Abstract
Insight into the regulation of complex physiological systems emerges from understanding how biological units communicate with each other. Recent findings show that mitochondria communicate at a distance with each other via nanotunnels, thin double-membrane protrusions that connect the matrices of non-adjacent mitochondria. Emerging evidence suggest that mitochondrial nanotunnels are generated by immobilized mitochondria and transport proteins. This review integrates data from the evolutionarily conserved structure and function of intercellular projections in bacteria with recent developments in mitochondrial imaging that permit nanotunnel visualization in eukaryotes. Cell type-specificity, timescales, and the selective size-based diffusion of biomolecules along nanotunnels are also discussed. The joining of individual mitochondria into dynamic networks of communicating organelles via nanotunnels and other mechanisms has major implications for organelle and cellular behaviors.
The Mitochondrion as a Signaling Organelle
Communication – the regulated exchange of information between biological compartments – is required for multicellular life. In mammals and lower organisms, specialized structures such as blood vessels and nerves facilitate the rapid transfer of signaling molecules between organs. At the tissue level, transmembrane receptors, gap junctions, and specialized synapses ensure efficient and selective molecular exchange between different cell types [1]. Likewise, at the intracellular level, molecular complexes regulating communication between different organelles have recently been defined (e.g., [2]) and are recognized to play important roles in regulating organelle function and lifespan [3].
Mitochondria, the only organelles in animal cells that contain their own genome, are an important hub of intracellular signaling. They exchange Ca2+ and reactive oxygen species (ROS) with the endoplasmic reticulum (ER) as well as with each other [4–6], and also communicate with the nucleus where they may regulate the transcription of important nuclear genes [7] via the release of metabolic intermediates and proteins acting as transcriptional regulators [8–10]. As a result, mitochondria impact on complex cellular processes including differentiation, stemness, and oncogenic behavior, and ultimately influence concerted physiological states that also contribute to aging and neurodegenerative disease [11,12].
This evidence has altered our view of mitochondria. Once thought of as powerhouses functioning in isolation from one another, a paradigm is now emerging that mitochondria constitute a dynamic network of signaling organelles (Box 1). Importantly, maintaining functional mitochondria requires mitochondrial content exchange (see Glossary). An evolutionarily conserved machinery enables the complete and sequential fusion of the outer and inner mitochondrial membranes [13]. As a result, a mitochondrion with a defective respiratory chain can be rescued by fusing with a respiration-competent mitochondrion [14]. Moreover, genetic disruption of such mitochondrial communication is a cause of human disease [15], demonstrating the physiological significance of intermitochondrial communication or exchange. However, several tissues including skeletal muscle have reduced mitochondrial motility, thus restricting such communication, but no detriment to function is observed. It is thus possible that alternative communication mechanisms can compensate for the lack of frequent fusion in vivo.
Box 1. Modes of Mitochondrial Communication.
Mitochondria communicate with each other via the release of soluble signaling molecules that can propagate through the cytoplasm. These mechanisms are driven by the diffusion of signals from source organelles to all surrounding organelles, and are limited by diffusion distances. For cell–cell communication, non-selective diffusible signals exert indiscriminate effects on multiple surrounding mitochondria rather than on a single receiver mitochondrion. For example, ROS disseminate by ROS-induced ROS-release (RIRR) [6], and Ca2+ is responsible for the propagation of apoptotic signals across the mitochondrial network through a regenerative mechanism [5].
Other mechanisms of mitochondrial communication involve physical contact and are enhanced by specialized structures to enable specific molecular exchanges. Adjacent mitochondria coordinate inner mitochondrial membrane cristae at intermitochondrial junctions (IMJs) [55]. Likewise, mitochondrial fusion is a form a ‘private’ communication because it leads to the mixing of matrix and intermembrane space content between two defined mitochondria [56]. Fusion is broad-acting in cells with unencumbered cytoplasm (e.g., in vitro) and relies on substantial microtubule-based motility, whereby mitochondria can collide with one another, kiss and run (i.e., transient fusion or hemifusion), or fuse completely [57]. However, in differentiated cells with a dense cytoarchitectural environment, such as skeletal and cardiac muscles, mitochondrial motility is restricted [29,54,58]. This, and possibly specific molecular anchors, precludes efficient movement and limits the frequency of potential fusion events. In muscle fibers, mitochondria form a lattice structure within the intermyofibrillar region [27] where mitochondria are tethered to the z-band by a protein complex containing desmin and plectin, preventing their free movement [36]. As a result, in these tissues mitochondrial fusion events and the observed exchange of contents are less frequent than in dividing cultured cells, and occur between immotile mitochondria sometimes over long distances [27], indicating that membrane protrusions are necessary to accomplish long-range interactions.
We review here recent evidence demonstrating that tubular protrusions, termed mitochondrial nanotunnels, are evolutionarily conserved structures enabling intermitochondrial communication (Figure 1). Specifically, we propose that mitochondrial nanotunnels are communicating structures arising from immobilized mitochondria ‘reaching out for help’. This interpretation is based on (i) imaging studies in mammalian systems including human tissues; (ii) nanotunnel-like structures that transport molecular information between bacteria, the mitochondrial ancestor; and (iii) an emerging literature regarding specialized cell protrusions that enable communication in mammalian cells. In particular, we discuss nanotunnel formation, ultrastructure and dimensions, growth rates, cargo selectivity, and potential regulatory mechanisms. Because nanotunnels have only recently been observed, several important questions remain unanswered, and we also outline the major gaps in our knowledge concerning the regulation and physiological significance of mitochondrial nanotunnels.
Figure 1. Specialized Membrane-Based Tubular Structures Enable Cell–Cell and Mitochondria–Mitochondria Information Transfer.
