Abstract
The DICER1 syndrome is associated with a variety of rare benign and malignant tumors, including pleuropulmonary blastoma (PPB), cystic nephroma (CN) and Sertoli-Leydig cell tumor (SLCT). The prevalence and penetrance of pathogenic DICER1 variation in the general population is unknown.
We examined three publicly-available germline whole exome sequence datasets: Exome Aggregation Consortium (ExAC), 1000 Genomes (1000G), and the Exome Sequencing Project (ESP). To avoid over-estimation of pathogenic DICER1 variation from cancer-associated exomes, we excluded The Cancer Genome Atlas (TCGA) variants from ExAC. All datasets were annotated with snpEFF and ANNOVAR and variants were classified into four categories: likely benign (LB), unknown significance (VUS), likely pathogenic (LP), or pathogenic (P).
The prevalence of DICER1 P/LP variants was 1:870 to 1:2,529 in ExAC-nonTCGA (53,105 exomes) estimated by metaSVM and REVEL/CADD, respectively. A more stringent prevalence calculation considering only loss-of-function and previously-published pathogenic variants detected in ExAC-nonTCGA, yielded a prevalence of 1:10,600.
Despite the rarity of most DICER1 syndrome tumors, pathogenic DICER1 variation is more common than expected. If confirmed, these findings may inform future sequencing-based newborn screening programs for PPB, CN and SLCT, in which early detection improves prognosis.
Keywords: DICER1, DICER1 syndrome, prevalence estimates, pleuropulmonary blastoma, Sertoli-Leydig cell tumor
INTRODUCTION
DICER1 is an RNaseIII endonuclease that is crucial for processing pre-microRNA into mature, active microRNA (miRNA)1. It is highly-conserved, with two functional enzymatic domains: RNaseIIIa and RNaseIIIb 2. Germline loss of function (LOF) mutations in DICER1 are associated with an increased risk of a neoplasm predisposition syndrome, typically affecting children and young adults 3. Somatic missense variation in five “hotspot” codons in the RNaseIIIb domain (E1705, D1709, G1809, D1810, E1813) are observed in virtually all DICER1-associated tumors; mosaicism for missense mutations in these hotspot codons confers a more severe phenotype 4. About 87% of individuals with the DICER1 syndrome have inherited a germline DICER1 pathogenic variant from one parent, illustrating an autosomal dominant inheritance pattern, with incomplete penetrance 4,5. Clinical features of the DICER1 syndrome include a variety of rare benign and malignant tumors, including pleuropulmonary blastoma (PPB), cystic nephroma, Sertoli-Leydig cell tumor, thyroid cancer, multinodular goiter (MNG), rhabdomyosarcoma, and pineoblastoma 3,6–10.
PPB, the most common pediatric primary lung cancer, is the most morbid manifestation of the DICER1 syndrome, and typically presents in children below age 7. DICER1 testing identifies carriers in at-risk families and provides an opportunity for early PPB detection (through chest imaging) in young children. This is especially important since early diagnosis of PPB is associated with improved clinical outcomes 11. In the future, sequence-based newborn screening may play an important role in early detection and diagnosis if the population prevalence of pathogenic variation and its penetrance in DICER1 is high enough to justify this approach. Today, early diagnosis in a child typically requires the previous recognition of a DICER1-associated tumor or DICER1-associated phenotype (e.g., early-onset MNG) in a family member 10. In our experience, the majority of DICER1 syndrome families are recognized after the diagnosis of PPB, cystic nephroma or Sertoli-Leydig cell tumor of the ovary. However, approximately 95% of children with a DICER1 pathogenic variant do not develop any DICER1-specific malignancy by age 10 years (unpublished data). Given this reduced penetrance, germline DICER1 pathogenic variants may be more common than the rare occurrence of PPB would suggest. Most pathogenic variation reported to date in DICER1 has been ascertained predominantly in children with rare DICER1-associated tumors. The majority of these are LOF variants, but their prevalence in the general population is unknown.
The availability of large-scale, whole-exome date from three publicly available datasets, the Exome Aggregation Consortium (ExAC)12, 1000 Genomes (1000G)13, and Exome Sequencing Project (ESP)14 permits the estimation of pathogenic variation within individual genes on a population basis 15–17. A better understanding of the prevalence of pathogenic DICER1 variants in the general population (and ultimately the penetrance of DICER1-associated phenotypes) is important in assessing the potential value of newborn screening.
