Abstract
Coordinated interactions between ovarian granulosa and theca cells are required for female endocrine function and fertility. To elucidate these interactions the regulation of the granulosa and theca cell transcriptomes during bovine antral follicle development were investigated. Granulosa cells and theca cells were isolated from small (<5 mm), medium (5–10 mm), and large (>10 mm) antral bovine follicles. A microarray analysis of 24,000 bovine genes revealed that granulosa cells and theca cells each had gene sets specific to small, medium and large follicle cells. Transcripts regulated (i.e., minimally changed 1.5-fold) during antral follicle development for the granulosa cells involved 446 genes and for theca cells 248 genes. Only 28 regulated genes were common to both granulosa and theca cells. Regulated genes were functionally categorized with a focus on growth factors and cytokines expressed and regulated by the two cell types. Candidate regulatory growth factor proteins mediating both paracrine and autocrine cell–cell interactions include macrophage inflammatory protein (MIP1 beta), teratocarcinoma-derived growth factor 1 (TDGF1), stromal derived growth factor 1 (SDF1; i.e., CXCL12), growth differentiation factor 8 (GDF8), glia maturation factor gamma (GMFG), osteopontin (SPP1), angiopoietin 4 (ANGPT4), and chemokine ligands (CCL 2, 3, 5, and 8). The current study examined granulosa cell and theca cell regulated genes associated with bovine antral follicle development and identified candidate growth factors potentially involved in the regulation of cell–cell interactions required for ovarian function.
Keywords: ovary, antral follicle, granulosa, theca, cow, bovine, microarray, cell–cell interactions, growth factors, folliculogenesis
INTRODUCTION
Antral follicle development is essential for maturation of the oocyte and to help control female endocrinology and fertility. The follicle is composed of the oocyte, granulosa cells, and theca cells. A rapid growth of granulosa cells and theca cells combined with an increase in the size of the antrum of the follicle is required for the transition from small to large pre-ovulatory follicles (Macklon and Fauser, 1999; Webb et al., 2004). The granulosa and theca cells are a site of action for the gonadotropins and a site for production of steroid hormones. The theca cells respond to luteinizing hormone (LH) and produce androgens, as well as progesterone in the pre-ovulatory large follicles (Magoffin, 2005; Wickenheisser et al., 2006). The granulosa cells respond to follicle stimulating hormone (FSH) and produce estrogen. Granulosa cells in the large pre-ovulatory follicle also respond to LH and produce progesterone (Webb et al., 2004; Skinner, 2005). The cow is a useful model for the investigation of ovarian physiology since the bovine follicle size, cell biology and endocrinology are similar to that of the human (Webb et al., 2004; Moore and Thatcher, 2006). The size of bovine antral follicles allows for isolation of purified theca and granulosa cell populations for the study of antral follicle development (Skinner and Osteen, 1988; Roberts and Skinner, 1990b).
Interactions between the theca cells and granulosa cells are required during antral follicle development (Parrott and Skinner, 1998b; Tajima et al., 2006). Theca–granulosa interactions are an example of a mesenchymal–epithelial cell interaction (Parrott et al., 1994; Kezele et al., 2005a). LH acts on the theca cells to promote the production of androgen that is then utilized by the granulosa cells to produce estrogen under the control of FSH. The estrogen can feedback on the theca cells to regulate theca cell function (Roberts and Skinner, 1990a,c). The local production of growth factors is important to mediate the cell–cell interactions required for antral follicle development (Webb et al., 2004; Skinner, 2005). Examples of growth factors that mediate cell–cell interactions include transforming growth factor-alpha (TGFα), transforming growth factor-beta (TGFβ), hepatocyte growth factor (HGF), and keratinocyte growth factor (KGF/FGF7) that are produced by theca cells and subsequently act on granulosa cells (Skinner et al., 1987; Roberts and Skinner, 1991; Parrott and Skinner, 1998a,b). Granulosa cells produce TGFβs, insulin-like growth factors (IGFs) and basic fibroblast growth factor (bFGF/FGF2) that can influence cell–cell interactions (Schams et al., 2002; Nilsson et al., 2003; Juengel et al., 2004a; Buratini et al., 2005; Drummond, 2005; Walters et al., 2006). Granulosa production of kit ligand (KITL) and leukemia inhibitory factor (LIF) influences the oocyte that in turn produces growth differentiation factor 9 (GDF9) and bone morphogenic protein 15 (BMP15; Elvin et al., 1999; Su et al., 2004; Juengel et al., 2004b). Other growth factors produced by granulosa and theca cells that mediate local cell–cell interactions include vascular endothelial growth factor (VEGF; Kaczmarek et al., 2005; Shimizu, 2006), nerve growth factor (NGF; Salas et al., 2006), bone morphogenic proteins (BMPs; Glister et al., 2004; Shimizu et al., 2006), and kit ligand (Ismail et al., 1996; Parrott and Skinner, 1997).
A genomic view of the granulosa and theca cell transcriptomes provides a much more global and efficient analysis of antral follicle development and growth factor mediated paracrine and autocrine cell–cell interactions than the analysis of individual growth factors. Previous analyses of granulosa and theca cell transcriptomes by microarray procedures have focused on treated cells in culture (Sasson et al., 2003; Sriraman et al., 2006) or isolated developmental time points (Chin et al., 2002; Wood et al., 2003; Ben-Ami et al., 2006; Hernandez-Gonzalez et al., 2006; Mihm et al., 2006). These studies have been useful to examine cell transcriptomes and specified gene sets (Amsterdam et al., 2003; Andreu-Vieyra et al., 2006), but do not examine antral follicle development or assess granulosa and theca cells isolated from the same follicles. The current study investigates the genes regulated during antral bovine follicle development to establish an understanding of the granulosa and theca cell transcriptome changes, with a focus on the growth factors expressed that may mediate ovarian follicle paracine and autocrine cell–cell interactions.
MATERIALS AND METHODS
Tissue Collection and Cell Isolation
Bovine ovaries were obtained from young nonpregnant cycling heifers less than 10 min after death. Ovaries were delivered fresh on ice by the Animal Reproduction Core Laboratory of the WSU/UI Center for Reproductive Biology. Only follicular phase ovaries were used as assessed by morphological criteria previously described (Ireland et al., 1980), that included the absence of a corpus luteum (CL) and the presence of developing large antral follicles. Follicles were dissected from the ovaries. Granulosa cells were isolated by dissection (i.e., scraped and flushed) from fresh tissue as previously described (Skinner and Osteen, 1988). Theca interna layers were then dissected (i.e., peeled) away in sheets from the follicle wall as previously described (Roberts and Skinner, 1990b). Cells were obtained from small (≤5 mm), medium (5–10 mm), and large (>10 mm) antral follicles (Skinner and Osteen, 1988; Roberts and Skinner, 1990b). Healthy vascularized follicles were identified and morphologically atretic follicles were not utilized. Pooled granulosa or theca cell samples from medium and large follicles may include cells from healthy subordinate, as well as future dominant follicles. Cell preparations obtained by this procedure have been characterized cytochemically to be greater than 98% pure and contain less than 1% contamination with endothelial cells (Skinner and Osteen, 1988; Roberts and Skinner, 1990b). RNA was extracted from freshly isolated cells using the TRIZOL reagent (Invitrogen, Grand Island, NY) per manufacturer’s instructions. About 20–40 different bovine follicles at each size were used for each individual RNA extraction, so that each pooled RNA sample represented 6–10 individual animals. All procedures were approved by the WSU Animal Care and Use Committee.
Microarray Analysis
An aliquot of each RNA sample was evaluated for quality by running on an agarose formaldehyde gel. Only samples with the expected proportions of 18S or 28S bands and which had no detectable (i.e., no extraneous smeared bands) genomic DNA contamination were used. RNA was hybridized to the Affymetrix (Affymetrix, Santa Clara, CA) bovine genome array chip. The Genomics Core in the Center for Reproductive Biology at Washington State University performed the analysis as previously described (McLean et al., 2002; Shima et al., 2004; Small et al., 2005; Kezele et al., 2005b). Briefly, RNA from individual granulosa and theca cell RNA preparations were reverse transcribed into cDNA and cDNA was transcribed into biotin labeled RNA. Biotin labeled RNA was then hybridized to the Affymetrix bovine genome array chips. Each gene set is composed of 11 pairs of 25-mer oligonucleotides, with one oligonucleotide being an antisense strand specific for the gene and one oligonucleotide being an antisense strand with a single point mutation for use as a comparative negative control. The oligonucleotides span the gene so 5′ and 3′ regions are contributing to the final signal obtained. Biotinylated RNA was then visualized by labeling with phycoerythrin-coupled avidin. The microarray was scanned on a Hewlett-Packard Gene Array Scanner (Hewlett-Packard Co., Palo Alto, CA). Two microarray chips were run from two different RNA samples from each of small, medium, and large bovine granulosa and theca cell preparations.
Bioinformatics and Microarray Statistics
Microarray output was examined visually for excessive background noise and physical anomalies. The default Microarray Suite (MAS) statistical values were used for all analyses. Probe sets were scaled to 125 signal for analysis. An absolute analysis using MAS was performed to assess the relative abundance of the transcripts based on signal and detection (present, absent, or marginal) for the 11 different oligonucleotides sets per gene. The absolute analysis from MAS was imported into GeneSpring 7.3 software (Silicon Genetics, Redwood City, CA). The developing follicle data were normalized within GeneSpring using the default/recommended normalization methods. These include setting of signal values below 0.01–0.01, total chip normalization to the 50th percentile, and normalization of each gene to the median. Therefore, normalization occurred on an individual gene level with the multiple probe sets rather than to a potentially irrelevant gene probe set. These normalizations allowed for the visualization of data based on relative abundance at any given time point rather than compared to a specific control value. Data restrictions and analytical tools in GeneSpring were applied to isolate noteworthy and possibly important patterns of gene expression. Transcripts expressed differentially with fold change 1.5 or higher (i.e., the majority were higher) between groups were determined separately for granulosa cell and theca cell samples. This was applied to all three follicle size samples and considered all transcripts represented on the arrays. Two repeats for each follicle development stage and cell type were performed and allowed for a 2×2 factorial comparison in the experiment. Subsequently, expression restrictions were applied to the transcripts expressed in a significant manner. These restrictions were designed so that the remaining transcripts met the following requirements in addition to being expressed in a significant manner: (1) each transcript must have a signal value of at least 75 in at least 1 of the 3 developmental stages; and (2) have an average fold change of 1.5 or greater in signal intensity between any 2 developmental stages. A signal of >75 was selected to avoid the false positive calls associated with a signal of >50 and false negative calls with a signal of >100. Genes were only selected if they had a statistically significant present call with a signal >75 and a change in expression (> 1.5-fold). The resulting transcripts were screened using Excel (Microsoft, Redmond, WA) for redundant UniGene entries. Transcripts that passed these restrictions were considered for further analysis. Cluster Analysis and patterns of gene expression were identified using unsupervised analysis within the set of differentially expressed transcripts that met the requirements detailed previously. Clustering algorithms allow for the separation of distinct patterns of expression based on the similarity of expression profiles between different genes (Eisen et al., 1998; Kezele et al., 2005b). In this analysis, a hierarchical clustering algorithm utilizing a smooth correlation with the default parameters was used in order to group genes with similar patterns of expression within the time course. The relationships of the various transcriptomes is depicted using an analysis that generates a dendrogram to illustrate major expression patterns within the differentially expressed transcripts (determined through statistical analysis) in a continuous fashion. This dendrogram analysis allows a visualization of the entire changing transcriptome for comparison between samples. Pathway Assist (Ariadne Genomics, Inc., Rockville, MD) software was used for detailed pathway analysis and gene associations. Previous studies have shown that microarray data correlates well with real-time quantitative PCR and Northern analysis (Sadate-Ngatchou et al.,2004; Kezele et al., 2005b). The microarray procedure involves 11 pairs of perfect match and mismatch oligonucleotides for each gene versus the single primer set used for the quantitative PCR analysis. Therefore, the microarray provides a more robust quantitation and more effective elimination of false positives and negatives in comparison to the PCR analysis. Therefore, global microarray data does not generally need to be confirmed as previously suggested and selected genes generally have been shown to confirm the analysis (McChlery and Clarke, 2003; Shima et al., 2004). However, three genes were selected in the current study for confirmation with real-time PCR (i.e., Vegf, TGFβ1, and aromatase Cyp19) as previously described (Kezele et al., 2005b), and the expression changes for all three genes were found to provide data similar to the microarray analysis (Supplemental Fig. S2).
