Engineering multicellular systems: using synthetic biology to control tissue self-organization
The control of multicellular systems in general and of tissue formation in particular is a frontier for regenerative medicine and basic biological research. Current manipulations of multicellular systems such as tissue engineering, in vitro organoid development, and stem cell differentiation are revolutionizing the field, yet remain confronted with difficulties controlling precision, complexity, and functional integration. New methodologies and tools are needed to address these issues before the ambitious goal of building complex, customizable organs and tissues can be achieved. One promising approach is starting to make gains in this area: the genetic engineering of cellular signaling to directly or indirectly affect cellular self-organization. This review will focus on genetic manipulations that make use of, and/or are modeled after, the self-organization programs that multicellular systems use during development and regeneration. In particular, current examples and future directions of the following three areas will be explored: (i) Engineering developmental trajectories in non-developmental systems, with an example for epithelial patterning; (ii) Engineering control in developmental systems, with an example of increasing cellular composition complexity in stem cell differentiation; (iii) Engineering regeneration in non-regenerating systems, with an example from limb regeneration with engineered cells.
The use of synthetic biology to control the genetic layer of these three areas will undoubtedly uncover important rules dictating cellular self-organization, putting us one step closer to a powerful approach for building multicellular systems, one we will call synthetic tissue development. In the future, we anticipate that convergence of this approach with more established approaches to multicellular system control will lead to improved functional tissue formation in vitro and the possibility of transformative advances in regenerative medicine.
Introduction
Building tissues is a fascination of modern biomedicine that is evident in the rapid expansion of tissue engineering for transplantation and regeneration, stem cell based therapies, advances in organoid development, and organ-on-a-chip disease modeling [1,2]. These revolutionary approaches face challenges in precision, complexity, control, and functional integration [3–5]. These are formidable challenges, as cells do not often behave in predictable or easily controllable ways. New approaches are therefore necessary to accomplish the lofty goal of building complex, customizable, self-organizing organs that could seamlessly replicate or augment endogenous organ function and integrate with host systems.
Yet, a technology that builds tissue with high precision already exists naturally: embryonic development. During embryonic development, undifferentiated progenitor cells are directed to build all the various tissue types of the mature organism. As revealed by developmental, regenerative, and stem cell biology, many of the programs that direct this self-organization are encoded at the genetic level in the form of gene regulatory networks and cell signaling cascades. It has been proposed that synthetic biology approaches at this scale could be transformative [6,7]. Until recently, the tools necessary for this kind of direct genetic engineering and control were unavailable, but recent technological advances, deepened scientific understanding, and social paradigm-shifts have created an opportunity for the growth of an entirely new engineering discipline focused on multicellular systems [8–10]. Specific synthetic biology tools such as synthetic receptors, synthetic transcription factors, and engineered communication pathways are described in more detail in the “Toolkit” section.
Recent technological advances in genetic manipulation have provided a favorable background for engineering multicellular systems. Of particular importance are genetic manipulations that allow for either the stable introduction of large exogenous signaling circuits in the genome or the ability to engineer changes directly in endogenous loci to rewire native signaling. Synthetic DNA of even very large size can now be produced relatively cheaply and landing pads can be used to integrate these larger synthetic constructs into the genome in site-specific ways [11]. Entirely artificial chromosomes can also be used as carriers of exogenous DNA, either alone or in combination with transposon-based technology [12]. In addition, advances in CRISPR based technologies have allowed for unprecedented manipulation of endogenous loci allowing both genetic replacement and nuanced gene control [13].
Concurrently, deepening biological knowledge of the principles relating to cellular differentiation have led to a greater understanding of the plasticity of cell fate via reprogramming [14] and to the development of protocols for in vitro organoid generation [15]. Many points of genetic control on cell differentiation have been identified and can be used as targets for the creation of synthetic circuits driving changes in cell fate. Recent breakthroughs in tumor immunotherapy and the FDA approval of CAR-T cells have changed the social landscape and have had the bystander effect of lowering perceived risks of engineered cells in the clinic [16].
Taken together we believe a new discipline is being defined. Described alternately as synthetic development, synthetic morphogenesis, or tissue development engineering, we will call it synthetic tissue development. Synthetic tissue development is based in the tools of synthetic biology and in the scientific underpinnings of developmental biology and it can be defined broadly as the use of synthetic biology tools to control tissue development. In a very general way, the goal is to control the developmental trajectory of a multicellular system by engineering synthetic genetic circuits in its component cells (Fig. 1). Developmental trajectory is itself broadly defined and encompasses self-organizational programs of information processing, patterning, morphogenesis, differentiation, and other such processes directed by cells and the matrix they inhabit.
Figure 1. Cell signaling networks as the key to engineering multicellular self-organization.
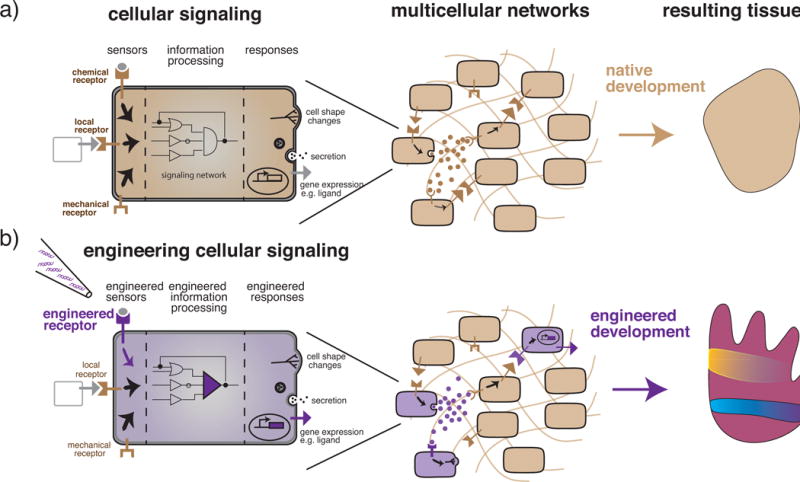
a) Representation of a native cell (represented as beige throughout this review) as a computational unit made up of sensing, processing, and response subunits. Receptors capable of sensing secreted chemicals, locally bound ligands, mechanical changes, or other environmental cues are an example of the sensing subunit of cells. An information processing subunit then uses signaling networks to transform the incoming signal into a response. Responses are computational outputs and can take many forms such as alterations of gene expression leading to morphological changes, molecule secretion, ligand production, or a variety of other behaviors. In a multicellular context, the computational output of any one cell both influences and is influenced by its neighbors. Each cell both senses and responds and the collection of cells creates its own signaling network. These complex networks generate, at the multicellular level, the emergent properties of self-organization, patterning, morphogenesis, differentiation, and decision-making that ultimately combine to build complex tissues. b) Engineering efforts (represented in purple throughout this review) can be directed towards modifications of cell sensing, processing, response or any combination of these subunits to suit the needs of the engineered system. In a multicellular context, the presence of even a few properly engineered cells can then change the computational output of the system as a whole. Engineering cell signaling at the level of the individual cell can thereby result in increased complexity and control in the multicellular context of tissues.
