Abstract
The CCN family of proteins plays important roles in development and homeostasis of bone and cartilage. To understand the role of CCN4 in chondrogenesis, human bone marrow stromal cells (hBMSCs) were transduced with CCN4 adenovirus (adCCN4) or siRNA to CCN4 (siCCN4) in the presence or absence of transforming growth factor-β3 (TGF-β3). Overexpression of CCN4 enhanced TGF-β3-induced SMAD2/3 phosphorylation and chondrogenesis of hBMSCs in an in vitro assay using a micromass culture model. On the other hand, knockdown of CCN4 inhibited the TGF-β3-induced SMAD2/3 phosphorylation and synthesis of cartilage matrix in micromass cultures of hBMSCs. Immunoprecipitation–western blot analysis revealed that CCN4 bound to TGF-β3 and regulated the ability of TGF-β3 to bind to hBMSCs. In vivo analysis confirmed there was a significant decrease in the gene expression levels of chondrocyte markers in cartilage samples from Ccn4-knock out (KO) mice, compared to those from wild type (WT) control. In order to investigate the regenerative properties of the articular cartilage in Ccn4-KO mice, articular cartilage defects were surgically performed in the knee joints of young mice, and the results showed that the cartilage was partially repaired in WT mice, but not in Ccn4-KO mice. In conclusion, these results show, for the first time, that CCN4 has a positive influence on chondrogenic differentiation by modulating the effects of TGF-β3.
Keywords: Bone marrow stromal cell (BMSC), Chondrogenesis, CCN4/WISP-1, TGF-β3, Articular cartilage
1. Introduction
Osteoarthritis (OA), which is characterized by the degeneration and loss of articular cartilage and subchondral bone, is one of the most common disorders of the musculoskeletal system, and is caused mainly by the effects of aging, trauma or injury and overloading, such as in the case of obesity. The main symptoms of OA of the hip or knee are pain and stiffness, causing loss of joint mobility. Articular cartilage consists basically of a single cell type, the chondrocyte, which can synthesize all components of the extracellular matrix (ECM), and the mechanical function and properties of articular cartilage is greatly dependent on the quality and quantity of ECM. However, adult cartilage has a poor capacity for repair and regeneration. Therefore, autologous chondrocyte transplantation has been widely used. Stem cell therapy, which employs the use of stem/progenitor cells from different tissue or induced-pluripotent stem cells (iPSCs), has gained particular attention in recent years. However, a deeper understanding of the detailed mechanisms of chondrogenic differentiation of stem/progenitor cells, including the interaction with growth factors, is necessary to eventually improve the current clinical outcomes for cartilage regeneration.
Transforming growth factor-beta (TGF-β) is a member of a family of dimeric polypeptide growth factors and is classified into four major subfamilies, TGF-βs, bone morphogenetic proteins (BMPs), activins/inhibins, and glia cell line-derived neurotrophic factors (GDNFs). Among the TGF-β superfamily, TGF-β1, β2 and β3 are known to play important roles not only in chondrocyte proliferation and differentiation but also in embryonic development of cartilage through activation of the SMAD2/3 signaling pathway; and alterations in TGF-β signaling are known to be involved in the pathogenesis of OA and osteoporosis [1,2]. In early stages of chondrogenesis from mesenchymal stem cell condensation to limb bud formation, TGF-β activates SMAD3, which binds to co-SMAD4, translocates to the nucleus and promotes the expression of Sox9, the master regulator of chondrogenesis [3].
The CCN family consists of six members, CCN1/Cyr61/Cef10, CCN2/CTGF/Fisp12, CCN3/Nov, CCN4/WISP-1/rCOP-1/Elm-1, CCN5/WISP-2, and CCN6/WISP-3. CCN family proteins are characterized by four distinct functional domains: an insulin-like growth factor binding protein like module (IGFBP), a von Willebrand factor type C repeat (VWC), a thrombospondin type 1 repeat (TSP1), and a cysteine-rich C-terminal module (CT). Because CCN family proteins have this unique modular structure, they can interact with various other proteins, cell membrane receptors and ECM components, and can regulate several cell functions, including adhesion, migration, proliferation, and differentiation both in vitro and in vivo. Interestingly, CCN family members can interact with and regulate the activity of members of the TGF-β superfamily and orchestrate tissue development during osteogenesis and chondrogenesis [4,5]. For instance, CCN2 was shown to promote the proliferation and differentiation of chondrocytes by interacting with BMP-2, FGF-2, and IGF-1 [4,6]. Additionally, our research group showed that CCN4 regulates osteoblast function by controlling the activity of TGF-β1 and BMP-2 [7,8].
Clinical and experimental evidence also suggest that CCN4 plays a role in OA [9,10]. Previous studies reported a high expression of CCN4 in the synovium and cartilage in an experimental OA model in mice as well as in human OA patients [10,11]. These findings suggest that CCN4 may be involved in the chondrocyte differentiation and/or cartilage regeneration. However, the specific function of CCN4 in chondrogenesis and cartilage regeneration remains unclear.
To understand the role of CCN4 in chondrogenesis, we investigated the effect of CCN4 on the chondrogenic differentiation of human bone marrow stromal cells (hBMSCs) using gain and loss of function models, and found that CCN4 is a positive regulator of TGF-β3-induced chondrogenic differentiation of hBMSCs in vitro. Moreover, in an experimental animal model of full-thickness articular cartilage defect using Ccn4-knock out (KO) mice, we showed that CCN4 is important for cartilage repair in vivo.
2. Materials and methods
2.1. Cells and culture medium
Human bone marrow stromal cells (hBMSCs) were purchased from Lonza (Walkersville, MD, USA). Cells were cultured in α-MEM containing 10% fetal bovine serum (FBS), 1% L-glutamine (Life Technologies, Rockville, MD, USA), and 1% penicillin and streptomycin (Sigma, St. Louis, MO, USA) [Basal medium].
For chondrogenic differentiation, hBMSCs at passages 4 to 5 were used for analysis of chondrogenic differentiation. hBMSCs were suspended (2 × 105 cells/10 μL) and cultured in micromasses, as previously reported [12,13]. Chondrogenic medium consisted of DMEM low glucose, supplemented with 6 μg/mL each of insulin, transferrin and selenous acid (ITS solution, BD, Bedford, MA, USA), 50 μg/mL of L-Ascorbic Acid Phosphate Magnesium Salt n-Hydrate (Wako Pure Chemical Industries, Osaka, Japan), 100 nM dexamethasone (Sigma), 10 ng/mL of rhTGF-β3 (R&D Systems, Inc., Minneapolis, MN, USA) and antibiotics. Medium was changed every 3–4 days and samples were cultured for 28 days prior to analysis.