RESULTS
The morphology of the bovine small, medium, and large antral follicles are shown in Supplemental Figure S1. The theca layer is composed of a theca externa and interna, which are vascularized. The granulosa cells adjacent to the theca cells are the mural granulosa cells while those adjacent to the oocyte are the cumulus granulosa cells, Supplemental Figure S1. Follicular phase healthy developing follicles having this morphology were used. As the follicle develops an increase in cell numbers and layers is in part responsible for the increased size of the antral follicle. Small (<5 mm), medium (5–10 mm), and large (>10 mm) antral follicles were isolated from freshly collected bovine ovaries as described in the Materials and Methods Section (Skinner and Osteen, 1988; Roberts and Skinner, 1990b). Granulosa cells were isolated and then theca cells microdissected from the ovary. For each experiment RNA was isolated from three different preparations of cells from different ovary collection sets and combined for analysis. Each pooled RNA sample represented 6–10 individual animals as described in the Materials and Methods Section. Two separate experiments were performed to provide duplicate Affymetrix microarray chips for each cell type/follicle size combination. The R2 values from the duplicate chip comparisons were greater than 0.98 for both theca cell and granulosa cell chips. Therefore, the variability of the chips and samples was negligible. A set of known ovarian cell specific genes was selected to verify the system and indicate the purity of the cell preparations. Table 1 demonstrates that the genes anticipated to be specific for granulosa cells [i.e., FSH receptor (Fshr), Anti-Müllerian hormone (Amh; i.e., Müllerian inhibitory substance), and aromatase (Cyp19)] are highly expressed in the granulosa cells with changes between the small, medium, and large samples, and have negligible expression in the small and medium follicle theca cells. The exception was the unexpected high level of aromatase in the large follicle theca cells. This large follicle theca cell aromatase gene expression was confirmed with a real-time PCR analysis and showed results similar to the microarray data, namely that there is negligible expression in small antral follicles and high levels of expression in large antral follicle theca cells, comparable to large antral follicle granulosa cells (Supplemental Fig. S2). A theca cell gene Lhr (i.e., LH receptor) was primarily expressed in the theca cells and not granulosa cells, Table 1. Expression of an oocyte gene, growth differentiation factor 9 (Gdf9), was not detected in either the granulosa or theca cell preparations, Table 1. Therefore, the cell population purity and isolation system appears appropriate for a transcriptome analysis.
TABLE 1.
Known Cell Specific Gene Expression Comparison
| Microarray signal
|
|||
|---|---|---|---|
| Gene | Small | Medium | Large |
| Granulosa cells | |||
| Fshr | 154 | 220 | 103 |
| Cyp19 (aromatase) | 120 | 2467 | 2477 |
| Lhr | ND | ND | ND |
| Gdf9 | ND | ND | ND |
| Amh | 644 | 373 | 186 |
| Theca cells | |||
| Fshr | ND | ND | ND |
| Cyp19 (aromatase) | ND | 548 | 2956 |
| Lhr | 32 | 45 | 123 |
| Gdf9 | ND | ND | ND |
| Amh | 150 | 89 | ND |
ND indicates a nondetectable signal or absent call on the microarray chip. Raw microarray signal from small (<5 mm), medium (5–10 mm), or large (> 10 mm) follicles.
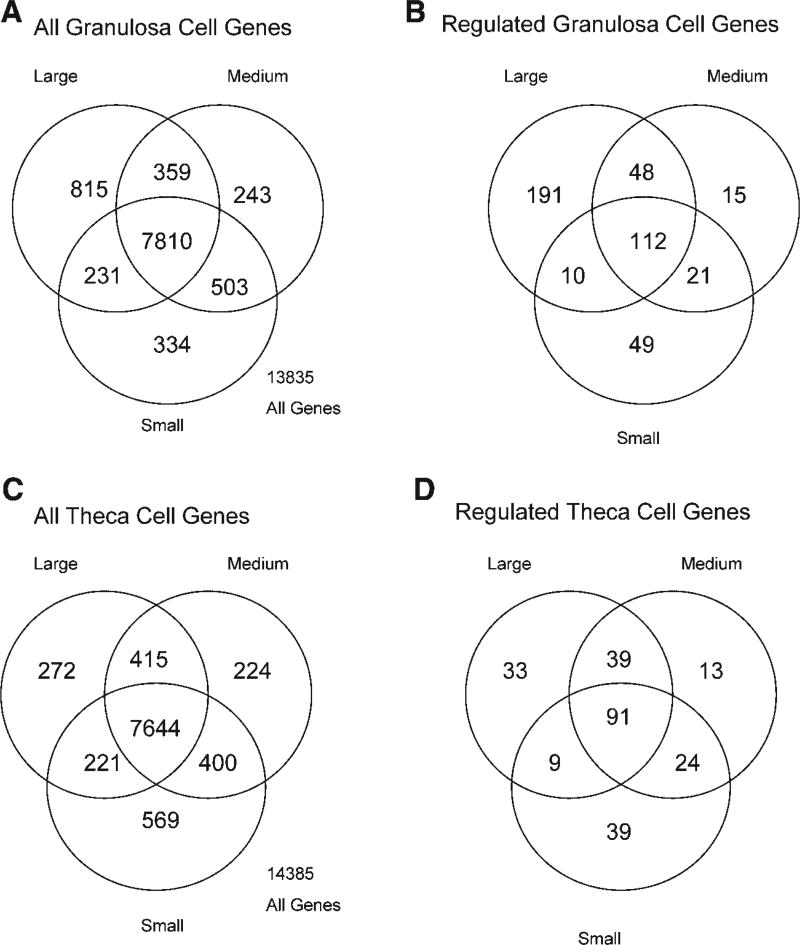
Analysis of the transcriptome was performed on an Affymetrix bovine genome microarray containing approximately 24,000 genes. Most genes are called absent with a signal that is approximately 50, but this varies between specific genes such that a signal cut-off of 75 was used. Therefore, the signal cut-off of 75 reduces the false positives observed with a cut-off of 50 and reduces the false negatives observed with a cut-off of >100. For granulosa cells a Venn diagram of all genes illustrates 13,835 genes detected with 7,810 common between small, medium, and large follicles and 334 specific to small, 243 specific to medium, and 815 specific to large follicle granulosa cells, Figure 1A. Regulated genes were defined as those with a minimum of 1.5-fold change between two (i.e., small to medium, medium to large, or small to large) of the small, medium, or large sample isolates. These regulated granulosa genes are shown in Figure 1B with overlap and specificity shown for 446 genes. The theca cell transcriptomes for all genes detected are shown in Figure 1C with 14,385 genes detected and 7,644 common between small, medium, and large follicles and 589 specific to small, 224 specific to medium, and 272 specific to large follicles. Regulated theca cell genes are shown in Figure 1D with overlap and specificity for 248 genes.
Fig. 1.
The microarray Venn diagram of expressed genes for bovine antral follicle (A,B) granulosa cells and (C,D) theca cells. The total number of expressed granulosa (A) and theca (C) genes are shown for small, medium and large antral follicles. The number of regulated genes (change > 1.5-fold) are shown for granulosa (B) and theca (D) cells from small, medium, and large antral follicles. Overlapping circles represent genes expressed in common.
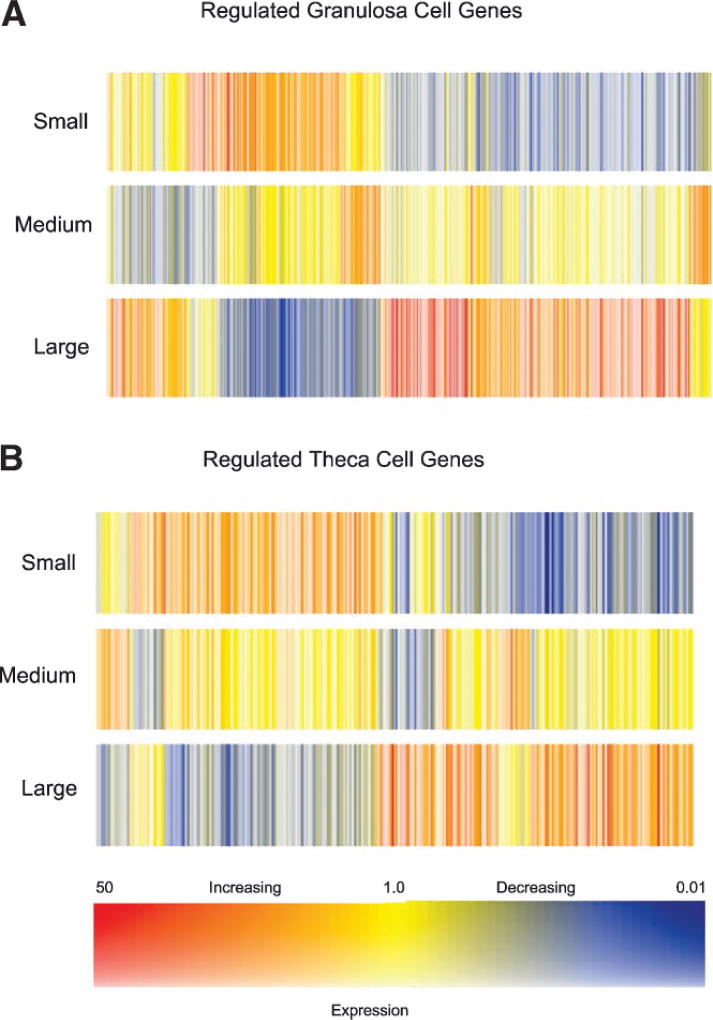
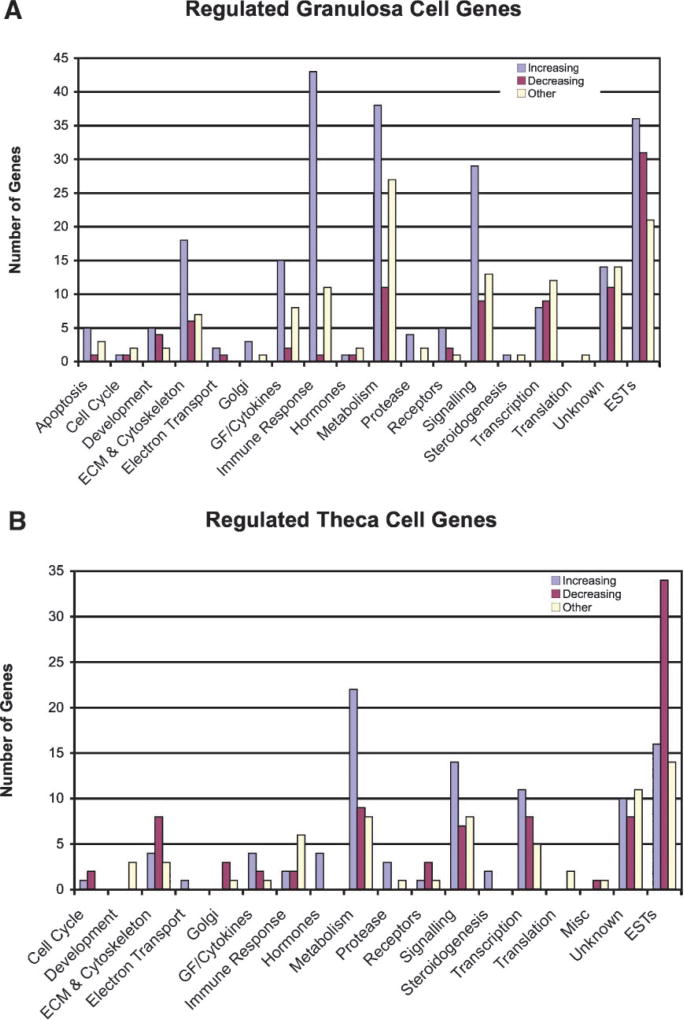
The dendrograms for the granulosa and theca cell regulated genes are shown in Figure 2 demonstrating the general changes during antral follicle development. This analysis allows the entire changing transcriptome differences to be compared between samples. Each gene has a specific colored line with an increase or decrease in expression as indicated. The regulated genes fall into five general clusters of expression patterns (i.e., color patterns) with the majority either increasing or decreasing between small, medium, and large antral follicles. Two clusters have highest or lowest levels of expression in the medium follicles, Supplemental Figure S3. The regulated gene lists for both the granulosa and theca cells were categorized into functionally related gene families or networks, Figure 3. For granulosa cells the major gene categories of immune regulation genes, metabolism genes, signaling genes and extracellular and cytoskeletal genes are predominant. For theca cells the major gene categories of metabolism genes, signaling genes, and transcription genes are predominant. The specific genes in these different categories are listed in the online Supplemental Tables S1 and S2. Both granulosa and theca cells had a large number of unknown and expressed sequence tags (ESTs), Figure 3. As shown in Figure 3 the majority of genes in specific categories have increased expression as the follicle develops. Genes that have an increased or decreased peak expression at the medium antral follicle stage (i.e., other) are also present in most categories, Figure 3.
