Abstract
Analysis of CT scans for studying Chronic Obstructive Pulmonary Disease (COPD) is generally limited to mean scores of disease extent. However, the evolution of local pulmonary damage may vary between patients with discordant effects on lung physiology. This limits the explanatory power of mean values in clinical studies. We present local disease and deformation distributions to address this limitation. The disease distribution aims to quantify two aspects of parenchymal damage: locally diffuse/dense disease and global homogeneity/heterogeneity. The deformation distribution links parenchymal damage to local volume change. These distributions are exploited to quantify inter-patient differences. We used manifold learning to model variations of these distributions in 743 patients from the COPDGene study. We applied manifold fusion to combine distinct aspects of COPD into a single model. We demonstrated the utility of the distributions by comparing associations between learned embeddings and measures of severity. We also illustrated the potential to identify trajectories of disease progression in a manifold space of COPD.
1 Introduction
Chronic Obstructive Pulmonary Disease (COPD) is a complex disorder arising from various pathological processes including emphysema and functional small airways disease (fSAD). The extent of emphysema and fSAD that make up overall disease burden can vary, which can affect lung physiology. Both disease processes can progress at different rates, complicating prognostication. Optimising the quantification of disease extent in COPD may improve the precision of disease staging and monitoring.
Analysis of lung disease from Computed Tomography (CT) has typically relied on the analysis of the lung using global averages. Such metrics cannot capture the anatomical distribution of disease. Methods have been proposed to quantify the contribution of various emphysema subtypes [5] or the distribution of image features [2]. Harmouche et al. [5] built an emphysema manifold by analysis of classified emphysema subtypes. A Severity Index (S) was derived from this space that is complimentary to the mean level of emphysema. In contrast, Bragman et al. [2] modelled local distributions of density and biomechanical features; exploiting them to investigate differences between subtypes of COPD whilst also classifying these subtypes.
2 Method
We present a new method to quantify the spread of parenchymal disease and measure its effect on lung deformation. It is based on locally quantifying tissue destruction and deformation to capture heterogeneity or homogeneity across the lung. The outcome is a distribution that quantifies various aspects of lung pathophysiology that can be modelled to test associations with various clinical hypotheses. The distributions can be exploited to quantify inter-patient differences in lung tissue pathology and deformation. A single model of tissue disease and deformation can be obtained by combining separate embeddings obtained from manifold learning with manifold fusion.
2.1 Lung deformation and tissue classification
The deformation between paired breath-hold CT scans acquired at forced residual capacity (ℐexp, Ω*) and total lung capacity (ℐins, Ω) can be obtained using nonrigid registration. The output is a transformation φ mapping each coordinate x ∈ Ω → x* ∈ Ω*. Local volume change is characterised by the Jacobian determinant J. It is calculated on a voxel-wise basis: J = det (∇xφ).
Parametric Response Mapping (PRM) [4] was used to classify voxels as emphysema (PRMemph) and functional small airways disease (PRMfSAD). For all voxels xi ∈ ℐins, the tissue class zi is based on Hounsfield Unit (HU) thresholds in ℐins and ℐexp. A voxel is classified as PRMemph if ℐins(xi) ≤ −950 and ℐexp(φ(xi)) ≤ −856. A voxel is classified as PRMfSAD if ℐins(xi) > −950 and ℐexp(φ(xi)) ≤ −856. The airways and vasculature are segmented by only considering voxels with an HU between −500HU and −1024HU in both scans.
2.2 Local disease and deformation distributions
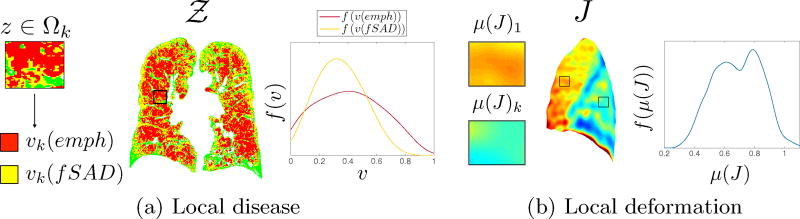
We present the concept of local feature distributions (Fig.1a and b). The aim is to quantify local abnormalities in lung physiology and pathology to define a signature unique to a patients disease state. We introduce two models: 1) local disease distributions and 2) local deformation distributions. The disease distributions model the spread of emphysema and fSAD whilst the deformation distribution characterises local volume change across the lung. They are created by locally sampling regions of 𝒵 and J in a Cartesian grid using local regions of interest Ωk (ROI) where k = 1 ⋯ K indexes the center voxel of the ROI. The size (r × r × r) of the ROI governs the scale of the sampling.
