Abstract
A cancer in the contralateral breast in a woman with a previous or synchronous breast cancer is typically considered to be an independent primary tumor. Emerging evidence suggests that in a small subset of these cases the second tumor represents a metastasis. We sought to investigate the issue using massively parallel sequencing targeting 254 genes recurrently mutated in breast cancer. We examined the tumor archives at Memorial Sloan Kettering Cancer Center for the period 1995–2006 to identify cases of contralateral breast cancer where surgery for both tumors was performed at the Center. We report results from 49 patients successfully analyzed by a targeted massively parallel sequencing assay. Somatic mutations and copy number alterations were defined by state-of-the-art algorithms. Clonal relatedness was evaluated by statistical tests specifically designed for this purpose. We found evidence that the tumors in contralateral breasts were clonally related in 3 cases (6%) on the basis of matching mutations at codons where somatic mutations are rare. Clinical data and the presence of similar patterns of gene copy number alterations were consistent with metastasis for all 3 cases. In 3 additional cases there was a solitary matching mutation at a common PIK3CA locus. The results suggest that a subset of contralateral breast cancers represent metastases rather than independent primary tumors. Massively parallel sequencing analysis can provide important evidence to clarify the diagnosis. However, given the inter-tumor mutational heterogeneity in breast cancer, sufficiently large gene panels need to be employed to define clonality convincingly in all cases.
Keywords: Clonality, bilateral breast cancer, next generation sequencing
Introduction
Women with unilateral breast cancer have a significantly elevated risk of developing a cancer in the contralateral breast.1 Contralateral breast cancer (CBC) is generally considered to be a second primary rather than a metastasis from the initial primary.2 This view is supported by studies showing frequent discrepancies in conventional histopathologic features such as histologic type, grade or estrogen receptor status between the two primaries, although assignment of histologic grade may vary among pathologists.3 However, a substantial body of evidence suggests that bilateral metachronous breast cancer is associated with poor prognostic features of the initial cancer.4–8 In particular, studies have shown that patients with bilateral cancer more frequently present with advanced disease than those presenting with unilateral cancer.9 These findings suggest that some CBCs may represent first site metastases.
Numerous studies have investigated clonal relationships between primary and contralateral breast cancers with most reaching the conclusion that the vast majority of CBCs have an independent origin.10–20 However, many of these studies were based on technologies with limited resolution for assessing clonality. The preponderance of the early studies involved assessment of a small number of markers of loss of heterozygosity at genetic regions that commonly exhibit allelic losses in breast cancer. Later studies employed genome-wide copy number arrays permitting, in principle, greater resolution and a more definitive test for clonal relatedness, but were often hampered by the use of deoxyribonucleic acid (DNA) obtained from paraffin-embedded tissues. 21,22 Three recent studies used mutational data derived from whole-exome sequencing for clonality analyses,23–25 and an additional study examined chromosomal rearrangements in 10 cases using low coverage whole genome sequencing.26 In the largest of these studies, Klevebring et al. examined 25 metachronous breast cancers and concluded that 3 (12%) were clonally related.25
In this study we used massively parallel sequencing targeting the 254 genes most frequently mutated in breast cancer or related to DNA repair to address the question of clonality in patients with synchronous and metachronous bilateral breast cancer. We also employed a novel statistical approach that has been developed to address the specific computational challenges in this setting.27
Material and Methods
Patients
After approval by the Memorial Sloan Kettering Cancer Center Institutional Review Board (WA0388-13), the medical records of patients previously identified as having bilateral breast cancer between 1995 and 2006 were reviewed to ascertain if both surgeries were performed at Memorial Sloan Kettering Cancer Center. Archival formalin-fixed paraffin-embedded (FFPE) tissue blocks from both tumors were obtained from the pathology department. Clinical information including patient age, tumor histology and hormone receptor status was obtained from the medical records.
DNA Extraction
Freshly cut hematoxylin and eosin sections were reviewed to ensure the adequacy of the specimen. Representative 8µm-thick FFPE sections were stained with nuclear fast red. Microdissection was performed using a sterile needle under a stereomicroscope (Olympus SZ61, Center Valley, PA) to ensure >80% of tumor cell content and that the normal tissue was devoid of any neoplastic cells as previously described.28,29 Genomic DNA extraction from each tumor and matched normal tissue was performed using the DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA), and quantified using the Qubit Fluorometer assay (Invitrogen, Thermo Fisher Scientific, Waltham, MA) following manufacturers’ instructions.
Targeted-Capture Massively Parallel Sequencing
DNA from all tumor and normal specimens were subjected to targeted capture massively parallel sequencing using a library of baits targeting exons of the 254 genes most frequently mutated in breast cancer and/or related to DNA repair.30 Genes harboring deleterious or potentially deleterious mutations in The Cancer Genome Atlas (TCGA) and the International Cancer Genome Consortium were included in the panel, in addition to a number of other genes that are of interest due to the presence of frequent copy number alterations mapping to their loci (e.g. CDKN2A, CDKN2B, RB1, FGFR1), germline mutations associated to hereditary cancer syndromes (e.g. BRCA1, BRCA2, PALB2, RAD51D, RAD51C, BRIP) or the presence of potentially actionable mutations (e.g. BRAF, KIT, EGFR, ERBB2 and ERBB3). Barcoded sequence libraries were prepared (New England Biolabs, KapaBiosystems) as previously described, and sequenced on an Illumina HiSeq 2000 (San Diego, CA) following validated protocols at the Memorial Sloan Kettering Cancer Center Integrated Genomics Operations.30
Reads were aligned to the human reference genome GRCh37 using the Burrows-Wheeler Aligner.31 Local realignment, duplicate removal and quality score recalibration were performed using Genome Analysis Toolkit.32 Somatic single nucleotide variants (SNVs) were identified using MuTect33 and small insertions and deletions (indels) were identified using Strelka and VarScan 2.34,35 SNVs and indels with mutant allelic fraction (MAF) <1% and/ or supported by <5 reads were disregarded.29,36 Variants found with >5% global minor allele frequency in dbSNP (Build 137) or that were covered by <10 reads in the tumor or <5 reads in the germline were disregarded. Variants for which the tumor variant allele fraction was <5 times than that of the normal variant allele fraction were disregarded. The cancer cell fractions of mutations were defined using ABSOLUTE as previously described.28,30
In cases in which multiple matching mutations were observed we performed validation by repeating the targeted capture massively parallel sequencing on a new DNA sample. In addition we repeated the sequencing for a few additional cases based on availability of tissue/DNA to examine the validation rate of observed mutations. The overall validation rate of observed mutations was 90% (35/39). In cases in which a single match was observed we resequenced the match using either amplicon sequencing or Sanger sequencing as previously described.28,30 The primers used for these methods are provided in Supplementary Table 1. For the amplicon sequencing mpileup files generated from samtools mpileup (version 1.2 htslib 1.2.1) were used to determine whether a mutation existed in the validation binary alignment/map file. Mutations that were found at <1% allele frequency in the germline and >1% in the tumor, where the MAF in the tumor was at least 5 times that in the normal (the same filter used in the targeted sequencing analysis) and had at least 50 reads in both the tumor and the normal were considered validated as somatic. Loci with <50 good-quality reads in either the tumor or normal were considered to have insufficient depth.
