Abstract
Objective
We investigated the effect and mechanism of hypoxic microenvironment and hypoxia-inducible factors (HIFs) on hepatocellular carcinoma (HCC) cancer stemness.
Design
HCC cancer stemness was analysed by self-renewal ability, chemoresistance, expression of stemness-related genes and cancer stem cell (CSC) marker-positive cell population. Specific small ubiquitin-like modifier (SUMO) proteases 1 (SENP1) mRNA level was examined with quantitative PCR in human paired HCCs. Immunoprecipitation was used to examine the binding of proteins and chromatin immunoprecipitation assay to detect the binding of HIFs with hypoxia response element sequence. In vivo characterisation was performed in immunocompromised mice and stem cell frequency was analysed.
Results
We showed that hypoxia enhanced the stemness of HCC cells and hepatocarcinogenesis through enhancing HIF-1α deSUMOylation by SENP1 and increasing stabilisation and transcriptional activity of HIF-1α. Furthermore, we demonstrated that SENP1 is a direct target of HIF-1/2α and a previously unrecognised positive feedback loop exists between SENP1 and HIF-1α.
Conclusions
Taken together, our findings suggest the significance of this positive feedback loop between HIF-1α and SENP1 in contributing to the increased cancer stemness in HCC and hepatocarcinogenesis under hypoxia. Drugs that specifically target SENP1 may offer a potential novel therapeutic approach for HCC.
Keywords: HEPATOCELLULAR CARCINOMA, MOLECULAR BIOLOGY, SIGNALING, MOLECULAR PATHOLOGY
Significance of this study.
What is already known on this subject?
Hepatocellular carcinoma (HCC) is a common cancer and leading cause of death worldwide.
Hypoxia is common in solid cancers and particularly in HCC, which is a fast growing cancer.
Hypoxic microenvironment is an important stem cell niche.
SUMOylation is an important post-translational protein modification and involved in a wide variety of cellular processes.
Specific SUMO proteases 1 (SENP1) is able to deSUMOylate hypoxia-inducible factor (HIF)-1α and increase its stability in hypoxia.
What are the new findings?
SENP1 is a direct target of HIF-1/2α.
A previously unrecognised positive feedback loop exists between SENP1 and HIF-1α.
This positive feedback loop between HIF-1α and SENP1 contributes to increased cancer stemness of HCC and hepatocarcinogenesis under hypoxia.
How might it impact on clinical practice in the foreseeable future?
Drugs that specifically target SENP1 may offer a potential novel therapeutic approach for HCC.
Introduction
Hepatocellular carcinoma (HCC) is a prevalent malignancy and ranks third in cancer mortality worldwide. Progression of HCC is believed to be partly driven by cancer stem cell (CSC) through their capacity of self-renewal, tumourigenicity, production of heterogeneous progenies, metastasis and resistance to chemotherapeutic drugs.1 Recently, liver CSCs have been identified by cell surface markers including CD133,2 CD90,3 epithelial cell adhesion molecule,4 CD245 and CD47.6
Hypoxia is a common phenomenon in solid cancers and is particularly frequent in HCC due to its rapid growth.7 Hypoxia-inducible factors (HIFs) are key transcription factors that allow cancer cells to survive in hypoxia and composed of the stable HIF-1β subunit and the oxygen-sensitive subunit HIF-1/2α.7 With O2 (normoxia), HIF-1/2α is hydroxylated and ubiquitin ligase Von Hippel-Lindau then targets HIF-1/2α for ubiquitin-proteasomal degradation. Without O2 (hypoxia), HIF-1/2α is no longer degraded and binds with HIF-1β to activate gene transcription and promote tumour progression and metastasis.7 HIF-1α is a master regulator contributing to the restoration of oxygen homeostasis.7 In HCC, HIF-1α is closely associated with poor prognosis of patients; HIF-1α expression is high in HCCs with venous and lymph node metastasis.8 Both HIF-1α and HIF-2α are induced by sorafenib in HCC cells, and this contributes to the resistance to sorafenib, the first-line molecular drug for advanced HCC.9 10
Hypoxic microenvironment is an important stem cell niche that promotes the persistence of CSCs in tumours.11 12 Increased levels of HIF-1α and HIF-2α were found in the stem cell-like populations of neuroblastomas12 13 and gliomas.14 Similarly, HIF-2α is preferentially expressed in immature neural crest-like neuroblastoma cells in vivo and may be required for the maintenance of undifferentiated neuroblastoma cells.13
Induced cancer stem-like sphere cells from HCC cells had higher HIF-1α mRNA levels and lower reactive oxygen species activity.15 Furthermore, hypoxia increased the proportion of HCC cells with stem-cell features, whereas echinomycin that inhibits HIF-1α DNA binding activity blocked this effect.15 It is unclear, however, how HIFs affect liver cancer stemness in hypoxic condition.
SUMOylation is an important post-translational protein modification by small ubiquitin-like modifier (SUMO) proteins, which belong to the growing family of ubiquitin-like proteins. SUMOylation is involved in a wide variety of cellular processes such as transcription, DNA repair, trafficking and signal transduction.16 SUMOylation is carried out by a multistep enzymatic cascade reaction facilitating the attachment of SUMO-1, SUMO-2 or SUMO-3 to the substrates.17 18 SUMOylation is a dynamic process and readily reversed by a family of specific SUMO protease (SENPs), in a process called deSUMOylation, in which SENPs remove SUMO conjugate from the conjugated proteins.19 Six SENP proteins (SENP1, SENP2, SENP3, SENP5, SENP6 and SENP7) have been identified in humans; each has distinct subcellular localisation and substrate specificity, suggesting that they are non-redundant.20–25
SENP1 has been shown to be essential for the stability and activation of HIF-1α and is able to deSUMOylate HIF-1α and increase its stability in hypoxia.26 SUMOylation plays an important role in the regulation of HIF-1α26–28 but the impact of SUMOylation on HIF-1α activity has been controversial. Moreover, how SENP1 modulates HIF-1α affecting cancer stemness is unknown. In this study, we report that SENP1 increases the stabilisation and transcriptional activity of HIF-1α in hypoxia via deSUMOylation in HCC. These enhance cancer stemness, increase liver CSC subpopulations and promote hepatocarcinogenesis. In addition, we demonstrate that SENP1 is a direct target gene of HIFs and a positive feedback loop exists between HIF-1α and SENP1 and contributes to HCC stemness and tumourigenesis.
Experimental procedures
Cell culture, cloning procedures and transfection
All cell lines were maintained in Dulbecco's modified Eagle's medium containing 1% penicillin and streptomycin, supplemented with 10% fetal bovine serum; 1% O2 was generated by flushing a 94% N2/5% CO2 mixture into the incubator. All expression plasmids and transfections are shown in the online supplementary experimental procedures.
gutjnl-2016-313264supp001.pdf (2.2MB, pdf)
Patient samples
Human HCCs and their paired non-tumourous liver (NT-L) tissues were collected during surgical resection at Queen Mary Hospital, Hong Kong. Use of human samples was approved by the Institutional Review Board (IRB) of University of Hong Kong/Hospital Authority Hong Kong West Cluster (IRB reference number: UW09-185). Demographic information of the patients is provided in online supplementary experimental procedures.
Chromatin immunoprecipitation assay
HCC cells were cross-linked with formaldehyde, lysed with sodium dodecyl sulfate buffer and sonicated. Sheared DNA was precleared with salmon sperm DNA/protein A agarose slurry (Merck Millipore) and immunoprecipitated with HIF-1α or HIF-2α antibody and IgG (Santa Cruz). Agarose beads were incubated with antibody/protein/DNA complex and washed with low-salt buffer, high-salt buffer and LiCl wash buffer according to manufacturer's protocol (Millipore). DNA was eluted in and extracted by phenol-chloroform.
