Genome Research 27: 677–685 (2017)
Due to a formula error, Table 2 in the above article displayed incorrect values of the observed structural variants. The authors would like to correct these values and the text referring to them (page 680), which should read as follows:
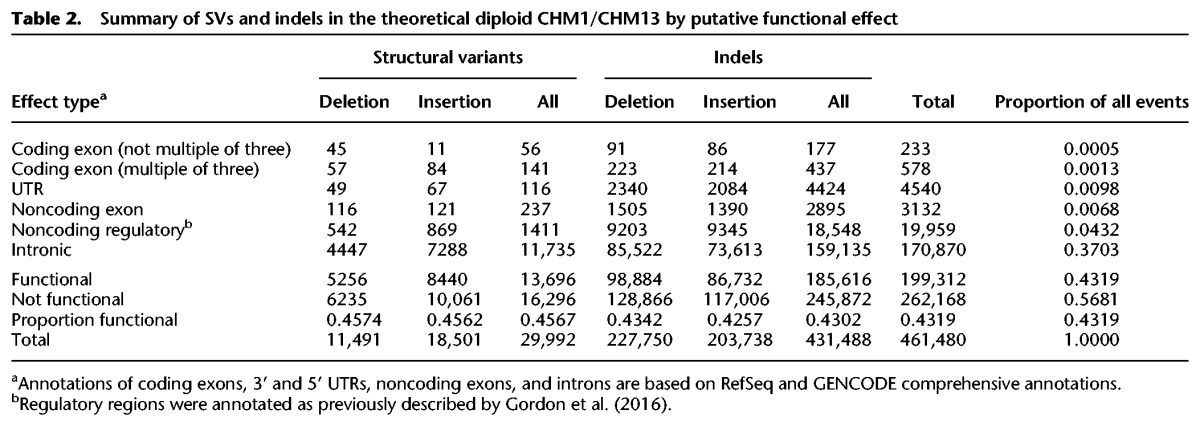
Of all 461,480 insertions and deletions detected as SVs or indels, only 1.8% of events occurred within a GENCODE or RefSeq coding exon, noncoding exon, or untranslated region (UTR) (Table 2). An additional 4.3% of events occurred in predicted noncoding regulatory regions, including DNase hypersensitivity sites, promoters (H3K27ac), and enhancers (H3K4me3), while 37.0% of events occurred in introns (Harrow et al. 2012).
Table 2.
Summary of SVs and indels in the theoretical diploid CHM1/CHM13 by putative functional effect
The authors apologize for any inconvenience. Table 2 and its associated text have been updated online to reflect the corrected values.
doi: 10.1101/gr.233007.117