Fig. 1.
Local disease and deformation distributions.
We modelled two properties of disease spread: 1) locally diffuse/dense disease and 2) global homogeneity/heterogeneity. For each ROI centered at zk where z ∈ Ωk, we computed the fraction of PRMemph and PRMfSAD voxels; defined as υk(emph) and υk(fSAD). Dense disease occurred when υk(·) → 1 whilst diffuse disease was present when υk(·) → 0. The deviation of diffuse and dense regions in the lung defined the heterogeneity/homogeneity of disease spread.
A distribution f(υ(·)) for each feature was built by sampling K regions. The shape of the distribution is governed by the two disease properties (Fig.1a). It provides information on the nature of local disease spread (diffuse or dense) and whether it is homogeneous or heterogeneous.
Expansion of the lung is dependent on local biomechanical properties (emphysema) and airway resistance (functional small airways disease), which will affect lung deformation locally. To capture volume change on a local basis, the Jacobian map (J) was sampled by calculating the mean Jacobian (μ(J)k) for all Ωk. A distribution f(μ(J)) of these measurements was built to capture local volume change throughout the lung using the same process as above (Fig.1b).
2.3 Manifold learning of COPD distributions
We hypothesised that the heterogeneity of COPD could be modelled by the local disease and deformation distributions. Manifold learning can be used to capture variability in the distributions and learn separate embeddings for emphysema, fSAD and lung deformation. Fusion of these embeddings can then be performed to create various models of COPD.
Distribution distance
Inter-patient differences are computed using the Earth Movers Distance (ℒEMD) [11]. It is a cross-bin distance metric, which measures the minimum amount of work needed to transform one distribution into another. The distributions are quantised into separate histograms hυ(emph), hυ(fSAD) and hJ using Nb bins. They are normalised to sum to 1 such that they have equal mass. A closed-form solution of the ℒEMD can be used for one-dimensional distributions with equal mass and bins [7]. It reduces to the ℒ1-norm between cumulative distributions (H) of two histograms h1,(·) and h2,(·): .
Manifold learning and fusion
Manifold learning is used to model emphysema, fSAD and Jacobian distributions. The aim is to capture variations in the distributions in a population of COPD patients. As emphysema and fSAD occur synchronously and both affect lung function, the manifold fusion framework of Aljabar et al. [1] is employed to create a single representation of these processes.
For P subjects, the PRM classified volumes are 𝒵1, ⋯, 𝒵P and their respective Jacobian determinant maps are J = J1, ⋯, JP. The distributions are quantised using Nb bins into their respective histograms hp,υ(emph), hp,υ(fSAD) and hp,J. Pairwise measures in the population are obtained with the ℒEMD yielding the pairwise matrices ℳemph, ℳfSAD and ℳJ. They can be visualised as connected graphs where each node represents a patient and the edge length is the ℒEMD. Isomap1 [12] is applied to each matrix. A K-nearest neighbour search is first performed to create a sparse representation of ℳ(·) where edges are restricted to the K-nearest neighbourhood of each node. A full pairwise geodesic distance matrix D(·) is then estimated by analysis of the K-nearest graph of ℳ(·) using Djikstra’s shortest-path algorithm [3]. The low-dimensional embedding , p = 1, ·, P is obtained by minimisation of
| (1) |
using Multi-Dimensional Scaling. The coordinate embeddings for ℳemph, ℳfSAD and ℳJ are ye, yf and yJ with dimensions de, df and dJ that are selected.
Fusion of the coordinates y(·) can be performed in any combination to investigate various processes. For simplicity, we consider all embeddings. The coordinates are uniformly scaled with the scale factors se, sf and sJ such that the first component of each embedding has a unit variance. These are concatenated to yield Y = (seye, sfyf, sJyJ) with dimension de + df + dJ. A distance matrix ℳc is obtained by calculating pairwise Euclidean distances of Y. Isomap is then applied to yield the combined coordinate embedding yc with dimension dc.
