Abstract
Objective(s):
Antibiotic resistance in Acinetobacter baumannii and outbreaks caused by this organism have been reported from several areas of the world. The present study aimed at determining the antibiotic susceptibility profiles and the distribution of OXA-type beta-lactamases among Iranian Acinetobacter baumannii isolates from Qom of Iran.
Materials and Methods:
For this study, 108 non-duplicate A. baumannii isolates were obtained from clinical specimens in four teaching hospitals in Qom in the central of Iran. The antimicrobial susceptibility of isolates was tested by standard disk diffusion and prevalence of bla OXA genes was investigated by PCR method.
Results:
Among 97 carbapenem non-susceptible isolates of A. baumannii, 90.72% (88 isolates) isolates showed extensive drug resistance to multiple antibiotics. Among carbapenem resistant isolates, 100% carried blaOXA-51-like, 82.47% carried blaOXA-23-like, 55.67% carried blaOXA-58-like, 22.68% carried blaOXA-40-like and 14.43% had blaOXA-143-like resistance genes.
Conclusion:
This study demonstrated high genetic diversity of OXA genes among isolates of A. baumannii in Qom, Iran.
Keywords: Acinetobacter baumanniiblaOXA-143, Carbapenem resistance, Iran, Polymerase chain-reaction
Introduction
Acinetobacter baumannii is a glucose non-fermentative, Gram-negative bacillus classified as a significant opportunistic pathogen and is usually involved in infectious outbreaks originating in intensive care units (1). The infections caused by A. baumannii include bacteremia, ventilator-associated pneumonia, meningitis, urinary tract and wound infections (2). The number of multidrug resistant (MDR) strains of A. baumannii has increased in recent years and therefore treatment of this organism is complicated (3). The terms of pan drug resistance (PDR), extensive drug resistance (XDR) and multidrug resistance (MDR) were explained as, resistance of an isolate to all antibiotics in all antimicrobial categories, resistance to at least one antibiotic in all expect one or two antimicrobial categories and resistance to at least one antibiotic in ≥3 antimicrobial categories, respectively.
The antimicrobial categories were included aminoglycosides, carbapenems, cephalosporins, fluoroq- uinolones, penicillins, monobactams and polymyxins (4).
Carbapenems are considered a last-line agent for the treatment of important Gram-negative bacilli, but resistance to these compounds in A. baumannii has been increased worldwide within the past decade (5, 6). Several mechanisms involved in resistance to carbapenem antibiotics in A. baumannii among them OXA-type beta-lactamases are the most widespread mechanism (7). At the present five main groups of OXA-type beta-lactamases have been identified in A. baumannii: blaOXA-23-like; blaOXA-40-like; blaOXA-51-like; blaOXA-58-like and blaOXA-143-like (8, 9). The OXA-type beta-lactamases contain between 243 and 260 amino acids residues, with a molecular mass ranging from 23 to 35.5 kDa (10). The first report of OXA-type beta-lactamases in A. baumannii was from Scotland in 1985 that named ARI-1 (Acinetobacter Resistant to Imipenem) (11) and later designated as OXA-23 (12). Nowadays, this gene has been discovered from many countries of the world (13). The second group of OXA-type beta-lactamases is blaOXA-40-like, originally called OXA-24, which was initially found in A. baumannii from Spain (14).
This gene encoded chromosomally or by plasmid and has 60% identity with OXA-23 (1). The largest group of OXA-type beta-lactamases is the blaOXA-51-like, which corresponds to chromosomal encoded enzymes and therefore naturally occurs in A. baumannii (15). The fourth group is blaOXA-58-like that was identified in France (16). These enzymes often located on plasmids, which may explain their wide distribution (1). Recently, Higgins et al. reported a new OXA-type beta-lactamase, the OXA-143, which isolated from a carbapenem resistant A. baumannii in Brazil. This gene has been identified in plasmid and so far was not reported from other countries of the world (8). The purpose of this study was to access the antibiotic susceptibility profile and the rate of MDR, XDR and PDR A. baumannii isolated from different hospitals in Qom in center of Iran and to investigate the distribution of OXA-type beta-lactamases among A. baumannii.