Fig. 2.
The microarray dendrogram for regulated granulosa (A) and theca (B) cell genes from small, medium, and large antral follicles. Each colored line represents a different gene. The color scale is presented with blue as decreasing expression and red as increasing expression between the different size antral follicles.
Fig. 3.
Functionally related gene categories for granulosa cell (A) and theca cell (B) regulated genes. The number of genes increasing in expression (L > M > S) or decreasing (S > M > L) are indicated. Those listed as Other have the highest or lowest level of expression at the medium follicle stage. Known genes without a specific category are listed as unknown and expressed sequence tags (ESTs) with unknown function are listed.
A pathway analysis revealed a number of signal transduction and cellular processes containing multiple (i.e., >5) regulated genes in developing antral granulosa cells, but fewer in theca cells, Table 2. Developing antral follicle granulosa cell regulated pathways included those for cell adhesion molecules, regulation of actin cytoskeleton, focal adhesion, the PPAR signal pathway, cytokine receptor interactions and the MAPK signaling pathway. The theca cell gene lists did not identify any specific pathways with greater than three regulated genes, but did contribute to regulation of several of the pathways that contained granulosa cell regulated genes, Table 2. These regulated pathways in the developing antral granulosa cells are anticipated to have a role in antral follicle development.
TABLE 2.
Regulated Cellular Pathways
| Pathway | Total pathway genes |
Granulosa regulated genes |
Theca regulated genes |
|---|---|---|---|
| Cell adhesion molecules (CAMs) | 58 | 10 | 0 |
| Regulated actin cytoskeleton | 82 | 7 | 0 |
| Focal adhesion | 103 | 6 | 2 |
| PPAR signaling pathway | 44 | 6 | 3 |
| Cytokine receptor interactions | 80 | 5 | 2 |
| MAPK signaling pathway | 106 | 5 | 0 |
Total pathway genes involves the total number of genes present in the listed pathway as compiled by KEGG and accessed through GeneSpring. Number of genes from granulosa or theca microarray analysis are defined as regulated according to the criteria in Methods and which are present in the listed pathway.
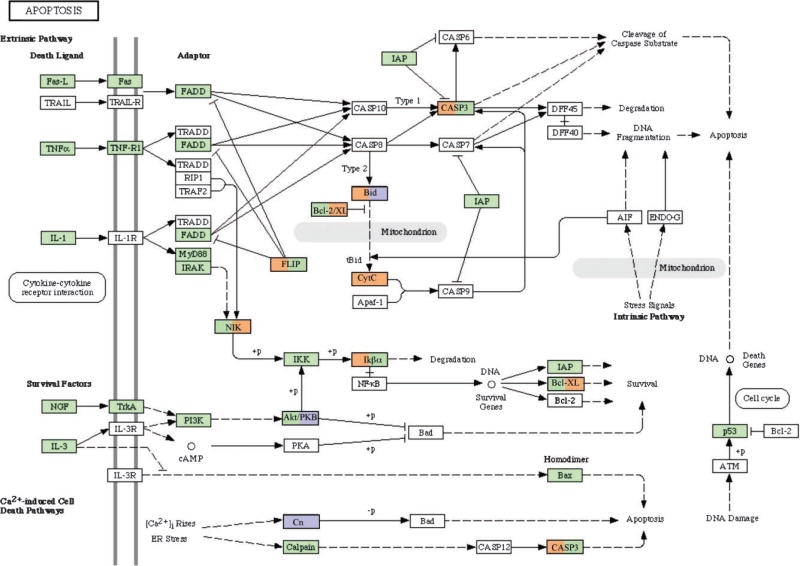
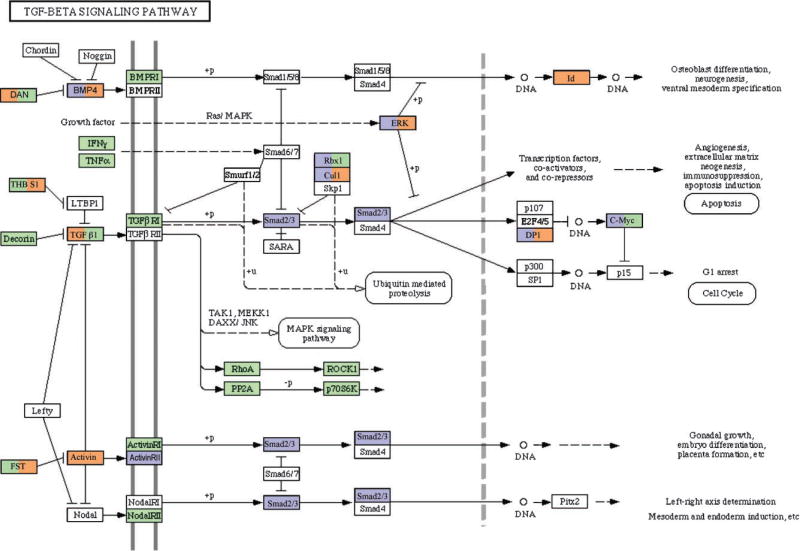
Analysis of specific cellular pathways was focused on extracellular regulation of antral follicle development. One signaling pathway critical for folliculogenesis is the apoptosis pathway. Developing follicles require a suppression of apoptosis while atretic follicles require a stimulation of this signaling pathway. A number of genes within the apoptosis pathway were regulated in both theca and granulosa cells, Figure 4. Genes associated with the induction of apoptosis [e.g., Bid, caspase 3 (Casp3)] were increased, but also those that inhibit apoptosis (e.g., Flip). Therefore, cells develop the capacity to undergo apoptosis, and regulation is independent of gene expression. Those apoptosis related genes with different expression patterns between theca and granulosa are indicated in Figure 4. Another signaling pathway regulated during antral follicle development was the TGFβ family signaling pathway, Figure 5. A large number of the genes were regulated with some differences between theca and granulosa cells. Due to the large number of TGFβ family growth factors having a role in the ovary (e.g., Gdf9, Tgfb1-3, Amh), this signaling pathway is expected to be regulated and influence both theca cell and granulosa cell growth and differentiation, Figure 5.
Fig. 4.
Apoptosis signaling pathway with green indicating genes with no change in expression between antral follicle stages, white indicating genes with no detectable expression or absent from the microarray chip, orange indicating genes with an increase in expression and blue indicating genes with a decrease in expression. Those genes with two colors have the granulosa expression on left and theca expression on right side of box.
Fig. 5.
Transforming growth factor-beta (TGFβ) signaling pathway with green indicating genes with no change in expression between antral follicle stages, white indicating genes with no detectable expression or absent from the microarray chip, orange indicating genes with an increase in expression and blue indicating genes with a decrease in expression. Those genes with two colors have the granulosa expression on left and theca expression on right side of box.
An analysis of growth factors and cytokines expressed and regulated during follicle development in either theca cells or granulosa cells was made, Table 3. Examples of genes that either decrease or increase during follicle development are observed. In granulosa cells a large number of cytokines have an increased expression, while the Igf family is prominent as well, Table 3. The number of theca cell candidate antral follicle regulatory growth factors is less than that observed for granulosa cells. Regulated gene expression in small, medium, and large antral follicles demonstrates a group of 16 growth factors that increase (e.g., Gmfg, Vegf, Tgfβ1, Ccl2, Ccl5, Ccl8), and a group of 4 that decrease (e.g., Tdgf1) in expression during antral follicle development, Table 3. Genes that regulate growth factor function are also regulated (i.e., Igfbp2). The increased expression of Vegf in theca cells between small and large antral cells was confirmed with a realtime PCR analysis (1.5-fold increase), as a selected gene to confirm the microarray analysis, Supplemental Figure S2. Two growth factors, Ccl2 and Spp1, increase in antral follicle development and are expressed by both granulosa and theca cells. Analysis of receptors associated with the growth factors identified is shown in Table 4. Not all growth factors have identified receptors and not all receptors are located on the microarray chips. Therefore, the list of receptors is a minimal list to be considered. The granulosa cells had 10 receptors associated with 8 of the growth factors shown to be expressed by granulosa cells, Table 4. The theca cells had one receptor (i.e., Ccr1), that was also expressed by granulosa cells, Table 4, and responds to the cytokine Ccl5 expressed by granulosa cells, Table 3. Four of the receptors had the highest level of expression in the large antral follicle granulosa cells (i.e., Cxcr4, Ccr1, Cxcr3, Ccr5), while most maintained relatively equal levels of expression in different follicle sizes, Table 4. These observations suggest both paracrine and autocrine growth factor signaling occurs. Therefore, a number of candidate regulatory growth factors were identified as being expressed by granulosa cells and theca cells that now require future studies to assess potential functional roles in antral follicle development.
TABLE 3.
Candidate Antral Follicle Regulatory Growth Factors
| Microarray signal
|
|||||
|---|---|---|---|---|---|
| Gene symbol | S | M | L | GenBank | Description |
| (A) Bovine granulosa cell growth factors and cytokines | |||||
| Gmfg | 40 | 81 | 435 | CK771942 | Glia maturation factor, gamma (GMFG) |
| Loc414347 | 26 | 57 | 338 | CB463807 | Putative MIP1-beta protein (LOC414347) |
| Sdf1 | 37 | 72 | 115 | CK771033 | Stromal cell derived factor 1 |
| Spp1 | 627 | 1,935 | 6,851 | NM_174187.2 | Secreted phosphoprotein 1 [osteopontin] |
| Notch1 | 8 | 26 | 90 | CK772450 | Similar to NP_032740.2 Notch gene homolog 1 |
| Vegf | 44 | 83 | 134 | NM_174216.1 | Vascular endothelial growth factor |
| Tgfβ1 | 65 | 107 | 231 | BM251237 | Transforming Growth Factor beta 1 |
| Ccl2 | 63 | 166 | 611 | NM_174006.2 | Chemokine (C-C motif) ligand 2 |
| Ccl3l1 | 50 | 77 | 305 | NM_174511.2 | Chemokine (C-C motif) ligand 3-like 1 |
| Ccl5 | 3 | 22 | 427 | NM_175827.2 | Chemokine (C-C motif) ligand 5 |
| Ccl8 | 8 | 15 | 105 | NM_174007.1 | Chemokine (C-C motif) ligand 8 |
| MGC137290 | 109 | 176 | 1054 | CK770974 | Similar to chemokine (C-X-C motif) ligand 16 |
| Tdgf1 | 75 | 9 | 15 | CB468433 | Teratocarcinoma-derived growth factor 1 (TDGF1) (Cripto) |
| MGC137324 | 101 | 46 | 21 | CK959718 | Similar to angiopoietin-4 precursor (ANG-4) |
| Loc504526 | 56 | 35 | 111 | CK771963 | Similar to growth arrest-specific 6 (GAS6) |
| Gdf8 | 81 | 13 | 34 | BF652431 | Growth differentiation factor 8 (Myostatin) |
| Igfbp2 | 293 | 151 | 76 | NM_174555.1 | Insulin-like growth factor binding protein 2, 36 kDa |
| Igfbp3 | 152 | 88 | 259 | NM_174556.1 | Insulin-like growth factor binding protein 3 |
| Igfbp4 | 74 | 259 | 167 | NM_174557.2 | Insulin-like growth factor-binding protein 4 |
| (B) Bovine theca cell growth factors and cytokines | |||||
| Ccl2 | 83 | 159 | 252 | NM_174006.2 | Chemokine (C-C motif) ligand 2 (Mcp-1) |
| Xcl1 | 269 | 855 | 1606 | NM_175716.2 | Chemokine (C motif) ligand 1 |
| Spp1 | 55 | 129 | 674 | NM_174187.2 | Secreted phosphoprotein 1 [osteopontin] |
| Ang | 93 | 269 | 501 | NM_173891.1 | Angiogenin |
| Loc508666 | 1193 | 697 | 2794 | CB465019 | Similar to Small inducible cytokine A23 (CCL23) (MIP3) |
| Loc522611 | 118 | 77 | 28 | CB431697 | Similar to Stem cell growth factor precursor (P47) |
| Vegf | 73 | 66 | 107 | CB450531 | Vascular endothelial growth factor |
Bold lines are genes that increase (S<M<L) during antral follicle development. Raw microarray signal for cells from small (S), medium (M), or large (L) follicles.