In the following pages, we will introduce examples that we believe underscore three categorically distinct approaches synthetic tissue development may take. They are based on the nature of the native, non-engineered system and the goal of the engineering:
Engineering developmental trajectories in non-developmental systems such as the bottom-up design of novel multicellular signaling networks. An example in this category could be engineering morphogenesis or patterning in simple epithelial layers in vitro.
Engineering control in developmental systems such as the modification of progenitor cells. An example in this category could be engineering stem cells to differentiate into multiple lineages with spatial precision.
Engineering regeneration in non-regenerating systems such as modifying degenerative trajectories to re-route them into regenerative healing. An example in this category could be engineering cells to direct scar-less healing in systems that normally undergo fibrosis.
Our hope is to share our excitement for and inspire consideration of a new approach to multicellular system engineering with synthetic biologists, tissue engineers, developmental, stem cell, and regenerative biologists. We would like to highlight how engineering cell-cell communication and signaling can provide a key knob for controlling multicellular systems and share what we see as the next steps in these areas. We feel that the synthetic tissue development angle could work in parallel with current tissue biology approaches to enrich our control over the structure and function of multicellular systems.
Engineering developmental trajectories in non-developmental systems
The goal here is to encode genetic programs for self-organized development in cellular systems that would not normally display patterning or morphogenesis on their own.
During embryonic development, cells self-organize through a combination of cell-cell and cell-matrix signaling interactions. Cells in tissues can be thought of as input-output units capable of concurrent communicate and response generation based on underlying genetic instructions (Fig. 1) [9,17]. Simple rules of communication along with their subsequent activation of functional genes provide the basic routine for emergence of more complex behaviors. It was speculated that manipulation of those rules with the introduction of synthetic input-output circuits should be able to generate developmental phenomena even in natively non-developmental systems [8,9,18].
In a recent example, the Lim Lab succeeded in generating a synthetic patterning circuit in a naïve epithelial cell line using synthetic cell-cell contact dependent pathways [19]. To achieve this, the authors designed simple rules of interaction in cells driven by a synthetic receptor system coined synNotch. SynNotch is a platform for engineering synthetic juxtacrine signaling circuits and works by altering both extracellular sensing specificity and intracellular response targets. The authors first generated a simple multicellular synthetic network where synthetic ligands on sender cells bind synthetic receptors on neighboring receiver cells, activating non-native target genes in the form of reporter genes. These simple rules generate a simple boundary detection pattern (Fig.2a–c). The network is then made more complex by further modifying and enriching the information flow by the addition of a second receptor and secondary response. Now, when the first ligand activates the receptor, transcription of a gene for a second ligand is also activated. This second ligand is itself sensed by neighboring cells that then activate a secondary reporter gene in a cascade fashion. This more complex network results in more complex rules (Fig.2e) and patterning (Fig. 2f). The initially uniform receiver cells are patterned into three different cell “types” (as defined by expression of different target genes) in concentric rings according to their distance from an island of sender cells (Fig. 2d–f). In another example, checkerboard patterns were generated by rewiring the Notch pathway to implement a controlled lateral-inhibition circuit [20].
Figure 2. Engineering cell-cell communications rules in epithelial cells produces precise single-cell thick concentric ring patterning.
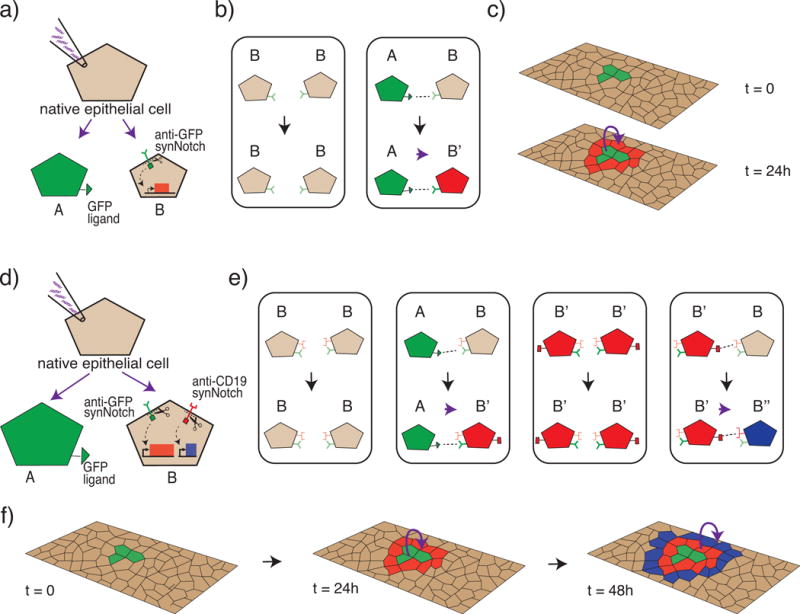
Shown is a schematic representations of results we wish to highlight from Morsut et al. [19]. a–c) Single ring patterning. a) Native epithelial cells are genetically engineered to create a sender cell (A) and receiver cell (B). The sender cell presents a GFP ligand on its surface while the receiver cell contains a synthetic receptor system (synNotch) with an anti-GFP sensing domain and a response domain capable of activating transcription of mCherry, a red fluorescent protein. b) A lookup table governing the rules of the engineered communication system, similar to those in a cellular automaton [62]. When B is near B, B remains unchanged. When A is near B, B changes state to B″. c) Schematic of how the system self-organizes in a multicellular context with an island of sender (A) cells in a field of receiver (B) cells. Receiver cells in the direct vicinity of sender cells change state and fluoresce red, while receiver cell further away remain unchanged. d–f) Double ring patterning. d) In a more complex example, the receiver cells are engineered with two synNotch pathways. The anti-GFP synNotch system from the first example is modified such that activation induces both red fluorescence and the production of a surface ligand (CD19) designed to serve as the ligand for a second synNotch system; the second synNotch system has an anti-CD19 sensing domain and a response domain capable of activating the transcription of tagBFP, a blue fluorescent protein. e) Look up table of rules governing this more complex system. When B is near B, B remains unchanged. When A is near B, B changes state to B′. When B′ is near B′, B′ remain unchanged. When B′ is near B, B changes state to B′. f) Schematic representation of the self-organization in a multicellular context with an island of sender cells (A) in a field of receiver cells (B). Receiver cells in the direct vicinity of sender cells change state, fluoresce red, and express the CD19 ligand. Receiver cells in the direct vicinity of cells expressing the CD19 ligand, change state and fluoresce blue.
