Abstract
Phosphorylation of histone H3 at serine 10 (H3S10ph) by Aurora kinases plays an important role in mitosis; however, H3S10ph also marks regulatory regions of inducible genes in interphase mammalian cells, implicating mitosis-independent functions. Using the fluorescent ubiquitin-mediated cell cycle indicator (FUCCI), we found that 30% of the genome in interphase mouse embryonic stem cells (ESCs) is marked with H3S10ph. H3S10ph broadly demarcates gene-rich regions in G1 and is positively correlated with domains of early DNA replication timing (RT) but negatively correlated with H3K9me2 and lamin-associated domains (LADs). Consistent with mitosis-independent kinase activity, this pattern was preserved in ESCs treated with Hesperadin, a potent inhibitor of Aurora B/C kinases. Disruption of H3S10ph by expression of nonphosphorylatable H3.3S10A results in ectopic spreading of H3K9me2 into adjacent euchromatic regions, mimicking the phenotype observed in Drosophila JIL-1 kinase mutants. Conversely, interphase H3S10ph domains expand in Ehmt1 (also known as Glp) null ESCs, revealing that H3S10ph deposition is restricted by H3K9me2. Strikingly, spreading of H3S10ph at RT transition regions (TTRs) is accompanied by aberrant transcription initiation of genes co-oriented with the replication fork in Ehmt1−/− and Ehmt2−/− ESCs, indicating that establishment of repressive chromatin on the leading strand following DNA synthesis may depend upon these lysine methyltransferases. H3S10ph is also anti-correlated with H3K9me2 in interphase murine embryonic fibroblasts (MEFs) and is restricted to intragenic regions of actively transcribing genes by EHMT2. Taken together, these observations reveal that H3S10ph may play a general role in restricting the spreading of repressive chromatin in interphase mammalian cells.
H3S10ph is a highly conserved histone post-translational modification (PTM); however, the influence of this mark on chromatin structure remains elusive, as H3S10ph is observed in two seemingly paradoxical contexts—broadly marking condensed chromosomes during mitosis and transiently marking the promoters of inducible genes in interphase. By early metaphase, histone H3 is extensively phosphorylated by Aurora kinase B (AURKB) at S10 and S28 (Wei et al. 1998), a process essential for the initiation of chromosome condensation and transition through mitosis (Van Hooser et al. 1998). Conversely, during interphase, activation of the MAPK pathway by mitogens or stress promotes rapid H3S10 phosphorylation at the promoter and enhancer regions of immediate early genes (Mahadevan et al. 1991; Cheung et al. 2000), stimulating the release of paused RNA POL II and P-TEFb-dependent transcription elongation (Zippo et al. 2009). While several nuclear kinases have been implicated as the downstream effectors of such inducible H3S10 phosphorylation at specific loci, including the JIL-1 homologs RPS6KA5 (also known as MSK1) and RPS6KA4 (also known as MSK2), RPS6KA3 (also known as RSK2), CHUK (also known as IKKα), and PIM1 (Sassone-Corsi et al. 1999; Thomson et al. 1999; Anest et al. 2003; Zippo et al. 2009), a genome-wide view of the dynamics of H3S10ph during the mammalian cell cycle has not been reported.
H3S10ph inhibits the lysine methyltransferase (KMT) activity of mammalian Su(var)3-9 family members in vitro, including SUV39H1, SETDB1, and EHMT2 (also known as G9a), and promotes displacement of Chromobox (CBX) proteins CBX1, CBX3, and CBX5 (also known as HP1β, HP1α, and HP1γ) from H3K9 methylated regions (Rea et al. 2000; Schultz 2002; Fischle et al. 2005; Rathert et al. 2008). While histone peptide binding studies have shown that H3K9me3 binding modules vary in their sensitivity to the presence of H3S10ph, the chromodomain of CBX1 and ankyrin repeats of EHMT1 and EHMT2 are inhibited by H3S10ph-marked peptides (Fischle et al. 2005; Rathert et al. 2008). Such antagonism between H3S10ph and methylation of the adjacent H3K9, termed the “phospho-methyl switch” (Fischle et al. 2005), has been best characterized in the context of mitosis; however, the extent of crosstalk between H3S10ph and H3K9me2/3 in interphase mammalian cells remains unexplored.
Interplay between H3S10ph and H3K9 methylation is well documented in Drosophila. The essential H3S10 kinase JIL-1, isolated originally as an anti-morph of position effect variegation (PEV), associates with chromatin throughout the cell cycle (Wang et al. 2001) and localizes to gene-dense regions, forming “blanket” domains over gene-bodies (Regnard et al. 2011). Notably, chromosome morphology is severely impacted in a JIL-1 hypomorph, with widespread loss of euchromatin interbands, despite normal mitotic H3S10ph (Jin et al. 1999). Furthermore, H3K9me2 and HP1 spread ectopically into euchromatin polytene bands, suggesting that H3S10ph functions to reinforce euchromatin boundaries in flies by antagonizing heterochromatin propagation (Zhang et al. 2006). Intriguingly, the lethality observed in JIL-1 null mutants is rescued in Su(var)3-9 double mutants, indicating that the aberrant gene expression observed in JIL-1 hypomorphs may be a consequence of ectopic H3K9 methylation. Indeed, depletion of interphase H3S10ph in the JIL-1 hypomorph leads to global redistribution of H3K9me2, with gain or loss of heterochromatin corresponding with down- and up-regulation of associated genes, respectively (Cai et al. 2014). Taken together, these observations reveal that crosstalk between H3K9me2 and H3S10ph plays a pivotal role in determining large-scale chromatin structure and, in turn, transcription potential in interphase Drosophila cells.
In mammals, H3K9me2 is deposited in euchromatic regions by EHMT2 and its obligate paralog, EHMT1 (Shinkai and Tachibana 2011). In mouse ESCs, H3K9me2 is enriched in megabase (Mb)-scale lamin-associated domains (LADs) that are gene-poor and late-replicating (Guelen et al. 2008; Yokochi et al. 2009; Lienert et al. 2011). Furthermore, EHMT2 and EHMT1 are required for repression of a number of late-replicating genes (Yokochi et al. 2009), including several gene clusters on the X Chromosome (Shinkai and Tachibana 2011), as well as specific families of retroelements (Maksakova et al. 2013). Interestingly, a recent report examining randomly integrated reporter constructs in ESCs revealed that transcription permissive integration sites cluster in large megabase-scale domains that are anti-correlated with H3K9me2 and LADs, indicating that the transcription potential of transgenes is dependent on chromatin features of the integration site (Akhtar et al. 2013). However, whether H3S10ph and H3K9me2 are mutually antagonistic in interphase mammalian cells has not been addressed, due largely to contaminating AURKB-dependent H3S10ph in mitotic cells. We employed the fluorescent ubiquitin-mediated cell cycle indicator (FUCCI) cell cycle reporter system or an Aurora kinase inhibitor, histone mutant overexpression, and genetic knockout approach to study the genome-wide distribution of H3S10ph in interphase ESCs and MEFs and the interplay between this mark and H3K9me2.
Results
H3S10ph in interphase ESCs
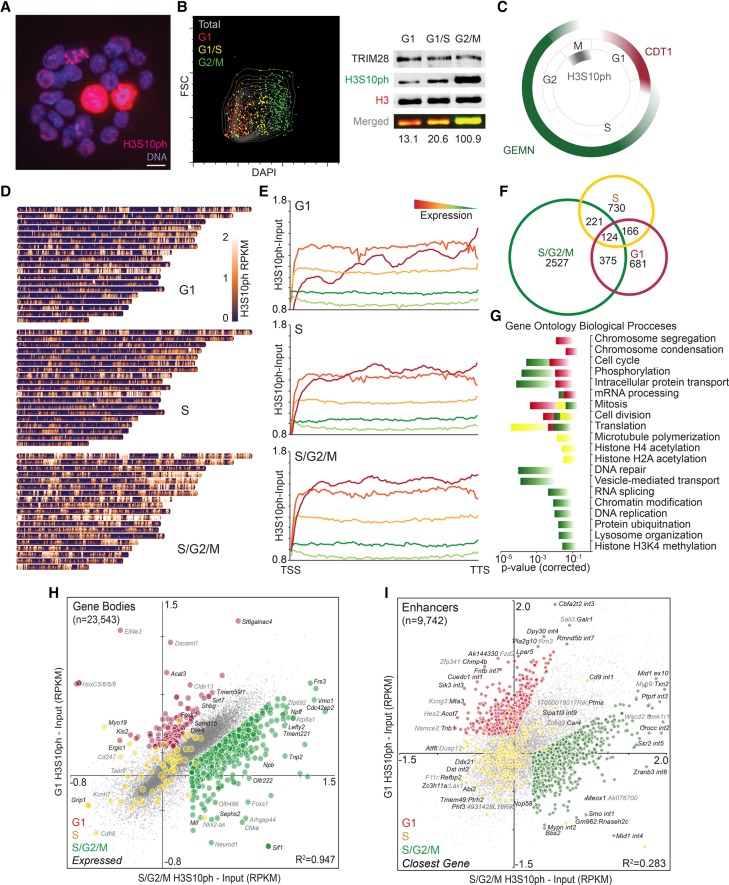
AURKB activity is recognized as a canonical marker of mitotic chromosomes, initiating phosphorylation of H3S10ph in G2/M at peri-centromeric heterochromatin and gradually modifying entire chromosomes as mitosis progresses. Indeed, staining of asynchronous ESCs cultures with a previously characterized (Hayashi-Takanaka et al. 2009) H3S10ph-specific monoclonal antibody (CMA311), which is insensitive to the presence of H3K9 mono- or dimethylation (Supplemental Table S1), reveals intense signal at chromocenters in G2 and condensed chromosomes in mitotic cells (Fig. 1A). However, abundant but faintly stained small H3S10ph foci are also clearly present in euchromatic and nucleolar compartments of interphase ESCs, consistent with previous studies using polyclonal or monoclonal H3S10ph antibodies (Sassone-Corsi et al. 1999; Fazzio et al. 2008; Hayashi-Takanaka et al. 2009). While considerable heterogeneity in interphase H3S10ph signal is apparent across ESC nuclei, presumably reflecting dynamic regulation of this labile PTM, these observations indicate that H3S10ph likely marks numerous genomic regions outside of mitosis in cycling ESCs. Indeed, quantitative Western blotting of extracts isolated from G1, G1/S, and G2/M populations (sorted by DNA content) clearly reveals the presence of H3S10ph in the G1-sorted fraction, albeit at lower levels than detected in G2/M cells (Fig. 1B).