Clinical Interpretations
For cases with matching mutations in both tumors we assigned a subjective clinical interpretation of whether or not the tumors were independent, based on tumor stage, the presence of in situ carcinoma, ER, PR, and HER2 status, and clinical course. Features considered suggestive of metastases included advanced stage of the initial tumor in the case of metachronous cancers or of one of the cancers in the case of synchronous cancers, the absence of in situ carcinoma in the proposed metastatic lesion, or the subsequent development of other sites of metastatic disease.
Statistical Methods
For each individual case we used a statistical test of the hypothesis that two tumors are of independent origin that was developed specifically for the purpose of comparing mutational profiles.27 The method relies strongly on the fact that somatic mutations at some loci are common in breast cancer while the preponderance of mutations are very rare. Matches at rare loci provide much stronger evidence for clonal relatedness than matches at common loci. These “marginal” loci-specific probabilities of observing a mutation were obtained from combining the empirical relative frequencies observed in the TCGA breast cancer dataset (downloaded from the Broad GDAC Firehose at https://gdac.broadinstitute.org) and the present study.37 Somatic mutations observed in our study that were not previously observed in TCGA were assigned a marginal probability of 1/1039, where 1039 is the combined number of patients in this study and in the TCGA breast study.
To derive copy number changes the number of reads that mapped to a locus and met base and mapping quality thresholds were obtained using the samtools mpileup function. Candidate loci were obtained from dbsnp build 137 along with pseudo- single nucleotide polymorphisms (SNPs) to obtain counts from regions that are sparse in SNPs. The genome was split into consecutive bins of size 200 bases and median count of candidate loci in a bin used for read depth of the bin. Only bins that had a count of at least 25 in the normal were considered. Guanine-cytosine (GC) normalization was accomplished by using locally weighted regression of the logarithm of the tumor to normal count ratio as a function of GC percentage of a 1000 base window around the locus. Loci with <25 reads in any of the normal samples were excluded. Log-ratios were then averaged in blocks of 5 consecutive markers to the final resolution of 2523 markers in order to smooth out the noise. Data were segmented using circular binary segmentation38 and gains and losses were called if the mean log-ratios were above or below 1 median absolute deviation of residuals from the sample’s median, where residuals were computed as a difference between log-ratios and the segmented value at each locus. We elected not to compare break points from full segmentation of these data because the targeted panel consisted of genes that are not spread uniformly in the genome. Consequently we elected to use a conservative approach whereby we limited attention to gains and losses that spanned an entire chromosome arm defined as a gain or loss covering at least 90% of the markers on the chromosome arm. We gauge and compare the similarities of these whole-arm copy number profiles using the log likelihood ratio measure proposed by Ostrovnaya et al. in which attention is restricted solely to the matching patterns of gains or losses that covered an entire chromosome arm.22
Results
The study population included 248 patients with bilateral invasive breast carcinoma in whom both surgeries were performed at Memorial Sloan Kettering Cancer Center, of which 97 were metachronous and 151 were synchronous. Tissue from both cancers was reported as archived in 98 of these cases. DNA of sufficient quality and quantity was obtained from both contralateral breast tumors and matched normal tissue and subjected successfully to targeted capture massively parallel sequencing for 49 of these cases. The clinical characteristics of these patients are described in Supplementary Table 2.
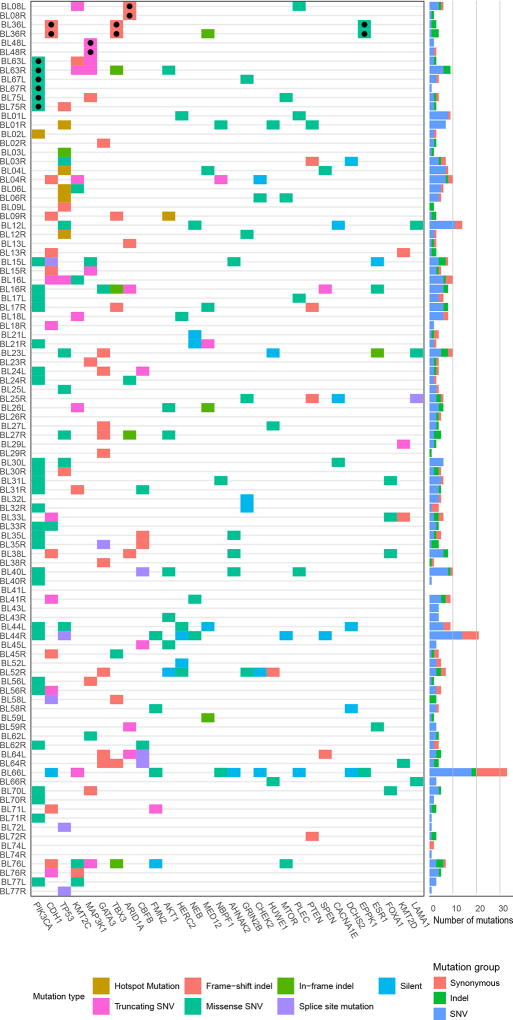
Targeted capture massively parallel sequencing of the tumors and matched normal tissue was performed to a median coverage of 544× (range 77×-1552×) in the tumors and 360× (range 117×-2069×) in the normal samples, and somatic mutations observed in both tumors from a patient were independently validated (see Methods). The numbers of somatic mutations per tumor observed in the 254 gene panel analyzed ranged from 0 to 33 with a median of 4 (the complete list of mutations is provided in Supplementary Table 3). Consistent with previous studies, the genes most frequently affected by somatic mutations in the breast cancers analyzed here included PIK3CA, TP53 and CDH1 (Figure 1). We note that 5 of the 7 genes found to be mutated and shared between the CBCs from a given patient (designated with a “●” in Figure 1) affected the top 8 most frequently mutated genes, specifically PIK3CA, ARID1A, CDH1, TBX3 and MAP3K1. Identical somatic mutations were identified in the tumors from both breasts in 6 of the 49 cases analyzed (Table 1). Importantly, these shared mutations were present at similar cancer cell fractions of tumor cells in the right and left breast cancers (Supplementary Table 3). For 3 of these cases our statistical test for clonality was significant. In case #36, 3 of 4 observed somatic mutations were matches and in case #48, 2 of 3 observed somatic mutations were matches. However, for case #8 only 1 of the 7 somatic mutations identified was shared between the two breast cancers. Notably, this ARID1A mutation was found to have a cancer cell fraction of 1 (i.e. bioinformatically inferred to be present in all tumor cells) in both the left and right invasive breast cancers, and to be the sole mutation in either left or right tumor of this case with a cancer cell fraction of 1 (Supplementary Table 3). The statistical test employed classified the two metachronous breast tumors of this case as clonal given that the shared mutation occurred at a very rare residue (e.g. ARID1A E250fs). In 3 additional cases (cases #63, #67 and #75) a single common PIK3CA H1047R hotspot mutation was found to be shared between the synchronously diagnosed left and right breast cancers. The statistical test employed was not significant for any of these cases. [We note that a validation experiment was not performed for the match in case #63.]