Cell sphere formation, proliferation, migration and chemoresistance assays
Details are provided in the online supplementary experimental procedures.
Immunohistochemistry, quantitative reverse transcription PCR, short hairpin RNA, luciferase reporter assay, immunoprecipitation and western blot analyses
Details are provided in the online supplementary experimental procedures.
Animal experiments
Animal care and experiments were performed in strict accordance with the ‘Guide for the Care and Use of Laboratory Animals’ and ‘Principles for the Utilisation and Care of Vertebrate Animals’ and were approved by the Experimental Animal Ethical Committee at University of Hong Kong. The detailed protocols are provided in the online supplementary experimental procedures.
Statistical analysis
All statistical analyses were performed by the SPSS Statistics SPSS 23.0. Student's t-test, χ2 test or Mann-Whitney U test were used for continuous data wherever appropriate. p Values <0.05 were considered to be statistically significant.
Results
Hypoxia enhances HCC stemness in HIF-1α-dependent and HIF-2α-dependent manner
Digoxin, a well-known HIF-1α inhibitor, inhibits the transcriptional activity of HIF-1α.29 First, we examined the effects of digoxin on the mRNA expression of the different liver CSC markers and sphere forming ability on two HCC cell lines (MHCC-97L and PLC/PRF/5) by treating them with digoxin or dimethyl sulfoxide for 24 hours under hypoxia. The mRNA levels of CD24 and CD133 were consistently upregulated in hypoxia, while digoxin treatment significantly abolished this upregulation (see online supplementary figure S1A). Similar results were observed in sphere formation assay (see online supplementary figure S1B).
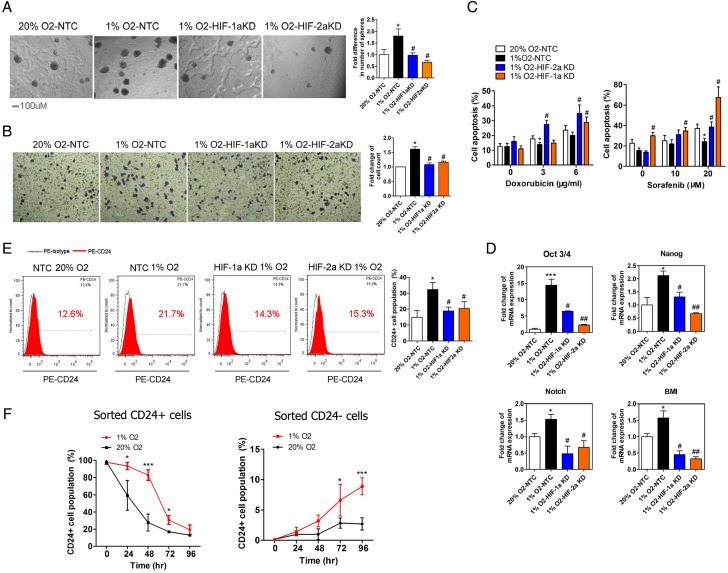
To determine whether HIF-1α and HIF-2α regulate stemness of HCC cells via specific molecular responses to hypoxia, we generated stable HIF-1α or HIF-2α knockdown HCC cells (MHCC-97L and PLC/PRF/5), as previously described.30 Knockdown of either HIF-1α or HIF-2α suppressed hypoxia-induced transcription of HIF-target genes including vascular endothelial growth factor (VEGF), lipoxygenase (LOX) and erythropoietin (EPO) (see online supplementary figure S2). Moreover, hypoxia enhanced the ability of self-renewal (in vitro using sphere formation assay5 6 31) (see figure 1A and online supplementary figure S3A), migration (see figure 1B and online supplementary figure S3B) and chemoresistance to sorafenib and doxorubicin (see figure 1C and online supplementary figure S3C). Upregulation of the mRNA levels of stemness-related genes (Oct3/4, Nanog, Notch1 and B cell-specific moloney murine leukemia virus integration site 1 (BMI-1)) was also observed in HCC cells with hypoxic treatment (see figure 1D and online supplementary figure S3D). Knockdown of HIF-1α or HIF-2α inhibited hypoxia-induced enhancement of stemness in HCC cells (see figure 1A–D and online supplementary figure S3A–D).
Figure 1.
The effect of hypoxia and hypoxia-inducible factor (HIF)-1/2α on the stemness of MHCC-97L cells. (A–C) The in vitro cell abilities for self-renewal (A), migration (B) and chemoresistance (C) to doxorubicin and sorafenib were enhanced under hypoxic condition (1% O2) in MHCC-97L cells and the knockdown of HIF-1α or HIF-2α suppressed these hypoxia-induced effects. (D) Hypoxia-induced increase of mRNA expression of stemness-related genes (Oct3/4, Nanog, BMI-1 and Notch1) was inhibited in HIF-1α or HIF-2α knockdown MHCC-97L cells. (E) CD24+ and CD133+ cell populations were increased in hypoxia-treated MHCC-97L cells, but the knockdown of HIF-1α or HIF-2α blunted the effects. (F) CD24+ population was analysed by FACS in sorted CD24+ (left) and CD24− (right) cells from MHCC-97L cells after cultured for indicated time under hypoxia or normoxia. (*p<0.05, **p<0.01, ***p<0.001, compared with the negative control in normoxia (20% O2); #p<0.05, ##p<0.01, ###p<0.001, compared with the negative control in hypoxia (1% O2)).
CD24+ tumour-initiating populations have been found in HCC,5 pancreatic,32 colorectal33 34 and bladder cancers.35 Promoter analysis demonstrated that a hypoxia response element (HRE) in the upstream promoter/enhancer region is required for both hypoxia-induced and HIF-1α-dependent CD24 expression.36 Previously, we reported that CD24 is a functional liver CSC marker that drives CSC through STAT3-mediated NANOG regulation.26 To determine the regulation of CD24 by hypoxia in HCC cells, we assessed the change of CD24+ cell population in hypoxia. After hypoxic treatment, the CD24+ cell population rose from 12.6% to 21.7% and 30.6% to 62.8% in MHCC-97L and PLC/PRF/5 cells, respectively (see figure 1E and online supplementary figure S3E). Similar but milder results were seen for CD133+ cells in the HCC cell lines after hypoxic treatment (see online supplementary figure S3E).
Furthermore, to address the influence of hypoxia on CD24+ and CD24− cells, we sorted CD24+ and CD24− subsets from MHCC-97L cells using fluorescence-activated cell sorting (FACS) with good sorting efficiency (see online supplementary figure S4A). High mRNA levels of CD24 and Nanog was observed in the sorted CD24+ cells (see online supplementary figure S4B). Next, we incubated CD24+ and CD24− cells in hypoxic or normoxic conditions (figure 1F). We observed that the CD24+ cell population was maintained at a significantly higher level under hypoxia than normoxia in both sorted CD24+ and CD24− (figure 1F) MHCC-97L cells, which was abolished with digoxin treatment (200 nM) in both CD24+ and CD24− HCC cells (see online supplementary figures S4C). These data suggest that HIF and/or hypoxia play an important role in the maintenance of the CD24+ CSC subpopulation.