Figure 1.
Genome-wide characterization of cell cycle-specific H3S10ph. (A) H3S10ph immunostaining of asynchronous WT TT2 ESCs, counterstained with Hoechst 33342. Scale bar, 10 µm. (B) G1, G1/S, and G2/M ESCs were isolated by FACS based on DNA content (DAPI-staining) and a flow cytometry plot showing the merge of each post-sort purity analysis is shown. Western analysis of whole-cell extract was conducted using H3S10ph- and H3-specific antibodies, and H3S10ph levels were quantified on the Odyssey infrared imager by normalizing to H3 fluorescence level. The percentage of H3S10ph/H3 is shown for each extract. TRIM28 served as a second loading control. (C) Schematic of the FUCCI reporter system for dissection of cell cycle-specific H3S10ph. CDT1-mKO and GEMN-Venus are fluorescent markers of G1 and S/G2/M, respectively. (D) Heat map depicting H3S10ph enrichment across all murine autosomes in G1, S, and S/G2/M sorted fractions. (E) Metagene analysis of normalized H3S10ph enrichment in ESCs (input-subtracted reads per kb mapped [RPKM]) over all ENSEMBL annotated gene bodies, clustered by expression levels in quintiles. (F) Venn diagram of genes enriched for H3S10ph (greater than twofold over input in gene bodies) in G1, S, and S/G2/M sorted fractions. (G) Gene Ontology of Biological Processes from genes identified as in E (Bonferroni corrected P < 0.05). (H) Pair-wise comparison of H3S10ph enrichment (input-subtracted RPKM) over all gene bodies in G1 vs. S/G2/M sorted fractions. Colored dots represent genes with cell cycle skewing (z > 0.5). R2 = Pearson correlation. (I) Pair-wise comparison of H3S10ph enrichment (input-subtracted RPKM) over annotated enhancer regions in G1 vs. S/G2/M sorted fractions. Colored dots represent sites with cell cycle skewing as in G. A subset of enhancers is labeled with the overlapping gene(s), exon (ex), intron (int), or flanking gene(s) with the closest gene in black.
To generate high-quality maps of H3S10ph in interphase cells in the absence of any confounding effects associated with chemical synchronization, we employed the dual-color FUCCI cell cycle reporter system (Fig. 1C), which allows for cell sorting based on fluorescent tagging of the labile cell cycle regulated proteins CDT1 and GEMN (Sakaue-Sawano et al. 2008). We derived a stable polyclonal ESC line expressing hCDT1-monomeric Kusabira Orange (mKO) and hGEMN-Venus from a multicistronic construct separated by T2A cleavage tag (Falconer et al. 2012) and validated the fidelity of these FUCCI fluorescent tags as cell cycle stage reporters in ESCs using Hoechst 33342 DNA counterstain (Supplemental Fig. S1A,B). Consistent with previous reports (Wilson et al. 2008), the mKO+Venus− subpopulation of these “FUCCI ESCs” is composed of cells predominantly in G1/early S, whereas the mKO−Venus+ subpopulation is enriched for cells in late S and G2/M phases.
We then sorted mKO+, mKO+Venus+, and Venus+ cells to >90% purity, and 106 nuclei, representing G1, S, S/G2/M stage fractions, respectively, were crosslinked and MNase-digested, followed by ChIP with the aforementioned H3S10ph antibody. Quantification of unamplified nucleosomal DNA recovered by ChIP revealed an approximately sixfold higher level of H3S10ph in the S/G2/M fraction (Supplemental Fig. S1C), reflecting the widespread deposition of this mark in mitotic cells. However, consistent with fluorescence microscopy and Western analysis of synchronized HeLa cells (Rothbart et al. 2015), the presence of nucleosomal DNA in chromatin isolated from G1- and S-phase cells indicates that H3S10ph is also present in interphase cells, albeit at lower levels. Indeed, ChIP-qPCR of several randomly chosen loci representing both genic and intergenic regions reveals that these regions are enriched for H3S10ph in G1-sorted cells (Supplemental Fig. S1D). Depending on the region analyzed, the percentage of input DNA immunoprecipitated ranged from ∼8% to 50%, which is well above the mitotic index of ESCs, ruling out the possibility that such enrichment is simply a result of contamination of the sorted G1- or S-phase populations with mitotic cells. Interestingly, G1-sorted cells showed higher levels of enrichment of H3S10ph than S-phase sorted cells, consistent with histone turnover during DNA replication. While H3S10ph enrichment is also detected in the S/G2/M-sorted cells by ChIP-qPCR at all tested loci, the absolute levels of enrichment are significantly lower than observed in G1, likely due to saturation of the antibody by the pervasive phosphorylation of H3S10 during mitosis.
Cell cycle dynamics of interphase H3S10ph
Evidence for the presence of H3S10ph in interphase prompted us to survey the landscape of H3S10ph genome-wide via ChIP-seq in G1, S, and S/G2/M purified ESCs. Consistent with bulk chromatin quantification, S/G2/M cells show widespread H3S10ph enrichment (Fig. 1D), reflecting the global accumulation of H3S10ph during mitosis. Notably, a significant fraction of the genome was also enriched with H3S10ph relative to input in G1 and S phases (Supplemental Fig. S1E), indicating that this mark may play a broader role in transcriptional regulation and/or other chromatin-related pathways in interphase than previously recognized. Input samples showed consistent recovery across the cell cycle, indicating that the dynamic profile of H3S10ph is unlikely to be an artifact of differential efficiency of nucleosome recovery in these cell cycle fractions.
To characterize the genomic distribution of H3S10ph in greater detail, we determined whether H3S10ph marks distinct DNA sequence features across cell cycle fractions by comparing input-normalized H3S10ph at genes and repetitive sequences. H3S10ph was relatively more enriched at genes than repetitive sequences across all cell cycle stages, as measured in 1-kb bins (Supplemental Fig. S1F). Metagene analysis of H3S10ph enrichment in FUCCI-sorted ESCs revealed that H3S10ph broadly marks gene bodies in interphase and is generally positively correlated with mRNA levels (Fig. 1E). However, toward the 5′ end of genes in G1-sorted cells, the most highly expressed quintile shows moderately lower levels of enrichment than the second and third quintile, perhaps due to a particularly high level of nucleosomal turnover and/or elevated H3K9ac in this region at highly expressed genes at this cell cycle stage. Indeed, the H3S10ph-specific antibody used in this experiment is inhibited by the presence of H3K9ac (Supplemental Table S1), a mark generally detected in the promoter regions of active genes. In addition, H3S10ph levels in the intragenic regions of expressed genes were modestly lower in S-phase relative to G1 and G2/M cells (Supplemental Fig. S1D,E), presumably due to replication-coupled deposition of nascent unmodified H3 during early S-phase and re-establishment of H3S10ph, in G2/M.
To identify genes that may be differentially marked at distinct cell cycle stages, we analyzed enrichment levels across ENSEMBL annotated genes in the G1 versus S/G2/M data sets. Notably, 77% of all annotated genes showed H3S10ph enrichment relative to the input control. While H3S10ph levels at most gene bodies were highly correlated across the cell cycle (R2= 0.947), a minority of genes show clear cell cycle-specific skewing of H3S10ph enrichment. To ascertain the function of those genes showing a cell cycle enrichment bias, genes with the highest levels of H3S10ph enrichment were identified (greater than or equal to twofold enriched over matched input reads per kb mapped [RPKM]), and Gene Ontology (GO) analysis was conducted. S/G2/M cells include a much larger subset of differentially marked genes (n = 3247) than G1 (n = 1346) or S (n = 1241) cells (Fig. 1F), as expected given that H3S10ph levels increase dramatically with the onset of mitosis. Across all cycle stages, heavily phosphorylated genes are enriched (Bonferroni-corrected P-value < 0.05) in housekeeping functions, specifically cell cycle-regulated biological processes (Fig. 1G). In contrast, genes enriched with H3S10ph in G1 were significantly overrepresented in mitosis-related GO terms, such as chromosome condensation and segregation, whereas genes with functions in DNA repair as well as DNA replication were prominent in S/G2/M. Furthermore, several GO terms were enriched in S compared to G1 and S/G2/M, including translation, microtubule polymerization, and histone acetylation. Taken together, these observations suggest a link between enrichment of H3S10ph in interphase and transcriptional activity, which for at least a subset of genes is apparently dynamically regulated during the cell cycle (Sasagawa et al. 2013).
To further characterize regions showing cell cycle-variable H3S10ph enrichment in ESCs, we first scored genomic bins that exhibit skewed H3S10 phosphorylation and found that 10.4% of these differentially phosphorylated regions are within 200 bp of a DNase I hypersensitive site (DHS), compared to 4.6% overlapping DHS in constitutively H3S10 phosphorylated regions (Supplemental Fig. S1G,H). To determine whether sites exhibiting dynamic H3S10ph include putative enhancers, ENCODE DHS coordinates were filtered for regions enriched for H3K4me1 and H3K27ac, a signature of active enhancers. Subsequent pair-wise comparisons of normalized H3S10ph in G1 vs. S/G2/M populations at the 9742 active enhancers identified in ESCs revealed much greater cell cycle variability in H3S10ph enrichment (R2= 0.283) than observed in gene bodies (Fig. 1H,I). Stage-specific H3S10ph enrichment (normalized RPKM > 0.5, z-score > 0.5) was observed at 1044 enhancer sites in G1, S, or S/G2/M, while 595 such sites were constitutively marked in all three fractions. To annotate gene targets of cell cycle-biased distal enhancers in ESCs, we parsed enhancers that physically contact promoters, as determined by Chromatin Interaction Analysis by Paired-End Tag Sequencing (Sahlén et al. 2015). Gene Ontology analysis of the predicted gene targets revealed enrichment for housekeeping functions, as observed for GO analysis of H3S10ph-marked gene bodies (Supplemental Fig. S1I), with the distinct emergence of fatty acid biosynthesis in G1, DNA replication and protein ubiquitination in S, and response to glucose, nutrients, and drugs in S/G2/M. Taken together, these observations reveal that, across the cell cycle, H3S10ph deposition and/or removal is dynamic in gene bodies as well as distal regulatory regions, perhaps reflecting transcriptional regulation at these stages.
