Abstract
To determine whether fruit bats in Singapore have been exposed to filoviruses, we screened 409 serum samples from bats of 3 species by using a multiplex assay that detects antibodies against filoviruses. Positive samples reacted with glycoproteins from Bundibugyo, Ebola, and Sudan viruses, indicating filovirus circulation among bats in Southeast Asia.
Keywords: virus surveillance, serology, Southeast Asia, Ebola virus, Bundibugyo virus, Sudan virus, virus envelope glycoprotein, viruses, filoviruses, fruit bats, Singapore
The genus Ebolavirus comprises 5 virus species: Zaire ebolavirus (EBOV), Sudan ebolavirus (SUDV), Bundibugyo ebolavirus (BDBV), Taï Forest ebolavirus (TAFV), and Reston ebolavirus (RESTV). The genus Marburgvirus comprises 1 species, Marburg marburgvirus, which includes 2 closely related virus strains: Marburg virus (MARV) and Ravn virus (RAVV). Viruses within the Ebolavirus and Marburgvirus genera are zoonotic; EBOV was the causative agent of the 2014–2016 Ebola virus disease epidemic in West Africa (1). Rousettus bats in Africa have been identified as Marburgvirus hosts (2), and viral nucleic acid and serologic evidence suggests that bats are also natural hosts of Ebolavirus spp. (3). Yet it remains unclear which species are the definitive reservoirs of filoviruses.
Ecologic models of Ebolavirus and Marburgvirus geographic distribution and habitat ranges of potential reservoir bat species suggest that both groups are distributed throughout Asia (3,4). Serologic evidence of filoviruses in frugivorous bats in Bangladesh, China, and the Philippines has been reported (5–7), and RESTV nucleic acid was detected in an insectivorous bat in the Philippines, where RESTV is considered endemic (8). We examined pteropodid bats of 3 species: Cynopterus brachyotis, Eonycteris spelaea, and Penthetor lucasi, which are widely distributed across Southeast Asia and share ecologic niches (9).
The Study
During 2011–2016, we collected serum from bats of the 3 aforementioned species in Singapore and screened samples for evidence of exposure to filoviruses. Samples were collected with permission from the National University of Singapore Institutional Animal Care and Use Committee (B01/12) and the National Parks Board (NP/RP11–011–3a). We diluted venous blood 1:10 in phosphate-buffered saline and then centrifuged, recovered, and heat-inactivated the serum at 56°C for 30 minutes and stored it at −80°C.
We developed a Bio-Plex (Bio-Rad, Hercules, CA, USA) bead-based multiplex assay that simultaneously probes serum for immunoglobulins specific to the viral envelope glycoproteins (GPs) from representative strains of all described Ebolavirus and Marburgvirus species (Table 1). A human FreeStyle 293-F stable cell-line expression system was used to produce the Ebolavirus and Marburgvirus spp. GPs as a soluble GP consisting of the entire ectodomain, sGP(1,2), which retains a native-like oligomeric conformation, as described previously with modifications (10). In brief, each GP(1,2) coding sequence was truncated at the C-terminus to remove the predicted transmembrane domain and cytoplasmic tail, then appended with the GCN trimerization peptide sequence (10) together with a factor Xa protease cleave site and a Twin-Strep-tag sequence (IBA Lifesciences, Göttingen, Germany). The sGP(1,2) proteins were produced in serum-free conditions and purified by Strep-Tactin XT technology (IBA Lifesciences). The Twin-Strep-tag was removed by factor Xa enzymatic cleavage; factor Xa was removed by Xarrest Agarose (Merck Millipore, Billerica, MA, USA); sGP(1,2) was purified further by S-200 size exclusion chromatography, concentrated, and stored frozen. These sGP(1,2)s were coupled to carboxylated beads (Bio-Rad). Screening was performed on a Bio-Rad Bio-Plex 200.
Table 1. Ebolavirus and Marburgvirus species soluble envelope glycoproteins conjugated Bio-Plex beads used in multiplex assay to detect antibodies against filoviruses*.
