Abstract
The majority of aneuploid fetuses are spontaneously miscarried. Nevertheless, some aneuploid individuals survive despite the strong genetic insult. Here, we investigate if the survival probability of aneuploid fetuses is affected by the genome-wide burden of slightly deleterious variants. We analyzed two cohorts of live-born Down syndrome individuals (388 genotyped samples and 16 fibroblast transcriptomes) and observed a deficit of slightly deleterious variants on Chromosome 21 and decreased transcriptome-wide variation in the expression level of highly constrained genes. We interpret these results as signatures of embryonic selection, and propose a genetic handicap model whereby an individual bearing an extremely severe deleterious variant (such as aneuploidy) could escape embryonic lethality if the genome-wide burden of slightly deleterious variants is sufficiently low. This approach can be used to study the composition and effect of the numerous slightly deleterious variants in humans and model organisms.
The majority of miscarriages are selective, i.e., contain chromosomal abnormalities or other severe mutations (Forbes 1997; Larsen et al. 2013). However, little is known about why fetuses with the same severe de novo variant can be either viable (at term) or not (miscarried). We hypothesize that the outcome is influenced by the burden of slightly deleterious variants (SDVs). Every human genome carries at least 1000 SDVs, including several loss-of-function variants (Kaiser et al. 2015), dozens of exon-intersecting copy number variants (Sudmant et al. 2015), hundreds of single-nucleotide missense coding substitutions (Xue et al. 2012; Henn et al. 2016), and thousands of single-nucleotide regulatory variants (Gulko et al. 2015). Numerous studies have demonstrated that this burden of SDVs is under purifying selection in the human population (Männik et al. 2015; Sulem et al. 2015; Narasimhan et al. 2016; Sohail et al. 2017) and thus might affect fetal viability.
In this study, we focus on trisomy 21 (T21). T21 fetuses have extremely high (up to 80%) miscarriage rates (Nussbaum et al. 2004), which may indicate strong embryonic selection. We hypothesize that as a result of this selection, live-born T21 individuals possess a reduced burden of SDVs compared to live-born, euploid control individuals. We hypothesized that a substantial fraction of the SDVs interacts with trisomy, so that their prevalence differs between T21 and euploid individuals. Although the SDVs on Chromosome 21 may directly affect the genes mapping on this chromosome, interactions may also involve genes elsewhere in the genome. We provisionally categorized SDVs into directly and indirectly interacting with trisomy on the basis of their genomic location: We assumed that Chromosome 21 SDVs are directly interacting, whereas the SDVs on all other autosomes may only be indirectly interacting.
In this study, we observed several lines of evidence, which is in line with embryonic selection of T21 individuals acting against a burden of SDVs. Based on our findings, we formulated the genetic handicap model, stating that an individual bearing an extremely severe deleterious variant (i.e., genetic handicap) might escape embryonic lethality if the genome-wide burden of SDVs is sufficiently low.
Results
Genomic and transcription variation of genes encoded on Chromosome 21 in T21 individuals
Functionally constrained Chromosome 21 genes demonstrate <1.5-fold increased expression in T21 individuals
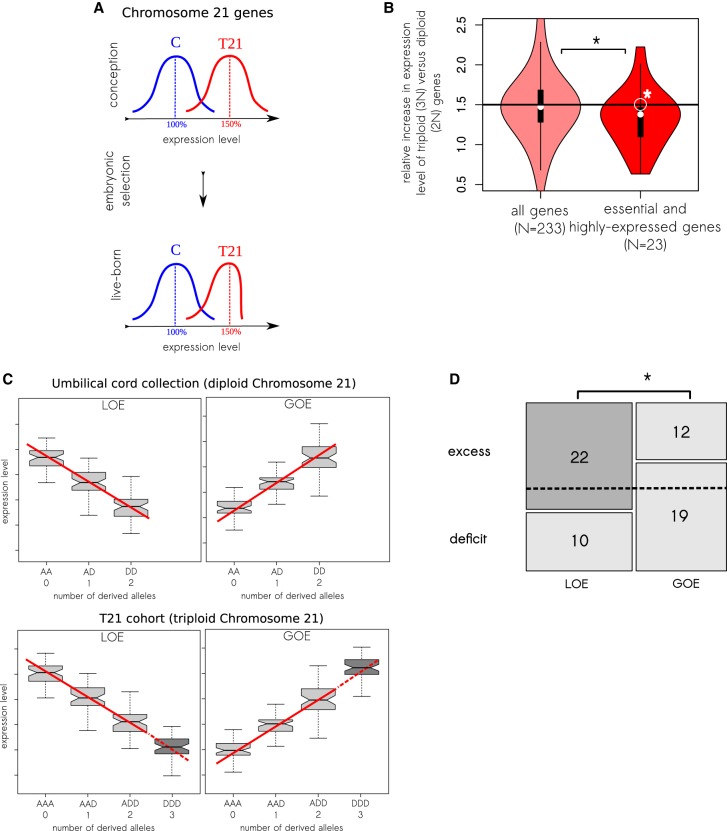
T21 fetuses with a relatively reduced expression of Chromosome 21 genes (<1.5-fold increase) might be favored by embryonic selection and thus have a higher probability of being live-born (Fig. 1A; Antonarakis et al. 2004; Aït Yahya-Graison et al. 2007; Prandini et al. 2007; Biancotti et al. 2010). We tested this by analyzing fibroblast transcriptomes of 16 T21 and 11 control live-born individuals. We used a set of Chromosome 21 genes (N = 233) that were expressed in all 27 samples. For each gene, we estimated the mean expression level in T21 and controls and computed a ratio of these values (Fig. 1B). The median of the ratios was 1.473, which does not significantly differ from the expected 1.5 (P-value = 0.78, one-sample Wilcoxon signed rank test with μ = 1.5). However, when we focused on a subset of functionally constrained genes (highly expressed and essential, N = 23) (Methods), the median of the ratios was significantly lower as compared to the whole set of Chromosome 21 genes (1.379 versus 1.473, P-value = 0.035, Mann-Whitney U test) (Fig. 1B), and the deviation from the expected 1.5 was also significant (P-value = 0.020, one-sample Wilcoxon signed rank test with μ = 1.5) (Fig. 1B).
Figure 1.
T21-specific variants: Expression level of Chromosome 21 genes is lower than expected. (A) Potential transcriptome signature of T21-specific selection: T21 fetuses with decreased expression level of Chromosome 21 genes are expected to have an advantage during embryogenesis. (B) Less than 1.5-fold increase in the expression level of highly constrained Chromosome 21 genes. Relative increase in the expression level of 233 Chromosome 21 genes in their trisomic (3N) state does not significantly differ from 1.5. However, for a subset of 23 highly functionally constrained genes, the increase in the expression level is lower than the expected 1.5. (*) P-value < 0.05. (C) A scheme defining loss of expression (LOE) and gain of expression (GOE) cis-eQTLs (upper) and our extrapolation of the effects of cis-eQTLs to the triploid case (dark gray box plots on lower panel). (D) Regulatory variants can compensate the overexpression of Chromosome 21 genes. The dotted line represents our expectation if the excess/deficit of genotypes is independent of the direction of expression changes (GOE or LOE). However, in the T21 live-born cohort, we observed an excess of homozygotes for the loss of expression derived alleles (LOE DDD) and a deficit of homozygotes for the gain of expression derived alleles (GOE DDD). (*) P-value < 0.05.
The <1.5-fold increase in the expression of functionally constrained genes is compatible with two explanations: (1) It might be the effect of embryonic selection against T21 fetuses with high overexpression of these genes; or (2) it might reflect a buffering of trisomy by a regulatory network. In order to distinguish between these two possibilities, we next analyzed the genetic component of the expression variation.