Interphase H3S10ph is not dependent on AURKB
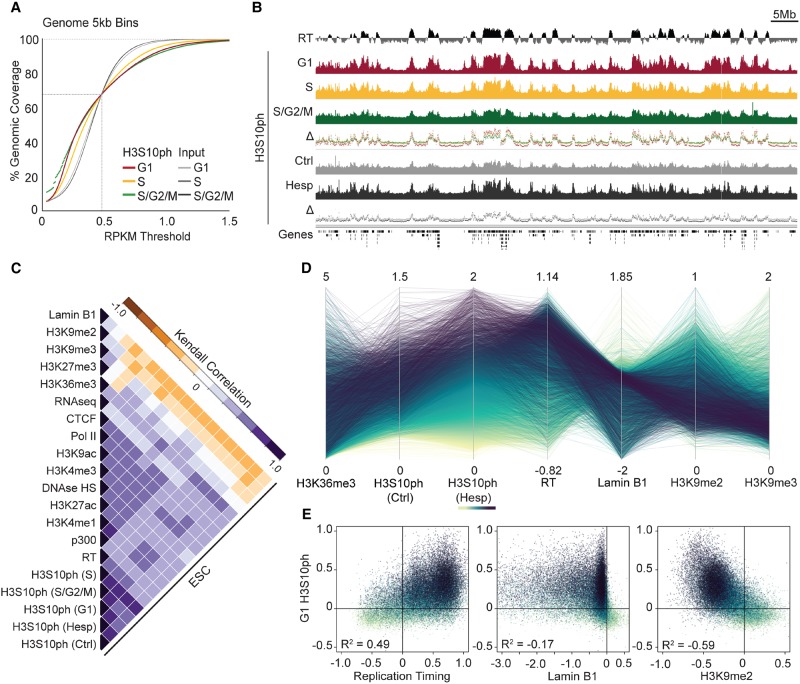
Curiously, as opposed to other PTMs positively associated with transcriptional activity, inspection of individual loci reveals that interphase H3S10ph is not limited to gene bodies or enhancers but also broadly marks intergenic regions (Supplemental Fig. S1G). Indeed, H3S10ph at gene-dense regions encompasses Mb-scale domains in interphase, covering ∼30% of the genome (Fig. 2A,B). To determine whether these broad domains reflect incomplete de-phosphorylation of H3S10ph deposited by AURKB during mitosis, we treated ESCs with Hesperadin (Hesp), an inhibitor of AURKB kinase (Fischle et al. 2005). As expected, a 3-h treatment with 200 nM Hesp effectively eliminated >98% of mitotic cells, as measured by DNA content and H3S10ph signal using flow cytometry (Supplemental Fig. S2A). Nevertheless, quantitative Western blotting revealed that Hesp-treated cells retain ∼60% of the level of H3S10ph in control asynchronous ESCs, likely reflecting AURKB-independent activity (Supplemental Fig. S2B). Furthermore, H3S10ph ChIP-seq on control and Hesp-treated ESCs revealed reduced global enrichment of H3S10ph following AURKB inhibition, as expected, but increased prominence of putative interphase H3S10ph domains, recapitulating the profile observed in G1-sorted cells (Fig. 2B; Supplemental Fig. S2C). Based on these observations, we conclude that the euchromatic H3S10ph domains observed in interphase ESCs are deposited by a serine kinase or kinases other than AURKB.
Figure 2.
Interphase H3S10ph forms domains highly concordant with replication timing. (A) Cumulative distribution frequency plot of ESC H3S10ph ChIP vs. input libraries in 5-kb bins. (B) Genome browser screenshot of H3S10ph enrichment and replication timing (RT) (Hiratani et al. 2010) in TT2 ESCs across Chr 18. (C) Kendall correlation matrix of H3S10ph with chromatin features (see Supplemental Table S2 for data sets analyzed) across 5-kb genomic bins in ESCs, following unsupervised hierarchical clustering. H3S10ph ESC data sets include sorted FUCCI subpopulations (S, S/G2/M, and G1), Hesp-treated, and vehicle control (Ctrl). (D) Parallel coordinate comparison of H3S10ph enrichment (RPKM) with histone marks H3K36me3, H3K9me2, and H3K9me3 as well as Lamin B1 and RT in randomly sampled 5-kb bins. Heat map color-coding is based on H3S10ph enrichment in Hesp-treated ESCs. Note the strong positive correlation between H3S10ph in interphase and RT and the negative correlation with H3K9me2. (E) Pair-wise comparisons of G1 H3S10ph enrichment with RT, LADs, and H3K9me2 (input subtracted RPKM) enrichment. Randomly sampled 5-kb bins are colored as in D. R2 = Pearson's correlation.
H3S10ph and H3K9me2 partition the genome into early- and late-replicating domains
To further characterize the chromatin state of regions marked with H3S10ph in interphase, we intersected the genome-wide distribution of this mark with additional chromatin features in ESCs, including those generated by the ENCODE Consortium (Supplemental Table S2; Yue et al. 2014). Given that H3S10ph domains in interphase ESCs were relatively broad, we used a tiling bin size of 5 kb. As expected, H3S10ph shows a positive correlation with RNA Pol II, transcript level (RNA-seq), and transcription-associated PTMs (H3K4me3, H3K9ac, H3K36me3) as well as enhancer PTMs (H3K4me1, H3K27ac) (Fig. 2C). However, consistent with the observation that H3S10ph is also enriched in intergenic regions of gene-rich genomic domains, 42% of interphase H3S10ph marked bins do not coincide with annotated genes. Notably, these intergenic H3S10ph-marked regions are almost universally replicated relatively early in S-phase, with boundaries of interphase H3S10ph domains closely aligning with replication timing (RT) transition regions (TTRs) (Fig. 2B). Indeed, RT was concordant with H3S10ph enrichment (R2= 0.49) genome-wide (Fig. 2D), and H3S10ph in G1 cells scales quantitatively with RT progression (Fig. 2E). To determine whether H3S10ph in interphase is specific to cells undergoing DNA replication, we costained proliferating ESCs with H3S10ph and the S-phase marker PCNA (Supplemental Fig. S2D). While replicating/PCNA+ nuclei were generally marked with punctate H3S10ph foci, PCNA− nuclei also harbored punctate H3S10ph foci, consistent with the observation that H3S10ph is present in G1 cells, i.e., prior to the onset of DNA replication.
In contrast, regions enriched for H3S10ph in G1 show a clear discordance with lamin-associated domains (Fig. 2C–E), shown previously to replicate relatively late in S-phase (Kind et al. 2015). Regions enriched for H3S10ph are also anti-correlated with H3K9me2, a mark deposited by the KMTases EHMT2 and EHMT1 at late-replicating genomic regions (Yokochi et al. 2009; Kind and van Steensel 2010) and, to a lesser extent, H3K9me3, which marks constitutive heterochromatin and specific retrotransposons in ESCs (Karimi et al. 2011). As H3K9me2 is specifically associated with LADs (Kind et al. 2013), we focused on the relationship between H3S10ph and H3K9me2. Consistent with the Mb-scale domains of H3K9me2 observed in ESCs via ChIP-chip (Lienert et al. 2011), ChIP-seq revealed that H3K9me2 demarcates large Mb domains, covering ∼40% of the mappable genome. As predicted, this mark is clearly anti-correlated with interphase H3S10ph (R2= 0.59, 5-kb bins) (Fig. 2E), indicating that H3S10ph may antagonize H3K9me2 deposition, and vice versa, in interphase ESCs. While the apparent anti-correlation may reflect epitope masking of the H3K9me2 antibody by H3S10ph (Duan et al. 2008; Rothbart et al. 2015), such a technical artifact is unlikely, as H3S10ph covers ∼30% of the genome and quantitative mass spectrometry analysis of WT ESCs revealed that under 20% of bulk H3 is dimethylated at H3K9 (Kubicek et al. 2007).
Expression of nonphosphorylatable H3.3 promotes aberrant accumulation of H3K9me2
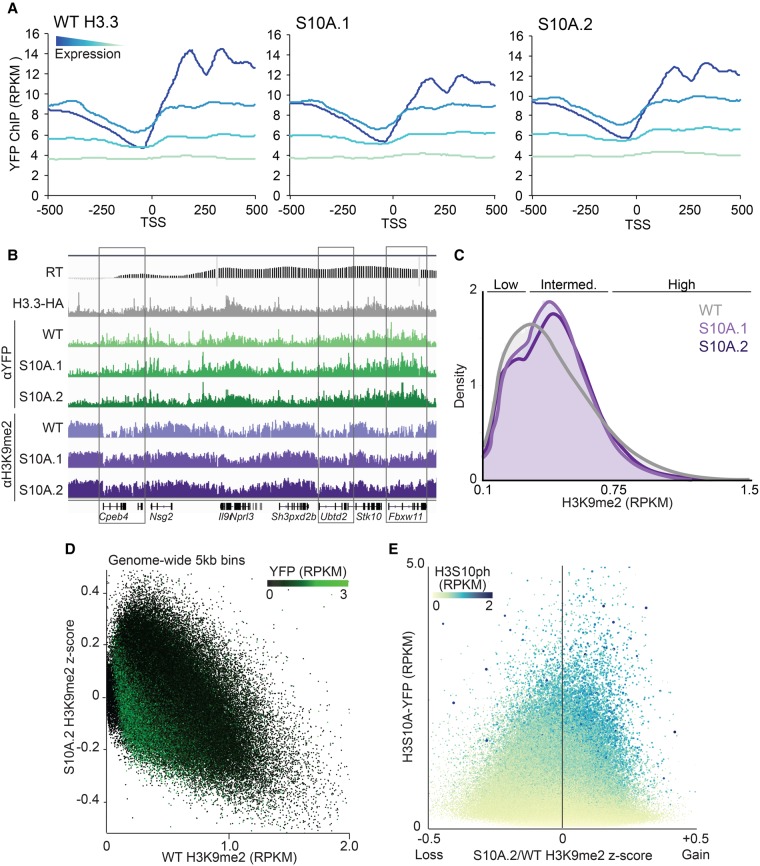
To determine whether H3S10ph influences H3K9me2 deposition, we generated ESC lines stably expressing tagged H3.3 and H3.3S10A, which mimics the loss of phosphoserine (Supplemental Fig. S3A). We reasoned that overexpression of the nonphosphorylatable H3.3 variant would mitigate the likelihood of mitotic defects, as H3.3 is deposited in a replication-independent pathway at specific genomic regions, including transcribed gene bodies (Ahmad and Henikoff 2002; Chen et al. 2013), which are enriched for interphase H3S10ph. Indeed, two independent clonal lines, “S10A.1” and “S10A.2,” which stably express the mutant H3.3-YFP transgene (Supplemental Fig. S3B,C), showed mitotic progression similar to that of the line expressing the wild-type (WT) H3.3-YFP transgene. To determine whether WT H3.3-YFP and H3.3S10A-YFP are targeted to intragenic regions of active genes, we performed ChIP-seq using a YFP-specific antibody. Metagene analysis revealed that the level of exogenous H3.3 enrichment in gene bodies increases with increasing transcription level (Fig. 3A), as expected. Importantly, WT and S10A mutant H3.3-YFP show similar enrichment over active gene bodies and genome-wide (Fig. 3A; Supplemental Fig. S3D), revealing that the incorporation of exogenous H3.3 is not dependent upon H3S10 per se. Furthermore, the genomic distribution of YFP-tagged H3.3 is similar to that of HA-tagged H3.3 (Elsässer et al. 2015), indicating that YFP-tagging does not perturb intragenic deposition of H3.3 (Fig. 3B).