| Virus | Isolation host/location | Bio-Plex bead no. | NCBI accession no. |
|---|---|---|---|
| Ebola virus/H.sapiens/COD/1976/Yambuku-Mayinga | Human/DRC | 33 | NC_002549.1 |
| Bundibugyo virus/H. sapiens/UGA/2007 | Human/Uganda | 64 | FJ217161.1 |
| Taï Forest virus/H. sapiens/COV/1994/Pauleoula-CI | Human/Côte d'Ivoire | 57 | NC_014372 |
| Sudan virus/H. sapiens/UGA/2000/Gulu-808892 | Human/Uganda | 77 | NC_006432.1 |
| Reston virus/M. fascicularis/USA/1989/Pennsylvania | Macaque/USA | 85 | AF522874.1 |
| Reston virus/S. domesticus/PHL/2008/Reston08-A | Swine/Philippines | 72 | FJ621583.1 |
| Marburg virus/H. sapiens/KEN/1980/Musoke | Human/Kenya | 37 | Z12132 S55429 |
| Marburg virus/H. sapiens/AGO/2005/Ang0126 | Human/Angola | 28 | DQ447656.1 |
| Ravn virus/H. sapiens/KEN/1987/Kitum cave-810040 | Human/Kenya | 49 | NC_024781.1 |
*Bio-Plex manufactured by Bio-Rad (Hercules, CA, USA). DRC, Democratic Republic of the Congo; NCBI, National Center for Biotechnology Information.
In the absence of confirmed filovirus-negative bat serum, we used methods developed by Peel et al. to establish a median fluorescence intensity (MFI) cutoff value (11). We confirmed a cutoff value of 200 MFI (Technical Appendix), as was previously used for Eidolon helvum bat serum in a Bio-Plex serologic assay (12). We screened 409 samples with our Ebolavirus and Marburgvirus spp. sGP(1,2) Bio-Plex assay modified from that described by Bossart et al. (13). Samples were diluted 1:100 and tested in duplicate; the sGP(1,2)-coupled beads were mixed with individual samples; and a 1:1 combination of recombinant biotinylated-protein A/protein G (1:500) (Pierce, Rockford, IL, USA) was added to the wells, followed by addition of streptavidin-phycoerythrin (1:1,000) (Bio-Rad) and determination of MFI.
Samples were positive for 17 (9.1%) of 186 E. spelaea, 13 (8.5%) of 153 C. brachyotis, and 3 (4.3%) of 70 P. lucasi bats (Figure 1). Positive samples reacted with EBOV, BDBV, SUDV, or TAFV sGP(1,2). However, no samples were positive for RESTV, MARV, or RAVV sGP(1,2). We further examined positive samples to determine cross-reactivity between the Ebolavirus spp. sGP(1,2) (Table 2). Twelve (71%) samples from E. spelaea bats cross-reacted with >2 Ebolavirus spp. sGP(1,2) (BDBV, EBOV, SUDV, or TAFV). In contrast, 8 (62%) C. brachyotis and 2 (66%) P. lucasi samples were positive for only 1 sGP(1,2) (BDBV or SUDV).
Figure 1.
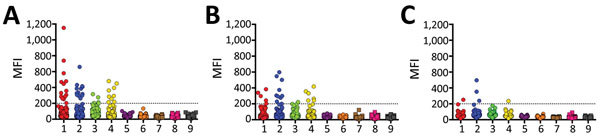
Mean fluorescence intensity (MFI) values obtained from Bio-Plex assay (Bio-Rad, Hercules, CA, USA) screening of individual serum samples from bats of 3 species with soluble filovirus glycoproteins. Dashed line indicates the cutoff value at 200 MFI. 1, Zaire ebolavirus; 2, Bundibugyo ebolavirus; 3, Taï Forest ebolavirus; 4, Sudan ebolavirus; 5, Reston ebolavirus–monkey; 6, Reston ebolavirus–pig; 7, Marburg virus–Musoke; 8, Marburg virus–Angola; 9, Ravn virus.
Table 2. Bio-Plex median fluorescence intensity values for bat serum positive for >1 filovirus antigen*.
