Abstract
CRISPR-Cas genome editing methods hold immense potential as therapeutic tools to fix disease-causing mutations at the level of DNA. In contrast to typical drug development strategies aimed at targets that are highly conserved among individual patients, treatment of disease at the genomic level must contend with significant inter-individual natural genetic variation. Here we analyze the recently released ExAC (http://exac.broadinstitute.org) and 1000 Genomes datasets (http://www.internationalgenome.org) to determine how human genetic variation impacts Cas endonuclease target choice in the context of therapeutic genome editing. We find that this variation confounds the target sites of certain Cas endonucleases more than others and we provide a compendium of guide RNAs predicted to have high efficacy in diverse patient populations. For further analysis, we focus on 12 therapeutically relevant genes and consider how genetic variation affects off-target candidates for these loci. Our analysis suggests that in large populations of individuals, most candidate off-target sites will be rare, underscoring the need for pre-screening of patients through whole genome sequencing to ensure safety. This information can be integrated with empirical methods for guide RNA selection into a framework for designing CRISPR-based therapeutics that maximize efficacy and safety across patient populations.
The development of CRISPR-based RNA-guided endonucleases such as Cas9 and Cpf1 for eukaryotic genome editing has sparked intense interest in the use of this technology for therapeutic applications1,2,3. In contrast to small molecule therapies, which target highly conserved protein active sites, therapies designed to target particular DNA sequences must take into account genetic variation among patient populations. If this variation disrupts the therapy target site it can affect the efficacy of a CRISPR-based therapeutic; if this variation generates off-target candidate sites it can affect the safety of a CRISPR-based therapeutic. Previously, it has been reported that genetic variation in cell lines can alter Cas9 targeting4, but there has been limited effort to comprehensively and systematically evaluate this phenomenon in large human populations.
As CRISPR-based therapies advance toward human clinical trials, it is important to consider how natural genetic variation in the human population may affect the results from these trials and may even affect patient safety. Recently, large scale sequencing datasets from the Exome Aggregation Consortium (ExAC) and 1000 Genomes Project have provided an unprecedented view of the landscape of human genetic variation5,6,7,8. These datasets have captured nearly all common variants in the human population and contain deep coverage of rare variants8,5, enabling evaluation of the effects of human variation on therapeutic genome editing in diverse human populations. Here we use these datasets to determine the impact of population genetic variation on therapeutic genome editing with Streptococcus pyogenes (Sp) Cas9, SpCas9 variants VQR and VRER, Staphylococcus aureus (Sa) Cas9, and Acidaminococcus sp. (As) Cpf11,2,9,10,3. We find extensive variation likely to impact the efficacy of these enzymes, and propose that unique, patient-specific off-target candidates will be one of the main challenges for ensuring the safety of these therapeutics. These results provide a framework for designing CRISPR-based therapeutics, highlight the need to develop multiple guide RNA-enzyme pairs for each target locus, and suggest that pre-therapeutic whole genome sequencing will be required to ensure uniform efficacy and safety for treatment across patient populations.
Results
Human genetic variation impacts choice of Cas enzyme
To date, two families of class 2 (single effector) CRISPR nucleases, Cas9 and Cpf1, have been harnessed for eukaryotic genome editing1,2,11,3. Both Cas9 and Cpf1 are programmed by RNA guides, which direct cleavage of DNA targets (protospacers) that are complementary to the RNA guide target and flanked by a short protospacer adjacent motif (PAM) specific to each endonuclease12,3 (Fig. 1a). Mismatches between the RNA guide and its DNA target have been shown to decrease RNA-guided endonuclease activity, and deviation from the canonical PAM sequence often completely abolishes nuclease activity13,14,15,16. The recently released ExAC dataset, based on 60,706 individuals, contains on average one variant per eight nucleotides in the human exome5. To assess the impact of this variation on RNA guide efficacy, we used the ExAC dataset to catalog variants present in all possible targets in the human reference exome that either (i) disrupt the target PAM sequence or (ii) introduce mismatches between the RNA guide and the genomic DNA, which we collectively term target variation (Fig. 1a).
Figure 1. Human genetic variation significantly impacts the efficacy of RNA-guided endonucleases.
a, Schematic illustrating the genomic target (consisting of spacer and adjacent PAM element), RNA guide, and target variation. b, Fraction of residues for individual nucleotides containing variation in the ExAC dataset. c, Fraction of 2-nt PAM motifs altered by variants in the ExAC dataset. d, Percent of target variants at different allele frequencies for each CRISPR endonuclease. e, Cumulative percent of targets containing variants for each CRISPR endonuclease. f, Fraction of targets containing homozygous variants at different allele frequencies. The Mean and SEM for 5 CRISPR endonucleases is shown.