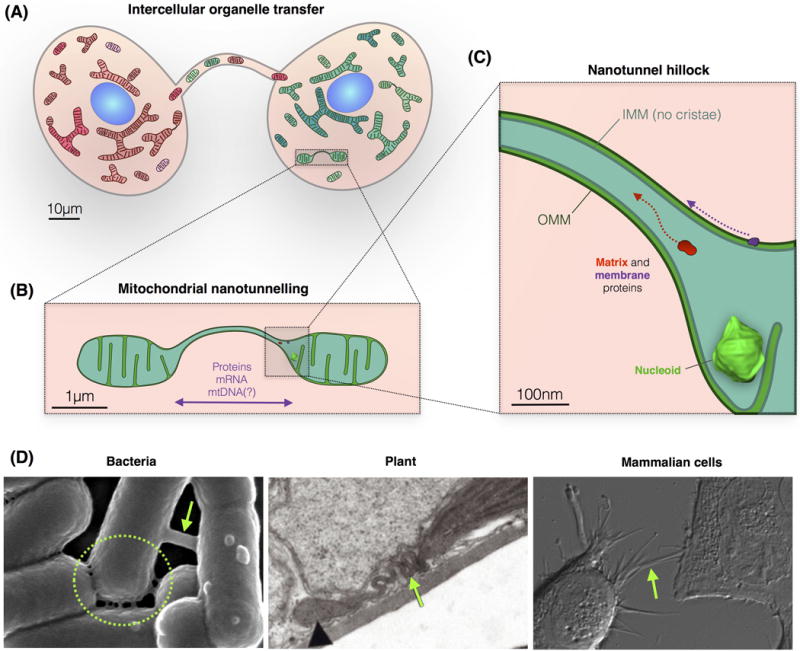
(A) Mammalian cell–cell exchange of organelles, vesicles, and soluble molecules occurs through cytonemes, nanotubes, and microtubules. (B) Within cells, mitochondria form similar tubular structures with contiguous outer and inner mitochondria membranes, and a continuous matrix space allowing the selective diffusion of specific molecular components. (C) Schematic of the nanotunnel junction, or ‘hillock’, showing the continuity of mitochondrial compartments. Nucleoid drawn to scale, see also Figure 2. (D) (Left) Scanning EM of intercellular nanotubes connecting PY79 bacteria [19]. (Center) Transmission EM of a tubular stromule extending from a chloroplast in a mesophyll cell of Arabidopsis thaliana [53]. (Right) Differential interference contrast (DIC) imaging of human HEK293 cells with cell–cell membrane protrusions. Abbreviations: EM, electron microscopy; IMM, inner mitochondrial membrane; mtDNA, mitochondrial DNA; OMM, outer mitochondrial membrane.
Ancestral Connections – Bacterial Membrane
Protrusions Mitochondria retain several structural and functional characteristics of their prokaryotic ancestors. Both harbor a double membrane, have a circular genome, and undergo population-level behavior akin to bacterial ‘quorum sensing’ [16] which can coordinate gene expression between bacteria to give rise to ‘complex’ behaviors [17]. Interestingly, bacteria also exchange molecular information with each other via membrane protrusions or bacterial nanotubes (Figure 1D).
Bacterial nanotubes are thin detergent-sensitive membrane projections that extend from the cell-wall surface and allow the transfer of small molecules and genetic material from one cell to another [18,19]. Protrusions extend from the surface of the donor bacterium within seconds to minutes, reaching lengths up to 1 μm – longer than the donor cell itself (Table 1). Standard electron microscopy (EM) imaging using gold coating indicates that bacterial nanotubes range from 30 to 130 nm in diameter [19]. In non-gold-coated samples, and when measured by transmission cryo-EM, nanotubes are smaller, ranging from 30 to 70 nm in diameter [18], which represents a more accurate estimate.
Table 1.
Summary of Findings from Nanotunnel Research Publications to Datea
| Tissue/cell type | Species | Condition | Diameter | Length | Elongation rate | Method | Refs |
|---|---|---|---|---|---|---|---|
| Kidney cells | African green monkey | In vitro | ~50 nm | <1–30 μm | 260 ± 20 nm/s | Confocal, TEM | [26] |
| Cardiomyocytes | Rat | Ex vivo | 90–120 nm | ~14 μm | N/A | Confocal, TEM | [27] |
| Kidney cells | Rat | In vitro | ~100 nm | ~6 μm | N/A | SIM, SEM, TEM | [31] |
| Skeletal muscle | Human | Biopsy | 62 ± 11 nm | 0.2–2.3 μm | N/A | TEM | [30] |
| Cardiomyocytes | Rat | In vivo, ex vivo | 40–200 nm | 0.7–3.6 μm | N/A | TEM, confocal | [28] |
| Skeletal muscle | Rat | Ex vivo | N/A | N/A | N/A | Confocal | [54] |
| Cardiomyocytes | Rat | Ex vivo | N/A | N/A | N/A | Confocal | [29] |
| Bacteria | B. subtilis | In vitro | ~40–60 nm | >50 μm | ~15–20 nm/s | TIRF-SIM | [18] |
| Bacteria | B. subtilis, S. aureus | In vitro | 30–130 nm | <1 μm | N/A | SEM | [19] |
Abbreviations: N/A, not available; SIM, structured illumination microscopy; TIRF, total internal reflection fluorescence microscopy.
Not unlike mitochondrial dynamics – that are regulated by substrate availability [20,21], bacterial nanotubes are regulated by nutrient availability and intracellular signaling pathways. Depletion of amino acids such as histidine and tryptophan via genetic ablation of key biosynthetic enzymes dramatically induced the growth of nanotubes and bidirectional cytoplasmic exchanges between cells [22]. Conversely, supplementing the growth medium with these amino acids was sufficient to prevent tubulation behavior and molecular exchanges [22]. Nanotubule formation in Bacillus subtilis is regulated by the cAMP-regulating phosphodiesterase enzyme YmdB [18]. Ablation of YmdB reduced nanotube formation by 95%, suggesting that bacterial nanotube formation is driven by cytoplasmic factors, and possibly by environmental cues, via modulation of cAMP signaling [18].
Functionally, bacterial nanotubes allow intercellular transfer of nutrients [22], small cytoplasmic molecules, and large proteins [19]. The evidence suggests that, in contrast to fast mixing of contents following cell fusion, nanotubes exchange molecules such as GFP with slow kinetics. In addition to proteins, small (6.6 Kb) non-conjugative genetic plasmids can also be exchanged, but not chromosomal genes, presumably because they are too large in size [19]. Decreasing bacterial nanotube formation by genetically ablating YmdB led to an ~25-fold reduction in the frequency of antibiotic-resistant colonies [18], underscoring the functional significance of bacteria-to-bacteria molecular exchanges through membrane protrusions.
Tubular structures physically connecting otherwise isolated units are evolutionarily conserved between bacteria and plant chloroplasts (Figure 1D). Among bacteria, cell protrusion-mediated genetic exchange occurs in both Gram-positive and Gram-negative bacteria [19], between evolutionary distinct bacterial species [19], and even in primitive archaebacteria [23]. Mammalian and invertebrate cells also exchange material and perform cell–cell signaling via membrane protrusions [24,25]. Therefore, membrane-based nanotubes likely represent an evolutionarily conserved mechanism for horizontal gene transfer. Given the bacterial origin of mitochondria, and that they have conserved several functional and structural features of their prokaryotic ancestry [16], the existence of tubular mitochondrial membrane protrusions allowing molecular exchanges is not unexpected.
Mitochondrial Nanotunnels
Nanotunnels are double-membrane protrusions that involve both the inner and outer mitochondrial membranes. Nanotunnels have been found to vary between 40 and 200 nm in diameter and between <1 and 30 μm in length, and have been observed in rat cardiomyocytes, human and rat skeletal muscle, and rat and African green monkey kidney cells (Table 1). In addition, timelapse imaging has demonstrated elongation rates of 260 ± 20 nm/sec. Examples of mitochondrial nanotunnels in human skeletal muscle imaged by TEM are presented in Figure 2A,B. When identifying nanotunnels one should consider the diameter, the double-membrane nature and length, and alternative structures such as tubular mitochondria (Figure 2C) and constricted mitochondria (Figure 2D) should not be confused with nanotunnels.
