Abstract
The incidence of squamous cell carcinoma of the oropharynx (SCCOP) continues to rise because of increasing rates of human papillomavirus (HPV) infection. Inherited polymorphisms in DNA repair pathways may influence the risk of SCCOP development and the prognosis of SCCOP. We sought to determine whether polymorphisms in microRNA (miRNA)-binding sites within 3' -untranslated regions (3’UTRs) of genes in DNA repair pathways modulate the risk of SCCOP recurrence. We evaluated the associations between nine such polymorphisms and SCCOP recurrence in 1008 patients with incident SCCOP using the log-rank test and multivariable Cox models. In an analysis of all the patients, patients with variant genotypes of BRCA1 rs12516 and RAD51 rs7180135 had better disease-free survival (log-rank, P = 0.0002 and P = 0.0003, respectively) and lower risk of SCCOP recurrence (hazard ratio [HR], 0.5, 95% confidence interval [CI], 0.2-0.8, and HR, 0.5, 95% CI, 0.3-0.9, respectively) than patients with common homozygous genotypes of the two polymorphisms after multivariable adjustment. Moreover, in tumor HPV16-positive patients, patients with variant genotypes of the same two polymorphisms also had better disease-free survival (log-rank, P = 0.004 and P = 0.003, respectively) and lower recurrence risk (HR, 0.2, 95% CI, 0.1-0.6, and HR, 0.2, 95% CI, 0.0-0.7, respectively) than patients with common homozygous genotypes of the two polymorphisms. No such significant associations were found for other polymorphisms. These findings support significant roles of BRCA1 rs12516 and RAD51 rs7180135 in modifying the risk of recurrence of SCCOP, particularly HPV16-positive SCCOP. However, these results must be validated in larger studies.
Keywords: miRNA, recurrence, genetic polymorphisms, squamous cell carcinoma of the oropharynx, double strand DNA repair
Introduction
The incidence of squamous cell carcinoma of the oropharynx (SCCOP), a subset of squamous cell carcinoma of the head and neck, is increasing, mainly because of the increasing prevalence of human papillomavirus (HPV) infection 1, 2. SCCOP is associated with poor long-term survival because of the high rate of recurrence even after multimodal treatment with combinations of surgery, radiotherapy, and chemotherapy 3. The recurrence rate might differ among patients with SCCOP, even though these patients received similar therapeutics and share similar clinical and pathological characteristics. Thus, genetic factors may affect an individual’s susceptibility to SCCOP recurrence 4.
Upon DNA damage, which can be caused by various endogenous and exogenous stimuli, damage repair responses activate specific signaling networks that are involved in DNA damage signaling, cell cycle regulation, and apoptosis to preserve genomic integrity 5, 6. Thus, genetic alterations, such as genetic polymorphisms, in DNA repair pathways may affect not only cancer risk but also the patient’s response to therapy, including chemotherapy and radiotherapy, leading to differences in recurrence and survival 7, 8.
Many studies have shown that microRNAs (miRNAs) play key roles in the regulation of DNA damage repair 9, 10. MiRNAs are single-stranded noncoding RNAs that pair with target mRNAs at the 3’-untranslated region (3’-UTR) and thereby cause cleavage of mRNA or suppression of translation; in this way, miRNAs regulate gene expression 11, 12. MiRNA-mediated gene silencing has been shown to modulate the activity of DNA repair pathways 7.
Studies have shown that polymorphisms in predicted miRNA-binding sites in genes implicated in DNA double strand break (DSB) repair can influence the risk of and prognosis of several types of cancer 13-16. However, to our knowledge, there are no large studies have examined the association between polymorphisms in miRNA-binding sites of DNA DSB repair genes and the risk of recurrence of SCCOP. In this study, we performed bioinformatics prediction and the determination of the Gibbs free energy for selection of SNPs in the DNA DSB repair pathway genes13. The identified SNPs were located within these putative miRNA-binding sites at the 3’-UTR of the DSB repair genes with potential functions, which might modify the binding ability of miRNAs, leading to different DSB gene expression or different DNA repair capacity. Moreover, these selected SNPs had a minor allele frequency (MAF) of ≥ 0.05 in European populations. The objective of the present study was to determine the extent to which polymorphism in miRNA-binding sites within 3’UTRs of genes involved in DNA DSB repair pathways modulate the risk of recurrence of SCCOP.
Materials and methods
Study subjects
This analysis was conducted in patients with SCCOP consecutively recruited between May 1995 and April 2010 as part of a molecular epidemiologic study at The University of Texas MD Anderson Cancer Center, as described previously 17. Briefly, the study was open to all patients with newly diagnosed, histopathologically confirmed, previously untreated SCCOP regardless of age, sex, ethnicity, or cancer stage; patients younger than 18 years of age or with distant metastasis were excluded. At initial presentation to MD Anderson, patients were interviewed regarding demographic, epidemiologic, and clinical characteristics, and blood samples were obtained for genetic testing. Approximately 95% of contacted patients consented to enrollment in the study. All subjects signed an informed consent form, and the study protocol was approved by the MD Anderson Institutional Review Board.
During and after treatment, patients had regularly scheduled clinical and radiographic examinations. The frequency of imaging studies and the specific imaging studies performed were at the discretion of each patient’s treatment team. Typically, patients had routine serial imaging supplemented as needed with extra imaging studies to investigate any symptoms or findings of concern on physical examination.
Assessment of baseline characteristics, treatment, and recurrence
Patient, tumor, and treatment characteristics for this analysis were obtained through review of medical records. Clinical stage at presentation of the index tumor was dichotomized into early stage (clinical stage I and II) and late stage (clinical stage III and IV). Treatment was dichotomized into treatment with surgery, radiotherapy, and chemotherapy or with just one or two of these modalities. Patients were classified as “ever drinkers” (those who had drunk at least one alcoholic beverage per week for at least 1 year) or “never drinkers” (those who had never had such a pattern of drinking). Patients were also classified as “ever smokers” (those who had smoked at least 100 cigarettes in their lifetime) or “never smokers” (those who had smoked fewer than 100 cigarettes in their lifetime).