3 Experiments
3.1 Data processing
A total of 1, 154 scans of COPD patients (GOLD ≥ 1) were downloaded from COPDGene [10]. They were acquired on various scanners (GE Medical Systems, Siemens and Philips) with the following reconstruction algorithms: STANDARD (GE), AS+ B31f and B31f (Siemens), and 64 B (Philips). The Pulmonary Toolkit2 was used for lung segmentation. Breath-hold scans were registered with NiftyReg [9] with a modified version of the EMPIRE10 pipeline [8]. The transformation was a stationary velocity field parameterised by a cubic B-spline and the similarity measure was MIND [6]. The constraint term was the bending energy of the velocity field, weighted at 1% for all stages of the pipeline. After manual inspection of the registrations, 743 patients were selected. Scans were rejected if there were major errors close to the fissures and the lung boundary.
The sampling size of the ROIs was r = 20mm, consistent with the size of the secondary pulmonary lobule. Sampling was performed with a Cartesian grid of center voxels spaced every 5mm. We chose a value of Nb = 60 as its effect on pairwise distances was minimal with increasing Nb when Nb > 50.
The dimensionality d of y and the parameter K for each embedding were determined by estimating the reconstruction quality of the lower-dimensional coordinates. The residual variance between the distances in ℳ(·) and the pairwise distances of y(·) was considered. For each embedding step (ye, yf and yJ), we determined the combination of K and d that minimised the residual variance. Grid-search parameters were set to d* ∈ [1, 5] and K* ∈ [5, 100]. Final parameters were K = [50, 30, 45] and d = [5, 5, 4] for ye, yf and yJ. We considered a model of the disease distributions (ye, yf → yc1) and a model also including the deformation (ye, yf, yJ → yc2). Parameters for both models were Kc1 = 55 and Kc2 = 60 with dc1 = 4 and dc2 = 4.
3.2 Associations with disease severity
Correlations between the embeddings and distribution moments were computed (Table 1). The first and second components of the embeddings had strong to moderate correlations with the distribution parameters, demonstrating that manifold learning of the distributions modelled the variation in the population.
Table 1.
Pearson correlation coefficient between the first three embedding coordinates and the distributions using the median (φ), median absolute deviation (ρ), skewness (γ1), kurtosis (γ2).
| PRMemph | PRMfSAD | J | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
||||||||||||||||
|
|
|
|
|
|
|
|
|
|
||||||||||
| φ | 0.96† | −0.19† | 0.01 | 0.97† | 0.07 | −0.01 | −0.48† | −0.06 | 0.04 | |||||||||
|
| ||||||||||||||||||
| ρ | 0.89† | 0.22† | −0.00 | 0.35† | −0.36† | −0.41† | −0.46† | 0.14* | −0.09 | |||||||||
|
| ||||||||||||||||||
| γ1 | −0.71† | −0.28† | 0.00 | −0.86† | 0.21† | 0.16† | −0.68† | −0.24† | 0.00 | |||||||||
|
| ||||||||||||||||||
| γ2 | −0.41† | −0.26† | −0.01 | −0.37† | 0.33† | 0.26† | −0.36† | −0.18† | −0.01 | |||||||||
[* = p < 0.05, † = p < 10−3]
We considered several models to predict COPD severity using FEV1%predicted and FEV1/FVC (Table 2). We considered three simple models (mean PRMemph, mean PRMfSAD and mean Jacobian μ(J)) and compared them to univariate and multivariate models of embedding coordinates (y). The univariate models showed moderate improvement over the simple mean models. However, the combined models ( and ) improved model prediction. The multivariate models demonstrated best performance, with model 2 (yc2 = ye + yf + yJ) performing best, even after adjusting for an increase in variables. It had a Bayesian Information Criterion (BIC) of 620 compared to 625 (yc1) and 633, 650 and 648 for PRMemph, PRMfSAD and μ(J) respectively. The increase in explanatory power was also seen when correlating the first component of the combined models with FEV1%predicted. The first components of the combined models had Pearson coefficients of r = 0.67, p < 0.001 and r = 0.70, p < 0.001 respectively. Coefficients for the mean models were r = −0.63, p < 0.001, r = −0.50, p < 0.001 and r = 0.52, p < 0.001 respectively. We also used manifold fusion to create a joint model between mean values of PRMemph and PRMfSAD and a second with PRMemph, PRMfSAD and μ(J). Pairwise mean differences were used to create ℳ(·). Correlation of the first component was r = 0.60, p < 0.001 and r = −0.65, p < 0.001 respectively. This corroborated the utility of combining embeddings based on the local distributions ( , p < 0.001).