Figure 3.
Overexpression of H3S10A in ESCs promotes H3K9me2 accumulation in early-replicating regions. (A) ChIP enrichment of YFP (RPKM) around transcription start sites (TSS ±500 bp) in WT and S10A H3.3-YFP-expressing lines, clustered by genic expression quartiles. (B) Genome browser screen shot spanning Cpeb4 to Fbxw11 loci on Chromosome 11 (31,696,800–32,668,200) with YFP and H3K9me2 ChIP enrichment in H3.3-YFP WT and S10A mutant lines presented, as well as previously published RT (Yokochi et al. 2009) and H3.3-HA (Elsässer et al. 2015) tracks. (C) Histogram showing the global density distribution of H3K9me2 in H3.3-YFP WT and S10A lines. (D) 2D scatterplot comparing the gain or loss (z-score) of H3K9me2 in the S10A.2 mutant line with WT H3K9me2 enrichment levels (RPKM). (E) Comparison of YFP incorporation levels with the change in H3K9me2 enrichment in H3.3-YFP S10A.2 relative to WT cells (z-score). Heat map and data-point size represent the range of WT H3S10ph enrichment levels. Note that gain of H3K9me2 occurs predominantly within regions that are enriched for H3S10ph in WT ESCs.
To determine whether incorporation of H3.3S10A influences the genomic distribution of H3K9me2, we performed H3K9me2 ChIP-seq, as above, on each H3.3-YFP-expressing line. Strikingly, differences in the distribution of H3K9me2 are clearly apparent in the H3.3S10A-YFP lines relative to the H3.3-YFP WT line (Fig. 3B). Analysis of the genome in 5-kb bins reveals a clear increase in the fraction of bins with intermediate levels of H3K9me2 in the mutant expressing lines (Fig. 3C), with 3.8% and 7.7% genome-wide bins showing increased H3K9me2 (z > 0.2) in S10A.1 and S10A.2 clones, respectively. In both H3.3S10A clonal lines, the ectopic gain of H3K9me2 at regions normally harboring low levels of H3K9me2 is accompanied by reduced levels of this mark at regions normally showing high enrichment (Fig. 3C,D; Supplemental Fig. S3E). The majority of regions showing such a gain in H3K9me2 enrichment may reflect indirect effects, since H3.3S10A-YFP signal was not detected over most of these regions. To identify regions that may accumulate H3K9me2 as a direct consequence of S10A-YFP incorporation, we filtered genomic bins showing a gain in this mark based on YFP enrichment level (>1 RPKM), which yielded 1846 and 1007 regions in S10A.1 and S10A.2 lines, respectively, with 40.2% overlap between these biological replicates. Importantly, in the regions identified as “direct targets,” the majority (92%) reside within H3S10ph-rich domains in WT cells (Fig. 3E; Supplemental Fig. S3F). Thus, the disruption of H3S10 phosphorylation via incorporation of ectopic H3.3S10A leads to accumulation of H3K9me2 at regions enriched for H3S10ph in WT cells, indicating that interphase H3S10ph may insulate gene-dense regions from EHMT2/EHMT1-mediated H3K9me2 deposition.
Interphase H3S10ph domains expand upon loss of H3K9me2
To investigate whether H3K9me2 restricts the expansion of H3S10ph domains at TTRs, we performed ChIP-seq on previously characterized Ehmt1−/− ESCs (Shinkai and Tachibana 2011; Maksakova et al. 2013). As expected, widespread depletion of H3K9me2 was observed (Fig. 4A), with 40% of genomic bins, which generally overlap with late-replicating domains, showing reduced enrichment (z < −0.2). Regions that retain H3K9me2 in Ehmt1−/− ESCs are generally enriched for H3K9me3 in WT ESCs (Fig. 4B), likely reflecting the independent deposition of this mark by SETDB1 at LTR retrotransposons (Karimi et al. 2011). Remarkably, 70% of genomic bins that lose H3K9me2 in Hesp-treated Ehmt1−/− cells show a gain of H3S10ph (Fig. 4A,C). Notably, ectopic H3S10ph is clearly observed in regions adjacent to previously identified TTRs (Fig. 4D; Pope et al. 2014), consistent with spreading of H3S10ph into gene-poor, relatively late-replicating regions normally marked by H3K9me2 in interphase.
Figure 4.
EHMT1 restricts spreading of H3S10ph and inhibits aberrant transcription near TTRs. (A) Genome browser screenshot of a region on Chromosome 1 showing H3K9me2, Hesp-treated (H+) H3S10ph, and strand-specific RNA-seq in WT and Ehmt1−/− ESCs. (B) Heat maps of the change in H3K9me2 enrichment in Ehmt1−/− vs. WT (z-score), H3K9me3 enrichment (RPKM) in WT, and differential expression in Ehmt1−/− vs. WT ESCs for all LTR transposable element subfamilies present at >100 copies in the reference mouse genome. (C) Scatterplot showing the changes in H3K9me2 and H3S10ph coverage in Hesp-treated Ehmt1−/− ESCs relative to WT, overlaid with ESC RT status of the genomic bins as a heat map (red: early; gray: late). In Ehmt1−/− ESCs, late-replicating regions concurrently lose H3K9me2 and gain H3S10ph. (D) Heat map of H3S10ph enrichment in Hesp-treated WT and Ehmt1−/− ESCs over 500 kb centered on all RT transition boundaries in WT ESCs (RT data from Yokochi et al. [2009]). (E) Cumulative density plot of RNA-seq coverage on the plus (top plot) and minus (bottom plot) strands at TTR boundaries (0–500 kb) in WT, Ehmt1−/−, and Ehmt2−/− ESCs. Note the increase in RNA coverage in the mutant lines on the plus strand in early to late transition regions and the minus strand in late to early transition regions. (F) 2D scatterplot comparing genic transcription in Ehmt1−/− and WT ESCs. Note that many of the genes up-regulated in Ehmt1−/− ESCs reside within RT transition boundaries (0–500 kb) and show a RT orientation strand-bias consistent with that described in E.
Asymmetrical transcriptional initiation at TTRs in Ehmt2−/− and Ehmt1−/− ESCs
To determine whether ectopic H3S10ph at TTRs in H3K9me2-depleted ESCs is associated with changes in transcription, we conducted strand-specific mRNA-seq in Ehmt1−/− and Ehmt2−/− ESCs as well as the control WT parent line. In Ehmt2−/− and Ehmt1−/− ESCs , 950 and 791 genes were up-regulated (|z| > 0.8), respectively. Up-regulated genes were significantly enriched within a TTR (χ2 test, P < 0.001), with 37.7% and 40.1% observed, versus 19% expected (Supplemental Fig. S4A). Furthermore, transcripts present exclusively in Ehmt2−/− and Ehmt1−/− ESCs are apparent in regions extending up to ∼500 kb beyond the boundaries of the H3S10ph/gene-rich domains observed in WT cells (Fig. 4A). These aberrant transcripts show strong strand bias depending on the orientation with respect to RT (Fig. 4A), with reads mapping to the minus and plus strands at TTRs on the L-E and E-L boundaries of early replicating regions, respectively. While meta-analysis of TTRs reveals that this asymmetry is present in WT ESCs, consistent with the previously described co-orientation of genes with replication forks in human cells (Huvet et al. 2007; Petryk et al. 2016), this phenomenon is greatly enhanced in the Ehmt1−/− and Ehmt2−/− lines (Fig. 4E). Transcripts up-regulated in the vicinity of TTRs (±250 kb centered on the inflection point) include both repetitive sequences and genes, such as MageA and Rhox gene clusters (Fig. 4F), shown previously to be up-regulated in Ehmt2−/− and Ehmt1−/− ESCs and in vivo (Shinkai and Tachibana 2011; Auclair et al. 2016). Taken together, these results indicate that within TTRs, EHMT2/EHMT1 play a role in regulating transcription units co-oriented with the gross direction of DNA replication.
As EHMT2-dependent H3K9me2 was reported to mediate Pol II transcriptional termination via R-loop formation (Skourti-Stathaki et al. 2014), we considered the possibility that regions showing aberrant transcription may also be the result of inefficient termination of the last transcribed gene oriented toward the replication timing boundary within the early replicating domain. Analysis of H3K36me3 ChIP-seq data generated from Ehmt2−/−, Ehmt1−/−, and WT ESCs, however, revealed similar profiles of reduced H3K36me3 downstream from annotated genic transcription termination sites (TTSs) (Supplemental Fig. S4B,C), indicating that aberrant Pol II processivity beyond the TTS is unlikely to account for such ectopic transcription. On the other hand, ChIP-seq analysis of H3K4me3 in Ehmt2−/− and Ehmt1−/− ESCs revealed numerous ectopic peaks of H3K4me3 at TTRs in the mutant lines, likely reflecting aberrant transcription initiation (Supplemental Fig. S4B,D). To identify candidate “de novo” initiation sites, we first called all H3K4me3 peaks in WT or mutant lines. Of 55,000 sites in total, 7878 gained H3K4me3 in both Ehmt2−/− and Ehmt1−/− ESCs (R2= 0.992 between Ehmt2−/− and Ehmt1−/−), with 1081 such regions lacking H3K4me3 in the WT control (Supplemental Fig. S4D,E). As expected, de novo H3K4me3 peaks are frequently found adjacent to regions of increased RNA-seq coverage on the plus or minus strand (Supplemental Fig. S4E), indicative of the presence of ectopic promoters in Ehmt2−/− and Ehmt1−/− ESCs. While most of the shared and gained H3K4me3 peaks were found at annotated promoters, de novo sites were also frequently found within LTR and LINE sequences (Supplemental Fig. S4F) and were significantly closer to TTRs than constitutive H3K4me3 peaks (Supplemental Fig. S4G). Taken together, these observations indicate that transcription units near TTRs may be particularly prone to aberrant initiation following loss of H3K9me2.