Figure 1. Somatic Mutations Observed.
Heatmap showing the 30 genes most frequently affected by mutations in 98 tumors subjected to targeted massively parallel sequencing from 49 patients with contralateral breast cancer. Samples are shown in rows, genes in columns. The top 6 cases are those with observed matching mutations, identified by ●. The histogram on the right displays the total numbers of mutations observed, per case, and the color code distinguishes different types of mutation, as indicated: indel, small insertion and deletion; SNV, single nucleotide variant.
Table 1.
Results Summary
| Case # | Somatic Mutations | Matches** | Copy Number Similarity/Rank*** |
|||
|---|---|---|---|---|---|---|
| Left | Right | Shared | P-Value* | |||
| 1 | 9 | 7 | 0 | 1.0 | −1.78 / 43 | |
| 2 | 3 | 3 | 0 | 1.0 | 0.76 / 20 | |
| 3 | 2 | 7 | 0 | 1.0 | 0.57 / 22 | |
| 4 | 8 | 10 | 0 | 1.0 | −1.75 / 42 | |
| 6 | 6 | 5 | 0 | 1.0 | 0.55/ 23 | |
| 8 | 6 | 2 | 1 | 0.004 | ARID1A E250fs | 3.74 / 2 |
| 9 | 2 | 3 | 0 | 1.0 | −1.16/ 35 | |
| 12 | 14 | 3 | 0 | 1.0 | −4.09 / 49 | |
| 13 | 3 | 3 | 0 | 1.0 | −0.22 / 28 | |
| 15 | 8 | 5 | 0 | 1.0 | −1.92 / 44 | |
| 16 | 10 | 8 | 0 | 1.0 | −2.65 / 47 | |
| 17 | 6 | 8 | 0 | 1.0 | −0.14/ 26 | |
| 18 | 8 | 2 | 0 | 1.0 | 2.37 / 5 | |
| 21 | 4 | 3 | 0 | 1.0 | 0.29 / 24 | |
| 23 | 10 | 4 | 0 | 1.0 | −1.47 / 40 | |
| 24 | 4 | 3 | 0 | 1.0 | 1.76/ 8 | |
| 25 | 4 | 6 | 0 | 1.0 | −0.32 / 30 | |
| 26 | 6 | 5 | 0 | 1.0 | 1.57 / 11 | |
| 27 | 4 | 5 | 0 | 1.0 | 1.38 / 13 | |
| 29 | 3 | 1 | 0 | 1.0 | −0.28 / 29 | |
| 30 | 6 | 5 | 0 | 1.0 | −1.18 / 36 | |
| 31 | 6 | 5 | 0 | 1.0 | −1.21 / 37 | |
| 32 | 5 | 4 | 0 | 1.0 | −0.98 / 34 | |
| 33 | 6 | 4 | 0 | 1.0 | 1.80 / 7 | |
| 35 | 5 | 4 | 0 | 1.0 | −0.01 / 25 | |
| 36 | 3 | 4 | 3 | <0.001 | CDH1 S111fs; TBX3 T267fs; EPPK1 R2337H | 4.46 / 1 |
| 38 | 8 | 2 | 0 | 1.0 | −1.60 / 41 | |
| 40 | 10 | 1 | 0 | 1.0 | 2.68 / 4 | |
| 41 | 0 | 9 | -- | 1.0 | 1.12 / 17 | |
| 43 | 4 | 4 | 0 | 1.0 | −0.69 / 33 | |
| 44 | 9 | 21 | 0 | 1.0 | −2.32 / 46 | |
| 45 | 3 | 4 | 0 | 1.0 | 1.24 / 15 | |
| 48 | 2 | 3 | 2 | <0.001 | MLH3 M346R; MAP3K1 R248* | 3.01 / 3 |
| 52 | 5 | 7 | 0 | 1.0 | 2.23 / 6 | |
| 56 | 2 | 5 | 0 | 1.0 | 1.51 / 12 | |
| 58 | 3 | 4 | 0 | 1.0 | −1.25 / 38 | |
| 59 | 2 | 3 | 0 | 1.0 | 0.83 / 19 | |
| 62 | 4 | 4 | 0 | 1.0 | −0.17 / 27 | |
| 63 | 3 | 9 | 1 | 0.08 | PIK3CA H1047R | 0.98 / 18 |
| 64 | 5 | 4 | 0 | 1.0 | 0.63 / 21 | |
| 66 | 33 | 3 | 0 | 1.0 | 1.34 / 14 | |
| 67 | 4 | 1 | 1 | 0.08 | PIK3CA H1047R | 1.75 / 9 |
| 70 | 5 | 2 | 0 | 1.0 | −2.95 / 48 | |
| 71 | 3 | 1 | 0 | 1.0 | 1.21 / 16 | |
| 72 | 1 | 3 | 0 | 1.0 | −2.19 / 45 | |
| 74 | 2 | 1 | 0 | 1.0 | −0.66 / 32 | |
| 75 | 4 | 3 | 1 | 0.08 | PIK3CA H1047R | −1.36 / 39 |
| 76 | 7 | 5 | 0 | 1.0 | 1.70 / 10 | |
| 77 | 3 | 1 | 0 | 1.0 | −0.60 / 31 | |
P-value for the test of clonal relatedness of the tumor pair.