Disease recurrence was defined as appearance of a new lesion of the same histology verified by biopsy (incisional, excisional, or needle biopsy) or reappearance of any lesion that had disappeared. Recurrences resulting from spread of tumor cells through the bloodstream (e.g., to the lung, esophagus, gastrointestinal, bone, or brain) were defined as distant recurrences, recurrences in the regional lymph nodes were defined as regional recurrences, and recurrences in the mucosa or soft tissue in or adjacent to the site of the primary tumor were defined as local recurrences. Freedom from disease was defined as absence of disease documented at the date of the last visit with the head and neck surgeon, head and neck radiation oncologist, or head and neck medical oncologist.
Selection and genotyping of study SNPs
Several single-nucleotide polymorphisms (SNPs) within miRNA-binding sites in the 3’UTRs of DNA DSB repair genes have been reported, and these SNPs were identified using the methods for identifying putative miRNA-binding sites, as previously described 13, 14. The miRNA target prediction was performed by either using available online bioinformatics tools or monitoring other SNP databases and related literature 13, 14. As part of the molecular genetic study in which patients in the present analysis were enrolled, genomic DNA was extracted from the buffy coat of whole blood samples before treatment using the Qiagen DNA Blood Mini Kit (Qiagen Inc.) per the manufacturer’s instructions. These DNA samples were used to genotype for nine potentially functional SNPs within miRNA-binding sites in the 3’UTRs of DNA DSB repair genes using TaqMan technology. The nine SNPs selected were ≥ 10% in our SCCOP patient population. Ten percent of samples were randomly selected for repeat assay, and results of initial and repeat assays were concordant in all cases.
Determination of tumor HPV16 status and BRCA1 and RAD51 protein expression
It is now standard clinical practice at MD Anderson that the pathology laboratory classifies all SCCOP specimens as being positive or negative for high-risk HPV by both in situ hybridization (ISH) and p16 immunohistochemistry (IHC). These data are now part of the patient’s clinical record and the pathology laboratory’s description of SCCOP biopsy specimens. Consequently, data on tumor HPV status of SCCOP patients are clinically available for most of our study patients. For SCCOP patients with missing data on tumor HPV status, in-house paraffin-embedded tissues were obtained or, where necessary, tissue blocks from outside institutions were requested, and these tissues were subjected to tumor HPV determination. Tumor HPV16 status was determined from paraffin-embedded tissue biopsy specimens or surgical specimens, when available. DNA was extracted from tissue specimens and analyzed for HPV16 using PCR and confirmed by ISH, as described previously 18. For quality control, 10% of randomly selected samples were re-assayed for tumor HPV16 status; the re-run results reached a 100% concordance with the original results.
Additionally, a total of 64 SCCOP patients were selected with tumor specimens available for the immunohistochemistry (IHC) as shown in Figure 1. Paraffin blocks from SCCOP specimens were sectioned into 4-μM-think sections. The IHC analysis followed previously described methodology with modification19. The slides were deparafinized with xylene and dehydrated with ethanol. Antigen retrieval was performed by microwave heating methods in pretreatment solution. Avidin/biotin blocking solution was used to prevent nonspecific binding of antibodies. These sections were incubated with anti-BRCA1 antibody (1:100 dilution, ab16780, Abcam, Cambridge, MA, USA) and anti-RAD51 antibody (1:50 clone 51RAD01, Abcam, Cambridge, MA, USA). The sections were then reviewed and categorized under a microscope as positively- or negatively-stained (section with ≥ 10% positively stained tumor cells were considered to be positively-stained)19.
Figure 1.
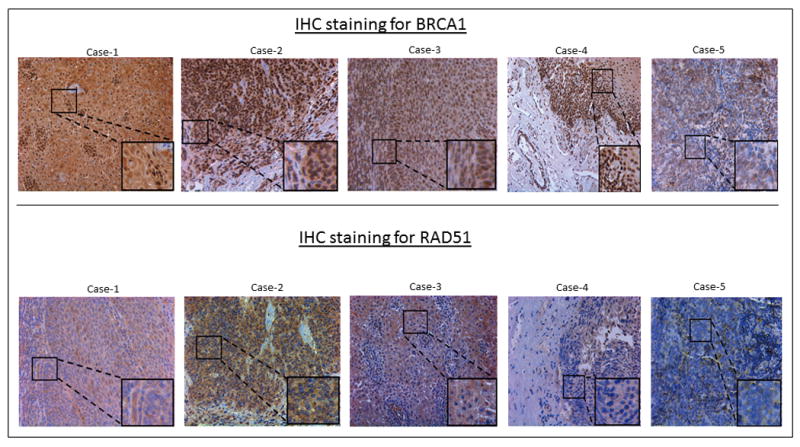
Immunohistochemistry staining for BRCA1 and RAD51.
Statistical analysis
Student’s t test was used to compare the mean age and follow-up time for patients with and without recurrence. The chi-squared test was used to evaluate differences in ethnicity, sex, smoking status, alcohol use status, index tumor site, disease stage, comorbidity, treatment, and genotype distributions between patients with and without recurrence.
Time to recurrence, the primary endpoint of the study, was computed from the date of presentation to the date of clinically detectable recurrence (local, regional, or distant). Participants who were recurrence free or lost to follow-up were considered censored. Kaplan-Meier survival curves and log-minus-log survival plots were also examined, and the data were confirmed to be consistent with the assumptions of the Cox proportional hazards regression model. The log-rank test was used to determine the associations between different variables and disease-free survival (DFS). Associations of SNPs in DNA DSB repair genes with SCCOP recurrence risk were estimated using hazard ratios (HRs) and 95% confidence intervals (CIs) among all patients in the study and among patients with HPV16-positive tumors. The multivariable Cox models were fully adjusted for major confounders, including age, sex, ethnicity, smoking and alcohol use status, stage, comorbidity and treatment. The chi-squared test or Z test was used for BRCA1 and RAD51 comparison of expression levels between the genotypes of the polymorphisms. For all analyses, statistical significance was set at P < 0.05, and all tests were two sided. SAS software (version 9.4; SAS Institute) was used to perform all statistical analyses.