These examples show the power of synthetic communication circuits, which can build on the endogenous way that cells self-organize. Contact dependent pathways such as the native Notch pathway are used during tissue formation for boundary establishment and precise cellular differentiation patterning [21]. Synthetic receptors engineered with different features, such as those capable of sensing soluble ligands, could extend the possible patterning configurations. Patterning circuits creating stripes or spots have been studied computationally [22] and could potentially be replicated in a multicellular context with these tools. Further enhancements to engineered behavior could be made by coupling functional gene activation with patterning circuits. Activating genes involved in differentiation, proliferation, and secretion [23] could contribute to the functional aspect of the engineered tissue. For example, differential expression of cadherin family adhesion molecules has been shown to enable formation of spatial patterns based, at least in part, on differential adhesion [24,25]. Implementation of this approach could lead to the creation of more complex morphogenesis and dynamically evolving structures.
Engineering and manipulation of non-developmental systems can also provide a useful model for studying pieces of more complex developmental trajectories in a more controlled manner. Along with other synthetic biology techniques such as optogenetics, and magnetogenetics, synthetic patterning circuits could be used to create genetic asymmetries capable of driving self-organization into desired geometries (see Toolkit section below for more on these techniques). These systems could have the potential to enhance our understanding of tissue development routines and principles by allowing us to ask questions like: What are the strengths and limitations of contact-dependent patterning as compared to soluble morphogen-like patterning? How much and what kind of morphogenesis can be designed with simple patterning rules when they are coupled with morphogenetic effectors? [9,23,26]. As these questions are explored, the principles they uncover could be then used to increase spatial precision at the cellular scale as, for example, in organoid differentiation contexts.
Engineering control in developmental systems
The goal here is to generate more complex and precise multicellular structures by controlling the developmental trajectories of stem cells using genetic manipulation. Multicellular systems are very generative and naturally self-organizing. In this case then, the challenge for the bioengineer is not reinventing self-organization, but rather redirecting endogenous routines [8,9,27].
Currently, there is a trade-off between complexity and control in stem cell differentiation protocols. When tight control over stem cell differentiation is desired only limited meaningful complexity in cellular composition is possible. Alternatively, when a high degree of complexity in cellular composition is desired, as in the case of organoids, direct control of individual cell fates is sacrificed [1,14]. Taken together, two recent examples show a path towards reconciling this trade-off.
In a powerful example of increasing the complexity of cellular composition, Patrick Guye in the Weiss Lab used simple genetic engineering in a stem cell population to seed spatial genetic asymmetries [28]. The authors generated a population of hiPSCs in which each cell had a different overexpression level of the transcription factor Gata6. Cell growth was followed for 18 days (Figure 3). The differentiation of this genetically asymmetric population generated a developmental trajectory much richer than one with native expression levels. Of particular note, the level of expression itself was not sufficient to predict the fate of each individual cell (Fig 3e). Instead, cell fate was in some cases influenced by the fate of neighboring cells. This highlights the importance of cell-cell signaling in translating genetic asymmetries into downstream cell behavior. This principle is evident during development, where initial asymmetries generated by various means serve as the starting point for more complex behaviors (e.g. primitive endoderm and epiblast differentiation and sorting in early mouse development [29,30]). It is interesting to note that it was sufficient to engineer differences in the starting point to influence the self-organization of the cellular ensemble. However, while this manipulation succeeded in increasing complexity, in order to increase the control of resultant cellular composition, circuits enabling spatial and temporal regulation of gene expression are necessary.
Figure 3. Increased complexity in cellular composition of stem cell differentiation as a function of initial genetic asymmetries.
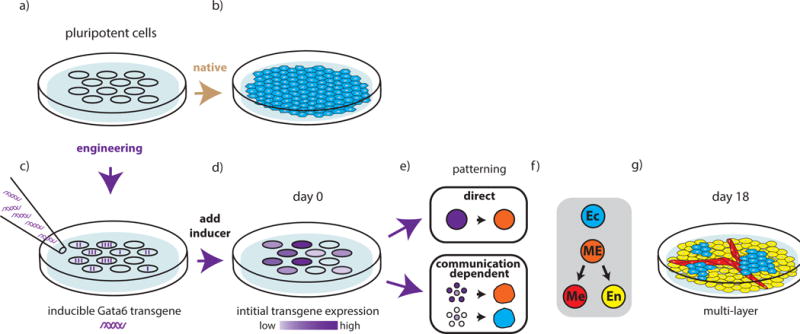
Shown is a schematic representation of the results we wish to highlight from Guye et al. [28]. a–b) Unengineered pluripotent cells undergo native developmental trajectories to build a sheet of ectodermal cells in a monolayer in media supporting pluripotency. c–g) Engineered pluripotent cells develop more complex multicellular structures c). An inducible GATA6 transgene is introduced into pluripotent cells such that individual cells take up variable copy numbers of the transgene. d) Once the inducer is added, the asymmetry introduced by variable copy number uptake is maintained as variable expression levels of the GATA6 protein. e) Two distinct types of patterning occur as a result of the engineering. Direct patterning leads cells with high levels of transgene expression to adopt a mesendodermal (ME) while cells with no transgene maintain ectodermal (Ec) fate. When transgene expression levels fall within a mid-range, patterning becomes communication-dependent and cell fate is largely determined by the expression levels of neighboring cells. f) With continued growth, and in the absence of specialized media or inducers, the cells continue to differentiate along ectodermal (Ec), mesodermal (Me), and endodermal (En) lineages and ultimately self-organize to create a complex and multilayer liver-bud-like structure.