Overexpression of SENP1 in human HCCs and HCC cells
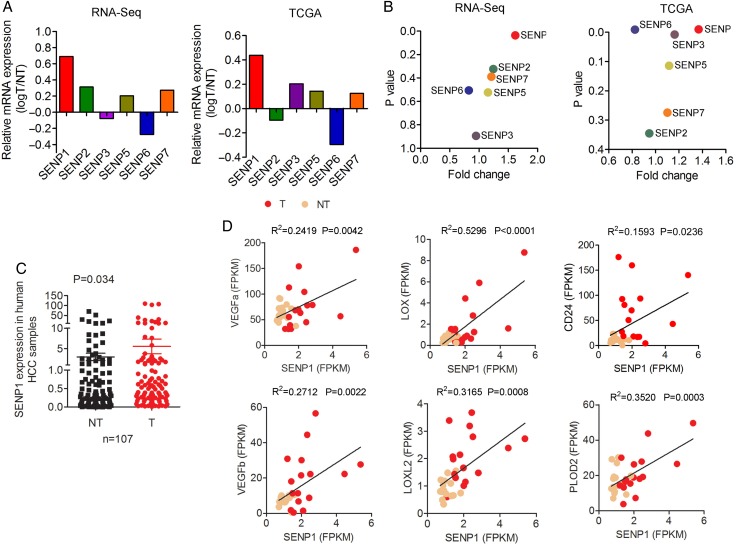
To delineate the roles of SENPs in hepatocarcinogenesis, we first assessed the mRNA levels of the six SENP family members in human HCC tissues. From our RNA-sequencing data (available in Sequence Read Archive of National Center for Biotechnology Information with the accession number SRP062885) on 16 pairs of human HCCs and their corresponding NT-Ls as well as The Cancer Genome Atlas (TCGA) data of National Cancer Institute, USA (202 HCC tumours of all aetiologies including ‘unspecified’ aetiologies), we observed a consistent upregulation of SENP1 (p=0.050 and p<0.001, respectively) (figure 2A, B). Of note, SENP1 was consistently and significantly upregulated among the six SENP family members examined in these two RNA-sequencing datasets, while this was not so for the other five SENPs. Using quantitative PCR (qPCR) on our larger cohort of 107 pairs of patients' HCCs and corresponding NT-Ls, we consistently observed significant SENP1 upregulation in the tumours (p=0.034) (figure 2C). Similarly, higher protein expression level of SENP1 in human HCC was shown by immunohistochemistry (IHC) (see online supplementary figure S5A).
Figure 2.
Upregulation of specific SUMO proteases 1 (SENP1) in hepatocellular carcinoma (HCC) tissues and the correlation between SENP1 and hypoxia-inducible factor (HIF)-α target genes. (A and B) SENP1 was consistently and significantly upregulated among the six SENP family members examined in our RNA-sequencing data on 16 pairs of human HCCs and their corresponding non-tumourous livers (NT-Ls) and data from The Cancer Genome Atlas (TCGA) of National Cancer Institute, USA (202 HCC tumours of all aetiologies including ‘unspecified’ aetiologies). (C) Using quantitative PCR on a larger cohort of 107 pairs of HCC tumour and corresponding NT-Ls, the similar result of SENP1 upregulation in HCC tumours was observed. (D) Correlation analysis of relative mRNA levels of SENP1 and VEGFa, VEGFb, LOX, LOXL2, PLOD2 or CD24 using our RNA-sequencing data on 16 pairs of human HCCs and their corresponding NT-Ls.
We also examined the SENP1 expression using qPCR and western blot analysis on a panel of HCC cell lines (Huh-7, PLC/PRF/5, MHCC-97L, MHCC-97H, BEL-7402 and SMMC-7721) and a non-tumourigenic immortalised normal liver cell line MIHA. These HCC cell lines showed a range of SENP1 mRNA and protein expression levels. In contrast, MIHA, which is incapable of tumour formation in vivo, had almost undetectable SENP1 protein level (see online supplementary figure S6).
Clinical significance of SENP1 and its correlation with HIF target gene expression in human HCCs
On clinicopathological correlation, SENP1 overexpression (OE) was significantly associated with more aggressive tumour behaviour, in terms of more frequent venous invasion (p=0.042), a feature of metastasis, and more advanced tumour stage (p=0.034) (see online supplementary table S1).
To address the relationship of SENP1 and HIF signals, we analysed the correlation between the mRNA levels of SENP1 and HIF-target genes using our RNA-sequencing data on 16 pairs of human HCCs. We observed a significant positive correlation between SENP1 and VEGFa, VEGFb, LOX, LOXL2 and PLOD2 (figure 2D). Furthermore, we also found a positive correlation between SENP1 and CD24 in this same cohort of patients with HCC (p=0.024; r2=0.159) (figure 2D). In addition, using IHC, we examined the expression of two HIF-1α-dependent genes, carbonic anhydrase 9 (CA9) and glucose transporter 1 (GLUT1), and observed good correlation among them in both mouse HCC xenograft (see online supplementary figure S5B) and human HCCs (see online supplementary figure S5C).
SENP1 OE enhances the expression of stemness-related genes in HCC cells in hypoxia
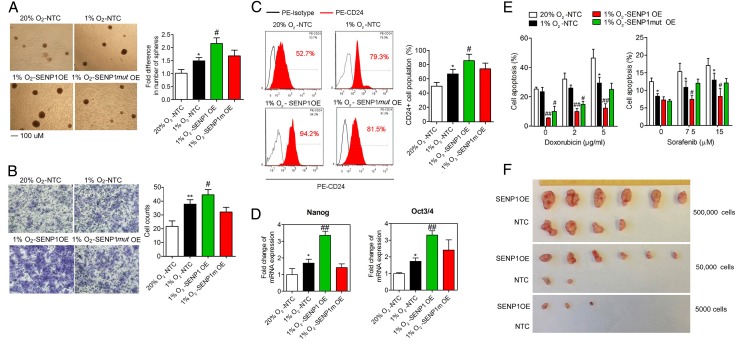
To address whether SENP1 regulated cancer stemness through its specific SENPs activity, we stably overexpressed SENP1 or SENP1 catalytic inactive mutant (SENP1mut) (in which a conserved amino acid, cysteine 603, in the catalytic domain of SENP1 was substituted with alanine)26 in Huh-7 and PLC/PRF/5 cells, to examine the functional roles of SENP1 in maintaining liver CSCs in vitro (see online supplementary figure S7). Huh-7 and PLC/PRF/5 cells were used in the OE experiment as they have a relatively lower SENP1 endogenous level (see online supplementary figure S6). The expression of SENP1, but not SENP1mut, enhanced stemness-related properties, including self-renewal ability (see figure 3A and online supplementary figure S8A), cell migration (see figure 3B and online supplementary figure S8B), CD24 cell population (see figure 3C and see online supplementary figure S8C), expression of stemness genes, Nanog and Oct4 (see figure 3D and online supplementary figure S8D) and chemoresistance to sorafenib and doxorubicin (figure 3E) under hypoxia. Increased cell proliferation was also observed (see online supplementary figure S7).
Figure 3.
Effect of specific SUMO proteases 1 (SENP1) overexpression on the stemness of hepatocellular carcinoma (HCC) cells. (A–E) The effects of overexpression of SENP1, SENP1mut and non-target control (NTC) on the stemness of HCC cells shown by in vitro abilities of self-renewal (A), migration (B), CD24+ cell population (C) and mRNA expression of stemness-related genes (D) and chemoresistance (E), in hypoxic condition. (F) Limiting dilution xenograft formation of Huh-7 cells with NTC or SENP1 overexpression. (*p<0.05, **p<0.01, compared with the negative control in normoxia (20% O2), #p<0.05; ##p<0.01, compared with the negative control in hypoxia (1% O2)).
