Abstract
DNA double-strand breaks (DSBs) are mainly repaired either by homologous recombination (HR) or by nonhomologous end-joining (NHEJ) pathways. Here, we showed that myeloid cell leukemia sequence 1 (Mcl-1) acts as a functional switch in selecting between HR and NHEJ pathways. Mcl-1 was cell cycle–regulated during HR, with its expression peaking in S/G2 phase. While endogenous Mcl-1 depletion reduced HR and enhanced NHEJ, Mcl-1 overexpression resulted in a net increase in HR over NHEJ. Mcl-1 directly interacted with the dimeric Ku protein complex via its Bcl-2 homology 1 and 3 (BH1 and BH3) domains, which are required for Mcl-1 to inhibit Ku-mediated NHEJ. Mcl-1 also promoted DNA resection mediated by the Mre11 complex and HR-dependent DSB repair. Using the Mcl-1 BH1 domain as a docking site, we identified a small molecule, MI-223, that directly bound to BH1 and blocked Mcl-1–stimulated HR DNA repair, leading to sensitization of cancer cells to hydroxyurea- or olaparib-induced DNA replication stress. Combined treatment with MI-223 and hydroxyurea or olaparib exhibited a strong synergy against lung cancer in vivo. This mechanism-driven combination of agents provides a highly attractive therapeutic strategy to improve lung cancer outcomes.
Keywords: Cell Biology
Keywords: Cancer
Introduction
DNA double-strand breaks (DSBs) are generated when the phosphodiester backbones of both DNA strands are broken at the same position or in sufficient proximity to allow physical dissociation of the double helix into 2 separate molecules (1). DSBs can be induced by ionizing radiation, chemotherapeutic drugs, oxidative stress, and replication fork collapse (2). Inappropriate repair of DSBs may result in propagation of deleterious mutations, genomic instability, immune deficiency, cancer predisposition, and accelerated aging or cell death (3–5).
Cells utilize 2 major pathways for DSB repair: nonhomologous end joining (NHEJ) and homologous recombination (HR). NHEJ facilitates DSB repair by direct ligation of broken DNA ends (6). To initiate NHEJ, the Ku70/Ku80 heterodimer binds to blunt or near-blunt DNA ends. DSB-bound Ku then recruits and activates the DNA-dependent protein kinase catalytic subunit (DNA-PKcs), which triggers a signaling cascade that orchestrates downstream repair processes that eventually seal the breaks (7). In the HR repair pathway, DSBs are recognized by the MRN (Mre11-Rad50-NBS1) complex and CtIP to initiate DSB end resection leading to generation of 3′ single-stranded DNA (ssDNA) overhangs through endonucleolytic cleavage followed by 3′–5′ exonucleolytic processing (6, 8, 9). The 3′ ssDNA overhangs are initially coated by the RPA complex to form an RPA-ssDNA nucleoprotein filament to allow extensive resection by the EXO and DNA2 nucleases, followed by displacement of RPA to allow assembly of the Rad51-ssDNA nucleoprotein filament (1). Rad51 loading promotes invasion onto the undamaged template and strand displacement, generating D-loop formation, which is necessary to generate a Holliday junction and a heteroduplex molecule. Repair ensues using the undamaged strand as a template, followed by ligation of the DNA ends (10).
The proper choice of DSB repair pathway has a profound impact on genomic integrity and the prevention of cancer (1). DSB repair via HR ensures fidelity and reduces the probability of mutation or genomic instability compared with NHEJ, because HR uses an undamaged template with homologous sequences to restore sequence information lost at the DSB site, whereas NHEJ does not require sequence homology for repair (1, 10). The choice between these pathways depends on the phase of the cell cycle and the nature of the DSB ends (11). NHEJ is active throughout all phases of the cell cycle but predominates in G1. HR is more active in S and G2 phases during DNA replication, since an identical sister chromatid is available as a template for repair (6, 12). A major restriction point in the choice between DSB repair pathways is the competition between Ku-mediated DNA end protection and MRN complex–initiated DNA end resection (13, 14). For example, initiation of 5′–3′ resection of DNA ends by the MRN complex commits cells to HR-dependent repair, and prevents repair by classical NHEJ. Once HR is initiated, NHEJ cannot be utilized in most instances. Conversely, once Ku binds dsDNA ends with high affinity, the DSB-bound Ku complex restricts the ability of the MRN complex to initiate DNA resection, which suppresses HR, while, at the same time, promoting NHEJ (1, 10, 15). A major, unanswered question is how MRN directly or indirectly counteracts NHEJ to promote HR. One possibility is that there is a pathway choice controller that allows NHEJ to predominate in G0/G1 and subsequently switch to the HR pathway in S/G2 during cell cycle progression through regulation of the core DSB repair machinery.
Myeloid cell leukemia sequence 1 (Mcl-1) is a unique antiapoptotic Bcl-2 family member that is overexpressed in many tumor types (16, 17). Mcl-1 expression is tightly regulated at multiple levels, including transcriptional, posttranscriptional, and posttranslational processes (18). In contrast to other antiapoptotic Bcl-2 family members (i.e., Bcl-2, Bcl-xL, Bcl-w, and Bfl-1A1), Mcl-1 has a considerably longer N-terminus that is intrinsically unstructured and therefore resistant to structural analyses (19). Mcl-1 is unique with respect to its short half-life (30 minutes to 3 hours) and short-term prosurvival function, which probably relates to the presence of a long proline-, glutamic acid–, serine-, and threonine-rich (PEST) region upstream of the Bcl-2 homology (BH) domain (17). Mcl-1 expression changes rapidly in response to cellular stresses or cell cycle progression, because its degradation is tightly regulated by three E3 ligases (Mule, FBW7, and β-TrCP) and two deubiquitinases (USP9X and Ku70) (17, 19, 20). In addition to its cell survival function, Mcl-1 has been demonstrated to regulate ATR-mediated CHK1 phosphorylation and localize to sites of DNA damage in response to DNA damage (21–23). One recent report reveals that Mcl-1 deficiency impairs DNA DSB repair and reinitiation of stalled DNA replication forks, leading to sensitization of cells to radiation (24).
Here we report the discovery that, during cell cycle progression, Mcl-1 peaks and selectively interacts with Ku in S/G2 phase, resulting in the suppression of NHEJ while simultaneously promoting DNA resection and activation of the HR pathway. This indicates that Mcl-1, in addition to its antiapoptotic function, also functions in mediating DSB repair pathway choice. Following our identification of this mechanism, we have discovered a novel small molecule, MI-223, that disrupts the Mcl-1/Ku interaction, inhibits HR activity, and strongly synergizes with DNA replication stress agents (hydroxyurea or olaparib) against lung cancer in vitro and in vivo.
Results
Mcl-1 accumulates in S/G2-phase cells.
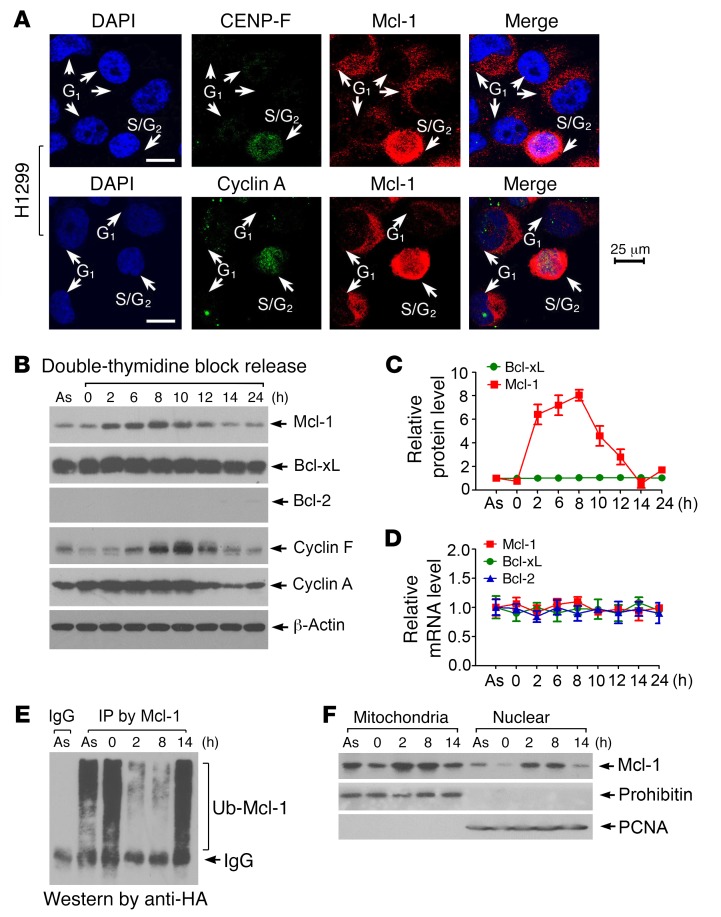
Mcl-1 is primarily localized in the outer mitochondrial membrane and in small quantities in the nucleus (16). To determine whether the cell cycle affects Mcl-1 expression and subcellular localization, we used the cell cycle markers CENP-F and cyclin A, which are absent in G1 phase and active during S and G2 phases (25, 26). Significantly increased levels of cytoplasmic and nuclear Mcl-1 were observed in S/G2 phase compared with G1 phase in H1299 human lung cancer cells (Figure 1A). The specificity of Mcl-1 antibody used in this experiment was further confirmed as shown in Supplemental Figure 1 (supplemental material available online with this article; https://doi.org/10.1172/JCI92742DS1 See complete unedited blots in the supplemental material). To verify the S/G2-specific nature of Mcl-1 accumulation, H1299 cells were synchronized at the G1/S boundary by double-thymidine block, followed by release into the cell cycle. Most cells entered S phase at 2 hours, reached G2 phase around 8 hours, and returned to G1 phase at 14 hours (Supplemental Figure 2A). Mcl-1 protein but not mRNA levels gradually increased from 2 hours and returned to a baseline level after 14 hours (Figure 1, B–D). As expected, cyclin A (an S/G2 marker) and cyclin F (a late G2 marker) were expressed in a cell cycle–dependent manner. In contrast, both protein and mRNA levels of Bcl-2 and Bcl-xL were not significantly changed during cell cycle progression (Figure 1, B–D), indicating that Mcl-1 but not Bcl-2 or Bcl-xL is a cell cycle–regulated molecule. A similar expression profile of Mcl-1 during cell cycle progression was also observed in H460 human lung cancer cells (Supplemental Figure 2B), indicating that this is a general response not limited to a single cell line. Since the cell cycle–dependent change in Mcl-1 occurs only at the protein but not the mRNA level, we tested whether ubiquitination regulates Mcl-1 expression during cell cycle progression. Intriguingly, significantly decreased levels of Mcl-1 ubiquitination were observed in S-phase (2-hour time point) and G2-phase (8-hour time point) cells compared with G1-phase cells (0 or 14-hour time point) (Figure 1E). Subcellular fractionation experiments revealed that Mcl-1 levels in both isolated mitochondria and nuclear fractions were significantly enhanced in S-phase (2 hours) and G2-phase (8 hours) cells (Figure 1F). These findings suggest that specific upregulation of Mcl-1 in S/G2 may play a role distinct from its antiapoptotic function.