For the overexpression of the CCN4 gene, hBMSCs were transfected with adenovirus that express human CCN4 (adCCN4) as reported previously [8]. Adenovirus CMV empty vector (adCMV) was used as the negative control. For the knockdown of Ccn4 gene, hBMSCs were cultured in penicillin and streptomycin-free basal medium, following the manufacturer’s instruction. Twenty four hours later, the medium was changed to Opti-MEM Reduced-Serum Medium (Life Technologies) supplemented with Lipofectamine RNAiMAX (Life Technologies) and 10 nM SiRNA to decrease the expression of human CCN4 (SiCCN4; Life Technologies) according to the manufacturer’s instructions. Stelth™ RNAi Negative Control High GC Duplex (Life Technologies) was used as the negative control.
For the isolation of chondrocytes, the growth plates of femur, which were collected from newborn WT and Ccn4-KO, were incubated with Trypsin-EDTA (Life Technologies) for 30 min and 86.5 U/mL of Type 1 collagenase (Worthington Biochemical Corporation, Lakewood, NJ, USA) overnight at 37 °C. Isolated chondrocytes were cultured with DMEM/F12 (Life Technologies) containing 10% fetal bovine serum (FBS), 1% L-glutamine, and 1% penicillin and streptomycin.
2.2. Reverse transcription and real time RT-PCR analysis
Total cellular RNA was extracted by RNA Minikit (Life Technologies), according to the manufacturer’s instructions. Possible residual DNA was removed by DNase (Purelink™DNase; Life Technologies). RNA samples were reverse-transcribed by using iScript cDNA synthesis kit (Bio-Rad; Hercules, CA, USA). Real time RT-PCR was performed to quantify the expression of the target gene by using KAPA SYBR FAST qPCR Master Mix (KAPA BIOSYSTEMS, Wilmington, MA, USA) and Chromo4 real-time detector. The levels of mRNAs of interest were normalized to that of the reference gene S29. Primer sequences are shown in Table 1.
Table 1.
Primers used for Real time RT-PCR analysis.
| Gene | Species | GenBank accession no. | Primer sequence | PCR product length (bp) |
|---|---|---|---|---|
| S29 | Human | BC032813 | 5′-T CTCGCT CTT GTCGT GT CT GTTC-3′(S) 5′-ACACTGGCGGCACATATTGAGG-3′(AS) |
75 |
| CCN1 | Human | AF003114 | 5′-GCCAATGAAGCAGCGTTTCCC-3′(S) 5′-CAAT GAGT CCCAT CACCCACACC-3′(AS) |
171 |
| CCN2 | Human | BT019794 | 5′-T GCGAGGAGT GGGTGTGTGAC-3′(S) 5′-TGGACCAGGCAGTTGGCTCTAATC-3′(AS) |
125 |
| CCN3 | Human | BC015028 | 5′-GAGCAGTGCCAATCTACAGCGAAG-3′(S) 5′-AGATGGAGAAGCAGGAAGGTCAGG-3′(AS) |
138 |
| CCN4 | Human | AB034725 | 5′-ACACTCATTAAGGCAGGGAAGAAG-3′(S) 5′-TCAGGACACTGGAAGGACACG-3′(AS) |
185 |
| CCN5 | Human | BC058074 | 5′-CACACCGAAGACCCACCTCCTG-3′(S) 5′-GCAGCAGCCACAGCCATCC-3′(AS) |
151 |
| CCN6 | Human | AY358350 | 5′-GGGTACAGGGCACTGGACCATTAG-3′(S) 5′-ACAGCATCCACAGCCATCTCTCAC-3′(AS) |
167 |
| COL2A1 | Human | BT007205 | 5′-TGGAGCAGCAAGAGCAAGGAGAAG-3′(S) 5′-CCGTGGACAGCAGGCGTAGG-3′(AS) |
136 |
| S29 | Mouse | NM_009093 | 5′-GGAGTCACCCACGGAAGTTCGG-3′(S) 5′-GGAAGCACTGGCGGCACATG-3′(AS) |
108 |
| Sox-9 | Mouse | NM_011448 | 5′-TCAGATGCAGTGAGGAGCAC-3′(S) 5′-CCAGCCACAGCAGTGAGTAA-3′(AS) |
208 |
| Acan | Mouse | L07049 | 5′-CCCGGTACCCTACAGAGACA-3′(S) 5′-ACAGTGACCCTGGAACTTGG-3′(AS) |
215 |
| Col2a1 | Mouse | BC082331 | 5′-GGGCAACAGCAGGTTCACATACAC-3′(S) 5′-TCAATAATGGGAAGGCGGGAGGTC-3′(AS) |
120 |
| Col10a1 | Mouse | NM_009925 | 5′-TTCTGCTGCTAATGTTCTTGA-3′(S) 5′-AATGCCTTGTTCTCCTCTTAC-3′(AS) |
164 |
| Bgn | Mouse | L20276 | 5′-ACCTGTCCCCTTCCATCTCT-3′(S) 5′-CCGTGTGTGTGTGTGTGTGT-3′(AS) |
201 |
| Dcn | Mouse | X53929 | 5′-CCAACATAACTGCGATCCCT-3′(S) 5′-TGTCCAAGTGGAGTTCCCTC-3′(AS) |
214 |
S, sense; AS, antisense.
2.3. Western blot analysis
Cells were lysed by M-PER (Mammalian Protein Extraction Reagent; Thermo, Waltham, Massachusetts, USA) supplemented with a Protease Inhibitor Cocktail (1 mg/mL; Roche, Indianapolis, IN, USA) and Phosphatase Inhibitor Signal-Use Cocktail (1:100; Thermo). Cell debris were removed from lysates by centrifugation at 12,000 rpm for 10 min at 4 °C. Protein concentration in the cell lysate was determined by using Pierce® BCA Protein assay kit (Thermo). Ten micrograms of the total protein lysate was separated in precast polyacrylamide gels (NuPage, Life Technologies) by electrophoresis and then transferred onto polyvinylidene fluoride membranes (PVDF; GE Healthcare Life sciences, Buckinghamshire, UK) at 30 V for 2 h. Blots were blocked and incubated in primary antibodies to SOX-9 (1:1000; Millipore, Billerica, MA, USA), phosphorylated SMAD2/3 (pSMAD2/3), total SMAD2/3 (1:1000; Cell Signaling Technologies, Danvers, MA, USA) and β-ACTIN as a loading control (1:5000; Sigma). Membranes were then washed, and incubated in HRP-labeled anti-rabbit IgG (1:2000; Santa Cruz Biotechnology, Santa Cruz, CA, USA) for 1 h at room temperature (RT). The blots were developed using Forte western HRP Substrate (Millipore), and visualized with Image Quant LAS 4000 mini (Fujifilm, Tokyo, Japan).