MATERIALS AND METHODS
Large, publicly available datasets
DICER1 variants were identified in the ExAC data set (53,105 exomes, accessed 10/16/16)12. Although it is not clear that there is an overall cancer predisposition in DICER1 pathogenic variant carriers outside of the few select organ sites/histologies, we decided to limit the dataset to non-The Cancer Genome Atlas (TCGA) cases. Since ExAC-nonTCGA comprises a subset of 1000G and ESP data, we also obtained DICER1 variation from 1000G phase 3 (2,504 exomes, accessed 12/28/16) 13 and ESP (6,503 exomes, accessed 12/28/16) to analyze datasets individually14. In addition to the publicly available databases, we utilized the whole exome sequencing controls (998 exomes) 18–20. To capture all variants found in general populations, we did not filter any variants by minor allele frequency (MAF).
Variant annotation, filtering, and classification
All exonic and splice-site region (<10 intronic base pairs from intron/exon boundary) variants from the canonical DICER1 transcript (NM_177438.2), including missense, frameshift, nonsense, and synonymous variants were included. Multi-allelic, deep intronic, and UTR variants were excluded in this analysis to focus on the protein-coding regions. SnpEff21 was used to annotate variants and ANNOVAR22 was used to predict pathogenicity in silico, obtain population allele frequencies from different databases, and obtain previously reported variants from ClinVar (2017-01-25 version). Annotation by ClinVar and the Human Gene Mutation Database (HGMD, Qiagen and Institute of Medical Genetics, Cardiff, Wales, UK; version Professional 2017.1) were used to identify previously recognized and interpreted variants. From our CLIA-certified DICER1 sequencing laboratory, we developed a database of pathogenic and non-pathogenic DICER1 variation from 391 subjects (233 families) with known or suspected DICER1-associated tumors. DICER1 was sequenced using next-generation methods 23.
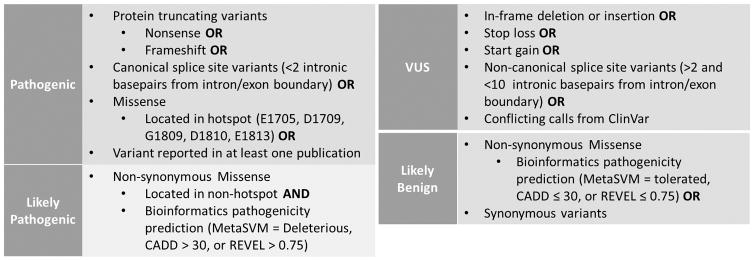
A scheme (Figure 1) to classify variants into pathogenic (P), likely pathogenic (LP), variant of uncertain significance (VUS), and likely benign (LB) categories was modelled after the guidelines proposed by the American College of Medical Genetics and Association for Molecular Pathology 24. In brief, variants that are LOF (e.g., nonsense and frameshift), located in the canonical splice site (≤2 intronic basepairs from the intron/exon boundary), missense located in DICER1 hotspots (e.g., E1705, D1709, G1809, D1810, E1813) or credibly reported as pathogenic in at least one publication are classified as P. Classification as LP required a non-synonymous missense variant to be outside a DICER1 hotspot locus and harbor a bioinformatics pathogenicity prediction of “Deleterious” by metaSVM, a REVEL25 score > 0.75 or a CADD26 score > 30 (top 0.01% of the predicted deleteriousness).
Figure 1. Schematic for DICER1 variant classification.
Variants are classified into pathogenic, likely pathogenic, variant of uncertain significance (VUS), or likely benign depending on type of variants, metaSVM, REVEL, and CADD prediction, and any previous, published report. Type of variant was based on the canonical transcript NM_177438.2.
Statistical Testing
Chi-square test was used to compare gender differences using SPSS version 21.
RESULTS
Classification of unique DICER1 variants from population databases
We identified 576 (ExAC-nonTCGA), 123 (1000G), and 150 (ESP) unique exonic and splice-site region DICER1 variants (Table 1, Tables S1, S2, and S3). Most of these variants (ExAC-nonTCGA = 92%; 1000G =94%; ESP = 96%), were categorized as LB. We found 7 in-frame variants (ExAC-nonTCGA = 5; ESP = 2) that we categorized as VUS, given the limited understanding of how in-frame DICER1 variants affect phenotype or protein function. There were 48 unique variants (ExAC-nonTCGA = 37; 1000G = 7; ESP = 4), 21 unique variants (ExAC-nonTCGA = 15; 1000G = 3; ESP = 3), and 20 unique variants (ExAC-nonTCGA = 17; 1000G = 0; ESP = 3) classified as P/LP because they were LOF or deleterious by metaSVM, REVEL, or CADD prediction, respectively (Table 1). One missense variant found in both ExAC and ESP (Gln1580His) was previously published and classified as LP, based on the metaSVM prediction 27. However, our review of the primary literature reporting the Gln1580His variant revealed that it was of questionable pathogenicity according to the author; this variant was thus re-classified for the current analysis as VUS (Table S4).