Next, we tested the in vivo tumour initiating capacity of SENP1. We injected SENP1-overexpressing Huh-7 cells or non-target control (NTC) into nonobese diabetic/severe combined immunodeficiency (NOD/SCID) mice subcutaneously at three dilutions (5×103, 5×104 and 5×105) and let them grow for 6 weeks. The tumour initiating capacity was analysed by the CIs for 1/(stem cell frequency) using extreme limiting dilution analysis37 (see figure 3F and see online supplementary tables S2 and S3). The estimated CI for the frequency of CSCs in SENP1-overexpressing group was 7121, as compared with 340 389 in NTC Huh-7 cells, indicating a much higher frequency of CSCs in SENP1-overexpressing HCC cells (p<0.001). These findings strongly suggest that SENP1 enhances hypoxia-induced cancer stemness in HCC cells, both in vitro and in vivo.
SENP1 knockdown suppresses stemness features in hypoxia
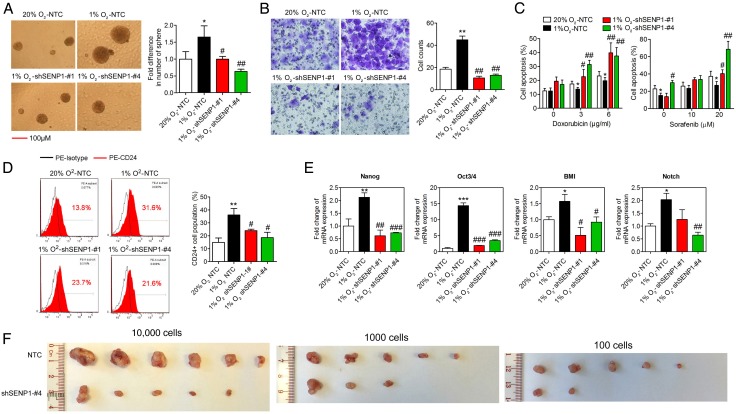
We used a lentiviral-based approach to establish stable SENP1-knockdown clones in MHCC-97L and BEL-7402 cells, which have a higher SENP1 endogenous expression level (see online supplementary figure S6). With successful SENP1 knockdown (see online supplementary figure S9A, sequences #1 and #4 had highest efficiency), we examined the stemness-associated features in vitro. First, we found the mRNA expression levels of liver CSC markers CD24, CD44 and CD133 were upregulated by hypoxia treatment and SENP1 knockdown abolished this response to hypoxia (see online supplementary figure S9B). By sphere formation assay, SENP1 knockdown resulted in the formation of fewer and smaller hepatospheres under hypoxia (figure 4A). Moreover, shSENP1 cells had significantly reduced migratory ability under hypoxia (figure 4B). In addition, knockdown of SENP1 suppressed the hypoxia-induced increase in chemosensitivity to sorafenib and doxorubicin in MHCC-97L cells (figure 4C). The CD24+ subpopulation, as detected using FACS assay, was also significantly reduced in SENP1-knockdown HCC cells under hypoxia (figure 4D). Finally, the stemness-related genes, Nanog, Notch1, Oct3/4 and BMI-1, were downregulated in the shSENP1 clones under hypoxia (figure 4E). These results were similarly observed in BEL-7402 cells (see online supplementary figure S10A–C).
Figure 4.
Effect of specific SUMO proteases 1 (SENP1) knockdown on the stemness of hepatocellular carcinoma (HCC) cells. (A–E) The effects of knockdown of SENP1 (shSENP1-#1 and shSENP1-#4) and non-target control (NTC) on the stemness of HCC cells shown by in vitro abilities of self-renewal (A), migration (B) and chemoresistance (C), CD24+ cell population (D) and mRNA expression of stemness-related genes (E) in hypoxic condition. (F) Limiting dilution xenograft formation of MHCC-97L cells with NTC or SENP1 knockdown. (*p<0.05, **p<0.01, ***p<0.001, as compared with the negative control in normoxia (20% O2); #p<0.05, ##p<0.01, ###p<0.001, compared with the negative control in hypoxia (1% O2)).
We examined the tumourigenicity of HCC cells on SENP1 knockdown in vivo by subcutaneous injection of SENP1-knockdown MHCC-97L cells or NTC into NOD/SCID mice at three dilutions (1×102, 1×103 and 1×104). The estimated CI for the frequency of CSCs in SENP1-knockdown group was 2722, compared with 249 in NTC MHCC-97L cells (p<0.0001) (see figure 4F, online supplementary tables S4 and S5). In addition, with in vivo tumour growth assay in nude mice, the size of the tumours was smaller in the shSENP1 BEL-7402 (see online supplementary figure S10D) and MHCC-97L (see online supplementary figure S11) clones when compared with the NTC group. These data suggest that SENP1 knockdown suppresses hypoxia-induced cell stemness in HCC cells and in vivo tumourigenicity. In addition, much reduced expression of CA9 and GLUT1, the 2 HIF-1α targets, was observed in the shSENP1 HCC cells in the xenografts in vivo (see online supplementary figure S5B).
SENP1 is a direct target of HIF-1α and HIF-2α
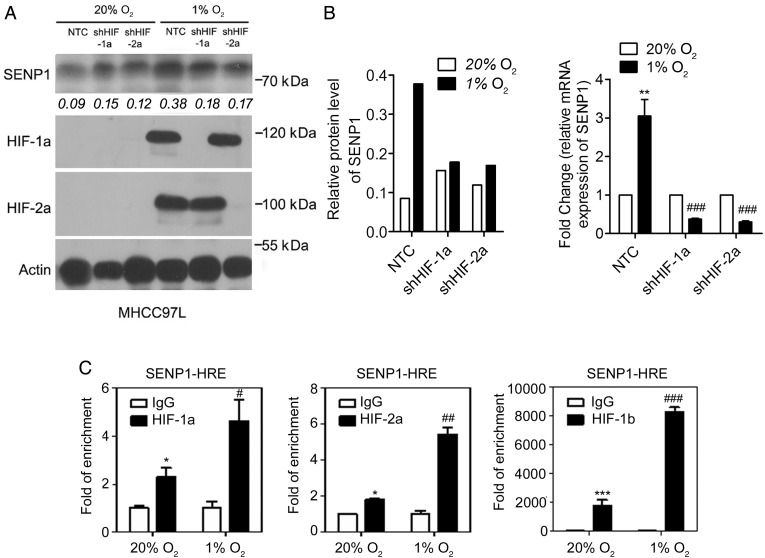
To investigate whether SENP1 was regulated by HIF-α, we examined the SENP1 mRNA and protein levels 24 hours after hypoxic treatment. SENP1 expression was markedly increased in MHCC-97L cells after hypoxic treatment (see online supplementary figure S2 and figure 5A, B). These effects were abolished when HIF-1α or HIF-2α was stably knocked down. Furthermore, chromatin immunoprecipitation assay30 was used to confirm the binding of HIF-1α or HIF-2α to SENP1 promoter. The HRE core sequence (A/G) CGTG was found at around −498 and −489 bp in the SENP1 promoter.38 Using the PCR primer sequences previously reported,38 we demonstrated that HIF-1α, HIF-2α and HIF-1β bound to the HRE sequence on the SENP1 promoter (figure 5C), strongly suggesting that HIF is a transcriptional factor that regulates SENP1 expression under hypoxia in HCC cells.