2.4. Immunoprecipitation
Cells were harvested using NP-40 buffer (1% NP-40, 0.15 M NaCl, 0.01 M sodium phosphate, and 1 mM ethylenediaminetetraacetic acid (EDTA)) containing a Complete Protease Inhibitor Mixture (Roche Diagnostics, Mannheim, Germany). Cleared cell lysates were incubated with magnetic beads conjugated with 4C5 anti-DDK mouse monoclonal antibody (OriGene, Rockville, MD, USA), or 1 μg mouse IgG1 kappa monoclonal [MOPC-21] isotype control (Abcam, Cambridge, UK) that was previously bound to Dynabeads protein G (Life Technologies). The beads were washed extensively, and the bound proteins were boiled in loading buffer for 10 min at 70 °C and then separated by SDS-PAGE and analyzed by immunoblot with 4C5 anti-DDK monoclonal antibody (OriGene) or anti-CCN4 antibody (LF-187). Details on the construction and specificity of LF-187 have been described previously [14,15].
2.5. Cell protein binding assay
To assess the effect of CCN4 on the cell binding affinity of rhTGF-β3, hBMSCs were seeded at a concentration of 1 × 104 cells/well in 96-well plates. After 24 h, cells were transfected with adCCN4 or siCCN4 for 48 h. Next, cells were fixed for 15 min at RT with 4% paraformaldehyde (PFA), washed and pre-blocked in 5% goat serum (Invitrogen) and 0.05% bovine serum albumin (BSA) for 1 h at 37 °C. Cells were incubated with 30 ng/mL of TGF-β3 for 4 h at RT. Bound TGF-β3 ligands were detected by incubating the cells with anti-TGF-β3 antibody (1:100; ProteinTech Group, Inc., Chicago, IL, USA) overnight at 4 °C. Cells were washed and incubated with the HRP-labeled anti-rabbit IgG (1:1000; Santa Cruz Biotechnology) for 1 h at RT. The amount of bound TGF-β3 was determined by tetramethylbenzidine (TMB) colorimetric substrate (Cell Signaling) after incubation for 10 min at RT. The reaction was terminated with an equal volume of 2 N H2SO4, and the optical absorbance was measured at 450 nm.
2.6. In vivo full-thickness articular cartilage defect model in Ccn4 knockout (KO) mice
Mice deficient in Ccn4 (Ccn4−/−) were generated using standard homologous recombination and gene targeting as described before [16]. Wild type (WT) and Ccn4-KO mice (C57BL/6) at 6 weeks of age were utilized in the experiments, according to the guidelines for Animal Care and Use at the NIDCR (ASP #13-700), NIH. Full-thickness articular cartilage defects were created according to a previously published report [17]. Briefly, three female WT and Ccn4-KO mice were anesthetized with isoflurane (2–5% in O2) and the joint capsule of the knee was opened, and the patella was dislocated laterally to expose the distal articular surface of the femur. Then, full-thickness articular cartilage defects of 400 μm diameter were created using 27 G needles, which has a diameter of 400 μm, in order to make the same size defect. Penetration to subchondral bone was confirmed by bleeding from the cartilage defect site. Muscles and patella were replaced on their original position and muscles and skin were sutured in separate layers. Four weeks after the surgery, animals were euthanized for histological analysis.
2.7. Isolation of articular cartilage from WT and Ccn4-KO mice
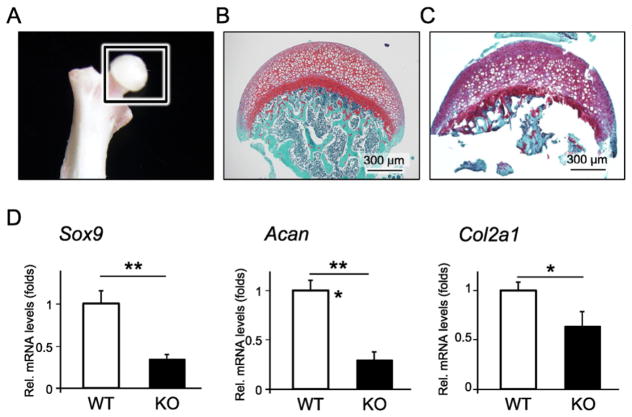
To compare the gene expression level of chondrocyte markers in the articular cartilage of WT and Ccn4-KO mice at 6 weeks of age, articular cartilage samples were carefully collected from the caput femur of WT and Ccn4-KO mice under a surgical microscope. Total RNA was isolated from the tissue samples using Trizol (Life Technologies), and gene expression levels of Sox9, aggrecan (Acan) and collagen type II, alpha 1 (Col2a1) were analyzed by real time RT-PCR.
2.8. Histological analysis and qualitative scoring
hBMSC pellets and mice samples were fixed in 4% PFA, dehydrated and embedded in paraffin. Serial sections of 5 μm were made, hydrated, and subsequently stained with hematoxylin and eosin (H&E), toluidine blue or safranin O for staining of glycosaminoglycans, as described previously [12]. For immunohistochemical staining of collagen type II, paraffin sections were deparaffinized, rehydrated, antigen-retrieved, blocked with 10% normal goat serum (Life Technologies) for 10 min, and incubated overnight at 4 °C with anti-collagen type II antibody (1:200; Millipore). Sections were then washed, incubated with secondary antibody and detected with 3-Amino-9-ethylcarbazole (AEC) color developing reagent (SuperPicture™ HRP Polymer Conjugate Broad Spectrum, Life Technologies), according to the manufacturer’s instructions. In a blinded fashion, an examiner used a modified ICRS score to evaluate the histological condition of the articular cartilage surface post-surgery, as reported previously [12]. In pellet culture experiments, quantitation of the positive area of collagen type II was performed using a BZ-X700 microscope equipped with a BZ analyzer (Keyence, Osaka, Japan).
2.9. Statistical analysis
The results obtained from quantitative experiments were reported as the mean values ± SD. Statistical differences were examined using a one-way factorial ANOVA followed by Tukey’s multiple comparison tests or Student’s unpaired t-tests.