Table 1. Number of DICER1 variants in each category.
Distribution of DICER1 variant classification in three publicly available datasets. Each category is separated into type of variants. Abbreviation: Exome Aggregation Consortium, non-The Cancer Genome Sequencing Atlas (TCGA; cancer) variants (ExAC-nonTCGA), Exome Sequencing Project (ESP), variant of uncertain significance (VUS). MetaSVM: Meta-Analytic Support Vector Machine, REVEL: Rare Exome Variant Ensemble Learner, CADD: Combined Annotation Dependent Depletion, syn: synonymous
| ExAC-nonTCGA (53,105 exome) | 1000 Genome (2,504 exome) | ESP (6,503 exome) | |||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Total Variants | 576 | 123 | 150 | ||||||
|
| |||||||||
| Likely Benign (LB) | metaSVM | REVEL | CADD | metaSVM | REVEL | CADD | metaSVM | REVEL | CADD |
|
| |||||||||
| 490 Total LB | 512 Total LB | 510 Total LB | 105 Total LB | 109 Total LB | 112 Total LB | 126 Total LB | 127 Total LB | 127 Total LB | |
| 217 syn | 217 syn | 217 syn | 52 syn | 52 syn | 52 syn | 59 syn | 59 syn | 59 syn | |
| 273 missense | 295 missense | 293 missense | 53 missense | 57 missense | 60 missense | 67 missense | 68 missense | 68 missense | |
|
| |||||||||
| Variant of Uncertain Significance (VUS) | 49 Total VUS | 11 Total VUS | 20 Total VUS | ||||||
| 5 in-frame deletion | 0 | 2 in-frame deletion | |||||||
| 43 splice-site | 11 splice-site | 17 splice-site | |||||||
| 1 missense | 0 | 1 missense | |||||||
|
| |||||||||
| Likely Pathogenic (LP) | metaSVM | REVEL | CADD | metaSVM | REVEL | CADD | metaSVM | REVEL | CADD |
|
| |||||||||
| 32 missense | 10 missense | 12 missense | 7 missense | 3 missense | 0 missense | 4 missense | 3 missense | 3 missense | |
|
| |||||||||
| Pathogenic (P) | 5 Total P | 0 Total P | 0 Total P | ||||||
| 1 splice donor | |||||||||
| 4 nonsense | |||||||||
|
| |||||||||
| Total P/LP | metaSVM | REVEL | CADD | metaSVM | REVEL | CADD | metaSVM | REVEL | CADD |
|
| |||||||||
| 37 Total P/LP | 15 Total P/LP | 17 Total P/LP | 7 Total P/LP | 3 Total P/LP | 0 Total P/LP | 4 Total P/LP | 3 Total P/LP | 3 Total P/LP | |
| 32 missense | 10 missense | 12 missense | 7 missense | 3 missense | 4 missense | 3 missense | 3 missense | ||
| 1 splice donor | 1 splice donor | 1 splice donor | |||||||
| 4 nonsense | 4 nonsense | 4 nonsense | |||||||
In addition to our own variant classification, we reviewed the ExAC constraint metrics for synonymous, missense and LOF DICER1 variants in ExAC-nonTCGA data (Table S5). The number of expected and observed synonymous DICER1 variants was approximately equal (Z-score = 0.083; median Z-score in ExAC (17,739 genes) = 0.054). There were significantly fewer observed DICER1 missense variants (Z-score = 4.24; median Z-score in ExAC (17,739 genes) = 0.542), consistent with DICER1 being intolerant to variation. Furthermore, there were many fewer DICER1 LOF variants than expected (probability of LOF intolerance, pLI = 1.00), also consistent with extreme intolerance to variation. In 17,739 genes in ExAC, 3,126 (17.6%) had pLI>0.9.