In addition to two orthologs of Cas9 (SpCas9 and SaCas9) and Cpf1, a number of SpCas9 variants have been engineered as tools for genome editing, each with a different PAM1,2,9,10,3 (Table 1). Consideration of multiple enzymes with different PAM requirements will increase the number of available genomic targets for therapeutic loci. We therefore assessed variation at each PAM in the human exome for SpCas9 (PAM = NGG), SpCas9-VQR (NGA), SpCas9-VRER (NGCG), SaCas9 (NNGRRT), and AsCpf1 (TTTN), all of which are currently being considered as candidate enzymes for CRISPR therapeutic development. The recently reported eSpCas9 and SpCas9-HF have the same NGG PAM as SpCas9, and are thus not considered separately here17,18. For each nuclease, we determined the fraction of exonic PAMs containing variants that alter PAM recognition either through abolishing or creating a new PAM relative to the reference genome. For the ExAC population, the total fraction of targets containing PAM-altering variants was similar for all enzymes (21 – 35%), except for SpCas9-VRER, which is impacted by PAM-altering variants in 80% of targets (Table 1, Fig. S1). The PAM for SpCas9-VRER contains a CpG motif, which has been shown to be highly mutable5. Consistent with these results, we find that CG is the most highly mutable 2-nt PAM motif in the human exome, and 66% of cytosine and guanine residues contained in CpG motifs show variation for the 60,706 ExAC individuals5 (Fig. 1b, c; Table S1). These results suggest that enzymes using PAMs containing CG motifs are significantly more affected by target variation in the human genome.
Table 1. Fraction of targets containing PAM-altering variants for five Cas enzymes.
n specifies the number of protein coding targets in the human exome for each enzyme. Orientation refers to the location of the PAM relative to the RNA guide complementary region of the target, and allele frequencies refer to the percentage of the ExAC population containing a particular PAM altering variant for a given target in the human exome.
| Whole-exome PAM variation by allele frequency (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Protein | PAM | Orientation | ≥10% | ≥1 | ≥0.1 | ≥0.01 | ≥0.001 | Total | n |
| AsCpf1-WT | TTTN | Left | 0.15 | 0.26 | 0.61 | 1.81 | 8.91 | 21.04 | 2702056 |
| SpCas9-VQR | NGA | Right | 0.11 | 0.25 | 0.69 | 2.28 | 11.39 | 23.19 | 9838603 |
| SpCas9-WT | NGG | Right | 0.16 | 0.37 | 1.13 | 3.82 | 17.46 | 32.61 | 10286445 |
| SaCas9-WT | NNGRRT | Right | 0.23 | 0.44 | 1.16 | 3.68 | 17.29 | 34.68 | 1938911 |
| SpCas9-VRER | NGCG | Right | 0.77 | 1.86 | 5.79 | 20.72 | 66.67 | 80.16 | 981524 |
Low-variation regions of the human exome are more reliably targeted
We extended our analysis to determine the fraction of all possible targets in the human exome for SpCas9, SpCas9-VQR, SaCas9, and AsCpf1 that contain variants. We find that 93 – 95% of targets contain variants in the ExAC dataset that are likely to alter the efficiency of target cleavage (Fig. 1d, e; Table S2), and most target variation occurring at frequencies less than 0.1% is heterozygous (Fig. 1d,f; Table S2).
The ExAC dataset is large enough that it provides near comprehensive coverage of variants at allele frequencies of greater than or equal to 0.01% in the population (i.e., variation will exist in at least 1 out of 10,000 alleles in the population)5. Hence, we used this dataset to compile a list of exome-wide target sites for SpCas9, Cas9-VQR, SaCas9, and AsCpf1 lacking variants occurring at ≥0.01% allele frequency (referred to as platinum targets; Whole exome platinum targets for each enzyme, Tables S4 – 7)). These platinum targets should be efficacious in ≥99.99% of the population (Fig. 2b; target variation less than or equal to 0.01%).
Figure 2. Selection of platinum targets maximizes population efficacy.
a, Schematic showing target variation within exon 2 of PCSK9, with regions containing high coverage in the ExAC dataset indicated by black lines below exons. Variants for a short region of PCSK9 exon 2 are highlighted along with 5 targets for SpCas9-WT. Top strand targets (PAM on the right) are shown above the region of PCSK9 exon 2, and bottom strand targets (PAM on the left) are shown below. The maximum frequency variant in the ExAC dataset intersecting each target is indicated colorimetrically and used as the “target variation frequency”. Variants that do not affect the target recognition by the endonuclease (such as a high frequency variant intersecting the ‘N’ in the NGG PAM of target 2) do not affect targeting efficiency and are excluded. Targets 2 and 4 show the lowest variation of the 5 targets shown. b, Frequency of target variation plotted by cut site position for targets spanning the start of PCSK9-001 exon 2, with the 5 targets shown in (a) indicated by arrows. The horizontal line at 0.01% separates platinum targets (grey) from targets with high variation (red). The classification for each target is depicted below for each enzyme (grey or red boxes). c, Classification of targets for each enzyme spanning exons 2 – 5 of PCSK9-001.