Figure 1. Mcl-1 is cell cycle–regulated with expression peaking in S/G2 phase.
(A) CENP-F and cyclin A as S/G2 markers were coimmunostained with Mcl-1 in H1299 cells. DAPI was used as nuclear marker. Scale bars: 25 μm. (B) H1299 cells were synchronized by double-thymidine block. After thymidine was washed off, cells were released to normal medium for a time course up to 24 hours. Mcl-1, Bcl-2, Bcl-xL, cyclin A, and cyclin F in total cell lysates were analyzed by Western blot. As, asynchronous cells. (C and D) Relative protein and mRNA levels of Mcl-1, Bcl-2, and Bcl-xL were quantified during cell cycle progression. (E) Mcl-1 ubiquitination was analyzed by anti–Mcl-1 co-IP and Western blot using anti-HA antibody following transfection with HA-tagged ubiquitin (HA-Ub) constructs during cell cycle progression. (F) Mitochondrial and nuclear fractions were isolated from cells at various time points during cell cycle progression. Mcl-1 was analyzed by Western blot. Prohibitin or PCNA was used as mitochondrial or nuclear marker, respectively.
Mcl-1 is required for HR-dependent DSB repair and clonogenic survival following DNA replication stress.
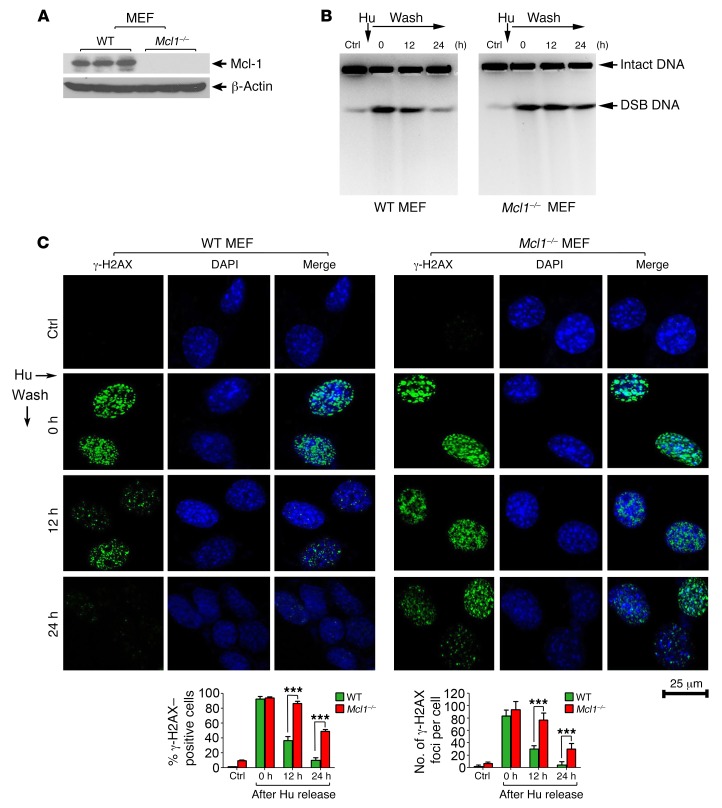
Hydroxyurea (Hu) is an inhibitor of ribonucleotide reductase, an enzyme essential for de novo deoxyribonucleotide triphosphate (dNTP) synthesis (27). Hu depletes cells of dNTPs, leading to DNA replication stress, which initially results in stalled replication forks that, after prolonged treatment, collapse into DSBs (28). To test whether Mcl-1 affects the repair of DNA replication stress–induced DSBs, WT and Mcl1–/– mouse embryonic fibroblasts (MEFs) were subjected to Hu (0.2 mM) treatment for 24 hours. Following Hu removal, cells were incubated in normal culture medium for various times up to 24 hours. Hu-induced DSBs were assessed by pulsed-field gel electrophoresis (a direct method for the measurement of DSBs) as previously described (29). After 24 hours of continuous treatment with Hu, DSBs were induced in MEF WT and MEF Mcl1–/– cells (Figure 2, A and B). DSBs were gradually reduced after removal of Hu from cell culture medium, indicating that some of the DSBs were repaired. Importantly, significantly more DSBs were observed in MEF Mcl1–/– cells compared with MEF WT cells 24 hours after removal of Hu, indicating that depletion of Mcl-1 via knockout suppresses the repair of Hu-induced DSBs (Figure 2B), possibly by retarding their repair. Formation of a chromatin-associated γ-H2AX focus is considered to be a sensitive and selective signal for the existence of DSBs (30). Therefore, Hu-induced DSBs were also evaluated by immunostaining with γ-H2AX antibody, and the percentage of γ-H2AX–positive cells and number of γ-H2AX foci per cell were determined as described previously (31). Knockout of Mcl-1 significantly delayed the disappearance of γ-H2AX foci following DNA replication stress (Figure 2, A and C). Because the repair of DNA replication stress–induced DSBs occurs through the HR DNA repair pathway (6), our results indicate a potential role for Mcl-1 in HR-dependent DSB repair, which is consistent with and/or extends recent findings from others (24). These data indicate that slower repair of Hu-induced DSBs in Mcl-1–deficient cells may result from inhibition of the HR repair pathway. The repair of x-ray–induced DSBs occurs mainly through the NHEJ pathway (10). Mcl-1 loss did not delay γ-H2AX disappearance and even slightly promoted the repair of x-ray radiation–induced DSB foci (Supplemental Figure 3). Collectively, these results indicate that Mcl-1 differentially regulates HR and NHEJ DNA repair pathways.
Figure 2. Knockout of Mcl-1 retards the repair of hydroxyurea-induced DSBs.
(A) Mcl-1 expression was analyzed by Western blot in WT and Mcl1–/– MEFs. (B and C) WT and Mcl1–/– MEFs were subjected to hydroxyurea (Hu, 0.2 mM) treatment for 24 hours. After Hu removal, cells were incubated in normal culture medium for various times up to 24 hours. Hu-induced DSBs were assessed by pulsed-field gel electrophoresis or immunofluorescence using γ-H2AX antibody at various time points. Scale bar: 25 μm. The percentage of γ-H2AX–positive cells (≥5 foci) and the number of γ-H2AX foci per cell were determined by counting of at least 100 cells from each sample. Data represent the mean ± SD, n = 3 per group. ***P < 0.001, by 2-tailed t test.
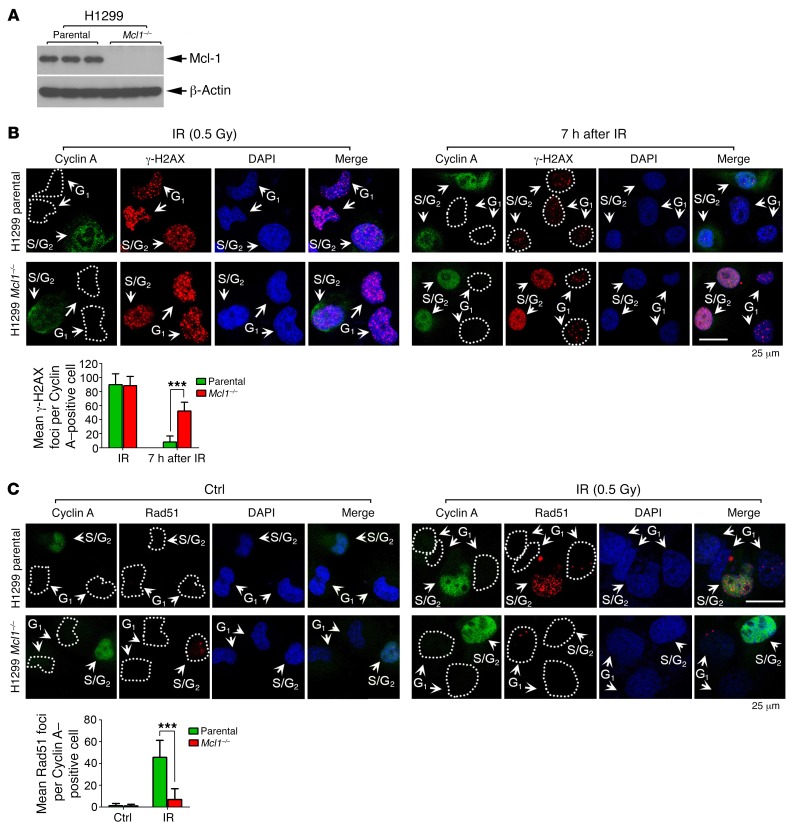
To better examine the effects of Mcl-1 on the rate of DNA repair and HR efficiency, endogenous Mcl-1 was knocked out from H1299 cells using the CRISPR/Cas9 system (Figure 3A). H1299 parental and H1299 Mcl1–/– cells were treated with irradiation (IR) (0.5 Gy), followed by costaining with cyclin A and γ-H2AX antibodies. IR induced clear γ-H2AX foci formation in both H1299 parental and H1299 Mcl1–/– cells (Figure 3B, left panel). By 7 hours after radiation, the majority of γ-H2AX foci had disappeared in H1299 parental cells, including cells in both S/G2 (i.e., cyclin A positive) and G1 (i.e., cyclin A negative) phases (Figure 3B, right panel), indicating that most DSBs were repaired. However, significantly more γ-H2AX foci were observed in the S/G2 but not the G1 population in H1299 Mcl1–/– cells by 7 hours after radiation (Figure 3B, right panel), indicating that Mcl-1 is essential for efficient DSB repair in S/G2-phase but not G1-phase cells. The formation of Rad51 foci is considered a marker of HR DNA repair (32, 33). Rad51 foci were compared in H1299 parental and H1299 Mcl1–/– cells following IR treatment. Rad51 foci were mainly observed in the S/G2 (cyclin A positive) population of H1299 parental cells after IR. Rad51 foci were significantly decreased in S/G2 cells in which Mcl-1 was knocked out following IR (Figure 3C). These findings provide additional evidence that Mcl-1 is required for HR repair in S/G2 cells.