3. Results
3.1. Expression of CCN family members in hBMSC cultures during chondrogenic differentiation in vitro
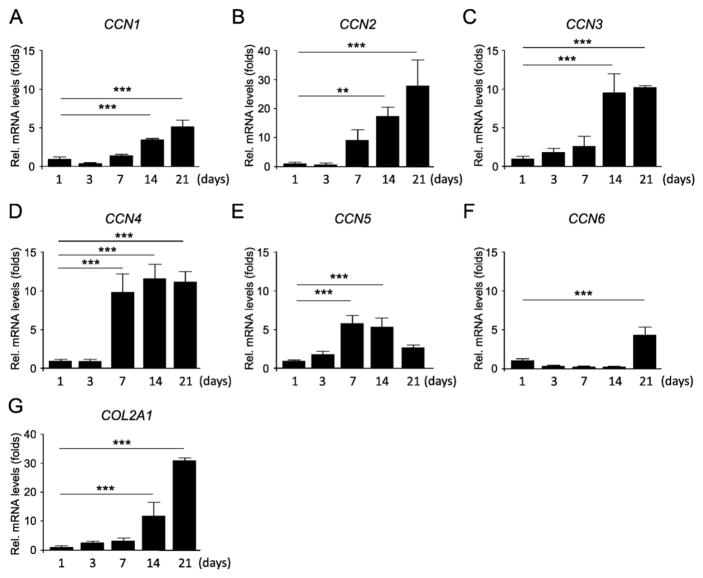
To obtain a better insight into the molecular mechanisms underlying the role of CCN family members in the process of chondrogenic differentiation of hBMSCs, we first investigated the expression levels of mRNA encoding genes of CCN family members in 21-day hBMSC micromass cultures. Although the transcript levels of CCN5 and CCN6 fluctuated during the culture period, there was a relatively high increase in the gene expression levels of CCN1, CCN2, CCN3 and CCN4 after the day 7, which followed a similar expression pattern of COL2A1, suggesting that these proteins could be playing important roles in modulating chondrogenesis (Fig. 1).
Fig. 1.
Expression pattern of CCN family genes during chondrogenesis. hBMSCs were cultured in a micromass culture system in chondrogenic induction medium for up to 21 days. Total RNA was collected at 1, 3, 7, 14, and 21 days, and the mRNA expression levels of CCN1 to CCN6 (A–F) and COL2A1 (G) were evaluated by real time RT-PCR. The expression of each gene was normalized to that of S29 ribosomal RNA. Data are reported as mean ± SD (n = 3). *p < 0.05, **p < 0.01, ***p < 0.001 versus day 1 (one-way ANOVA/Tukey). Results are representative data of at least three independent experiments.
3.2. CCN4 regulates TGF-β3-induced chondrogenesis of hBMSCs
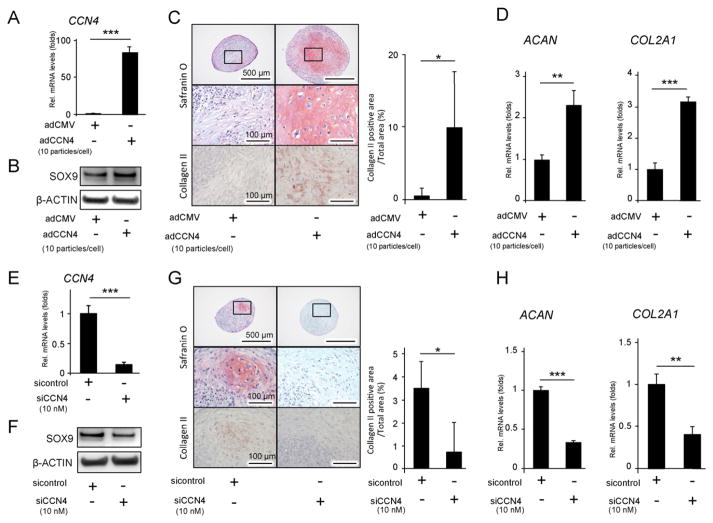
Since CCN1, CCN3 and mainly CCN2 had already been reported to be important factors regulating chondrocyte differentiation [3,18,19], we evaluated the influence of CCN4 on the chondrogenic process of hBMSCs by inducing CCN4 over-expression using adenovirus encoding CCN4 (adCCN4) as well as by silencing CCN4 gene using siCCN4. AdCCN4 was transduced into hBMSCs [8], and the increase in CCN4 gene expression level was confirmed by real time RT-PCR, in comparison with adCMV-transduced cells (Fig. 2A). Conversely, CCN4 gene expression level was decreased with a targeted gene knock down using siCCN4 (Fig. 2E). First, we confirmed that overexpression and down regulation of CCN4 did not affect hBMSC proliferation by MTS assay (data not shown). Next, since CCN4 is considered to be a downstream target of TGF-β [7], we tested the effect of CCN4 over expression and down regulation on TGF-β3-induced chondrogenic differentiation of hBMSCs in a micromass culture system. After 24 h of culture, we observed that adCCN4-transduced cells presented a notable increase in SOX9 protein levels (Fig 2B). In accordance, after 28 days of culture, we could observe an intense staining with safranin O and Alcian blue for detection of glycosaminoglycans, as well as an intense immunostaining for collagen type II in adCCN4-transduced hBMSC cultures (Figs. 2C, S1). A quantitative analysis of the major chondrocyte marker genes, ACAN and COL2A1, confirmed a marked increase in the levels of the two genes in adCCN4-transduced hBMSC cultures, compared to adCMV (Fig. 2D). By contrast, knockdown of CCN4 suppressed chondrogenic differentiation of hBMSCs, as demonstrated by a decrease in SOX9 levels in 24 h hBMSC pellets (Fig. 2F). Additionally, chondrocyte-like cells were nearly absent, and ACAN and COL2A1 transcript levels were also significantly down regulated in siCCN4-transduced hBMSC micromasses, after 28 days of culture (Figs. 2G, H, S1). Taken together, these results demonstrate that CCN4 plays an important role in enhancing chondrogenic differentiation of hBMSCs in vitro.
Fig. 2.
The effect of CCN4 on the chondrogenic differentiation of hBMSCs in vitro. hBMSCs were transduced with adCCN4 (A–D) or siCCN4 (E–H) for two days, and cultured in micromasses. (A, E) Two days after the transduction, mRNA expression levels of CCN4 gene were measured by real-time RT-PCR. (B, F) The cellular proteins were collected 24 h after chondrogenic induction, and protein levels of SOX9 were detected by western blot. β-ACTIN was used as protein loading control. (C, G) Twenty eight days after chondrogenic induction, histological analysis of hBMSC micromass cultures was performed for detection of glycosaminoglycans with safranin O staining, or collagen type II by immunohistochemistry. Graphs show the quantitation of the positive area of collagen type II. (D, H) mRNA expression level of ACAN COL2 gene was measured by real time RT-PCR. The expression of each gene was normalized to that of S29 ribosomal RNA. Bars represent the mean values and standard deviation (+/−SD) (n = 3). **p < 0.01, ***p < 0.001 (Student’s t-tests). Results are representative data of at least three independent experiments.