Regulatory variants of Chromosome 21 genes tend to diminish their overexpression in T21 individuals
Inter-individual gene expression variation is associated with single-nucleotide regulatory variants—expression quantitative trait loci in cis (cis-eQTLs) (Montgomery and Dermitzakis 2011). Therefore, regulatory variants that increase expression of Chromosome 21 genes (gain of expression [GOE] cis-eQTLs) may aggravate the effect of trisomy 21; in contrast, regulatory variants that decrease the expression of Chromosome 21 genes (loss of expression [LOE] cis-eQTLs) may partially compensate the trisomy 21 effect and be favorable. The embryonic selection eliminating such GOE and retaining LOE cis-eQTLs would result in a <1.5-fold increase in the expression of Chromosome 21 genes.
To functionally annotate GOE and LOE alleles on Chromosome 21, we analyzed the cis-eQTLs detected in three cell types (fibroblasts, lymphoblastoid cell lines, and T-cells) derived from umbilical cords of 195 unrelated healthy European newborns (The GenCord collection) (Dimas et al. 2009; Gutierrez-Arcelus et al. 2013). Since the umbilical cord is a fetal tissue and we are interested in embryonic selection, this collection of cis-eQTLs is relevant for our analyses. For each Chromosome 21 cis-eQTL, we polarized alleles as ancestral (A) or derived (D) using ancestral allele information from The 1000 Genomes Project Consortium (2010) and annotated derived alleles as GOE and LOE cis-eQTLs (Fig. 1C, upper). We assumed that the direction of the derived allele in trisomic cases is the same as in disomic, i.e., if D is annotated in the GenCord collection as GOE, the expression levels would be ordered as AAA < AAD < ADD < DDD (Fig. 1C, lower). We only used concordant (i.e., with the same direction) cis-eQTLs detected in more than one cell type.
We then analyzed the genotype frequencies of the annotated cis-eQTLs in a population of unrelated live-born T21 individuals (N = 338). For the 63 annotated cis-eQTLs on Chromosome 21 (32 GOE and 31 LOE), we compared the observed and predicted (Methods) frequencies of the DDD genotypes. Due to the small sample size, we did not split the cis-eQTLs according to the properties (functionally constrained and functionally nonconstrained) of associated genes. Our null expectation is that excess and deficit of different genotypes is independent of the effect of regulatory alleles (Fig. 1D, dotted line). However, we observed that the majority of LOE DDD genotypes (22 of 32) have a higher frequency than expected, whereas the majority of GOE DDD genotypes (19 of 31) have a lower frequency than expected (N = 63, Fisher's odds ratio = 3.41, P = 0.023) (Fig. 1D). Interestingly, if we further restrict our analysis only to those cis-eQTLs that were concordant in all three cell types of the GenCord collection, the signal becomes stronger, although less significant due to the small sample size (N = 16, Fisher's odds ratio = 7.63, P = 0.118). We conclude that the distribution of LOE and GOE cis-eQTLs (Fig. 1D) in live-born T21 individuals is compatible with embryonic selection favoring decreased gene expression level of trisomic genes.
Deviation of Chromosome 21 alleles from the Hardy–Weinberg Equilibrium uncovers signatures of selection
We next aimed to estimate the effect of all potentially deleterious variants on Chromosome 21. Rare derived alleles are enriched in deleterious variants (MacArthur et al. 2012; Fu et al. 2013), allowing us to use them as a proxy for SDVs. Correspondingly, strong selection driven by trisomy 21 will result in a deficit of rare derived alleles.
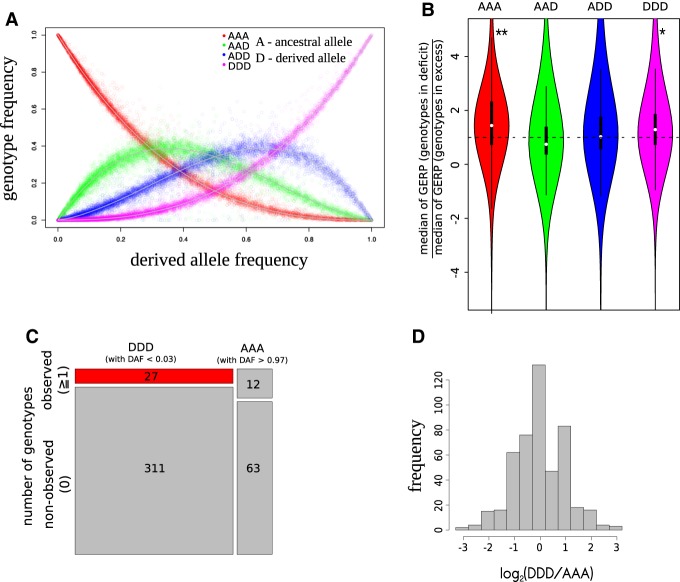
To test this, we analyzed a European cohort of 338 unrelated genotyped individuals with trisomy 21 (Sailani et al. 2013). If the parents of T21 individuals represent a random subset from a population in the Hardy–Weinberg equilibrium, trisomic genotype frequencies of Chromosome 21 single-nucleotide polymorphisms (SNPs) can be predicted from allele frequencies observed in the T21 cohort and the type of nondisjunction of Chromosome 21. Nondisjunction of Chromosome 21 can occur at meiosis I or meiosis II, leading to different relationships between allele and genotype frequencies (Methods). Fitting observed and expected numbers of genotypes in our analyzed population (Methods), we estimated that the fraction of T21 individuals resulting from nondisjunction in meiosis I is 74%, which is in agreement with a previously published ratio (Methods; Antonarakis et al. 1992). With the estimation of this fraction, we were able to reconstruct the number of expected genotypes (AAA, AAD, ADD, DDD, where “A” denotes the ancestral allele and “D” denotes the derived allele) for each allele based on its frequency (Fig. 2A).
Figure 2.
Hardy–Weinberg distribution of Chromosome 21 alleles and signatures of selection in the live-born T21 cohort. (A) Hardy–Weinberg distribution for trisomic alleles. The four curves represent the expected relationships between allele frequencies and genotype frequencies, assuming that the fraction of T21 individuals coming from nondisjunction in meiosis I is 74%. All dots are empirical observations. (B) The deficit of homozygous genotypes AAA and DDD is associated with increased evolutionary constraints of the corresponding nucleotide positions. Taking into account the genotypes in deficit and in excess (deviating by >5% from the expectation), we analyzed the GERP scores of the corresponding nucleotide positions. For each percentile of the derived allele frequency, we obtained the ratio of the median GERP scores for nucleotides associated with a deficit or an excess of genotypes. Violin plots reflect the distribution of these ratios for each genotype. In the case of AAA and DDD, the distribution of ratios has medians higher than the expected one: (*) P-value < 0.05; (**) P-value < 0.01. (C) Homozygotes for rare derived alleles (DDD) are rarer than the homozygotes for rare ancestral alleles (AAA) with the similar allele frequency: analysis of the two opposite tails of the Hardy–Weinberg distribution. (D) Observed homozygotes for rare derived alleles (DDD) are rarer than the observed homozygotes for rare ancestral alleles (AAA) with the same allele frequency: analysis of the 500 allele pairs, matched by allele frequency and characterized by the nonzero number of the observed homozygous genotypes. The distribution of the log2(DDD/AAA) has the median (−0.193) which is lower than the expected zero.
First, for each of the 8929 SNPs on Chromosome 21, we compared the number of observed and expected genotypes. We did not find any SNPs with a significant difference between observed and expected genotype frequencies (Bonferroni-corrected P-values > 0.01, Fisher's exact test), indicating that the potential compensatory selection, if it occurs, is not based on a single SNP with a strong effect, but rather on many SNPs with small effect. Thus, there are no strong outliers from the expected distribution shown in Figure 2A.