Figure 2. Anatomy of Mitochondrial Nanotunnels.
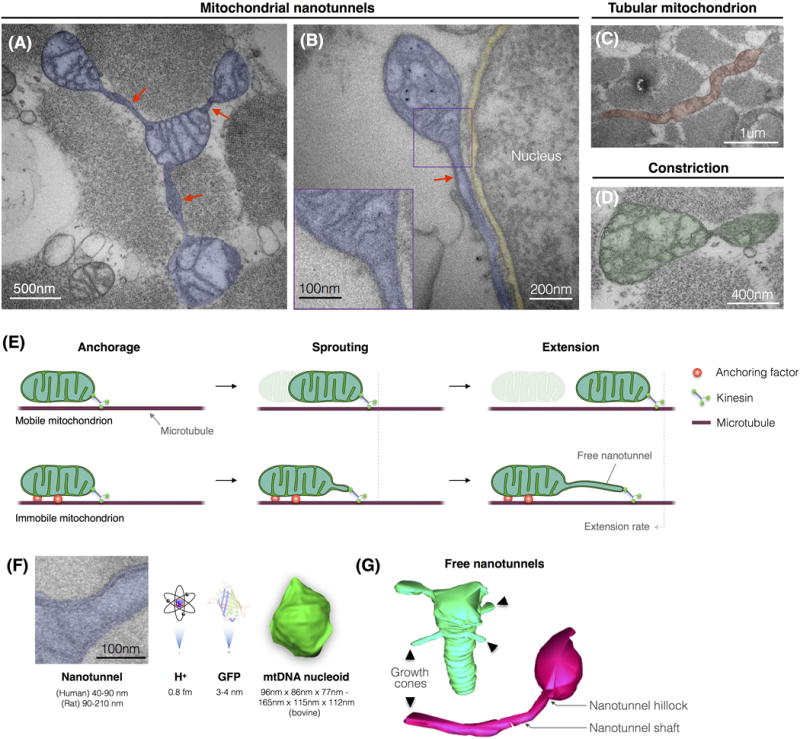
(A) Four mitochondria connected by three nanotunnels (arrows) in human skeletal muscle. (B) A mitochondrion with a nanotunnel running adjacent to the nuclear envelope (yellow) in human skeletal muscle. The high magnification inset shows the nanotunnel double membrane with an internal lumen devoid of cristae. (C) Elongated tubular mitochondrion with variable diameter harboring cristae and a localized mitochondrial constriction with concave membrane curvature consistent with mitochondrial fission. (D) Mitochondria undergoing membrane constriction. Structures in (C,D) are not nanotunnels. (E) Hypothetical model of mitochondrial nanotunnels arising from immobilized mitochondria through the action of motor proteins. (Top) A free mitochondrion pulled by kinesin along a microtubule. (Bottom) A mitochondrion immobilized by anchoring proteins but pulled by the same kinesin protein, resulting in the production of a free nanotunnel. See text for discussion. (F) Human mitochondrial nanotunnel drawn to scale with a proton, GFP, and an mtDNA nucleoid [42]. (G) 3D reconstructions of free mitochondrial nanotunnels in human skeletal muscle showing blunt-end protrusions consistent with an autonomous mode of nanotunnel growth. The nanotunnel growth cones are shown with arrowheads.
At different stages of growth they can be observed to be blunt-ended (Figure 3, step 3A), ‘free nanotunnels’, or to connect two mitochondria (Figure 3, step 4). However, the proportion of nanotunnels that are in the free versus connected state at any one time remains to be determined.
Figure 3. Life Cycle of Mitochondrial Nanotunnels.
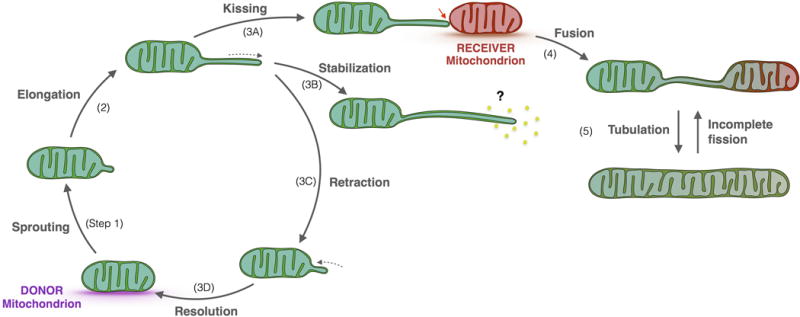
This model proposes that initial nanotunnel sprouting starts with a membrane protrusion from a donor mitochondrion (step 1), subsequently elongating into a free nanotunnel (2). Nanotunnels then either contact a recipient mitochondrion for subsequent fusion (3A), stabilize, and further extend towards a signaling molecule (3B), or retract (3C) and resolve (3D). Fusion of mitochondrial nanotunnels with a recipient mitochondrion (4) leads to connecting nanotunnels, which can expand to accommodate cristae and generate tubular mitochondria (5). Incomplete mitochondrial fission of mitochondrial tubules may generate anatomically similar structures to nanotunnels.
The first report of mitochondrial nanotunnels, almost a decade ago, used EM and confocal imaging of GFP-labeled mitochondria in cultured African green monkey kidney cells [26]. Thin mitochondrial ‘extensions’ with diameters near the diffraction limit of light microscopy were observed to emerge from tubular mitochondria, particularly after the addition of a cysteine alkylating agent (N-ethylmaleimide) that inhibits mitochondrial motility [26]. A subsequent study [27] demonstrated the existence and elongation of mitochondrial tubular structures, coined nanotunnels, in primary cardiomyocytes, and made the observation that matrix-located GFP could be transferred from the donor mitochondrion to receiver mitochondria through nanotunnels. Imaging of adult ventricular cardiomyocytes [28,29], isolated mouse skeletal muscle fibers, and human skeletal muscle biopsies of patients with mitochondrial DNA (mtDNA) disease [30] has since also identified nanotunnels.
Formation and Resolution of Nanotunnels
Live-cell imaging has demonstrated that mitochondrial nanotunnels form and elongate in a kinesin (KIF5B)- and microtubule-dependent manner within seconds [31]. This process can even be recapitulated in a cell-free system by the addition of polymerized microtubules, KIF5B, ATP, and isolated mitochondria. The microtubule-dependent mechanism of nanotunnel growth suggests a model whereby molecular motors pull on the ‘elastic’ membrane of an immobilized mitochondrion [26] (Figure 2E). If this were so, the growth rate of nanotunnels would be lower than the speed at which motor proteins can pull an untethered ‘free’ cargo. Accordingly, in a live-cell model of mitochondrial arrest, nanotunnel growth rate was found to be ~32% slower than the most rapid movement of whole mitochondria [26]. In cultured cells [31], EM tomography in cardiomyocytes showed nanotunnel alignment with microtubules [28], and depolymerization of microtubules with nocodazole also prevents nanotunnel formation [26,31].