Results
A total of 1226 patients with SCCOP were enrolled during the study period. Of these patients, 218 were excluded from our analysis because follow-up or treatment data were missing or no blood sample was available for genotyping. Therefore, a total of 1008 patients with incident SCCOP were included in the final analysis. The follow-up of these patients occurred from May 1995 to October 2013. The median follow-up duration for all 1008 patients was 44.7 months (range, 1.7-170.9 months). During follow-up, 181 patients had disease recurrence. The median follow-up duration for the patients with recurrence was 11.6 months; the median follow-up duration for the patients without recurrence was 50.9 months.
Patient characteristics and 5-year recurrence rates by patient subgroup were shown in Table 1. There were significant differences in DFS between the subgroups defined on the basis of age, ethnicity, smoking status, alcohol use status, comorbidity, and treatment (log-rank, all P < 0.05); DFS did not differ significantly between men and women or between the two groups of treatment. In our study population, the majority of patients were male and non-Hispanic white.
Table 1.
Characteristics of patients with SCCOP (N = 1008)
| Characteristic | No. (%) of patients | No. of patients with recurrence | 5-year recurrence rate | P valuea |
|---|---|---|---|---|
| No. of patients | 1008 (100) | 181 | 0.20 | |
| Age, years | ||||
| ≤57 | 621 (61.6) | 85 | 0.15 | < 0.0001 |
| > 57 | 387 (38.4) | 96 | 0.27 | |
| Sex | ||||
| Male | 872 (86.5) | 161 | 0.20 | 0.3110 |
| Female | 136 (13.5) | 20 | 0.19 | |
| Ethnicity | ||||
| Non-Hispanic white | 913 (90.6) | 146 | 0.17 | < 0.0001 |
| Other | 95 (9.4) | 35 | 0.41 | |
| Smoking | ||||
| Never | 388 (38.5) | 51 | 0.14 | 0.0004 |
| Ever | 620 (61.5) | 130 | 0.23 | |
| Alcohol use | ||||
| Never | 247 (24.5) | 26 | 0.10 | 0.0005 |
| Ever | 761 (75.5) | 155 | 0.23 | |
| Comorbidity | ||||
| None or mild | 913 (90.6) | 157 | 0.19 | 0.0370 |
| Moderate to severe | 95 (9.4) | 24 | 0.27 | |
| Index cancer stage | ||||
| 1 or 2 | 72 (7.1) | 11 | 0.19 | 0.5280 |
| 3 or 4 | 936 (92.9) | 170 | 0.20 | |
| Treatment | ||||
| X/XC/X/S | 947 (93.9) | 166 | 0.19 | 0.0030 |
| XC | 61 (6.1) | 15 | 0.32 | |
| HPV status | ||||
| HPV(+) | 324 | 45 | 0.12 | 0.0187 |
| HPV(-) | 51 | 10 | 0.21 | |
| HPV (not available) | 633 | 126 | 0.24 |
P: Log-rank test for DFS between the two groups.
X, radiotherapy
C, chemotherapy
S, surgery (for treatment purpose)
As shown in Figure 2, patients with the BRCA1 rs12516 and RAD51 rs7180135 variant genotypes had significantly better DFS than patients with the corresponding common homozygous genotypes among all SCCOP patients (log-rank, P = 0.0002 and P = 0.0003, respectively) and among SCCOP patients with HPV16-positive tumors (log-rank, P = 0.004 and P = 0.003, respectively).
Figure 2.
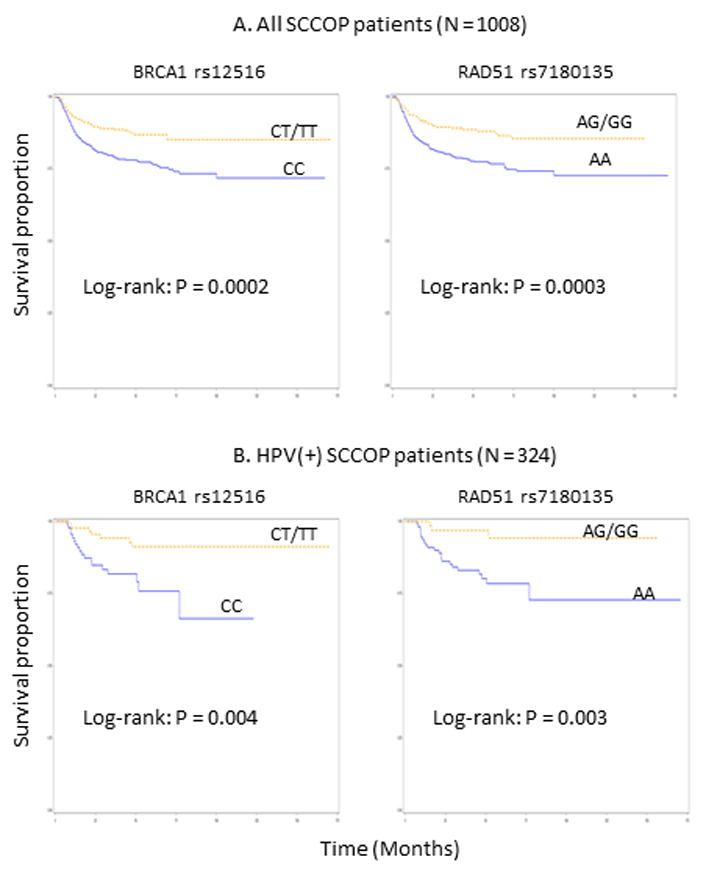
Kaplan-Meier estimates for the cumulative recurrence rates of patients according to the genotypes.
As shown in Table 2, patients with the BRCA1 rs12516 and RAD51 rs7180135 variant genotypes had a significantly lower risk of disease recurrence than patients with the corresponding common homozygous genotypes (HR = 0.5, 95% CI 0.2-0.8, and HR = 0.5, 95% CI 0.3-0.9, respectively). No such significant associations were observed for the other seven polymorphisms in DNA DSB repair genes.
Table 2.