Prevalence of pathogenic DICER1 variants
To calculate a prevalence rate of all pathogenic (P/LP) variants, we determined the total allele count (AC) of the unique P/LP variants (Table 2), as determined by metaSVM. Using these, we estimated P/LP variant prevalence to range from 1:310 (1000G) through 1:870 (ExAC-nonTCGA) to 1:1,300 (ESP). However, in our classification scheme, non-hotspot, unpublished missense variants deemed “Deleterious” by the metaSVM score were classified as LP, potentially inflating the prevalence of damaging variation. Therefore, we used two independent in silico meta-score prediction tools, REVEL25 and CADD26, to validate our findings using stringent criteria. We, compared the overlap of LP variants from the three independent bioinformatic tools (metaSVM, REVEL, and CADD); in all three databases (ExAC-nonTCGA, 1000G, and ESP), REVEL overlapped 100% and CADD overlapped 50% with metaSVM (Figure S1), suggesting that the most deleteriousness variants were identified, regardless of method. REVEL and CADD found identical P/LP prevalence in ESP (1:2168) and ExAC-nonTCGA (1: 2529), about 2–3X less frequent than the estimate from metaSVM (Table 2).
Table 2. Allele count of pathogenic (P) and likely pathogenic (LP) variation.
Table shows the number of unique variants and the allele count (AC) of each type of variant. Prevalence was calculated by dividing allele count by the total number of exomes for a given study. “P/LP Prevalence” is sum of all LP and P variants; “Loss of function (LOF) Prevalence” includes only truncating variants.
| ExAC-nonTCGA (53,105 exomes) | 1000 Genome (2,504 exomes) | ESP (6,503 exomes) | ||||
|---|---|---|---|---|---|---|
| # of unique variants | AC | # of unique variants | AC | # of unique variants | AC | |
| Likely Pathogenic metaSVM | 32 missense | 56 | 7 missense | 8 | 4 missense | 5 |
| Likely Pathogenic REVEL | 10 missense | 16 | 3 missense | 3 | 3 missense | 3 |
| Likely Pathogenic CADD | 12 missense | 16 | 0 missense | 0 | 3 missense | 3 |
| Pathogenic | 1 canonical splice 4 stop gained |
1 4 |
0 canonical splice 0 stop gained |
0 0 |
0 canonical splice 0 stop gained |
0 0 |
| Pathogenic Total AC | 5 | 0 | 0 | |||
| P/LP Prevalence metaSVM | 1:870 | 1:310 | 1:1,300 | |||
| P/LP Prevalence REVEL | 1:2,529 | 1:835 | 1:2,168 | |||
| P/LP Prevalence CADD | 1:2,529 | N/A | 1:2,168 | |||
| LOF (P) Prevalence | 1:10,600 | N/A | N/A | |||
To better categorize unpublished missense variants, we examined our in-house DICER1 variant database. Since the DICER1 syndrome is autosomal dominant, we assumed that in individuals with a known pathogenic or LOF DICER1 allele, additional co-segregating germline DICER1 variants were non-pathogenic. However, of 35 unique LP missense variants from ExAC-nonTCGA and ESP, none overlapped with 107 missense variants in our DICER1 database (data not shown). Therefore, we conducted a more stringent prevalence calculation by only considering LOF (e.g., truncating, canonical splice sites) and previously published pathogenic variants. With these criteria, 1000G and ESP did not contain any P variants, potentially due to their small population size. For ExAC-nonTCGA, the prevalence rate was 1:10,600 (Table 2). We next examined the 998 whole exome sequencing controls but did not find any P/LP variants (data not shown). Although we did not filter by MAF, review of P/LP variants identified in each database showed that all had MAF < 0.1%.
Ethnicity and gender distribution of pathogenic DICER1 variants
Next, we evaluated the ethnicity and gender distribution of P/LP variants (determined by metaSVM only). The DICER1 syndrome is not known to exclusively affect any particular ethnic or racial group. However, in ExAC-nonTCGA, there were more P variants (by AC) in non-Finnish Europeans, resulting in a higher AF for this group compared with other ethnic groups. This may be due to a larger sample size in this group. Only limited observations on ethnic distribution can be made in ESP, due to its small number of variants and inclusion of only two ethnic groups (white and African-American) (Table S6). In ExAC-nonTCGA P variants, there was a significantly higher AC in males versus females (X2=5.000, p=0.0253); however, the absolute counts are small. LP variants in both ExAC-nonTCGA and 1000G showed no significant differences (ExAC-nonTCGA: X2=1.143, p=0.285; 1000G: X2=0.500, p=0.4795) (Table S7). ESP was excluded from this analysis due to its lack of gender information.