TABLE 4.
Candidate Antral Follicle Regulatory Growth Factor Receptors
| Microarray signal
|
||||||
|---|---|---|---|---|---|---|
| Gene symbol | S | M | L | Genbank | Receptor description |
Ligand |
| (A) Bovine granulosa cell growth factor receptors | ||||||
| Cxcr4 | 96 | 53 | 460 | NM_174301.3 | Chemokine (C-X-C motif) receptor 4 | SDF1 |
| Ccr1 | 35 | 53 | 264 | CK773226 | Chemokine (C-C motif) receptor 1 | CCL5 |
| Cxcr3 | ND | ND | 63 | CK774970 | Chemokine receptor 3 | CCL3L1 |
| Ccr5 | ND | ND | 70 | AW658666 | Chemokine receptor 5 | MIP1 |
| Flk1 | 22 | 24 | 19 | CB166172 | Tyrosine kinase receptor | VEGF |
| Cxcr6 | ND | ND | 158 | CB170970 | Chemokine (C-X-C motif) receptor 6 | CXC16 |
| TgfbR1 | 18 | 24 | 40 | NM174621.2 | Transforming growth factor beta receptor 1 | TGFB1 |
| Loc504429 | 704 | 534 | 797 | AW345638 | Similar to MER receptor tyrosine kinase | GAS6 |
| Loc516598 | 656 | 715 | 837 | BF440555 | Similar to AXL receptor tyrosine kinase | GAS6 |
| Loc788224 | 813 | 507 | 710 | AW313997 | Similar to RSE receptor tyrosine kinase | GAS6 |
| (B) Bovine theca cell growth factor receptors | ||||||
| Ccr1 | 57 | 43 | 59 | CK773226 | Chemokine (C-C motif) receptor 1 | CCL5 |
Bold lines are genes that increase (S < M < L) during antral follicle development. Raw microarray signal for cells from small (S), medium (M), or large (L) follicles.
DISCUSSION
Regulation of the granulosa and theca cell transcriptomes during bovine ovarian antral follicle development was investigated. Bovine granulosa and theca cells were isolated from small (≤5 mm), medium (5–10 mm), and large (> 10 mm) antral follicles. The purity of the isolated cells has been shown to be greater than 98% in cell culture (Skinner and Osteen, 1988; Roberts and Skinner, 1990b). The microarray data for specific granulosa and theca cell genes demonstrated a high level of purity of the cells, Table 1. FSH receptor (Fshr) was only observed in granulosa, while LH receptor (Lhr) was in theca cells. A marker for oocytes, Gdf9, was not present in either cell preparation. Therefore, as previously described (Skinner and Osteen, 1988; Roberts and Skinner, 1990b) the purity of freshly isolated antral granulosa and theca cells is appropriate for the subsequent analysis. Interestingly, a high level of aromatase (Cyp19) gene expression was observed in large and medium follicle theca cells, but not small theca cells, Table 1. Previously, theca cells from small, medium or large follicles were shown to have no detectable aromatase bioactivity in the ability to aromatize androgen (Roberts and Skinner, 1990a,b). Therefore, the functional role of this aromatase gene expression by the large follicle theca cells is unclear and remains to be investigated. In contrast to the rodent CL that only produces progesterone, the bovine and human CL produces both progesterone and estrogen. Although not rigorously investigated, the ability of the luteinized theca cells to cooperate in the CL estrogen production has been suggested (Inkster and Brodie, 1991; Watson, 2000; Okuda et al., 2001; Lenz et al., 2004). Future studies will need to investigate this further along with the bioactivity of any aromatase expressed. The hypothesis would be that large antral follicle theca cells express aromatase, but do not have aromatase bioactivity until in the CL when the luteinized theca develop the capacity to produce estrogen. Since the purity of the isolated cells has been shown to be >98% (Skinner and Osteen, 1988; Roberts and Skinner, 1990b), the level of aromatase (Cyp19) gene expression is too high to suggest a contamination issue, Table 1, but this issue needs to be further investigated. Although this bovine theca cell aromatase expression is an interesting observation, it does not influence the current study nor the conclusion that the cell populations are adequate for further investigation. The lack of detectable expression of LH receptor (LHR) in the large follicle granulosa cells was also unexpected. The theca cell expression of LHR is higher than granulosa in most mammals, but the raw signal value in large follicle theca was only 123, Table 1. Therefore, either the steady state levels of LHR mRNA are low compared to protein, or the array signal sensitivity for LHR is low. The lack of LHR detection in large follicle granulosa cells is likely due to one of these parameters.
The morphological criteria described in the Materials and Methods Section used to select follicles to pool for each RNA sample allow for the possibility that both healthy subordinate follicles as well as future dominant follicles may be included in the medium and possibly large follicle samples. The high R2 values for the duplicate chips from each treatment group indicate that the proportions of subordinate and future dominant follicles are similar between duplicate samples. Therefore, comparing the genes expressed in small, medium and large follicles will not apply to dominant versus subordinate follicle development, rather will reflect those changes in gene expression that are common to dominant and subordinate follicles. The developmental processes that occur within the small, medium and large follicle sub-categories will need to be more thoroughly investigated in future analyses.
A large number of studies have investigated the expression of various individual genes during antral follicle development. To provide a more systems biology approach to understanding ovarian physiology a genome wide view of the different cells transcriptomes is required. The current study used a microarray approach to study granulosa cell and theca cell gene expression during antral follicle development. The bovine Affymetrix microarray chip has approximately 24,000 gene transcripts arrayed that represent the majority of the cow genome. Approximately 14,000 genes were expressed by either theca or granulosa cells, with approximately 7,000 of these in common. Unique cell specific sets of genes were expressed at each of the small, medium, and large stages of antral follicle development, Figure 1. Those genes found to be regulated (altered greater than 1.5-fold in expression between follicle stages) included some that were specific to a particular cell type and stage of development. Some genes decreased while others increased during antral follicle development, Figure 2 and Supplemental Figure S3. The bovine genome sequence is not fully annotated, so it was not surprising a large number of genes fell into the unknown or EST categories. Further identification of these genes and characterization of the bovine genome will provide additional insights into these regulated genes.
Previous microarray analyses of follicle development has primarily focused on isolated cell types from a specific developmental stage (Chin et al., 2002; Wood et al., 2003; Ben-Ami et al., 2006; Hernandez-Gonzalez et al., 2006; Mihm et al., 2006; Xu et al., 2006). Granulosa cells have been analyzed in cell culture (Sasson et al., 2003; Sriraman et al., 2006), at ovulation (Hernandez-Gonzalez et al., 2006), and in disease states (Chin et al., 2002). Theca cells have not been rigorously investigated (Wood et al., 2003, 2004; Mihm et al., 2006). Whole ovary analysis has also been useful to elucidate early stage follicle assembly and primordial follicle development (Kezele et al., 2005b), normal follicle development (Liu et al., 2001; Jo et al., 2004; De et al., 2006) and the impacts of disease states (Burns et al., 2003). The ovarian follicle transcriptomes from several species have been examined including primate (Xu et al., 2006), bovine (Mihm et al., 2006), pig (Caetano et al., 2004), and human (Wood et al., 2004). The analysis of the oocyte at various stages of follicle development has also been accomplished (Yao et al., 2004; Acevedo and Smith, 2005). The global analysis of the granulosa and theca cell transcriptomes during antral follicle development has not been previously investigated, and is the focus of the current study.
The microarray analysis of granulosa and theca cell transcriptomes during antral follicle development revealed sets of regulated genes that are specific to a particular cell type and follicle stage. Analysis of cellular pathways with multiple regulated genes present identified a number of pathways in granulosa cells, Table 2. Although several of these pathways have been shown to have a role in the ovary (Shiota et al., 2003; Tamura et al., 2004; Froment et al., 2006; Peluso, 2006), the role they have in granulosa cell antral follicle development remains to be elucidated. Pathways emphasized were the apoptosis and TGFβ signaling pathways. Apoptosis is required for both granulosa and theca cells during follicle atresia, so both cell types have the capacity to respond to cell death or survival signals. A large number of apoptosis pathway genes were not regulated, Figure 4, but several key genes were altered. The hormonal regulation of apoptosis (Elvin et al., 1999) is critical throughout antral follicle development. The TGFβ signaling pathway is also active in both cell types and regulated in a similar manner. Since a number of TGFβ family members such as BMPs, TGFB1-3, inhibin, activin, follistatin and AMH all are known to be produced locally in the ovary, the TGFβ signaling pathway has functional relevance to the expression data provided (Cook et al., 2004; Drummond, 2005; Mazerbourg and Hsueh, 2006). The current study focused on this and other growth factor mediated paracrine and autocrine cell–cell interactions to identify potential regulatory candidates for future analysis.
The ability of theca cells to influence granulosa cells initially was observed with steroid interactions (Roberts and Skinner, 1990a,b; Webb et al., 2004), and then a number of growth factors were shown to be produced by theca cells that regulate granulosa cells (Skinner et al., 1987; Roberts and Skinner, 1990a,c, 1991; Parrott et al., 1994; Parrott and Skinner, 1998a,b; Kezele et al., 2005a; Tajima et al., 2006). Theca cells were shown to produce the proteins TGFα (Roberts and Skinner, 1991), TGFβ (Skinner et al., 1987; Roberts and Skinner, 1991; Nilsson et al., 2003; Juengel et al., 2004a; Drummond, 2005), insulin-like growth factor (IGF; Schams et al., 2002; Walters et al., 2006), HGF (Parrott et al., 1994; Parrott and Skinner, 1998a) and keratocyte growth factor (KGF; Parrott et al., 1994; Parrott and Skinner, 1998b; Kezele et al., 2005a) that can all influence granulosa cell growth. Granulosa cells also produce secreted regulatory factors that can influence theca cells including kit ligand (KITL; Ismail et al., 1996; Parrott and Skinner, 1997; Shimizu et al., 2006), FGF2 (Buratini et al., 2005), and VEGF (Kaczmarek et al., 2005; Shimizu, 2006). Granulosa cells and oocytes also interact in the production of secretory factors such as GDF9 and BMP15 (Elvin et al., 1999; Su et al., 2004; Juengel et al., 2004b). Therefore, paracrine and autocrine cell–cell interactions are thought to be essential for the growth and development of the antral follicle (Webb et al., 2004; Tajima et al., 2006). Characterization of additional growth factors that can mediate ovarian cell–cell interactions on a genome wide level with microarray analysis is an objective of the current study.
A number of candidate regulatory growth factors were found to be expressed by both granulosa and theca cells, Table 3. Interestingly, some increased during antral follicle development, while others decreased. For granulosa cells several groups of functionally related genes were identified. The first is the insulin-like growth factor (Igf) family. Igf1 has been shown to be important for both granulosa and theca cell function, in particular for cell growth regulation, as observed for most cell types (Edmondson et al., 2003; Romano, 2003; Oksbjerg et al., 2004; Kurmasheva and Houghton, 2006; Ye and D’Ercole, 2006). The Igf binding proteins, Igfbp2, 3, and 4, were found to be expressed by granulosa cells. Interestingly, Igfbp2 was highest in small follicles, Igfbp4 highest in medium follicles and Igfbp3 highest in large follicles, Table 3. Potential differential actions and functions should be considered. The interplay of Igf1 and the IGFBPs has been previously implicated in regulating antral follicle development (Fortune et al., 2004; Gerard et al., 2004; Voge et al., 2004; Santiago et al., 2005; Canty et al., 2006; Walters et al., 2006).