For further analysis, we selected 12 therapeutically relevant genes, including those that are currently the focus of therapeutic development; CEP290, CFTR, DMD, G6PC, HBB, IDUA, IL2RG, PCSK9, PDCD1, SERPINA1, TTR, VEGFA (Fig. S2; platinum targets for each enzyme for these 12 genes, Tables S8 - 11). For these 12 genes, approximately two-thirds of possible exonic targets meet our platinum criteria, with PCSK9 containing the smallest fraction of platinum targets (50%) (Table S3). This suggests that for most genomic regions, there will exist ample platinum targets that can be considered when beginning the process of therapeutic target selection.
We observed that both high variation targets and platinum targets cluster along exons for each of the 12 genes examined. For example, all targets in the 5′ half of PCSK9 exon 4 are platinum, whereas very few platinum targets exist for exon 5 (Fig. 2c). However, even for regions of high frequency variation, such as PCSK9 exons 1-4, it is still possible to find small numbers of platinum targets for some enzymes (Fig. 2c). This observation for PCSK9 is representative of the other genes investigated in this study and suggests that considering multiple enzymes with distinct PAM requirements increases the likelihood of finding a platinum target. In the event that a genomic region of interest contains variation that cannot be avoided, it will be necessary to design multiple RNA guides, each tailored to accommodate the presence of high frequency (≥0.01% allele frequency) variants.
Low frequency off-target candidates predominate in large populations
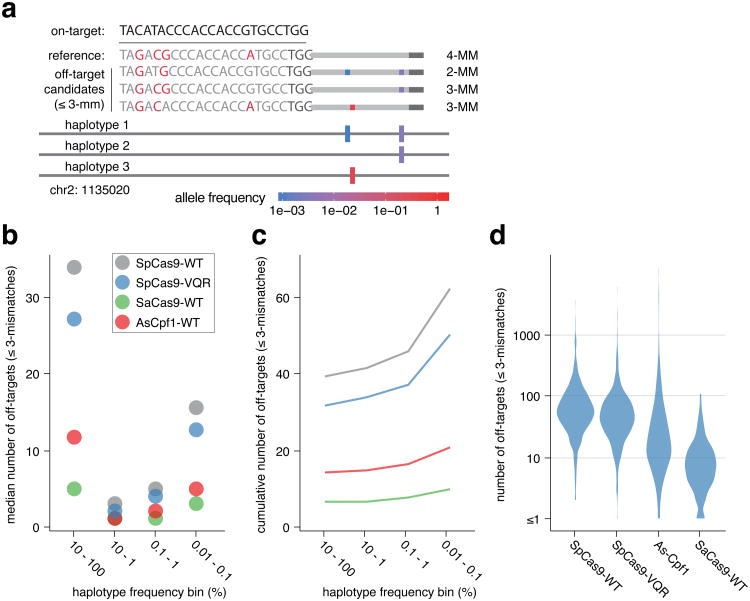
A second major consideration for CRISPR-based therapeutics is safety, which can be improved by designing RNA guides with minimal potential off-target activity. Unbiased investigation of genome-wide CRISPR nuclease activity suggests that most off-target activity occurs at loci with ≤3 mismatches to the RNA guide13,19,20,21,9,22,23,24. Current approaches for Cas9 target selection rank off-target candidates found in the reference human genome by both the number and position of RNA guide mismatches, with the assumption that loci containing ≤3 mismatches or containing PAM distal mismatches are more likely to be cleaved13,14,15. However, in a population of individuals, this strategy is complicated by the existence of multiple haplotypes (sets of variants that co-occur), which will contain different positions or numbers of mismatches at candidate off-target sites (Fig. 3a). To assess the predicted safety of an RNA guide within a population, we turned to the 1000 Genomes dataset, which contains phased single nucleotide variant calls for 2504 individuals25. From this data, we reconstructed allele-specific whole genome sequences for each individual. In contrast to the much larger ExAC dataset, which collapses all variants, the 1000 Genomes dataset contains information about haplotypes, enabling us to identify off-target sites in the population arising from single or multiple variants in an individual haplotype. For platinum targets in the 12 genes considered here, we quantified off-target candidates (defined as genomic loci with ≤3 mismatches to a given RNA guide) arising from all 1000 Genomes haplotypes. In this relatively small population of 2504 individuals more than half of the haplotypes containing off-target candidates are present in ≥10% of individuals (Fig. 3b). However, for haplotypes present in <10% of individuals, the number of off-target candidates for each RNA guide increases with decreasing haplotype frequency (Fig. 3b). This trend indicates that for large populations most unique off-target candidates for a given RNA guide will differ between individuals as shown by the rise in cumulative off-targets for an individual guide RNA accompanying decreasing allele frequency (Fig. 3c).