Synthetic circuits providing sophisticated temporal control of genes driving differentiation of stem cells into single cell types have been described. Saxena et al. describe a synthetic network that executes a preprogrammed sequential differentiation agenda coordinating the timely induction and repression of multiple genes [31]. During development, tight temporal control of gene expression profiles is what creates coherent differentiation and maturation of a tissue containing multiple types. Integrating this sort of temporal control into synthetic patterning circuits driving differentiation of multiple cell types concurrently is a frontier of the field of synthetic tissue development.
We think that these first results demonstrate the potential power of engineering in natively self-organizing developmental systems. The combination of transcription-factor mediated differentiation (e.g. with Gata6 or others) with cell-cell communication engineering could provide a more rational design for concurrent differentiation of multiple cell types. The next generation of this kind of intervention could benefit from greater control of initial spatial asymmetries using the techniques of synthetic patterning, optogenetics, or magnetogenetics (see “Toolkit” section) and could be used, among other things, to develop more precisely constructed multicellular disease modeling systems.
Engineering regeneration in non-regenerating systems
The goal here is to re-route a degenerative response into a regenerative one. In many biological systems, it is thought that an endogenous regeneration potential is present, but impeded by competing blocking mechanisms [32–34]. Currently, the combination of stem cells, growth factors, and scaffolds is being explored as a possible therapeutic intervention in degenerative contexts. The potential and limitations of this approach have been widely evaluated [35,36]. One major limitation is the difficulty of integrating new growth with existing tissue, especially in complex organs. Cell therapy with engineered cells is a promising avenue, since cells are able to provide very complex and reliable therapeutic inputs [37]. A striking example of the power of engineered cells to increase complex tissues regeneration is reported by Lin et al. [38] in the context of limb regeneration (Figure 4).
Figure 4. Using engineered cells to enhance limb regeneration in a non-regenerative system.
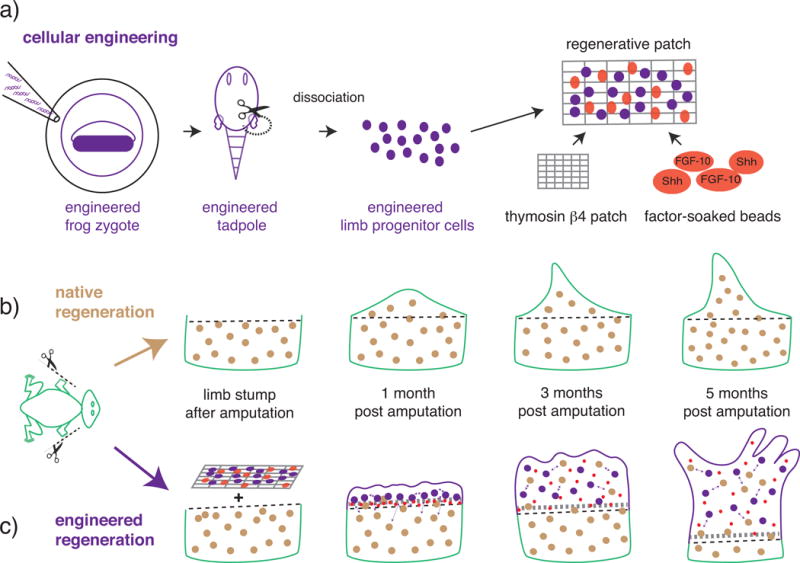
Shown is a schematic representation of the results we wish to highlight from Lin et al.[38]. a) Steps to the construction of a regenerative patch. X. laevis zygotes are engineered to express a heat inducible construct that constitutively activates the β-catenin gene (β-catenin*). A tadpole is then generated with a decondensed sperm nuclei transfer technique. Limb progenitor cells containing the engineered construct are combined with a thymosin β4 soaked fibrin scaffold, or patch, and Fibroblast Growth Factor 10 (FGF-10) and Sonic Hedgehog (Shh) factor-soaked affi-gel beads. b–c) Limb regeneration in presence or absence of the regenerative patch. b) Post amputation, native regeneration in post-metamorphic juvenile X. laevis adults is minimal: five months post-amputation the regenerate is an unsegmented, disorganized, cartilaginous spike. c) Post-amputation treatment with the regenerative patch containing both engineered cells and growth factors led to enhanced regeneration (patches containing only factor-soaked beads or only engineered cells preformed less optimally). Five months post-amputation, treatment with the full regenerative patch created a regenerate with partial segmentation and morphology more closely resembling that of a normal limb.
In juvenile frogs, the native response to amputation is a very limited form of regeneration with no resultant patterning of the regenerated tissue (Fig 4b). Lin et al successfully improved this response with a “synthetic blastema” construct made of engineered cells, growth factors beads, and an underlying scaffold. Engineered cells overexpressed beta-catenin, a signaling molecule known to be important during limb development (Fig 4a). When the authors used the components parts of the construct separately, they saw minimal regeneration. When combined, the engineered cells, through unknown mechanisms, communicated amongst themselves and with the host cells triggering a regenerative response. Strikingly, the resulting limb structures were made of both engineered donor cells and host cells (Fig 4c). This supports the idea that host cells, even in non-regenerative contexts, can be induced to regenerate and also suggests that engineered cells, with their enhanced communications, could be a suitable medium to overcome the block to regeneration. In another example, mesenchymal stem cells were engineered to overexpress the growth factor BMP-2 to increase bone regeneration responses [39]. These studies represent validation of the idea that engineered cells can be used as the basis for regenerative therapeutic interventions, and lays the foundation for more complex and rational interventions.