Despite the fact that there are no significantly deviating individual alleles, we have observed a substantial variation: Some genotypes are in deficit (below the curve), whereas others are in excess (above the curve) as compared to the expectation (Fig. 2A). In order to uncover a potential effect of selection acting on many low-effect-size variants, we focused on this variation: If selection is responsible for some of these deviations, we expect that the deficit of certain genotypes would be associated with increased constraints of the nucleotides. We used the Genomic Evolutionary Rate Profiling (GERP) score (Davydov et al. 2010) to approximate the evolutionary constraints on each nucleotide and binned SNPs into percentiles according to their derived allele frequencies, yielding 100 bins with 89 SNPs in each. Next, in each bin we characterized all genotypes in deficit and genotypes in excess by the median values of the corresponding GERP scores. Finally, for each bin we computed the ratio of medians [median(GERP score of alleles with a deficit of observed genotypes)/median(GERP score of alleles with an excess of observed genotypes)] which is expected to be one under the null hypothesis. For the AAD, ADD, and DDD genotypes, the ratios were indistinguishable from 1 (P-value > 0.2, one-sample Wilcoxon signed rank test with μ = 1; P-value is adjusted for multiple tests using Bonferroni correction). However, for the AAA genotype the ratio was greater than 1 (AAA: Bonferroni adjusted P-value = 0.0026, median = 1.26; one-sample Wilcoxon signed rank test with μ = 1), suggesting that more constrained SNPs (with high GERP scores) might be associated with deficient genotypes. If the observed effect is a result of selection, we expect that the stronger the deviation from the expectation, the higher the GERP score in deficient genotypes. Thus, we repeated our analysis taking into account only SNPs with genotypes deviating (decreased or increased) by >5% from the expectation. Indeed, we observed a stronger effect for both homozygous genotypes (AAA: Bonferroni adjusted P-value = 0.005, median = 1.44; DDD: Bonferroni adjusted P-value = 0.027, median = 1.29; one-sample Wilcoxon signed rank test with μ = 1), but did not observe this for heterozygous genotypes (Bonferroni adjusted P-values > 0.4; median = 1.02 [ADD] and 0.74 [DDD]; one-sample Wilcoxon signed rank test with μ = 1). The observed deficit of homozygous genotypes corresponding to highly constrained nucleotide positions is compatible with purifying selection against many slightly deleterious alleles. More pronounced selection against triploid homozygous genotypes might be the result of dosage effect, whereas in the heterozygotic state, the increased ploidy might be buffered by the presence of two different alleles. Interestingly, when we performed the same analysis for the rest of the autosomes (all autosomes but Chromosome 21), we did not observe any significant GERP differences between genotypes in deficit and in excess (all P-values for AA, AD, and DD genotypes > 0.15; one-sample Wilcoxon signed rank test with μ = 1). This result suggests that SNPs on Chromosome 21 are under the most stringent purifying selection due to the trisomy.
In the previous analysis, we did not observe any associations between the derived allele frequency and the ratio of GERP medians for all four genotypes (AAA, AAD, ADD, DDD; all P-values > 0.1, rank correlation). Because rare derived alleles are enriched in deleterious variants, another test of a potential selection might be a comparison of the expected and observed genotypes from the point of view of the derived allele frequency. To do this, we used the symmetry of the Hardy–Weinberg equilibrium (Fig. 2A), in which the number of DDD genotypes was expected to be similar to the number of AAA genotypes when comparing alleles of the same frequency [freq(D) = freq(A)]. First, we compared the fractions of DDD and AAA genotypes among rare derived (with DAF < 0.03) and rare ancestral (with DAF > 0.97) alleles (derived and ancestral alleles have similar distributions of frequencies, P-value > 0.8, Mann-Whitney U test). As expected, we did not observe DDD and AAA genotypes for the majority of these alleles in our T21 cohort (Fig. 2C, mosaic-plot); however, the fraction of the observed DDD genotypes was lower as compared to the AAA genotypes (P-value = 0.047, odds ratio = 0.46, Fisher's exact test). Second, to focus the analysis on the observed DDD and AAA genotypes, we analyzed 500 allele-frequency matched DDD–AAA genotype pairs (Methods). We found a significant deficit of DDD versus AAA genotypes (P-value = 5.409 × 10−5, paired Mann-Whitney U test) (Fig. 2D), confirming the deficit of homozygotes for rare derived alleles in the T21 cohort. Interestingly, using the same approach for all autosomes except Chromosome 21, we did not detect significant differences between DD and AA genotypes, emphasizing once more the most important role of the Chromosome 21 variability in the live-born trisomy. The Hardy–Weinberg equilibrium on Chromosome 21 alleles (Fig. 2A) allowed us to uncover nonuniform distribution of GERP scores (Fig. 2B) as well as a deficit of homozygotes for rare derived alleles (Fig. 2C,D). Both of these findings uncover signatures of potential purifying selection targeting many low-effect-size variants. Altogether, the preceding three analyses are consistent with selection against T21 fetuses with an excess of directly interacting deleterious variants.
Whole-transcriptome perturbations in T21 individuals
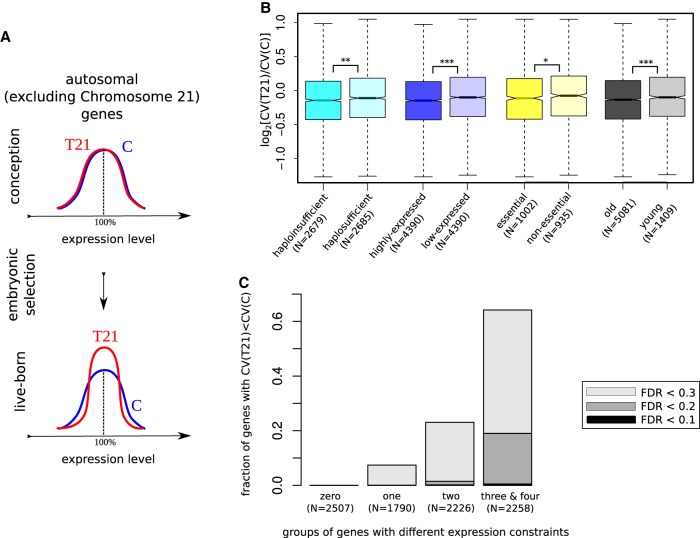
The deviations from the median gene expression might be deleterious, so that individuals whose gene expression level is too high or too low are less fit. This assumption is consistent with several lines of evidence: Both overexpression and underexpression of genes have been associated with pathological conditions (Carmona-Mora and Walz 2010; Jacquemont et al. 2011; Adamo et al. 2015; McCoy et al. 2015); variation in gene expression affects the severity of different mutations (Vu et al. 2015); increased variation in gene expression is associated with aging (Bahar et al. 2006; Végh et al. 2014) and low fitness (Wang and Zhang 2011); single-nucleotide regulatory variants have a genome-wide distribution and population genetic properties similar to slightly deleterious variants (Popadin et al. 2013, 2014). If T21 fetuses with nonoptimal (too low or too high) gene expression patterns are preferentially eliminated through miscarriages, we expect decreased variation in gene expression among live-born T21 individuals as compared to euploid controls (Fig. 3A). The transcriptome variation is expected to be deleterious for all fetuses, including euploid, but the effect might be exacerbated in T21 individuals. We hypothesize that the variation of gene expression outside Chromosome 21 is involved in such indirect interactions with trisomy.
Figure 3.