DISCUSSION
The hallmark tumors of the DICER1 syndrome (PPB, cystic nephroma, Sertoli-Leydig cell tumor) are rare. The prevalence of these tumors is unknown but collectively are estimated to occur in a few hundred individuals each year in the United States. Because of this phenotypic rarity, the prevalence of damaging DICER1 variants in the general population was also assumed to be rare. The current investigation, to our knowledge, is the first to quantify such variation. In this study, we classified germline DICER1 variants from three large, publicly available datasets and found the estimated prevalence of pathogenic DICER1 variants ranged from 1:310 – 1:10,600. The latter, our most conservative estimate, is based on the frequency of truncating and previously-published variants (P only; excludes LP) in the non-cancer portion of the ExAC database (53,105 exomes), and it predicts that approximately 30,000 Americans harbor pathogenic DICER1 variants. It is important to distinguish between the prevalence of pathogenic DICER1 variants and the penetrance of a phenotype associated with those variants. Although our analysis did not include any pediatric cancer cases, our results support the observation that DICER1-associated pediatric tumors are of low-to-moderate penetrance in DICER1 families since the prevalence of pathogenic DICER1 variants is more frequent than expected. The penetrance of other DICER1-associated features such as MNG (~75% by age 40 in women) 10 and macrocephaly (larger head circumference; ~40% in males and females) 28 is much higher in clinically- ascertained populations compared with non-DICER1 carriers; it may be lower in on a population-ascertained basis. The tumor risk associated with pathogenic DICER1 variants among older adults needs additional study. (This is a goal of the NCI DICER1 syndrome study; more information can be found at http://ppb.cancer.gov).
The >30-fold difference in our prevalence estimates is driven by different dataset sample sizes and whether we include LP variants, a category entirely comprised of in silico-predicted missense variation which lack published or publicly available functional or clinical data. To refine our estimates, we used two additional in silico-prediction tools, REVEL and CADD and applied stringent criteria for deleteriousness. With these, we observed a P/LP DICER1 prevalence in the larger datasets (ExAC-nonTCGA and ESP) of 1: 2,168 – 1: 2,529. Using these lower estimates, the DICER1 P/LP prevalence ranges from 1: 2,168 – 1: 10,600, a ~5-fold difference.
Since the DICER1 syndrome is relatively recently recognized, the number of curated, annotated variants is limited and mainly comprised of LOF variation. To address this, we reviewed our in-house database of known pathogenic and co-segregating, presumed non-pathogenic DICER1 variants from our CLIA-certified sequencing laboratory. However, there was no overlap between the 35 unique missense variants from ExAC, 1000G and ESP and the 107 missense DICER1 variants observed by our lab. The proportion of germline missense variants among all mutation classes varies across tumor suppressor genes, from 73% (TP53) 29 to ~20% (NF1) 30. In our published comprehensive analysis of clinically significant DICER1 variation (children presenting with PPB), only 4% of germline pathogenic variation was missense 4. It is possible that non-LOF DICER1 (missense) carriers are under-ascertained due to a milder phenotype or because they develop features later in life. To develop a better understanding of non-LOF DICER1 variants, population-based sequence data linked with comprehensive phenotyping will likely enhance our ability to characterize the range of syndrome-associated clinical features. Furthermore, reporting of co-segregating, presumed benign DICER1 variation by commercial testing laboratories would assist in classification of missense variants.
The widespread availability of population-based exome sequencing has prompted a revision in our understanding of tumor-predisposition disorders; there is much more apparently non-penetrant, pathogenic variation in BRCA1/2 (1:300–1:500)31, TP53 (1:500) (de Andrade, under review) and mismatch repair genes (Lynch syndrome 1:440)32 than previously expected. A revision of the cancer penetrance of these disorders is needed. Well-known, highly penetrant genetic disorders with population prevalence frequencies spanning our DICER1 pathogenic variation estimates include sickle cell anemia (1/500 African American births)33, Down syndrome (1/700 live births)34, velo-cardio-facial syndrome (1/2,000)35, cystic fibrosis (1/2,500 white Americans)36, neurofibromatosis type 1 (1/3,000)37, and Duchenne muscular dystrophy (1/5,000 – 1/7,500 boys) 38, hemophilia A (factor VIII deficiency; 1/6,500 live male births) 39, Williams syndrome (1/7,500)40 and Marfan syndrome (1/7,500 – 1/10,000) 41. As noted, the penetrance related to the pathogenic DICER1 variants we have reported here needs to be determined.
There are limitations to our analyses. Most prominently, the phenotype data available from publicly-available population databases are limited. We have previously shown that thyroid cancer prevalence is higher in DICER1 carriers versus non-carriers 10. Since ExAC does not report the types of cancer that are included in the dataset, we excluded the TCGA component of ExAC to avoid inflation of cancer-associated variation. In addition, ExAC-nonTCGA and ESP do not share individual genotype data; therefore, we cannot exclude the possibility of one person possessing multiple P/LP DICER1 variants. However, in 1000G, we were able to confirm that no subject harbored more than one P or LP alleles.