Moving forward the goal would be to generate sensing-and-response mechanisms, similar to those explored in tumor immunotheraphy [37,40], that could detect degeneration and re-route cells towards regeneration. Recently identified genetic elements from organisms with high regeneration potential show activation at the site of degeneration and could be used as sensors of degeneration in synthetic systems [41]. Other sensors could include synthetic receptors designed to bind specific signals produced at the site of degeneration. Responses could include the production of growth factors or communication factors for resident stem cells or immune cells. Other responses could be informed by existing limb regeneration mechanisms in organisms capable of regeneration [42–45]. Stabilization and maintenance of regeneration could require additional patterning and could benefit from the combination of optogenetics, magnetogenetics, and synthetic patterning.
Synthetic biology toolkit for synthetic tissue development
Here we report some of the tools we think will be useful in the future of synthetic tissue development. As we discussed earlier (see Introduction and Engineering development) these synthetic biology tools are derived from the template that nature uses for self-organization during embryonic development, i.e. cell communication networks. These natural networks are implemented by signaling pathways that process the information from input to output, sensing information from the extracellular world with receptors, processing that information intracellularly, and producing appropriate cellular outputs (Fig. 1). On a molecular level, the information largely flows through protein-protein interactions cascades. The direction of the flow is dictated by the specific signaling proteins and their partners, which are in turn dictated by modular protein-protein interaction domains [46]. This means that you can direct information flow across multicellular communication networks by adding, removing, swapping or otherwise modifying the protein domains of the proteins that transduce cellular signals. We can generate in this way synthetic networks that are either completely orthogonal to endogenous signaling or that rewire it. This concept, and the interchangeability of protein domains, underlies all the techniques at the heart of the synthetic biology tools for cellular signaling engineering [47,48]. More recently, this same approach has been used to build synthetic receptors, exploiting in this case the modularity present in both the extracellular/sensing domains and the intracellular/transduction domains of native receptors [49].
At the level of the sensing, one class of synthetic receptors sense synthetic/controllable factors such as exogenous compounds in anti-GFP synNotch receptors [19], light in optogenetics [50], and magnetic fields in magnetogenetics [51]. In optogenetics, receptors or signaling proteins with light-sensitive domains are under the control of light activation. Similarly, in magnetogenetics, magnetosensitive receptors have been generated that can activate target genes upon magnetic stimulation. These synthetic sensors can be used to trigger specific responses with high temporal and spatial precision by either controlling the inputs (light and magnetic fields) or via implementation of synthetic patterning rules (synNotch). In multicellular systems, this type of synthetic sensors has proven useful for probing endogenous systems [52], and could conceivably be used to control the development of organoids or the release of therapeutics in vivo.
Another class of synthetic receptors can sense endogenous factors: tumor antigens like CD19 can be sensed with CARs [53] or synNotch receptors [19,54]; growth factors like VEGF and/or small peptides can be sensed with MESA receptors [55], Tango [56] and dCas9-synR [57]. These receptors couple sensing of endogenous factors with synthetic transduction, usually using a synthetic intracellular domain. This type of synthetic receptor is ideally suited for disease state sensing (e.g. sensing molecules produced at a site of tissue degeneration), or for sensing the differentiation status of neighbor cells to trigger downstream programs.
At the level of the responses, the transduction of the signaling usually leads to the induction or repression of target genes through activation or repression of transcription factors. Both synthetic and endogenous transcription factors and target genes can be used as output of synthetic pathways. Synthetic exogenous cassettes with responsive promoters and custom-designed target genes can be introduced in the genome as in the synNotch examples, to introduce synthetic genetic responses. The advantage of synthetic loci is the flexibility of what can be created in terms of promoters and chimeric proteins. Endogenous genes can be activated or repressed in endogenous loci using microRNAs [31] or synthetic transcription factors like CRISPR/Cas9 [58] variants. The advantage of targeting the endogenous genetic loci is more stable and long-term impact on the cellular phenotype. To this end, interesting tools for manipulating the genetic information at the epigenetic level have been developed using dCAS9 fusion with enzymes that can write different epigenetic modifications in a targeted way [13]; an interesting frontier for synthetic tissue development will be generating synthetic pathways that would result in epigenome editing.
The cellular responses produced by the synthetic signaling pathways can be of different kinds. They can change cell fate, for example with master transcription factors like myoD for a myogenic cell fate, or snail for epithelial to mesenchymal transition [19]. Or the response can be a change in signaling properties. In an example of altered signaling properties, negative feedback on endogenous TNF-alfa signaling was introduced such that TNF-alfa target genes were replaced in the genome with TNF-alfa inhibitors [59]. More complex intracellular signal processing like memory and recombinase-based networks are available [60], and will be interesting to see if they can be coupled to extracellular sensing in multicellular contexts as they have been implicated in driving tissue development. Finally, responses could also result in changes to cell shape or to mechanical properties such as cell adhesion; in a recent example, drug-inducible expression of cadherin molecules in epithelial monolayer has been used to generate 2D and 3D synthetic patterning based on phase separation [24]. The implementation of these responses under control of synthetic pathways will be important for the implementation and the study of more complex morphogenetic trajectories.
The modularity of the various components of synthetic signaling pathways means that this toolkit could be combined in a very open-ended fashion. Synthetic sensing can be linked to activation of endogenous target genes, or sensing of endogenous factors can be linked to synthetic therapeutic responses. Alternatively, both strategies can be combined so that synthetic sensing is linked with activation of exogenous target genes for a completely synthetic pathway. Each approach will have different benefits. Completely synthetic pathways will be well suited to engineering development as by controlling both the inputs and outputs we can explore bottom-up designs. Synthetic pathways could be useful in these systems to set up patterning and asymmetries. Synthetic pathways designed to sense endogenous factors, could be particularly useful for triggering temporally controlled synthetic programs by sensing the differentiation status of neighboring cells. Systems sensing endogenous degeneration factors seem particularly relevant for engineering regeneration, as they could be coupled with responses dictating precise temporal and spatial patterning and morphogenesis.