Figure 5.
Specific SUMO proteases 1 (SENP1) play a role as the direct target gene of hypoxia-inducible factor (HIF)-1/2α in hepatocellular carcinoma (HCC) cells. (A and B) Protein (A) and mRNA (B) levels of SENP1 were increased under hypoxia in HIF-1α-dependent and HIF-2α-dependent manner in MHCC-97L cells. (C) Chromatin immunoprecipitation assay was used to determine the bind of HIF-1α, HIF-2α and HIF-1β to the hypoxia response element (HRE) sequence in SENP1 promoter. (*p<0.05, **p<0.01, ***p<0.001, as compared with the negative control in normoxia (20% O2); #p<0.05, ##p<0.01, ###p<0.001, as compared with the negative control in hypoxia (1% O2)).
SENP1 enhances the stability and transcriptional activity of HIF-1α through deSUMOylation
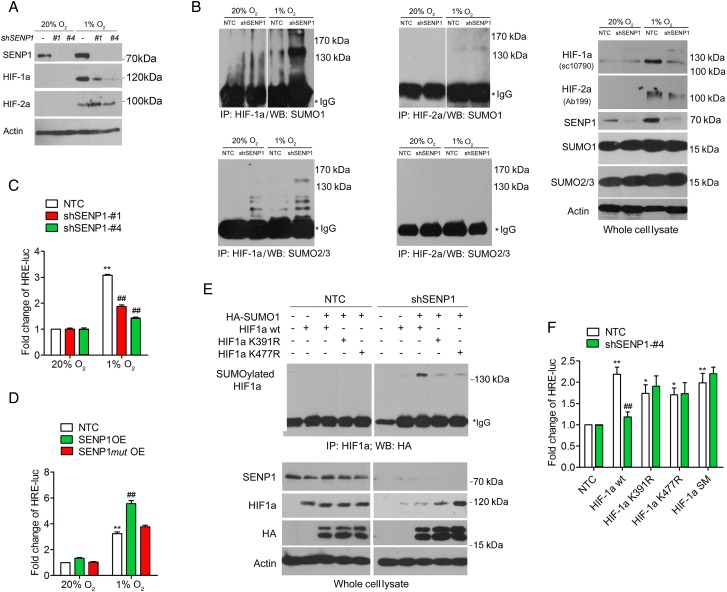
The correlation of SENP1 and hypoxia-induced HCC cell stemness prompted us to investigate how SENP1 regulates HIF-α stability and transcriptional activity. As previously shown,26 SENP1 was able to deSUMOylate HIF-1α and increase its stability in hypoxia. To this end, we examined the effect of SENP1 on HIF-1/2α SUMO modification, stability and transcriptional activity in HCC cells. The protein level of HIF-1α but not HIF-2α was reduced in SENP1-knockdown HCC cells in hypoxia (figure 6A). Next, using immunoprecipitation (IP) assay, we observed that in hypoxia, increased amounts of SUMO1-conjugated or SUMO2/3-conjugated HIF-1α accumulated in SENP1-knockdown HCC cells, as compared with NTC (figure 6B). Consistent with the results of figure 6A, we found very little detectable level of SUMOylated HIF-2α in both NTC and SENP1-knockdown cells whether in hypoxia or normoxia (figure 6B).
Figure 6.
Increased stability and transcriptional activity of hypoxia-inducible factor (HIF)-1α through deSUMOylation by specific SUMO proteases 1 (SENP1) in hepatocellular carcinoma (HCC) cells under hypoxic condition. (A) Western blot analyses were used to examine the protein level of HIF-1α and HIF-2α in SENP1-knockdown and non-target control (NTC) HCC cells under hypoxia or normoxia. (B) immunoprecipitation (IP) assay was used to detect the SUMOylation of HIF-1/2α by SUMO-1 or SUMO-2/3 in HCC cells. IP was conducted to obtain the complex protein with anti-HIF-1α or anti-HIF-2α antibody in SENP1-knockdown or vehicle-infected SMMC-7721 cells, which were incubated under normoxia or hypoxia for 24 hours. Western blot analyses were then carried out on the whole cell lysates and the IP complex with anti-SUMO-1 or SUMO-2/3 antibody. Inputs are presented in the right-most panel. (C) Fold change of the relative luciferase activity was examined by luciferase-reporter assay in SENP1-knockdown and NTC SMMC-7721 cells which were incubated under normoxia or hypoxia for 24 hours. (D) Fold change of the relative luciferase activity was examined in NTC, SENP1 overexpression (SENP1 OE) and SENP1mut overexpression (SENP1mut OE) Huh-7 cells which were incubated under normoxia or hypoxia for 36 hours. (E) IP assay was used to detect the binding of exogenous HIF-1α or the mutants with SUMO-1 in SMMC-7721 cells in normoxia. First, IP was conducted to obtain the complex protein with anti-HIF-1α antibody in SENP1-knockdown or vehicle-infected SMMC-7721 cells, in which HA-SUMO-1 was cotransfected with RGS-HIF-1α wild type (WT), RGS-HIF-1α K391R, RGS-HIF-1α K477R, or RGS-HIF-1α SM. Western blot analyses for indicated proteins in the whole cell lysates and the IP complex with anti-HA antibody. (F) Fold of the relative luciferase activity was examined in SENP1-knockdown and NTC SMMC-7721 cells (in normoxia), in which HA-SUMO-1 was cotransfected with RGS-HIF-1α WT, RGS-HIF-1α K391R, RGS-HIF-1α K477R or RGS-HIF-1α SM. (*p<0.05, **p<0.01, ***p<0.001, compared with the negative control in normoxia (20% O2); #p<0.05, ##p<0.01, ###p<0.001, compared with the negative control in hypoxia (1% O2)).
Then we assessed the transcriptional activity of HIF-1α on VEGF using 6×HRE VEGF promoter driven-luciferase reporter assay and qPCR. pGL3−6×VEGF HRE was cotransfected with pSV40 into SMMC-7721 HCC cells with high efficiency. Hypoxia-induced VEGF transcription was abolished in SENP1-knockdown SMMC-7721 cells (see figure 6C and online supplementary figure S12A). On the other hand, Huh-7 cells with SENP1 OE displayed increased HIF-1α transcriptional activity in hypoxia, as compared with the NTC and SENP1-mut OE controls (see figure 6D and online supplementary figure S12B). Altogether, these results suggest that under hypoxia, SENP1 increases the stability and transcriptional activity of HIF-1α in a SENPs-dependent manner.
Then we asked whether SUMO-mediated HIF-1α degradation was dependent on proteasome signalling. We examined the hypoxia-induced SUMOylation of HIF-1α in HCC cells with or without treatment with MG132, a specific cell-permeable proteasome inhibitor. In hypoxia, HIF-1α protein level was increased in SENP1-knockdown HCC cells with MG132 treatment, when compared with untreated cells (see online supplementary figure S12C). SUMOylated HIF-1α was easily detected in both NTC and SENP1-knockdown cells exposed to hypoxia and MG132 treatment. In contrast, without MG132 treatment, this was undetectable and only weakly detectable in NTC and SENP1-knockdown cells, respectively. These results suggest that SUMOylated HIF-1α is degraded through a proteasome-dependent mechanism in HCC cells.