Figure 3. Mcl-1 is essential for DSB repair via HR in S/G2 cells.
(A) Endogenous Mcl-1 was knocked out from H1299 using CRISPR/Cas9 system. (B) H1299 parental and H1299 Mcl1–/– cells were treated with IR (0.5 Gy), followed by costaining with cyclin A and γ-H2AX antibodies immediately or after 7 hours. Cyclin A is a well-known S/G2 marker; therefore, cyclin A–positive cells are considered as S/G2-phase cells. Scale bar: 25 μm. The γ-H2AX foci per cell were determined by counting of at least 100 cells from each sample. Data represent the mean ± SD, n = 3 per group. ***P < 0.001, by 2-tailed t test. (C) H1299 parental and H1299 Mcl1–/– cells were treated with IR (0.5 Gy), followed by costaining with cyclin A and Rad51 antibodies. Scale bar: 25 μm. The Rad51 foci per cell were determined by counting of at least 100 cells from each sample. Data represent the mean ± SD, n = 3 per group. ***P < 0.001, by 2-tailed t test.
To test whether Mcl-1 loss sensitizes cells to DNA replication stress, WT and Mcl1–/– MEFs were treated with increasing doses of Hu, which is known to induce cell killing via DNA replication stress (34). Clonogenic survival assays revealed that knockout of Mcl-1 significantly enhanced Hu-induced cell killing (Supplemental Figure 4A). Furthermore, Mcl-1 was knocked down using Mcl-1 shRNA or knocked out using CRISPR/Cas9 from H1299 cells, followed by treatment with Hu or the PARP inhibitor olaparib (35). Disruption of Mcl-1 significantly sensitized H1299 cells to the DNA replication stress agents Hu and olaparib (Supplemental Figure 4, B and C).
Mcl-1 enhances HR and suppresses NHEJ.
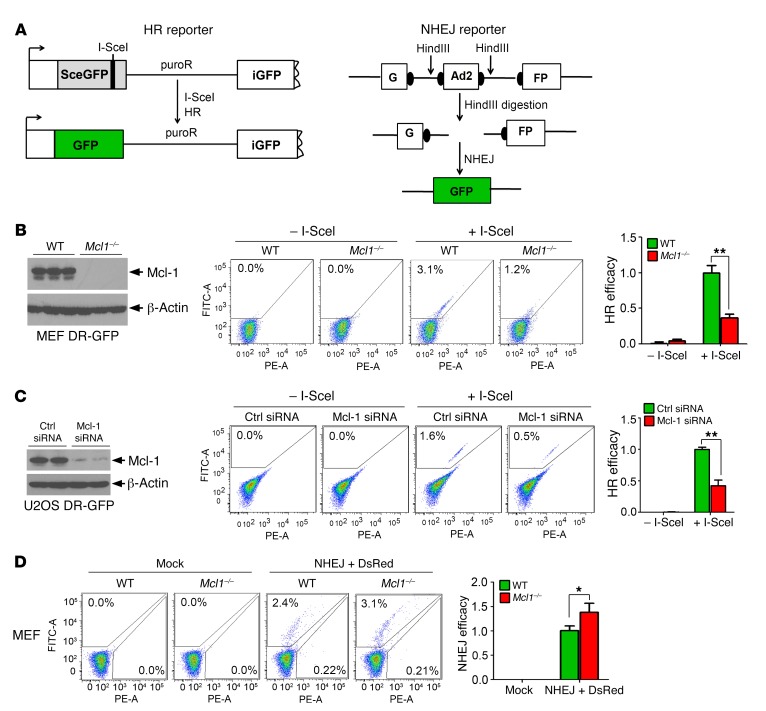
An HR reporter plasmid (pDR-GFP) that contains a GFP gene inactivated by insertion of the I-SceI endonuclease recognition site was used to measure HR activity (Figure 4A, left panel, and ref. 36). Active GFP can be restored only after I-SceI–induced DSB is repaired by HR. We observed a significant reduction in HR activity in Mcl1–/– MEFs compared with WT MEFs (Figure 4B). To provide a more physiologically relevant test of HR efficiency, U2OS DR-GFP cells carrying a chromosomally integrated single copy of the GFP HR reporter with I-SceI site were also used to measure HR activity (37, 38). Accordingly, depletion of Mcl-1 from U2OS DR-GFP cells using Mcl-1 siRNA also significantly reduced HR efficiency (Figure 4C). NHEJ activity was also compared in WT versus Mcl1–/– MEFs using the NHEJ substrate pGFP-Pem1-Ad2 system, in which the GFP sequence is interrupted by an inserted adenovirus exon 2 (Ad2) sequence (39). Following removal of Ad2 by HindIII, active GFP can be restored in the resulting plasmid only through NHEJ repair (Figure 4A, right panel). We found that NHEJ activity was increased in the absence of Mcl-1 (Figure 4D). These findings suggest that Mcl-1 positively regulates HR and negatively regulates NHEJ, thus controlling DSB repair pathway choice. Intriguingly, depletion of Mcl-1 by knockout or knockdown did not significantly affect the percentage of cells in S and G2 phases of the cell cycle or proliferation rate (Supplemental Figure 5, A–D). Similar effects of Mcl-1 on HR, NHEJ, cell cycle, and proliferation rate were also observed in H1299 cells expressing Mcl-1 siRNA versus control siRNA (Supplemental Figure 6). Additionally, knockout of Mcl-1 from H1299 or MEF cells did not significantly affect plating efficiency (Supplemental Tables 1 and 2).
Figure 4. Depletion of Mcl-1 downregulates HR and upregulates NHEJ.
(A) Schematic diagram of HR and NHEJ reporter systems. HR reporter is composed of 2 defective GFP genes that can be rescued only by HR, resulting in GFP fluorescence. In the NHEJ reporter, GFP is interrupted by an adenoviral exon (Ad2) and can be restored upon HindIII digestion and NHEJ repair. (B) HR activity was compared in WT and Mcl1–/– MEFs. Data represent the mean ± SD, n = 3 per group. **P < 0.01, by 2-tailed t test. (C) Mcl-1 was knocked down using Mcl-1 siRNA from U2OS DR-GFP cells, followed by analysis of HR activity. Data represent the mean ± SD, n = 3 per group. **P < 0.01, by 2-tailed t test. (D) NHEJ activity was compared in WT and Mcl1–/– MEFs. Data represent the mean ± SD, n = 3 per group. *P < 0.05, by 2-tailed t test.
Mcl-1 facilitates recruitment of Mre11 to DSBs via direct interaction with Ku.
To test whether Mcl-1 affects the recruitment of factors regulating HR (i.e., Mre11) or NHEJ (i.e., Ku70/Ku80) to DSB sites, a ChIP assay was performed following induction of DSBs with I-SceI in DR-GFP MEFs or DR-GFP U2OS cells. Two pairs of primers, located 200 bp upstream or downstream of the I-SceI site, were used to determine the relative abundance of Mre11 and Ku70 at the induced break site (Supplemental Figure 7A). Levels of Ku70 at DSBs induced by I-SceI were lower in WT MEFs compared with Mcl1–/– MEFs. Conversely, levels of Mre11 recruited to DSBs were significantly higher in WT MEFs compared with Mcl1–/– MEFs (barely detectable in Mcl1–/– MEFs) (Supplemental Figure 7B). Similar results were also observed in U2OS DR-GFP cells when Mcl-1 was knocked down with Mcl-1 siRNA (Supplemental Figure 7C). These results indicate that Mcl-1 is essential for recruitment of Mre11 to the DSB site. Thus, the negative effect of Mcl-1 on Ku70 recruitment, and its positive effect on Mre11 recruitment following DSB induction, may be a critical factor for pathway choice in DSB repair.
Mcl-1 was enhanced in S/G2, but total levels of Ku70/Ku80 and Mre11 did not change during cell cycle progression (Supplemental Figure 7D). To test whether Mcl-1 associates with factors regulating NHEJ and HR, coimmunoprecipitation (co-IP) was performed in H1299 cells during cell cycle progression. Mcl-1 selectively interacted with Ku70/Ku80 but not Mre11, peaking at S/G2 phase (Supplemental Figure 7E). To further assess whether cell cycle progression affects subcellular localization of Mcl-1 and Ku, first, G1- and S/G2-phase H1299 cells were isolated from double-thymidine block synchronization. Subcellular fractionation experiments were then performed to isolate heavy membrane–containing mitochondria, light membrane–containing endoplasmic reticulum, and nuclear (Nuc) fractions from G1- or S/G2-phase H1299 cells as we previously described (17). Levels of Ku70 and Mcl-1 in each fraction were quantified from Western blot by ImageJ software (NIH). In G1-phase cells, the majority of Mcl-1 (86%) was localized in mitochondria and only a small portion of Mcl-1 was localized in the endoplasmic reticulum (8%) and nucleus (6%), while Ku70 and Ku80 were localized only in the nucleus (Supplemental Figure 7F). When cells entered S/G2 phase, the distribution of Mcl-1 in the nuclear fraction was significantly enhanced (i.e., from 6% to 40%) (Supplemental Figure 7F), thus having great potential to colocalize and/or interact with Ku proteins in the nucleus.