Materials and Methods
Bacterial isolation and identification
A total of l08 non-duplicate isolates of A. baumannii were collected from clinical specimens of hospitalized patients in (four teaching hospitals in Qom, Iran) Shahid-beheshti hospital (a 530-bed referral hospital with three 12-bed ICUs), Nekoei Hospital (a 170-bed referral hospital), Kamkar Hospital (a 150-bed referral hospital) and Valiasr Hospital (a 158-bed referral hospital) between November 2012 and October 2013. The isolates were obtained from clinical specimens, including tracheal aspirate, urine, blood, wounds and cerebrospinal fluid. The identification of the isolates was performed using standard microbiological and biochemical tests such as Gram stain, colony morphology, glucose oxidation, citrate utilization, oxidase tests and growth at 44 °C (17). In addition, identification of A. baumannii was confirmed using blaOXA-51 PCR (18).
Antibiotic susceptibility testing
Susceptibility to a panel of 18 antimicrobial agents was determined by Kirby-Bauer disc diffusion according to Clinical and Laboratory Standard Institution (CLSI) guideline (19). Antibiotics used were ceftriaxon (30 µg), ceftazidime (30 µg), gentamicin (10 µg), amikacin (30 µg), ciprofloxacin (5 µg), piperacillin (100 µg), piperacillin–tazobactam (100/10 µg), imipenem (10 µg), meropenem (10 µg), aztreonam (30 µg), cefepime (30 µg), levofloxacin (5 µg), ampicillin-sulbactam (10/10 µg), trimethoprime-sulfamethoxazole (25 µg), ticarcillin-clavulanic acid (75/10 µg), tobramycin (10 µg), colistin (10 µg), polymyxin B (30 unit) (MAST, Merseyside, United Kingdom). Escherichia coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 were used as controls. Intermediate sensitivity was considered as resistance.
DNA extraction and PCR assay
All A. baumannii isolates were grown for 24 hr at 37 °C in MacConkey agar then pure colonies were isolated and cultured in the LB broth medium and DNA was extracted by boiling method (20). Detection of blaOXA-23-like, blaOXA-40-like, blaOXA-51-like, blaOXA-58-like and blaOXA-143-like in clinical isolates of A. baumannii was carried out by PCR with specific primers (8, 21-23). PCR amplification conditions were as follows: Initial denaturation at 94 °C for 4 min, followed by 35 cycles of denaturation for 1 min at 94 °C, annealing at specific temperature for each gene for 1 min, and extension at 72 °C for 1 min with a final extension at 72 °C for 7 min. PCR products were analyzed by gel electrophoresis in 1.2 % agarose gel at 85 V. A. baumannii reference strain NCTC 12516, NCTC 13305, NCTC 13304 and NCTC 13302 were used as positive control for the blaOXA-51-like, blaOXA-58-like, blaOXA-23-like and blaOXA-40-like respectively. PCR product of blaOXA-143-like gene was extracted from the gel, purified and sequenced using an automated sequencer. DNA sequence was compared to the national center for biotechnology information (NCBI) database.
Results
A total of 108 A. baumannii isolates were recovered from clinical specimens collected in four teaching hospitals in Qom, Iran. Clinical samples were tracheal aspirate (81 isolates, 75%), urine (17 isolates, 15.74%), blood (7 isolates, 6.48%), wounds (2 isolates, 1.85 %) and cerebrospinal fluid (1 isolate, 0.93%).
The age ranges of the patients were from 20 to 90 years old with a median of 52 years. Seventy-five (69.44%) patients were male and thirty-three (30.56%) were female. A. baumannii isolates were collected from different hospital wards as follows; ICU wards (92 isolates, 85.19%), burn wards (2 isolates, 1.85%), trauma and emergency wards (7 isolates, 6.48%), general surgery wards (2 isolates, 1.85%) and internal wards (5 isolates, 4.63%). Analysis for presence of blaOXA-51-like gene with PCR method showed that all isolates were positive for this gene and confirmed them as A. baumannii. A. baumannii isolates showed 100% resistance to aztreonam and ticarcillin-clavulanic. In addition, 41.6% and 12% of the isolates were resistant to colistin and polymixin B, respectively (Table 1). In addition, among 108 A. baumannii isolates screened 12 isolates (11.11%) were multidrug resistant (MDR), 89 isolates (82.4%) were extensive drug resistant (XDR) and 6 isolates (5.55%) were pandrug resistant (PDR). Of the total 108 A. baumannii collected in this study, 97 isolates (89.81%) were found non-susceptible to imipenem and meropenem.