Associations between miRNA-binding-site polymorphisms in DNA DSB repair genes and recurrence risk of patients with SCCOP (N = 1008)
| Genotype | No. of recurrences/no. of patients | 5-year recurrence rate | Log-rank P value | Adjusted HR* (95% CI) |
|---|---|---|---|---|
| ATM rs227091 | 0.494 | |||
| TT† | 90/508 | 0.18 | 1.0 | |
| TC/CC | 91/500 | 0.20 | 1.2 (0.8-1.6) | |
| BRCA1 rs12516 | 0.0002 | |||
| CC† | 130/592 | 0.23 | 1.0 | |
| CT/TT | 51/416 | 0.14 | 0.5 (0.2-0.8) | |
| BRCA1 rs8176318 | 0.393 | |||
| GG† | 123/709 | 0.19 | 1.0 | |
| GT/TT | 58/299 | 0.22 | 1.1 (0.8-1.6) | |
| PARP1 rs8679 | 0.499 | |||
| TT | 152/860 | 0.19 | 1.0 | |
| TC/CC | 29/148 | 0.22 | 1.0 (0.6-1.6) | |
| LIG3 rs4796030 | 0.298 | |||
| CC† | 45/281 | 0.16 | 1.0 | |
| CA/AA | 3/727 | 0.21 | 1.2 (0.8-1.7) | |
| NBS1 rs2735383 | 0.685 | |||
| GG† | 88/516 | 0.19 | 1.0 | |
| GC/CC | 93/492 | 0.20 | 1.0 (0.8-1.4) | |
| NBS1 rs1063054 | 0.818 | |||
| AA† | 95/573 | 0.19 | 1.0 | |
| AC/CC | 86/435 | 0.20 | 1.2 (0.8-1.7) | |
| NBS1 rs1063053 | 0.914 | |||
| CC† | 111/619 | 0.19 | 1.0 | |
| CT/TT | 70/389 | 0.18 | 1.0 (0.7-1.4) | |
| RAD51 rs7180135 | 0.0003 | |||
| AA† | 139/642 | 0.23 | 1.0 | |
| AG/GG | 42/366 | 0.13 | 0.5 (0.3-0.9) |
HR, hazard ratio.
Adjusted for age, sex, ethnicity, smoking status, alcohol use status, stage, comorbidity, and treatment.
Reference group.
Patient stratification by both HPV status and genetic alterations will be central to appropriate treatment selection. Therefore, we further evaluated the associations between these polymorphisms and risk of recurrence in SCCOP patients with HPV16-positive tumors. As shown in Table 3, patients with HPV16-positive SCCOP with BRCA1 rs12516 and RAD51 rs7180135 variant genotypes had a significantly lower risk of disease recurrence than patients with the corresponding common homozygous genotypes (HR = 0.2, 95% CI 0.1-0.6, and HR = 0.2, 95% CI 0.0-0.7, respectively). No such significant associations were observed for the other seven polymorphisms in DNA DSB repair genes.
Table 3.
Associations between miRNA-binding-site polymorphisms in DNA DSB repair genes and recurrence risk of patients with HPV16-positive SCCOP (N = 324)
| Genotype | No. of recurrences/no. of patients | 5-year recurrence rate | Log-rank P value | Adjusted HR* (95% CI) |
|---|---|---|---|---|
| ATM rs227091 | 0.570 | |||
| TT† | 29/183 | 0.15 | 1.0 | |
| TC/CC | 16/141 | 0.13 | 0.7 (0.3-1.7) | |
| BRCA1 rs12516 | 0.004 | |||
| CC† | 33/161 | 0.26 | 1.0 | |
| CT/TT | 12/163 | 0.10 | 0.2 (0.1-0.6) | |
| BRCA1 rs8176318 | 0.372 | |||
| GG† | 27/223 | 0.15 | 1.0 | |
| GT/TT | 18/101 | 0.18 | 1.3 (0.6-3.0) | |
| PARP1 rs8679 | 0.430 | |||
| TT | 29/264 | 0.17 | 1.0 | |
| TC/CC | 16/60 | 0.22 | 1.4 (0.6-3.7) | |
| LIG3 rs4796030 | 0.613 | |||
| CC† | 12/76 | 0.16 | 1.0 | |
| CA/AA | 33/248 | 0.15 | 0.8 (0.3-2.0) | |
| NBS1 rs2735383 | 0.738 | |||
| GG† | 22/168 | 0.17 | 1.0 | |
| GC/CC | 23/156 | 0.15 | 1.2 (0.6-2.7) | |
| NBS1 rs1063054 | 0.547 | |||
| AA† | 27/233 | 0.16 | 1.0 | |
| AC/CC | 18/91 | 0.18 | 1.4 (0.6-3.5) | |
| NBS1 rs1063053 | 0.529 | |||
| CC† | 33/254 | 0.15 | 1.0 | |
| CT/TT | 12/70 | 0.16 | 1.4 (0.6-3.5) | |
| RAD51 RS7180135 | 0.003 | |||
| AA† | 40/202 | 0.26 | 1.0 | |
| AG/GG | 5/122 | 0.10 | 0.2 (0.0-0.7) |
HR, hazard ratio.
Adjusted for age, sex, ethnicity, smoking status, alcohol use status, stage, comorbidity, and treatment.
Reference group.
To further characterize the potentially functional relevance of these 2 significant polymorphisms, we performed correlation analyses between tumor BRCA1 and RAD51 protein expression by IHC and genotypes of BRCA1 rs12516 and RAD51 rs7180135 polymorphisms among a subset of 64 SCCOP patients. As shown in Figure 3, among 64 SCCOP patients, we found that 50 cases were HPV16-positive tumors and 14 had HPV16-negative tumors. In all patients, we found that the BRCA1 rs12516 CC genotype had a significantly higher BRCA1 protein expression than BRCA1 rs12516 CT/TT variant genotypes (P = 0.036), while RAD51 rs7180135 AA genotype had a borderline significantly higher RAD51 protein expression than RAD51 rs7180135 AG/GG variant genotypes (P = 0.057) (Figure 3). Furthermore, in 50 HPV16-positive patients only, the patterns of above significant associations of these 2 SNPs with their protein expression remained. BRCA1 rs12516 CC genotype had a significantly higher BRCA1 protein expression than BRCA1 rs12516 CT/TT genotypes (P = 0.021); and RAD51 rs7180135 AA genotype had a borderline significantly higher RAD51 protein expression than RAD51 rs7180135 AG/GG genotypes (P = 0.053) (Figure 3).
Figure 3.