To test whether replication stress affects the Mcl-1/Ku interaction, time course experiments were carried out. H1299 cells were treated with Hu or olaparib for various times (0, 0.5, 1, 2, 8, or 24 hours). The Mcl-1/Ku interaction and cell cycle were analyzed simultaneously at each time point. Mcl-1/Ku interaction was increased at 2, 8, and 24 hours following treatment of cells with Hu or olaparib (Supplemental Figure 8). However, Hu treatment did not alter the percentage of cells in S phase at short time points (i.e., 2 and 8 hours), and the percentage of cells in S phase was enhanced only at the 24-hour time point, indicating that increased Mcl-1/Ku interaction also occurs before cells enter S phase following Hu treatment (Supplemental Figure 8A). Intriguingly, treatment of H1299 cells with olaparib did not significantly affect the percentage of S phase cells at time points tested, but increased Mcl-1/Ku interactions were observed at 2, 8, and 24 hours (Supplemental Figure 8B). These findings indicate that, in addition to S/G2 cell cycle phase, Hu- or olaparib-induced DNA replication stress can also promote Mcl-1/Ku interaction.
Mcl-1 directly interacts with Ku via BH1 and BH3 domains, leading to suppression of Ku-DNA binding activity.
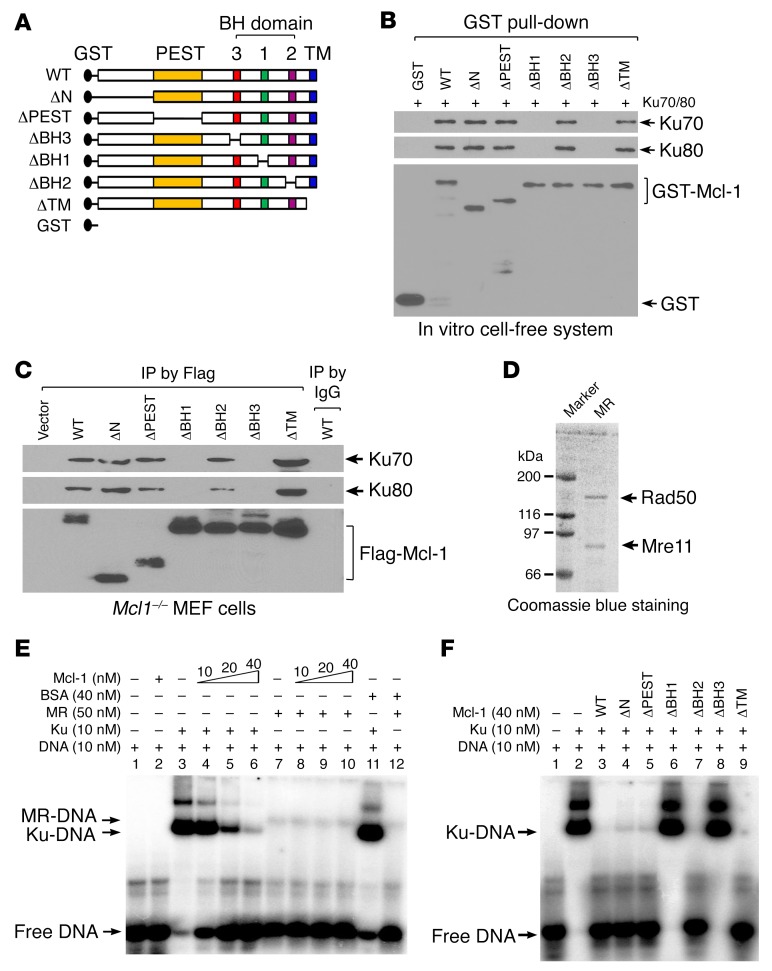
Mcl-1 contains multiple functional domains, including N-terminal, PEST, BH1, BH2, BH3, and transmembrane (TM) domains (40, 41). To identify the binding region of Mcl-1 to Ku, a panel of Mcl-1 deletion mutants, including ΔN (aa 10–120), ΔPEST (aa 120–200), ΔBH1 (aa 256–265), ΔBH2 (aa 305–315), ΔBH3 (aa 213–221), and ΔTM (aa 329–346), were generated (Figure 5A). Purified recombinant glutathione S-transferase–fused (GST-fused) Mcl-1 WT or deletion mutants (Supplemental Figure 9) were incubated with purified Ku70/Ku80 complex. GST pull-down experiments revealed that WT, ΔN, ΔPEST, ΔBH2, and ΔTM, but not ΔBH1 or ΔBH3, Mcl-1 mutants directly interact with Ku70/Ku80 heterodimer (Figure 5B), indicating that the BH1 and BH3 domains comprise the Ku binding sites on Mcl-1 protein. To further test this, FLAG-tagged Mcl-1 WT and deletion mutants were exogenously expressed in Mcl1–/– MEFs, followed by co-IP using a FLAG antibody. Similarly, deletion of the BH1 or BH3 domain resulted in loss of Mcl-1’s ability to interact with Ku proteins in cells (Figure 5C).
Figure 5. Mcl-1 directly interacts with Ku via BH1 and BH3 domains, which are required for Mcl-1 dissociation of Ku/DNA complex.
(A) Schematic representation of various Mcl-1 deletion mutants. (B) GST beads coated with purified recombinant GST-tagged WT or individual Mcl-1 deletion mutants were incubated with recombinant Ku70/Ku80 heterodimer. Mcl-1–associated Ku70 or Ku80 and GST–Mcl-1 were analyzed by Western blot. (C) Mcl1–/– MEFs were transfected with FLAG-tagged Mcl-1 WT or individual Mcl-1 deletion mutants using Amaxa electroporation system. Co-IP experiments were performed using anti-FLAG M2 beads, followed by Western blot analysis of Ku70, Ku80, and FLAG–Mcl-1. (D) Mre11-Rad50 (MR) complex was expressed in Sf9 insect cells and purified using an anti-FLAG M2 affinity column. (E and F) The 5′-end-labeled overhang DNA was incubated with Ku or MR complex in the absence or presence of increasing concentrations of Mcl-1 (E) or individual Mcl-1 deletion mutant proteins (F). BSA was used as negative control. The Ku/DNA or MR/DNA complexes were analyzed by EMSA.
Since Mcl-1 selectively binds to Ku but not Mre11 (Supplemental Figure 7E), the effect of Mcl-1 on Ku-DNA binding or Mre11-DNA binding was compared by EMSA. Recombinant human Mre11-Rad50 (MR) complex was expressed and purified from baculovirus-infected Sf9 insect cells as previously described (42) (Figure 5D). As previously reported (43), Ku70/Ku80 heterodimer displayed binding to a 32P-labeled, 3′ overhang DNA substrate with high affinity, while the MR complex displayed low binding affinity (Figure 5E, lane 3 vs. lane 7). Addition of purified Mcl-1 suppressed Ku-DNA binding in a dose-dependent manner (Figure 5E, lane 3 vs. lanes 4–6) but had no significant effect on MR-DNA binding (Figure 5E, lane 7 vs. lanes 8–10). These results indicate that purified Mcl-1 protein can directly disrupt Ku-DNA binding but not MR-DNA binding. To further address whether the Mcl-1/Ku interaction is essential for Mcl-1 disruption of Ku-DNA binding, similar experiments using purified recombinant Mcl-1 WT or deletion mutants were carried out. Notably, WT, ΔN, ΔPEST, ΔBH2, and ΔTM, but not ΔBH1 and ΔBH3, Mcl-1 mutants suppressed Ku-DNA binding (Figure 5F), suggesting that the Ku binding site(s) on Mcl-1 (i.e., BH1 or BH3) is required for Mcl-1–mediated dissociation of the Ku/DNA complex in a cell-free system. The sequence of addition in the above experiments was DNA→Ku→Mcl-1. To test whether the effect of purified Mcl-1 on Ku binding to DNA depends on the order of addition, we also performed experiments with the following order of addition: DNA→Mcl-1→Ku, or DNA→Ku→Mcl-1. Similar results were observed (Supplemental Figure 10), indicating that the effect of purified Mcl-1 on Ku-DNA binding is independent of the order of addition. These findings suggest that Mcl-1 protein may not only block the interaction of Ku with DNA but may also have the capacity to dissociate Ku from the Ku/DNA complex.
To further test the effect of ΔBH1 and ΔBH3 Mcl-1 mutants on the recruitment of Ku to DSBs in cells, first, WT, ΔBH1, and ΔBH3 mutant Mcl-1 was transfected into Mcl-1–knockout DR-GFP H1299 (Mcl1–/– H1299 DR-GFP) cells (Supplemental Figure 11A), followed by transfection of I-SceI into cells to produce DSBs. ChIP experiments to measure the level of Ku recruitment to DSBs were performed using anti-Ku70 antibody. Results indicated that knockout of endogenous Mcl-1 resulted in increased Ku70 recruitment to DSBs (Supplemental Figure 11B, lane 2 vs. lane 4). Intriguingly, expression of endogenous or exogenous WT Mcl-1 but not the Ku binding–deficient Mcl-1 ΔBH1 and ΔBH3 mutants in Mcl-1–knockout H1299 DR-GFP cells reduced Ku recruitment to DSBs (Supplemental Figure 11B, lane 2 vs. lane 4, lane 6 vs. lane 8 or 10), indicating that the Ku binding–deficient ΔBH1 and ΔBH3 Mcl-1 mutants failed to prevent Ku recruitment to DSBs.
Mcl-1 molecules can be recruited to DSB sites following DNA DSBs.
To address how Mcl-1 has the capacity to inhibit Ku even when there are relatively low levels of Mcl-1 in the nucleus, we first assessed whether Mcl-1 can be recruited to DNA DSBs. I-SceI was transfected into U2OS DR-GFP cells to induce DSBs, followed by ChIP using Mcl-1 antibody and PCR to detect DR-GFP break DNA fragment. Results indicated that Mcl-1 was associated with I-SceI–induced DSBs (Supplemental Figure 12A). To further test whether Mcl-1 could be recruited to DSB sites following radiation, S/G2-phase H1299 cells harvested at 6 hours after double-thymidine block were exposed to IR (5 Gy), followed by costaining with Mcl-1 and γ-H2AX. Intriguingly, Mcl-1 molecules were mainly enriched on DSB sites to form foci and colocalized with γ-H2AX (i.e., a classic DSB marker) (Supplemental Figure 12B). The accumulation of Mcl-1 proteins on DSBs could yield a sufficiently high number of Mcl-1 molecules to inhibit Ku at DNA break sites.
BH1 and BH3 domains of Mcl-1 are required for its enhancement of HR and suppression of NHEJ.