Table 1.
Antibiotic resistance of Acinetobacter baumannii isolates
| Antimicribial agents | Resistance (%) |
|---|---|
| Ceftriaxone | 97.22 |
| Ceftazidime | 94.44 |
| Gentamicin | 81.48 |
| Amikacin | 93.51 |
| Ciprofloxacin | 93.51 |
| Piperacillin | 97.22 |
| Piperacillin-Tazobactam | 94.44 |
| Imipenem | 89.81 |
| Meropenem | 89.81 |
| Colistin | 41.66 |
| Polymyxin B | 12.03 |
| Azotreonam | 100 |
| Cefepime | 93.51 |
| Levofloxacin | 91.66 |
| Tobramycin | 47.22 |
| Ampicillin-Sulbactam | 92.59 |
| Ticarcillin-Clavulanic acid | 100 |
| Cotrimoxazole | 95.37 |
The results of PCR assays for 97 carbapenem non-susceptible isolates of A. baumannii showed that 80 isolates (82.47%) carried blaOXA-23-like, 54 isolates (55.67%) carried blaOXA-58-like, 22 isolates (22.68%) carried blaOXA-40-like and 14 isolates (14.43%) had blaOXA-143-like genes (Figure 1). The sequencing and alignment results for PCR product of blaOXA-143-like gene was showed that sequence of this gene was 100% similar to sequences of blaOXA-143-like genes recorded in the GenBank and registered in GenBank with accession number KX349208. Table 2 shows the genotypic profiles of OXA-type beta-lactamase genes in carbapenem resistant A. baumannii isolates.
Figure 1.
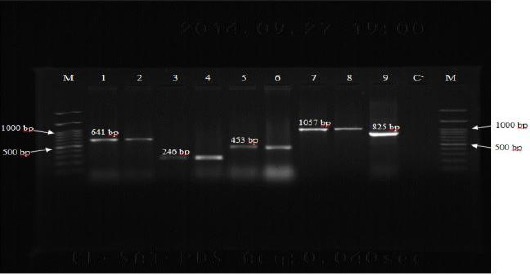
PCR amplification for the detection of OXA-type beta-lactamase genes among Acinetobacter baumannii isolates. Lane M: DNA size marker (100 bp plus), Lane 1: OXA-51 positive control (A. baumannii NCTC12516), Lane 2: OXA-51 (641 bp) positive isolate, Lane 3: OXA-40 positive control (A. baumannii NCTC 13302), Lane 4: OXA-40 (246 bp) positive isolate, Lane 5: OXA-58 positive control (A. baumannii NCTC 13305), Lane 6: OXA-58 (453 bp) positive isolate, Lane 7: OXA-23 positive control (A. baumannii NCTC 13304), Lane 8: OXA-23 (1057 bp) positive isolate, Lane 9: OXA-143 (825 bp) positive isolate, Lane C-: negative control
Table 2.