H3S10ph domains are restricted to gene bodies in interphase MEFs
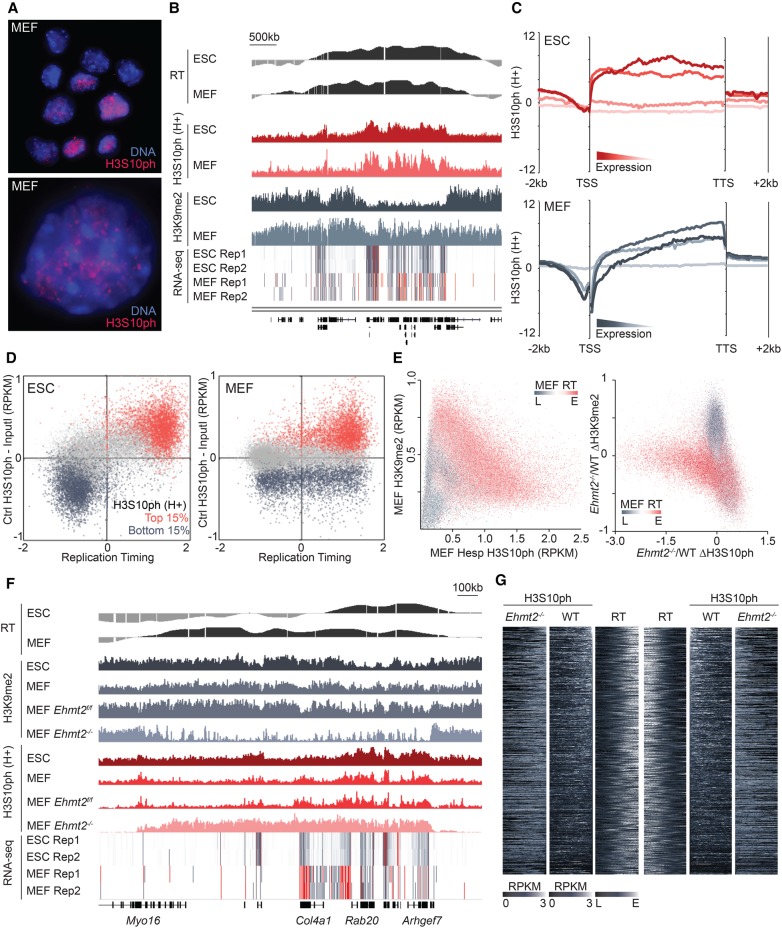
As somatic cells exhibit chromatin marks and replication timing profiles distinct from ESCs (Mikkelsen et al. 2007; Yue et al. 2014), we next determined whether early-replicating regions are also enriched in H3S10ph in interphase MEFs. As observed in ESCs, MEFs show heterogeneity in H3S10ph staining in interphase, with a subset of cells showing small but abundant foci exclusive of DAPI-dense regions (Fig. 5A). However, ChIP-seq of Hesp-arrested MEFs revealed distinct differences in H3S10ph distribution in early-replicating regions relative to ESCs, with extended domains in ESCs resolved into smaller domains in MEFs (Fig. 5B). Metagene analysis reveals prominent enrichment of H3S10ph in gene bodies, with a bias toward the 3′ end of transcribed genes (Fig. 5C). As in ESCs, gene body H3S10ph enrichment was generally higher in Hesp-treated than untreated control MEFs (Supplemental Fig. S5A), indicating that such regions are maintained by an AURKB-independent H3S10 kinase in interphase. However, consistent with ChIP-seq on untreated MEFs, H3S10ph in Hesp-treated MEFs does not mark broad domains. While H3S10ph covers ∼30% of the genome in interphase ESCs, H3S10ph is enriched at only 11% of all genomic bins (H3S10ph RPKM – input > 0.1) in Hesp-arrested MEFs. Inspection of the distribution of H3S10ph in relation to replication timing reveals that, unlike in ESCs, this mark is not uniformly distributed across early-replicating regions in MEFs (Fig. 5B). While H3S10ph-enriched regions are typically replicated early in MEFs, H3S10ph-depleted regions can be early- or late-replicating (Fig. 5D). Using 10-kb tiled genomic bins, H3S10ph is 94.0% sensitive and 97.9% specific as a predictor of early RT in ESCs; in stark contrast, in MEFs, sensitivity of H3S10ph to predict early RT is 91.7%, but specificity is only 46.7%. Notably, the subset of genomic regions that are late-replicating in ESCs but early-replicating in MEFs show relatively low levels of H3S10ph in the latter (Supplemental Fig. S5B), indicating that H3S10ph is not a universal mark of early RT.
Figure 5.
H3S10ph is restricted to genic regions in interphase MEFs. (A) H3S10ph immunostaining of interphase WT MEFs, counterstained with Hoechst 33342 for DNA, as in Figure 1A. (B) Genome browser screenshot showing RT, H3K9me2, and biological replicates of RNA-seq coverage in ESCs and MEFs as well as H3S10ph in Hesp-treated cells. (C) Metagene analysis of normalized H3S10ph enrichment (input subtracted RPKM) over all murine gene bodies (−2 kb-TSS-TTS+2 kb), clustered by RNA-seq expression levels into quartiles in the respective cell type. (D) Pair-wise comparison of H3S10ph with replication timing in asynchronous ESCs and MEFs using genome-wide 10-kb tiling bins. Red and blue data points correspond to bins that were in the top and bottom 15% of H3S10ph enrichment, respectively, in Hesp-treated cells. (E) Pair-wise comparison of H3S10ph (Hesp-treated) with H3K9me2 genome-wide (5-kb bins) in WT and Ehmt2−/− MEFs, overlaid with a heat map of MEF RT data with red and gray data points representing early- and late-replicating bins, respectively. (F) Genome browser snapshot of a MEF-specific early-replicon showing the loss of intergenic H3K9me2 and concurrent gain of H3S10ph in Ehmt2−/− MEFs. (G) Heat map of H3S10ph enrichment in Hesp-treated WT and Ehmt2−/− MEFs over 500 kb, centered on all RT transition boundaries in WT MEFs.
EHMT2-mediated H3K9me2 restricts H3S10ph to intergenic regions in MEFs
Meta-epigenomic comparisons in MEFs reveals many of the same trends observed in ESCs, with regions marked with H3S10ph in Hesp-treated cells showing a positive correlation with RT and other features of transcriptionally active regions and a negative correlation with H3K9me2, H3K27me3, and LADs (Supplemental Fig. S5C,D). Notably, unlike in ESCs, many intergenic early-replicating regions are depleted of H3S10ph in MEFs. As H3K9me2 and H3S10ph are anti-correlated in ESCs as well as in Drosophila, we surmised that the absence of H3S10ph in such regions in MEFs could be coincident with a reciprocal enrichment of H3K9me2. Indeed, early-replicating regions depleted of H3S10ph in MEFs are generally enriched for H3K9me2 (Fig. 5E) but depleted of H3K9me2 in ESCs (Supplemental Fig. S5E). The trend toward increased abundance of H3K9me2 in MEFs relative to ESCs is consistent with previous mass spectrometry data, which revealed a 10% greater abundance of H3K9me2 in MEFs (Kubicek et al. 2007). These results indicate that EHMT2 and/or EHMT1 mark a subset of intergenic regions in MEFs, leading to an apparent “fragmentation” of the broad interphase H3S10ph domains observed in ESCs.
Utilizing a previously characterized Ehmt2−/− MEF line (Leung et al. 2011), we tested whether EHMT2-mediated H3K9me2 restricts H3S10ph into localized peaks in MEFs. H3K9me2 ChIP-seq confirmed that EHMT2 ablation in MEFs causes global loss of H3K9me2, predominantly at early-replicating regions (Fig. 5E,F). Furthermore, Hesp-arrested Ehmt2−/− cells showed widespread expansion of H3S10ph into mega-base domains (Fig. 5F), with new boundaries aligning to MEF TTRs (Fig. 5G). The ectopic H3S10ph in Ehmt2−/− MEFs frequently marks regions beyond the boundaries of TTRs as identified in WT cells, however, indicating that the extent of H3S10ph spreading may be restricted by other chromatin features, such as H3K9me3. Taken together, these results indicate that H3S10ph and H3K9me2 are deposited over broad domains in interphase pluripotent and somatic mammalian cells in a mutually exclusive manner, likely as a consequence of direct reciprocal antagonism of the enzymes responsible.
Discussion
H3S10ph marks Mb-scale domains in interphase ESCs
H3S10ph has generally been recognized as a transient modification in interphase mammalian cells, marking specific gene promoter and enhancer regions following induction with external growth factors or stress (Bode 2005; Wang and Higgins 2013). Our genome-wide analysis of H3S10ph reveals that this PTM is not limited to transcriptional regulatory regions in interphase ESCs but rather broadly marks gene-rich regions, encompassing ∼30% of the genome. The majority of gene bodies are marked with H3S10ph in interphase ESCs, a pattern similar to that observed for Drosophila JIL-1 occupancy (Regnard et al. 2011). The broad distribution of interphase H3S10ph in euchromatic regions is consistent with the “hyperdynamic plasticity” of chromatin reported in ESCs (Meshorer and Misteli 2006) and may play a role in the relatively short chromatin residency time of chromatin-associated proteins observed in ESCs in interphase and/or the exclusion of these regions from LADs. Regardless, a subset of intragenic regions as well as enhancers embedded in these broad euchromatic domains show varying levels of H3S10ph enrichment in G1 vs. S vs. S/G2/M data sets, perhaps reflecting the dynamics of transcriptional regulation during the cell cycle. Indeed, genes enriched preferentially with H3S10ph in specific cell stages are overrepresented in distinct GO biological processes associated with housekeeping functions.