Table 2.
Regression of models versus various clinical measures of COPD severity. Model performance quoted as adjusted-r2.
| Mean features | univariate | multivariate | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
||||||||||||||||
| Y | PRMe | PRMf | μ(J) |
|
|
|
|
|
yc1 | yc2 | ye | yf | yJ | |||||
| FEV1%p | 0.40† | 0.25† | 0.26† | 0.45† | 0.49† | 0.42† | 0.29† | 0.13† | 0.48† | 0.51† | 0.43† | 0.34† | 0.14† | |||||
| FEV1/FVC | 0.51† | 0.30† | 0.22† | 0.54† | 0.53† | 0.54† | 0.32† | 0.09† | 0.59† | 0.60† | 0.55† | 0.38† | 0.10† | |||||
[† = p < 10−3]
3.3 Trajectories of emphysema and fSAD progression
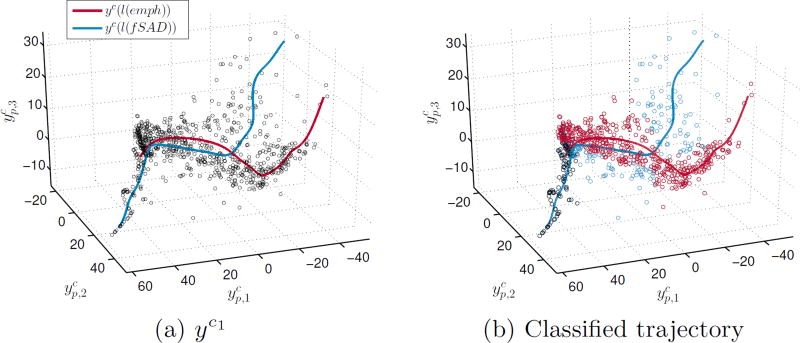
It is likely that trajectories of disease progression in COPD vary depending on the dominant disease phenotype. We assessed whether we can model these in the tissue disease model (yc1). We parameterised yc1 using the emphysema and fSAD distributions as covariates (l) with kernel regression: where K is a Gaussian kernel and υ is a normalisation constant. The covariate was the ℒEMD between the distributions and an idealised healthy distribution (distribution peak at υ = 0). The outcome is two trajectories in the manifold space (Fig.3a). The emphysema trajectory can be considered as the path taken when emphysema progression is dominant and vice-versa for fSAD. We classified patients based on these trajectories. A patient is seen to follow an emphysema progression trajectory if it is closest to yc(l(emph)). At the baseline, patients are classified as both emphysema and fSAD subtypes. When considering two sets of patients stratified by trajectory, the explanatory power of the embeddings improved in comparison to yc1 (Table 2). The emphysema regression produced an adjusted-r2 of 0.52 and 0.63 when predicting FEV1%predicted and FEV1/FVC respectively whilst fSAD was 0.45 and 0.62.
Fig. 3.
a) Three-dimensional projection of yc1 and b) classified trajectories of yc1.
4 Discussion and Conclusion
We have presented a method to parameterise distributions of various local features implicated in COPD progression. The disease distributions model local aspects of tissue destruction whilst modelling global properties of heterogeneity and homogeneity. The deformation distribution quantifies the local effect of disease on lung function. Patients exhibiting different mechanisms of tissue destruction can have identical global averages yet can display different disease distributions. These differences are likely to cause differences in local biomechanical properties, which are captured by the deformation distribution.
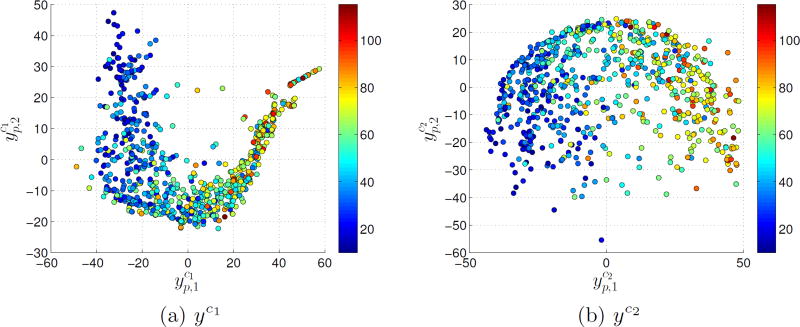
We have shown that models of the proposed distributions better predict COPD severity than conventional metrics (Table 2). We have shown that embeddings based on distribution dissimilarities have stronger correlations with FEV1%predicted than those learned from mean differences. Both these results suggest that the position of a patient in the manifold space of yc1 or yc2 is critical for assessing COPD. This was observed in the trajectory classification (Fig.3). Determining the trajectory that a patient is following may help inform therapeutic decisions and improve our understanding of COPD progression.