3.3. CCN4 enhances TGF-β3-induced phosphorylation of SMADs
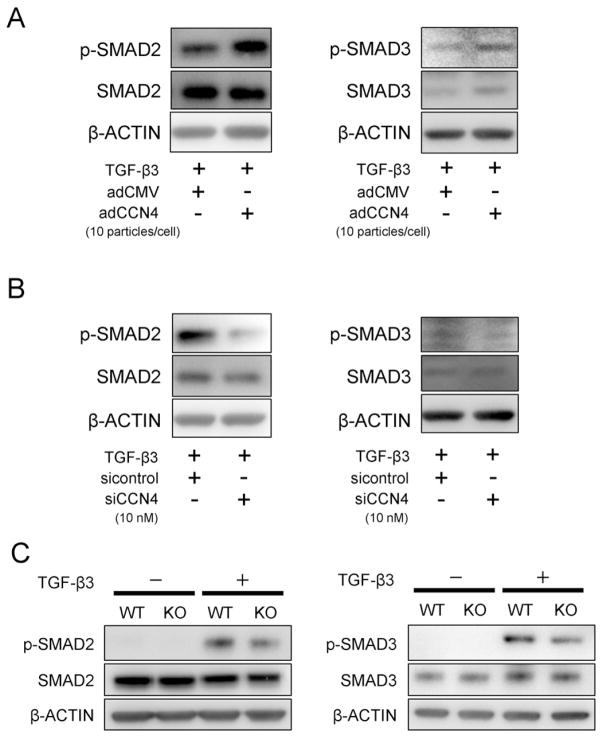
Next, in order to investigate the mechanism of how CCN4 interacts with TGF-β3 during chondrogenic differentiation of hBMSCs, we cultured the cells in monolayer, and transduced the cells with adCCN4. Forty eight hours after adCCN4 transduction, stimulation with rhTGF-β3 (10 ng/mL) for 10 min significantly increased the phosphorylation of SMAD2 and SMAD3 as compared to adCMV or TGF-β3-treated control cells (Fig. 3A). In contrast, as a response to the CCN4 knockdown, TGF-β3-induced activation of SMAD2 and SMAD3 was repressed in hBMSCs transduced with siCCN4 (Fig. 3B). In order to confirm the effect of CCN4 on the TGF-β signaling pathway, we isolated chondrocytes from growth plate of newborn WT and Ccn4-KO mice, and observed that TGF-β3-induced phosphorylation of SMAD2/3 was also reduced in chondrocytes derived from Ccn4-KO mice (Fig. 3C). These results explain in part the effect of CCN4 in enhancing TGF-β3-induced chondrogenic differentiation of hBMSCs by promoting a stronger activation of SMAD2 and SMAD3.
Fig. 3.
The effect of CCN4 on TGF-β3-SMAD signaling pathway in vitro. hBMSCs were transduced with adCCN4 (A) or siCCN4 (B) for 2 days. (A, B) Transduced-hBMSCs in monolayer culture were treated with 10 ng/mL of TGF-β3 for 10 min, and cellular proteins were collected for detection of SMAD2/3 activity. β-ACTIN was used as protein loading control. (C) Chondrocytes were collected from mandibular condylar cartilage of newborn WT and Ccn4-KO mice, and stimulated with TGF-β3 for 10 min. The cellular proteins were collected for analysis of TGF-β3-induced phosphorylation of SMAD2/3. SMAD2/3 activity was decreased in chondrocytes derived from Ccn4-KO mice, compared to WT mice.
3.4. Interaction between CCN4 and TGF-β3
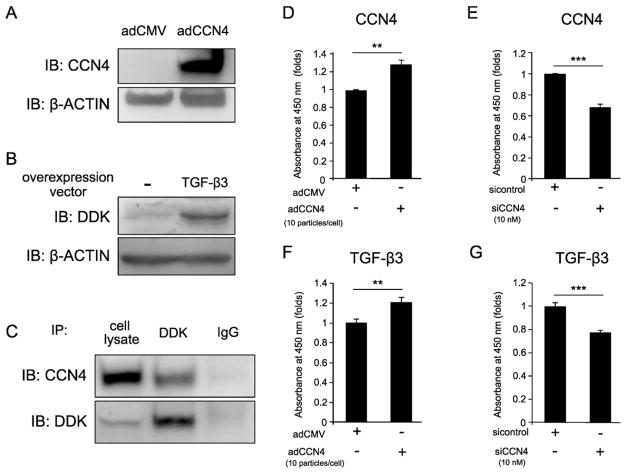
It has been reported that CCN family members bind to the members of the TGF-β superfamily and regulate their function, thus we speculated that CCN4 could bind to TGF-β3. To test this hypothesis, hBMSCs were transduced with adCCN4 and/or TGF-β3 overexpression vector for 2 days and the protein levels of CCN4 or TGF-β3 were detected by western blot (Fig. 4A and B). Immunoprecipitation (IP)–western blot analysis showed a binding between DDK-tagged TGF-β3 and CCN4 (Fig. 4C). The direct binding between the two proteins was also evaluated using recombinant proteins (rhCCN4 and rhTGF-β3); however, the IP-western results did not confirm that there was a direct binding between the two recombinant proteins (data not shown). Thus in cell extracts our data shows that CCN4 binds to TGF-β3, however we cannot conclude definitively whether this binding is direct or, rather, requires additional intermediating factors.
Fig. 4.
Interaction between CCN4 and TGF-β3. hBMSCs were transduced with adCCN4 and/or TGF-β3-DDK-over expression vector, and incubated for 2 days, before collection of cell lysates. (A, B) Western blotting was performed using anti-CCN4 antibody or anti-DDK antibody, respectively. (C) Immunoprecipitation assay was performed using an anti-DDK antibody, and CCN4 or TGF-β3/DDK was detected by immunoblotting using an anti-CCN4 or anti-DDK antibodies. (D, E) hBMSCs were transduced with adCCN4 or siCCN4, respectively. After two days, cells were fixed and, the amount of CCN4 bound to cells was measured by its immunoreactivity to anti-CCN4 antibody. (F, G) To analyze the interaction of CCN4 and TGF-β3, fixed cells were incubated with TGF-β3 for 2 h. The amount of TGF-β3 bound to cells was measured by its immunoreactivity using anti-DDK antibody. Bars represent the mean values and standard deviation (+/−SD) (n = 3). **p < 0.01, ***p < 0.001 (Student’s t-tests). Results are representative data of at least three independent experiments.
Since both CCN4 and TGF-β3 are secreted proteins, we then hypothesized that they could interact to each other at the cell membrane or pericellularly at the extracellular level. Therefore, to determine whether CCN4 could regulate TGF-β3-to-cell binding affinity, we performed an on-cell protein binding assay with CCN4 overexpression and knockdown conditions. First, we performed a quantitative analysis of the amount of CCN4 bound to the surface of cells using anti-CCN4 antibody. As shown in Fig. 4D and E, the amount of CCN4 on the surface of cells was increased in the adCCN4-transduced hBMSCs; and on the other hand, it was decreased in siCCN4-transduced hBMSCs.