To further test whether Mcl-1/Ku binding influences NHEJ and HR activities, WT and the panel of Mcl-1 deletion mutants were stably expressed in Mcl1–/– MEFs. Expression levels of the endogenous Mcl-1 in intact WT MEF cells and exogenously expressed Mcl-1 in Mcl1–/– MEF cells were simultaneously analyzed by Western blot using Mcl-1 antibody (Supplemental Figure 13A). Results revealed that knockout of endogenous Mcl-1 resulted in downregulation of HR activity and upregulation of NHEJ activity (Supplemental Figure 13, B and C, panel 1 vs. panel 2). Intriguingly, expression of exogenous WT, ΔN, ΔPEST, ΔBH2, and ΔTM, but not ΔBH1 and ΔBH3, Mcl-1 mutants restored HR and suppressed NHEJ activity (Supplemental Figure 13, B and C), but did not significantly affect the percentage of cells in S and G2 phases of the cell cycle or the proliferation rate (Supplemental Figure 13, D–F). These findings suggest that Mcl-1/Ku binding via the BH1 and BH3 domains is an essential component for Mcl-1 enhancement of HR activity via inhibition of NHEJ. Importantly, these data also provided information on the relative levels of endogenous Mcl-1 in intact WT MEF cells compared with exogenously expressed Mcl-1 in Mcl1–/– MEF cells, which could efficiently regulate HR or NHEJ in cells.
Mcl-1 reverses the inhibitory effect of Ku on Mre11 complex–induced DNA end resection.
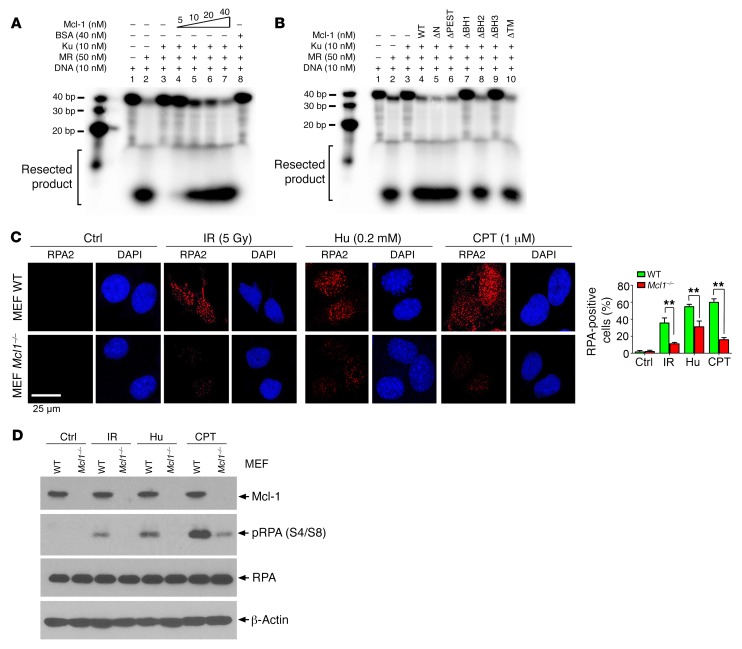
It is known that HR repair is initiated by DSB end resection that is mediated by the Mre11 complex (6), whereas Ku tightly bound to DNA ends blocks Mre11 DNA resection (43). Since our findings demonstrate that Mcl-1 negatively regulates Ku function via direct binding, Mcl-1 may reverse the Ku-mediated inhibitory effect on Mre11 complex–mediated DNA end resection. Mre11-Rad50 complex–mediated exonuclease activity was analyzed in the presence of increasing concentrations of Mcl-1 protein using 5′-32P-end-labeled fork DNA as a substrate (43). The Mre11 complex exhibited potent DNA resection activity (Figure 6A, lane 2 vs. lane 1). Ku heterodimer inhibited MR-mediated DNA resection (Figure 6A, lane 3 vs. lane 2), which was gradually reversed by the addition of increasing concentrations of purified Mcl-1 protein in the presence of a constant amount of Ku protein (Figure 6A, lane 3 vs. lanes 4–7). BSA was used as control and had no effect on DNA resection. However, deletion of BH1 or BH3 resulted in the failure of Mcl-1 to reverse the inhibitory effect of Ku on Mre11 complex–mediated DNA resection (Figure 6B, lanes 3 and 4 vs. lanes 7 and 9), indicating that Mcl-1/Ku binding is required for Mcl-1 to promote DNA resection. In addition, RPA2 is an established reporter of DNA end resection during HR repair in cells (1, 44, 45). To further test the effect of Mcl-1 on DNA resection in cells, RPA2 foci formation following treatment with Hu, camptothecin (CPT), or IR was compared in WT and Mcl1–/– MEFs. WT and Mcl1–/– MEF cells were treated with Hu (0.2 mM) for 24 hours or CPT (1 μM) for 1 hour, or exposed to IR (5 Gy), followed by immunostaining with anti-RPA2 antibody. Results indicated that IR and CPT as well as Hu induced RPA2 foci formation in WT MEF cells. Knockout of Mcl-1 significantly decreased IR-, CPT-, or Hu-induced RPA2 foci (Figure 6C). Furthermore, RPA2 phosphorylation at Ser4 and Ser8 has also been extensively used as a surrogate marker for DNA end resection (44–47). RPA2 phosphorylation at Ser4 and Ser8 was analyzed by Western blot using the S4/S8 dual-site phosphospecific RPA2 antibody following exposure of MEF WT or MEF Mcl1–/– cells to IR, CPT, and Hu. Results indicate that IR, CPT, and Hu stimulated RPA2 phosphorylation at S4 and S8, and knockout of Mcl-1 reduced RPA2 phosphorylation (Figure 6D), indicating that depletion of Mcl-1 suppresses DNA end resection. Thus, Mcl-1 may also promote DNA resection in cells.
Figure 6. Mcl-1 promotes DNA resection in cell-free system and in cells.
(A and B) 5′-End-labeled forked DNA substrate was incubated with MR complex in the absence or presence of Ku and/or increasing concentrations of WT Mcl-1 protein (A) or individual Mcl-1 deletion mutant proteins (B). Resected DNA product was run on 16% urea-PAGE gel and analyzed by phosphoimager. (C) MEF WT and MEF Mcl1–/– cells were treated with Hu (0.2 mM) for 24 hours or CPT (1 μM) for 1 hour or exposed to IR (5 Gy), followed by immunostaining with anti-RPA2 antibody. Data represent the mean ± SD, n = 3 per group. **P < 0.01, by 2-tailed t test. Scale bar: 25 μm. (D) RPA2 phosphorylation at Ser4 and Ser8 was analyzed by Western blot using the S4/S8 dual-site phosphospecific RPA2 antibody following exposure of MEF WT or MEF Mcl1–/– cells to IR (5 Gy), Hu (0.2 mM) for 24 hours, or CPT (1 μM) for 1 hour.
Expression of Mcl-1 accelerates repair of DNA replication stress–induced DSBs leading to prolonged clonogenic survival, requiring BH1 and BH3 domains.
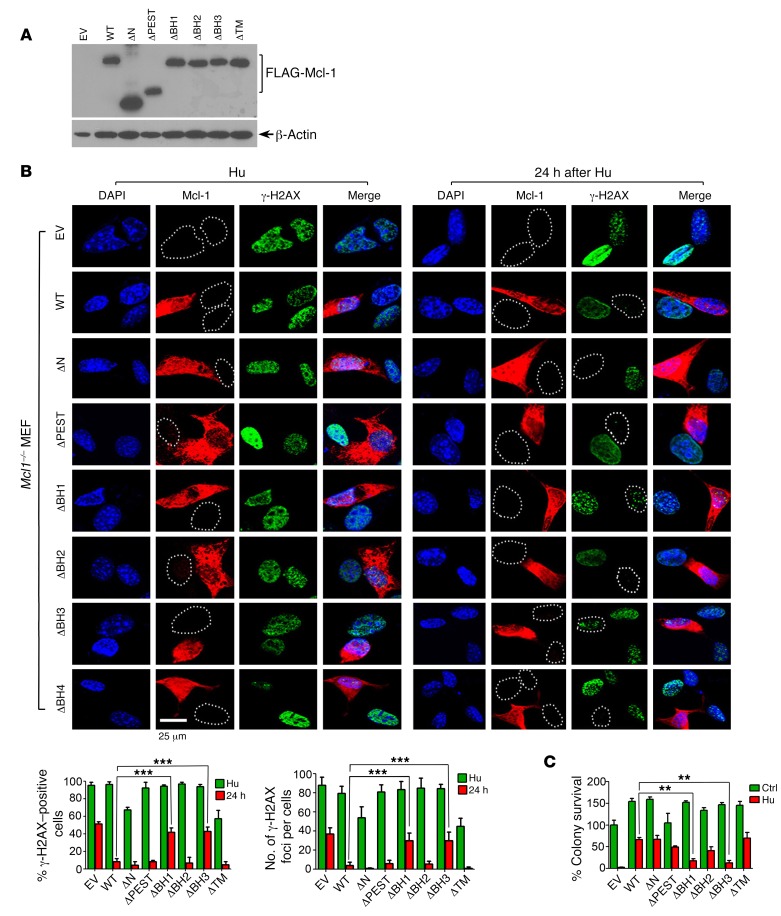
Our findings reveal that knockout of Mcl-1 resulted in impairment of HR-mediated DSB repair (Figures 2–4). To test whether expression of exogenous Mcl-1 restores HR-mediated DSB repair capacity, FLAG-tagged WT and a panel of Mcl-1 deletion mutants were expressed in Mcl1–/– MEFs (Figure 7A). As expected, Hu-induced DNA replication stress led to formation of γ-H2AX DSB foci in MEFs expressing Mcl-1 WT or various deletion mutants (Figure 7B, left panel). After removal of Hu from the medium for 24 hours, most DSB foci disappeared in MEFs expressing WT, ΔN, ΔPEST, ΔBH2, and ΔTM Mcl-1 deletion mutants (Figure 7B, right panel), indicating that Hu-induced DSBs were repaired within 24 hours. However, a significant number of DSB foci persisted in cells expressing ΔBH1, ΔBH3, or vector-only control (Figure 7B, right panel). Since the repair of DNA replication stress–induced DSBs mainly occurs through the HR pathway (6), these results indicate that Mcl-1 can restore HR activity to repair Hu-induced DSBs, which requires its BH1 and BH3 domains. Furthermore, expression of exogenous WT, ΔN, ΔPEST, ΔBH2, or ΔTM Mcl-1 deletion mutants restored clonogenic survival to different extents following Hu treatment, whereas deletion of the BH1 or BH3 domain resulted in failure of Mcl-1 to restore clonogenic survival (Figure 7C and Supplemental Figure 14).