The second functionally related growth factor family of genes expressed by granulosa cells was the cytokines, Table 3. Granulosa cells expressed chemokines Ccl2, Ccl3l1, Ccl5, Ccl8, and Cxcl16. All cytokines increased with antral follicle development and all had relatively high levels of expression in the large antral follicle, with Cxcl16 being highest. These cytokines have been shown to be expressed by cells during viral infection and to induce activation and migration of immune cells to combat infection (Salazar-Mather and Hokeness, 2006). A previous granulosa cell mircroarray analysis identified cytokines and other immune related genes as being prevalent in cumulus–oocyte complexes (Hernandez-Gonzalez et al., 2006). Combined observations of the granulosa cell transcriptome supports a potentially important role for these immune related genes in the antral follicle. Although immune cell inter-communication may be a factor (e.g., immune cells in pre-ovulatory follicles; Castilla et al., 1990; Loukides et al., 1990), cytokine actions on granulosa cells, theca cells and the oocyte need to be considered. The receptors for a number of these cytokines were found to be expressed by granulosa cells, Table 4. Therefore, autocrine actions of these cytokines are speculated to be important for antral follicle development.
A number of other granulosa cell expressed growth factors were identified. Growth factors that increased in expression during antral follicle development included glial maturation factor gamma (Gmfg). Gmfg is related to Gmfb which affects growth and differentiation of neurons and glia (Asai, 2001). Gmfg is expressed in endothelial and inflammatory cells and localizes with cytoskeletal elements to modulate migration and differentiation (Ikeda et al., 2006). Macrophage inducing protein beta (Mip1), also known as Ccl4, increased during antral development and has been implicated in a variety of chemokine-mediated actions including hematopoesis, cancer-mediated bone disease, asthma, and macrophage and lymphocyte function (Maurer and von Stebut, 2004; Bisset and Schmid-Grendelmeier, 2005; Sezer, 2005; Ramkissoon et al., 2006). Stromal cell derived factor-1 (Sdf1), also known as Cxcl12, mediates a wide variety of chemokine actions including regulation of hematopoesis (Lataillade et al., 2004), cancer progression (Egeblad et al., 2005), and embryogenesis (Nagasawa et al., 1998). Previously Sdf1a has been implicated in inhibiting the initiation of growth of primordial follicles in the ovary (Holt et al., 2006). The receptor for SDF1 (i.e., CXCR4) is expressed by granulosa cells, with highest levels in the large follicles, Table 4. Therefore, an autocrine action of Sdf1 is proposed on antral follicle granulosa cells. Osteopontin is a secreted glycoprotein (Denhardt and Guo, 1993)that is expressed by granulosa cells at high levels and has been previously shown to be involved in mammary gland development and function (Nemir et al., 2000). Osteopontin (SSP1) may mediate cell matrix interactions in the ovarian antral follicle (Denhardt and Guo, 1993). Two growth factors, Vegf and Tgfb1, were shown in the current experiment to have their highest levels of expression in the granulosa cells of large antral follicles (Table 3). The identification of these factors helps confirm the microarray approach. Vegf has been shown to have a role in vascularization of the developing large follicles (Wulff et al., 2002; Fraser, 2006), while Tgfb1 has been shown to regulate cell proliferation and differentiation of both granulosa and theca cells (Dorrington et al., 1993; Attia et al., 2000; Saragueta et al., 2002; Johnson et al., 2004; Woods et al., 2005).
Growth factors expressed by granulosa cells that declined during antral follicle development or peaked at the medium antral follicle stage, were also identified, Table 3. The teratocarcinoma-derived growth factor 1 (Tdgf1; also known as cripto) was expressed at its highest levels in the small antral follicle. Tdgf1 acts as both a ligand for activation of the src-Akt pathway and in the regulation of the nodal signaling pathway. Tdgf1 is involved in embryogenesis and tumorigenesis (Kenney et al., 2004; Strizzi et al., 2005). Granulosa cells from small antral follicles expressed the angiogenin (Ang) family member (Lee et al., 1999; Koga et al., 2000; Kawano et al., 2003; Horikawa et al., 2005) angiopoietin-4 (Ang4), a modulator of angiogenesis through binding of the endothelial Tie2 receptor (Lee et al., 2004). A granulosa expressed growth factor with high levels of expression in the large and small antral follicles is growth arrest-specific 6 (Gas6),that has previously been implicated in kidney function, blood clot formation and in regulating cell apoptosis (Yanagita, 2004; Lafdil et al., 2006; Saller et al., 2006; Shankar et al., 2006). The GAS6 and associated receptors Mer, Axl, and Rse have been shown to be important in testis function (Lu et al., 1999). The GAS6 receptors, MER, AXL, and RSE, were found to be expressed by granulosa cells at high levels throughout antral follicle development, Table 3. Therefore, GAS6 appears to have the potential to mediate autocrine granulosa cell–cell interactions during antral follicle development. Granulosa cells from small antral follicles also expressed growth differentiation factor 8 (GDF8) (also known as myostatin) that is a member of the TGFβ family of growth factors and has been shown to negatively regulate muscle growth (Tsuchida, 2004). Although the GDF8 knockout mice do not have fertility defects (McCroskery et al., 2005), GDF8 may have other compensatory growth factors present, such as TGFβ family members, that mask important functions or phenotypes in the null mutant experiments. Similar observations have been made with other ovarian regulatory factors such as LIF that do not have an ovarian null mutant phenotype, but are known to influence germ cell development (Cheng et al., 2002; Molyneaux et al., 2003). These candidate antral follicle regulatory growth factors from the granulosa now need to be investigated on a functional level.
Candidate antral follicle regulatory growth factors were also identified in theca cells, although there were fewer than were found in granulosa. As found with granulosa cells, theca cells produced several immune related genes and cytokines. Theca cells expressed Ccl2 with the highest levels found in large antral follicles, Table 3. Therefore, both theca and granulosa cells express Ccl2. Previous literature has also reported expression of CCL2 (i.e., monocyte chemoattractant protein-1, MCP1), from both granulosa and theca/ stroma cells of pre-ovulatory follicles (Arici et al., 1997; Kawano et al., 2001, 2004). Another cytokine expressed by large antral follicle theca cells at relatively high levels is chemokine (Cmotif) ligand 1 (Cxcl1; also known as lymphotactin) that has previously been shown to be a chemoattractant for lymphocytes (Hedrick and Zlotnik, 1998; Stievano et al., 2004). A theca cell cytokine expressed at high levels in the large and small follicle stages is small inducible cytokine A23 (Ccl23; also known as Mpif1) previously shown to be chemotactic for lymphocytes, but inhibitory to myeloid precursor cells (Patel et al., 1997). As found with granulosa cells, the major group of theca cell growth factors identified was also cytokines, providing additional support for these factors in antral follicle development. Cytokines are known for their roles in attracting and regulating white blood cells, and such leukocytes can comprise a significant proportion of the cells present in pre-ovulatory follicles (Castilla et al., 1990; Loukides et al., 1990). Inflammatory processes mediated by leukocytes may contribute to pre-ovulatory follicle development (Wong et al., 2002). The bovine granulosa and theca cell preparations have previously been shown to be >98% pure, with negligible cell contamination (Skinner and Osteen, 1988; Roberts and Skinner, 1990b). Although we cannot exclude immune cell involvement, the potential that these signaling molecules have a direct role in regulation of follicle development, outside of this role as leukocyte mediators, now needs to be considered. An example of such local cytokine mediated cellular interaction potentially identified involves the expression of Ccl5 by granulosa cells and associated receptor Ccr1 expression by both theca cells and granulosa cells, Table 4. CCL5 may have both an autocrine and paracrine role in mediating theca and granulosa cell interactions during antral follicle development. The functional role of Ccl5 in antral follicle development now needs to be investigated. The final growth factor found to be expressed by the small antral follicle theca cells is stem cell growth factor (Scgf) that has previously been shown to stimulate growth of primitive hematopoetic progenitor cells (Hiraoka et al., 1997). These candidate antral follicle regulatory growth factors now need to be functionally investigated.
The analysis of the bovine granulosa and theca cell transcriptomes during antral follicle development has provided a resource to correlate gene expression to follicle development and identify novel regulatory factors. The database for this microarray study is available at www.skinner.wsu.edu or NCBI GEO. The candidate antral follicle regulatory growth factors identified suggest novel roles for a number of cytokines, as previously suggested (Machelon and Emilie, 1997; Bukulmez and Arici, 2000; Brannstrom and Enskog, 2002), and provides candidates for future functional analyses. These observations support a critical role for the local production and action of regulatory factors to mediate the cell–cell interactions essential for antral follicle development and ovarian function.
Supplementary Material
Acknowledgments
We acknowledge the expert technical assistance of Ms. Nada Cummings for the bovine tissue collection and the Center for Reproductive Biology (CRB) Animal Reproduction Core Laboratory. We acknowledge the expert technical assistance of Mr. Derek Pouchnik for microarray analyses and the CRB Genomics Core Laboratory. We thank Ms. Jill Griffin for assistance in preparation of this manuscript. This study was supported by a National Institutes of Health, NIH NICHD, to MKS.
Footnotes
The authors have no conflicts of interest to disclose.