Genotypic profile of Acinetobacter baumannii isolates according to OXA-type beta-lactamase genes in carbapenem resistant
| Resistance determinants | Cases (no.) | Percent |
|---|---|---|
| blaOXA-51-like only | 10 | 10.30 |
| blaOXA-51-like+blaOXA-23-like | 24 | 24.74 |
| blaOXA-51-like +blaOXA-58-like | 6 | 6.18 |
| blaOXA-51-like+blaOXA-23-like+blaOXA-40-like | 6 | 6.18 |
| blaOXA-51-like+blaOXA-23-like+blaOXA-58-like | 27 | 27.83 |
| blaOXA-51-like+blaOXA-23-like+blaOXA-143-like | 2 | 2.06 |
| blaOXA-51-like+blaOXA-23-like+blaOXA-40-like+blaOXA-143-like | 1 | 1.03 |
| blaOXA-51-like+blaOXA-23-like+blaOXA-40-like+blaOXA-58-like | 10 | 10.30 |
| blaOXA-51-like+blaOXA-23-like+blaOXA-58-like+blaOXA-143-like | 6 | 6.18 |
| blaOXA-51-like+blaOXA-40-like+blaOXA-58-like+blaOXA-143-like | 1 | 1.03 |
| blaOXA-51-like+blaOXA-23-like+blaOXA-40-like+blaOXA-58-like+blaOXA-143-like | 4 | 4.12 |
| Total | 97 | 100 |
Discussion
Nosocomial infections are an important origin of mortality and morbidity in hospitalized patients. A. baumannii plays a significant role in these infections especially in ICU wards due to its resistance against multiple classes of antibiotics (24). The finding of this study showed that 85.18% of A. baumannii isolates were recovered from hospitalized patients in ICU wards. This result is in line with previous report about the role of A. baumannii in ICU infections (25, 26). Findings of this study showed that 89.81% of A. baumannii isolates were resistant to imipenem and meropenem. Previous reports from Iran showed the resistant rate to imipenem in 50.7% (25), 52% (27) and 62% (28) of A. baumannii isolates which indicates an increase in the rate of resistance to carbapenems. Results of other studies also show that resistance rate to carbapenems in recent years for detection of blaOXA-51-like revealed that all A. baumannii isolates had blaOXA-51-like gene. This finding supports the proposal that detection of blaOXA-51-like gene can be used as a simple and valid method to differentiate A. baumannii strains from other species (18). In addition, we found considerable levels of multiple-resistance to antimicrobials tested in our study. The distribution of XDR and PDR among our isolates was found to be 82.4% and 5.55% which was much lower than the rate of XDR (89%) and PDR (11%) reported from Arak in central part of Iran (31). These results explain that the appropriate choices for the treatment of infection caused by A. baumannii are limited. Our data indicated that 82.47% (80 isolates) of the carbapenem non-susceptible isolates of A. baumannii contained blaOXA-23-like gene. The distribution of blaOXA-23-like gene in A. baumannii isolates from the Iran (central part of Iran) was shown to be 25% in 2008 (25). In other studies, increasing level of blaOXA-23-like gene was reported so that, 84% of the isolates were found to have the blaOXA-23-like gene in the capital of Iran in 2011(32) and 88.7% of the isolates had this gene in Northwest of Iran on 2012 (28). The comparison in the rate of blaOXA-23-like gene in these three regions could be explained by an increased distribution of blaOXA-23-like gene. Outbreaks of blaOXA-23 producing carbapenem resistant A. baumannii isolates have been reported from many countries and this gene has found a worldwide dissemination (33). In studies conducted by Mak et al. and Zhou et al. 87.5% and 94% of carbapenem resistant isolates had blaOXA-23-like gene, respectively (34, 21). In our study, 22.68% of carbapenem non-susceptible A. baumannii isolates had blaOXA-40-like gene. This result is similar to the rate of blaOXA-40-like gene reported from central part of Iran (17.9%) (25), but is considerably higher than the rate of this gene reported from northwest of Iran (1.6%) (28). The highest prevalence of blaOXA-40-like gene has been reported from European countries such as Spain (23) and Portugal (35). In this study, we identified blaOXA-58-like gene in 55.67% of carbapenem non-susceptible A. baumannii isolates. The distribution of blaOXA-58-like gene in A. baumannii isolates from the northwest and the capital of Iran was shown to be 3.2% (28) and 21.2% (36), respectively. These rates are significantly lower than the rate found in current study. Ruiz et al. in Spain (23) and Stoeva et a.l in Bulgaria (30) reported that 19% and 27.27% of carbapenem resistant A. baumannii isolates contained blaOXA-58-like gene, respectively. To date, blaOXA-143-like gene has been reported only for A. baumannii isolates in Brazil (8, 37). Our study displayed, 14.43% (14 isolates) of the carbapenem non-susceptible isolates of A. baumannii carried blaOXA-143-like gene. This result is similar to the rates of blaOXA-143-like gene reported from Brazil (8.4%) (37) and 18.6% (38). However, our results are consistent with the data presented in some other studies documenting high distribution of blaOXA-23-like gene (84%, 88.7%), which followed by a lower occurrence of blaOXA-58-like gene (21.2%, 3.2%) and blaOXA-40-like gene (17.9%, 1.6%) (25, 28, 32). In the present study, we found that among carbapenem non-susceptible A. baumannii isolates 90.72% (88 isolates) of isolates were extensive drug resistance (XDR), 80.68% (71 isolates), 54.54% (48 isolates), 23.86% (21 isolates) and 13.63% (12 isolates) of them carried blaOXA-23-like, blaOXA-58-like, blaOXA-40-like and blaOXA-143 genes, respectively. The current study found 10 (10.3%) isolates of carbapenem resistant A. baumannii isolates that contained only the blaOXA-51-like gene. Therefore, it implies that the association between blaOXA-51-like gene and resistance of A. baumannii to carbapenem is low and other blaOXA genes considerably involved in resistance of A. baumannii to carbapenems.