| Bat species, ID | EBOV | BDBV | TAFV | SUDV | RESTVm | RESTVp | MARV(Mus) | MARV(Ang) | RAVV |
|---|---|---|---|---|---|---|---|---|---|
| Eonycteris spelaea, n = 186 | |||||||||
| 0805149† | 738 | 124 | 68 | 40 | 44 | 22 | 23 | 21 | 24 |
| 080814 | 86 | 318 | 105 | 258 | 26 | 12 | 17 | 16 | 20 |
| 082154 | 143 | 161 | 113 | 214 | 35 | 41 | 21 | 31 | 39 |
| 052313 | 284 | 408 | 177 | 285 | 89 | 72 | 29 | 23 | 30 |
| 052335 | 203 | 191 | 124 | 219 | 42 | 21 | 38 | 38 | 24 |
| 052339 | 357 | 306 | 141 | 293 | 54 | 31 | 26 | 26 | 42 |
| 071839 | 330 | 299 | 164 | 480 | 65 | 44 | 28 | 33 | 45 |
| 071842 | 446 | 327 | 202 | 362 | 65 | 49 | 42 | 38 | 57 |
| 110733 | 126 | 416 | 166 | 95 | 58 | 42 | 34 | 42 | 58 |
| 011603† | 1151 | 130 | 91 | 69 | 36 | 32 | 51 | 35 | 39 |
| 011616 | 252 | 294 | 168 | 175 | 32 | 49 | 47 | 29 | 50 |
| 011656 | 306 | 386 | 204 | 394 | 89 | 73 | 18 | 39 | 37 |
| 012309† | 579 | 659 | 315 | 69 | 35 | 31 | 27 | 33 | 35 |
| 021303 | 478 | 431 | 188 | 450 | 52 | 37 | 24 | 30 | 47 |
| 111903 | 469 | 384 | 276 | 113 | 52 | 57 | 37 | 69 | 54 |
| 111907 | 285 | 336 | 213 | 158 | 39 | 36 | 29 | 50 | 30 |
| 042722 |
260
|
262
|
174 |
167 |
75 |
31 |
54 |
24 |
42 |
| Cynopterus brachyotis, n = 153 | |||||||||
| 051253 | 121 | 133 | 59 | 242 | 40 | 41 | 19 | 25 | 68 |
| 0516613 | 146 | 293 | 127 | 73 | 47 | 36 | 25 | 29 | 22 |
| 0516632 | 138 | 139 | 86 | 356 | 35 | 25 | 28 | 34 | 34 |
| 0726122† | 119 | 501 | 100 | 60 | 40 | 46 | 25 | 19 | 29 |
| 1103241 | 84 | 141 | 128 | 241 | 50 | 47 | 66 | 38 | 34 |
| 100903 | 148 | 201 | 71 | 108 | 42 | 33 | 18 | 16 | 36 |
| 100914 | 74 | 228 | 70 | 55 | 39 | 38 | 30 | 27 | 26 |
| 100925 | 166 | 304 | 109 | 116 | 43 | 18 | 33 | 30 | 28 |
| 021357 | 201 | 299 | 179 | 264 | 65 | 44 | 25 | 55 | 47 |
| 050804 | 242 | 276 | 140 | 124 | 41 | 30 | 34 | 33 | 44 |
| 050818 | 383 | 374 | 198 | 332 | 60 | 55 | 29 | 26 | 68 |
| 040807† | 297 | 597 | 194 | 192 | 40 | 38 | 122 | 95 | 32 |
| 042701† |
339
|
547
|
222
|
417
|
60 |
78 |
54 |
25 |
62 |
| Penthetor lucasi, n = 70 | |||||||||
| 062590† | 34 | 496 | 93 | 39 | 36 | 18 | 23 | 17 | 23 |
| 070409† | 95 | 238 | 129 | 89 | 62 | 27 | 34 | 36 | 37 |
| 112112† | 251 | 352 | 148 | 235 | 51 | 29 | 23 | 23 | 29 |
*Bio-Plex manufactured by Bio-Rad (Hercules, CA, USA). Boldface indicates positive results. BDBV, Bundibugyo virus; EBOV, Ebola virus; ID, specimen identification number; MARV(Mus), Marburg virus–Musoke; MARV(Ang), Marburg virus–Angola; RESTVm, Reston virus–monkey; RESTVp, Reston virus–pig; SUDV, Sudan virus; RAVV, Ravn virus; TAFV, Taï Forest virus. †Sample screened by Western blot and shown in Figure 2.
To further determine the cross-reactivity of positive samples and to corroborate Bio-Plex assay results for a selected number of samples, we performed Western blot (WB) assays (Figure 2). The filovirus GP(1,2) is a trimer of heterodimeric GP1 and GP2 subunits. The trimeric-like sGP(1,2) is the antigen in the multiplex Bio-Plex assay, whereas linearized monomeric sGP1 and sGP2 subunits are the antigens in WBs. Reduced and denatured EBOV or BDBV unconjugated sGP(1,2) was loaded on 8% sodium dodecyl sulfate–polyacrylamide electrophoresis gels, transferred to a polyvinylidene difluoride membrane, and probed with 1:100 dilutions of positive and negative bat serum, as previously determined by the Bio-Plex assay. All 3 E. spelaea bat samples and 2 of 3 C. brachyotis bat samples that were Bio-Plex positive were also positive by WB and displayed reactivity with EBOV and BDBV GP1 and GP2 antigens; no P. lucasi bat samples positive by Bio-Plex were positive by WB.
Figure 2.