SUMO sites K391 and K477 in HIF-1α are pivotal in SENP1-regulated HIF-1α deSUMOylation
We assessed the effect of deSUMOylation on HIF-1α activity in HCC cells using HIF-1α SUMOylated site mutants, K391R and K477R.26 SUMOylation of exogenous HIF-1α was detected through cotransfection of haemagglutinin (HA)-SUMO-1 and regulation of G-protein signaling domain (RGS)-HIF-1α wild-type (WT) or its mutants (RGS-HIF-1α K391R; RGS-HIF-1α K477R) in HCC cells. As shown in figure 6E, SUMOylated HIF-1α accumulated only in SENP1-knockdown HCC cells but not in NTC cells, and the SUMO-1 conjugated HIF-1α was markedly decreased in SENP1 knockdown cells transfected with HIF-1α mutant (K391R and K477R). Furthermore, mutation of the SUMOylation sites of HIF-1α significantly increased the transcriptional activity of HIF-1α in SENP1-knockdown cells (see figure 6F and online supplementary figure S12D).
Mutation of the SUMO sites in HIF-1α rescues the loss of hypoxia-induced stemness in SENP1-knockdown HCC cells
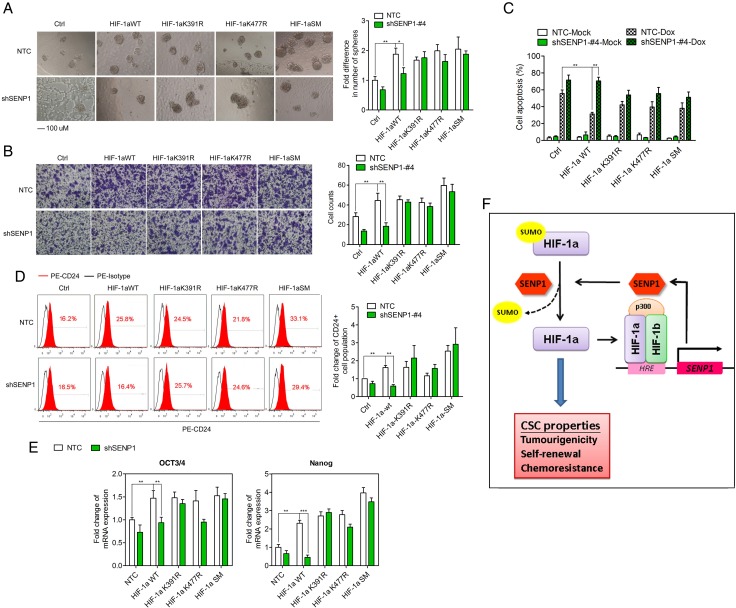
Since K391 and K477 SUMO sites are required by SENP1-regulated HIF-1α deSUMOylation, we wondered if the mutations of SUMO site in HIF-1α resisted the effect of SENP1 knockdown in HCC cells. To address this, we generated MHCC-97L cells which stably overexpressed HIF-1α WT, HIF-1α K391R, HIF-1α K477R or HIF-1α K391R/K477R (HIF-1α SM, simultaneous double mutant). Western blot analysis confirmed the overexpression of HIF-1α WT and mutants in SENP1-knockdown cells or NTC (see online supplementary figure S13). We found that stable transfection of HIF-1α WT or HIF-1α mutants enhanced the stemness features including self-renewal (figure 7A), cell migration (figure 7B), chemoresistance (figure 7C), CD24+ cell population (figure 7D) and mRNA expression of Oct3/4 and Nanog (figure 7E). However, SENP1 knockdown suppressed the effects of HIF-1α WT, while the three HIF-1α mutants partially to almost completely rescued HIF-1α-induced enhancement of HCC cell stemness in shSENP1 cells.
Figure 7.
Mutation of SUMO sites rescues the loss of hypoxia-induced stemness in specific SUMO proteases 1 (SENP1)-knockdown hepatocellular carcinoma (HCC) cells. The effects of SENP1 knockdown on the stemness of HCC cells with overexpressing hypoxia-inducible factor (HIF)-1α wild type (WT), HIF-1α K391R, HIF-1α K477R or HIF-1α SM. In vitro abilities of self-renewal (A), migration (B) and chemoresistance (C), CD24+ cell population (D) and mRNA expression of stemness-related genes (E) were determined in hypoxic condition. SENP1 knockdown suppressed the effects of HIF-1α WT, while the three HIF-1α mutants partially to almost completely rescued HIF-1α-induced enhancement of HCC cell stemness in shSENP1 HCC cells. (F) A cartoon summarising our findings. HIF-1/2α induced by the hypoxic microenvironment increases the transcription of SENP1 in HCC cells. SENP1 represses the SUMOylation of HIF-1α at K391 and K477 by its SUMO protease activity, thus enhancing the stability and transcriptional activity of HIF-1α. These in turn upregulate the expression of HIF target genes, including SENP1 and stemness-related genes, and contribute to the increased stemness properties. (*p<0.05, **p<0.01, ***p<0.001, as compared with the negative control).
HIF-1α knockdown suppresses SENP1-enhanced cancer stemness in hypoxia
To further determine if the functional roles of SENP1 in hypoxia were dependent on the activity of HIF-1α, we knocked down the expression of HIF-1α or HIF-2α using lentivirus-mediated short hairpin RNA in Huh-7 cells which overexpressed SENP1 (see online supplementary figure S14A). The blockage of HIF-1α signal partly repressed the enhancement of cell migration (see online supplementary figure S14B). Furthermore, FACS results showed that HIF-1α knockdown inhibited the increase of CD24+ cells induced by SENP1 in hypoxia (see online supplementary figure S14C). There was also chemoresistance to doxorubicin induced by SENP1 OE (see online supplementary figure S14D). In contrast, these stemness-associated features induced by SENP1 were unchanged in the HIF-2α-knockdown HCC cells under hypoxia (see online supplementary figure S14A–D).
Discussion
In this study, our results show that SENP1 increases the stabilisation and transcriptional activity of HIF-1α under hypoxic condition via deSUMOylation. In addition, we have demonstrated that SENP1 is a direct target gene of HIFs, and a previously unrecognised positive feedback loop exists between HIF-1α and SENP1 and contributes to HCC stemness and tumourigenesis.
Our results have provided evidence of the effects of hypoxia and HIF-1/2α on liver cancer stemness. The stemness-associated features were significantly enhanced in HCC cells in hypoxia, while knockdown of HIF-1α or HIF-2α suppressed these hypoxia-induced effects. Previously, we reported that CD24 is a functional liver CSC marker that drives CSC through STAT3-mediated NANOG regulation. Promoter analysis demonstrates that a HRE located in the promoter/enhancer region is required for both hypoxia-induced and HIF-1α-dependent CD24 expression.36 Here, we consistently observed that hypoxia maintained the CD24+ subpopulation in HCC cells in a HIF-1/2α-dependent manner. Furthermore, CD24+ cell populations were maintained at a significantly higher level under hypoxia than normoxia in both sorted CD24+ and CD24− HCC cells, while exposure to digoxin abolished these hypoxia-induced effects in both CD24+ and CD24− HCC cells.
SUMOylation is known to regulate the functional activity of some transcription factors associated with the acquisition of CSC properties.39 So far, there is no report on whether SUMO signalling is involved in the regulation of liver CSC properties and hepatocarcinogenesis. We showed that in human HCCs, OE of SENP1 was significantly associated with more aggressive tumour behaviour in terms of more advanced tumour stage and presence of venous invasion. Functionally, SENP1 was able to enhance the liver CSC properties. Furthermore, we revealed that mechanistically, the regulation of SENP1 on hypoxia-induced enhancement of liver CSC properties was dependent on its catalytic activity, as inactivation of its catalytic activity by specific mutants resulted in loss of such ability in enhancing cancer stemness in hypoxia.