Matches were validated either by re-sampling the tumor and resequencing if sufficient tumor was available or by targeted sequencing (MiSeq), or by Sanger sequencing.
Log likelihood ratio measure of similarity of copy number profiles and ranking
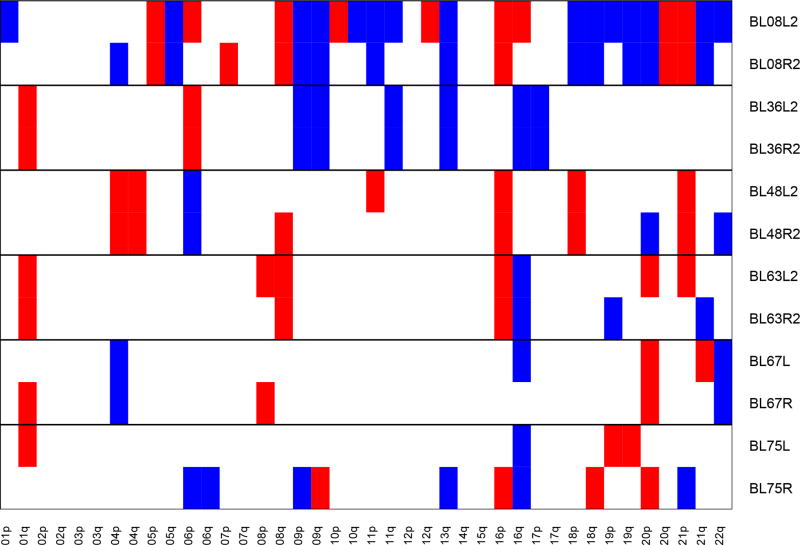
We further evaluated the evidence for clonal relatedness by examining the copy number alterations. These are displayed for the 6 cases with matches in Figure 2. For the 3 cases designated as “clonally related” by mutational analysis (cases #8, #36 and #48) the copy number profiles of the tumor pairs displayed multiple common gains and losses. These 3 cases had the 3 highest values of our log likelihood measure characterizing the similarity of the copy number profiles (Table 1). However several other cases demonstrated multiple matching gains and/or losses, indicating that there may be cases which are clonal but where no matches were observed on the panel (Supplementary Plot 1).
Figure 2. Copy Number Alterations.
Each row represents whole chromosome arm copy number gains (in blue) and losses (in red) for an individual case. Pairs of tumors for all 6 cases with matching mutations are presented. Copy number plots for all cases included in this study are presented in Supplementary Plot 1.
Key clinical details and interpretations of the 6 cases with mutational matches are provided in Table 2 (additional clinical information on all cases is provided in Supplementary Table 2). For cases #8, #36 and #48 the clinical features are consistent with the possibility of the second breast cancer being a metastasis, supporting the evidence from both matching mutations and similar copy number patterns, although distant metastases were not observed after longer-term follow-up for 2 of these 3 cases. By contrast, the 3 cases with single matching PIK3CA H1047R mutations all have clinical features consistent with independent primary tumors.
Table 2.
Clinical/Pathological Details of Potentially Clonal Cases
| Case# | Initial Histology | Interval to 2nd Primary |
2nd Tumor Histology |
Outcome | Clinical Interpretation |
|---|---|---|---|---|---|
| 8 | T= 6.8cm, 22 positive nodes, extensive LVI, DCIS present, ER+/PR−/HER2− | 1yr | Inflammatory, 12 positive nodes, DCIS present, ER−/PR−/HER2− | Distant metastases 11 months after second cancer | Consistent with metastases |
| 36 | T>5cm, 19 positive nodes, extensive LVI, no DCIS, ER+/PR−/HER2− | 3yrs | T-diffuse, >5cm, 12 positive nodes, extensive LVI, no DCIS, ER+/PR−/HER2− | NED years after initial diagnosis | Consistent with metastases |
| 48 | Inflammatory, 5 positive nodes, no DCIS, ER+/PR+/HER2− | Synchronous | T= 5cm, 5 positive nodes, extensive LVI, no DCIS, ER+/PR+/HER2− | Chest wall recurrence 12 yrs later | Consistent with metastases |
| 63 | T= 1.0cm, 0/5 nodes, DCIS present, no LVI, ER+/PR+/HER2− | Synchronous | T= 1.8cm, 0/2 nodes, DCIS present, no LVI, ER+/PR+/HER2− | NED 4 years 9months | Independent primaries |
| 67 | T= 1.6cm, 0/1 nodes, DCIS present, no LVI, ER+/PR+/HER2− | Synchronous | T= 1.4cm, 1/3 positive nodes, DCIS present, no LVI, ER+/PR+/HER2− | NED 3 years | Independent primaries |
| 75 | T= 1.1cm, 0/1 nodes, DCIS present, no LVI, ER+/PR+/HER2− | Synchronous | T= 1.4cm, 0/1 nodes, DCIS present, no LVI, ER+/PR+/HER2− | NED 1year | Independent primaries |
Abbreviation: DCIS, ductal carcinoma in situ; LVI, lymphovascular invasion; NED, no evidence of disease.
Discussion
Our findings are consistent with the hypothesis that a small subset of CBCs are clonally related and constitute metastatic dissemination from one breast to another. This phenomenon can occur regardless of whether the two breast tumors occur synchronously or metachronously. We found strong molecular evidence in this study to support the clonal relationship between the two invasive breast cancers in 3 out of 49 (6%) CBCs. In these 3 cases, we identified identical mutations at rare residues, all of which were validated, and the copy number alterations and clinical information supported the interpretation that these tumor pairs are clonally related. In 3 additional patients a shared single PIK3CA hotspot mutation was observed. However this evidence for clonality alone is weak due to the high frequency of mutations at this residue in breast cancers generally and the fact that the clinical data suggest that the tumors are independent in all 3 patients.