Western blot results of individual bat serum samples probed against Zaire ebolavirus and Bundibugyo ebolavirus glycoproteins 1 and 2 (GP1, GP2). Boldface indicates positivity by Western blot and underlining indicates positivity by Bio-Plex (Bio-Rad, Hercules, CA, USA). 1, soluble GP1 and GP2 blotted with control anti–Ebola virus nonhuman primate polyclonal serum that demonstrates cross-reactivity with Bundibugyo ebolavirus soluble GP. Other numbers along baseline correspond to the following sample identifiers, also used in Table 2: 2, 0805149; 3, 012309; 4, 011603; 5, 0116048; 6, 0719036; 7, 1128015; 8, 0726122; 9, 042701; 10, 040807; 11, 0512540; 12, 1009010; 13, 0408029; 14, 070409; 15, 112112; 16, 062590; 17, 0228004; 18, 0919025; 19, 0625095. BDBV, Bundibugyo virus; EBOV, Ebola virus.
Conclusions
We present evidence of antibodies specific to filoviruses antigenically related to Ebolavirus spp. in 3 species of fruit bats widely distributed throughout Southeast Asia. We detected seroreactivity with Ebolavirus spp. but not Marburgvirus spp. GP. Despite the close relatedness of the viruses, we detected samples reacting with only SUDV, not RESTV, GP. This finding contrasts with previous reports of bat serum cross-reactivity with RESTV nucleoprotein (5,7,14). Possible explanations include 1) the fact that our customized Bio-Plex assay is based on conformational sGP(1,2), which can differentiate antibody specificity better than the more sequence conserved nucleoprotein, and 2) the lack of evidence of RESTV GP positivity with Cynopterus and Eonycteris bat serum samples, which is in line with previous findings (both species were negative while only Rousettus amplexicaudatus bats were positive) (7). E. spelaea bats were previously predicted to be filovirus hosts (15), and sequences of novel filoviruses have been discovered in E. spelaea bat populations in Yunnan, China (14). Our data provide additional empirical evidence that populations of C. brachyotis, E. spelaea, and P. lucasi bats in Southeast Asia are hosts of filoviruses, which seem antigenically more closely related to EBOV, BDBV, and SUDV than to RESTV.
Examination of cross-reactivity of positive samples from E. spelaea, C. brachyotis, and P. lucasi bats revealed no clear patterns of preferential reactivity with EBOV, BDBV, or SUDV GP. Factors that might contribute to the lack of P. lucasi positivity by WB include sensitivity differences between Bio-Plex and WB assays paired with the change in sGP(1,2) conformation. Two Bio-Plex EBOV-positive samples (E. spelaea samples 0805149 and 011603) reacted with EBOV sGP2 and BDBV sGP1 in the WB. Bio-Plex and WB data strongly suggest the presence of yet-undetected batborne filoviruses, which are antigenically related to but distinct from BDBV, EBOV, and SUDV circulating in local bat populations. Reasons why these filoviruses have remained undetected include their inability to cross the species barrier, the rarity of spillovers into humans or domestic animals, or the fact that spillover events cause mild or no disease. We suggest that a yet-undescribed diversity of filoviruses exists in Southeast Asia bat populations, a hypothesis supported by the recent identification of filovirus sequences in E. spelaea and R. leschenaulti bats in China (14,16). Comprehensive surveillance including serology and detection of viral nucleic acid, along with virus isolation, will help elucidate the characteristics of filoviruses endemic to Asia and identify bat species that function as maintenance populations and reservoirs.
Median fluorescence intensity cutoff value determination and results for filoviruses in serum from fruit bats, Singapore, 2011–2016.
Acknowledgment
We thank Alison J. Peel for assistance with determination of the median fluorescence intensity cutoff and statistical advice.
This study was supported by the Duke-National University of Singapore Signature Research Program funded by the Agency of Science, Technology and Research, and the Ministry of Health, Singapore, and by grants from National University of Singapore–Global Asia Institute (NIHA-2011-1-005), the National Medical Research Council (NMRC/BNIG/2005/2013), the Ministry of Health (CDPHRG/0006/2014) in Singapore, and the US Department of Defense, Defense Threat Reduction Agency. C.C.B., E.D.L., L.Y., and S.L.S. were supported by funding from the Biological Defense Research Directorate of the Naval Medical Research Center. E.D.L. was also supported by the National Science Foundation, an East Asia and Pacific Summer Institutes Fellowship award (1515304), with collaborative support from the National University of Singapore.
Biography
Dr. Laing is a postdoctoral fellow at the Uniformed Services University and performed this work while a National Science Foundation EAPSI fellow at Duke-National University of Singapore Medical School. His research focuses on biosurveillance, batborne viruses, and antiviral immunity.