While our ChIP-seq analyses are consistent with the model that AURKB promiscuously phosphorylates all nucleosomes in mitotic cells, AURKB is unlikely to be responsible for euchromatic H3S10ph in G1 ESCs, as treatment with the specific inhibitor Hesperadin did not result in a loss of H3S10ph in gene-dense regions. RPS6KA5 and RPS6KA4 (also known as MSK1 and MSK2), the two orthologs of JIL-1 in mammalian genomes, phosphorylate H3S10 upon MAPK-mediated signal transduction (Thomson et al. 2001); however, both RPS6KA4/5 are expressed at low levels in ESCs relative to MEFs, and Rps6ka4/5 double knockout MEF showed no appreciable loss of basal H3S10ph (Soloaga et al. 2003). RPS6KA3, a member of the related kinase family, RSKs, was implicated in EGF-induced H3S10ph in interphase ESCs (Sassone-Corsi et al. 1999), but like MSKs, RPS6KA3 acts only at a subset of inducible promoters/enhancers. Indeed, we have found that H3S10ph in gene-rich regions is maintained in Rps6ka4, Rps6ka5, and Rps6ka3 knockout ESCs (C Chen, C Fan, H Kimura, M Lorincz, in prep). Identifying the kinase(s) responsible for H3S10ph deposition in interphase will require systematic depletion of individual candidates and perhaps combinatorial knockouts.
Reciprocal H3K9me2/H3S10ph antagonism
H3K9me2 was previously recognized as a broadly distributed modification (Lienert et al. 2011). Our analyses reveal that H3S10ph and H3K9me2 are essentially mutually exclusive in both MEFs and ESCs, with the majority of the mappable genome covered by one or the other mark. H3K9me2 accumulates over transcribed genic regions in ESCs overexpressing H3.3S10A, indicating that H3S10ph may directly inhibit deposition of H3K9me2 in mammalian cells, consistent with previous reports of heterochromatin spreading in JIL-1 hypomorphs in Drosophila (Zhang et al. 2006; Cai et al. 2014). Notably, antagonism between H3K9me2 and H3S10ph in interphase ESCs seems to be reciprocal, as EHMT1 prevents the spreading of H3S10ph into gene-poor regions at most TTRs (Fig. 6A). Such expansion of H3S10ph domains is unlikely to be a consequence of transcription, as this mark spreads over ∼100 kb beyond TTRs in Ehmt1−/− ESCs regardless of the presence of aberrant transcripts. As few new H3S10ph domains appear in Ehmt1−/− ESCs, ectopic H3S10ph at TTRs may depend upon spreading of this mark via mass action into regions previously marked with H3K9me2. Whether H3K9me2 and/or readers of this mark directly inhibit the catalytic activity of the relevant H3S10 kinase(s) remains to be determined.
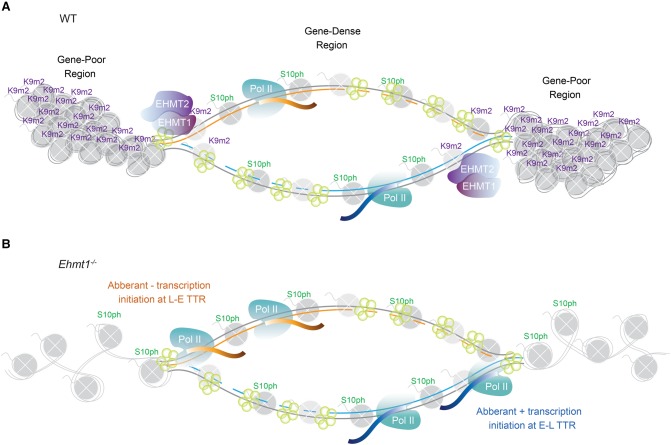
Figure 6.
Model of the interplay between H3S10ph and H3K9me2 during DNA synthesis in ESCs and the influence of perturbing H3K9me2 deposition on strand-biased transcription. (A) Schematic of a hypothetical replicon in WT ESCs, with AURKB-independent H3S10ph marking early-replicating, gene-dense regions and EHMT2/EHMT1-dependent H3K9me2 marking late-replicating, gene-poor regions. Replication timing transition regions, present at the edges of the replicon, coincide with the interface of H3S10ph and H3K9me2 domains in WT ESCs. In the interest of clarity, a single replication fork encompassing the entire replicon is depicted; multiple adjacent replicons may coexist within TTRs (Petryk et al. 2016). (B) Schematic of the same replicon in Ehmt1−/− ESCs showing that, in the absence of H3K9me2, H3S10ph spreads into late-replicating regions, coincident with aberrant transcription initiating asymmetrically at TTRs; showing a plus-strand bias at early-to-late (E to L) and a minus-strand bias at late-to-early (L to E) TTRs. Thus, in both cases, transcription is co-oriented with leading strand synthesis.
Asymmetrical transcription at TTRs in the absence of H3K9me2
We observed a strong directional bias in de novo initiated transcripts in Ehmt2−/− and Ehmt1−/− ESCs, specifically in TTRs. Minus-stranded transcription was favored at late-early TTRs, whereas plus-stranded transcription was favored at early-late TTRs (Figs. 4E, 6B). Presuming that TTRs are generally replicated unidirectionally from a single origin firing in an adjacent earlier replicating region, this observation is consistent with co-orientation of transcription with the replication fork (Huvet et al. 2007). Alternatively, if the TTR is replicated by multiple sequentially firing origins (Petryk et al. 2016), transcription may be biased to leading strands oriented in the net direction of early to late replicating regions, minimizing the likelihood of collision between RNA POL II and the replication machinery emanating from an adjacent sequentially activated replicon.
Notably, many of the genes previously reported to be up-regulated in the absence of EHMT2 or EHMT1, including the MageA and Rhox gene clusters (Tachibana et al. 2002), are within TTRs and are co-oriented with the gross direction of DNA replication (Fig. 4F), revealing that this phenomenon may be extended to endogenous genes. Regardless, as replication timing in EHMT2-deficient ESCs is largely unchanged (Yokochi et al. 2009), such aberrant transcription is unlikely to be a consequence of a shift in replication timing. The strand bias in transcriptional up-regulation observed at the boundaries of H3K9me2- and H3S10ph-marked domains in EHMT1- and EHMT2-deficient cells may thus reflect an intrinsic asymmetry in the “transcription competency,” either initiation or productive elongation, of leading versus lagging strand templates in the wake of replication within a TTR. A difference in the kinetics of chromatin maturation of the leading versus lagging strand following replication could render the former more sensitive to the establishment of an open chromatin state in the absence of H3K9me2, potentiating transcription behind the replication fork.
H3S10ph domains in ESCs versus MEFs
While replication timing profiles are generally conserved in ESCs and MEFs, the broad uninterrupted domains of H3S10ph observed in early-replicating, gene-dense regions in ESCs are frequently interspersed with smaller domains of H3K9me2 in MEFs. Notably, in the absence of EHMT2, H3K9me2 is lost within early-replicating domains, accompanied by expansion of H3S10ph into these regions. However, in contrast to ESCs, H3K9me2 is not diminished at late-replicating regions in Ehmt2−/− MEFs, likely reflecting deposition of this mark by other KMTs such as SUV39H1/2 or SETDB1. The accumulation of H3K9me2 may be a general feature of somatic cells, as a global increase of EHMT2/EHMT1-dependent H3K9me2 was also observed in the E5.5 epiblast after implantation, specifically within gene bodies (Zylicz et al. 2015). EHMT2 is essential for mid-gestation development, with protracted expression of pluripotency markers such as Nanog and Pou5f1 observed at E7.5 and lethality observed at E10.5 in Ehmt2−/− embryos (Shinkai and Tachibana 2011; Yamamizu et al. 2012). Whether the broader domains of H3S10ph in ESCs reflects the distribution of this mark in the epiblast and/or plays a role in restricting H3K9me2 in vivo remains to be determined.
Reconciling the role of mitotic and interphase H3S10ph
To reconcile the paradoxical association of H3S10ph with condensed chromatin in mitosis and de-condensed chromatin in interphase, Johansen and Johansen proposed that this mark promotes the detachment of chromatin from the nuclear scaffold (Johansen and Johansen 2006), facilitating chromosome segregation and transcription. Consistent with this model, following stimulation in B lymphocytes, immediate early genes physically shift from the periphery of the nucleus to constitutively active transcription factories prior to the onset of transcription (Osborne et al. 2007). Furthermore, the stable radial positioning of chromosomes is established de novo within the first 2 h of G1 (Walter et al. 2003), coinciding with establishment of self-associating topological associating domains (TADs) and RT (Dileep et al. 2015). We show that interphase H3S10ph domains are most prominent in G1, are not associated with the nuclear lamina, and are universally early-replicating (Fig. 3E). Thus, H3S10ph-mediated nuclear positioning may enhance both transcription and access to the replication machinery during interphase.
As H3S10ph exerts little influence on physical packing of nucleosome arrays in vitro (Shogren-Knaak 2003; Fry et al. 2004), this histone mark may promote untethering through repulsion of lamin-associated proteins. Consistent with this model, disruption of mitotic H3S10ph through AURKB inhibition results in retention of CBX1/3/5 and the lamin resident protein PRR14 on metaphase chromosomes (Fischle et al. 2005; Poleshko and Katz 2014). Intriguingly, EHMT2 itself is physically tethered to the nuclear lamina (Kind et al. 2013), possibly via interaction with the lamin-associated protein Barrier to Auto-integration Factor (Montes de Oca et al. 2014), and genes repressed by EHMT2 are localized to the nuclear periphery in ESCs (Yokochi et al. 2009). Determining whether interphase H3S10ph is a driver or bystander in nuclear compartmentalization of marked euchromatic regions in mammals will depend upon identification of the kinase(s) involved.