References
- Acevedo N, Smith GD. Oocyte-specific gene signaling and its regulation of mammalian reproductive potential. Front Biosci. 2005;10:2335–2345. doi: 10.2741/1702. [DOI] [PubMed] [Google Scholar]
- Amsterdam A, Keren-Tal I, Aharoni D, Dantes A, Land-Bracha A, Rimon E, Sasson R, Hirsh L. Steroidogenesis and apoptosis in the mammalian ovary. Steroids. 2003;68:861–867. doi: 10.1016/j.steroids.2003.09.003. [DOI] [PubMed] [Google Scholar]
- Andreu-Vieyra C, Lin YN, Matzuk MM. Mining the oocyte transcriptome. Trends Endocrinol Metab. 2006;17:136–143. doi: 10.1016/j.tem.2006.03.010. [DOI] [PubMed] [Google Scholar]
- Arici A, Oral E, Bukulmez O, Buradagunta S, Bahtiyar O, Jones EE. Monocyte chemotactic protein-1 expression in human preovulatory follicles and ovarian cells. J Reprod Immunol. 1997;32:201–219. doi: 10.1016/s0165-0378(97)82476-x. [DOI] [PubMed] [Google Scholar]
- Asai K. Review of the research of glia maturation factor and cloning of human and rat glia maturation factor-gamma (GMFG) cDNA. Nihon Shinkei Seishin Yakurigaku Zasshi. 2001;21:15–20. [PubMed] [Google Scholar]
- Attia GR, Dooley CA, Rainey WE, Carr BR. Transforming growth factor beta inhibits steroidogenic acute regulatory (StAR) protein expression in human ovarian thecal cells. Mol Cell Endocrinol. 2000;170:123–129. doi: 10.1016/s0303-7207(00)00335-x. [DOI] [PubMed] [Google Scholar]
- Ben-Ami I, Freimann S, Armon L, Dantes A, Ron-El R, Amsterdam A. Novel function of ovarian growth factors: Combined studies by DNA microarray, biochemical and physiological approaches. Mol Hum Reprod. 2006;12:413–419. doi: 10.1093/molehr/gal045. [DOI] [PubMed] [Google Scholar]
- Bisset LR, Schmid-Grendelmeier P. Chemokines and their receptors in the pathogenesis of allergic asthma: Progress and perspective. Curr Opin Pulm Med. 2005;11:35–42. doi: 10.1097/01.mcp.0000144502.50149.e0. [DOI] [PubMed] [Google Scholar]
- Brannstrom M, Enskog A. Leukocyte networks and ovulation. J Reprod Immunol. 2002;57:47–60. doi: 10.1016/s0165-0378(02)00009-8. [DOI] [PubMed] [Google Scholar]
- Bukulmez O, Arici A. Leukocytes in ovarian function. Hum Reprod Update. 2000;6:1–15. doi: 10.1093/humupd/6.1.1. [DOI] [PubMed] [Google Scholar]
- Buratini J, Jr, Teixeira AB, Costa IB, Glapinski VF, Pinto MG, Giometti IC, Barros CM, Cao M, Nicola ES, Price CA. Expression of fibroblast growth factor-8 and regulation of cognate receptors, fibroblast growth factor receptor-3c and −4, in bovine antral follicles. Reproduction. 2005;130:343–350. doi: 10.1530/rep.1.00642. [DOI] [PubMed] [Google Scholar]
- Burns KH, Owens GE, Ogbonna SC, Nilson JH, Matzuk MM. Expression profiling analyses of gonadotropin responses and tumor development in the absence of inhibins. Endocrinology. 2003;144:4492–4507. doi: 10.1210/en.2003-0476. [DOI] [PubMed] [Google Scholar]
- Caetano AR, Johnson RK, Ford JJ, Pomp D. Microarray profiling for differential gene expression in ovaries and ovarian follicles of pigs selected for increased ovulation rate. Genetics. 2004;168:1529–1537. doi: 10.1534/genetics.104.029595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canty MJ, Boland MP, Evans AC, Crowe MA. Alterations in follicular IGFBP mRNA expression and follicular fluid IGFBP concentrations during the first follicle wave in beef heifers. Anim Reprod Sci. 2006;93:199–217. doi: 10.1016/j.anireprosci.2005.06.033. [DOI] [PubMed] [Google Scholar]
- Castilla JA, Sampalo A, Molina R, Samaniego F, Mozas J, Vergara F, Garrido F, Herruzo AJ. Mononuclear cell subpopulations in human follicular fluid from stimulated cycles. Am J Reprod Immunol. 1990;22:127–129. doi: 10.1111/j.1600-0897.1990.tb00655.x. [DOI] [PubMed] [Google Scholar]
- Cheng JG, Rodriguez CI, Stewart CL. Control of uterine receptivity and embryo implantation by steroid hormone regulation of LIF production and LIF receptor activity: Towards a molecular understanding of “the window of implantation”. Rev Endocr Metab Disord. 2002;3:119–126. doi: 10.1023/a:1015402811650. [DOI] [PubMed] [Google Scholar]
- Chin KV, Seifer DB, Feng B, Lin Y, Shih WC. DNA microarray analysis of the expression profiles of luteinized granulosa cells as a function of ovarian reserve. Fertil Steril. 2002;77:1214–1218. doi: 10.1016/s0015-0282(02)03114-x. [DOI] [PubMed] [Google Scholar]
- Cook RW, Thompson TB, Jardetzky TS, Woodruff TK. Molecular biology of inhibin action. Semin Reprod Med. 2004;22:269–276. doi: 10.1055/s-2004-831902. [DOI] [PubMed] [Google Scholar]
- De A, Park JI, Kawamura K, Chen R, Klein C, Rauch R, Mulders SM, Sollewijn Gelpke MD, Hsueh AJ. Intraovarian tumor necrosis factor-related weak inducer of apoptosis/fibroblas factor-inducible-14 ligand-receptor system limits ovarian preovulatory follicles from excessive luteinization. Mol Endocrinol. 2006;20:2528–2538. doi: 10.1210/me.2006-0028. [DOI] [PubMed] [Google Scholar]
- Denhardt DT, Guo X. Osteopontin: A protein with diverse functions. FASEB J. 1993;7:1475–1482. [PubMed] [Google Scholar]
- Dorrington JH, Bendell JJ, Khan SA. Interactions between FSH, estradiol-17 beta and transforming growth factor-beta regulate growth and differentiation in the rat gonad. J Steroid Biochem Mol Biol. 1993;44:441–447. doi: 10.1016/0960-0760(93)90248-u. [DOI] [PubMed] [Google Scholar]
- Drummond AE. TGFbeta signalling in the development of ovarian function. Cell Tissue Res. 2005;322:107–115. doi: 10.1007/s00441-005-1153-1. [DOI] [PubMed] [Google Scholar]
- Edmondson SR, Thumiger SP, Werther GA, Wraight CJ. Epidermal homeostasis: The role of the growth hormone and insulin-like growth factor systems. Endocr Rev. 2003;24:737–764. doi: 10.1210/er.2002-0021. [DOI] [PubMed] [Google Scholar]
- Egeblad M, Littlepage LE, Werb Z. The fibroblastic coconspirator in cancer progression. Cold Spring Harb Symp Quant Biol. 2005;70:383–388. doi: 10.1101/sqb.2005.70.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elvin JA, Clark AT, Wang P, Wolfman NM, Matzuk MM. Paracrine actions of growth differentiation factor-9 in the mammalian ovary. Mol Endocrinol. 1999;13:1035–1048. doi: 10.1210/mend.13.6.0310. [DOI] [PubMed] [Google Scholar]
- Fortune JE, Rivera GM, Yang MY. Follicular development: The role of the follicular microenvironment in selection of the dominant follicle. Anim Reprod Sci. 2004;82–83:109–126. doi: 10.1016/j.anireprosci.2004.04.031. [DOI] [PubMed] [Google Scholar]
- Fraser HM. Regulation of the ovarian follicular vasculature. Reprod Biol Endocrinol. 2006;4:18. [Google Scholar]
- Froment P, Gizard F, Defever D, Staels B, Dupont J, Monget P. Peroxisome proliferator-activated receptors in reproductive tissues: From gametogenesis to parturition. J Endocrinol. 2006;189:199–209. doi: 10.1677/joe.1.06667. [DOI] [PubMed] [Google Scholar]
- Gerard N, Delpuech T, Oxvig C, Overgaard MT, Monget P. Proteolytic degradation of IGF-binding protein (IGFBP)-2 in equine ovarian follicles: Involvement of pregnancy-associated plasma protein-A (PAPP-A) and association with dominant but not subordinated follicles. J Endocrinol. 2004;182:457–466. doi: 10.1677/joe.0.1820457. [DOI] [PubMed] [Google Scholar]
- Glister C, Kemp CF, Knight PG. Bone morphogenetic protein (BMP) ligands and receptors in bovine ovarian follicle cells: Actions of BMP-4, −6 and −7 on granulosa cells and differential modulation of Smad-1 phosphorylation by follistatin. Reproduction. 2004;127:239–254. doi: 10.1530/rep.1.00090. [DOI] [PubMed] [Google Scholar]
- Hedrick JA, Zlotnik A. Lymphotactin. Clin Immunol Immunopathol. 1998;87:218–222. doi: 10.1006/clin.1998.4546. [DOI] [PubMed] [Google Scholar]
- Hernandez-Gonzalez I, Gonzalez-Robayna I, Shimada M, Wayne CM, Ochsner SA, White L, Richards JS. Gene expression profiles of cumulus cell oocyte complexes during ovulation reveal cumulus cells express neuronal and immune-related genes: Does this expand their role in the ovulation process? Mol Endocrinol. 2006;20:1300–1321. doi: 10.1210/me.2005-0420. [DOI] [PubMed] [Google Scholar]
- Hiraoka A, Sugimura A, Seki T, Nagasawa T, Ohta N, Shimonishi M, Hagiya M, Shimizu S. Cloning, expression, and characterization of a cDNA encoding a novel human growth factor for primitive hematopoietic progenitor cells. Proc Natl Acad Sci USA. 1997;94:7577–7582. doi: 10.1073/pnas.94.14.7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt JE, Jackson A, Roman SD, Aitken RJ, Koopman P, McLaughlin EA. CXCR4/SDF1 interaction inhibits the primordial to primary follicle transition in the neonatal mouse ovary. Dev Biol. 2006;293:449–460. doi: 10.1016/j.ydbio.2006.02.012. [DOI] [PubMed] [Google Scholar]
- Horikawa M, Kirkman NJ, Mayo KE, Mulders SM, Zhou J, Bondy CA, Hsu SY, King GJ, Adashi EY. The mouse germ-cell-specific leucine-rich repeat protein NAL P14: A member of the NACHT nucleoside triphosphatase family. Biol Reprod. 2005;72:879–889. doi: 10.1095/biolreprod.104.033753. [DOI] [PubMed] [Google Scholar]
- Ikeda K, Kundu RK, Ikeda S, Kobara M, Matsubara H, Quertermous T. Glia maturation factor-gamma is preferentially expressed in microvascular endothelial and inflammatory cells and modulates actin cytoskeleton reorganization. Circ Res. 2006;99:424–433. doi: 10.1161/01.RES.0000237662.23539.0b. [DOI] [PubMed] [Google Scholar]
- Inkster SE, Brodie AM. Expression of aromatase cytochrome P-450 in premenopausal and postmenopausal human ovaries: An immunocytochemical study. J Clin Endocrinol Metab. 1991;73:717–726. doi: 10.1210/jcem-73-4-717. [DOI] [PubMed] [Google Scholar]
- Ireland JJ, Murphee RL, Coulson PB. Accuracy of predicting stages of bovine estrous cycle by gross appearance of the corpus luteum. J Dairy Sci. 1980;63:155–160. doi: 10.3168/jds.S0022-0302(80)82901-8. [DOI] [PubMed] [Google Scholar]
- Ismail RS, Okawara Y, Fryer JN, Vanderhyden BC. Hormonal regulation of the ligand for c-kit in the rat ovary and its effects on spontaneous oocyte meiotic maturation. Mol Reprod Dev. 1996;43:458–469. doi: 10.1002/(SICI)1098-2795(199604)43:4<458::AID-MRD8>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Jo M, Gieske MC, Payne CE, Wheeler-Price SE, Gieske JB, Ignatius IV, Curry TE, Jr, Ko C. Development and application of a rat ovarian gene expression database. Endocrinology. 2004;145:5384–5396. doi: 10.1210/en.2004-0407. [DOI] [PubMed] [Google Scholar]
- Johnson AL, Bridgham JT, Woods DC. Cellular mechanisms and modulation of activin A- and transforming growth factor beta-mediated differentiation in cultured hen granulosa cells. Biol Reprod. 2004;71:1844–1851. doi: 10.1095/biolreprod.104.032573. [DOI] [PubMed] [Google Scholar]
- Juengel JL, Bibby AH, Reader KL, Lun S, Quirke LD, Haydon LJ, McNatty KP. The role of transforming growth factor-beta (TGF-beta) during ovarian follicular development in sheep. Reprod Biol Endocrinol. 2004a;2:78. doi: 10.1186/1477-7827-2-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juengel JL, Bodensteiner KJ, Heath DA, Hudson NL, Moeller CL, Smith P, Galloway SM, Davis GH, Sawyer HR, McNatty KP. Physiology of GDF9 and BMP15 signalling molecules. Anim Reprod Sci. 2004b;82–83:447–460. doi: 10.1016/j.anireprosci.2004.04.021. [DOI] [PubMed] [Google Scholar]