Increasing evidence has shown that HIF-1α is an important SUMO substrate, although HIF-1α SUMOylation and its effects vary among cell types.27 28 40 41 42 43 With regard to deSUMOylation, here, we show that SENP1 increased the stability and transcriptional activity of HIF-1α by deSUMOylation at K391 or K477 residues of HIF-1α in HCC cells. SENP1 knockdown significantly increased the accumulation of SUMO-1-conjugated HIF-1α and decreased the HIF-1α protein level under hypoxia in HCC cells. Consistently, hypoxia-induced transcriptional activity of HIF-1α on VEGF was inhibited by SENP1 knockdown in HCC cells. On the contrary, mutation of the SUMO sites at K391 and K477 of HIF-1α, either singly or combined, alleviated the inhibitory effect of SENP1 knockdown on HIF-1α activity and liver CSC properties. However, we failed to detect significant conjugation of SUMO with HIF-2α in HCC cells, although it was reported that HIF-2α was also regulated by SUMO and SENP1 in HeLa cells.43 Functionally, knockdown of HIF-1α, but not HIF-2α, partially suppressed SENP1-enhanced stemness of HCC cells in hypoxia. From our results, SENP1 is able to enhance HCC stemness properties by reducing the SUMOylation and increasing the stability and transcriptional activity of HIF-1α in hypoxia. In previous studies, SENP1 has been identified as an erythropoiesis regulator as well as being essential for the stability and activity of HIF-1α by deSUMOylation.26 SENP1 also contributes to the progression of prostate cancer through stabilising HIF-1α and enhancing VEGF production and angiogenesis.38 Nonetheless, it is the first time that SENP1 is reported to be involved in liver CSC properties and hepatocarcinogenesis via regulation of HIF-1α deSUMOylation in hypoxia.
Interestingly, we found that SENP1 was induced by hypoxia as a direct target of HIF-1α and HIF-2α in HCC cells, in line with a previous report on endothelial cell model.38 Of significance, we have demonstrated a previously unrecognised positive feedback loop between HIF-1α and SENP1, which contributes to the maintenance of HCC stemness and tumourigenesis under hypoxia. Overall, as a summary of our findings in this study (figure 7F), HIF-1/2α, as induced by hypoxic microenvironment, increases the transcription of SENP1 in HCC cells. SENP1 represses the SUMOylation of HIF-1α at K391 and K477 by its SENPs activity, which decreases the stability and transcriptional activity of HIF-1α, thus increasing the expression of HIF target genes, including SENP1 and stemness-related genes (Nanog, Oct4 and CD24), and contributing to the enhancement of stemness properties. Developing new inhibitors that specifically target SENP1 may offer a novel therapeutic approach to block HCC growth, metastasis and recurrence.
Acknowledgments
We thank Professor ETH Yeh of the University of Texas, Houston Health Science Center for providing plasmids. We also thank LKS Faculty of Medicine at The University of Hong Kong for the Faculty Core Facility. IOL Ng is Loke Yew Professor in Pathology.
Footnotes
Contributors: C-PC and IOLN provided study concept and design. C-PC, CC-LW, AK-LK, DW-HH, EY-TL, Y-MT, TK-WL and IOLN collected and analysed the data. CPC, CCW, DWH, LKC, TKL and IOLN interpreted the data. C-PC, CC-LW, AK-LK, EY-TL, Y-MT, JM-FL and TK-WL performed the experiments. T-TC, KS-HC, ACYC, RC-LL and IOLN collected the patients' samples. C-PC and IOLN wrote the manuscript. All authors approved the final version of manuscript.
Funding: This work was supported by the Hong Kong Research Grants Council (RGC) Theme-based Research Scheme (T12-704116-R), RGC General Research Fund (17111315), Hong Kong Scholars programme (81572373), SK Yee Medical Research Fund 2011 and Lee Shiu Family Foundation.
Competing interests: None declared.
Patient consent: Obtained.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Lee TK, Cheung VC, Ng IO. Liver tumor-initiating cells as a therapeutic target for hepatocellular carcinoma. Cancer Lett 2013;338:101–9. 10.1016/j.canlet.2012.05.001 [DOI] [PubMed] [Google Scholar]
- 2.Ma S, Chan KW, Hu L, et al. Identification and characterization of tumorigenic liver cancer stem/progenitor cells. Gastroenterology 2007;132:2542–56. 10.1053/j.gastro.2007.04.025 [DOI] [PubMed] [Google Scholar]
- 3.Yang ZF, Ho DW, Ng MN, et al. Significance of CD90+ cancer stem cells in human liver cancer. Cancer Cell 2008;13:153–66. 10.1016/j.ccr.2008.01.013 [DOI] [PubMed] [Google Scholar]
- 4.Yamashita T, Ji J, Budhu A, et al. EpCAM-positive hepatocellular carcinoma cells are tumor-initiating cells with stem/progenitor cell features. Gastroenterology 2009;136:1012–24. 10.1053/j.gastro.2008.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee TK, Castilho A, Cheung VC, et al. CD24(+) liver tumor-initiating cells drive self-renewal and tumor initiation through STAT3-mediated NANOG regulation. Cell stem cell 2011;9:50–63. 10.1016/j.stem.2011.06.005 [DOI] [PubMed] [Google Scholar]
- 6.Lee TK, Cheung VC, Lu P, et al. Blockade of CD47-mediated cathepsin S/protease-activated receptor 2 signaling provides a therapeutic target for hepatocellular carcinoma. Hepatology 2014;60:179–91. 10.1002/hep.27070 [DOI] [PubMed] [Google Scholar]
- 7.Keith B, Johnson RS, Simon MC. HIF1α and HIF2α: sibling rivalry in hypoxic tumour growth and progression. Nat Rev Cancer 2011;12:9–22. 10.1038/nrc3183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng SS, Chen XH, Yin X, et al. Prognostic significance of HIF-1α expression in hepatocellular carcinoma: a meta-analysis. PLoS ONE 2013;8:e65753 10.1371/journal.pone.0065753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liang Y, Zheng T, Song R, et al. Hypoxia-mediated sorafenib resistance can be overcome by EF24 through Von Hippel-Lindau tumor suppressor-dependent HIF-1α inhibition in hepatocellular carcinoma. Hepatology 2013;57:1847–57. 10.1002/hep.26224 [DOI] [PubMed] [Google Scholar]
- 10.Zhao D, Zhai B, He C, et al. Upregulation of HIF-2α induced by sorafenib contributes to the resistance by activating the TGF-α/EGFR pathway in hepatocellular carcinoma cells. Cell Signal 2014;26:1030–9. 10.1016/j.cellsig.2014.01.026 [DOI] [PubMed] [Google Scholar]
- 11.Lin Q, Yun Z. Impact of the hypoxic tumor microenvironment on the regulation of cancer stem cell characteristics. Cancer Biol Ther 2010;9:949–56. 10.4161/cbt.9.12.12347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pietras A, Gisselsson D, Ora I, et al. High levels of HIF-2alpha highlight an immature neural crest-like neuroblastoma cell cohort located in a perivascular niche. J Pathol 2008;214:482–8. 10.1002/path.2304 [DOI] [PubMed] [Google Scholar]