Methods
Cell culture, plasmids, and transfection
TT2 (WT), 2–3 Ehmt2−/−, and Cd12 Ehmt1−/− ESCs were cultured in Dulbecco Modified Eagle's Medium (DMEM), supplemented with 15% fetal bovine serum (FBS) (HyClone Laboratories), 20 mM HEPES, 1 mM L-glutamine, 1 mM nonessential amino acids, 1 mM sodium pyruvate, 0.1 mM betamercaptoethnol, 100 U/mL penicillin-streptomycin, and recombinant leukemia inhibitory factor on gelatinized plates. For AURKB inhibition experiments, ESCs were cultured for 3 h in complete media with 200 nM Hesperadin (stock 1 mM in DMSO; Millipore). To derive ESC lines stably expressing the FUCCI reporters, 1.5 µg of CAG-hCDT1:mKO-T2A-hGEMN:Venus plasmid (kind gift from U. Naumann) were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's recommendation. Stable mKO+ and Venus+ cells were isolated by FACS at day 5 and 13 post-transfection. To generate ESCs expressing H3.3 mutated at S10, a Ser10Ala point mutation was introduced in the CMV-H3.3-YFP vector (Addgene) using the QuikChange Lightning site-directed mutagenesis kit (Cat.# 210518; Agilent). WT and S10A CMV-H3.3-YFP were transfected using Lipofectamine, as above. Clones were isolated by puromycin selection followed by FACS for YFP+ 24 h after Dox induction. WT MEFs were grown in DMEM supplemented with 10% FBS, 1 mM L-glutamine, and 100 U/mL penicillin-streptomycin.
Immunofluorescence and flow cytometry
Indirect immunofluorescence staining was performed using standard methods. Briefly, cells were trypsinized and crosslinked with 1% formaldehyde, permeabilized with 0.25% Triton X-100 and 1% bovine serum albumin (Sigma-Aldrich). Cells were then incubated with H3S10ph- (CMA311, 1:100) and PCNA- (ab18197, 1:100; Abcam) specific antibodies at 37°C for 1 h and subsequently incubated with Alexa Fluor 488 and 594-labeled secondary antibodies (Life Technologies). DNA was counter-stained with 500 ng/mL Hoechst 33342 (Sigma-Aldrich). Flow cytometry data were acquired using a BD LSRII561 flow cytometer using BD FACSDIVA software. Cells were sorted on BD Influx.
Protein extraction and Western blotting
For Western analysis of histones, cell extracts were prepared by lysis in RIPA buffer (50 mM Tris pH 8.0, 150 mM NaCl, 1% NP-40, 0.25% deoxycholate, 0.1% SDS). Extracts were boiled in SDS-PAGE loading buffer and blotting was performed as previously described using anti-TRIM28 (ab22553, 1:5000; Abcam), anti-H3S10ph (H. Kimura, CMA311, 1:5000), and anti-pan H3 (H0164, 1:5000; Sigma). Following addition of IRDYE-conjugated secondary antibodies, fluorescence was quantified on an Odyssey imager (LiCOR Biosciences).
ChIP-seq and RNA-seq
NChIP-seq of H3K9me2, H3K4me3, and H3K36me3 was performed as previously described (Karimi et al. 2011). ChIP of H3S10ph was performed by crosslink MNase ChIP, as described previously (Hiragami-Hamada et al. 2016). See Supplemental Methods for detailed protocol. Construction of Illumina ChIP and RNA sequencing libraries were as described previously (Brind'Amour et al. 2015; Morrissy et al. 2016). Paired-end sequencing (75 or 100 bp) was performed on an Illumina HiSeq 2000 following the recommended protocol. Reads were aligned to mm9 using the Burrows–Wheeler Aligner using default parameters (Li and Durbin 2009). PCR duplicates and multimatch reads (Q = 0) were removed from all subsequent statistical analysis. Calculations of RPKM and z-score were performed as per Karimi et al. (2011) using SeqMonk (https://www.bioinformatics.babraham.ac.uk/projects/seqmonk/) and VisRSeq (Younesy et al. 2015). Data visualization and plots were generated in VisRSeq, Excel, or RStudio.
Data access
The ChIP-seq and RNA-seq data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE97947.
Supplementary Material
Acknowledgments
We thank Julie Brind'Amour and Louis Lefebvre for critical reading of the manuscript, Irina Maksakova for technical assistance, Yoichi Shinkai for the Ehmt2−/− and Ehmt1−/− ESCs, Fabio Rossi for the Ehmt2−/− MEFs, Ulrike Naumann for the modified FUCCI plasmid, and members of the Lorincz Lab for helpful discussions. We also thank the UBCFlow facility for FACS services and the BC Genome Science Centre and UBC Biomedical Research Centre for Illumina sequencing. This work was supported by Canadian Institutes of Health Research (CIHR) grant MOP133417 and Natural Sciences and Engineering Research Council of Canada (NSERC) grant 05228 to M.C.L. C.C.L.C. is supported by an NSERC doctoral fellowship. H.K. was supported by the Platform Project for Supporting Drug Discovery and Life Science Research from the Japan Agency for Medical Research and Development, and JSPS KAKENHI (JP25116005, JP26291071, and JP17H01417).
Author contributions: C.C.L.C., P.G., and M.H.A. conducted the experiments. C.C.L.C., M.M.K., and M.C.L. analyzed the data. H.K. provided reagents and contributed to experimental procedure optimization. C.C.L.C. and M.C.L. designed the experiments and wrote the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.224717.117.
References
- Ahmad K, Henikoff S. 2002. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell 9: 1191–1200. [DOI] [PubMed] [Google Scholar]
- Akhtar W, de Jong J, Pindyurin AV, Pagie L, Meuleman W, de Ridder J, Berns A, Wessels LFA, van Lohuizen M, van Steensel B. 2013. Chromatin position effects assayed by thousands of reporters integrated in parallel. Cell 154: 914–927. [DOI] [PubMed] [Google Scholar]
- Anest V, Hanson JL, Cogswell PC, Steinbrecher KA, Strahl BD, Baldwin AS. 2003. A nucleosomal function for IκB kinase-α in NF-κB-dependent gene expression. Nature 423: 659–663. [DOI] [PubMed] [Google Scholar]
- Auclair G, Borgel J, Sanz LA, Vallet J, Guibert S, Dumas M, Cavelier P, Girardot M, Forné T, Feil R, et al. 2016. EHMT2 directs DNA methylation for efficient gene silencing in mouse embryos. Genome Res 26: 192–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode AM. 2005. Inducible covalent posttranslational modification of histone H3. Sci STKE 2005: pre4. [DOI] [PubMed] [Google Scholar]
- Brind'Amour J, Liu S, Hudson M, Chen C, Karimi MM, Lorincz MC. 2015. An ultra-low-input native ChIP-seq protocol for genome-wide profiling of rare cell populations. Nat Commun 6: 6033. [DOI] [PubMed] [Google Scholar]
- Cai W, Wang C, Li Y, Yao C, Shen L, Liu S, Bao X, Schnable PS, Girton J, Johansen J, et al. 2014. Genome-wide analysis of regulation of gene expression and H3K9me2 distribution by JIL-1 kinase mediated histone H3S10 phosphorylation in Drosophila. Nucleic Acids Res 42: 5456–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Zhao J, Wang Y, Wang M, Long H, Liang D, Huang L, Wen Z, Li W, Li X, et al. 2013. H3.3 actively marks enhancers and primes gene transcription via opening higher-ordered chromatin. Genes Dev 27: 2109–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung P, Tanner KG, Cheung WL, Sassone-Corsi P, Denu JM, Allis CD. 2000. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol Cell 5: 905–915. [DOI] [PubMed] [Google Scholar]
- Dileep V, Ay F, Sima J, Vera DL, Noble WS, Gilbert DM. 2015. Topologically associating domains and their long-range contacts are established during early G1 coincident with the establishment of the replication-timing program. Genome Res 25: 1104–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Q, Chen H, Costa M, Dai W. 2008. Phosphorylation of H3S10 blocks the access of H3K9 by specific antibodies and histone methyltransferase. Implication in regulating chromatin dynamics and epigenetic inheritance during mitosis. J Biol Chem 283: 33585–33590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsässer SJ, Noh K-M, Diaz N, Allis CD, Banaszynski LA. 2015. Histone H3.3 is required for endogenous retroviral element silencing in embryonic stem cells. Nature 522: 240–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconer E, Hills M, Naumann U, Poon SSS, Chavez EA, Sanders AD, Zhao Y, Hirst M, Lansdorp PM. 2012. DNA template strand sequencing of single-cells maps genomic rearrangements at high resolution. Nat Meth 9: 1107–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazzio TG, Huff JT, Panning B. 2008. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell 134: 162–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W, Tseng BS, Dormann HL, Ueberheide BM, Garcia BA, Shabanowitz J, Hunt DF, Funabiki H, Allis CD. 2005. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature 438: 1116–1122. [DOI] [PubMed] [Google Scholar]
- Fry CJ, Shogren-Knaak MA, Peterson CL. 2004. Histone H3 amino-terminal tail phosphorylation and acetylation: synergistic or independent transcriptional regulatory marks? Cold Spring Harb Symp Quant Biol 69: 219–226. [DOI] [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, et al. 2008. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453: 948–951. [DOI] [PubMed] [Google Scholar]
- Hayashi-Takanaka Y, Yamagata K, Nozaki N, Kimura H. 2009. Visualizing histone modifications in living cells: spatiotemporal dynamics of H3 phosphorylation during interphase. J Cell Biol 187: 781–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiragami-Hamada K, Soeroes S, Nikolov M, Wilkins B, Kreuz S, Chen C, De La Rosa-Velázquez IA, Zenn HM, Kost N, Pohl W, et al. 2016. Dynamic and flexible H3K9me3 bridging via HP1β dimerization establishes a plastic state of condensed chromatin. Nat Commun 7: 11310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I, Ryba T, Itoh M, Rathjen J, Kulik M, Papp B, Fussner E, Bazett-Jones DP, Plath K, Dalton S, et al. 2010. Genome-wide dynamics of replication timing revealed by in vitro models of mouse embryogenesis. Genome Res 20: 155–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huvet M, Nicolay S, Touchon M, Audit B, d'Aubenton-Carafa Y, Arneodo A, Thermes C. 2007. Human gene organization driven by the coordination of replication and transcription. Genome Res 17: 1278–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Wang Y, Walker DL, Dong H, Conley C, Johansen J, Johansen KM. 1999. JIL-1: a novel chromosomal tandem kinase implicated in transcriptional regulation in Drosophila. Mol Cell 4: 129–135. [DOI] [PubMed] [Google Scholar]
- Johansen KM, Johansen J. 2006. Regulation of chromatin structure by histone H3S10 phosphorylation. Chromosome Res 14: 393–404. [DOI] [PubMed] [Google Scholar]