Live-born T21 individuals are closer to expression optimum in autosomal genes encoded outside Chromosome 21. (A) A schematic representation illustrating a potential effect of purifying selection against T21 fetuses with a nonoptimal pattern of expression during embryogenesis. The wide distribution of genes with similar mean expression levels in T21 and control (C) cohort in early stages of embryogenesis (top) becomes narrower (bottom) as a result of selection that eliminates low-fitness fetuses with excessively low or high expression level of genes. (B) The expression variation in T21 is decreased more in old versus young, haploinsufficient versus haplosufficient, essential versus nonessential, and highly expressed versus low-expressed genes. (*) P-value < 0.05; (**) P-value < 0.01; (***) P-value < 0.001. (C) The expression variation in the T21 cohort is decreased more in highly constrained genes. The number of constraints, from zero to four: zero if the gene is neither old nor essential nor highly expressed nor haploinsufficient, and four if a given gene has all four metrics for constrained genes.
To test this, we used previously described transcriptome data (fibroblast transcriptomes from 16 live-born T21 individuals and 11 controls). We selected 8781 genes that are expressed in all 27 samples, located on autosomes other than Chromosome 21, and have similar expression level in T21 and control cohorts (Methods). For each of these genes, we calculated the log ratio of the coefficients of variation (CV) in expression level in T21 versus control (Methods). We then performed 10,000 permutations, randomly assigning 16 “T21” and 11 “controls” out of 27 samples, and defined a test statistic (P-value) as the fraction of permuted log ratios deviating from the observed value. Using vectors of gene-specific P-values (N = 8781), we estimated the π1 statistics (Supplemental Fig. S1A), which reflect the proportion of truly significant P-values (Storey and Tibshirani 2003). Using π1 statistics with different lambdas, we demonstrated that there is a group of genes with significantly decreased expression variation in T21 versus controls (Methods; Supplemental Fig. S1). As the most conservative estimation of the number of genes with decreased expression variation in “T21” versus “controls,” we used π1 statistics equal to 3% (Supplemental Fig. S1B, left) that corresponds to 250 genes. Importantly, when we estimated the opposite effect (genes with increased expression variation in “T21” versus “controls”) using the same approaches, we did not observe any such genes (π1 = 0).
Fluctuations in gene expression affect the fitness according to gene-specific expression constraints, which are related to gene age (Popadin et al. 2014), gene essentiality (Georgi et al. 2013), haploinsufficiency (Steinberg et al. 2015), and expression level (Wolf et al. 2009). Thus, a subset of constrained genes might be the primary target of selection and show a more extreme reduction in expression variation in T21 versus controls. To test this, we grouped all analyzed genes into categories defined by gene age, essentiality, haploinsufficiency status, and expression level (Methods), and compared the corresponding log ratios. Indeed, the constrained groups of genes—old versus young, essential versus nonessential, haploinsufficient versus haplosufficient, highly expressed versus low-expressed—demonstrated a stronger reduction in expression variation in T21 versus control (all P-values < 0.05, Mann-Whitney U test) (Fig. 3B). Next, we grouped all genes according to their number of constraints, from zero to four: zero if a gene is neither old, essential, highly expressed, or haploinsufficient; four if a given gene is in all four categories for constrained genes. We observed that the more constrained groups are indeed characterized by a higher fraction of genes with decreased expression variation in T21 versus controls (Fig. 3C). For example, among the most constrained 2258 genes (with three and four levels of constraint), 11 genes have significantly lower variation in their expression level in T21 versus control with FDR < 0.1, 121 genes with FDR < 0.15, and 429 genes with FDR < 0.2 (Fig. 3C).
Recently, Sullivan et al. (2016) compared transcriptomes of T21 individuals with sex- and age-matched controls using fibroblasts (six versus six) and lymphoblastoid cell lines (three versus three). We repeated our gene expression variation analysis on this independent data set. For each gene, we estimated the ratio of the coefficients of variation between T21 and controls and derived the P-values of the decreased variation in T21. Analyzing the distribution of P-values, we observed a very similar trend to that described in our main data set (N = 6069 genes; π1 statistics is higher than zero, with lambdas changing from 0.05 to 1). Moreover, for a subset of genes expressed in both our original data set and in the data set of Sullivan et al. (2016) (N = 3536), we observed a significant positive correlation between gene-specific log ratios (Spearman's ρ = 0.20, P-value < 2.2 × 10−16). Thus, genes with decreased variance in T21 relative to controls tend to show the same trend in both data sets. For the lymphoblastoid cell lines, we did not observe any signals: The π1 statistic equals zero, meaning that all P-values follow the Uniform (0,1) distribution.
The decreased variation in gene expression level in T21 versus control individuals is compatible with a reduced burden of regulatory variants (both rare and common), which affect gene expression. This observation suggests that severe mutations, such as trisomy 21, might be at least partially compensated by a decreased genome-wide burden of indirectly interacting deleterious variants.
Genetic handicap hypothesis
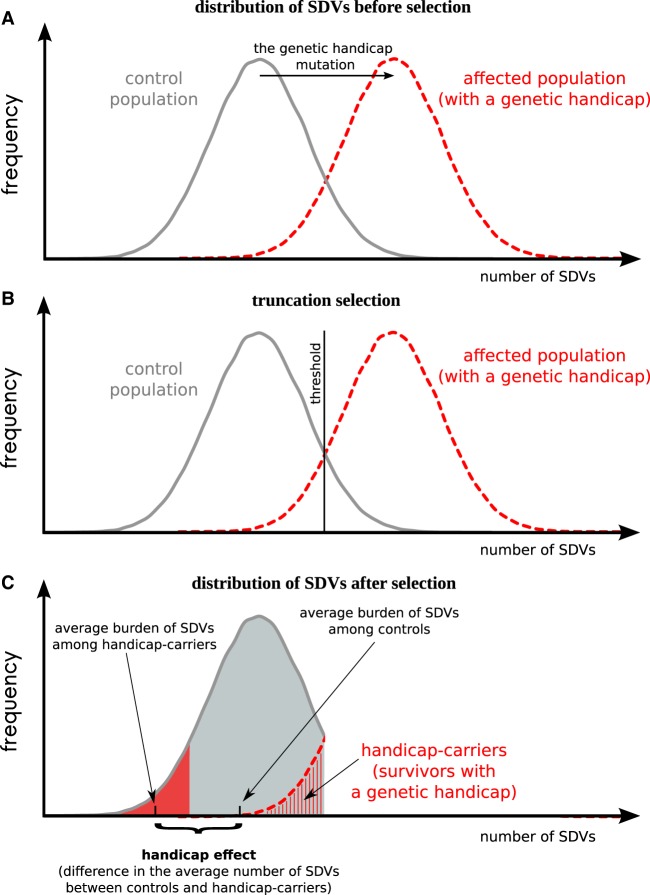
If the biological fitness of an organism is determined by its mutational burden (the total effect of all slightly, moderately, and severely deleterious variants), we expect to observe a trade-off between the presence of a severely deleterious mutation and the number of SDVs. The term handicap, defined in the English Oxford Living Dictionaries (2017) as “a circumstance that makes progress or success difficult,” has been used by Zahavi (1975) to refer to a phenotypically disadvantageous trait such that its presence is the marker of a high genome quality of the carrier. We propose the genetic handicap model, whereby an organism bearing a severely deleterious mutation (a “genetic handicap” in Zahavi's sense) is only fit (i.e., viable at birth) if its genome-wide burden of SDVs is sufficiently low (Fig. 4). The rationale for this hypothesis is that only highly fit organisms, i.e., those with a sufficiently low burden of SDVs, are able to tolerate the effects of severely deleterious mutations and survive. Our approach resembles the “liability” introduced by Falconer (1965) as a single continuous normally distributed factor representing a mixture of environmental and genetic traits and determining the probability of acquiring a complex disease. Here, we define genetic handicap as a severely deleterious mutation with very broad effect on cellular metabolism. The broad effect assumes that the genetic handicap is unlikely to be compensated/modified by a few variants, and therefore might only be compensated by the genome-wide decreased burden of slightly deleterious variants.