Conclusion
This study showed that the rate of resistance to most of the available antibiotics for treatment of infection caused by A. baumannii was high in the study region. This study also revealed that suceptibility to carbapenems in the population of A. baumannii isolates in Iran reduced and the blaOXA-23-like gene was the most prevalent types of carbapenemase among carbapenem resistant A. baumannii isolates in this area. The rapid identification of carbapenemase-producing isolates is important and the use of specific methods are necessary to control the further transmission of infection by these organisms.
Acknowledgment
This study is part of MSc thesis for Zohreh Sarikhani that approved in the Qom Branch, Islamic Azad University. The authors thank Dr Morovat Taheri Kalani for gift of the OXA-type positive controls of A. baumannii. The authors declare no conflicts of interests.
References
- 1.Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii:emergence of a successful pathogen. Clin Microbiol Rev. 2008;21:538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moradi J, Hashemi B, Bahador A. Antibiotic resistance of Acinetobacter baumannii in Iran:A systemic review of the published literature. Osong Public Health Res Perspect. 2015;6:79–86. doi: 10.1016/j.phrp.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sadeghi P, Khosravi A, Hashemi A, Beiranvand M. Identification of clinical isolates of Acinetobacter baumannii from Iran and study of their heterogeneity. J Chin Med Assoc. 2016;79:382–386. doi: 10.1016/j.jcma.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 4.Magiorakos AP, Srinivassan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pan drug-resistant bacteria:an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 5.Helfand MS, Bonomo RA. Current challenges in antimicrobial chemotherapy:the impact of extended-spectrum beta-lactamases and metallo-beta-lactamases on the treatment of resistant Gram-negative pathogens. Curr Opin Pharmacol. 2005;5:452–458. doi: 10.1016/j.coph.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 6.Farajnia S, Azhari F, Alikhani MY, Hosseini MK, Peymani A, Sohrabi N. Prevalence of PER and VEB type extended spectrum betalactamases among multidrug resistant Acinetobacter baumannii isolates in North-West of Iran. Iran J Basic Med Sci. 2013;16:751–755. [PMC free article] [PubMed] [Google Scholar]
- 7.Poirel L, Nordmann P. Carbapenem resistance in Acinetobacter baumannii:mechanism and epidemiology. Clin Microbiol Infect. 2006;12:826–836. doi: 10.1111/j.1469-0691.2006.01456.x. [DOI] [PubMed] [Google Scholar]
- 8.Higgins PG, Poirel L, Lehmann M, Nordmann P, Seifert H. OXA-143, a novel carbapenem-hydroltoyzing class D beta-lactamase in Acinetobacter baumannii. Antimicrob Agents Chemother. 2009;53:5035–5038. doi: 10.1128/AAC.00856-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Higgins PG, Dammhayn C, Hackel M, Seifert H. Global spread of carbapenem-resistant Acinetobacter baumannii. J Antimicrob Chemother. 2010;65:233–238. doi: 10.1093/jac/dkp428. [DOI] [PubMed] [Google Scholar]
- 10.Walther-Rasmussen J, Hoiby N. OXA-type carbapenemases. J Antimicrob Chemother. 2006;57:373–383. doi: 10.1093/jac/dki482. [DOI] [PubMed] [Google Scholar]
- 11.Paton R, Miles RS, Hood J, Amyes SGB. ARI-1:Beta lactamase-mediated imipenem resistance in Acinetobacter baumannii. Int J Antimicrob Agents. 1993;2:81–88. doi: 10.1016/0924-8579(93)90045-7. [DOI] [PubMed] [Google Scholar]