Next, to perform the TGF-β3 affinity assay, adCCN4 or siCCN4-transduced hBMSCs were treated with rhTGF-β3 (1 μg/mL) for 4 h and the amount of TGF-β3 bound to the cell membrane was detected by using anti-TGF-β3 antibody. As predicted, increased amounts of TGF-β3 were detected in hBMSCs transduced with adCCN4 (Fig. 4F). Conversely, the binding affinity of TGF-β3 to hBMSCs was decreased under CCN4 knockdown condition (Fig. 4G). Taken together, these data support the concept that CCN4 can enhance TGF-β3 function by facilitating its binding to the surface of hBMSCs and subsequently enhancing the activation of TGF-β downstream signaling.
3.5. Ccn4 deficiency decreases the chondrogenic differentiation related markers in articular cartilage and delays articular cartilage regeneration in vivo
Next, we isolated articular cartilage samples from the caput femur of WT and Ccn4-KO mice (Fig. 5A–C), and found that the gene expression levels of chondrocyte-related markers, Sox9, Acan and Col2a1, cartilage ECM including Biglycan (Bgn) and Decorin (Dcn), were significantly decreased in Ccn4-KO mice (Figs. 5D, S2), indicating that CCN4 is necessary for the maintenance of articular cartilage homeostasis. Of note, however, gene expression levels of collagen type X, alpha 1 (Col10a1), which is hypertrophic chondrocyte marker and highly expressed in articular cartilage of OA, were not detected in articular cartilage of either WT or Ccn4-KO mice (data not shown). Moreover, histological analysis of the joints showed that there was no notable difference in morphology between WT and Ccn4-KO mice in the articular cartilage of the knees (Fig. S3A–C).
Fig. 5.
Comparison of gene expression level of chondrogenic markers in articular cartilage between WT and Ccn4-KO mice. Articular cartilage samples of caput femoris for analysis of gene expression level of chondrogenic markers in healthy articular cartilage. Photograph and histological image of safranin O staining before the isolation of the articular cartilage are shown in (A) and (B), respectively; and the histological image after isolation is shown in (C). (D) Total RNA was collected from the articular cartilage separated from the caput femoris, and the mRNA expression levels of chondrogenic markers were measured by real time RT-PCR. The expression of each gene was normalized to that of S29 ribosomal RNA. Bars represent the mean values and standard deviation (+/−SD) (n = 3). *p < 0.05, **p < 0.01, ***p < 0.001 (Student’s t-tests). Results are representative data of at least three independent experiments.
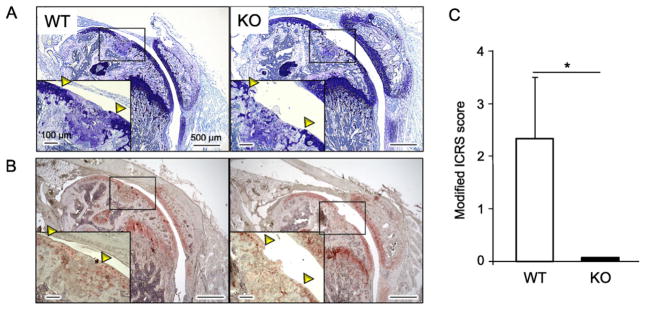
Finally, we performed full-thickness articular cartilage defects in the knee joints of WT and Ccn4-KO mice, and investigated whether CCN4 could work as a positive regulator of articular cartilage repair. One month after the surgical defect, histological analysis of the joints revealed the formation of a newly-formed cartilage-like tissue positively stained with toluidine blue in WT mice (Fig. 6A). Moreover, immunostaining analysis showed that the newly-formed articular cartilage-like tissue was positive for collagen type II (Fig. 6B). Conversely, no cartilage-like tissue could be observed in the defect region of Ccn4-KO (Fig. 6A), and a histopathology-based scoring of the repair cartilage showed a significant difference between the WT mice group and Ccn4-KO mice group, indicating a substantial decrease in the ability of the articular cartilage to repair in Ccn4-KO mice (Fig. 6C).
Fig. 6.
Comparison of the articular cartilage healing between WT and KO mice. The full-thickness articular cartilage defects of 400 μm in diameter were surgically created in the knee joints of WT and KO mice to evaluate the healing ability of articular cartilage. Four weeks after the surgery, samples were collected and specimens were stained with toluidine blue (A), and immunostained for collagen type II (B). (C) Evaluation of the histological condition of the articular cartilage surface was performed using a modified ICRS score. Bars represent the mean values and standard deviation (+/−SD) (n = 3). *p < 0.05 (Student’s t-tests).
4. Discussion
CCN family members are expressed in development of growth plate cartilage and, because of their unique structural domain, they can interact with various growth factors and ECM components [20]. CCN2 for example, is expressed dominantly in the pre-hypertrophic zone of growth plate and interacts with BMP-2 and ACAN, and regulates chondrocyte proliferation and differentiation [4,21]. Furthermore, CCN2 was shown to promote the regeneration of articular cartilage [22]; and aging-related cartilage degradation could be prevented by cartilage-specific overexpression of CCN2 [23]. Using the targeted mutation mice model of CCN3, Roddy K.A. et al. have also reported that CCN3 is required for maintenance of articular cartilage and for joint homeostasis [24].
CCN4 is also expressed in the hypertrophic zone [25], and our previous results show that CCN4 can regulate osteogenesis by controlling the affinity of BMP-2 to hBMSCs [8]. Despite the implications of our previous reports that elucidated the effect of CCN4 on osteoblast differentiation and function [8,26], no study has yet determined a decisive role of CCN4 in chondrogenesis and cartilage regeneration. In this study, we demonstrated that CCN4 overexpression significantly increased the TGF-β3-induced chondrogenic differentiation of hBMSCs, and that CCN4 plays important roles in joint homeostasis and in the process of articular cartilage repair. Nevertheless, the articular cartilage defect model used in this study did not enable a complete repair of the articular surface in WT mice (Fig. 6). A possible reasoning for these results could be the age of the mice, since Matsuoka et al. demonstrated that age is one of the crucial factors for the articular cartilage repair, with superior results observed in young (3 week-old) or juvenile (4 week-old) mice [17]; and thus, the 6 week-old mice used in this study could be relatively aged, with a limited capacity for natural healing of the articular cartilage. Despite the incomplete repair of the articular cartilage in WT mice, our results show that CCN4 may be playing important roles in cartilage metabolism, as we could detect a significant decrease in the gene expression levels of Sox9, Acan, and Col2a1 in the articular cartilage of Ccn4-KO mice, which eventually, could also contribute to the observed impaired healing capacity of chondrocytes.