Figure 4.
The genetic handicap model. (A) The distribution of the number of SDVs in control (gray) and affected (red) populations. The genetic handicap mutation (black arrow) is an equivalent of many SDVs. (B) Truncation selection eliminates all organisms with the number of SDVs higher than the given threshold (vertical black line) from both control and affected populations. (C) Handicap carriers have a decreased number of SDVs (SDVs do not include the genetic handicap per se) than controls; this difference represents the handicap effect.
We assume here that embryonic viability is an important component of fitness, so that the genetic handicap splits the affected population into two groups: survivors at birth (handicap carriers) and nonsurvivors (Fig. 4). If the increased number of SDVs leads to a disproportional decline in fitness, which would be the case, e.g., under truncation selection (Kondrashov 1988; Crow and Kimura 2009), then handicap carriers will have a decreased number of SDVs as compared to controls (live-born organisms without handicap) (Fig. 4). Moreover, we predict that the more severe the genetic handicap, the higher the handicap effect (difference in the mean numbers of SDVs between controls and handicap carriers) (Fig. 4).
Discussion
In this report, we observed genomic and transcriptomic signatures of embryonic selection in live-born individuals with Down syndrome. In these individuals, we found that SDVs are underrepresented, and/or have a weaker phenotypic effect, suggesting that more deleterious variants are being eliminated. These results are consistent with the liability threshold/truncation selection model involving synergistic epistasis between the genome-wide burden of slightly deleterious variants (SDVs) and trisomy 21. Thus we propose the genetic handicap model, which states that a severe mutation (i.e., genetic handicap) might be partially compensated by a reduced genome-wide burden of SDVs. In this case, the presence of a severe mutation in a viable (fit) organism might indicate a reduced burden of SDVs.
From the medical point of view, future comparative investigations of the burden of SDVs in human genomes carrying a severe mutation might shed light on (1) the difference between miscarried and live-born individuals carrying the same genetic handicap; (2) the extensive clinical heterogeneity often observed among live-born individuals carrying identical severe mutations; and (3) the relative impact of directly versus indirectly interacting SDVs. From the evolutionary genetics point of view, one of the important applications of the genetic handicap approach is the possibility of establishing qualitative and quantitative links between the burden of SDVs and fitness. Within a given population, individuals differ in their fitness, defined as their relative reproductive success (Haldane 1937). It has been suggested that the genetic component of this variation is predominantly due to slightly deleterious variants (Muller 1950), which, contrary to strongly deleterious and beneficial variants, can segregate within a population for a long time and reach both relatively high frequencies in a population and high numbers in individual genomes (Crow 1958). The functional relationship between the burden of SDVs and fitness is of considerable importance for basic evolutionary and medical genetics concepts, such as the maintenance of intra-population variation in fitness (Haldane 1937), evolution of recombination and sexual selection (Kondrashov 1988; Agrawal 2001), fitness landscapes, speciation, and species extinction (Ohta 1992; Popadin et al. 2007; Meer et al. 2010; Polishchuk et al. 2015), inbreeding depression (Charlesworth and Willis 2009), and the etiology of complex diseases (Cortopassi 2002).
Despite the fundamental importance of the relationship between SDVs and fitness, there is no solid empirical evidence regarding whether intra-population fitness variation is driven by variation in the burden of SDVs. This is because both the burden of SDVs is difficult to quantify, and fitness is one of the most complex phenotypes to measure. All apparently healthy people probably carry thousand(s) of deleterious variants in their genome; the unknown selection coefficients associated with SDVs and their epistatic interactions make it challenging to reconstruct fitness from genetic data. For example, it is unknown whether there is a difference in the burden of SDVs between human populations (see the controversy in the estimation of the burden in African and non-African populations) (Lohmueller et al. 2008; Fu et al. 2014; Simons et al. 2014; Do et al. 2015; Henn et al. 2016).
Important data on associations between SDVs and fitness in humans come from genetic studies of complex diseases, in which an increased burden of certain types of variants in affected versus unaffected individuals has been demonstrated (Cooper et al. 2011; Girirajan et al. 2011; Krumm et al. 2015). Recently, the genome-wide burden of copy number variants (CNVs) (Männik et al. 2015) and runs of homozygosity (ROHs) (Joshi et al. 2016) have been linked to fitness-related phenotypes (educational attainment) among healthy individuals. More direct effects of ongoing purifying selection in the healthy human population have been shown recently as a deficit of loss-of-function (LoF) variants transmitted from heterozygous parents to homozygous offspring (Sulem et al. 2015) and of underrepresentation of individuals with large numbers of LoF variants (Sohail et al. 2017). Although these studies provide a first empirical correlation between the burden of SDVs and fitness-related traits in human populations, they are still restricted to a few types of variants/genes and would require large sample sizes to uncover the effects of slightly deleterious variants.
Additionally to direct inference of SDVs, fitness might be approximated through transcriptomic data (Marigorta et al. 2017). The transcriptome can be considered as an intermediate molecular phenotype between DNA and organism-level phenotype. Leveraging the transcriptome may have advantages over DNA sequencing because gene expression reflects complex interactions between numerous DNA coding, DNA regulatory, and chromatin variants. Gene expression level was shown to reveal minor differences between individuals and distinguish noncarriers, heterozygous carriers, and patients homozygous for autosomal recessive disorders (Cheung and Ewens 2006; Smirnov and Cheung 2008), and the first two groups are phenotypically indistinguishable at the organism level.
The genetic handicap approach described in this paper can provide an a priori expectation for the difference in the burden of SDVs between handicap carriers and controls, which might be tested in empirical or experimental studies, and ultimately improve our understanding and functional annotation of the numerous slightly deleterious variants in humans and model organisms.
Methods
Transcriptome analyses
The primary skin fibroblast of 16 European unrelated T21 and 11 European unrelated control individuals stratified by sex and age were described in Letourneau et al. (2014). To avoid batch effects, T21 and control samples were randomized prior to culturing, RNA extraction, and sequencing. All fibroblasts were grown in DMEM media containing high glucose, GlutaMAX, and pyruvate (Life Technologies 31966), and supplemented with 10% fetal bovine serum (Life Technologies 10270) and 1% penicillin/streptomycin/fungizone mix (Amimed, BioConcept 4-02F00-H) at 37°C in a 5% CO2 atmosphere. Total RNA was collected using TRIzol reagent (Life Technologies 15596) according to the manufacturer's instructions. mRNA-seq libraries were prepared using the Illumina mRNA-seq Sample Preparation kit (RS-100-0801) and TruSeq RNA sample preparation kit, according to the manufacturer's instructions. Libraries were sequenced on the Illumina HiSeq 2000 using paired-end sequencing 2 × 100 bp. Reads were mapped against the human (hg19) genome using the default parameters of the TopHat mapper (Trapnell et al. 2009), filtering out multiple mapping reads, and FPKMs (fragments per kilobase of exon per million fragments mapped) were calculated for each gene. Because sequence content is largely unchanged between hg19 and GRCh38, especially for protein-coding genes, we argue that realigning reads to GRCh38 would not significantly affect our results. No further normalization procedures were applied (such as quantile normalization), because all analyses were run in the gene-by-gene manner (however, see below for a test confirming that quantile normalization does not affect results).