Our findings, if validated in other large, population-based datasets, suggest that pathogenic DICER1 variants are more common than previously expected. The DICER1-associated tumor with the greatest mortality is PPB11, although there is also significant potential morbidity associated with cystic nephroma, Sertoli-Leydig cell tumor, renal sarcoma, pineoblastoma and thyroid cancer. Malignant PPB types II and III arise from the precursor type I PPB, which is readily detected on routine chest computed tomography and potentially by radiograph as a lung cyst. Given the limited window of development of type I PPB (95% by age 2.5 years)11, availability of good radiographic screening tools and the efficacy of complete resection, PPB may be amenable to a newborn screening program. Historically, newborn screening has been performed to detect very rare, highly-penetrant biochemical genetic disorders with devastating phenotypes that, once identified, can be ameliorated with dietary and medical management. However, there are significant differences between the DICER1 syndrome and the metabolic disorders detected by traditional state screening programs. In the DICER1 syndrome, the effectiveness of a screening program will likely be driven by several factors. These include the population prevalence of pathogenic DICER1 variation (as first estimated here and in need of validation), penetrance of PPB type I (precursor, pre-malignant) cysts, rate of malignant transformation of those cysts and parental and physician tolerance for a screening regimen which with current technology requires ionizing radiation. The penetrance of type I PPB and their rate of malignant transformation are unknown. In screening the general population for rare conditions, a small decrease in specificity has a significant impact on positive predictive value (PPV). As PPV decreases, the number of false positives increases. However, depending on the condition, the tolerance for false-positives will likely vary. For disorders without readily apparent precursor lesions or symptoms and thus a pre-emptive requirement for surgery or device implantation (e.g., BRCA1/2, Lynch syndrome, long QT syndrome), the tolerance for false positives is likely low or very low. For disorders with readily available, low-cost, low-morbidity screening or exposure-avoidance (e.g., familial hypercholesterolemia, Marfan syndrome, hemochromatosis, alpha-1-anti-trypsin deficiency), the tolerance for false positives is likely higher42. The tolerance for false positives screening results in the DICER1 syndrome is unknown, but likely falls between these two extremes, given the current need to use ionizing radiation to detect the precursor PPB type I. A better understanding of the penetrance of PPB type I cysts and the rate of, and risk factors for, malignant transformation of those cysts is needed to inform these considerations. The early detection of other DICER1-associated tumors has other benefits: prompt diagnosis of cystic nephroma can result in partial rather than total nephrectomy; stage 1a Sertoli-Leydig cell tumor is associated with improved survival and avoidance of chemotherapy. Other screening strategies under development (e.g., MRI, blood-based biomarkers) may influence the tolerance for screening.
We are first to report a range of DICER1 pathogenic variant prevalence in the general population. Such variation appears to be more common than expected, based on the known prevalence of DICER1-associated tumors. Our results, if confirmed, open the door to the possible development of sequence-based screening programs for the DICER1 syndrome.
Supplementary Material
Novelty and impact.
DICER1 syndrome is a cancer-predisposition syndrome that is associated with rare benign and malignant tumors. As a recently identify syndrome, the general population prevalence is unknown. In this paper, utilizing publically available datasets, we identified that the prevalence of pathogenic/likely pathogenic DICER1 variation ranges from 1:870–1:1300, and if only considering loss of function variants, 1:10,600. As DICER1 variation is more common than expected, if confirmed, it could potentially inform sequencing-based newborn screening programs.
Acknowledgments
This work was supported by the Intramural Research Program of the Division of Cancer Epidemiology and Genetics of the National Cancer Institute, Bethesda, MD and National Cancer Institute R01CA143167 (to DAH). Support for the International Pleuropulmonary Blastoma Registry is derived in part from the Pine Tree Apple Tennis Classic.
Clinical Genetics Branch Support Services Contract with Westat, Inc., Contract Number HHSN261201300003C. We thank patients, families and referring physicians. This work utilized the computational resources of the NIH High Performance Computing Biowulf cluster.
Footnotes
Conflict of Interest: The authors declare no relevant conflicts of interest.