Additionally, our recent article showed that the body length of Ccn4-KO mice is slightly but significantly reduced compared to wild type mice [16]. We also analyzed the growth plates of growing mice at embryonic day E18.5 and showed that the hypertrophic zone of growth plate was expanded in the Ccn4-KO compared to wild type mice. From these data, we also suspect that CCN4 could regulate the long bone growth by controlling the apoptosis of hypertrophic chondrocytes. Further experiments will be required to elucidate this issue.
Of note, in vitro experiments showed that CCN4 alone does not have any function for osteo-chondrogenesis. Therefore, CCN4 action may be mediated by interacting with other growth factors associated with chondrogenesis, and regulating their function. Here we show that CCN4 can interact with and control TGF-β3 function; however, additional experiments are necessary to clarify the exact mechanisms of action, since the direct binding between the two factors could not be determined. Other cellular mechanisms, independent of TGF-β3, could also be involved in the CCN4’s role in chondrogenesis. For instance, it has been reported that tumor necrosis factor-α (TNF-α), a pro-inflammatory cytokine, induces focal loss of cartilage in OA knee joints [27]. Additionally, in the cardiac fibroblast, TNF-α up-regulates the expression level of CCN4 via its cyclic AMP response element-binding protein (CREB) site, and that CCN4 protects from TNF-α induced cell death [28]. Indeed, elevated levels of both TNF-α and CCN4 have been found in the cartilage of OA patients [9,10,29,30] and, CCN4 could be not only a key component in the process of cartilage differentiation but also could be playing important roles in organizing the inflammatory condition in OA, and promoting cartilage repair.
In summary, for the first time, we showed that CCN4 exerts a positive effect on TGF-β3-induced chondrogenesis by modulating TGF-β3 binding to the surface of hBMSCs and enhancing its downstream signaling. Further investigations are necessary for a better understanding of the underlying molecular and cellular mechanisms involved in effect of CCN family proteins in the process of chondrogenesis and/or cartilage repair, as well as for advancing new therapeutic modalities for OA to increase current clinical outcomes.
Supplementary Material
Acknowledgments
This study was supported in part by JSPS KAKENHI Grant Numbers #23890122 and #26861639 and the Intramural Program of the NIH, NIDCR.
Appendix A. Supplementary data
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.bone.2015.11.007.
Footnotes
Conflict of interest
The authors declare no competing interests.
Author contributions
Study design: MO, MFY and TK. Study conduct: MO and MFY. Data collection: YY, AM, TMK, ESH and EA. Data analysis: YY, AM, HK, and JU. Data interpretation: MO, TO, MT, and TK. Drafting manuscript: YY, MO and ESH. Revising manuscript content: MFY, and TK. Approving final version of manuscript: TO, MT and TK.
References
- 1.Yamada Y, Okuizumi H, Miyauchi A, Takagi Y, Ikeda K, Harada A. Association of transforming growth factor beta1 genotype with spinal osteophytosis in Japanese women. Arthritis Rheum. 2000;43:452–460. doi: 10.1002/1529-0131(200002)43:2<452::AID-ANR28>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 2.Lau HH, Ho AY, Luk KD, Kung AW. Transforming growth factor-beta1 gene polymorphisms and bone turnover, bone mineral density and fracture risk in southern Chinese women. Calcif Tissue Int. 2004;74:516–521. doi: 10.1007/s00223-004-0163-4. [DOI] [PubMed] [Google Scholar]
- 3.Lorda-Diez CI, Montero JA, Diaz-Mendoza MJ, Garcia-Porrero JA, Hurle JM. Defining the earliest transcriptional steps of chondrogenic progenitor specification during the formation of the digits in the embryonic limb. PLoS One. 2011;6:e24546. doi: 10.1371/journal.pone.0024546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maeda A, Nishida T, Aoyama E, Kubota S, Lyons KM, Kuboki T, Takigawa M. CCN family 2/connective tissue growth factor modulates BMP signalling as a signal conductor, which action regulates the proliferation and differentiation of chondrocytes. J Biochem. 2009;145:207–216. doi: 10.1093/jb/mvn159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abreu JG, Ketpura NI, Reversade B, De Robertis EM. Connective-tissue growth factor (CTGF) modulates cell signalling by BMP and TGF-beta. Nat Cell Biol. 2002;4:599–604. doi: 10.1038/ncb826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aoyama E, Kubota S, Takigawa M. CCN2/CTGF binds to fibroblast growth factor receptor 2 and modulates its signaling. FEBS Lett. 2012;586:4270–4275. doi: 10.1016/j.febslet.2012.10.038. [DOI] [PubMed] [Google Scholar]
- 7.Inkson CA, Ono M, Kuznetsov SA, Fisher LW, Robey PG, Young MF. TGF-beta1 and WISP-1/CCN-4 can regulate each other’s activity to cooperatively control osteoblast function. J Cell Biochem. 2008;104:1865–1878. doi: 10.1002/jcb.21754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ono M, Inkson CA, Kilts TM, Young MF. WISP-1/CCN4 regulates osteogenesis by enhancing BMP-2 activity. J Bone Miner Res. 2011;26:193–208. doi: 10.1002/jbmr.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Urano T, Narusawa K, Shiraki M, Usui T, Sasaki N, Hosoi T, Ouchi Y, Nakamura T, Inoue S. Association of a single nucleotide polymorphism in the WISP1 gene with spinal osteoarthritis in postmenopausal Japanese women. J Bone Miner Metab. 2007;25:253–258. doi: 10.1007/s00774-007-0757-9. [DOI] [PubMed] [Google Scholar]
- 10.Blom AB, Brockbank SM, van Lent PL, van Beuningen HM, Geurts J, Takahashi N, van der Kraan PM, van de Loo FA, Schreurs BW, Clements K, Newham P, van den Berg WB. Involvement of the Wnt signaling pathway in experimental and human osteoarthritis: prominent role of Wnt-induced signaling protein 1. Arthritis Rheum. 2009;60:501–512. doi: 10.1002/art.24247. [DOI] [PubMed] [Google Scholar]