Figure 3. Human genetic variation significantly impacts CRISPR endonuclease therapeutic safety.
a, Schematic illustrating off-target candidates arising due to multiple different haplotypes. MM, mismatch. b, Number of off-target candidates present in the 1000 Genomes dataset for each CRISPR endonuclease for 12 therapeutically relevant genes at different allele frequencies. c, Distribution of the number of off-target candidates per platinum target for each CRISPR endonuclease. d, Distribution of the number of off-target candidates per enzyme for the 4 CRISPR endonucleases studied here.
Avoiding high-frequency off-target candidates should maximize population safety
For individual RNA guides in these 12 genes, we find that the number of off-target candidates for SpCas9, SpCas9-VQR, SaCas9, and AsCpf1 varies from 0 to greater than 10,000 in the 1000 Genomes population (Fig. 3d). Much of this disparity reflects how unique or repetitive an individual target sequence is within the human genome. For instance SaCas9, which has a longer PAM and hence fewer genomic targets, has on average fewer off-target candidates per RNA guide. Of the 12 genes we considered, some contain more repetitive regions relative to the rest of the human genome, as reflected by increased numbers of targets with high candidate off-target counts in the 12 genes studied (Fig. 4a). For example, within PCSK9 exons 2 – 5, we observed that platinum targets with high or low numbers of off-target candidates tend to cluster in regions of sequence that are either repetitive or unique within the genome, respectively (Fig. 4b) This pattern holds true for all 12 genes studied. Interestingly, within repetitive regions of exons, we did identify small numbers of platinum targets with significantly reduced quantities of off-target candidates. These findings further support the notion that utilizing multiple enzymes with distinct PAM requirements should enhance both safety and efficacy by increasing the number of available targets for therapeutically relevant genomic loci.
Figure 4. Gene- and population-specific variation informs therapeutic design.
a, Distribution of the number of off-target candidates per platinum target for the 12 therapeutically relevant genes studied here. b, Total off-target candidates for platinum targets spanning exons 2 – 5 of PCSK9-001 are shown for each CRISPR endonuclease. c, Principal component analysis (PCA) separating 1000 Genomes individuals into superpopulations based on patient-specific off-target profiles for platinum targets spanning 12 therapeutically relevant genes. PC2 and PC3 are shown. AFR, African; AMR, Ad mixed American; EAS, East Asian; EUR, European; SAS, South Asian. d, Proposed framework for identifying therapeutic guides that maximize efficacy and safety.
Additionally, in a population, the number of off-target candidates at a given locus is compounded by the existence of multiple haplotypes, the number of which will increase with the size of the population. Hence, for each off-target candidate present in a high frequency haplotype, in a large population, multiple lower frequency haplotypes are likely to exist that may lead to different gene editing outcomes. Thus, minimizing the number of off-target candidates occurring in high frequency haplotypes is of critical importance for the selection of therapeutic RNA guides. The current 1000 genomes dataset provides comprehensive coverage of alleles occurring at up to 0.1% in the population (considered to be the lower bound of high frequency variants), allowing us to identify platinum targets with minimal off-target candidates occurring in high frequency haplotypes in the human population25,5. Use of the enhanced specificity enzymes eSpCas9 and Cas9-HF1 will further reduce the likelihood of cleavage at off-target candidate sites, but it will still remain important to avoid target regions that are repetitive or have candidate off-targets in high frequency haplotypes17,18.
Consideration of patient populations
Genome editing therapies are currently being designed for a range of applications including treatment of rare genetic diseases (e.g., Leber's Congenital Amaurosis), common conditions (e.g., high cholesterol), and therapeutic augmentation whereby a genetic change increases the efficacy of a treatment (e.g., PDCD1 knockout for enhanced immunotherapy). Each of these applications will have a unique patient population with its own landscape of genetic variation, and this can be considered when choosing therapeutic targets. For example, Tay-Sachs disease occurs in Ashkenazi populations at more than ten times the rate of occurrence in the general population26; because of the shared genetic heritage among Tay-Sachs patients, there will be fewer variants and those that are present will occur at higher frequency in the patient population. On the other hand, diverse patient populations will contain large numbers of variants occurring at high frequencies in a subset of patients but low frequencies overall in the patient population. Because the 1000 Genomes project provides demographic information, including gender and ethnicity, for each individual, we used this data to explore how much off-target candidate variation for a given individual is explained by population demographics. For all off-target candidates for RNA guides targeting the 12 genes considered here, we performed principal component analysis (PCA) and found that the first five principal components separate individuals most effectively by continent, and also by sub-continent and sex (Fig. 4c, Fig. S3 – 5). We found that these first 5 principle components account for 12% of the variation in off-target candidates occurring at less than 100% frequency for the population, indicating that safety and efficacy of therapeutics can be enhanced by designing therapeutic targets for specific patient subpopulations.