Another possibility for the biogenesis of nanotunnels is autonomous growth relying on exclusively endogenous processes. For example, mammalian cells generate membrane protrusions through the coordinated polymerization of endogenous cytoskeletal proteins (microtubules, microfilaments) which push and extend thin stretches of plasma membrane from the inside [24]. This produces nanotubes, filopodia, and other types of communicating membrane protrusions of different lengths. However, given the known protein composition of mitochondria, this process seems unlikely to underlie the generation of nanotunnels.
Regulation of Mitochondrial Nanotunnels
It is intriguing to consider what signaling mechanisms initiate and regulate mitochondrial nanotunnel formation. Evolutionarily related bacterial and mammalian cell-membrane protrusions both have known regulatory mechanisms, such as cAMP-dependent signaling in bacteria [18], but similar regulatory mechanisms have not been identified for mitochondria.
Ca2+ dysregulation, which causes mitochondrial stress when prolonged, may represent an important trigger for nanotunnel formation. In several cell types, and particularly in muscle cells, Ca2+ is a major physiological regulator of mitochondrial oxidative phosphorylation [32], as well as of the mitochondrial calcium uniporter (MCU) that governs mitochondrial calcium dynamics and influences cytoplasmic calcium regulation [33]. Indeed, ryanodine receptor dysfunction causing Ca2+ dysregulation induced a dramatic increase in the number of mitochondrial nanotunnels in cardiomyocytes [28].
The link between Ca2+ dynamics and nanotunnels could be explained by a few non-mutually exclusive processes. One possible explanation is that disruption of Ca2+ dynamics prevents fusion because normal Ca2+ spiking is necessary to maintain normal fusion [29]. Inhibition of fusion would prohibit the molecular exchanges that are necessary for functional complementation between mitochondria. This in turn would either lead to mitochondrial dysfunction or activate a putative sensor for the absence of fusion. The limited evidence available thus far suggests that absence of movement might trigger nanotunnel formation as a compensatory response, possibly to maintain some degree of intermitochondrial exchange. Another possibility is that Ca2+ dysregulation and other abnormal signals within the cell trigger a general mitochondrial stress response. In the absence of mitochondrial motility/fusion that would normally be initiated as an initial compensatory mechanism [34], membrane protrusions may be formed in cell types where mitochondria are immobilized. Finally, we cannot exclude the possibility that a Ca2+-dependent machinery for nanotunnel formation, perhaps analogous to the bacterial phosphodiesterase YmdB [18], might initiate and promote the growth of mitochondrial nanotunnels. In human cells we have also observed a higher abundance of mitochondrial nanotunnels in the presence of mtDNA mutations. More work will be necessary to determine the mechanisms that regulate nanotunnel formation and their involvement in disease.
Nanotunnel Life Cycle
In addition to nanotunnels that continuously elongate to eventually fuse with a recipient mitochondrion, short-lived membrane protrusions also emerge from mitochondria. Visualization of cardiac and skeletal muscle by electron tomography and serial EM reveals that protrusions are often blunt-ended [28,35] (Figure 2G). In vivo monitoring of mitochondrial fusion dynamics in adult cardiomyocytes showed occasional emerging tunneling structures that may remain unconnected (Figure 3, step 3A) or complete linkage between two distant mitochondria (Figure 3, step 4, and Video S1 in the supplemental material online). These could represent actively growing or retracting nanotunnels, or possibly stable nanotunnels undergoing some form of ‘sensing’ (Figure 3, step 3B), similarly to some mammalian cell protrusions [25]. Live-cell imaging of GFP-labeled mitochondria also reveals fusion of thin mitochondrial nanotunnels with a receiver mitochondrion (see Figure I in Box 2).
Figure I. Live-Cell Imaging and 3D EM Imaging of Mitochondrial Nanotunnels.
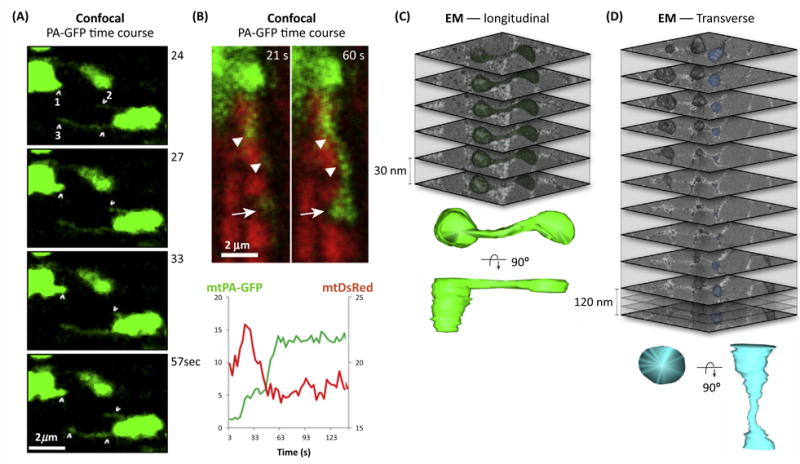
(A) Dynamic mitochondrial nanotunnels in a freshly isolated adult ventricular cardiomyocytes (AVCM) expressing mito-targeted photoactivatable GFP (mtPA-GFP). Timelapse confocal imaging with time since photoconversion. (1) Early protrusion emerging from a globular mitochondrion. (2,3) Thin mitochondrial protrusions, likely representing free mitochondrial nanotunnels, emerging and retracting from a donor mitochondrion. Note that image contrast is enhanced (and mitochondria overexposed) to enable visualization of nanotunnels. (B) mtPA-GFP live-cell timelapse confocal imaging of an AVCM showing the relatively slow exchange kinetics of PA-GFP to a receiver mitochondrion over ~40 s. The bottom plot represents the diffusion kinetics of the receiver mitochondrion: an increase of mtPA-GFP fluorescence and a simultaneous decrease of mtDsRed (mitochondrial matrix targeted Discosoma sp. red fluorescent protein) that replenished the PA-GFP donor organelle which suffered photobleaching upon GFP photoconversion (adapted from [28]). (C) Serial block-face scanning electron microscopy (SBF-SEM; Gatan 3 view) showing pseudocolored mitochondrial nanotunnels running through the image plane in longitudinal and (D) transverse orientations. A 3D surface reconstruction of nanotunnels is shown below. In (D) every fourth image is shown where the actual section thickness is 30 nm.<stream name=“fig_1365_gr1b2” position=“4” desc=“1”/
Box 2. Detecting Nanotunnels by EM and Light Microscopy.
Why have nanotunnels remained elusive and have only recently been described? The answer likely lies in their narrow diameter (<100 nm) and the limitations of microscopy techniques. Light microscopy is largely limited by diffraction, which places the resolution limit at around 200 nm for confocal microscopy. Super-resolution fluorescent light microscopy approaches addresses this difficulty and has allowed sub-diffraction limit (50–100 nm) imaging of fixed mitochondrial structures [59] and of structures within live cells [60]. Each approach is associated with specific limitations and cellular toxicity that should be considered in experimental design, including imaging duration and fluorophore intensity [61]. Examples from confocal imaging of life cardiomyocytes following two-photon photoconversion of PA-GFP demonstrating nanotunnel-mediated mitochondrial content exchange are shown in Figure IA,B.