Footnotes
Suggested citation for this article: Laing ED, Mendenhall IH, Linster M, Low DHW, Chen Y, Yan L, et al. Serologic evidence of fruit bat exposure to filoviruses, Singapore, 2011–2016. Emerg Infect Dis. 2018 Jan [date cited]. https://doi.org/10.3201/eid2401.170401
These authors contributed equally to this article.
References
- 1.Baize S, Pannetier D, Oestereich L, Rieger T, Koivogui L, Magassouba N, et al. Emergence of Zaire Ebola virus disease in Guinea. N Engl J Med. 2014;371:1418–25. 10.1056/NEJMoa1404505 [DOI] [PubMed] [Google Scholar]
- 2.Towner JS, Amman BR, Sealy TK, Carroll SA, Comer JA, Kemp A, et al. Isolation of genetically diverse Marburg viruses from Egyptian fruit bats. PLoS Pathog. 2009;5:e1000536. 10.1371/journal.ppat.1000536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Olival KJ, Hayman DT. Filoviruses in bats: current knowledge and future directions. Viruses. 2014;6:1759–88. 10.3390/v6041759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peterson AT, Bauer JT, Mills JN. Ecologic and geographic distribution of filovirus disease. Emerg Infect Dis. 2004;10:40–7. 10.3201/eid1001.030125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olival KJ, Islam A, Yu M, Anthony SJ, Epstein JH, Khan SA, et al. Ebola virus antibodies in fruit bats, bangladesh. Emerg Infect Dis. 2013;19:270–3. 10.3201/eid1902.120524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yuan J, Zhang Y, Li J, Zhang Y, Wang LF, Shi Z. Serological evidence of ebolavirus infection in bats, China. Virol J. 2012;9:236. 10.1186/1743-422X-9-236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taniguchi S, Watanabe S, Masangkay JS, Omatsu T, Ikegami T, Alviola P, et al. Reston Ebolavirus antibodies in bats, the Philippines. Emerg Infect Dis. 2011;17:1559–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jayme SI, Field HE, de Jong C, Olival KJ, Marsh G, Tagtag AM, et al. Molecular evidence of Ebola Reston virus infection in Philippine bats. Virol J. 2015;12:107. 10.1186/s12985-015-0331-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mendenhall IH, Borthwick S, Neves ES, Low D, Linster M, Liang B, et al. Identification of a lineage D betacoronavirus in cave nectar bats (Eonycteris spelaea) in Singapore and an overview of lineage D reservoir ecology in SE Asian bats. Transbound Emerg Dis. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan YP, Yan L, Feng YR, Broder CC. Preparation of recombinant viral glycoproteins for novel and therapeutic antibody discovery. [xiii.]. Methods Mol Biol. 2009;525:31–58, xiii. 10.1007/978-1-59745-554-1_2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peel AJ, McKinley TJ, Baker KS, Barr JA, Crameri G, Hayman DT, et al. Use of cross-reactive serological assays for detecting novel pathogens in wildlife: assessing an appropriate cutoff for henipavirus assays in African bats. J Virol Methods. 2013;193:295–303. 10.1016/j.jviromet.2013.06.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayman DT, Suu-Ire R, Breed AC, McEachern JA, Wang L, Wood JL, et al. Evidence of henipavirus infection in West African fruit bats. PLoS One. 2008;3:e2739. 10.1371/journal.pone.0002739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bossart KN, McEachern JA, Hickey AC, Choudhry V, Dimitrov DS, Eaton BT, et al. Neutralization assays for differential henipavirus serology using Bio-Plex protein array systems. J Virol Methods. 2007;142:29–40. 10.1016/j.jviromet.2007.01.003 [DOI] [PubMed] [Google Scholar]
- 14.Yang XL, Zhang YZ, Jiang RD, Guo H, Zhang W, Li B, et al. Genetically diverse filoviruses in Rousettus and Eonycteris spp. bats, China, 2009 and 2015. Emerg Infect Dis. 2017;23:482–6. 10.3201/eid2303.161119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han BA, Schmidt JP, Alexander LW, Bowden SE, Hayman DT, Drake JM. Undiscovered bat hosts of filoviruses. PLoS Negl Trop Dis. 2016;10:e0004815. 10.1371/journal.pntd.0004815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He B, Feng Y, Zhang H, Xu L, Yang W, Zhang Y, et al. Filovirus RNA in fruit bats, China. Emerg Infect Dis. 2015;21:1675–7. 10.3201/eid2109.150260 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Median fluorescence intensity cutoff value determination and results for filoviruses in serum from fruit bats, Singapore, 2011–2016.