Discussion
Ideally, personalized genomic medicine would tailor RNA-guided endonuclease therapeutics for each patient. However, in most cases, the cost and time required to obtain regulatory approval for each individualized therapeutic would be prohibitive given current regulatory framework. Instead, a small number of carefully chosen combinations of enzyme and guide may be developed and tested to provide a suite of potential therapeutics for a particular patient population. Current methods for selecting targets and guides typically rely solely on sequence information from the human reference genome and criteria obtained from empirical tests of efficacy. However, while RNA guide efficacy does vary and can be difficult to predict, if therapeutic targets are selected based on efficacy alone, those therapies run the risk of being mired in a clinical trial with confounding results and/or undesirable outcomes due to human genetic variation.
Our findings on the impact of genetic variation can be integrated with empirical methods to streamline the design and testing of genome editing therapeutics in a consolidated framework (Fig. 4d). First, when possible, regions of low variation should be targeted, which will ensure maximal efficacy across a patient population with diverse genetic backgrounds. Second, RNA guides need to be selected to minimize the number of off-target candidates occurring on high frequency haplotypes in the patient population, as to reduce the likelihood of transpositions or off-target mutations resulting in oncogenic events. Third, assessing the amount of low frequency variation present in the patient population can be helpful for estimating the number of RNA guide-enzyme combinations required to effectively and safely treat the anticipated patient population. This will be particularly important when designing targets for use within specific populations. For example, for treatment of common diseases, more guide-enzyme combinations will need to be developed given the breadth of the natural genetic variation in the patient population. Fourth, in silico screening and empirical assays9,19,20,21,27,28,29 to assess target efficacy and genome-wide specificity should be used to identify the optimal RNA guide-enzyme combinations from the pool of selected RNA guides. In the event that no high efficacy guides are found, the number of RNA guide-enzyme combinations should be increased and tailored to the presence of multiple independent high frequency haplotypes. The safety of selected combinations of RNA guide and enzyme should be evaluated through unbiased whole-genome off-target detection in relevant cell lines (ideally patient specific). Combinations that pass all of these filters should then be moved forward for regulatory approval. Finally, pre-therapeutic whole genome sequencing of individual patients will be needed to select a single approved RNA guide-enzyme combination for treatment that is a perfect match to the patient's genome and free of patient-specific off-target candidates.
The selection of specific targets to pursue for therapeutic development will also depend on the type of gene edit desired. For example, gene knockout strategies (which are being pursued for PDCD1 for immunotherapy and PCSK9 for cardiovascular disease) have many guide choices, and researchers can choose from a range of low frequency target regions within or near the gene of interest and then select the guide within that subset that provides the most efficient gene knockdown. Other diseases can be addressed by removal of single or multiple causal variants, and therapies are being developing that aim to remove mutated segments of genes to restore function (e.g., CEP290 for Leber's Congenital Amaurosis and DMD for Duchenne Muscular Dystrophy). While this strategy for therapeutic intervention also affords some flexibility in target selection, because two gRNAs are needed for each gene, researchers will be limited to working with a single enzyme, reducing the number of target options; this strategy carries the risk of doubling the potential off-target candidates as well. Finally, homology directed repair (HDR) is being used for correction of disease causing mutations affecting mitotically active cells in the body (such as SERPINA1 for Alpha-1 Antitrypsin and CFTR for Cystic Fibrosis). For HDR strategies, Cas nucleases are used to cleave the target gene typically within 10 – 20 nucleotides of the desired integration site, greatly restricting the targetable range. However, considering SpCas9, SpCas9-VQR, SaCas9, and AsCpf1, a target is present approximately every 4 nucleotides in the human exome, which should allow selection of a low-variation region even in situations with a narrow target range, such as would be required by HDR.
Continued technological development will deliver more powerful and precise systems for therapeutic genome editing. Beyond nuclease-based strategies, new approaches that leverage the programmable DNA binding activity of CIRSPR-based enzymes to direct DNA base-modifying enzymes such as base editing30 also promises to further expand therapeutic options. Finally, well-designed clinical trials that are carried out efficiently and smoothly will be central to addressing the regulatory and ethical challenges facing therapeutic genome editing. Failure to anticipate the genetic diversity in patient populations will confound clinical trials and may lead to adverse outcomes. Our analysis of the impact of human genetic variation on CRISPR-Cas-based therapeutics provides a toolset and resources that will increase the efficacy and safety of these therapies, ultimately moving them more quickly toward the clinic.