Figure 7. BH1 and BH3 domains are required for Mcl-1 promotion of HR-dependent DSB repair and clonogenic survival.
(A) FLAG-tagged WT or individual Mcl-1 deletion mutants were transfected into Mcl1–/– MEFs. Mcl-1 expression was analyzed by Western blot using FLAG antibody. (B) Mcl1–/– MEFs expressing exogenous WT or individual Mcl-1 deletion mutants were treated with 0.2 mM Hu for 24 hours. After washing, cells were cultured in normal medium for another 24 hours. DSBs were analyzed by immunofluorescence using γ-H2AX antibody. Scale bar: 25 μm. The percentage of γ-H2AX–positive cells (left panel) and the number of γ-H2AX foci per cell (right panel) were determined by counting of at least 100 cells from each sample. Data represent the mean ± SD, n = 3 per group. ***P < 0.001, by 2-tailed t test. (C) Mcl1–/– MEFs expressing exogenous WT or individual Mcl-1 deletion mutants were treated with 0.2 mM Hu for 24 hours. After washing, cells were cultured in normal medium, followed by colony formation analysis. EV, empty vector. Data represent the mean ± SD, n = 3 per group. **P < 0.01, by 2-tailed t test.
Nuclear Mcl-1 promotes DSB repair and prolongs clonogenic survival following DNA replication stress.
Our findings reveal that, in addition to mitochondria, Mcl-1 levels are also elevated in nuclei of S- and G2-phase cells (Figure 1). However, the exact role of nuclear Mcl-1 remains unknown. A nuclear-targeted Mcl-1 construct (Nuc–Mcl-1) was created and transfected into H1299 Mcl-1–knockout cells (Supplemental Figure 15A). Expression of Nuc–Mcl-1 was analyzed by immunostaining. As expected, Nuc–Mcl-1 was exclusively expressed in nuclei (Supplemental Figure 15B, positive cell 1 vs. negative cells 2 and 3). To test the role of Nuc–Mcl-1 in the repair of DNA replication stress–induced DSBs, H1299 Mcl-1–knockout cells expressing exogenous Nuc-WT, Nuc-ΔBH1, or Nuc-ΔBH3 mutant Mcl-1 were treated with Hu (0.2 mM) for 24 hours. Following removal of Hu, cells were released into normal culture medium for an additional 6 hours. DSBs were analyzed by immunostaining with γ-H2AX. As expected, Hu induced significant DSBs in H1299 Mcl-1–knockout cells expressing Nuc-WT, Nuc-ΔBH1, or Nuc-ΔBH3 mutant Mcl-1 or empty vector control (Supplemental Figure 15C, left panel). Following depletion of Hu, a number of γ-H2AX foci were still observed in H1299 Mcl-1–knockout cells transfected with the empty vector control (Supplemental Figure 15C, right panel, cell 4, 5, or 6). Intriguingly, γ-H2AX foci were almost undetectable in cells that expressed Nuc-WT Mcl-1 (Supplemental Figure 15C, right panel, Nuc-WT Mcl-1–positive cells 8 and 9 vs. –negative cells 7 and 10). These results indicate that nuclear WT Mcl-1 significantly promotes the repair of DNA replication stress–induced DSBs. However, a significant number of DSB foci persisted in cells expressing Nuc-ΔBH1 or Nuc-ΔBH3 6 hours after Hu depletion (Supplemental Figure 15, C–E), suggesting that the BH1 and BH3 domains are essential for Nuc–Mcl-1 to promote DSB repair. To further test the effect of Nuc–Mcl-1 on cell survival, clonogenic survival experiments were carried out following treatment of cells with Hu or staurosporine. Expression of Nuc-WT but not Nuc-ΔBH1 or Nuc-ΔBH3 mutant Mcl-1 prolonged clonogenic cell survival after Hu treatment but had no effect following staurosporine exposure (Supplemental Figure 15, F and G), indicating that Nuc–Mcl-1 enhances clonogenic survival through promoting repair of Hu-induced DSBs but has no antiapoptotic function in staurosporine-induced mitochondria-dependent apoptosis.
Small molecule MI-223 targets the BH1 binding pocket of Mcl-1, reduces HR efficiency, and inhibits HR-mediated DNA repair, leading to synergism with DNA replication stress agents against lung cancer in vitro and in vivo.
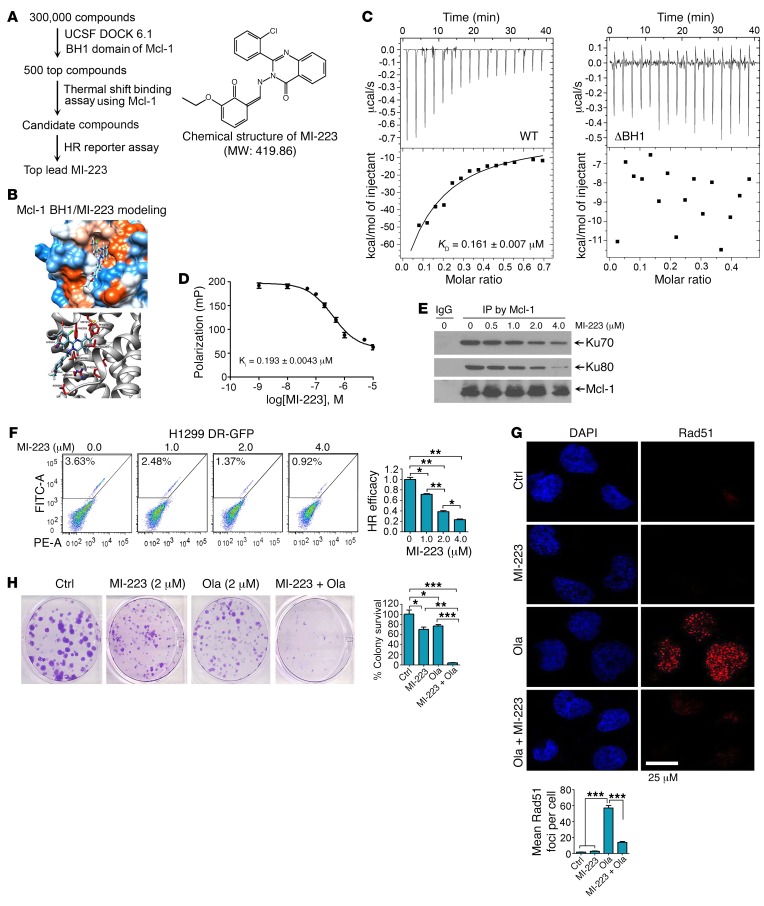
Our findings reveal that the BH1 and BH3 domains of Mcl-1 are required to enhance HR and prolong clonogenic survival following DNA replication stress (Figure 7 and Supplemental Figures 14 and 15), indicating that BH1 or BH3 is an attractive target for screening small molecules to interfere with HR DNA repair and potentially sensitize cancer cells to DNA replication stress. A National Cancer Institute (NCI) database library of 300,000 small molecules was docked into the Mcl-1 BH1 structure pocket (aa 256–265) identified by the UCSF DOCK 6.1 program suite for a first round of screening as we previously described (ref. 48 and Figure 8A). The small molecules were ranked according to their energy scores. The top 500 small molecules were then selected for a second round of screening by thermal shift binding assay using Mcl-1 protein (49) and HR reporter assay (Figure 8A). One lead compound (NSC320223, C23H18ClN3O3, MW: 419.86032) was identified as shown in Figure 8, A and B, and was termed Mcl-1 inhibitor-223 (MI-223). To further confirm the binding of MI-223 with Mcl-1, we used isothermal titration calorimetry (ITC) to measure MI-223/Mcl-1 binding. ITC is a direct, label- and immobilization-free technique that measures the binding affinity between proteins and small-molecule ligands that interact with each other, and can analyze binding constant (KD) values in the millimolar and nanomolar range (50, 51). We performed ITC experiments to assess MI-223/Mcl-1 binding using an auto-iTC200 instrument as previously described (52). Results revealed that MI-223 directly bound WT Mcl-1 protein with good binding affinity (KD value: 0.161 ± 0.007 μM) (Figure 8C, left panel). In contrast, MI-223 failed to bind to the BH1 deletion Mcl-1 mutant protein (ΔBH1) in ITC assay (Figure 8C, right panel), suggesting that the BH1 domain is essential for Mcl-1 to interact with MI-223. In addition to the binding constant (KD) value measured by ITC, we also used fluorescence polarization (FP) to measure the inhibitory constant (KI) value using a fluorescence-labeled PUMA BH3 peptide. We chose the PUMA BH3 peptide for this FP assay to evaluate Mcl-1/MI-223 binding because it has been reported to specifically bind to the BH1 domain of Mcl-1 (53). Results indicated that the KI value of MI-223/Mcl-1 binding in the FP assay was 0.193 ± 0.0043 μM (Figure 8D). Based on findings from both ITC (i.e., KD) and FP (i.e., KI), we conclude that MI-223 may directly bind to Mcl-1 with good affinity.
Figure 8. Discovery of small molecule MI-223 as a lead compound that specifically binds to Mcl-1, inhibits HR DNA repair, and sensitizes cancer cells to DNA replication agents.