We believe that the clinical features of the 3 cases classified as clonal are consistent with this diagnosis. The 3-year interval between the initial cancer and the contra-lateral diagnosis for case #36 is congruent with recent studies that support the premise that cancer cells can disseminate to distant sites early in tumor development but may remain dormant for long periods before clinically detectable metastases emerge.39,40 This prolonged period of dormancy is particularly common in ER positive tumors, where more than 50% of distant recurrences occur more than 5 years post diagnosis, particularly in the setting of adjuvant endocrine therapy.41 Case #36 was still in remission one year later. In case #48 the patient had synchronous inflammatory cancers and developed a chest wall recurrence 12 years post-diagnosis. Primary therapy for both of these cases consisted of modified radical mastectomy, chemotherapy with an anthracycline, cyclophosphamide and a taxane (ACT), post mastectomy radiotherapy to the chest wall and node fields, and tamoxifen. The same treatment strategy was used for the subsequent contra-lateral tumor in case #36 in addition to an aromatase inhibitor. The application of what was in effect cytoreductive surgery for a solitary metastasis for both these cases may have proved to be effective treatment in providing the long disease-free interval for case #48. Indeed, prolonged survival after aggressive local and systemic therapy is well documented in stage IV breast cancers. In an analysis of 21,372 patients with stage IV cancer and an intact primary, survival at 10 years was observed in 10% of patients who received surgery on the primary tumor.42
In contrast to these two cases, in case #8, after treatment with modified radical mastectomy, ACT and post mastectomy radiotherapy to the chest wall and node fields, the contralateral lesion developed 1 year post-diagnosis and despite treatment with neoadjuvant ACT followed by modified radical mastectomy, chest wall recurrence developed 2 months postoperatively followed by distant metastases 11 months later. The finding of DCIS in the second cancer in this case is somewhat puzzling since the presence of DCIS is generally regarded as an indicator of a primary tumor. However, a recent large observational study identified a small but significant number of patients who died of metastatic breast cancer without local recurrence following diagnosis and treatment of DCIS, suggesting that DCIS itself represents very early stage breast cancer that nonetheless has metastatic potential.43 It is notable that the preponderance of observed mutations in this case were not matches.
The knowledge that a proportion of CBCs are in fact metastases is clinically important to allow individualization of the use of surgery on the contralateral breast and axilla based on a diagnosis of metastatic disease versus a new primary cancer, and to direct the choice and duration of systemic therapy. The management of the breast and axilla in patients presenting with stage IV cancer and an intact primary tumor remains a subject of debate. Although multiple retrospective studies suggest a survival benefit following surgery of the primary tumor in these patients,44 a prospective randomized trial45 and a prospective registry study45 have failed to confirm this. Until this issue is resolved with the reporting of the on-going Eastern Cooperative Oncology Group randomized trial (NCT01242800), it is unclear if the finding that a subset of women with CBC actually have metastases to the contralateral breast rather than a new primary tumor has the potential to alter clinical management of CBC.
To implement the approach employed here in a clinical setting when a new contralateral breast tumor is diagnosed would require the availability of tumor tissue from both tumors as well as matched normal tissue. This may be challenging for metachronous CBCs if the first tumor was diagnosed several years previously or at a different hospital. In the era of precision medicine, however, and with the implementation of routine panel mutation testing such as the Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT)47 at our institution and other similar assays at other institutions, genomic analysis of tumors is likely to become increasingly common. Our success rate in extracting DNA from archived tissue samples and successfully sequencing the samples was 75% in this study. Whole-exome and/ or targeted capture massively parallel sequencing analyses have been proven to constitute a successful strategy to resolve the clonality of synchronous neoplasms in other contexts,48–50 and to define the origin of metastatic deposits in the presence of independent primaries in distinct anatomical sites.51 Given the increasing robustness of sequencing assays based on the analysis of DNA extracted from archival FFPE tissue samples, genotyping approaches may constitute a useful ancillary method to distinguish independent primary cancers from metastatic disease.
Our study has important limitations. When the DNA samples from both tumors are successfully sequenced there remain challenges in making the differential diagnosis of independence versus metastasis. Our targeted sequencing panel contained 254 genes, including all the known driver genes in breast cancer. Yet the number of mutations observed in any given tumor was frequently small. The median number of somatic mutations per tumor was 4, and in 1 of the 98 tumors no mutation was observed at all. As a result there will frequently be considerable uncertainty in calling cases as clonal versus independent. For example, even if no shared genetic alterations are observed between two tumors using a given targeted capture massively parallel sequencing assay, there may be clonal mutations present in both breast cancers in genomic locations that were not included in the sequencing panel or in non-coding regions of the genome. Indeed for a few cases in our study in which no mutational matches were observed the copy number profiles showed considerable similarities (Supplementary Plot 1), suggesting that CBCs may be determined to be clonally related based on their patterns of gene copy number alterations rather than on their mutational profiles if no matches are observed after testing a limited panel of genes. Conversely, the presence of a mutation shared between two tumors does not necessarily indicate a clonal origin for the tumors, since mutations at the same locus could occur by chance in independent tumors, especially if the mutation is common, such as the PIK3CA hotspot mutation that occurred in both breasts in three cases in this study, #63, #67 and #75. Whole-exome sequencing has the potential to greatly reduce the diagnostic uncertainty, but it would not necessarily eliminate it. Indeed in an earlier study by our group that used whole-exome sequencing to assess the evidence of clonal relationships between pre-malignant lesions (lobular carcinoma in situ) and invasive breast cancers several cases were observed with a single shared somatic mutation, rendering the evidence for clonality equivocal.52 Finally, our statistical testing is based on the premise that the probability of a matching mutation in two independently occurring tumors is the square of the relative frequiency of the given mutation in breast cancers generally. However, matches are more likely if host characteristics of the patient influence these probabilities, a phenomenon that will occur if somatic mutational patterns are influenced by the germline. While there are, to our knowledge, no published studies addressing this issue, there has been an anecdotal report of a pair of monozygotic twins with breast cancer whose tumors had unusually similar copy number patterns.53
Despite these limitations, our study confirms that a small but substantial proportion of CBCs may actually represent metastases from the original primary cancers. Formal clonality testing of tissue samples from both tumors has the potential to alter the clinical management for these cases and represents a paradigm shift in our thinking about CBC.
Supplementary Material
Acknowledgments
We are grateful to the following individuals who provided assistance with specimen processing or data management:- Nicola Fusco, Elena Guerini-Rocco, Caterina Marchio, Russell Towers.