Because transmission electron microscopy (TEM) and scanning electron microscopy (SEM) only allow single-plane imaging, the likelihood that an ultrathin (~70 nm) section or sample is perfectly orientated to capture such a thin structure along its length is relatively low. If captured in the longitudinal plane, the donor/receiver mitochondria are rarely visualized (Figure IC); if caught in cross-section, nanotunnels appear as small electron-dense vesicle-like structures (Figure ID). 3D imaging by electron tomography [28,35] allows nanotunnel structures to be visualized with an optimal spatial resolution of <1 nm, but has a limited imaging depth of 200–500 nm. Because nanotunnels can be >1 μm and distributed in 3D within the cell, electron tomography cannot be reliably used to quantify and discriminate between free and connecting nanotunnels. Recent EM methods including SBF-SEM and FIB-SEM have lower absolute spatial resolution but allow imaging of substantially larger biological volumes, making it possible to track nanotunnels at EM resolution through the complex cytoarchitectural environment.
Overall, the only approach currently available to validate the presence of mitochondrial nanotunnels is ultrastructure analysis by EM imaging of fixed samples. High-resolution light microscopy may eventually overcome this technical limitation and offer opportunities to precisely probe nanotunnel growth and molecular exchanges. Further developments will be necessary to define the molecular composition of the nanotunnel hillock, shaft, and growth cone.
Mitochondrial Nanotunnels Arise from Immobilized Mitochondria
Mitochondrial nanotunnels have only been observed in immotile mitochondria, and an absence of movement/motility appears to promote the formation of membrane protrusions in different systems. One study reported a 20-fold induction of nanotunnel growth following inhibition of mitochondrial motility in vitro [26]. In tissues, nanotunnels are observed in cell types where mitochondrial motility is prevented by physical constraints, such as in skeletal and cardiac muscle cells that are densely packed with myofibrils [27,28,30]. In skeletal myofibers there are three populations of mitochondria: (i) intermyofibrillar (IMF) mitochondria that are physically constrained by surrounding myofibrils and are tethered by cytoskeletal components at the z-line [36]; (ii) perinuclear mitochondria, adjacent to the nuclei; and (iii) subsarcolemmal (SS) mitochondria which exist as a pool of organelles that are loosely bound only by the plasma membrane and the myofibrillar compartment. The motility of SS and perinuclear mitochondria is not well characterized, but IMF mitochondria are largely immobile. Possibly as a result, nanotunnels are observed in the constrained IMF mitochondria but not in the SS mitochondria; however, they have been observed in perinuclear mitochondria (Figure 2B) [27]. In bacteria, tubular membrane protrusions are also promoted by low motility because tunneling membrane protrusions only form when grown on solid medium [19], consistent with the requirement for organelle immobilization for nanotunnel formation.
Thin Membranous Structures Arise From Stalled Fusion
An alternative to de novo growth of free nanotunnels may be that nanotunnels result from stalled or incomplete fission of an existing mitochondrion [37]. This idea is mainly supported by the similar diameters of mitochondrial nanotunnels and restriction rings caused by dynamin-related protein 1 (Drp1) during fission in yeast [38]. Drp1 can constrict mitochondrial membranes to generate tubules of 60 ± 12 nm (similar to nanotunnels); these can be constricted further to 39 ± 9 nm upon GTP binding but are incapable of completing the fission process alone [39]. The final step of constriction may be performed by dynamin 2 (DNM2) [40]. Upon DNM2 knockdown in cultured cells, tubular structures of ~55 ± 12 nm in diameter have been observed to form [40]. Likewise, in mouse brain exposed to hypoxia, long tubular nanotunnel-like connections between strings of mitochondria have been observed, possibly as a result of arrested or incomplete fission, although there is no direct evidence [37]. Thus, based on static EM images, it is not possible to discount the possibility that at least some nanotunnels linking two mitochondria result from incomplete fission.
Both Failed Fission and De Novo Synthesis Produce Nanotunnels
Free nanotunnels that emerge from single mitochondria cannot be explained by this mechanism. In human skeletal muscle, 3D reconstruction of mitochondrial networks reveals several mitochondria with nanotunnel protrusions that are blunt-ended (Figure 2F), similar to the blunt-ended bacterial protrusions [19]. In the case of bacteria, tubular extensions arising from a single bacterium also cannot be the result of failed or incomplete constriction, and must therefore represent de novo protrusions. This same conclusion is consistent with live-cell imaging showing extension of nanotunnels from existing organelles and subsequent fusion with distant organelles [26,28,29,31], as shown in Video S1.
Both de novo mitochondrial nanotunnels that grow from single organelles, and constricted mitochondrial tubules that result from failed fission, may coexist in various cell types. It may not be possible to distinguish between these etiologies by EM. However, based on limited evidence, it is possible that failed fission yields organelles with a relatively minimal membrane curvature and that are shorter in length. By contrast, mitochondrial nanotunnels often exhibit more pronounced curvature at the nanotunnel hillock, and generally extend over considerably longer distances that can exceed 2 μm (Table 1). Blunt-end mitochondrial membrane protrusions defined by 3D EM imaging are therefore unlikely to result from failed fission, and the most logical mechanism for mitochondrial nanotunnel biogenesis is growth from a donor organelle.
Nanotunnels Allow Molecular Exchange
Dynamic distribution of fluorescent proteins targeted to the mitochondrial matrix (or other mitochondrial compartments) among individual mitochondria has provided clues to molecular transfer mediated by nanotunnels. Fluorescence and confocal imaging do not have sufficient resolution to identify structures of the size of nanotunnels, but the distinctive fluorescence distribution patterns in cardiomyocytes that are uniquely rich in nanotunnels are instructive. First, fusion-mediated exchange of soluble matrix contents often appears between mitochondria separated by ≥1 μm, together with the emergence of narrow connectors, and occurs with slower kinetics than in any previously characterized paradigm of full fusion [28,29]. The slower diffusion kinetics through nanotunnels has been ascribed to the narrow diameter of nanotunnel lumen (matrix). Furthermore, a ryanodine receptor 2 mutation that is associated with a substantial increase in nanotunnels in cardiac muscle, as validated by EM, also increases the fraction of slow content-mixing events, indicating that nanotunnels mediate slow matrix exchange between mitochondria [28]. The slow kinetics of mitochondrial fusion also displays a clear stepwise pattern in many cases, suggesting intermittent fusion-pore formation between the nanotunnel growth cone and the receiver mitochondrion [29] (Figure 3, oscillations between steps 3A and 4). Thus, the initiation of a diffusion event must involve some stochastic transition in the relationship between the two interacting mitochondria.