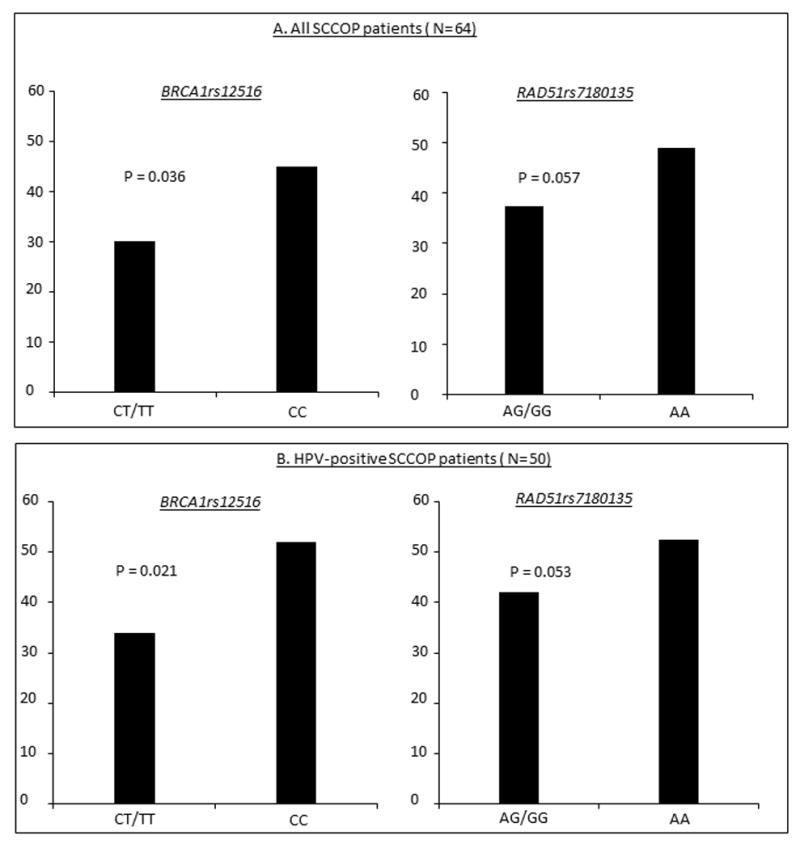
Comparison of BRCA1 and RAD51 expression levels by immunohistochemistry staining among SCCOP patients (A: all 64 SCCOP patients and B: Fifty HPV-positive SCCOP patients).
Discussion
In this study, we assessed the roles of polymorphisms in miRNA-binding sites within 3’UTRs of genes specifically involved in DNA DSB repair pathways in modulating the recurrence of SCCOP. We found that genotypic variations at BRCA1 rs12516 and RAD51 rs7180135 were significantly associated with a reduced risk of disease recurrence, particularly in patients with HPV16-positive SCCOP.
BRCA1 is required for efficient repair of DSBs through the homology-directed repair pathway in DNA damage response20, 21. It plays an mutually exclusive role in selecting between the homology-directed repair and nonhomologous end joining DSB repair pathways 22. Deregulation of BRCA1 reduces DNA repair capacity and increases the susceptibility of cancer cells to DNA-damaging agents, including chemotherapy and radiotherapy 23, 24. Although many studies have explored the relationships between BRCA1 and miRNAs 25, few have focused on the roles of genetic variants in the miRNA-binding sites of the BRCA1 3’UTR. A Thai study in women found a reduced breast cancer recurrence rate in association with two polymorphic sites alleles in the BRCA1 3’UTR: 5711+421 (G or T) and 5711+1286 (C or T) 26. Another study showed that the BRCA1 rs12516 polymorphism resulted in increased risk of early-onset and/or familial breast cancer in the Turkish population. These results indicated that this polymorphism in BRCA1 may function in the development of breast cancer 27. In contrast, in this study, SCCOP patients with the variant genotypes of BRCA1 rs12516 had a significantly reduced risk of disease recurrence. Our finding may support that this polymorphism in the 3’UTR of BRCA1 alters the ability of miRNA to bind to the BRCA1 3’ UTR and leads to loss of certain functions of BRCA1.
Recently, BRCA1 rs12516 has been shown to have several potential miRNAs for binding. Bioinformatics analysis and SNP function prediction demonstrated that the site of BRCA1 rs12516 C-to-T transition had six potential binding miRNAs: hsa-miR-188-5p, hsa-miR-502-5p, hsa-miR-557, hsa-miR-623, hsa-miR-637, and hsa-miR-639 28. Among these miRNAs, miRNA-188 was downregulated in oral squamous cell carcinoma and inhibited proliferation and invasion by targeting SIX1 29. Similar functions of miRNA-188 were found in other types of cancer, including prostate cancer, hepatocellular carcinoma, and acute myeloid leukemia 30-32. Another miRNA, miR-639, was found to have an oncogenic role in human tongue cancer and breast cancer 33, 34. Further studies are recommended to explore the roles of these miRNAs in outcome and to elucidate the mechanisms underlying the associations.
RAD51, like BRCA1, is essential for catalyzing homologous recombination. Recruitment of RAD51 to DSBs is mediated by many factors, including BRCA1. Overexpression of RAD51 is associated with a more aggressive cancer phenotype and treatment resistance in a variety of tumors, while downregulation of RAD51 reduces the efficiency of DNA repair and thus improves sensitivity to radiotherapy and chemotherapy 35, 36. Our studies showed that SCCOP patients with the RAD51 rs7180135 variant genotypes had a significantly reduced risk of disease recurrence. Similar findings have been reported in other types of cancer. Improved survival was associated with RAD51 rs7180135 G allele in human bladder cancer 13. This study suggested that the binding of miR-197 to RAD51 rs7180135 might alter miR-197-mRNA-binding and might predict sensitivity of tumors to radiotherapy, thus affecting outcomes in bladder cancer 13. Several other miRNAs have previously been shown to target RAD51 and inhibit its role in homologous recombination repair, including miRNA-182, miR-155, miR-103, and miR-107 in several types of human cancer 37-39. In addition, miR-182 can impede DNA repair and may increase the susceptibility of breast cancer to radiotherapy and chemotherapy through downregulation of BRCA1 40.