(A) Schematic illustration of screening strategies used to identify the lead compound Mcl-1 inhibitor MI-223 and its chemical structure. (B) Structural modeling of MI-223 in the BH1 domain binding pocket of Mcl-1 protein. (C) The binding affinity of MI-223 with WT Mcl-1 or ΔBH1 Mcl-1 deletion mutant protein was examined by isothermal titration calorimetry assay. The binding constant (KD) value was determined by fitting of the titration curve to a 1-site binding mode. Data represent the mean ± SD, n = 3 per group. (D) Fluorescence polarization assay was performed to measure the inhibitory constant (KI) value using purified Mcl-1 protein, MI-223, and fluorescence-labeled PUMA BH3 peptide. Data represent the mean ± SD, n = 3 per group. (E) H1299 cells were treated with increasing concentrations of MI-223 for 24 hours, followed by co-IP using Mcl-1 antibody. (F) HR repair efficiency was measured in H1299 DR-GFP cells in the absence or presence of increasing concentrations of MI-223. Data represent the mean SD, n = 3 per group. *P < 0.05, **P < 0.01, by 2-tailed t test. (G) H1299 cells were treated with olaparib (Ola, 20 μM), MI-223 (4 μM), or the combination for 24 hours, followed by immunostaining with Rad51 antibody. Rad51 foci were quantified by counting of at least 100 cells from each sample. Data represent the mean ± SD, n = 3 per group. ***P < 0.001, by 2-tailed t test. Scale bar: 25 μm. (H) H1299 cells were treated with Ola (2 μM), MI-223 (2 μM), or the combination, followed by colony formation assay. Data represent the mean ± SD, n = 3 per group. *P < 0.05, **P < 0.01, ***P < 0.001, by 2-tailed t test.
MI-223 not only disrupted the Mcl-1/Ku complex (Figure 8E) but also potently inhibited HR activity in both H1299 and U2OS DR-GFP cells in a dose-dependent manner (Figure 8F and Supplemental Figure 16A). To further assess whether Mcl-1 is essential for the effect of MI-223 on HR activity, Mcl-1 was knocked out from H1299 cells using CRISPR/Cas9 to generate Mcl-1–deficient cells. WT Mcl-1, Ku binding–deficient mutant ΔBH1, and empty vector control were exogenously expressed in Mcl-1–knockout H1299 (Mcl1–/– H1299) cells (Supplemental Figure 16B), followed by treatment with MI-223 for 24 hours and analysis of HR efficacy. Results indicated that MI-223 induced a dose-dependent reduction of HR efficacy in H1299 parental cells, and knockout of endogenous Mcl-1 from H1299 cells resulted in a significant decrease in HR efficacy. MI-223 had no significant further effect on HR efficacy in Mcl-1–deficient H1299 cells. Intriguingly, expression of exogenous WT Mcl-1 but not the Ku binding–deficient ΔBH1 Mcl-1 mutant in Mcl-1–deficient H1299 cells restored the inhibitory effect of MI-223 on HR activity (Supplemental Figure 16, C and D). These findings suggest that Mcl-1 expression is essential for the inhibitory effect of MI-223 on HR activity, which requires its BH1 domain.
To test whether inhibition of HR by MI-223 sensitizes cancer cells to DNA replication stress, H1299 cells were treated with olaparib or Hu in the absence or presence of MI-223, followed by analysis of Rad51 foci and clonogenic survival. Olaparib or Hu induced Rad51 foci and MI-223 strongly inhibited Rad51 foci formation (Figure 8G and Supplemental Figure 16E), suggesting that MI-223 suppresses HR-dependent DNA repair. Importantly, MI-223 suppression of HR DNA repair activity synergizes with olaparib or Hu to kill H1299 lung cancer cells (Figure 8H and Supplemental Figure 16F). Annexin V binding assays revealed that MI-223 did not induce significant apoptotic cell death (Supplemental Figure 16G), indicating that MI-223–induced cell killing occurs mainly through suppression of Mcl-1–enhanced HR DNA repair activity, which occurs independently of apoptosis.
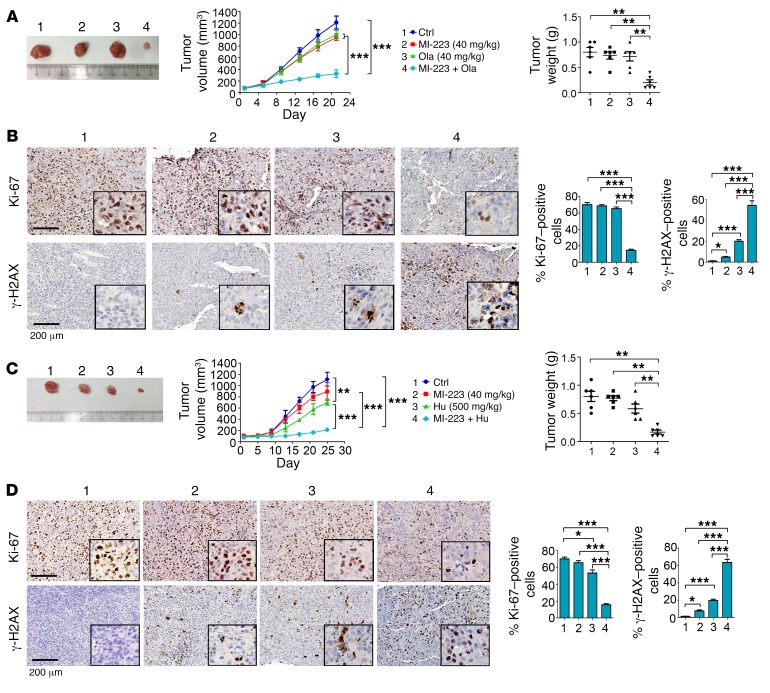
To further test whether MI-223 and DNA replication stress agents synergistically inhibit cancer growth in vivo, nude mice with non–small cell lung carcinoma (i.e., H1299) xenografts were treated with MI-223 (40 mg/kg/d), olaparib (40 mg/kg/d), Hu (500 mg/kg), and the combination of MI-223 with olaparib or Hu for 3 weeks. Importantly, the combination of MI-223 with olaparib or Hu exhibited significantly greater efficacy than a single agent alone in suppressing lung tumor growth in vivo (Figure 9, A and C), leading to sustained tumor repression. Compared with olaparib or Hu alone, there was significant reduction of Ki-67 in association with increased levels of γ-H2AX in tumor tissues from animals treated with the combination (Figure 9, B and D), indicating that MI-223 blocks the HR-dependent repair of olaparib- or Hu-induced DNA damage, leading to increased cell killing in tumor tissue. There was no significant weight loss, increase in alanine transaminase, aspartate transaminase, or blood urea nitrogen, nor reduction in wbcs, rbcs, hemoglobin, or platelets, in mice treated with MI-223 alone or in combination with olaparib or Hu. Histopathologic evaluation of harvested normal tissues (brain, heart, lung, liver, spleen, kidney, and intestine) revealed no evidence of normal tissue toxicity (Supplemental Figure 17).
Figure 9. MI-223 synergizes with DNA replication stress agents against lung cancer in vivo.
(A) nu/Nu nude mice with H1299 lung cancer xenografts were treated with MI-223 (40 mg/kg), olaparib (Ola, 40 mg/kg), or the combination for 3 weeks. Tumor volume was measured once every 4 days. After 21 days, mice were sacrificed and tumors were removed and analyzed. Data represent the mean ± SD, n = 6 per group. **P < 0.01, ***P < 0.001, by 2-tailed t test. (B) Ki-67 and γ-H2AX in tumor tissues were analyzed by IHC staining at the end of experiments and quantified. Scale bars: 200 μm. Data represent the mean ± SD, n = 6 per group. *P < 0.05, ***P < 0.001, by 2-tailed t test. (C and D) H1299 lung cancer xenografts were treated with MI-223 (40 mg/kg), Hu (500 mg/kg), or the combination for 25 days. Tumor volume and weight, Ki-67, and γ-H2AX in tumor tissues were analyzed as above. Scale bars: 200 μm. Data represent the mean ± SD, n = 6 per group. *P < 0.05, **P < 0.01, ***P < 0.001, by 2-tailed t test.
Discussion
Competition between NHEJ and HR, which stems from the molecular interplay between Ku and the Mre11 complex at DSB ends, decides the choice of DSB repair (1). The first control point for this pathway choice is the process of DNA resection. Multiple proteins or protein complexes influence the HR/NHEJ pathway choice by regulating DNA end resection directly or indirectly, including the MRN complex, CtIP, EXO1, BLM, DNA2, BRAC1, 53BP1, RIF1, CDKs, RNF138, chromatin remodeling factors, and others (14, 54–57). Initiation of DNA end resection by the MRN complex and CtIP removes Ku from DNA ends to generate ssDNA overhangs that not only inhibit NHEJ but also provide a platform to recruit proteins involved in HR repair (47). DNA end resection is a 2-step process in which the MRN complex and CtIP are involved in the initial step, and EXO1 and DNA2-BLM are involved in the second step (54, 58). CDKs promote these 2 steps in the process of DNA resection by phosphorylating CtIP, NBS1, or EXO1, respectively (14, 55). Ku ubiquitination mediated by the E3 ligase RNF138 and subsequent degradation promote DNA resection (57). Intriguingly, BRAC1 promotes, whereas RIF1 acts as the effector of 53BP1 to inhibit, DNA resection and HR (55, 56).
Several issues remain, in particular how cell cycle phase influences pathway choice and repair outcome. A cell cycle activation step is required to initiate the process of DNA end resection, but the mechanism remains unclear (59). Mcl-1 has been reported to play an important role in DNA repair/DNA damage response (21, 23, 24). However, the exact mechanism is not fully understood. Here we have demonstrated that Mcl-1, in addition to its well-known antiapoptotic function, plays an unexpected role in DSB repair pathway choice during cell cycle progression.
NHEJ, which does not require DNA sequence homology, is active throughout the cell cycle in vertebrate cells (60, 61). In contrast, HR occurs largely during the S/G2 phase of the cell cycle, when a replicated sister chromatid is present and can be used as a homologous template to copy and restore the missing DNA sequence on the damaged chromatid (1). This indicates that cell cycle transition from G1 to S/G2 is a critical check point for switching the predominant DSB repair pathway from NHEJ to HR. We have demonstrated that Mcl-1 is a positive regulator of HR DNA repair, and that accumulation of Mcl-1 via reduction of its ubiquitination in S/G2 may tip the balance toward HR, consistent with HR being restricted to S and G2 phases of the cell cycle (47). For example, Mcl-1 depletion by knockout or knockdown can change the balance of repair toward an increase in NHEJ. Conversely, Mcl-1 overexpression shifts the balance toward HR, suggesting that Mcl-1 is able to direct DSB repair pathway choice by altering the NHEJ/HR ratio during cell cycle progression.