Conclusions
Since its inception, arguably around 2000 in the modern sense [61], synthetic biology has been used in many ways, from metabolic rewiring in bacteria to tumor cell immunotherapy. In general, synthetic biology approaches have provided tools to control and analyze the biological systems at hand, from bacterial cells to T-cells. The time has come to turn this approach to tissue biology through the control of information flow in multicellular contexts. As previously discussed, a tissue can be thought of as the computational output of individual sensing-processing-response units (Fig. 1). With synthetic receptors providing the key to connecting sensing to information processing and genome editing providing the key to connecting information processing to responses, we believe that synthetic biology is poised to become a foundational technology for multicellular systems control.
In particular, the expectation is development of more control over cellular behaviors (see Engineering control), development of novel properties (see Engineering development), and increased applicability for cell therapy (see Engineering regeneration). For example, synthetic tissue development efforts could contribute to the development of complex model organs that more closely mirror endogenous organ structure and function. More physiologically relevant systems for screening therapeutic molecules and testing novel disease interventions could reduce the time from basic research to clinical application. Advances in tissue engineering could also be translated into therapies augmenting tissue regeneration or even tissue transplantation. Other explorations could expand normal tissue function by introducing novel behaviors and shapes. Once a more complete toolkit is in place, the variety of tissues and multicellular structures that could be created might only be limited by the imagination of the engineer.
Moreover, while we artificially divided the field into categories based on the nature of the system and the goal of engineering, such categories need not remain distinct. For instance, engineered cells derived from non-developmental systems could be used in concert with unengineered stem cells or iPSCs to drive and guide developmental programs in spatially or temporally controlled ways. The flexibility of the approach underlines a methodological advantage of synthetic tissue development. Its investigative process has much in common with iterative engineering processes. Engineered multicellular systems can become model systems on their own, and can be subjected to study alongside natural tissue systems, to discover their mechanisms and (mal)functions; the engineered systems will now have the added advantage of an increased access to their manipulation and improvement. The design-build-test-learn-validate approach of engineering has been very successful in driving progress and building quickly on successes. This is of particular value in the age of translational research where it is desirous to turn the truths uncovered by basic science into applications relevant to treating human disease and dysfunction.
For the first time, we find ourselves equipped with the tools and basic knowledge necessary for engineering self-organization in multicellular systems. We believe that this could enable “biological moonshot” projects, such as complete limb regeneration, that could energize the field. Looking to tissue engineers for better scaffolds, stem cell biologists for improved differentiation targets, developmental biologists for basic principles and mechanisms, and computational scientists for modeling and planning, this effort will necessarily span many disciplines. The incipient field of synthetic tissue development holds the potential to enhance our control of the development and functional behavior of complex multicellular systems and to serve as a valuable testing ground for the nature and importance of cell signaling in multicellular contexts. We believe it will play a meaningful role in uncovering basic biological principles and in creating the next generation of therapies for regenerative medicine.
Cell signaling pathways within and between cells dictate form and function in multicellular systems.
Synthetic cell signaling pathways can be engineered due to technological advances in synthetic biology.
Synthetic signaling pathways can be used to engineer developmental processes (e.g. patterning) in non-developmental systems (e.g. epithelial cell lines)
Possible uses of synthetic pathways to engineer stem cells differentiation into complex and controlled multicellular systems are examined.
The potential for using synthetic signaling to enhance regeneration in non-regenerative systems is explored.
Acknowledgments
The authors want to thank Cristy Lytal for revising the text. Jonathan Brunger, Satoshi Toda, Alex Ng, the Editor and an anonymous reviewer for the feedback on an earlier draft of the manuscript. This work was supported by an R00 grant from the National Institute of Biomedical Imaging and Bioengineering to LM (4R00EB021030-03), and from a Startup Fund from the Department of Stem Cell Biology and Regenerative Medicine at USC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Yin X, Mead BE, Safaee H, Langer R, Karp JM, Levy O. Engineering Stem Cell Organoids. Stem Cell. 2016;18:25–38. doi: 10.1016/j.stem.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Langer R, Vacanti J. Advances in tissue engineering. Journal of Pediatric Surgery. 2016;51:8–12. doi: 10.1016/j.jpedsurg.2015.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matthys OB, Hookway TA, McDevitt TC. Design Principles for Engineering of Tissues from Human Pluripotent Stem Cells. Curr Stem Cell Rep. 2016;2:43–51. doi: 10.1007/s40778-016-0030-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mao AS, Mooney DJ. Regenerative medicine: Current therapies and future directions. Proc Natl Acad Sci U S A. 2015;112:14452–14459. doi: 10.1073/pnas.1508520112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peloso A, Dhal A, Zambon JP, Li P, Orlando G, Atala A, Soker S. Current achievements and future perspectives in whole-organ bioengineering. Stem Cell Research & Therapy. 2015;6:107. doi: 10.1186/s13287-015-0089-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davies JA. Synthetic morphology: prospects for engineered, self-constructing anatomies. J Anat. 2008;212:707–719. doi: 10.1111/j.1469-7580.2008.00896.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sprinzak D, Elowitz MB. Reconstruction of genetic circuits. Nature. 2005;438:443–448. doi: 10.1038/nature04335. [DOI] [PubMed] [Google Scholar]