- Kaczmarek MM, Schams D, Ziecik AJ. Role of vascular endothelial growth factor in ovarian physiology—An overview. Reprod Biol. 2005;5:111–136. [PubMed] [Google Scholar]
- Kawano Y, Kawasaki F, Nakamura S, Matsui N, Narahara H, Miyakawa I. The production and clinical evaluation of macrophage colony-stimulating factor and macrophage chemoattractant protein-1 in human follicular fluids. Am J Reprod Immunol. 2001;45:1–5. doi: 10.1111/j.8755-8920.2001.450101.x. [DOI] [PubMed] [Google Scholar]
- Kawano Y, Zeineh Hasan K, Fukuda J, Mine S, Miyakawa I. Production of vascular endothelial growth factor and angiogenic factor in human follicular fluid. Mol Cell Endocrinol. 2003;202:19–23. doi: 10.1016/s0303-7207(03)00056-x. [DOI] [PubMed] [Google Scholar]
- Kawano Y, Fukuda J, Itoh H, Takai N, Nasu K, Miyakawa I. The effect of inflammatory cytokines on secretion of macrophage colony-stimulating factor and monocyte chemoattractant protein-1 in human granulosa cells. Am J Reprod Immunol. 2004;52:124–128. doi: 10.1111/j.1600-0897.2004.00198.x. [DOI] [PubMed] [Google Scholar]
- Kenney NJ, Adkins HB, Sanicola M. Nodal and Cripto-1: Embryonic pattern formation genes involved in mammary gland development and tumorigenesis. J Mammary Gland Biol Neoplasia. 2004;9:133–144. doi: 10.1023/B:JOMG.0000037158.91940.1c. [DOI] [PubMed] [Google Scholar]
- Kezele P, Nilsson EE, Skinner MK. Keratinocyte growth factor acts as a mesenchymal factor that promotes ovarian primordial to primary follicle transition. Biol Reprod. 2005a;73:967–973. doi: 10.1095/biolreprod.105.043117. [DOI] [PubMed] [Google Scholar]
- Kezele PR, Ague JM, Nilsson E, Skinner MK. Alterations in the ovarian transcriptome during primordial follicle assembly and development. Biol Reprod. 2005b;72:241–255. doi: 10.1095/biolreprod.104.032060. [DOI] [PubMed] [Google Scholar]
- Koga K, Osuga Y, Tsutsumi O, Momoeda M, Suenaga A, Kugu K, Fujiwara T, Takai Y, Yano T, Taketani Y. Evidence for the presence of angiogenin in human follicular fluid and the up-regulation of its production by human chorionic gonadotropin and hypoxia. J Clin Endocrinol Metab. 2000;85:3352–3355. doi: 10.1210/jcem.85.9.6837. [DOI] [PubMed] [Google Scholar]
- Kurmasheva RT, Houghton PJ. IGF-I mediated survival pathways in normal and malignant cells. Biochim Biophys Acta. 2006;1766:1–22. doi: 10.1016/j.bbcan.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Lafdil F, Chobert MN, Couchie D, Brouillet A, Zafrani ES, Mavier P, Laperche Y. InductionofGas6proteininCCl4-induced rat liver injury and anti-apoptotic effect on hepatic stellate cells. Hepatology. 2006;44:228–239. doi: 10.1002/hep.21237. [DOI] [PubMed] [Google Scholar]
- Lataillade JJ, Domenech J, Le Bousse-Kerdiles MC. Stromal cell-derived factor-1 (SDF-1)\CXCR4 couple plays multiple roles on haematopoietic progenitors at the border between the old cytokine and new chemokine worlds: Survival, cell cycling and trafficking. Eur Cytokine Netw. 2004;15:177–188. [PubMed] [Google Scholar]
- Lee HS, Lee IS, Kang TC, Jeong GB, Chang SI. Angiogenin is involved in morphological changes and angiogenesis in the ovary. Biochem Biophys Res Commun. 1999;257:182–186. doi: 10.1006/bbrc.1999.0359. [DOI] [PubMed] [Google Scholar]
- Lee HJ, Cho CH, Hwang SJ, Choi HH, Kim KT, Ahn SY, Kim JH, Oh JL, Lee GM, Koh GY. Biological characterization of angiopoietin-3 and angiopoietin-4. FASEB J. 2004;18:1200–1208. doi: 10.1096/fj.03-1466com. [DOI] [PubMed] [Google Scholar]
- Lenz S, Pohland R, Becker F, Vanselow J. Expression of the bovine aromatase cytochrome P450 gene (Cyp19) is primarily regulated by promoter 2 in bovine follicles and by promoter 1.1 in corpora lutea. Mol Reprod Dev. 2004;67:406–413. doi: 10.1002/mrd.20000. [DOI] [PubMed] [Google Scholar]
- Liu HC, He Z, Rosenwaks Z. Application of complementary DNA microarray (DNA chip) technology in the study of gene expression profiles during folliculogenesis. Fertil Steril. 2001;75:947–955. doi: 10.1016/s0015-0282(01)01706-x. [DOI] [PubMed] [Google Scholar]
- Loukides JA, Loy RA, Edwards R, Honig J, Visintin I, Polan ML. Human follicular fluids contain tissue macrophages. J Clin Endocrinol Metab. 1990;71:1363–1367. doi: 10.1210/jcem-71-5-1363. [DOI] [PubMed] [Google Scholar]
- Lu Q, Gore M, Zhang Q, Camenisch T, Boast S, Casagranda F, Lai C, Skinner MK, Klein R, Matsushima GK, Earp HS, Goff SP, Lemke G. Tyro-3 family receptors are essential regulators of mammalian spermatogenesis. Nature. 1999;398:723–728. doi: 10.1038/19554. [DOI] [PubMed] [Google Scholar]
- Machelon V, Emilie D. Production of ovarian cytokines and their role in ovulation in the mammalian ovary. Eur Cytokine Netw. 1997;8:137–143. [PubMed] [Google Scholar]
- Macklon NS, Fauser BC. Aspects of ovarian follicle development throughout life. Horm Res. 1999;52:161–170. doi: 10.1159/000023456. [DOI] [PubMed] [Google Scholar]
- Magoffin DA. Ovarian theca cell. Int J Biochem Cell Biol. 2005;37:1344–1349. doi: 10.1016/j.biocel.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Maurer M, von Stebut E. Macrophage inflammatory protein-1. Int J Biochem Cell Biol. 2004;36:1882–1886. doi: 10.1016/j.biocel.2003.10.019. [DOI] [PubMed] [Google Scholar]
- Mazerbourg S, Hsueh AJ. Genomic analyses facilitate identification of receptors and signalling pathways for growth differentiation factor 9 and related orphan bone morphogenetic protein/ growth differentiation factor ligands. Hum Reprod Update. 2006;12:373–383. doi: 10.1093/humupd/dml014. [DOI] [PubMed] [Google Scholar]
- McChlery SM, Clarke SC. The use of hydrolysis and hairpin probes in real-time PCR. Mol Biotechnol. 2003;25:267–274. doi: 10.1385/MB:25:3:267. [DOI] [PubMed] [Google Scholar]
- McCroskery S, Thomas M, Platt L, Hennebry A, Nishimura T, McLeay L, Sharma M, Kambadur R. Improved muscle healing through enhanced regeneration and reduced fibrosis in myostatin-null mice. J Cell Sci. 2005;118:3531–3541. doi: 10.1242/jcs.02482. [DOI] [PubMed] [Google Scholar]
- McLean DJ, Friel PJ, Pouchnik D, Griswold MD. Oligonucleotide microarray analysis of gene expression in follicle-stimulating hormone-treated rat Sertoli cells. Mol Endocrinol. 2002;16:2780–2792. doi: 10.1210/me.2002-0059. [DOI] [PubMed] [Google Scholar]
- Mihm M, Baker PJ, Ireland JL, Smith GW, Coussens PM, Evans AC, Ireland JJ. Molecular evidence that growth of dominant follicles involves a reduction in follicle-stimulating hormone dependence and an increase in luteinizing hormone dependence in cattle. Biol Reprod. 2006;74:1051–1059. doi: 10.1095/biolreprod.105.045799. [DOI] [PubMed] [Google Scholar]
- Molyneaux KA, Schaible K, Wylie C. GP130, the shared receptor for the LIF/IL6 cytokine family in the mouse, is not required for early germ cell differentiation, but is required cell-autonomously in oocytes for ovulation. Development. 2003;130:4287–4294. doi: 10.1242/dev.00650. [DOI] [PubMed] [Google Scholar]
- Moore K, Thatcher WW. Major advances associated with reproduction in dairy cattle. J Dairy Sci. 2006;89:1254–1266. doi: 10.3168/jds.S0022-0302(06)72194-4. [DOI] [PubMed] [Google Scholar]
- Nagasawa T, Tachibana K, Kishimoto T. A novel CXC chemokine PBSF/SDF-1 and its receptor CXC R4: Their functions in development, hematopoiesis and HIV infection. Semin Immunol. 1998;10:179–185. doi: 10.1006/smim.1998.0128. [DOI] [PubMed] [Google Scholar]
- Nemir M, Bhattacharyya D, Li X, Singh K, Mukherjee AB, Mukherjee BB. Targeted inhibition of osteopontin expression in the mammary gland causes abnormal morphogenesis and lactation deficiency. J Biol Chem. 2000;275:969–976. doi: 10.1074/jbc.275.2.969. [DOI] [PubMed] [Google Scholar]
- Nilsson EE, Doraiswamy V, Skinner MK. Transforming growth factor-beta isoform expression during bovine ovarian antral follicle development. Mol Reprod Dev. 2003;66:237–246. doi: 10.1002/mrd.10350. [DOI] [PubMed] [Google Scholar]
- Oksbjerg N, Gondret F, Vestergaard M. Basic principles of muscle development and growth in meat-producing mammals as affected by the insulin-like growth factor (IGF) system. Domest Anim Endocrinol. 2004;27:219–240. doi: 10.1016/j.domaniend.2004.06.007. [DOI] [PubMed] [Google Scholar]
- Okuda K, Uenoyama Y, Berisha B, Lange IG, Taniguchi H, Kobayashi S, Kobayashi S, Miyamoto A, Schams D. Estradiol-17beta is produced in bovine corpus luteum. Biol Reprod. 2001;65:1634–1639. doi: 10.1095/biolreprod65.6.1634. [DOI] [PubMed] [Google Scholar]
- Parrott JA, Skinner MK. Direct actions of kit-ligand on theca cell growth and differentiation during follicle development. Endocrinology. 1997;138:3819–3827. doi: 10.1210/endo.138.9.5368. [DOI] [PubMed] [Google Scholar]
- Parrott JA, Skinner MK. Developmental and hormonal regulation of hepatocyte growth factor expression and action in the bovine ovarian follicle. Biol Reprod. 1998a;59:553–560. doi: 10.1095/biolreprod59.3.553. [DOI] [PubMed] [Google Scholar]
- Parrott JA, Skinner MK. Developmental and hormonal regulation of keratinocyte growth factor expression and action in the ovarian follicle. Endocrinology. 1998b;139:228–235. doi: 10.1210/endo.139.1.5680. [DOI] [PubMed] [Google Scholar]
- Parrott JA, Vigne JL, Chu BZ, Skinner MK. Mesenchymal-epithelial interactions in the ovarian follicle involve keratinocyte and hepatocyte growth factor production by thecal cells and their action on granulosa cells. Endocrinology. 1994;135:569–575. doi: 10.1210/endo.135.2.8033804. [DOI] [PubMed] [Google Scholar]
- Patel VP, Kreider BL, Li Y, Li H, Leung K, Salcedo T, Nardelli B, Pippalla V, Gentz S, Thotakura R, Parmelee D, Gentz R, Garotta G. Molecular and functional characterization of two novel human C-C chemokines as inhibitors of two distinct classes of myeloid progenitors. J Exp Med. 1997;185:1163–1172. doi: 10.1084/jem.185.7.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peluso JJ. N-cadherin mediated cell contact inhibits germinal vesicle breakdown in mouse oocytes maintained in vitro. Reproduction. 2006;131:429–437. doi: 10.1530/rep.1.00863. [DOI] [PubMed] [Google Scholar]
- Ramkissoon SH, Patel HJ, Taborga M, Rameshwar P. G protein-coupled receptors in haematopoietic disruption. Expert Opin Biol Ther. 2006;6:109–120. doi: 10.1517/14712598.6.2.109. [DOI] [PubMed] [Google Scholar]
- Roberts AJ, Skinner MK. Estrogen regulation of thecal cell steroidogenesis and differentiation: Thecal cell-granulosa cell interactions. Endocrinology. 1990a;127:2918–2929. doi: 10.1210/endo-127-6-2918. [DOI] [PubMed] [Google Scholar]
- Roberts AJ, Skinner MK. Hormonal regulation of thecal cell function during antral follicle development in bovine ovaries. Endocrinology. 1990b;127:2907–2917. doi: 10.1210/endo-127-6-2907. [DOI] [PubMed] [Google Scholar]
- Roberts AJ, Skinner MK. Mesenchymal-epithelial cell interactions in the ovary: Estrogen-induced theca cell steroidogenesis. Mol Cell Endocrinol. 1990c;72:R1–5. doi: 10.1016/0303-7207(90)90242-z. [DOI] [PubMed] [Google Scholar]