Online Methods
Datasets
Our target variation analysis was performed using the Exome Aggregation Consortium (ExAC) dataset from 60,706 globally diverse individuals5. The ExAC data was downloaded from the following ftp site: ftp://ftp.broadinstitute.org/pub/ExAC_release/release0.3.1.
Our investigation of off-target candidates was performed using the 1000 Genomes Project phase 3 dataset containing phased whole genome sequences from 2504 globally diverse individuals25. The 1000 Genomes data was acquired from: ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502.
Whole-exome target variation analysis
We included all targets containing the canonical Protospacer Adjacent Motif (PAM) sequences for the for CRISPR enzymes SpCas9-WT (NGG), SpCas9-VQR (NGA), SpCas9-VRER (NGCG), SaCas9 (NNGRRT), and AsCpf1 (TTTN) for all exons in Gencode release 19 (GRCh37.p13) annotated as protein coding and having an average coverage of at least 20 reads per ExAC sample (See Table S2). For analysis of variation in these targets, we included all missense or synonymous variants passing quality filtering in the ExAC dataset as described previously5. Because the publicly available ExAC dataset includes only summary information for each variant, it was not possible to determine if multiple variants occurring in a single genomic target occur on different haplotypes. Hence, we calculated target variation frequency as the maximum frequency of variants in an individual target. While accurately approximating the variation of most targets in the population, this approach does underestimate the variation frequency for rare targets containing multiple high frequency variants existing on separate haplotypes. Platinum targets were defined as those with a maximum variant frequency of less than 0.01% in the ExAC population.
Off-target candidate analysis
Phased haplotypes included in the 1000 Genomes phase 3 dataset were used to create whole genome allele-specific references for 2504 individuals. We included in our analysis all single nucleotide polymorphisms passing quality filtering in the 1000 Genomes phase 3 dataset as described previously25. Up to 100 protein-coding platinum targets for each therapeutically relevant gene (as available, see Table S3), CEP290, CFTR, DMD, G6PC, HBB, IDUA, IL2RG, PCSK9, PDCD1, SERPINA1, TTR, VEGFA were selected for proteins SpCas9-WT, SpCas9-VQR, SaCas9, and AsCpf1. Targets for each gene were searched against the references for each of the 2504 1000 genomes individuals to profile candidate off-target sites specific to each individual. All PAM sequences associated with nuclease activity were included in the off-target analysis for each enzyme as follows: SpCas9-WT (NGG, NAG), SpCas9-VQR (NGAN, NGNG), SaCas9 (NNGRRT), and AsCpf1 (TTTN). For the purpose of this study, off-target candidates are defined as unintended genome-wide targets for a specific guide RNA-enzyme combination with less than or equal to 3-mismatches with the guide RNA protospacer.
Demographics Analysis
We performed principal component analysis (PCA) taking into account for each 1000 Genomes individual, the presence or absence of off-target candidates for each target included in our analysis of 12 therapeutically relevant genes present in less than 100% of the 1000 Genomes individuals (n= 46,362 off-target candidates; PCA computed using the R prcomp function). Superpopulation groups included: AFR, African; AMR, Ad mixed American; EAS, East Asian; EUR, European; SAS, South Asian. Population groups included: CHB, Han Chinese in Bejing, China; JPT, Japanese in Tokyo, Japan; CHS, Southern Han Chinese; CDX, Chinese Dai in Xishuangbanna, China; KHV, Kinh in Ho Chi Minh City, Vietnam; CEU, Utah Residents (CEPH) with Northern and Western Ancestry; TSI, Toscani in Italia; FIN, Finnish in Finland; GBR, British in England and Scotland; IBS, Iberian Population in Spain; YRI, Yoruba in Ibadan, Nigeria; LWK, Luhya in Webuye, Kenya; GWD, Gambian in Western Divisions in the Gambia; MSL, Mende in Sierra Leone; ESN, Esan in Nigeria; ASW, Americans of African Ancestry in SW USA; ACB, African Caribbeans in Barbados; MXL, Mexican Ancestry from Los Angeles USA; PUR, Puerto Ricans from Puerto Rico; CLM, Colombians from Medellin, Colombia; PEL, Peruvians from Lima, Peru; GIH, Gujarati Indian from Houston, Texas; PJL, Punjabi from Lahore, Pakistan; BEB, Bengali from Bangladesh; STU, Sri Lankan Tamil from the UK; ITU, Indian Telugu from the UK.
Supplementary Material
Figure S1: a – e, Left, fraction of PAMs altered by variants in the ExAC dataset; center, distribution of PAM-altering variant frequencies; right, fraction of homozygous variants by frequency. Data shown for AsCpf1 (a), SpCas9-VQR (b), SpCas9 (c), SaCas9 (d), and SpCas9-VRER (e).