- Karimi MM, Goyal P, Maksakova IA, Bilenky M, Leung D, Tang JX, Shinkai Y, Mager DL, Jones S, Hirst M, et al. 2011. DNA methylation and SETDB1/H3K9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell Stem Cell 8: 676–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind J, van Steensel B. 2010. Genome–nuclear lamina interactions and gene regulation. Curr Opin Cell Biol 22: 320–325. [DOI] [PubMed] [Google Scholar]
- Kind J, Pagie L, Ortabozkoyun H, Boyle S, de Vries SS, Janssen H, Amendola M, Nolen LD, Bickmore WA, van Steensel B. 2013. Single-cell dynamics of genome-nuclear lamina interactions. Cell 153: 178–192. [DOI] [PubMed] [Google Scholar]
- Kind J, Pagie L, de Vries SS, Nahidiazar L, Dey SS, Bienko M, Zhan Y, Lajoie B, de Graaf CA, Amendola M, et al. 2015. Genome-wide maps of nuclear lamina interactions in single human cells. Cell 163: 134–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubicek S, O'Sullivan RJ, August EM, Hickey ER, Zhang Q, Teodoro ML, Rea S, Mechtler K, Kowalski JA, Homon CA, et al. 2007. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol Cell 25: 473–481. [DOI] [PubMed] [Google Scholar]
- Leung DC, Dong KB, Maksakova IA, Goyal P, Appanah R, Lee S, Tachibana M, Shinkai Y, Lehnertz B, Mager DL, et al. 2011. Lysine methyltransferase G9a is required for de novo DNA methylation and the establishment, but not the maintenance, of proviral silencing. Proc Natl Acad Sci 108: 5718–5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lienert F, Mohn F, Tiwari VK, Baubec T, Roloff TC, Gaidatzis D, Stadler MB, Schübeler D. 2011. Genomic prevalence of heterochromatic H3K9me2 and transcription do not discriminate pluripotent from terminally differentiated cells. PLoS Genet 7: e1002090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan LC, Willis AC, Barratt MJ. 1991. Rapid histone H3 phosphorylation in response to growth factors, phorbol esters, okadaic acid, and protein synthesis inhibitors. Cell 65: 775–783. [DOI] [PubMed] [Google Scholar]
- Maksakova IA, Thompson PJ, Goyal P, Jones SJ, Singh PB, Karimi MM, Lorincz MC. 2013. Distinct roles of KAP1, HP1 and G9a/GLP in silencing of the two-cell-specific retrotransposon MERVL in mouse ES cells. Epigenetics Chromatin 6: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshorer E, Misteli T. 2006. Chromatin in pluripotent embryonic stem cells and differentiation. Nat Rev Mol Cell Biol 7: 540–546. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim T-K, Koche RP, et al. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montes de Oca R, Andreassen PR, Wilson KL. 2014. Barrier-to-Autointegration Factor influences specific histone modifications. Nucleus 2: 580–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissy AS, Garzia L, Shih DJH, Zuyderduyn S, Huang X, Skowron P, Remke M, Cavalli FMG, Ramaswamy V, Lindsay PE, et al. 2016. Divergent clonal selection dominates medulloblastoma at recurrence. Nature 529: 351–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne CS, Chakalova L, Mitchell JA, Horton A, Wood AL, Bolland DJ, Corcoran AE, Fraser P. 2007. Myc dynamically and preferentially relocates to a transcription factory occupied by Igh. PLoS Biol 5: e192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petryk N, Kahli M, d'Aubenton-Carafa Y, Jaszczyszyn Y, Shen Y, Silvain M, Thermes C, Chen C-L, Hyrien O. 2016. Replication landscape of the human genome. Nat Commun 7: 10208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poleshko A, Katz RA. 2014. Specifying peripheral heterochromatin during nuclear lamina reassembly. Nucleus 5: 32–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope BD, Ryba T, Dileep V, Yue F, Wu W, Denas O, Vera DL, Wang Y, Hansen RS, Canfield TK, et al. 2014. Topologically associating domains are stable units of replication-timing regulation. Nature 515: 402–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathert P, Dhayalan A, Murakami M, Zhang X, Tamas R, Jurkowska R, Komatsu Y, Shinkai Y, Cheng X, Jeltsch A. 2008. Protein lysine methyltransferase G9a acts on non-histone targets. Nat Chem Biol 4: 344–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, et al. 2000. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406: 593–599. [DOI] [PubMed] [Google Scholar]
- Regnard C, Straub T, Mitterweger A, Dahlsveen IK, Fabian V, Becker PB. 2011. Global analysis of the relationship between JIL-1 kinase and transcription. PLoS Genet 7: e1001327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbart SB, Dickson BM, Raab JR, Grzybowski AT, Krajewski K, Guo AH, Shanle EK, Josefowicz SZ, Fuchs SM, Allis CD, et al. 2015. An interactive database for the assessment of histone antibody specificity. Mol Cell 59: 502–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahlén P, Abdullayev I, Ramsköld D, Matskova L, Rilakovic N, Lötstedt B, Albert TJ, Lundeberg J, Sandberg R. 2015. Genome-wide mapping of promoter-anchored interactions with close to single-enhancer resolution. Genome Biol 16: 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, Osawa H, Kashiwagi S, Fukami K, Miyata T, Miyoshi H, et al. 2008. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 132: 487–498. [DOI] [PubMed] [Google Scholar]
- Sasagawa Y, Nikaido I, Hayashi T, Danno H, Uno KD, Imai T, Ueda HR. 2013. Quartz-Seq: a highly reproducible and sensitive single-cell RNA sequencing method, reveals non-genetic gene-expression heterogeneity. Genome Biol 14: R31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassone-Corsi P, Mizzen CA, Cheung P, Crosio C, Monaco L, Jacquot S, Hanauer A, Allis CD. 1999. Requirement of Rsk-2 for epidermal growth factor-activated phosphorylation of histone H3. Science 285: 886–891. [DOI] [PubMed] [Google Scholar]
- Schultz DC. 2002. SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev 16: 919–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkai Y, Tachibana M. 2011. H3K9 methyltransferase G9a and the related molecule GLP. Genes Dev 25: 781–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shogren-Knaak MA. 2003. A native peptide ligation strategy for deciphering nucleosomal histone modifications. J Biol Chem 278: 15744–15748. [DOI] [PubMed] [Google Scholar]
- Skourti-Stathaki K, Kamieniarz-Gdula K, Proudfoot NJ. 2014. R-loops induce repressive chromatin marks over mammalian gene terminators. Nature 516: 436–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soloaga A, Thomson S, Wiggin GR, Rampersaud N, Dyson MH, Hazzalin CA, Mahadevan LC, Arthur JSC. 2003. MSK2 and MSK1 mediate the mitogen- and stress-induced phosphorylation of histone H3 and HMG-14. EMBO J 22: 2788–2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, Ohki M, Fukuda M, Takeda N, Niida H, Kato H, et al. 2002. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev 16: 1779–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson S, Mahadevan LC, Clayton AL. 1999. MAP kinase-mediated signalling to nucleosomes and immediate-early gene induction. Semin Cell Dev Biol 10: 205–214. [DOI] [PubMed] [Google Scholar]
- Thomson S, Clayton AL, Mahadevan LC. 2001. Independent dynamic regulation of histone phosphorylation and acetylation during immediate-early gene induction. Mol Cell 8: 1231–1241. [DOI] [PubMed] [Google Scholar]
- Van Hooser A, Goodrich DW, Allis CD, Brinkley BR, Mancini MA. 1998. Histone H3 phosphorylation is required for the initiation, but not maintenance, of mammalian chromosome condensation. J Cell Sci 111: 3497–3506. [DOI] [PubMed] [Google Scholar]
- Walter J, Schermelleh L, Cremer M, Tashiro S, Cremer T. 2003. Chromosome order in HeLa cells changes during mitosis and early G1, but is stably maintained during subsequent interphase stages. J Cell Biol 160: 685–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Higgins JMG. 2013. Histone modifications and mitosis: countermarks, landmarks, and bookmarks. Trends Cell Biol 23: 175–184. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhang W, Jin Y, Johansen J, Johansen KM. 2001. The JIL-1 tandem kinase mediates histone H3 phosphorylation and is required for maintenance of chromatin structure in Drosophila. Cell 105: 433–443. [DOI] [PubMed] [Google Scholar]
- Wei Y, Mizzen CA, Cook RG, Gorovsky MA, Allis CD. 1998. Phosphorylation of histone H3 at serine 10 is correlated with chromosome condensation during mitosis and meiosis in Tetrahymena. Proc Natl Acad Sci 95: 7480–7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A, Laurenti E, Oser G, van der Wath RC, Blanco-Bose W, Jaworski M, Offner S, Dunant CF, Eshkind L, Bockamp E, et al. 2008. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell 135: 1118–1129. [DOI] [PubMed] [Google Scholar]
- Yamamizu K, Fujihara M, Tachibana M, Katayama S, Takahashi A, Hara E, Imai H, Shinkai Y, Yamashita JK. 2012. Protein kinase A determines timing of early differentiation through epigenetic regulation with G9a. Cell Stem Cell 10: 759–770. [DOI] [PubMed] [Google Scholar]
- Yokochi T, Poduch K, Ryba T, LU J, Hiratani I, Tachibana M, Shinkai Y, Gilbert DM. 2009. G9a selectively represses a class of late-replicating genes at the nuclear periphery. Proc Natl Acad Sci 106: 19363–19368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younesy H, Möller T, Lorincz MC, Karimi MM, Jones SJM. 2015. VisRseq: R-based visual framework for analysis of sequencing data. BMC Bioinformatics 16: pS2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue F, Cheng Y, Breschi A, Vierstra J, Wu W, Ryba T, Sandstrom R, Ma Z, Davis C, Pope BD, et al. 2014. A comparative encyclopedia of DNA elements in the mouse genome. Nature 515: 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Deng H, Bao X, Lerach S, Girton J, Johansen J, Johansen KM. 2006. The JIL-1 histone H3S10 kinase regulates dimethyl H3K9 modifications and heterochromatic spreading in Drosophila. Development 133: 229–235. [DOI] [PubMed] [Google Scholar]
- Zippo A, Serafini R, Rocchigiani M, Pennacchini S, Krepelova A, Oliviero S. 2009. Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation. Cell 138: 1122–1136. [DOI] [PubMed] [Google Scholar]
- Zylicz JJ, Dietmann S, Günesdogan U, Hackett JA. 2015. Chromatin dynamics and the role of G9a in gene regulation and enhancer silencing during early mouse development. eLife 10.7554/eLife.09571. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.