It is possible that an exchange is initiated by an actual fusion event which permits direct mixing of matrices (fusion hypothesis). An alternative is that the membranes at kissing junctions become permissive to direct movements of proteins from one organelle to the other (gating hypothesis). This might be analogous to the fusion of neurotransmitter-filled vesicles with the plasma membrane that occurs through partial opening of a fusion pore as a form of exocytosis [41]. In mammalian cells this behavior leads to so-called ‘kiss-and-run’ between the two membranes which remain connected via a nanotube and open up to a larger pore, and then reclose to a nanotube [41]. Both hypotheses have some intrinsic weaknesses. In the case of fusion, the problem is that matrix exchange is relatively slow, even considering the possible negative effect of matrix space complexity. In the case of kissing junctions, the mechanism that coordinates the opening of pores in the outer and inner mitochondrial membranes remains unknown.
Live-cell imaging demonstrates the ability of nanotunnels not only to transfer matrix-targeted GFP from one mitochondrion to another via a transient nanotunnel connection [27]. Given this and the range of nanotunnel diameters measured, it would appear possible that small proteins, RNA, and free mtDNA could be transported in this manner, although nucleoid-packaged mtDNA may be too large (Figure 2F). However, data from cardiomyocytes indicate that ‘nanotunnels’ can be up to 200 nm in diameter, which would be large enough for a nucleoid to be transported. Nevertheless, these larger nanotunnels tend to contain cristae, which are not present in thinner nanotunnels. It is therefore likely be that these larger nanotunnel-like structures are nanotunnels in the process of expanding to form cristae-bearing tubular mitochondria (i.e., a tubulation process), as proposed by Wang et al. [31] (Figure 3, step 5). Moreover, mammalian mtDNA is tightly associated in a protein complex as a nucleoid that is significantly larger in size and presumably less malleable than individual DNA molecules [42]. Therefore, whether nucleoid-bound mtDNA can be transported remains to be determined.
Concluding Remarks
The view of mitochondria as individual powerhouses is expired. Mitochondria are dynamic living organelles that move, fuse, and divide in response to biochemical cues. They also generate signals that influence a wide spectrum of cellular and physiological functions. The discovery that mitochondria grow membrane protrusions to engage in private and selective communication with other mitochondria under conditions of stress raises a new set of questions about their behavior (see Outstanding Questions). We especially need to understand the molecular drivers for nanotunnel formation, the selectivity for donor and receiver mitochondria, and the physiological significance of nanotunnels for the cell and the organism as a whole.
Outstanding Questions.
The existence of mitochondrial nanotunnels in cells and human tissues highlights their potential relevance to mitochondrial pathophysiology. However, several questions remain.
What are the molecular mechanisms supporting mitochondrial membranes curvature and extension that initiate and promote the extension of nanotunnels? Do intrinsic processes within mitochondria cooperate with cytoskeletal and motor proteins to guide nanotunnel initiation and elongation?
In the same way that the ER marks sites of mitochondrial division, is the ER involved in determining the initiation or elongation of mitochondrial nanotunnels?
What signals precede the formation of nanotunnels and determine the receiver mitochondrion? Is there an ‘SOS’ stress signal that is released from the receiver mitochondrion?
Can nanotunnels transport genetic material? If mtDNA nucleoids are selectively excluded based on their size, could the sharing of gene products and membrane potential provide an alternative mechanism for functional complementation between dysfunctional mitochondria?
Is there a selective filter that regulates molecular exchanges along nanotunnels and the rate at which this exchange occurs?
Do tubular structures formed during failed or stalled fission play similar roles as nanotunnels? What proportion of nanotunnels in tissues arise from de novo nanotunnel biogenesis compared to failed fission events?
Is the formation of nanotunnels dependent on mitochondria-to-microtubule interaction in muscle cells?
There is evidence for transport of matrix and outer mitochondrial membrane proteins, but can inner mitochondrial membrane proteins also be transported? Could functional complementation occur through nanotunnel-mediated transfer of respiratory chain subunits?
Tubular connections between whole organisms, cells, and organelles are ubiquitous in biology. Tubular connections are conserved across numerous branches of the evolutionary tree – from unicellular organisms such as bacteria to complex multicellular mammalian organisms. Such fractal-like or scale-free properties are common in biology [43,44]. These epistemological observations say little about the specific function of mitochondrial nanotunnels, the mechanisms regulating their behavior, or their relevance to disease, but underscore their widespread biological significance.
Examining the function of tubular membrane protrusions at the cellular level may provide insight into the functional significance of nanotunnels. Using tunneling nanotubes (TNTs), neurons transfer dysfunctional mitochondria to astrocytes, possibly to ‘outsource’ mitophagy and quality control [45]. Alternatively, cell-to-cell membrane protrusions enable coordination of cytoplasmic signals between cells and cellular rescue via the transfer of organelles such as mitochondria [46] and lysosomes [47]. Although the transfer of dysfunctional components is less intuitive in mitochondrial nanotunnels, selective quality-control mechanisms in the form of mitochondria-derived vesicles (MDVs) have been identified [48]. Nanotunnels could provide a means of functional complementation similar to that achieved by mitochondrial fusion [14,49]. This may be even more likely in cells with highly organized cytoarchitectures, such as skeletal and cardiac muscle cells, where mitochondrial movement is restricted, limiting the opportunities for mitochondria to encounter potential fusion partners.
Some mammalian cells also use cellular protrusions to ‘screen’ and sense the environment, for example in the stem cell niche [25]. Similarly, amino acid starvation can promote the formation of thin nanotubes in bacteria, leading to metabolic sharing between connected cells, which in turn directs colony growth, consistent with a role in environmental sensing [22]. Mitochondrial nanotunnels that do not result in fusion with a recipient mitochondrion could possibly serve a similar function.
Discovering how different parts of a system communicate with one another can lead to insights into the function and regulation of the system as a whole. The field of neuroscience is a good example, and our understanding of brain function has been transformed by mapping the mechanisms that enable and regulate communication between neurons [50]. Likewise, resolving outstanding questions about mitochondrial communication generally, and about mitochondrial nanotunnels more specifically, should bring us closer to understanding the factors that orchestrate the complex network behavior of mitochondria. This should in turn enlighten us regarding potential new roles for mitochondria in regulating cellular stress responses that define health and disease states.
Supplementary Material
Trends.
Nanotunnels are communicating double-membrane tubular protrusions 40–200 nm in diameter, and up to 30 μm in length, that emerge primarily from the surface of immobilized mitochondria or from mitochondria in tissues with restricted mitochondrial motility.
Nanotunnels transport matrix and membrane proteins between mitochondria, and probably also transport smaller molecules such as ions, RNA, and metabolites.
In a cell-free system, microtubules, mitochondria, ATP, and kinesin 5b are sufficient to produce mitochondrial protrusions, whereas disruption of microtubules hinders nanotunnel formation, implicating a motor-driven microtubule-dependent mechanism of nanotunnel formation.
Disruption of calcium dynamics in muscle cells and genetic mitochondrial defects are associated with greater abundance of mitochondrial nanotunnels, suggesting that nanotunnels arise as a compensatory mechanism to promote mitochondrial communication in stress conditions.