Table S1: Fraction of total residues and fraction of residues containing variation for individual nucleotides (nt) in the human exome.
Table S2: Fraction of targets containing target variation for each CRISPR endonuclease (n specifies the number of protein coding targets in the human exome for each enzyme).
Figure S2: a – d, Top, distribution of target variation for therapeutically relevant genes. Targets with frequencies of variation less than 0.01% (red line) are considered platinum. Bottom, fraction of all targets in these genes containing variation. Data shown for AsCpf1 (a), SpCas9-VWR (b), SpCas9-WT (c), SaCas9-WT (d).
Table S3: Number of platinum and high variation frequency targets for 12 therapeutically relevant genes for AsCpf1, SpCas9, SpCas9-VQR, or SaCas9.
Figure S3: Separation of 1000 Genomes individuals into super populations based on patient specific off-target profiles for targets spanning 12 therapeutically relevant genes. Principal components 1 – 5 shown. AFR, African; AMR, Ad mixed American; EAS, East Asian; EUR, European; SAS, South Asian.
Figure S4: Separation of 1000 Genomes individuals into populations based on patient specific off-target profiles for targets spanning 12 therapeutically relevant genes. Principal components 1 – 5 shown. CHB, Han Chinese in Bejing, China; JPT, Japanese in Tokyo, Japan; CHS, Southern Han Chinese; CDX, Chinese Dai in Xishuangbanna, China; KHV, Kinh in Ho Chi Minh City, Vietnam; CEU, Utah Residents (CEPH) with Northern and Western Ancestry; TSI, Toscani in Italia; FIN, Finnish in Finland; GBR, British in England and Scotland; IBS, Iberian Population in Spain; YRI, Yoruba in Ibadan, Nigeria; LWK, Luhya in Webuye, Kenya; GWD, Gambian in Western Divisions in the Gambia; MSL, Mende in Sierra Leone; ESN, Esan in Nigeria; ASW, Americans of African Ancestry in SW USA; ACB, African Caribbeans in Barbados; MXL, Mexican Ancestry from Los Angeles USA; PUR, Puerto Ricans from Puerto Rico; CLM, Colombians from Medellin, Colombia; PEL, Peruvians from Lima, Peru; GIH, Gujarati Indian from Houston, Texas; PJL, Punjabi from Lahore, Pakistan; BEB, Bengali from Bangladesh; STU, Sri Lankan Tamil from the UK; ITU, Indian Telugu from the UK.
Figure S5: Separation of 1000 Genomes individuals by sex based on patient specific off-target profiles for targets spanning 12 therapeutically relevant genes. Principal components 1 – 5 shown.
Acknowledgments
We would like to thank R. Macrae, L. Francioli, S. Jones, J. Strecker, D. Cox, I. Slaymaker, and W. Yan for helpful discussions and insights. F.Z. is a New York Stem Cell Foundation-Robertson Investigator. F.Z. is supported by the NIH through NIMH (5DP1-MH100706 and 1R01-MH110049); NSF; the New York Stem Cell Foundation; the Howard Hughes Medical Institute; the Simons, Paul G. Allen Family, and Vallee Foundations; the Skoltech-MIT Next Generation Program; James and Patricia Poitras; Robert Metcalfe; and David Cheng. F.Z. is a New York Stem Cell Foundation-Robertson Investigator. The computer code and resources related to this work are available through the Zhang lab website (www.genome-engineering.org) and GitHub (github.com/fengzhanglab).
Footnotes
Competing Financial Interest: F.Z. is a founder of Editas Medicine and a scientific advisor for Editas Medicine and Horizon Discovery.
Authorship contributions: D.A.S. and F.Z. conceived the study; D.A.S. performed all experiments and analyses; D.A.S. and F.Z. wrote the manuscript.
Data Availability: Tables including all Platinum Targets in the human exome are freely available as a supplement to this manuscript.
Code Availability: All computer code used in this work is freely available from https://github.com/fengzhanglab/CRISPR-Human_Variation_Nature_Medicine_manuscript.