- 12.Donald HM, Scaife W, Amyes SG, Young HK. Sequence analysis of ARI-1, a novel OXA beta-lactamase, responsible for imipenem resistance in Acinetobacter baumannii6B92. Antimicrob Agents Chemother. 2000;44:196–199. doi: 10.1128/aac.44.1.196-199.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mugnier PD, Poirel L, Naas T, Nordmann P. Worldwide dissemination of the blaOXA-23-likecarbapenemase gene of Acinetobacter baumannii. Emerg Infect Dis. 2010;16:35–40. doi: 10.3201/eid1601.090852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bou G, Oliver A, Martinez-Beltran J. OXA-24, a novel class D beta-lactamase with carbapenemase activity in an Acinetobacter baumannii clinical strain. Antimicrob Agents Chemother. 2000;44:1556–1561. doi: 10.1128/aac.44.6.1556-1561.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Evans BA, Hamouda A, Towner KJ, Amyes SG. OXA-51-like beta-lactamases and their association with particular epidemic lineages of Acinetobacter baumannii . Clin Microbiol Infect. 2008;14:268–275. doi: 10.1111/j.1469-0691.2007.01919.x. [DOI] [PubMed] [Google Scholar]
- 16.Poirel L, Marque S, Heritier C, Segonds C, Chabanon G, Nordmann P. OXA-58, a novel class D beta-lactamase involved in resistance to carbapenems in Acinetobacter baumannii. Antimicrob Agents Chemother. 2005;49:202–208. doi: 10.1128/AAC.49.1.202-208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bergogne-Berezin E, Towner KJ. Acinetobacter spp as nosocomial pathogens:microbiological, clinical, and epidemiological features. Clin Microbiol Rev. 1993;9:148–165. doi: 10.1128/cmr.9.2.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turton JF, Woodford N, Glover J, Yarde S, Kaufmann ME, Pitt TL. Identification of Acinetobacter baumannii by detection of the blaOXA-51-likecarbapenemase gene intrinsic to this species. J Clin Microbiol. 2006;44:2974–2976. doi: 10.1128/JCM.01021-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial disk susceptibility tests. 11th ed. Wayne, PA: Approved standards, CLSI Document M02-A11; 2012. [Google Scholar]
- 20.Pitout JD, Gregson DB, Poirl L, McClure JA, Le P, Church DL. Detection of Pseudomonas aeruginosa producing metallo-beta-lactamases in a large centralized laboratory. J Clin Microbiol. 2005;43:3129–3135. doi: 10.1128/JCM.43.7.3129-3135.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou H, Pi BR, Yang Q, Yu YS, Chen YG, Li LJ, et al. Dissemination of imipenem resistant Acinetobacter baumannii strains carrying the ISAba1bla OXA-23 genes in a Chinese hospital. J Med Microbiol. 2007;56:1076–1080. doi: 10.1099/jmm.0.47206-0. [DOI] [PubMed] [Google Scholar]
- 22.Woodford N, Ellington MJ, Coelho JM, Turton JF, Ward ME, Brown S, et al. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int J Antimicrob Agents. 2006;27:351–353. doi: 10.1016/j.ijantimicag.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 23.Ruiz M, Marti S, Fernandez-Cuenca F, Pascual A, Vila J. High prevalence of carbapenem-hydrolysing oxacillinases in epidemiologically related and unrelated Acinetobacter baumannii clinical isolates in Spain. Clin Microbiol Infect. 2007;13:1192–1198. doi: 10.1111/j.1469-0691.2007.01825.x. [DOI] [PubMed] [Google Scholar]
- 24.Garcia-Garmendia JL, Ortiz-Leyba C, Garnacho-Montero J, Jimenez-Jimenez FJ, Perez-Paredes C, Barrero-Almodovar AE, et al. Risk factors for Acinetobacter baumannii bacteremia in critically III patients:A cohort study. Clin Infect Dis. 2001;33:939–946. doi: 10.1086/322584. [DOI] [PubMed] [Google Scholar]
- 25.Feizabadi MM, Fathollahzadeh B, Taherikalani M, Rasoolinejad M, Sadeghifard N, Aligholi M, et al. Antimicrobial susceptibility patterns and distribution of bla OXA genes among Acinetobacter spp. isolated from patients at Tehran hospitals. Jpn J Infect Dis. 2008;61:274–278. [PubMed] [Google Scholar]