Grant Sponsor: The study was funded in part by the Alan and Sandra Gerry Metastasis Research Initiative at Memorial Sloan Kettering Cancer Center, by award CA08748 from the National Cancer Institute, and in part by a Cancer Center Support Grant of the National Institutes of Health/National Cancer Institute (grant No P30CA008748). J.S. Reis-Filho is funded in part by the Breast Cancer Research Foundation. S. Piscuoglio is funded by Swiss National Science Foundation (Ambizione grant PZ00P3_168165)
Abbreviations
- CBC
contralateral breast cancer
- FFPE
formalin-fixed paraffin-embedded
- TCGA
The Cancer Genome Atlas
- SNV
single nucleotide variants
- MAF
mutant allelic fraction
- GC
guanine-cytosine
- DNA
deoxyribonucleic acid
- SNP
Single nucleotide polymorphism
Footnotes
Novelty and Impact: Contralateral occurrences of breast cancer have been considered by convention to be independent primaries. The results of our study support a growing literature that in a small proportion of these cases one of the tumors is a metastasis of the other. Panel sequencing is highly informative in diagnosing metastases, but not always definitive. This study is the largest study to date of this issue.
References
- 1.Hayat MJ, Howlader N, Reichman ME, Edwards BK. Cancer statistics, trends, and multiple primary cancer analyses from the Surveillance, Epidemiology, and End Results (SEER) Program. Oncologist. 2007;12:20–37. doi: 10.1634/theoncologist.12-1-20. [DOI] [PubMed] [Google Scholar]
- 2.Banelli B, Casciano I, Di Vinci A, Gatteschi B, Levaggi A, Carli F, Bighin C, Salvi S, Allemanni G, Ghiorzo P, Pronzato P, Venturini M, Romani M, Del Mastro L. Pathological and molecular characteristics distinguishing contralateral metastatic from new primary breast cancer. Ann Oncol. 2010;21:1237–1242. doi: 10.1093/annonc/mdp470. [DOI] [PubMed] [Google Scholar]
- 3.Pusztai L, Viale G, Kelly CM, Hudis CA. Estrogen and HER-2 receptor discordance between primary breast cancer and metastasis. Oncologist. 2010;15:1164–1168. doi: 10.1634/theoncologist.2010-0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beckmann KR, Buckingham J, Craft P, Dahlstrom JE, Zhang Y, Roder D, Stuart-Harris R. Clinical characteristics and outcomes of bilateral breast cancer in an Australian cohort. Breast. 2011;20:158–164. doi: 10.1016/j.breast.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 5.Vichapat V, Garmo H, Holmqvist M, Liljegren G, Wärnberg F, Lambe M, Fornander T, Adolfsson J, Lüchtenborg M, Holmberg L. Tumor stage affects risk and prognosis of contralateral breast cancer: results from a large Swedish-population-based study. J Clin Oncol. 2012;30:3478–3485. doi: 10.1200/JCO.2011.39.3645. [DOI] [PubMed] [Google Scholar]
- 6.Vichapat V, Garmo H, Holmberg L, Fentiman IS, Tutt A, Gillett C, Lüchtenborg M. Patterns of metastasis in women with metachronous contralateral breast cancer. Br J Cancer. 2012;107:221–223. doi: 10.1038/bjc.2012.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuo WH, Yen AM, Lee PH, Chen KM, Wang J, Chang KJ, Chen TH, Tsau HS. Cumulative survival in early-onset unilateral and bilateral breast cancer: an analysis of 1907 Taiwanese women. Br J Cancer. 2009;100:563–570. doi: 10.1038/sj.bjc.6604898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quan G, Pommier SJ, Pommier RF. Incidence and outcomes of contralateral breast cancers. Am J Surg. 2008;195:645–650. doi: 10.1016/j.amjsurg.2008.01.007. discussion 650. [DOI] [PubMed] [Google Scholar]
- 9.Shi YX, Xia Q, Peng RJ, Yuan ZY, Wang SS, An X, Cao Y, Tan YT, Jin Y, Cai XY, Sun YL, Teng XY, Liu DG, Jiang WQ. Comparison of clinicopathological characteristics and prognoses between bilateral and unilateral breast cancer. J Cancer Res Clin Oncol. 2012;138:705–714. doi: 10.1007/s00432-011-1141-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Imyanitov EN, Suspitsin EN, Grigoriev MY, Togo AV, Kuligina ESh, Belogubova EV, Pozharisski KM, Turkevich EA, Rodriquez C, Cornelisse CJ, Hanson KP, Theillet C. Concordance of allelic imbalance profiles in synchronous and metachronous bilateral breast carcinomas. Int J Cancer. 2002;100:557–564. doi: 10.1002/ijc.10530. [DOI] [PubMed] [Google Scholar]
- 11.Chunder N, Roy A, Roychoudhury S, Panda CK. Molecular study of clonality in multifocal and bilateral breast tumors. Pathol Res Pract. 2004;200:735–741. doi: 10.1016/j.prp.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 12.Janschek E, Kandioler-Eckersberger D, Ludwig C, Kappel S, Wolf B, Taucher S, Rudas M, Gnant M, Jakesz R. Contralateral breast cancer: molecular differentiation between metastasis and second primary cancer. Breast Cancer Res Treat. 2001;67:1–8. doi: 10.1023/a:1010661514306. [DOI] [PubMed] [Google Scholar]
- 13.Kollias J, Man S, Marafie M, Carpenter K, Pinder S, Ellis IO, Blamey RW, Cross G, Brook JD. Loss of heterozygosity in bilateral breast cancer. Breast Cancer Res Treat. 2000;64:241–251. doi: 10.1023/a:1026575619155. [DOI] [PubMed] [Google Scholar]
- 14.Regitnig P, Ploner F, Maderbacher M, Lax SF. Bilateral carcinomas of the breast with local recurrence: analysis of genetic relationship of the tumors. Mod Pathol. 2004;17:597–602. doi: 10.1038/modpathol.3800089. [DOI] [PubMed] [Google Scholar]
- 15.Schlechter BL, Yang Q, Larson PS, Golubeva A, Blanchard RA, de las Morenas A, Rosenberg CL. Quantitative DNA fingerprinting may distinguish new primary breast cancer from disease recurrence. J Clin Oncol. 2004;22:1830–1838. doi: 10.1200/JCO.2004.05.123. [DOI] [PubMed] [Google Scholar]