For whole-transcriptome analyses, we eliminated genes located on sex chromosomes, Chromosome 21, and the mitochondrial genome. We used a subset of genes expressed in all 27 samples with FPKM > 0. In order to avoid any effects of differentially expressed genes, we only kept genes with a similar expression level between T21 and controls, defined as 20% range of mean expression levels in control cohort: for each gene, mean(C) × 0.9 ≤ mean(Ts) ≤ mean(C) × 1.1.
For each gene, we estimated the coefficient of variation (CV) as the ratio of the standard deviation to the mean separately for T21 and control cohorts. Thereafter, we obtained the log ratio of these coefficients as log2[CV(T21)/CV(C)].
To assess the significance of decreased expression variation in T21 versus controls (P-values), we ran 10,000 permutations for each gene. For a given gene, we compared the observed log ratios obtained after randomly assigning 16 “T21” and 11 “control” labels in a shuffled vector containing 27 expression levels of this gene. Counting the fraction of permuted values, which are less than the observed value, we approximated the P-value of decreased expression variation of this gene in T21 cohort. It is important to note, that this permutation analysis takes into account potential outliers in our data set and thus assures that our results are robust. Using our vector of gene-specific P-values, we estimated the π0 statistic and Q-values using the R package “qvalue” (Storey and Tibshirani 2003). Thereafter, we obtained π1 as 1 − π0.
The coefficient of variation and the sample size are inversely proportional. Thus, our log ratio can be partially affected by an increased sample size of T21 cohort. Our permutation approach overcomes this limitation by retaining the asymmetrical sample size during the permutations (16 randomly assigned “Ts” versus 11 “control” labels), thus eliminating this potential bias. This permutation analysis also refuses the possibility that our results might be driven by outliers (for example, by several too variably expressed control individuals).
To corroborate robustness of our results to normalization steps, we reran our variation analysis using quantile normalized FPKMs of 16 T21 and 11 controls (normalize.quantiles function in the R package preprocessCore) (Bolstad et al. 2003). First, the log ratio of the coefficients of variation of normalized data highly correlated with the log ratio based on non-normalized data (Spearman's ρ = 0.91, P-value <2.2 × 10−16). Second, on normalized data, we ran 10,000 permutations randomly assigning 16 “T21” and 11 “Controls” out of 27 samples, and defined a test statistic as the fraction of permuted log ratios less than or equal to the observed value. Using this vector of gene-specific P-values (N = 8781), we estimated the π1 statistic as 0.435, which is near identical to our value 0.448 obtained for non-normalized data.
Gene age was used from Zhang et al. (2010), in which old or young genes were defined as genes originating before or after the divergence of zebrafish. Lists of essential and nonessential genes have been used from Georgi et al. (2013). Haploinsufficiency score was used according to Steinberg et al. (2015). Genes with a haploinsufficiency score higher or lower than the median were deemed haploinsufficient or haplosufficient. The expression level of each gene was derived as the mean expression (FPKM) across all 27 samples. Genes with an expression level higher or lower than median were called highly or lowly expressed genes. For analysis of Chromosome 21 genes, those genes with an expression level higher than the Chromosome 21–specific median were called highly expressed genes.
Genotype analyses
The analyzed population, as well as the procedure for detecting four clusters of genotypes (AAA, AAB, ABB, and BBB) for each triploid SNP from Chromosome 21, were as in Sailani et al. (2013).
Hardy–Weinberg principle for trisomic cases
Nondisjunction of Chromosome 21 can occur at meiosis I or meiosis II, leading to different relationships between allele and genotype frequencies. In the case of meiosis I, the nondisjoining parent may contribute gametes AA, AB, or BB with frequencies p2, 2pq, and q2, leading to the next distribution of AAA, AAB, ABB, and BBB genotypes as follows:
| (1) |
In the case of meiosis II, the nondisjoining parent contributes gametes AA and BB with frequencies p and q, leading to the next distribution of AAA, AAB, ABB, and BBB genotypes as follows:
| (2) |
Combining both Equations (1) and (2) yields
| (3) |
where X is the fraction of T21 individuals, resulting from nondisjunction at meiosis I. In order to estimate X in our data set of 338 T21, we estimated the differences in the observed and predicted numbers of AAA, AAB, ABB, and BBB genotypes with X, changing from 0.60 to 0.90 with step 0.01. We found that the predicted genotype frequencies are closer to the observed values when X is 0.72–0.76. For our analyses, we used X = 0.74; however, all results with X equal to 0.72, 0.73, 0.75, and 0.76 are qualitatively similar: Figures 1D, and 2C and D, remain unchanged, whereas Figure 2, A and B, remain very similar (there are no significantly deviating alleles on Fig. 2A; there is the same trend for AAA and DDD genotypes on Fig. 2B).
It has been shown that 77.1% of maternal nondisjunctions (128 out of 166) occurred in meiosis I; however, only 22.2% of paternal nondisjunctions (two of nine) occurred in meiosis I (Antonarakis et al. 1992). Taking into account the prevalence of maternal nondisjunctions, the average probability of nondisjunctions in meiosis I is 74% (130 out of 175), which is consistent with our estimation.
Paired test of symmetry
Due to the small number of rare ancient (ancestral) alleles with which to match pairs of alleles (one derived and once ancestral with the same frequency), we started from the ancestral allele. Each SNP was used only once, and the frequencies of alleles D and A in each matched pair were the same (P > 0.2, paired Mann-Whitney U test). This analysis is deterministic and consists of the next steps: (1) We subset all rare ancestral alleles A [freq(A) < 0.1] with at least one observed homozygous genotype AAA and sort them according to the frequency of allele A; (2) we subset all rare derived alleles D [freq(D) < 0.1] with at least one observed homozygous genotype DDD and sort them according to the frequency of allele D; (3) starting from the most rare ancestral allele we create matched pairs: Find the derived allele with the same or the most similar allele frequency; if there are several alleles D with identical frequencies (this occurs in <10% of pairs) we take the most distant one from the allele A, the matched allele D is excluded from further pairing; and (4) analysis stops at 500 matched pairs.
Direction of cis-eQTL derived alleles
We calculated the direction of effect of each cis-eQTL as previously described (Popadin et al. 2014). We estimated the slope of a linear model between the number of derived alleles (AA = 0, AD = 1, DD = 2) and the expression level of the exon used for the cis-eQTL call. If a given cis-eQTL affected more than one exon, we used the direction of the strongest effect (with the highest absolute value of the effect).
The congruent cis-eQTLs were defined as “+,+,+”; “+,+,0”; “−,−,−”; and “−,−,0”; where “+,” “−,” and “0” means GOE, LOE, and no detected cis-eQTLs in three cell type of the umbilical cord collection.
Supplementary Material
Acknowledgments
We thank Amotz Zahavi and Alexey Kondrashov for discussion of the interpretation of handicap and Christopher Brown for statistical comments and suggestions. K.P. was supported by the 5 Top 100 Russian Academic Excellence Project at the Immanuel Kant Baltic Federal University. This work was also supported by SNF grant 163180 and ERC grant 249968 to S.E.A., and the Swiss National Science Foundation (grant 31003A_160203) to A.R. S.P. was partly supported by a Swiss NSF grant No. 31003A-143393 to L.E.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.228411.117.