- 8.Teague BP, Guye P, Weiss R. Synthetic Morphogenesis. Cold Spring Harb Perspect Biol. 2016 doi: 10.1101/cshperspect.a023929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morsut L. Programming cells to build tissues with synthetic biology: A new pathway toward engineering development and regeneration. In: Gardiner DM, editor. Regenerative Engineering and Developmental Biology: Principles and Applications. CRC Press; 2017. pp. 555–593. [Google Scholar]
- 10.Kicheva A, Rivron NC. Creating to understand - developmental biology meets engineering in Paris. Development. 2017;144:733–736. doi: 10.1242/dev.144915. [DOI] [PubMed] [Google Scholar]
- 11.Duportet X, Wroblewska L, Guye P, Li Y, Eyquem J, Rieders J, Rimchala T, Batt G, Weiss R. A platform for rapid prototyping of synthetic gene networks in mammalian cells. Nucleic Acids Research. 2014;42:13440–13451. doi: 10.1093/nar/gku1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jung CJ, Ménoret S, Brusselle L, Tesson L, Usal C, Chenouard V, Remy S, Ouisse L-H, Poirier N, Vanhove B, et al. Comparative Analysis of piggyBac, CRISPR/Cas9 and TALEN Mediated BAC Transgenesis in the Zygote for the Generation of Humanized SIRPA Rats. Sci Rep. 2016 doi: 10.1038/srep31455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dominguez AA, Lim WA, Qi LS. Beyond editing: repurposing CRISPR–Cas9 for precision genome regulation and interrogation. Nature Publishing Group. 2015;17:5–15. doi: 10.1038/nrm.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu J, Belmonte JCI. Stem Cells: A Renaissance in Human Biology Research. Cell. 2016;165:1572–1585. doi: 10.1016/j.cell.2016.05.043. [DOI] [PubMed] [Google Scholar]
- 15.Huch M, Koo B-K. Modeling mouse and human development using organoid cultures. Development. 2015;142:3113–3125. doi: 10.1242/dev.118570. [DOI] [PubMed] [Google Scholar]
- 16.Miller JFAP, Sadelain M. The Journey from Discoveries in Fundamental Immunology to Cancer Immunotherapy. Cancer Cell. 2015;27:439–449. doi: 10.1016/j.ccell.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 17.Vila AOX, Duran-Nebreda S, Conde-Pueyo NXR, ez RXLMX, xe9 RS A morphospace for synthetic organs and organoids: the possible and the actual. Integrative Biology. 2016;8:485–503. doi: 10.1039/c5ib00324e. [DOI] [PubMed] [Google Scholar]
- 18.Davies JA, Cachat E. Synthetic biology meets tissue engineering. Biochemical Society Transactions. 2016;44:696–701. doi: 10.1042/BST20150289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morsut L, Roybal KT, Xiong X, Gordley RM, Coyle SM, Thomson M, Lim WA. Engineering Customized Cell Sensing and Response Behaviors Using Synthetic Notch Receptors. Cell. 2016;164:780–791. doi: 10.1016/j.cell.2016.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsuda M, Koga M, Woltjen K, Nishida E, Ebisuya M. Synthetic lateral inhibition governs cell-type bifurcation with robust ratios. Nat Commun. 2015;6:6195. doi: 10.1038/ncomms7195. [DOI] [PubMed] [Google Scholar]
- 21.Hori K, Sen A, Artavanis-Tsakonas S. Notch signaling at a glance. J Cell Sci. 2013;126:2135–2140. doi: 10.1242/jcs.127308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meinhardt H, Gierer A. Pattern formation by local self-activation and lateral inhibition. BioEssays. 2000;22:753–760. doi: 10.1002/1521-1878(200008)22:8<753::AID-BIES9>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 23.Davies J. Using synthetic biology to explore principles of development. Development. 2017;144:1146–1158. doi: 10.1242/dev.144196. [DOI] [PubMed] [Google Scholar]
- 24.Cachat E, Liu W, Martin KC, Yuan X, Yin H, Hohenstein P, Davies JA. 2-and 3-dimensional synthetic large-scale de novo patterning by mammalian cells through phase separation. Sci Rep. 2016 doi: 10.1038/srep20664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steinberg MS. Differential adhesion in morphogenesis: a modern view. Curr Opin Genet Dev. 2007;17:281–286. doi: 10.1016/j.gde.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 26.Elowitz M, Lim WA. Build life to understand it. Nature. 2010;468:889–890. doi: 10.1038/468889a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prochazka L, Benenson Y, Zandstra PW. Synthetic gene circuits and cellular decision-making in human pluripotent stem cells. Current Opinion in Systems Biology. 2017;5:93–103. [Google Scholar]
- 28.Guye P, Ebrahimkhani MR, Kipniss N, Velazquez JJ, Schoenfeld E, Kiani S, Griffith LG, Weiss R. Genetically engineering self-organization of human pluripotent stem cells into a liver bud-like tissue using Gata6. Nat Commun. 2016;7:10243. doi: 10.1038/ncomms10243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chazaud C, Yamanaka Y. Lineage specification in the mouse preimplantation embryo. Development. 2016;143:1063–1074. doi: 10.1242/dev.128314. [DOI] [PubMed] [Google Scholar]
- 30.Wennekamp S, Mesecke S, Nédélec F, Hiiragi T. A self-organization framework for symmetry breaking in the mammalian embryo. Nature Publishing Group. 2013;14:452–459. doi: 10.1038/nrm3602. [DOI] [PubMed] [Google Scholar]
- 31.Saxena P, Heng BC, Bai P, Folcher M, Zulewski H, Fussenegger M. A programmable synthetic lineage-control network that differentiates human IPSCs into glucose-sensitive insulin-secreting beta-like cells. Nat Commun. 2016;7:11247. doi: 10.1038/ncomms11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tanaka EM. Regeneration: if they can do it, why can't we? Cell. 2003;113:559–562. doi: 10.1016/s0092-8674(03)00395-7. [DOI] [PubMed] [Google Scholar]
- 33.Laurencin CT, Nair LS. The Quest toward limb regeneration: a regenerative engineering approach. Regen Biomater. 2016;3:123–125. doi: 10.1093/rb/rbw002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quijano LM, Lynch KM, Allan CH, Badylak SF, Ahsan T. Looking Ahead to Engineering Epimorphic Regeneration of a Human Digit or Limb. Tissue Engineering Part B: Reviews. 2016 doi: 10.1089/ten.teb.2015.0401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han I, Ropper AE, Konya D, Kabatas S, Toktas Z, Aljuboori Z, Zeng X, Chi JH, Zafonte R, Teng YD. Biological Approaches to Treating Intervertebral Disk Degeneration: Devising Stem Cell Therapies. Cell Transplantation. 2015;24:2197–2208. doi: 10.3727/096368915X688650. [DOI] [PubMed] [Google Scholar]