- 16.Stenmark-Askmalm M, Gentile M, Wingren S, Ståhl O. Protein accumulation and gene mutation of p53 in bilateral breast cancer. South-East Sweden Breast Cancer Group. Acta Oncol. 2001;40:56–62. doi: 10.1080/028418601750071064. [DOI] [PubMed] [Google Scholar]
- 17.Tse GM, Kung FY, Chan AB, Law BK, Chang AR, Lo KW. Clonal analysis of bilateral mammary carcinomas by clinical evaluation and partial allelotyping. Am J Clin Pathol. 2003;120:168–174. doi: 10.1309/6YEP-MCHA-CPG2-BD15. [DOI] [PubMed] [Google Scholar]
- 18.Noguchi S, Motomura K, Inaji H, Imaoka S, Koyama H. Differentiation of primary and secondary breast cancer with clonal analysis. Surgery. 1994;115:458–462. [PubMed] [Google Scholar]
- 19.Brommesson S, Jonsson G, Strand C, Grabau D, Malmström P, Ringnér M, Fernö M, Hedenfalk I. Tiling array-CGH for the assessment of genetic similarities among synchronous unilateral and bilateral invasive breast cancer tumor pairs. BMC Clin Pathol. 2008;8:6. doi: 10.1186/1472-6890-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masuda S, Kadowaki T, Kumaki N, Tang X, Tokuda Y, Yoshimura S, Takekoshi S, Osamura RY. Analysis of gene alterations of mitochondrial DNA D-loop regions to determine breast cancer clonality. Br J Cancer. 2012;107:2016–2023. doi: 10.1038/bjc.2012.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ostrovnaya I, Begg CB. Testing clonal relatedness of tumors using array comparative genomic hybridization: a statistical challenge. Clin Cancer Res. 2010;16:1358–1367. doi: 10.1158/1078-0432.CCR-09-2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ostrovnaya I, Olshen AB, Seshan VE, Orlow I, Albertson DG, Begg CB. A metastasis or a second independent cancer? Evaluating the clonal origin of tumors using array copy number data. Stat Med. 2010;29:1608–1621. doi: 10.1002/sim.3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song F, Li X, Song F, Zhao Y, Li H, Zheng H, Gao Z, Wang J, Zhang W, Chen K. Comparative genomic analysis reveals bilateral breast cancers are genetically independent. Oncotarget. 2015;6:31820–31829. doi: 10.18632/oncotarget.5569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bao L, Messer K, Schwab R, Harismendy O, Pu M, Crain B, Yost S, Frazer KA, Rana B, Hasteh F, Wallace A, Parker BA. Mutational profiling can establish clonal or independent origin in synchronous bilateral breast and other tumors. PLoS One. 2015;10:e0142487. doi: 10.1371/journal.pone.0142487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klevebring D, Lindberg J, Rockberg J, Hilliges C, Hall P, Sandberg M, Czene K. Exome sequencing of contralateral breast cancer identifies metastatic disease. Breast Cancer Res Treat. 2015;151:319–324. doi: 10.1007/s10549-015-3403-6. [DOI] [PubMed] [Google Scholar]
- 26.Alkner S, Tang MHE, Brueffer C, Dahlgren M, Chen Y, Olsson E, Winter C, Baker S, Ehinger A, Ryden L, Saal LH, Ferno M, Gruvberger-Saal SK. Contralateral breast cancer can represent a metastatic spread of the first primary tumor: determination of clonal relationship between contralateral breast cancers using next generation whole genome sequencing. Breast Cancer Research. 2015;17:102. doi: 10.1186/s13058-015-0608-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ostrovnaya I, Seshan VE, Begg CB. Using somatic mutation data to test tumors for clonal relatedness. Ann Appl Stat. 2015;9:1533–1548. doi: 10.1214/15-AOAS836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Piscuoglio S, Ng CK, Murray M, Burke KA, Edelweiss M, Geyer FC, Macedo GS, Inagaki A, Papanastasiou AD, Martelotto LG, Marchio C, Lim RS, Ioris RA, Nahar PK, Bruijn ID, Smyth L, Akram M, Ross D, Petrini JH, Norton L, Solit DB, Baselga J, Brogi E, Ladanyi M, Weigelt B, Reis-Filho JS. Massively parallel sequencing of phyllodes tumours of the breast reveals actionable mutations, and TERT promoter hotspot mutations and TERT gene amplification as likely drivers of progression. J Pathol. 2016;238:508–518. doi: 10.1002/path.4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martelotto LG, De Filippo MR, Ng CK, Natrajan R, Fuhrmann L, Cyrta J, Piscuoglio S, Wen HC, Lim RS, Shen R, Schultheis AM, Wen YH, Edelweiss M, Mariani O, Stenman G, Chan TA, Colombo PE, Norton L, Vincent-Salomon A, Reis-Filho JS, Weigelt B. Genomic landscape of adenoid cystic carcinoma of the breast. J Pathol. 2015;237:179–189. doi: 10.1002/path.4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guerini-Rocco E, Hodi Z, Piscuoglio S, Ng CK, Rakha EA, Schultheis AM, Marchiò C, da Cruz Paula A, De Filippo MR, Martelotto LG, De Mattos-Arruda L, Edelweiss M, Jungbluth AA, Fusco N, Norton L, Weigelt B, Ellis IO, Reis-Filho JS. The repertoire of somatic genetic alterations of acinic cell carcinomas of the breast: an exploratory, hypothesis-generating study. J Pathol. 2015;237:166–178. doi: 10.1002/path.4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA, Mardis ER, Ding L, Wilson RK. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saunders CT, Wong WS, Swamy S, Becq J, Murray LJ, Cheetham RK. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics. 2012;28:1811–1817. doi: 10.1093/bioinformatics/bts271. See comment in PubMed Commons below. [DOI] [PubMed] [Google Scholar]
- 36.De Mattos-Arruda L, Weigelt B, Cortes J, Won HH, Ng CK, Nuciforo P, Bidard FC, Aura C, Saura C, Peg V, Piscuoglio S, Oliveira M, Smolders Y, Patel P, Norton L, Tabernero J, Berger MF, Seoane J, Reis-Filho JS. Capturing intra-tumor genetic heterogeneity by de novo mutation profiling of circulating cell-free tumor DNA: a proof-of-principle. Ann Oncol. 2014;25:1729–1735. doi: 10.1093/annonc/mdu239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Venkatraman ES, Olshen AB. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics. 2007;23:657–663. doi: 10.1093/bioinformatics/btl646. [DOI] [PubMed] [Google Scholar]