HPV-positive SCCOP is more sensitive to radiotherapy than HPV-negative SCCOP 3, 41. Impaired DNA DSB repair in both the nonhomologous end joining and homologous recombination pathways may underlie the enhanced sensitivity of HPV-positive head and neck cancers to treatment 42. Our findings from this study showed that SCCOP patients, particularly patients with HPV16-positive SCCOP, with BRCA1 rs12516 and RAD51 rs7180135 variant genotypes had significantly better DFS and reduced risk of disease recurrence. These findings are biologically plausible since almost every SCCOP patient in our study underwent definitive radiotherapy or chemoradiotherapy and such treatments lead to DNA DSB damage, which is repaired by DNA DSB repair pathways. HPV16-positive tumors, particularly in never smokers, rarely exhibit somatic mutations of TP53. Tumors harboring intact p53 might activate DNA repair pathways, and these patients who received radiotherapy or chemoradiotherapy seem to be correlated with response to radiotherapy partially because of activation of the p53-mediated repair pathway. Therefore, genetic variants of BRCA1 rs12516 and RAD51 rs7180135 may cause different expression of these two genes and lead to inter-individual differences in DNA repair capacity phenotype. Such differences in turn may result in different susceptibility to the genotoxic effects of radiation, resulting in different clinical outcomes.
To date, no investigations on functional relevance of BRCA1 rs12516 and RAD51 rs7180135 polymorphisms have been reported. Since these 2 polymorphisms are within the functional regions of the 3’-UTR of these two genes, the variant genotypes of these 2 polymorphisms might have potentially function to decrease expression levels of these 2 DNA repair proteins, thus leading to better outcome. In fact, in this study, we found that BRCA1 rs12516 and RAD51 rs7180135 polymorphisms significantly or borderline significantly affected expression of their protein levels in SCCOP patients, particularly HPV16-positive patients. Thus, our results could partially suggest a functional correlation between these 2 polymorphism and their protein expressions, which may provide preliminary evidence of biological plausibility for the associations.
Although the exact molecular mechanisms of these 2 variants in the double-strand break (DSB) repair pathway is still largely uncertain, the biological plausibility of significance for these 2 SNPs could be because 1) the expression level or biological function of these 2 genes could be determinant for the entire DNA repair process they are belonging to the process; 2) these 2 SNPs might affect either biological function or expression level of the gene of interest; and 3) these 2 SNPs might not directly affect the DNA repair function, while it may be in linkage disequilibrium with other known or unknown functional polymorphisms or adjacent susceptibility loci of the gene, thereby affecting this gene expression and activity. For example, in this study, the BRCA1 rs12516 and RAD51 rs7180135 variant genotypes might have an increased miRNA-mRNA binding ability and a decreased BRCA1 and RAD51 expression. Such lower levels of DNA repair proteins would cause reduced DNA repair and increased cancer risk. However, in tumors with radiotherapy or chemoradiotherapy, we speculate that the reduced DNA repair proteins may provide a favorable advantage for tumor cells to die, leading to reduced risk of cancer recurrence. Therefore, the altered BRCA1 and RAD51 expression caused by BRCA1 rs12516 and RAD51 rs7180135 might modify the susceptibility to radiotherapy or chemoradiotherapy. This modification effect thus influence risk of SCCOP recurrence.
Our study has several limitations. First, because the study was hospital based and retrospective, it is subject to selection bias. Second, results may not be generalizable to other ethnic groups since most of patients in the study were non-Hispanic whites. Third, the small sample size, limited number of outcome events, and short follow-up time might cause chance findings. Finally, some HPV16-positive SCCOP may be negative for p16 expression, It is likely that the some HPV16-positive SCCOP that failed to express p16 was not driven by HPV, but HPV was just an incidental bystander in that tumor43. This discordance could occur between HPV status and p16 expression since some events (e.g., mutation, deletion, other epigenetic changes, etc) may impact the p16, leading to biased estimation of the associations.
In conclusion, we believe this is the first comprehensive study investigating the roles of genetic variants in miRNA-binding sites of DNA DSB repair genes in recurrence of SCCOP. We found that genetic variants of BRCA1 rs12516 and RAD51 rs7180135 significantly reduced the risk of recurrence of SCCOP, particularly HPV16-positive SCCOP. Although our findings require validation in further studies, these miRNA-related SNPs might serve as prognostic biomarkers for patients with SCCOP.
Novelty and impact.
Our study found that BRCA1 rs12516 and RAD51 rs7180135, polymorphisms in miRNA-binding sites within 3’UTRs of genes specifically involved in DNA DSB repair pathways, were significantly associated with a reduced risk of SCCOP recurrence, particularly in patients with HPV16-positive SCCOP. If these associations are confirmed, clinicians could use these functional polymorphisms to identify subgroups of patients with different risks of recurrence and offer them appropriately tailored treatment.
Acknowledgments
The authors thank Margaret Lung, Kathryn L. Tipton, Liliana Mugartegui, and Angeli Fairly for their help with participant recruitment and Li-E Wang for laboratory management.
Funding support: This work was supported by start-up funds from The University of Texas MD Anderson Cancer Center (to E.M.S.), a National Institutes of Health Head and Neck Specialized Program of Research Excellence Career Development Award (P50CA097007 to E.M.S.), an Institutional Research Grant from The University of Texas MD Anderson Cancer Center (to E.M.S.), and other National Institutes of Health grants: grants ES011740 and CA131274 (to Q.W.), a Clinician Investigator Award (K12CA88084 to E.M.S.), a Cancer Center Support Grant to The University of Texas MD Anderson Cancer Center (CA16672), and grants CA135679, CA133099, and 1R03CA186261-01A1 (to G.L.).
Abbreviations
- 3' -UTR
3' -untranslated region
- miRNA
microRNA
- CI
confidence interval
- HR
hazard ratio
- SCCOP
squamous cell carcinoma of the oropharynx
- HPV
human papillomavirus
- SNP
single-nucleotide polymorphism
- DSB
double strand break
Footnotes
Conflict of Interest: The authors declare no conflict of interest.