References
- The 1000 Genomes Project Consortium. 2010. A map of human genome variation from population-scale sequencing. Nature 467: 1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamo A, Atashpaz S, Germain PL, Zanella M, D'Agostino G, Albertin V, Chenoweth J, Micale L, Fusco C, Unger C, et al. 2015. 7q11.23 dosage-dependent dysregulation in human pluripotent stem cells affects transcriptional programs in disease-relevant lineages. Nat Genet 47: 132–141. [DOI] [PubMed] [Google Scholar]
- Agrawal AF. 2001. Sexual selection and the maintenance of sexual reproduction. Nature 411: 692–695. [DOI] [PubMed] [Google Scholar]
- Aït Yahya-Graison E, Aubert J, Dauphinot L, Rivals I, Prieur M, Golfier G, Rossier J, Personnaz L, Creau N, Bléhaut H, et al. 2007. Classification of human chromosome 21 gene-expression variations in Down syndrome: impact on disease phenotypes. Am J Hum Genet 81: 475–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonarakis SE, Petersen MB, McInnis MG, Adelsberger PA, Schinzel AA, Binkert F, Pangalos C, Raoul O, Slaugenhaupt SA, Hafez M. 1992. The meiotic stage of nondisjunction in trisomy 21: determination by using DNA polymorphisms. Am J Hum Genet 50: 544–550. [PMC free article] [PubMed] [Google Scholar]
- Antonarakis SE, Lyle R, Dermitzakis ET, Reymond A, Deutsch S. 2004. Chromosome 21 and Down syndrome: from genomics to pathophysiology. Nat Rev Genet 5: 725–738. [DOI] [PubMed] [Google Scholar]
- Bahar R, Hartmann CH, Rodriguez KA, Denny AD, Busuttil RA, Dollé ME, Calder RB, Chisholm GB, Pollock BH, Klein CA, et al. 2006. Increased cell-to-cell variation in gene expression in ageing mouse heart. Nature 441: 1011–1014. [DOI] [PubMed] [Google Scholar]
- Biancotti JC, Narwani K, Buehler N, Mandefro B, Golan-Lev T, Yanuka O, Clark A, Hill D, Benvenisty N, Lavon N. 2010. Human embryonic stem cells as models for aneuploid chromosomal syndromes. Stem Cells 28: 1530–1540. [DOI] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19: 185–193. [DOI] [PubMed] [Google Scholar]
- Carmona-Mora P, Walz K. 2010. Retinoic acid induced 1, RAI1: a dosage sensitive gene related to neurobehavioral alterations including autistic behavior. Curr Genomics 11: 607–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D, Willis JH. 2009. The genetics of inbreeding depression. Nat Rev Genet 10: 783–796. [DOI] [PubMed] [Google Scholar]
- Cheung VG, Ewens WJ. 2006. Heterozygous carriers of Nijmegen Breakage Syndrome have a distinct gene expression phenotype. Genome Res 16: 973–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper GM, Coe BP, Girirajan S, Rosenfeld JA, Vu TH, Baker C, Williams C, Stalker H, Hamid R, Hannig V, et al. 2011. A copy number variation morbidity map of developmental delay. Nat Genet 43: 838–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortopassi GA. 2002. A neutral theory predicts multigenic aging and increased concentrations of deleterious mutations on the mitochondrial and Y chromosomes. Free Radic Biol Med 33: 605–610. [DOI] [PubMed] [Google Scholar]
- Crow JF. 1958. Some possibilities for measuring selection intensities in man. Hum Biol 30: 1–13. [PubMed] [Google Scholar]
- Crow JF, Kimura M. 2009. An introduction to population genetics theory. The Blackburn Press, Caldwell, NJ. [Google Scholar]
- Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S. 2010. Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS Comput Biol 6: e1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimas AS, Deutsch S, Stranger BE, Montgomery SB, Borel C, Attar-Cohen H, Ingle C, Beazley C, Gutierrez Arcelus M, Sekowska M, et al. 2009. Common regulatory variation impacts gene expression in a cell type–dependent manner. Science 325: 1246–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do R, Balick D, Li H, Adzhubei I, Sunyaev S, Reich D. 2015. No evidence that selection has been less effective at removing deleterious mutations in Europeans than in Africans. Nat Genet 47: 126–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English Oxford Living Dictionaries. 2017. Oxford University Press, Oxford: http://www.oxforddictionaries.com/definition/english/handicap. [Google Scholar]
- Falconer DS. 1965. The inheritance of liability to certain diseases, estimated from the incidence among relatives. Ann Hum Genet 29: 51–76. [Google Scholar]
- Forbes LS. 1997. The evolutionary biology of spontaneous abortion in humans. Trends Ecol Evol 12: 446–450. [DOI] [PubMed] [Google Scholar]
- Fu W, O'Connor TD, Jun G, Kang HM, Abecasis G, Leal SM, Gabriel S, Rieder MJ, Altshuler D, Shendure J, et al. 2013. Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature 493: 216–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu W, Gittelman RM, Bamshad MJ, Akey JM. 2014. Characteristics of neutral and deleterious protein-coding variation among individuals and populations. Am J Hum Genet 95: 421–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgi B, Voight BF, Bućan M. 2013. From mouse to human: evolutionary genomics analysis of human orthologs of essential genes. PLoS Genet 9: e1003484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girirajan S, Brkanac Z, Coe BP, Baker C, Vives L, Vu TH, Shafer N, Bernier R, Ferrero GB, Silengo M, et al. 2011. Relative burden of large CNVs on a range of neurodevelopmental phenotypes. PLoS Genet 7: e1002334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulko B, Hubisz MJ, Gronau I, Siepel A. 2015. A method for calculating probabilities of fitness consequences for point mutations across the human genome. Nat Genet 47: 276–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Arcelus M, Lappalainen T, Montgomery SB, Buil A, Ongen H, Yurovsky A, Bryois J, Giger T, Romano L, Planchon A, et al. 2013. Passive and active DNA methylation and the interplay with genetic variation in gene regulation. eLife 2: e00523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane JBS. 1937. The effect of variation on fitness. Am Nat 71: 337–386. [Google Scholar]
- Henn BM, Botigué LR, Peischl S, Dupanloup I, Lipatov M, Maples BK, Martin AR, Musharoff S, Cann H, Snyder MP, et al. 2016. Distance from sub-Saharan Africa predicts mutational load in diverse human genomes. Proc Natl Acad Sci 113: E440–E449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemont S, Reymond A, Zufferey F, Harewood L, Walters RG, Kutalik Z, Martinet D, Shen Y, Valsesia A, Beckmann ND, et al. 2011. Mirror extreme BMI phenotypes associated with gene dosage at the chromosome 16p11.2 locus. Nature 478: 97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi PK, Esko T, Mattsson H, Eklund N, Gandin I, Nutile T, Jackson AU, Schurmann C, Smith AV, Zhang W, et al. 2016. Directional dominance on stature and cognition in diverse human populations. Nature 523: 459–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser VB, Svinti V, Prendergast JG, Chau YY, Campbell A, Patarcic I, Barroso I, Joshi PK, Hastie ND, Miljkovic A, et al. 2015. Homozygous loss-of-function variants in European cosmopolitan and isolate populations. Hum Mol Genet 24: 5464–5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov AS. 1988. Deleterious mutations and the evolution of sexual reproduction. Nature 336: 435–440. [DOI] [PubMed] [Google Scholar]
- Krumm N, Turner TN, Baker C, Vives L, Mohajeri K, Witherspoon K, Raja A, Coe BP, Stessman HA, He ZX, et al. 2015. Excess of rare, inherited truncating mutations in autism. Nat Genet 47: 582–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen EC, Christiansen OB, Kolte AM, Macklon N. 2013. New insights into mechanisms behind miscarriage. BMC Med 11: 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letourneau A, Santoni FA, Bonilla X, Sailani MR, Gonzalez D, Kind J, Chevalier C, Thurman R, Sandstrom RS, Hibaoui Y, et al. 2014. Domains of genome-wide gene expression dysregulation in Down's syndrome. Nature 508: 345–350. [DOI] [PubMed] [Google Scholar]