Acknowledgments
A.E.V. and D.M.T. are supported by the Wellcome Centre for Mitochondrial Research (203105), Newcastle University Centre for Ageing and Vitality (supported by the Biotechnology and Biological Sciences Research Council and the Medical Research Council), and a UK National Institute for Health Research (NIHR) Biomedical Research Centre for Ageing and Age-related disease award to the Newcastle upon Tyne Foundation Hospitals National Health Service (NHS) Trust. V.E. is supported by a FONDECYT grant (1150677). G.H. is supported by National Institutes of Health(NIH) grant DK051526. M. P. is supported by NIH grant GM119793 and the Wharton Fund.
Glossary
- Donor mitochondrion
the mitochondrion from which the nanotunnel originates
- Mitochondrial content exchange
diffusion of molecular content (proteins, nucleic acids, ions, and other small molecules) from the donor to the receiver mitochondria, over periods ranging from seconds to minutes [28]
- Mitochondrial nanotunnel
a thin double-membrane protrusion of the mitochondrial outer and inner membranes containing matrix, and capable of transporting proteins. Nanotunnels can be found as either ‘free nanotunnel’ with a blunt end, or as a ‘connecting nanotunnel’ fused on both ends with mitochondria
- Nanotunnel hillock
a conical-shaped connecting segment with high membrane curvature and continuous matrix between the donor mitochondrion and the nanotunnel shaft
- Nanotunnel growth cone
the tip of the outer and inner mitochondrial membranes protrusion as it extends from the donor mitochondrion
- Nucleoid
the packaged form of the mitochondrial DNA (mtDNA) and associated proteins localized in the mitochondrial matrix [51], which typically contain 1–2 copies of mtDNA [42]
- Receiver mitochondrion
the mitochondrion with which a free mitochondrial nanotunnel fuses, forming a connecting nanotunnel with the donor mitochondrion
- Serial block-face scanning electron microscopy (SBF-SEM)
a technique which enables 3D automated imaging at sub-micron resolution of large sample volumes [52]. Similar results are obtained with focused ion-beam SEM (FIB-SEM)
Footnotes
Supplemental Information
Supplemental information associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.tcb.2017.08.009.
References
- 1.Lerner TN, et al. Communication in neural circuits: tools, opportunities, and challenges. Cell. 2016;164:1136–1150. doi: 10.1016/j.cell.2016.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Phillips MJ, Voeltz GK. Structure and function of ER membrane contact sites with other organelles. Nat Rev Mol Cell Biol. 2016;17:69–82. doi: 10.1038/nrm.2015.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gottschling DE, Nystrom T. The upsides and downsides of organelle interconnectivity. Cell. 2017;169:24–34. doi: 10.1016/j.cell.2017.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Booth DM, et al. Redox nanodomains are induced by and control calcium signaling at the ER–mitochondrial interface. Mol Cell. 2016;63:240–248. doi: 10.1016/j.molcel.2016.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pacher P, Hajnóczky G. Propagation of the apoptotic signal by mitochondrial waves. EMBO J. 2001;20:4107–4121. doi: 10.1093/emboj/20.15.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zorov DB, et al. Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol Rev. 2014;94:909–950. doi: 10.1152/physrev.00026.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Picard M, et al. Progressive increase in mtDNA 3243A>G heteroplasmy causes abrupt transcriptional reprogramming. Proc Natl Acad Sci U S A. 2014;111:E4033–E4042. doi: 10.1073/pnas.1414028111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chandel NS. Evolution of mitochondria as signaling organelles. Cell Metab. 2015;22:204–206. doi: 10.1016/j.cmet.2015.05.013. [DOI] [PubMed] [Google Scholar]
- 9.Klecker T, et al. Making connections: interorganelle contacts orchestrate mitochondrial behavior. Trends Cell Biol. 2014;24:537–545. doi: 10.1016/j.tcb.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 10.Quiros PM, et al. Mitonuclear communication in homeostasis and stress. Nat Rev Mol Cell Biol. 2016;17:213–226. doi: 10.1038/nrm.2016.23. [DOI] [PubMed] [Google Scholar]
- 11.Latorre-Pellicer A, et al. Mitochondrial and nuclear DNA matching shapes metabolism and healthy ageing. Nature. 2016;535:561–565. doi: 10.1038/nature18618. [DOI] [PubMed] [Google Scholar]
- 12.Picard M, McManus MJ, et al. Mitochondrial signaling and neurodegeneration. In: Reeve AK, editor. Mitochondrial Dysfunction in Neurodegenerative Disorders. Springer International; 2016. pp. 107–137. [Google Scholar]
- 13.Friedman JR, Nunnari J. Mitochondrial form and function. Nature. 2014;505:335–343. doi: 10.1038/nature12985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang L, et al. Mitochondrial fusion provides an ‘initial metabolic complementation’ controlled by mtDNA. Cell Mol Life Sci. 2015;72:2585–2598. doi: 10.1007/s00018-015-1863-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Archer SL. Mitochondrial dynamics – mitochondrial fission and fusion in human diseases. N Eng J Med. 2013;369:2236–2251. doi: 10.1056/NEJMra1215233. [DOI] [PubMed] [Google Scholar]
- 16.Picard M, Burelle Y. Mitochondria: starving to reach quorum? Insight into the physiological purpose of mitochondrial fusion. Bioessays. 2012;34:272–274. doi: 10.1002/bies.201100179. [DOI] [PubMed] [Google Scholar]
- 17.Goo E, et al. Control of bacterial metabolism by quorum sensing. Trends Microbiol. 2015;23:567–576. doi: 10.1016/j.tim.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 18.Dubey GP, et al. Architecture and characteristics of bacterial nanotubes. Dev Cell. 2016;36:453–461. doi: 10.1016/j.devcel.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 19.Dubey GP, Ben-Yehuda S. Intercellular nanotubes mediate bacterial communication. Cell. 2011;144:590–600. doi: 10.1016/j.cell.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 20.Gomes LC, et al. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat Cell Biol. 2011;13:589–598. doi: 10.1038/ncb2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rambold AS, et al. Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proc Natl Acad Sci. 2011;108:10190–10195. doi: 10.1073/pnas.1107402108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pande S, et al. Metabolic cross-feeding via intercellular nanotubes among bacteria. Nat Commun. 2015;6:6238. doi: 10.1038/ncomms7238. [DOI] [PubMed] [Google Scholar]
- 23.Rosenshine I, et al. The mechanism of DNA transfer in the mating system of an archaebacterium. Science. 1989;245:1387–1389. doi: 10.1126/science.2818746. [DOI] [PubMed] [Google Scholar]
- 24.Inaba M, et al. Nanotubes mediate niche-stem-cell signalling in the Drosophila testis. Nature. 2015;523:329–332. doi: 10.1038/nature14602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buszczak M, et al. Signaling by cellular protrusions: keeping the conversation private. Trends Cell Biol. 2016;26:526–534. doi: 10.1016/j.tcb.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bowes T, Gupta RS. Novel mitochondrial extensions provide evidence for a link between microtubule-directed movement and mitochondrial fission. Biochem Biophys Res Commun. 2008;376:40–45. doi: 10.1016/j.bbrc.2008.08.120. [DOI] [PubMed] [Google Scholar]