References
- 1.D’Souza G, Kreimer AR, Viscidi R, Pawlita M, Fakhry C, Koch WM, Westra WH, Gillison ML. Case-control study of human papillomavirus and oropharyngeal cancer. N Engl J Med. 2007;356:1944–56. doi: 10.1056/NEJMoa065497. [DOI] [PubMed] [Google Scholar]
- 2.Chaturvedi AK, Engels EA, Pfeiffer RM, Hernandez BY, Xiao W, Kim E, Jiang B, Goodman MT, Sibug-Saber M, Cozen W, Liu L, Lynch CF, Wentzensen N, Jordan RC, Altekruse S, Anderson WF, Rosenberg PS, Gillison ML. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol. 2011;29:4294–301. doi: 10.1200/JCO.2011.36.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ang KK, Harris J, Wheeler R, Weber R, Rosenthal DI, Nguyen-Tân PF, Westra WH, Chung CH, Jordan RC, Lu C. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363:24–35. doi: 10.1056/NEJMoa0912217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seiwert TY, Zuo Z, Keck MK, Khattri A, Pedamallu CS, Stricker T, Brown C, Pugh TJ, Stojanov P, Cho J. Integrative and comparative genomic analysis of HPV-positive and HPV-negative head and neck squamous cell carcinomas. Clin Cancer Res. 2015;21:632–641. doi: 10.1158/1078-0432.CCR-13-3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iyama T, Wilson DM. DNA repair mechanisms in dividing and non-dividing cells. DNA repair. 2013;12:620–636. doi: 10.1016/j.dnarep.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bottai G, Pasculli B, Calin GA, Santarpia L. Targeting the microRNA-regulating DNA damage/repair pathways in cancer. Expert Opin Biol Ther. 2014;14:1667–1683. doi: 10.1517/14712598.2014.950650. [DOI] [PubMed] [Google Scholar]
- 8.Raleigh DR, Haas-Kogan DA. Molecular targets and mechanisms of radiosensitization using DNA damage response pathways. Future Oncology. 2013;9:219–233. doi: 10.2217/fon.12.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei W, Ba Z, Gao M, Wu Y, Ma Y, Amiard S, White CI, Danielsen JMR, Yang Y-G, Qi Y. A role for small RNAs in DNA double-strand break repair. Cell. 2012;149:101–112. doi: 10.1016/j.cell.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 10.d’Adda di Fagagna F. A direct role for small non-coding RNAs in DNA damage response. Trends Cell Biol. 2014;24:171–178. doi: 10.1016/j.tcb.2013.09.008. [DOI] [PubMed] [Google Scholar]
- 11.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 12.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 13.Teo MT, Landi D, Taylor CF, Elliott F, Vaslin L, Cox DG, Hall J, Landi S, Bishop DT, Kiltie AE. The role of microRNA-binding site polymorphisms in DNA repair genes as risk factors for bladder cancer and breast cancer and their impact on radiotherapy outcomes. Carcinogenesis. 2012;33:581–586. doi: 10.1093/carcin/bgr300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu H, Gao F, Dahlstrom KR, Li G, Sturgis EM, Zevallos JP, Wei Q, Liu Z. A variant at a potentially functional microRNA-binding site in BRIP1 was associated with risk of squamous cell carcinoma of the head and neck. Tumor Biology. 2015:1–10. doi: 10.1007/s13277-015-4682-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naccarati A, Rosa F, Vymetalkova V, Barone E, Jiraskova K, Di Gaetano C, Novotny J, Levy M, Vodickova L, Gemignani F, Buchler T, Landi S, Vodicka P, Pardini B. Double-strand break repair and colorectal cancer: gene variants within 3’ UTRs and microRNAs binding as modulators of cancer risk and clinical outcome. Oncotarget. 2016;7:23156–23169. doi: 10.18632/oncotarget.6804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang X, Yang H, Lee JJ, Kim E, Lippman SM, Khuri FR, Spitz MR, Lotan R, Hong WK, Wu X. MicroRNA-related genetic variations as predictors for risk of second primary tumor and/or recurrence in patients with early-stage head and neck cancer. Carcinogenesis. 2010;31:2118–2123. doi: 10.1093/carcin/bgq177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang C, Sturgis EM, Zheng H, Zafereo ME, Wei Q, Li G. TNF-α promoter polymorphisms and risk of recurrence in patients with squamous cell carcinomas of the nonoropharynx. Int J Cancer. 2014;135:1615–1624. doi: 10.1002/ijc.28793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ji X, Sturgis EM, Zhao C, Etzel CJ, Wei Q, Li G. Association of p73 G4C14-to-A4T14 polymorphism with human papillomavirus type 16 status in squamous cell carcinoma of the head and neck in non-Hispanic whites. Cancer. 2009;115:1660–1668. doi: 10.1002/cncr.24184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang ZD, Lippman SM, Wu TT, Lotan R, Xu XC. RRIG1 mediates effects of retinoic acid receptor beta2 on tumor cell growth and gene expression through binding to and inhibition of RhoA. Cancer Res. 2006;66:7111–7118. doi: 10.1158/0008-5472.CAN-06-0812. [DOI] [PubMed] [Google Scholar]
- 20.Moynahan ME, Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nature reviews Molecular cell biology. 2010;11:196–207. doi: 10.1038/nrm2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huen MS, Sy SM, Chen J. BRCA1 and its toolbox for the maintenance of genome integrity. Nat Rev Mol Cell Biol. 2010;11:138–148. doi: 10.1038/nrm2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aparicio T, Baer R, Gautier J. DNA double-strand break repair pathway choice and cancer. DNA repair. 2014;19:169–175. doi: 10.1016/j.dnarep.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roy R, Chun J, Powell SN. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nature Reviews Cancer. 2012;12:68–78. doi: 10.1038/nrc3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosen EM. BRCA1 in the DNA damage response and at telomeres. Frontiers in genetics. 2013;4:85. doi: 10.3389/fgene.2013.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang S, Sharan SK. BRCA1 and microRNAs: emerging networks and potential therapeutic targets. Mol Cells. 2012;34:425–432. doi: 10.1007/s10059-012-0118-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pongsavee M, Yamkamon V, Dakeng S, Oc P, Smith DR, Saunders GF, Patmasiriwat P. The BRCA1 3’-UTR: 5711+421T/T_5711+1286T/T genotype is a possible breast and ovarian cancer risk factor. Genet Test Mol Biomarkers. 2009;13:307–317. doi: 10.1089/gtmb.2008.0127. [DOI] [PubMed] [Google Scholar]