- Lohmueller KE, Indap AR, Schmidt S, Boyko AR, Hernandez RD, Hubisz MJ, Sninsky JJ, White TJ, Sunyaev SR, Nielsen R, et al. 2008. Proportionally more deleterious genetic variation in European than in African populations. Nature 451: 994–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur DG, Balasubramanian S, Frankish A, Huang N, Morris J, Walter K, Jostins L, Habegger L, Pickrell JK, Montgomery SB, et al. 2012. A systematic survey of loss-of-function variants in human protein-coding genes. Science 335: 823–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Männik K, Mägi R, Macé A, Cole B, Guyatt AL, Shihab HA, Maillard AM, Alavere H, Kolk A, Reigo A, et al. 2015. Copy number variations and cognitive phenotypes in unselected populations. JAMA 313: 2044–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marigorta UM, Denson LA, Hyams JS, Mondal K, Prince J, Walters TD, Griffiths A, Noe JD, Crandall WV, Rosh JR, et al. 2017. Transcriptional risk scores link GWAS to eQTLs and predict complications in Crohn's disease. Nat Genet 49: 1517–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy RC, Demko Z, Ryan A, Banjevic M, Hill M, Sigurjonsson S, Rabinowitz M, Fraser HB, Petrov DA. 2015. Common variants spanning PLK4 are associated with mitotic-origin aneuploidy in human embryos. Science (80-) 348: 235–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meer MV, Kondrashov AS, Artzy-Randrup Y, Kondrashov FA. 2010. Compensatory evolution in mitochondrial tRNAs navigates valleys of low fitness. Nature 464: 279–282. [DOI] [PubMed] [Google Scholar]
- Montgomery SB, Dermitzakis ET. 2011. From expression QTLs to personalized transcriptomics. Nat Rev Genet 12: 277–282. [DOI] [PubMed] [Google Scholar]
- Muller HJ. 1950. Our load of mutations. Am J Hum Genet 2: 111–176. [PMC free article] [PubMed] [Google Scholar]
- Narasimhan VM, Hunt KA, Mason D, Baker CL, Karczewski KJ, Barnes MR, Barnett AH, Bates C, Bellary S, Bockett NA, et al. 2016. Health and population effects of rare gene knockouts in adult humans with related parents. Science (80-) 352: 474–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussbaum RL, McInnes RR, Willard HF. 2004. Thompson & Thompson genetics in medicine, revised reprint, 6th ed Saunders, Philadelphia, PA. [Google Scholar]
- Ohta T. 1992. The nearly neutral theory of molecular evolution. Annu Rev Ecol Syst 23: 263–286. [Google Scholar]
- Polishchuk LV, Popadin KY, Baranova MA, Kondrashov AS. 2015. A genetic component of extinction risk in mammals. Oikos 124: 983–993. [Google Scholar]
- Popadin K, Polishchuk LV, Mamirova L, Knorre D, Gunbin K. 2007. Accumulation of slightly deleterious mutations in mitochondrial protein-coding genes of large versus small mammals. Proc Natl Acad Sci 104: 13390–13395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popadin K, Gutierrez-Arcelus M, Dermitzakis ET, Antonarakis SE. 2013. Genetic and epigenetic regulation of human lincRNA gene expression. Am J Hum Genet 93: 1015–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popadin KY, Gutierrez-Arcelus M, Lappalainen T, Buil A, Steinberg J, Nikolaev SI, Lukowski SW, Bazykin GA, Seplyarskiy VB, Ioannidis P, et al. 2014. Gene age predicts the strength of purifying selection acting on gene expression variation in humans. Am J Hum Genet 95: 660–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prandini P, Deutsch S, Lyle R, Gagnebin M, Delucinge Vivier C, Delorenzi M, Gehrig C, Descombes P, Sherman S, Dagna Bricarelli F, et al. 2007. Natural gene-expression variation in Down syndrome modulates the outcome of gene-dosage imbalance. Am J Hum Genet 81: 252–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sailani MR, Makrythanasis P, Valsesia A, Santoni FA, Deutsch S, Popadin K, Borel C, Migliavacca E, Sharp AJ, Duriaux Sail G, et al. 2013. The complex SNP and CNV genetic architecture of the increased risk of congenital heart defects in Down syndrome. Genome Res 23: 1410–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons YB, Turchin MC, Pritchard JK, Sella G. 2014. The deleterious mutation load is insensitive to recent population history. Nat Genet 46: 220–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnov DA, Cheung VG. 2008. ATM gene mutations result in both recessive and dominant expression phenotypes of genes and microRNAs. Am J Hum Genet 83: 243–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohail M, Vakhrusheva OA, Sul JH, Pulit SL, Francioli LC, van den Berg LH, Veldink JH, de Bakker PIW, Bazykin GA, Kondrashov AS, et al. 2017. Negative selection in humans and fruit flies involves synergistic epistasis. Science 356: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg J, Honti F, Meader S, Webber C. 2015. Haploinsufficiency predictions without study bias. Nucleic Acids Res 43: e101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. 2003. Statistical significance for genomewide studies. Proc Natl Acad Sci 100: 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudmant PH, Mallick S, Nelson BJ, Hormozdiari F, Krumm N, Huddleston J, Coe BP, Baker C, Nordenfelt S, Bamshad M, et al. 2015. Global diversity, population stratification, and selection of human copy number variation. Science 349: aab3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulem P, Helgason H, Oddson A, Stefansson H, Gudjonsson SA, Zink F, Hjartarson E, Sigurdsson GT, Jonasdottir A, Jonasdottir A, et al. 2015. Identification of a large set of rare complete human knockouts. Nat Genet 47: 448–452. [DOI] [PubMed] [Google Scholar]
- Sullivan KD, Lewis HC, Hill AA, Pandey A, Jackson LP, Cabral JM, Smith KP, Liggett LA, Gomez EB, Galbraith MD, et al. 2016. Trisomy 21 consistently activates the interferon response. eLife 5: e16220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Végh MJ, Rausell A, Loos M, Heldring CM, Jurkowski W, van Nierop P, Paliukhovich I, Li KW, del Sol A, Smit AB, et al. 2014. Hippocampal extracellular matrix levels and stochasticity in synaptic protein expression increase with age and are associated with age-dependent cognitive decline. Mol Cell Proteomics 13: 2975–2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu V, Verster AJ, Schertzberg M, Chuluunbaatar T, Spensley M, Pajkic D, Hart GT, Moffat J, Fraser AG. 2015. Natural variation in gene expression modulates the severity of mutant phenotypes. Cell 162: 391–402. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zhang J. 2011. Impact of gene expression noise on organismal fitness and the efficacy of natural selection. Proc Natl Acad Sci 108: E67–E76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf YI, Novichkov PS, Karev GP, Koonin EV, Lipman DJ. 2009. The universal distribution of evolutionary rates of genes and distinct characteristics of eukaryotic genes of different apparent ages. Proc Natl Acad Sci 106: 7273–7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Chen Y, Ayub Q, Huang N, Ball EV, Mort M, Phillips AD, Shaw K, Stenson PD, Cooper DN, et al. 2012. Deleterious- and disease-allele prevalence in healthy individuals: insights from current predictions, mutation databases, and population-scale resequencing. Am J Hum Genet 91: 1022–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahavi A. 1975. Mate selection—a selection for a handicap. J Theor Biol 53: 205–214. [DOI] [PubMed] [Google Scholar]
- Zhang YE, Vibranovski MD, Krinsky BH, Long M. 2010. Age-dependent chromosomal distribution of male-biased genes in Drosophila. Genome Res 20: 1526–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.