- 26.Papa A, Koulourida V, Souliou E. Molecular epidemiology of carbapenem-resistant Acinetobacter baumannii in a newly established Greek hospital. Microb Drug Resist. 2009;15:257–260. doi: 10.1089/mdr.2009.0060. [DOI] [PubMed] [Google Scholar]
- 27.Rahbar M, Monnavar KM, Vatan KK, Fadaei-haq A, Shakerian F. Carbapenem resistance in gram negative bacilli isolates in an Iranian 1000-bed Tertiary hospital. Pak J Med Sci. 2008;24:537–540. [Google Scholar]
- 28.Sohrabi N, Farajnia S, Akhi MT, Nahaei MR, Naghili B, Peymani A, et al. Prevalence of OXA-type beta-lactamases among Acinetobacter baumannii isolates from Northwest of Iran. Microb Drug Resist. 2012;18:385–389. doi: 10.1089/mdr.2011.0077. [DOI] [PubMed] [Google Scholar]
- 29.Gur D, Korten V, Unal S, Deshpande LM, Castanheira M. Increasing carbapenem resistance due to the clonal dissemination of oxacillinase (OXA-23 and OXA-58) producing Acinetobacter baumannii:report from the Turkish Sentry Program Site. J Med Microbiol. 2008;57:1529–1532. doi: 10.1099/jmm.0.2008/002469-0. [DOI] [PubMed] [Google Scholar]
- 30.Stoeva T, Higgins PG, Savov E, Markovska R, Mitov I, Seifert H. Nosocomial spread of OXA-23 and OXA-58 beta-lactamase producing Acinetobacter baumannii in a Bulgarian hospital. J Antimicrob Chemother. 2009;63:618–620. doi: 10.1093/jac/dkn537. [DOI] [PubMed] [Google Scholar]
- 31.Japoni-Nejad A, Sofian M, VanBelkum A, Ghaznavi-Rad E. Nosocomial outbreak of extensively and pandrug resistant Acinetobacter baumannii in tertiary hospital in Central part of Iran. Jundishapur J Microbiol. 2013;6:1–5. [Google Scholar]
- 32.Shahcheraghi F, Abbasalipour M, Feizabadi M, Ebrahimipour G, Akbari N. Isolation and genetic characterization of metallo-beta-lactamase and carbapenemase producing strains of Acinetobacter baumannii from patients at Tehran hospitals. Iran J Microbiol. 2011;3:68–74. [PMC free article] [PubMed] [Google Scholar]
- 33.Mugnier PD, Poirel L, Nass T, Nordmann P. Worldwide dissemination of the bla OXA-23 carbapenemase gene of Acinetobacter baumannii. Emerg Infect Dis. 2010;16:35–40. doi: 10.3201/eid1601.090852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mak JK, Kim MJ, Pham J, Tapsall J, White PA. Antibiotic resistance determinants in nosocomial strains of Multidrug-resistant Acinetobacter baumannii. J Antimicrob Chemother. 2009;63:47–54. doi: 10.1093/jac/dkn454. [DOI] [PubMed] [Google Scholar]
- 35.Quinteira S, Grosso F, Ramos H, Peixe L. Molecular epidemiology of imipenem resistant Acinetobacter haemolyticus and Acinetobacter baumannii isolates carrying plasmid mediated OXA-40 from a Portuguese hospital. Antimicrob Agents Chemother. 2007;51:3465–3466. doi: 10.1128/AAC.00267-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taherikalani M, Fatolahzadeh B, Emaneini M, Soroush S, Feizabadi MM. Distribution of different carbapenem resistant clones of Acinetobacter baumannii in Tehran hospitals. New Microbiol. 2009;32:265–271. [PubMed] [Google Scholar]
- 37.Werneck JS, Picao RC, Girardello R, Cayo R, Marguti V, Dalla-Costa L, et al. Low prevalence of bla OXA-143 in private hospitals in Brazil. Antimicrob Agents Chemother. 2011;55:4494–4495. doi: 10.1128/AAC.00295-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neves F, Clemente W, Lincopan N, Paiao I, Neves P, Romanelli R, et al. Clinical and microbiological characteristics of OXA-23 and OXA-143 producing Acinetobacter baumannii in ICU patients at a teaching hospital, Brazil. Braz J Infect Dis. 2016;20:556–563. doi: 10.1016/j.bjid.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]