- 39.Hosseini H, Obradovic MMS, Hoffman M, Harper KL, Sosa MS, Werner-Klein M, Nanduri LK, Werno C, Ehrl C, Maneck M, Patwary N, Haunschild G, Guzvic M, Reimelt C, Grauvog M, Eichner N, Weber F, Hartkopf AD, Taran FA, Brucker SY, Fehm T, Rack B, Buchholz S, Spang R, Meister G, Aguirre-Ghiso JA, Klein CA. Early dissemination seeds metastasis in breast cancer. Nature. 2016;540:552–558. doi: 10.1038/nature20785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harper KL, Sosa MS, Entenberg D, Hosseini H, Cheung JF, Nobre R, Avivar-Valderas A, Nagi C, Girnius N, Davis RJ, Farias EF, Condeelis J, Klein CA, Aguirre-Ghiso JA. Mechanism of early dissemination and metastasis in Her2+ mammary cancer. Nature. 2016;540:588–592. doi: 10.1038/nature20609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Colleoni M, Gray KP, Gelber S, Láng I, Thürlimann B, Gianni L, Abdi EA, Gomez HL, Linderholm BK, Puglisi F, Tondini C, Kralidis E, Eniu A, Cagossi K, Rauch D, Chirgwin J, Gelber RD, Regan MM, Coates AS, Price KN, Viale G, Goldhirsch A. Low-Dose Oral Cyclophosphamide and Methotrexate Maintenance for Hormone Receptor-Negative Early Breast Cancer: International Breast Cancer Study Group Trial 22-00. J Clin Oncol. 2016 Oct 1;34:3400–8. doi: 10.1200/JCO.2015.65.6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thomas A, Khan SA, Chrischilles EA, Schroeder MC. Initial Surgery and Survival in Stage IV Breast Cancer in the United States, 1988–2011. JAMA Surg. 2016 May 1;151:424–31. doi: 10.1001/jamasurg.2015.4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Narod SA, Iqbal J, Giannakeas V, Sopik V, Sun P. Breast cancer mortality after a diagnosis of ductal carcinoma in situ. JAMA Oncol. 2015;1:888–896. doi: 10.1001/jamaoncol.2015.2510. [DOI] [PubMed] [Google Scholar]
- 44.Khan SA. Surgical management of de novo stage IV breast cancer. Semin Radiat Oncol. 2016;26:79–86. doi: 10.1016/j.semradonc.2015.08.004. [DOI] [PubMed] [Google Scholar]
- 45.Badwe R, Hawaldar R, Nair N, Kaushik R, Parmar V, Siddique S, Budrukkar A, Mittra I, Gupta S. Locoregional treatment versus no treatment of the primary tumour in metastatic breast cancer: an open-label randomised controlled trial. Lancet Oncol. 2015;16:1380–1388. doi: 10.1016/S1470-2045(15)00135-7. [DOI] [PubMed] [Google Scholar]
- 46.King TA, Lyman J, Gonen M, Reyes S, Hwang E-S, Rugo HS, Liu MC, Boughey JC, Jacobs LK, McGuire KP, Storniolo AM, Isaacs C, Meszoely IM, Van Poznak CH, Babiera G, Norton L, Morrow M, Wolff AC, Winer EP, Hudis CA Translational Breast Cancer Research Consortium (TBCRC) A prospective analysis of surgery and survival in stage IV breast cancer (TBCRC 013) J Clin Oncol. 2016;34 (suppl; abstr 1006) [Google Scholar]
- 47.Cheng DT, Mitchell TN, Zehir A, Shah RH, Benayed R, Syed A, Chandramohan R, Liu ZY, Won HH, Scott SN, Brannon AR, O'Reilly C, Sadowska J, Casanova J, Yannes A, Hechtman JF, Yao J, Song W, Ross DS, Oultache A, Dogan S, Borsu L, Hameed M, Nafa K, Arcila ME, Ladanyi M, Berger MF. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn. 2015;17:251–264. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schultheis AM, Ng CK, De Filippo MR, Piscuoglio S, Macedo GS, Gatius S, Perez Mies B, Soslow RA, Lim RS, Viale A, Huberman KH, Palacios JC, Reis-Filho JS, Matias-Guiu X, Weigelt B. Massively parallel sequencing-based clonality analysis of synchronous endometrioid endometrial and ovarian carcinomas. J Natl Cancer Inst. 2016;108:djv427. doi: 10.1093/jnci/djv427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anglesio MS, Wang YK, Maassen M, Horlings HM, Bashashati A, Senz J, Mackenzie R, Grewal DS, Li-Chang H, Karnezis AN, Sheffield BS, McConechy MK, Kommoss F, Taran FA, Staebler A, Shah SP, Wallwiener D, Brucker S, Gilks CB, Kommoss S Huntsman DG2. Synchronous endometrial and ovarian carcinomas: evidence of clonality. J Natl Cancer Inst. 2016;108:djv428. doi: 10.1093/jnci/djv428. [DOI] [PubMed] [Google Scholar]
- 50.Murphy SJ, Aubry MC, Harris FR, Halling GC, Johnson SH, Terra S, Drucker TM, Asiedu MK, Kipp BR, Yi ES, Peikert T, Yang P, Vasmatzis G, Wigle DA. Identification of independent primary tumors and intrapulmonary metastases using DNA rearrangements in non-small-cell lung cancer. J Clin Oncol. 2014;32:4050–4058. doi: 10.1200/JCO.2014.56.7644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.De Mattos-Arruda L, Bidard FC, Won HH, Cortes J, Ng CK, Peg V, Nuciforo P, Jungbluth AA, Weigelt B, Berger MF, Seoane J, Reis-Filho JS. Establishing the origin of metastatic deposits in the setting of multiple primary malignancies: the role of massively parallel sequencing. Mol Oncol. 2014;8:150–158. doi: 10.1016/j.molonc.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Begg CB, Ostrovnaya I, Carniello JV, Sakr RA, Giri D, Towers R, Schizas M, De Brot M, Andrade VP, Mauguen A, Seshan VE, King TA. Clonal relationships between lobular carcinoma in situ and other breast malignancies. Breast Cancer Res. 2016;18:66. doi: 10.1186/s13058-016-0727-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wistuba II, Tomlinson GE, Behrens C, Virmani A, Geradts J, Blum JL, Minna JD, Gazdar AF. Two identical triplet sisters carrying a germline BRCA1 gene mutation acquire very similar breast cancer somatic mutations at multiple other sites throughout the genome. Genes, Chromosomes & Cancer. 2000;28:359–369. doi: 10.1002/1098-2264(200008)28:4<359::aid-gcc1>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.