- 11.Liu JF, Hou SM, Tsai CH, Huang CY, Hsu CJ, Tang CH. CCN4 induces vascular cell adhesion molecule-1 expression in human synovial fibroblasts and promotes monocyte adhesion. Biochim Biophys Acta. 1833;2013:966–975. doi: 10.1016/j.bbamcr.2012.12.023. [DOI] [PubMed] [Google Scholar]
- 12.Hara ES, Ono M, Hai PT, Sonoyama W, Kubota S, Takigawa M, Matsumoto T, Young MF, Olsen BR, Kuboki T. Fluocinolone acetonide is a potent synergistic factor of TGF-beta3-associated chondrogenesis of bone marrow-derived mesenchymal stem cells for articular surface regeneration. J Bone Miner Res. 2015 doi: 10.1002/jbmr.2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woods A, Wang G, Dupuis H, Shao Z, Beier F. Rac1 signaling stimulates N-cadherin expression, mesenchymal condensation, and chondrogenesis. J Biol Chem. 2007;282:23500–23508. doi: 10.1074/jbc.M700680200. [DOI] [PubMed] [Google Scholar]
- 14.Xu L, Corcoran RB, Welsh JW, Pennica D, Levine AJ. WISP-1 is a Wnt-1- and beta-catenin-responsive oncogene. Genes Dev. 2000;14:585–595. [PMC free article] [PubMed] [Google Scholar]
- 15.Ono M, Inkson CA, Sonn R, Kilts TM, de Castro LF, Maeda A, Fisher LW, Robey PG, Berendsen AD, Li L, McCartney-Francis N, Brown AC, Crawford NP, Molinolo A, Jain A, Fedarko NS, Young MF. WISP1/CCN4: a potential target for inhibiting prostate cancer growth and spread to bone. PLoS One. 2013;8:e71709. doi: 10.1371/journal.pone.0071709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maeda A, Ono M, Holmbeck K, Li L, Kilts TM, Kram V, Noonan ML, Yoshioka Y, McNerny EM, Tantillo MA, Kohn DH, Lyons KM, Robey PG, Young MF. WNT1-induced Secreted Protein-1 (WISP1), a novel regulator of bone turnover and Wnt signaling. J Biol Chem. 2015;290:14004–14018. doi: 10.1074/jbc.M114.628818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsuoka M, Onodera T, Sasazawa F, Momma D, Baba R, Hontani K, Iwasaki N. An articular cartilage repair model in common C57Bl/6 mice. Tissue Eng Part C Methods. 2015 doi: 10.1089/ten.tec.2014.0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lafont J, Jacques C, Le Dreau G, Calhabeu F, Thibout H, Dubois C, Berenbaum F, Laurent M, Martinerie C. New target genes for NOV/CCN3 in chondrocytes: TGF-beta2 and type X collagen. J Bone Miner Res. 2005;20:2213–2223. doi: 10.1359/JBMR.050818. [DOI] [PubMed] [Google Scholar]
- 19.Ivkovic S, Yoon BS, Popoff SN, Safadi FF, Libuda DE, Stephenson RC, Daluiski A, Lyons KM. Connective tissue growth factor coordinates chondrogenesis and angiogenesis during skeletal development. Development. 2003;130:2779–2791. doi: 10.1242/dev.00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawaki H, Kubota S, Suzuki A, Lazar N, Yamada T, Matsumura T, Ohgawara T, Maeda T, Perbal B, Lyons KM, Takigawa M. Cooperative regulation of chondrocyte differentiation by CCN2 and CCN3 shown by a comprehensive analysis of the CCN family proteins in cartilage. J Bone Miner Res. 2008;23:1751–1764. doi: 10.1359/jbmr.080615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aoyama E, Hattori T, Hoshijima M, Araki D, Nishida T, Kubota S, Takigawa M. N-terminal domains of CCN family 2/connective tissue growth factor bind to aggrecan. Biochem J. 2009;420:413–420. doi: 10.1042/BJ20081991. [DOI] [PubMed] [Google Scholar]
- 22.Nishida T, Kubota S, Kojima S, Kuboki T, Nakao K, Kushibiki T, Tabata Y, Takigawa M. Regeneration of defects in articular cartilage in rat knee joints by CCN2 (connective tissue growth factor) J Bone Miner Res. 2004;19:1308–1319. doi: 10.1359/JBMR.040322. [DOI] [PubMed] [Google Scholar]
- 23.Itoh S, Hattori T, Tomita N, Aoyama E, Yutani Y, Yamashiro T, Takigawa M. CCN family member 2/connective tissue growth factor (CCN2/CTGF) has anti-aging effects that protect articular cartilage from age-related degenerative changes. PLoS One. 2013;8:e71156. doi: 10.1371/journal.pone.0071156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roddy KA, Boulter CA. Targeted mutation of NOV/CCN3 in mice disrupts joint homeostasis and causes osteoarthritis-like disease. Osteoarthr Cartil. 2015;23:607–615. doi: 10.1016/j.joca.2014.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yanagita T, Kubota S, Kawaki H, Kawata K, Kondo S, Takano-Yamamoto T, Tanaka S, Takigawa M. Expression and physiological role of CCN4/Wnt-induced secreted protein 1 mRNA splicing variants in chondrocytes. FEBS J. 2007;274:1655–1665. doi: 10.1111/j.1742-4658.2007.05709.x. [DOI] [PubMed] [Google Scholar]
- 26.Maeda A, Ono M, Holmbeck K, Li L, Kilts TM, Kram V, Noonan ML, Yoshioka Y, McNerny E, Tantillo M, Kohn D, Lyons KM, Robey PG, Young MF. WNT1 induced secreted protein-1 (WISP1): a novel regulator of bone turnover and Wnt signaling. J Biol Chem. 2015 doi: 10.1074/jbc.M114.628818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Westacott CI, Barakat AF, Wood L, Perry MJ, Neison P, Bisbinas I, Armstrong L, Millar AB, Elson CJ. Tumor necrosis factor alpha can contribute to focal loss of cartilage in osteoarthritis. Osteoarthr Cartil. 2000;8:213–221. doi: 10.1053/joca.1999.0292. [DOI] [PubMed] [Google Scholar]
- 28.Venkatachalam K, Venkatesan B, Valente AJ, Melby PC, Nandish S, Reusch JE, Clark RA, Chandrasekar B. WISP1, a pro-mitogenic, pro-survival factor, mediates tumor necrosis factor-alpha (TNF-alpha)-stimulated cardiac fibroblast proliferation but inhibits TNF-alpha-induced cardiomyocyte death. J Biol Chem. 2009;284:14414–14427. doi: 10.1074/jbc.M809757200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stannus O, Jones G, Cicuttini F, Parameswaran V, Quinn S, Burgess J, Ding C. Circulating levels of IL-6 and TNF-alpha are associated with knee radiographic osteoarthritis and knee cartilage loss in older adults. Osteoarthr Cartil. 2010;18:1441–1447. doi: 10.1016/j.joca.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 30.Xue J, Wang J, Liu Q, Luo A. Tumor necrosis factor-alpha induces ADAMTS-4 expression in human osteoarthritis chondrocytes. Mol Med Rep. 2013;8:1755–1760. doi: 10.3892/mmr.2013.1729. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.