References
- 1.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409(6818):363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 2.Wilson RC, Tambe A, Kidwell MA, Noland CL, Schneider CP, Doudna JA. Dicer-TRBP complex formation ensures accurate mammalian microRNA biogenesis. Mol Cell. 2015;57(3):397–407. doi: 10.1016/j.molcel.2014.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hill DA, Ivanovich J, Priest JR, et al. DICER1 mutations in familial pleuropulmonary blastoma. Science. 2009;325(5943):965. doi: 10.1126/science.1174334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brenneman M, Field A, Yang J, et al. Temporal order of RNase IIIb and loss-of-function mutations during development determines phenotype in DICER1 syndrome: a unique variant of the two-hit tumor suppression model. F1000Res. 2015;4:214. doi: 10.12688/f1000research.6746.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doros L, Schultz KA, Stewart DR, et al. DICER1-Related Disorders. In: Pagon RA, Adam MP, Ardinger HH, et al., editors. GeneReviews(R) Seattle (WA): 1993. [Google Scholar]
- 6.de Kock L, Sabbaghian N, Druker H, et al. Germ-line and somatic DICER1 mutations in pineoblastoma. Acta Neuropathol. 2014;128(4):583–595. doi: 10.1007/s00401-014-1318-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bahubeshi A, Bal N, Rio Frio T, et al. Germline DICER1 mutations and familial cystic nephroma. J Med Genet. 2010;47(12):863–866. doi: 10.1136/jmg.2010.081216. [DOI] [PubMed] [Google Scholar]
- 8.Rio Frio T, Bahubeshi A, Kanellopoulou C, et al. DICER1 mutations in familial multinodular goiter with and without ovarian Sertoli-Leydig cell tumors. JAMA. 2011;305(1):68–77. doi: 10.1001/jama.2010.1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doros L, Yang J, Dehner L, et al. DICER1 mutations in embryonal rhabdomyosarcomas from children with and without familial PPB-tumor predisposition syndrome. Pediatr Blood Cancer. 2012;59(3):558–560. doi: 10.1002/pbc.24020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khan NE, Bauer AJ, Schultz KA, et al. Quantification of thyroid cancer and multinodular goiter risk in the DICER1 syndrome: a family-based cohort study. J Clin Endocrinol Metab. 2017 doi: 10.1210/jc.2016-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Messinger YH, Stewart DR, Priest JR, et al. Pleuropulmonary blastoma: a report on 350 central pathology-confirmed pleuropulmonary blastoma cases by the International Pleuropulmonary Blastoma Registry. Cancer. 2015;121(2):276–285. doi: 10.1002/cncr.29032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Genomes Project C. Auton A, Brooks LD, et al. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Exome Variant Server. [Accessed Nov 15, 2016];NHLBI GO Exome Sequencing Project (ESP) http://evs.gs.washington.edu/EVS/
- 15.Akinrinade O, Koskenvuo JW, Alastalo TP. Prevalence of Titin Truncating Variants in General Population. PLoS One. 2015;10(12):e0145284. doi: 10.1371/journal.pone.0145284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cousminer DL, Arkader A, Voight BF, Pacifici M, Grant SF. Assessing the general population frequency of rare coding variants in the EXT1 and EXT2 genes previously implicated in hereditary multiple exostoses. Bone. 2016;92:196–200. doi: 10.1016/j.bone.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flannick J, Beer NL, Bick AG, et al. Assessing the phenotypic effects in the general population of rare variants in genes for a dominant Mendelian form of diabetes. Nat Genet. 2013;45(11):1380–1385. doi: 10.1038/ng.2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Landi MT, Consonni D, Rotunno M, et al. Environment And Genetics in Lung cancer Etiology (EAGLE) study: an integrative population-based case-control study of lung cancer. BMC Public Health. 2008;8:203. doi: 10.1186/1471-2458-8-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prorok PC, Andriole GL, Bresalier RS, et al. Design of the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial. Control Clin Trials. 2000;21(6 Suppl):273S–309S. doi: 10.1016/s0197-2456(00)00098-2. [DOI] [PubMed] [Google Scholar]
- 20.Calle EE, Rodriguez C, Jacobs EJ, et al. The American Cancer Society Cancer Prevention Study II Nutrition Cohort: rationale, study design, and baseline characteristics. Cancer. 2002;94(2):500–511. doi: 10.1002/cncr.10197. [DOI] [PubMed] [Google Scholar]