References
- 1.Cong L, et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mali P, et al. RNA-Guided Human Genome Engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zetsche B, et al. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang L, et al. Targeted and genome-wide sequencing reveal single nucleotide variations impacting specificity of Cas9 in human stem cells. Nat Commun. 2014;5:5507. doi: 10.1038/ncomms6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Consortium, T. 1000 G. P. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Consortium, T. 1000 G. P. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.1000 Genomes Project Consortium et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ran FA, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kleinstiver BP, et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Makarova KS, et al. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol. 2015;13:722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garneau JE, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 13.Hsu PD, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pattanayak V, et al. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol. 2013;31:839–843. doi: 10.1038/nbt.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu Y, et al. High frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Slaymaker IM, et al. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kleinstiver BP, et al. High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsai SQ, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. 2015;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frock RL, et al. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat Biotechnol. 2015;33:179–186. doi: 10.1038/nbt.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim D, et al. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat Methods. 2015;12:237–243. doi: 10.1038/nmeth.3284. [DOI] [PubMed] [Google Scholar]
- 22.Lin Y, et al. CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic Acids Res. 2014;42:7473–7485. doi: 10.1093/nar/gku402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kleinstiver BP, et al. Genome-wide specificities of CRISPR-Cas Cpf1 nucleases in human cells. Nat Biotechnol. 2016;34:869–874. doi: 10.1038/nbt.3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim D, et al. Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat Biotechnol. 2016;34:863–868. doi: 10.1038/nbt.3609. [DOI] [PubMed] [Google Scholar]
- 25.The 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu YF, Goldstein DB, Angrist M, Cavalleri G. Personalized Medicine and Human Genetic Diversity. Cold Spring Harb Perspect Med. 2014;4:a008581. doi: 10.1101/cshperspect.a008581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cameron P, et al. SITE-Seq: A Genome-wide Method to Measure Cas9 Cleavage. Protoc Exch. 2017 doi: 10.1038/protex.2017.043. [DOI] [Google Scholar]
- 28.Tsai SQ, et al. CIRCLE-seq: a highly sensitive in vitro screen for genome-wide CRISPR-Cas9 nuclease off-targets. Nat Methods. 2017;14:607–614. doi: 10.1038/nmeth.4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan WX, et al. BLISS is a versatile and quantitative method for genome-wide profiling of DNA double-strand breaks. Nat Commun. 2017;8 doi: 10.1038/ncomms15058. ncomms15058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: a – e, Left, fraction of PAMs altered by variants in the ExAC dataset; center, distribution of PAM-altering variant frequencies; right, fraction of homozygous variants by frequency. Data shown for AsCpf1 (a), SpCas9-VQR (b), SpCas9 (c), SaCas9 (d), and SpCas9-VRER (e).
Table S1: Fraction of total residues and fraction of residues containing variation for individual nucleotides (nt) in the human exome.
Table S2: Fraction of targets containing target variation for each CRISPR endonuclease (n specifies the number of protein coding targets in the human exome for each enzyme).
Figure S2: a – d, Top, distribution of target variation for therapeutically relevant genes. Targets with frequencies of variation less than 0.01% (red line) are considered platinum. Bottom, fraction of all targets in these genes containing variation. Data shown for AsCpf1 (a), SpCas9-VWR (b), SpCas9-WT (c), SaCas9-WT (d).
Table S3: Number of platinum and high variation frequency targets for 12 therapeutically relevant genes for AsCpf1, SpCas9, SpCas9-VQR, or SaCas9.
Figure S3: Separation of 1000 Genomes individuals into super populations based on patient specific off-target profiles for targets spanning 12 therapeutically relevant genes. Principal components 1 – 5 shown. AFR, African; AMR, Ad mixed American; EAS, East Asian; EUR, European; SAS, South Asian.
Figure S4: Separation of 1000 Genomes individuals into populations based on patient specific off-target profiles for targets spanning 12 therapeutically relevant genes. Principal components 1 – 5 shown. CHB, Han Chinese in Bejing, China; JPT, Japanese in Tokyo, Japan; CHS, Southern Han Chinese; CDX, Chinese Dai in Xishuangbanna, China; KHV, Kinh in Ho Chi Minh City, Vietnam; CEU, Utah Residents (CEPH) with Northern and Western Ancestry; TSI, Toscani in Italia; FIN, Finnish in Finland; GBR, British in England and Scotland; IBS, Iberian Population in Spain; YRI, Yoruba in Ibadan, Nigeria; LWK, Luhya in Webuye, Kenya; GWD, Gambian in Western Divisions in the Gambia; MSL, Mende in Sierra Leone; ESN, Esan in Nigeria; ASW, Americans of African Ancestry in SW USA; ACB, African Caribbeans in Barbados; MXL, Mexican Ancestry from Los Angeles USA; PUR, Puerto Ricans from Puerto Rico; CLM, Colombians from Medellin, Colombia; PEL, Peruvians from Lima, Peru; GIH, Gujarati Indian from Houston, Texas; PJL, Punjabi from Lahore, Pakistan; BEB, Bengali from Bangladesh; STU, Sri Lankan Tamil from the UK; ITU, Indian Telugu from the UK.
Figure S5: Separation of 1000 Genomes individuals by sex based on patient specific off-target profiles for targets spanning 12 therapeutically relevant genes. Principal components 1 – 5 shown.