- 27.Huang X, et al. Kissing and nanotunneling mediate inter-mitochondrial communication in the heart. Proc Natl Acad Sci U S A. 2013;110:2846–2851. doi: 10.1073/pnas.1300741110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lavorato M, et al. Increased mitochondrial nanotunneling activity, induced by calcium imbalance, affects intermitochondrial matrix exchanges. PNAS. 2017;114:E849–E858. doi: 10.1073/pnas.1617788113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eisner V, et al. Mitochondrial fusion dynamics is robust in the heart and depends on calcium oscillations and contractile activity. Proc Natl Acad Sci. 2017;114:E859–E868. doi: 10.1073/pnas.1617288114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vincent AE, et al. The spectrum of mitochondrial ultrastructural defects in mitochondrial myopathy. Sci Rep. 2016;6:30610. doi: 10.1038/srep30610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang C, et al. Dynamic tubulation of mitochondria drives mitochondrial network formation. Cell Res. 2015;25:1108–1120. doi: 10.1038/cr.2015.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Glancy B, Balaban RS. Role of mitochondrial Ca2+ in the regulation of cellular energetics. Biochemistry. 2012;51:2959–2973. doi: 10.1021/bi2018909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paillard M, et al. Tissue-specific mitochondrial decoding of cytoplasmic Ca2+ signals is controlled by the stoichiometry of MICU1/2 and MCU. Cell Rep. 2017;18:2291–2300. doi: 10.1016/j.celrep.2017.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shutt TE, McBride HM. Staying cool in difficult times: mitochondrial dynamics, quality control and the stress response. Biochim Biophys Acta. 2013;1833:417–424. doi: 10.1016/j.bbamcr.2012.05.024. [DOI] [PubMed] [Google Scholar]
- 35.Lavorato M, et al. Electron tomography of mitochondrial nanotunnels in a CPVT model with RyR2 loss-of-function mutation. Biophys J. 2016;110:367a. [Google Scholar]
- 36.Milner DJ, et al. Desmin cytoskeleton linked to muscle mitochondrial distribution and respiratory function. J Cell Biol. 2000;150:1283–1298. doi: 10.1083/jcb.150.6.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang L, et al. Altered brain energetics induces mitochondrial fission arrest in Alzheimer’s Disease. Sci Rep. 2016;6:18725. doi: 10.1038/srep18725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mears JA, et al. Conformational changes in Dnm1 support a contractile mechanism for mitochondrial fission. Nat Struct Mol Biol. 2011;18:20–26. doi: 10.1038/nsmb.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Francy CA, et al. The mechanoenzymatic core of dynamin-related protein 1 comprises the minimal machinery required for membrane constriction. J Biol Chem. 2015;290:11692–11703. doi: 10.1074/jbc.M114.610881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee JE, et al. Multiple dynamin family members collaborate to drive mitochondrial division. Nature. 2016;540:139–143. doi: 10.1038/nature20555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mellander LJ, et al. Two modes of exocytosis in an artificial cell. Sci Rep. 2014;4:3847. doi: 10.1038/srep03847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kukat C, et al. Cross-strand binding of TFAM to a single mtDNA molecule forms the mitochondrial nucleoid. Proc Natl Acad Sci U S A. 2015;112:11288–11293. doi: 10.1073/pnas.1512131112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aon MA, et al. The scale-free dynamics of eukaryotic cells. PLoS One. 2008;3:e3624. doi: 10.1371/journal.pone.0003624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barabasi AL, Oltvai ZN. Network biology: understanding the cell’s functional organization. Nat Rev Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 45.Davis CHO, et al. Transcellular degradation of axonal mitochondria. Proc Natl Acad Sci. 2014;111:9633–9638. doi: 10.1073/pnas.1404651111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Torralba D, et al. Mitochondria know no boundaries: mechanisms and functions of intercellular mitochondrial transfer. Front Cell Dev Biol. 2016;4:107. doi: 10.3389/fcell.2016.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yasuda K, et al. Tunneling nanotubes mediate rescue of prematurely senescent endothelial cells by endothelial progenitors: exchange of lysosomal pool. Aging (Milano) 2011;3:597–608. doi: 10.18632/aging.100341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sugiura A, et al. A new pathway for mitochondrial quality control: mitochondrial-derived vesicles. EMBO J. 2014;33:2142–2156. doi: 10.15252/embj.201488104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen H, et al. Mitochondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell. 2010;141:280–289. doi: 10.1016/j.cell.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lerner TN, et al. Communication in neural circuits: tools, opportunities, and challenges. Cell. 2016;164:1136–1150. doi: 10.1016/j.cell.2016.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gilkerson R, et al. The mitochondrial nucleoid: integrating mitochondrial DNA into cellular homeostasis. Cold Spring Harb Perspect Biol. 2013;5:a011080. doi: 10.1101/cshperspect.a011080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Denk W, Horstmann H. Serial block-face scanning electron microscopy to reconstruct three-dimensional tissue nanostructure. PLoS Biol. 2004;2:e329. doi: 10.1371/journal.pbio.0020329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Holzinger A, et al. Effects of arc3, arc5 and arc6 mutations on plastid morphology and stromule formation in green and nongreen tissues of Arabidopsis thaliana. Photochem Photobiol. 2008;84:1324–1335. doi: 10.1111/j.1751-1097.2008.00437.x. [DOI] [PubMed] [Google Scholar]
- 54.Eisner V, et al. Mitochondrial fusion is frequent in skeletal muscle and supports excitation-contraction coupling. J Cell Biol. 2014;205:179–195. doi: 10.1083/jcb.201312066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Picard M, et al. Trans-mitochondrial coordination of cristae at regulated membrane junctions. Nat Commun. 2015;6:6259. doi: 10.1038/ncomms7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Twig G, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu X, et al. Mitochondrial ‘kiss-and-run’: interplay between mitochondrial motility and fusion-fission dynamics. EMBO J. 2009;28:3074–3089. doi: 10.1038/emboj.2009.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Glancy B, et al. Power grid protection of the muscle mitochondrial reticulum. Cell Rep. 2017;19:487–496. doi: 10.1016/j.celrep.2017.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jans DC, et al. STED super-resolution microscopy reveals an array of MINOS clusters along human mitochondria. Proc Natl Acad Sci U S A. 2013;110:8936–8941. doi: 10.1073/pnas.1301820110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shelden EA, et al. Focusing super resolution on the cytoskeleton. F1000Research. 2016;5:998. doi: 10.12688/f1000research.8233.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Godin AG, et al. Super-resolution microscopy approaches for live cell imaging. Biophys J. 2014;107:1777–1784. doi: 10.1016/j.bpj.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