- 21.Cingolani P, Platts A, Wang le L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6(2):80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doros LA, Rossi CT, Yang J, et al. DICER1 mutations in childhood cystic nephroma and its relationship to DICER1-renal sarcoma. Mod Pathol. 2014;27(9):1267–1280. doi: 10.1038/modpathol.2013.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ioannidis NM, Rothstein JH, Pejaver V, et al. REVEL: An Ensemble Method for Predicting the Pathogenicity of Rare Missense Variants. Am J Hum Genet. 2016;99(4):877–885. doi: 10.1016/j.ajhg.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46(3):310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slade I, Bacchelli C, Davies H, et al. DICER1 syndrome: clarifying the diagnosis, clinical features and management implications of a pleiotropic tumour predisposition syndrome. J Med Genet. 2011;48(4):273–278. doi: 10.1136/jmg.2010.083790. [DOI] [PubMed] [Google Scholar]
- 28.Khan NE, Bauer AJ, Doros L, et al. Macrocephaly associated with the DICER1 syndrome. Genet Med. 2017;19(2):244–248. doi: 10.1038/gim.2016.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ognjanovic S, Olivier M, Bergemann TL, Hainaut P. Sarcomas in TP53 germline mutation carriers: a review of the IARC TP53 database. Cancer. 2012;118(5):1387–1396. doi: 10.1002/cncr.26390. [DOI] [PubMed] [Google Scholar]
- 30.Messiaen LMWK. NF1 mutational spectrum. In: DK, editor. Neurofibromatoses. Monographs in Human Genetics. Vol. 16. Karger; 2008. pp. 63–77. [Google Scholar]
- 31.King MC, Levy-Lahad E, Lahad A. Population-based screening for BRCA1 and BRCA2: 2014 Lasker Award. JAMA. 2014;312(11):1091–1092. doi: 10.1001/jama.2014.12483. [DOI] [PubMed] [Google Scholar]
- 32.Rubenstein JH, Enns R, Heidelbaugh J, Barkun A Clinical Guidelines C. American Gastroenterological Association Institute Guideline on the Diagnosis and Management of Lynch Syndrome. Gastroenterology. 2015;149(3):777–782. doi: 10.1053/j.gastro.2015.07.036. quiz e716–777. [DOI] [PubMed] [Google Scholar]
- 33.Hassell KL. Population estimates of sickle cell disease in the U.S. Am J Prev Med. 2010;38(4 Suppl):S512–521. doi: 10.1016/j.amepre.2009.12.022. [DOI] [PubMed] [Google Scholar]
- 34.Parker SE, Mai CT, Canfield MA, et al. Updated National Birth Prevalence estimates for selected birth defects in the United States, 2004–2006. Birth Defects Res A Clin Mol Teratol. 2010;88(12):1008–1016. doi: 10.1002/bdra.20735. [DOI] [PubMed] [Google Scholar]
- 35.Shprintzen RJ. Velo-cardio-facial syndrome: 30 Years of study. Dev Disabil Res Rev. 2008;14(1):3–10. doi: 10.1002/ddrr.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scotet V, Dugueperoux I, Saliou P, et al. Evidence for decline in the incidence of cystic fibrosis: a 35-year observational study in Brittany, France. Orphanet J Rare Dis. 2012;7:14. doi: 10.1186/1750-1172-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Evans DG, Howard E, Giblin C, et al. Birth incidence and prevalence of tumor-prone syndromes: estimates from a UK family genetic register service. Am J Med Genet A. 2010;152A(2):327–332. doi: 10.1002/ajmg.a.33139. [DOI] [PubMed] [Google Scholar]
- 38.Romitti PA, Zhu Y, Puzhankara S, et al. Prevalence of Duchenne and Becker muscular dystrophies in the United States. Pediatrics. 2015;135(3):513–521. doi: 10.1542/peds.2014-2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Srivastava A, Brewer AK, Mauser-Bunschoten EP, et al. Guidelines for the management of hemophilia. Haemophilia. 2013;19(1):e1–47. doi: 10.1111/j.1365-2516.2012.02909.x. [DOI] [PubMed] [Google Scholar]
- 40.Stromme P, Bjornstad PG, Ramstad K. Prevalence estimation of Williams syndrome. J Child Neurol. 2002;17(4):269–271. doi: 10.1177/088307380201700406. [DOI] [PubMed] [Google Scholar]
- 41.Groth KA, Hove H, Kyhl K, et al. Prevalence, incidence, and age at diagnosis in Marfan Syndrome. Orphanet J Rare Dis. 2015;10:153. doi: 10.1186/s13023-015-0369-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adams MC, Evans JP, Henderson GE, Berg JS. The promise and peril of genomic screening in the general population. Genet Med. 2016;18(6):593–599. doi: 10.1038/gim.2015.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.