- 36.Reynolds J, Lamba DA. Human embryonic stem cell applications for retinal degenerations. Experimental Eye Research. 2014;123:151–160. doi: 10.1016/j.exer.2013.07.010. [DOI] [PubMed] [Google Scholar]
- 37.Fischbach MA, Bluestone JA, Lim WA. Cell-based therapeutics: the next pillar of medicine. Sci Transl Med. 2013;5:179ps7–179ps7. doi: 10.1126/scitranslmed.3005568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin G, Chen Y, Slack JMW. Imparting Regenerative Capacity to Limbs by Progenitor Cell Transplantation. Developmental Cell. 2013;24:41–51. doi: 10.1016/j.devcel.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pensak M, Hong S, Dukas A, Tinsley B, Drissi H, Tang A, Cote M, Sugiyama O, Lichtler A, Rowe D, Lieberman JR. The Role of Transduced Bone Marrow Cells Overexpressing BMP-2 in Healing Critical-Sized Defects in a Mouse Femur. Gene Therapy. 2015;22:467–75. doi: 10.1038/gt.2015.14. [DOI] [PubMed] [Google Scholar]
- 40.Roybal KT, Lim WA. Synthetic Immunology: Hacking Immune Cells to Expand Their Therapeutic Capabilities. Annu Rev Immunol. 2017;35:229–253. doi: 10.1146/annurev-immunol-051116-052302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kang J, Hu J, Karra R, Dickson AL, Tornini VA, Nachtrab G, Gemberling M, Goldman JA, Black BL, Poss KD. Modulation of tissue repair by regeneration enhancer elements. Nature. 2016;532:201–206. doi: 10.1038/nature17644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bryant SV, Gardiner DM. The relationship between growth and pattern formation. Regeneration. 2016;3:103–122. doi: 10.1002/reg2.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McCusker C, Bryant SV, Gardiner DM. The axolotl limb blastema: cellular and molecular mechanisms driving blastema formation and limb regeneration in tetrapods. Regeneration. 2015;2:54–71. doi: 10.1002/reg2.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murawala P, Tanaka EM, Currie JD. Regeneration: The ultimate example of wound healing. Seminars in Cell and Developmental Biology. 2012;23:954–962. doi: 10.1016/j.semcdb.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 45.Simkin J, Sammarco MC, Dawson LA, Schanes PP, Yu L, Muneoka K. The mammalian blastema: regeneration at our fingertips. Regeneration. 2015;2:93–105. doi: 10.1002/reg2.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lim WA. Designing customized cell signalling circuits. 2010 doi: 10.1038/nrm2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gordley RM, Bugaj LJ, Lim WA. Modular engineering of cellular signaling proteins and networks. Curr Opin Struct Biol. 2016;39:106–114. doi: 10.1016/j.sbi.2016.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ausländer S, Fussenegger M. Engineering Gene Circuits for Mammalian Cell-Based Applications. Cold Spring Harb Perspect Biol. 2016;8:a023895–18. doi: 10.1101/cshperspect.a023895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brenner M, Cho JH, Wong WW. Synthetic biology: Sensing with modular receptors. Nature Publishing Group. 2017;13:131–132. doi: 10.1038/nchembio.2290. [DOI] [PubMed] [Google Scholar]
- 50.Kolar K, Weber W. ScienceDirect Synthetic biological approaches to optogenetically control cell signaling. Curr Opin Biotechnol. 2017;47:112–119. doi: 10.1016/j.copbio.2017.06.010. [DOI] [PubMed] [Google Scholar]
- 51.Stanley SA, Sauer J, Kane RS, Dordick JS, Friedman JM. Remote regulation of glucose homeostasis in mice using genetically encoded nanoparticles. Nat Med. 2014;21:92–98. doi: 10.1038/nm.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Johnson HE, Goyal Y, Pannucci NL, Schüpbach T, Shvartsman SY, Toettcher JE. The Spatiotemporal Limits of Developmental Erk Signaling. Developmental Cell. 2017;40:185–192. doi: 10.1016/j.devcel.2016.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sadelain M, Brentjens R, Rivière I. The Basic Principles of Chimeric Antigen Receptor Design. Cancer Discovery. 2013;3:388–398. doi: 10.1158/2159-8290.CD-12-0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roybal KT, Williams JZ, Morsut L, Rupp LJ, Kolinko I, Choe JH, Walker WJ, McNally KA, Lim WA. Engineering T Cells with Customized Therapeutic Response Programs Using Synthetic Notch Receptors. Cell. 2016;167:419–432.e16. doi: 10.1016/j.cell.2016.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schwarz KA, Daringer NM, Dolberg TB, Leonard JN. Rewiring human cellular input–output using modular extracellular sensors. Nature Publishing Group. 2016 doi: 10.1038/nchembio.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barnea G, Strapps W, Herrada G, Berman Y, Ong J, Kloss B, Axel R, Lee KJ. The genetic design of signaling cascades to record receptor activation. Proc Natl Acad Sci U S A. 2008;105:64–69. doi: 10.1073/pnas.0710487105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Baeumler TA, Ahmed AA, Fulga TA. Engineering Synthetic Signaling Pathways with Programmable dCas9-Based Chimeric Receptors. CellReports. 2017;20:2639–2653. doi: 10.1016/j.celrep.2017.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zalatan JG, Lee ME, Almeida R, Gilbert LA, Whitehead EH, La Russa M, Tsai JC, Weissman JS, Dueber JE, Qi LS, et al. Engineering Complex Synthetic Transcriptional Programs with CRISPR RNA Scaffolds. Cell. 2015;160:339–350. doi: 10.1016/j.cell.2014.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brunger JM, Zutshi A, Willard VP, Gersbach CA, Guilak F. Genome Engineering of Stem Cells for Autonomously Regulated, Closed-Loop Delivery of Biologic Drugs. Stem Cell Reports. 2017;8:1202–1213. doi: 10.1016/j.stemcr.2017.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Purcell O, Lu TK. Synthetic analog and digital circuits for cellular computation and memory. Curr Opin Biotechnol. 2014;29:146–155. doi: 10.1016/j.copbio.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cameron DE, Bashor CJ, Collins JJ. A brief history of synthetic biology. Nature Publishing Group. 2014;12:381–390. doi: 10.1038/nrmicro3239. [DOI] [PubMed] [Google Scholar]
- 62.Bilotta E, Pantano P. Emergent patterning phenomena in 2D cellular automata. Artif Life. 2005;11:339–362. doi: 10.1162/1064546054407167. [DOI] [PubMed] [Google Scholar]