Complexity of the modelling may be increased to model more specific information about lung pathophysiology. Separate manifolds can be produced on a lobar basis. This is likely to further increase the explanatory power of the models since inter-lobar disease metrics correlate with different aspects of physiology. The detection of regional differences in local deformation may add further important information regarding the pathophysiology of a patient.
Fig. 2.
Projection of embeddings a) yc1 and b) yc2 with FEV1%predicted overlayed.
Acknowledgments
This work was supported by the EPSRC under Grant EP/H046410/1 and EP/K502959/1, and the UCLH NIHR RCF Senior Investigator Award under Grant RCF107/DH/2014. It used data (phs000179.v3.p2) from the COPDGene study, supported by NIH Grant U01HL089856 and U01HL089897.
Footnotes
References
- 1.Aljabar P, Wolz R, Srinivasan L, Counsell SJ, Rutherford MA, Edwards AD, Hajnal JV, Rueckert D. A combined manifold learning analysis of shape and appearance to characterize neonatal brain development. IEEE transactions on medical imaging. 2011;30(12):2072–86. doi: 10.1109/TMI.2011.2162529. [DOI] [PubMed] [Google Scholar]
- 2.Bragman F, McClelland J, Modat M, Ourselin S, Hurst JR, Hawkes DJ. Multi-scale Analysis of Imaging Features and Its Use in the Study of COPD Exacerbation Susceptible Phenotypes. MICCAI. 2014:417–424. doi: 10.1007/978-3-319-10443-0_53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dijkstra EW. A note on two problems in connexion with graphs. Numerische Mathematik. 1959;1(1):269–271. [Google Scholar]
- 4.Galbán CJ, Han MK, Boes JL, Chughtai KA, Charles R, Johnson TD, Galbán S, Rehemtulla A, Kazerooni EA, Martinez FJ, Ross BD. CT-based biomarker provides unique signature for diagnosis of COPD phenotypes and disease progression. Nature Medicine. 2013;18(11):1711–1715. doi: 10.1038/nm.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harmouche R, Ross JC, Diaz AA, Washko GR, Estepar RSJ. A Robust Emphysema Severity Measure Based on Disease Subtypes. Academic Radiology. 2016;23(4):421–428. doi: 10.1016/j.acra.2015.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heinrich MP, Jenkinson M, Bhushan M, Matin T, Gleeson FV, Brady M, Schnabel JA. MIND: Modality Independent Neighbourhood Descriptor for Multi-Modal Deformable Registration. Medical Image Analysis. 2012;16(7):1423–1435. doi: 10.1016/j.media.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 7.Levina E, Bickel P. The earth mover’s distance is the Mallows distance: some insights from statistics. Eighth IEEE International Conference on Computer Vision. 2001;2:251–256. [Google Scholar]
- 8.Modat M, McClelland J, Ourselin S. Lung Registration Using the NiftyReg Package. Medical Image Analysis for the Clinic: A Grand Challenge EMPIRE. 2010;10:33–42. [Google Scholar]
- 9.Modat M, Ridgway GR, Taylor ZA, Lehmann M, Barnes J, Hawkes DJ, Fox NC, Ourselin S. Fast free-form deformation using graphics processing units. Computer methods and programs in biomedicine. 2010;98(3):278–84. doi: 10.1016/j.cmpb.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 10.Regan EA, Hokanson JE, Murphy JR, Make B, Lynch DA, Beaty TH, Curran-Everett D, Silverman EK, Crapo JD. Genetic epidemiology of COPD (COPDGene) study design. COPD. 2010;7(1):32–43. doi: 10.3109/15412550903499522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rubner Y, Tomasi C, Guibas LJ. The Earth Mover's Distance as a Metric for Image Retrieval. International Journal of Computer Vision. 2000;40(2):99–121. [Google Scholar]
- 12.Tenenbaum JB, de Silva V, Langford JC. A global geometric framework for nonlinear dimensionality reduction. Science. 2000;290(5500):2319–23. doi: 10.1126/science.290.5500.2319. [DOI] [PubMed] [Google Scholar]