- Roberts AJ, Skinner MK. Transforming growth factor-alpha and -beta differentially regulate growth and steroidogenesis of bovine thecal cells during antral follicle development. Endocrinology. 1991;129:2041–2048. doi: 10.1210/endo-129-4-2041. [DOI] [PubMed] [Google Scholar]
- Romano G. The complex biology of the receptor for the insulinlike growth factor-1. Drug News Perspect. 2003;16:525–531. doi: 10.1358/dnp.2003.16.8.829351. [DOI] [PubMed] [Google Scholar]
- Sadate-Ngatchou PI, Pouchnik DJ, Griswold MD. Identification of testosterone-regulated genes in testes of hypogonadal mice using oligonucleotide microarray. Mol Endocrinol. 2004;18:422–433. doi: 10.1210/me.2003-0188. [DOI] [PubMed] [Google Scholar]
- Salas C, Julio-Pieper M, Valladares M, Pommer R, Vega M, Mastronardi C, Kerr B, Ojeda SR, Lara HE, Romero C. Nerve growth factor-dependent activation of trkA receptors in the human ovary results in synthesis of follicle-stimulating hormone receptors and estrogen secretion. J Clin Endocrinol Metab. 2006;91:2396–2403. doi: 10.1210/jc.2005-1925. [DOI] [PubMed] [Google Scholar]
- Salazar-Mather TP, Hokeness KL. Cytokine and chemokine networks: Pathways to antiviral defense. Curr Top Microbiol Immunol. 2006;303:29–46. doi: 10.1007/978-3-540-33397-5_2. [DOI] [PubMed] [Google Scholar]
- Saller F, Burnier L, Schapira M, Angelillo-Scherrer A. Role of the growth arrest-specific gene 6 (gas6) product in thrombus stabilization. Blood Cells Mol Dis. 2006;36:373–378. doi: 10.1016/j.bcmd.2005.12.038. [DOI] [PubMed] [Google Scholar]
- Santiago CA, Voge JL, Aad PY, Allen DT, Stein DR, Malayer JR, Spicer LJ. Pregnancy-associated plasma protein-A and insulin-like growth factor binding protein mRNAs in granulosa cells of dominant and subordinate follicles of preovulatory cattle. Domest Anim Endocrinol. 2005;28:46–63. doi: 10.1016/j.domaniend.2004.06.002. [DOI] [PubMed] [Google Scholar]
- Saragueta PE, Lanuza GM, Baranao JL. Autocrine role of transforming growth factor beta1 on rat granulosa cell proliferation. Biol Reprod. 2002;66:1862–1868. doi: 10.1095/biolreprod66.6.1862. [DOI] [PubMed] [Google Scholar]
- Sasson R, Dantes A, Tajima K, Amsterdam A. Novel genes modulated by FSH in normal and immortalized FSH-responsive cells: New insights into the mechanism of FSH action. FASEB J. 2003;17:1256–1266. doi: 10.1096/fj.02-0740com. [DOI] [PubMed] [Google Scholar]
- Schams D, Berisha B, Kosmann M, Amselgruber WM. Expression and localization of IGF family members in bovine antral follicles during final growth and in luteal tissue during different stages of estrous cycle and pregnancy. Domest Anim Endocrinol. 2002;22:51–72. doi: 10.1016/s0739-7240(01)00116-3. [DOI] [PubMed] [Google Scholar]
- Sezer O. Myeloma bone disease. Hematology. 2005;10:19–24. doi: 10.1080/10245330512331389782. [DOI] [PubMed] [Google Scholar]
- Shankar SL, O’Guin K, Kim M, Varnum B, Lemke G, Brosnan CF, Shafit-Zagardo B. Gas6/Axl signaling activates the phosphatidylinositol 3-kinase/Akt1 survival pathway to protect oligodendrocytes from tumor necrosis factor alpha-induced apoptosis. J Neurosci. 2006;26:5638–5648. doi: 10.1523/JNEUROSCI.5063-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima JE, McLean DJ, McCarrey JR, Griswold MD. The murine testicular transcriptome: Characterizing gene expression in the testis during the progression of spermatogenesis. Biol Reprod. 2004;72:492–501. doi: 10.1095/biolreprod.103.026880. [DOI] [PubMed] [Google Scholar]
- Shimizu T. Promotion of ovarian follicular development by injecting vascular endothelial growth factor (VEGF) and growth differentiation factor 9 (GDF-9) genes. J Reprod Dev. 2006;52:23–32. doi: 10.1262/jrd.17072. [DOI] [PubMed] [Google Scholar]
- Shimizu T, Jayawardana BC, Nishimoto H, Kaneko E, Tetsuka M, Miyamoto A. Involvement of the bone morphogenetic protein/ receptor system during follicle development in the bovine ovary: Hormonal regulation of the expression of bone morphogenetic protein 7 (BMP-7) and its receptors (ActRII and ALK-2) Mol Cell Endocrinol. 2006;249:78–83. doi: 10.1016/j.mce.2006.01.015. [DOI] [PubMed] [Google Scholar]
- Shiota M, Tanihiro T, Nakagawa Y, Aoki N, Ishida N, Miyazaki K, Ullrich A, Miyazaki H. Protein tyrosine phosphatase PTP20 induces actin cytoskeleton reorganization by dephosphorylating p190 RhoGAP in rat ovarian granulosa cells stimulated with follicle-stimulating hormone. Mol Endocrinol. 2003;17:534–549. doi: 10.1210/me.2002-0187. [DOI] [PubMed] [Google Scholar]
- Skinner MK. Regulation of primordial follicle assembly and development. Hum Reprod Update. 2005;11:461–471. doi: 10.1093/humupd/dmi020. [DOI] [PubMed] [Google Scholar]
- Skinner MK, Osteen KG. Developmental and hormonal regulation of bovine granulosa cell function in the preovulatory follicle. Endocrinology. 1988;123:1668–1675. doi: 10.1210/endo-123-3-1668. [DOI] [PubMed] [Google Scholar]
- Skinner MK, Keski-Oja J, Osteen KG, Moses HL. Ovarian thecal cells produce transforming growth factor-beta which can regulate granulosa cell growth. Endocrinology. 1987;121:786–792. doi: 10.1210/endo-121-2-786. [DOI] [PubMed] [Google Scholar]
- Small CL, Shima JE, Uzumcu M, Skinner MK, Griswold MD. Profiling gene expression during the differentiation and development of the murine embryonic gonad. Biol Reprod. 2005;72:492–501. doi: 10.1095/biolreprod.104.033696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriraman V, Rudd MD, Lohmann SM, Mulders SM, Richards JS. Cyclic guanosine 5’-monophosphate-dependent protein kinase II is induced by luteinizing hormone and progesterone receptor-dependent mechanisms in granulosa cells and cumulus oocyte complexes of ovulating follicles. Mol Endocrinol. 2006;20:348–361. doi: 10.1210/me.2005-0317. [DOI] [PubMed] [Google Scholar]
- Stievano L, Piovan E, Amadori A. C and CX3C chemokines: Cell sources and physiopathological implications. Crit Rev Immunol. 2004;24:205–228. doi: 10.1615/critrevimmunol.v24.i3.40. [DOI] [PubMed] [Google Scholar]
- Strizzi L, Bianco C, Normanno N, Salomon D. Cripto-1: A multifunctional modulator during embryogenesis and oncogenesis. Oncogene. 2005;24:5731–5741. doi: 10.1038/sj.onc.1208918. [DOI] [PubMed] [Google Scholar]
- Su YQ, Wu X, O’Brien MJ, Pendola FL, Denegre JN, Matzuk MM, Eppig JJ. Synergistic roles of BMP15 and GDF9 in the development and function of the oocyte-cumulus cell complex in mice: Genetic evidence for an oocyte-granulosa cell regulatory loop. Dev Biol. 2004;276:64–73. doi: 10.1016/j.ydbio.2004.08.020. [DOI] [PubMed] [Google Scholar]
- Tajima K, Orisaka M, Yata H, Goto K, Hosokawa K, Kotsuji F. Role of granulosa and theca cell interactions in ovarian follicular maturation. Microsc Res Tech. 2006;69:450–458. doi: 10.1002/jemt.20304. [DOI] [PubMed] [Google Scholar]
- Tamura M, Nakagawa Y, Shimizu H, Yamada N, Miyano T, Miyazaki H. Cellular functions of mitogen-activated protein kinases and protein tyrosine phosphatases in ovarian granulosa cells. J Reprod Dev. 2004;50:47–55. doi: 10.1262/jrd.50.47. [DOI] [PubMed] [Google Scholar]
- Tsuchida K. Activins, myostatin and related TGF-beta family members as novel therapeutic targets for endocrine, metabolic and immune disorders. Curr Drug Targets Immune Endocr Metabol Disord. 2004;4:157–166. doi: 10.2174/1568008043339901. [DOI] [PubMed] [Google Scholar]
- Voge JL, Aad PY, Santiago CA, Goad DW, Malayer JR, Allen D, Spicer LJ. Effect of insulin-like growth factors (IGF), FSH, and leptin on IGF-binding-protein mRNA expression in bovine granulosa and theca cells: Quantitative detection by real-time PCR. Peptides. 2004;25:2195–2203. doi: 10.1016/j.peptides.2004.07.008. [DOI] [PubMed] [Google Scholar]
- Walters KA, Binnie JP, Campbell BK, Armstrong DG, Telfer EE. The effects of IGF-I on bovine follicle development and IGFBP-2 expression are dose and stage dependent. Reproduction. 2006;131:515–523. doi: 10.1530/rep.1.00682. [DOI] [PubMed] [Google Scholar]
- Watson ED. Compartmentalization of steroidogen esis by the equine corpus luteum. Theriogenology. 2000;53:1459–1466. doi: 10.1016/S0093-691X(00)00288-0. [DOI] [PubMed] [Google Scholar]
- Webb R, Garnsworthy PC, Gong JG, Armstrong DG. Control of follicular growth: Local interactions and nutritional influences. J Anim Sci. 2004;82:E63–E74. doi: 10.2527/2004.8213_supplE63x. [DOI] [PubMed] [Google Scholar]
- Wickenheisser JK, Nelson-DeGrave VL, McAllister JM. Human ovarian theca cells in culture. Trends Endocrinol Metab. 2006;17:65–71. doi: 10.1016/j.tem.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Wong KH, Negishi H, Adashi EY. Expression, hormonal regulation, and cyclic variation of chemokines in the rat ovary: Key determinants of the intraovarian residence of representatives of the white blood cell series. Endocrinology. 2002;143:784–791. doi: 10.1210/endo.143.3.8699. [DOI] [PubMed] [Google Scholar]
- Wood JR, Nelson VL, Ho C, Jansen E, Wang CY, Urbanek M, McAllister JM, Mosselman S, Strauss JF., III The molecular phenotype of polycystic ovary syndrome (PCOS) theca cells and new candidate PCOS genes defined by microarray analysis. J Biol Chem. 2003;278:26380–26390. doi: 10.1074/jbc.M300688200. [DOI] [PubMed] [Google Scholar]
- Wood JR, Ho CK, Nelson-Degrave VL, McAllister JM, Strauss JF., III The molecular signature of polycystic ovary syndrome (PCOS) theca cells defined by gene expression profiling. J Reprod Immunol. 2004;63:51–60. doi: 10.1016/j.jri.2004.01.010. [DOI] [PubMed] [Google Scholar]
- Woods DC, Haugen MJ, Johnson AL. Opposing actions of TGFbeta and MAP kinase signaling in undifferentiated hen granulosa cells. Biochem Biophys Res Commun. 2005;336:450–457. doi: 10.1016/j.bbrc.2005.08.107. [DOI] [PubMed] [Google Scholar]
- Wulff C, Wilson H, Wiegand SJ, Rudge JS, Fraser HM. Prevention of thecal angiogenesis, antral follicular growth, and ovulation in the primate by treatment with vascular endothelial growth factor Trap R1 R2. Endocrinology. 2002;143:2797–2807. doi: 10.1210/endo.143.7.8886. [DOI] [PubMed] [Google Scholar]
- Xu J, Hennebold JD, Stouffer RL. Dynamic expression and regulation of the corticotropin-releasing hormone/urocortin-receptor-binding protein system in the primate ovary during the menstrual cycle. J Clin Endocrinol Metab. 2006;91:1544–1553. doi: 10.1210/jc.2005-2776. [DOI] [PubMed] [Google Scholar]
- Yanagita M. The role of the vitamin K-dependent growth factor Gas6 in glomerular pathophysiology. Curr Opin Nephrol Hypertens. 2004;13:465–470. doi: 10.1097/01.mnh.0000133981.63053.e9. [DOI] [PubMed] [Google Scholar]
- Yao J, Ren X, Ireland JJ, Coussens PM, Smith TP, Smith GW. Generation of a bovine oocyte cDNA library and microarray: Resources for identification of genes important for follicular development and early embryogenesis. Physiol Genomics. 2004;19:84–92. doi: 10.1152/physiolgenomics.00123.2004. [DOI] [PubMed] [Google Scholar]
- Ye P, D’Ercole AJ. Insulin-like growth factor actions during development of neural stem cells and progenitors in the central nervous system. J Neurosci Res. 2006;83:1–6. doi: 10.1002/jnr.20688. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.