- 27.Erturk E, Cecener G, Polatkan V, Gokgoz S, Egeli U, Tunca B, Tezcan G, Demirdogen E, Ak S, Tasdelen I. Evaluation of genetic variations in miRNA-binding sites of BRCA1 and BRCA2 genes as risk factors for the development of early-onset and/or familial breast cancer. Asian Pac J Cancer Prev. 2014;15:8319–8324. doi: 10.7314/apjcp.2014.15.19.8319. [DOI] [PubMed] [Google Scholar]
- 28.Yang F, Chen F, Xu J, Guan X. Identification and frequency of the rs12516 and rs8176318 BRCA1 gene polymorphisms among different populations. Oncol Lett. 2016;11:2481–2486. doi: 10.3892/ol.2016.4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang L, Liu H. microRNA-188 is downregulated in oral squamous cell carcinoma and inhibits proliferation and invasion by targeting SIX1. Tumor Biology. 2016;37:4105–4113. doi: 10.1007/s13277-015-4246-9. [DOI] [PubMed] [Google Scholar]
- 30.Zhang H, Qi S, Zhang T, Wang A, Liu R, Guo J, Wang Y, Xu Y. miR-188-5p inhibits tumour growth and metastasis in prostate cancer by repressing LAPTM4B expression. Oncotarget. 2015;6:6092. doi: 10.18632/oncotarget.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fang F, Chang R-m, Yu L, Lei X, Xiao S, Yang H, Yang L-Y. MicroRNA-188-5p suppresses tumor cell proliferation and metastasis by directly targeting FGF5 in hepatocellular carcinoma. J Hepatol. 2015;63:874–885. doi: 10.1016/j.jhep.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 32.Jinlong S, Lin F, Yonghui L, Li Y, Weidong W. Identification of let-7a-2-3p or/and miR-188-5p as prognostic biomarkers in cytogenetically normal acute myeloid leukemia. PLoS One. 2015;10:e0118099. doi: 10.1371/journal.pone.0118099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin Z, Sun L, Chen W, Liu B, Wang Y, Fan S, Li Y, Li J. miR-639 regulates transforming growth factor beta-induced epithelial–mesenchymal transition in human tongue cancer cells by targeting FOXC1. Cancer Sci. 2014;105:1288–1298. doi: 10.1111/cas.12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li L, Qiu XG, Lv PW, Wang F. miR-639 promotes the proliferation and invasion of breast cancer cell in vitro. Cancer Cell Int. 2014;14:39. doi: 10.1186/1475-2867-14-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ward A, Khanna KK, Wiegmans AP. Targeting homologous recombination, new pre-clinical and clinical therapeutic combinations inhibiting RAD51. Cancer Treat Rev. 2015;41:35–45. doi: 10.1016/j.ctrv.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 36.Le Scodan R, Cizeron-Clairac G, Fourme E, Meseure D, Vacher S, Spyratos F, de la Lande B, Cvitkovic F, Lidereau R, Bieche I. DNA repair gene expression and risk of locoregional relapse in breast cancer patients. Int J Radiat Oncol Biol Phys. 2010;78:328–336. doi: 10.1016/j.ijrobp.2009.07.1735. [DOI] [PubMed] [Google Scholar]
- 37.Lai TH, Ewald B, Zecevic A, Liu C, Sulda M, Papaioannou D, Garzon R, Blachly JS, Plunkett W, Sampath D. HDAC Inhibition Induces MicroRNA-182, which Targets RAD51 and Impairs HR Repair to Sensitize Cells to Sapacitabine in Acute Myelogenous Leukemia. Clin Cancer Res. 2016;22:3537–3549. doi: 10.1158/1078-0432.CCR-15-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gasparini P, Lovat F, Fassan M, Casadei L, Cascione L, Jacob NK, Carasi S, Palmieri D, Costinean S, Shapiro CL, Huebner K, Croce CM. Protective role of miR-155 in breast cancer through RAD51 targeting impairs homologous recombination after irradiation. Proc Natl Acad Sci U S A. 2014;111:4536–4541. doi: 10.1073/pnas.1402604111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang J-W, Wang Y, Dhillon KK, Calses P, Villegas E, Mitchell PS, Tewari M, Kemp CJ, Taniguchi T. Systematic screen identifies miRNAs that target RAD51 and RAD51D to enhance chemosensitivity. Mol Cancer Res. 2013;11:1564–1573. doi: 10.1158/1541-7786.MCR-13-0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moskwa P, Buffa FM, Pan Y, Panchakshari R, Gottipati P, Muschel RJ, Beech J, Kulshrestha R, Abdelmohsen K, Weinstock DM. miR-182-mediated downregulation of BRCA1 impacts DNA repair and sensitivity to PARP inhibitors. Mol Cell. 2011;41:210–220. doi: 10.1016/j.molcel.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marur S, D’Souza G, Westra WH, Forastiere AA. HPV-associated head and neck cancer: a virus-related cancer epidemic. The lancet oncology. 2010;11:781–789. doi: 10.1016/S1470-2045(10)70017-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dok R, Kalev P, Van Limbergen EJ, Asbagh LA, Vázquez I, Hauben E, Sablina A, Nuyts S. p16INK4a Impairs Homologous Recombination–Mediated DNA Repair in Human Papillomavirus–Positive Head and Neck Tumors. Cancer Res. 2014;74:1739–1751. doi: 10.1158/0008-5472.CAN-13-2479. [DOI] [PubMed] [Google Scholar]
- 43.Garnaes E, Frederiksen K, Kiss K, Andersen L, Therkildsen MH, Franzmann MB, Specht L, Andersen E, Norrild B, Kjaer SK, von Buchwald C. Double positivity for HPV DNA/p16 in tonsillar and base of tongue cancer improves prognostication: Insights from a large population-based study. Int J Cancer. 2016;139:2598–2605. doi: 10.1002/ijc.30389. [DOI] [PubMed] [Google Scholar]