- 13.Pietras A, Hansford LM, Johnsson AS, et al. HIF-2alpha maintains an undifferentiated state in neural crest-like human neuroblastoma tumor-initiating cells. Proc Natl Acad Sci USA 2009;106:16805–10. 10.1073/pnas.0904606106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Z, Bao S, Wu Q, et al. Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer Cell 2009;15:501–13. 10.1016/j.ccr.2009.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muramatsu S, Tanaka S, Mogushi K, et al. Visualization of stem cell features in human hepatocellular carcinoma reveals in vivo significance of tumor-host interaction and clinical course. Hepatology 2013;58:218–28. 10.1002/hep.26345 [DOI] [PubMed] [Google Scholar]
- 16.Flotho A, Melchior F. Sumoylation: a regulatory protein modification in health and disease. Annu Rev Biochem 2013;82:357–85. 10.1146/annurev-biochem-061909-093311 [DOI] [PubMed] [Google Scholar]
- 17.Johnson ES. Protein modification by SUMO. Annu Rev Biochem 2004;73:355–82. 10.1146/annurev.biochem.73.011303.074118 [DOI] [PubMed] [Google Scholar]
- 18.Hay RT. SUMO: a history of modification. Mol Cell 2005;18:1–12. 10.1016/j.molcel.2005.03.012 [DOI] [PubMed] [Google Scholar]
- 19.Kim JH, Baek SH. Emerging roles of desumoylating enzymes. Biochim Biophys Acta 2009;1792:155–62. 10.1016/j.bbadis.2008.12.008 [DOI] [PubMed] [Google Scholar]
- 20.Deng R, Zhao X, Qu Y, et al. Shp2 SUMOylation promotes ERK activation and hepatocellular carcinoma development. Oncotarget 2015;6:9355–69. 10.18632/oncotarget.3323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang QF, Tian YW, Shen Q, et al. SENP2 regulated the stability of β-catenin through WWOX in hepatocellular carcinoma cell. Tumour Biol 2014;35:9677–82. 10.1007/s13277-014-2239-8 [DOI] [PubMed] [Google Scholar]
- 22.Liu J, Sha M, Wang Q, et al. Small ubiquitin-related modifier 2/3 interacts with p65 and stabilizes it in the cytoplasm in HBV-associated hepatocellular carcinoma. BMC cancer 2015;15:675 10.1186/s12885-015-1665-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mukhopadhyay D, Dasso M. Modification in reverse: the SUMO proteases. Trends Biochem Sci 2007;32:286–95. 10.1016/j.tibs.2007.05.002 [DOI] [PubMed] [Google Scholar]
- 24.Yang ST, Yen CJ, Lai CH, et al. SUMOylated CPAP is required for IKK-mediated NF-κB activation and enhances HBx-induced NF-κB signaling in HCC. J Hepatol 2013;58:1157–64. 10.1016/j.jhep.2013.01.025 [DOI] [PubMed] [Google Scholar]
- 25.Tomasi ML, Tomasi I, Ramani K, et al. S-adenosyl methionine regulates ubiquitin-conjugating enzyme 9 protein expression and sumoylation in murine liver and human cancers. Hepatology 2012;56:982–93. 10.1002/hep.25701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng J, Kang X, Zhang S, et al. SUMO-specific protease 1 is essential for stabilization of HIF1alpha during hypoxia. Cell 2007;131:584–95. 10.1016/j.cell.2007.08.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berta MA, Mazure N, Hattab M, et al. SUMOylation of hypoxia-inducible factor-1α reduces its transcriptional activity. Biochem Biophys Res Commun 2007;360:646–52. 10.1016/j.bbrc.2007.06.103 [DOI] [PubMed] [Google Scholar]
- 28.Carbia-Nagashima A, Gerez J, Perez-Castro C, et al. RSUME, a small RWD-containing protein, enhances SUMO conjugation and stabilizes HIF-1α during hypoxia. Cell 2007;131:309–23. 10.1016/j.cell.2007.07.044 [DOI] [PubMed] [Google Scholar]
- 29.Parhira S, Zhu GY, Jiang RW, et al. 2′-Epi-uscharin from the latex of Calotropis gigantea with HIF-1 inhibitory activity. Sci Rep 2014;4:4748 10.1038/srep04748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wong CC, Tse AP, Huang YP, et al. Lysyl oxidase-like 2 is critical to tumor microenvironment and metastatic niche formation in hepatocellular carcinoma. Hepatology 2014;60:1645–58. 10.1002/hep.27320 [DOI] [PubMed] [Google Scholar]
- 31.Ma S, Lee TK, Zheng BJ, et al. CD133+ HCC cancer stem cells confer chemoresistance by preferential expression of the Akt/PKB survival pathway. Oncogene 2008;27:1749–58. 10.1038/sj.onc.1210811 [DOI] [PubMed] [Google Scholar]
- 32.Li C, Heidt DG, Dalerba P, et al. Identification of pancreatic cancer stem cells. Cancer Res 2007;67:1030–7. 10.1158/0008-5472.CAN-06-2030 [DOI] [PubMed] [Google Scholar]
- 33.Ke J, Wu X, Wu X, et al. A subpopulation of CD24+ cells in colon cancer cell lines possess stem cell characteristics. Neoplasma 2012;59:282–8. 10.4149/neo_2012_036 [DOI] [PubMed] [Google Scholar]
- 34.Vermeulen L, Todaro M, de Sousa Mello F, et al. Single-cell cloning of colon cancer stem cells reveals a multi-lineage differentiation capacity. Proc Natl Acad Sci USA 2008;105:13427–32. 10.1073/pnas.0805706105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ishizawa K, Rasheed ZA, Karisch R, et al. Tumor-initiating cells are rare in many human tumors. Cell Stem Cell 2010;7:279–82. 10.1016/j.stem.2010.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomas S, Harding MA, Smith SC, et al. CD24 is an effector of HIF-1-driven primary tumor growth and metastasis. Cancer Res 2012;72:5600–12. 10.1158/0008-5472.CAN-11-3666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods 2009;347:70–8. 10.1016/j.jim.2009.06.008 [DOI] [PubMed] [Google Scholar]
- 38.Xu Y, Zuo Y, Zhang H, et al. Induction of SENP1 in endothelial cells contributes to hypoxia-driven VEGF expression and angiogenesis. J Biol Chem 2010;285:36682–8. 10.1074/jbc.M110.164236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Du L, Li YJ, Fakih M, et al. Role of SUMO activating enzyme in cancer stem cell maintenance and self-renewal. Nat Commun 2016;7:12326 10.1038/ncomms12326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Antico Arciuch VG, Tedesco L, Fuertes M, et al. Role of RSUME in inflammation and cancer. FEBS Lett 2015;589:3330–5. 10.1016/j.febslet.2015.07.048 [DOI] [PubMed] [Google Scholar]
- 41.Li J, Xu Y, Long XD, et al. Cbx4 governs HIF-1α to potentiate angiogenesis of hepatocellular carcinoma by its SUMO E3 ligase activity. Cancer Cell 2014;25:118–31. 10.1016/j.ccr.2013.12.008 [DOI] [PubMed] [Google Scholar]
- 42.Mei Z, Jiao H, Wang W, et al. Polycomb chromobox 4 enhances migration and pulmonary metastasis of hepatocellular carcinoma cell line MHCC97L. Sci China Life Sci 2014;57:610–17. 10.1007/s11427-014-4663-9 [DOI] [PubMed] [Google Scholar]
- 43.van Hagen M, Overmeer RM, Abolvardi SS, et al. RNF4 and VHL regulate the proteasomal degradation of SUMO-conjugated Hypoxia-Inducible Factor-2α. Nucleic Acids Res 2010;38:1922–31. 10.1093/nar/gkp1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gutjnl-2016-313264supp001.pdf (2.2MB, pdf)