Because DNA end resection is considered a major control point between NHEJ and HR choice (47), DSB end resection must be appropriately restricted to S/G2, as HR requires the presence of an intact sister chromatid to promote repair (6). Cells favor DSB repair by NHEJ if the DNA ends are suitable for joining, while DNA resection is activated if joining fails, particularly when DNA ends are not suitable for NHEJ (1). Since Mcl-1 molecules were recruited to DSB sites following the induction of DSBs by IR or I-SceI, this could provide potential for Mcl-1 to be involved in the process of DNA resection. Intriguingly, Mcl-1 facilitates Mre11 complex–mediated DNA resection in a mechanism involving inhibition of Ku/DSB binding via direct interaction with Ku proteins, suggesting that Mcl-1 binding to Ku may release Ku from DSBs to initiate Mre11 complex–mediated DNA resection leading to promotion of HR. The positive effect of Mcl-1 on DNA resection and HR accelerates the repair of DNA replication stress–induced DSBs. Selective promotion of HR-dependent DSB repair by Mcl-1 might play an important role in prolonging cell survival following DNA replication stress.
Domain-mapping studies reveal that Mcl-1 directly interacts with the Ku70/Ku80 dimer via its BH1 and BH3 domains. Importantly, these 2 Ku binding sites on Mcl-1 are essential not only for its inhibitory effect on NHEJ but also for its positive effects on DNA end resection and HR-mediated DSB repair, which reveals the mechanism of action of Mcl-1 in directing DSB repair pathway choice and increasing clonogenic survival following treatment with DNA replication stress agents.
Inhibition of apoptosis by Mcl-1 occurs through its heterodimerization with multiple proapoptotic Bcl-2 family proteins (i.e., Bim, Bak, or Bax) within the mitochondrial membranes (62). In addition to this well-characterized antiapoptotic mechanism, we have discovered that Mcl-1 not only regulates the choice between HR and NHEJ, but also supports clonogenic cell survival through promotion of HR-dependent DSB repair following Hu- or olaparib-induced DNA replication stress. This activity could also contribute to therapeutic resistance in human cancers.
In addition to its mitochondrial localization, Mcl-1 has also been shown to be localized in the nucleus (21, 63) with levels peaking in S/G2. The exact role of nuclear Mcl-1 remains unclear. We have shown that selective expression of Mcl-1 in the nucleus facilitates HR-dependent DSB repair leading to increased clonogenic survival following DNA replication stress–induced DSBs. However, nuclear Mcl-1 does not have an antiapoptotic function, as evidenced by the inability of the nuclear-targeted Mcl-1 to support survival following treatment with the apoptotic agent staurosporine. We propose that nuclear Mcl-1 promotes HR-dependent DSB repair while mitochondrial Mcl-1 may inhibit apoptosis.
Using the NCI small-molecule library and the UCSF DOCK 6.1 screen program, we identified MI-223 as a lead compound that directly binds to Mcl-1 protein via its BH1 domain and disrupts the interaction between Mcl-1 and Ku. MI-223 induces robust cancer cell killing via inhibition of HR-mediated DNA repair. Importantly, MI-223–mediated reduction of HR activity renders cancer cells highly sensitive to DNA replication stress agents (Hu or olaparib). This observation helps to explain why the combination of MI-223 with Hu or olaparib displayed strong synergism against lung cancer in mouse xenografts.
In conclusion, our findings have demonstrated that Mcl-1 is directly involved in the regulation of NHEJ- and HR-mediated DNA repair pathway choice. Physiologic accumulation of Mcl-1 in S/G2 phase renders a net increase of HR over NHEJ. Direct interaction of Mcl-1 with Ku via its BH1 and BH3 domains is required for Mcl-1 inhibition of Ku-DNA binding, promotion of Mre11 complex–mediated DNA resection, and enhancement of HR activity, resulting in HR-dependent DSB repair and increased cell survival. Therefore, Mcl-1, in addition to its antiapoptotic function, appears to be a driver of the mechanism utilized for choice of DSB repair pathway. Specifically targeting this novel function of Mcl-1 by using small molecules such as MI-223 represents a potentially new, effective strategy for cancer therapy.
Methods
Supplemental Methods are available online with this article (supplemental material available online with this article; https://doi.org/10.1172/JCI92742DS1).
Cell lines, plasmids, and transfections.
WT and Mcl-1–knockout (Mcl1–/–) mouse embryonic fibroblasts (MEFs) were obtained from Joseph Opferman (St. Jude Children’s Research Hospital, Memphis, Tennessee, USA) and maintained in DMEM with 10% FBS. H1299 cells were obtained from the American Type Culture Collection and grown in RPMI 1640 medium supplemented with 10% FBS. U2OS DR-GFP cells carrying a chromosomally integrated single copy of HR reporter were obtained from Maria Jasin (Memorial Sloan-Kettering Cancer Center, New York, New York, USA) (37). Sf9 cells were cultured in Sf-900 III SFM medium (Invitrogen). FLAG-tagged full-length Mcl-1 in pCMV-Tag2A, pDR-GFP, and pCBASce-I was purchased from Addgene. NHEJ substrate pGFP-Pem1-Ad2 and pDsRed2-N1 constructs were provided by Vera Gorbunova (University of Rochester, Rochester, New York, USA). Baculoviruses harboring FLAG-hMre11 and His6-hRad50 were provided by Tanya Paull (University of Texas at Austin, Austin, Texas, USA). The pShooter-pCMV/Myc/Nuc vector was purchased from Invitrogen. Transfection of DNA plasmid into MEF cells was performed using Amaxa electroporation system with program A23 (Lonza). H1299 cells were transfected using NanoJuice (EMD Millipore) according to the manufacturer’s instructions.
Cell synchronization and fractionation.
Cells were synchronized at G1/S boundary by double-thymidine block as previously described (20). Growing cells were incubated with thymidine (2 mM) for 16 hours. After washing, cells were cultured in normal medium for 9 hours, followed by addition of thymidine for another 16 hours. After the double-thymidine block, cells were released to fresh media and analyzed at various time points.
HR and NHEJ assays.
HR activity was measured as previously described (36). HR reporter pDR-GFP plasmid was stably transfected into cells. Cells with single copy number were identified by quantitative PCR as previously described (64), then transfected with I-SceI expression plasmid (pCBASce-I), followed by flow cytometry analysis for GFP recovery.
NHEJ assay in cells was analyzed as previously reported (39). First, NHEJ substrate GFP-Pem1-Ad2 plasmids were linearized by restriction enzyme HindIII (Thermo Fisher Scientific). After purification, the linearized GFP-Pem1-Ad2 and internal control pDsRed2-N1 plasmids were cotransfected into cells. After 72 hours, cells were harvested for analysis of GFP (green) and DsRed (red) expression by flow cytometry. NHEJ activity was calculated by the ratio between GFP-positive and DsRed-positive cells.
Measurement of DNA resection.
To study the DNA end resection of Mre11-Rad50, its DNA exonuclease activity was examined using 5′-32P-labeled fork DNA (F-DNA, long strand: 5′-CGCGCCCAGCTTTCCCAGCTAATAAACTAAAAACTCCTAAGG-3′; short strand: 5′-CCTTAGGAGTTTTTAGTTTATTGGGCGCG-3′) as described previously (43). Ten nanomolar F-DNA was incubated with 50 nM Mre11-Rad50 in digestion buffer (25 mM MOPS [pH 7.0], 50 mM KCl, 1 mM DTT, and 1 mM MnCl2) at 37°C for 1.5 hours. Ten nanomolar Ku and various concentrations of Mcl-1 were added to study the effect of Ku and Mcl-1 on Mre11-Rad50–mediated DNA end resection. After the reaction, 10 μl of formamide dye (95% formamide, 0.025% bromophenol blue, 0.025% xylene cyanol FF, and 5 mM EDTA) was added and separated by 16% urea-PAGE gel and analyzed by Typhoon 9210 phosphoimager (GE Healthcare). To measure DNA resection in cells, RPA2 foci were analyzed by immunofluorescence as previously described (14).
Knockout of Mcl-1 by CRISPR/Cas9.
H1299 cells were transfected with U6-Mcl-1gRNA-Cas9-2A-GFP plasmid (Sigma-Aldrich) using NanoJuice. The sequence of Mcl-1–targeting guide RNA (gRNA) was 5′-GATTACCGCGTTTCTTTTGAGG-3′. After transfection, GFP-positive cells were sorted by flow cytometry and plated at a density of 1 cell per well in a 96-well plate. Mcl-1 expression of cells from a single clone was confirmed by Western blot.
Treatment of lung cancer xenografts.
Lung cancer xenografts were generated as previously described (48). Six-week-old male nude mice were purchased from Harlan and housed under pathogen-free conditions. H1299 cells (1 × 107) were implanted s.c. into mouse flanks. Tumor-bearing mice were randomly grouped, and tumors were allowed to grow to an average volume of 100 mm3 before treatment. Mice were treated with olaparib, MI-223, Hu, or the combination through i.p. injection at the indicated dose. During treatment, tumor volumes were measured by caliper once every 4 days and calculated with the formula V = (L × W2)/2 (L, length; W, width) as previously described (48).
Statistics.
The statistical significance of differences between groups was analyzed by 2-tailed unpaired Student’s t test. P less than 0.05 was considered statistically significant. All data are presented as the mean ± SD.
Study approval.
All mouse experiments were conducted with approval from the Institutional Animal Care and Use Committee of Emory University (Atlanta, Georgia, USA).
Author contributions
XD conceived the project. GC, DP, KX, SWS, and GLS performed experiments. ATM performed screening of small molecules. DSY provided technical support for HR assay. TKO, SSR, WJC, and PWD analyzed data. XD and GC wrote the manuscript. TKO, SSR, WJC, and PWD edited the manuscript.
Supplementary Material
Acknowledgments
This work was supported by NIH/NCI grants R01CA193828, R01CA136534, and R01CA200905 (to XD) and R01CA178999 (to DSY); by the Winship Research Pathology and Integrated Cellular Imaging shared resource and the Emory Comprehensive Glycomics Core supported by the Winship Cancer Institute of Emory University (P30CAJ 38292); by the Winship Fashion a Cure Research Scholar Award (to XD), a philanthropic award provided by the Winship Cancer Institute of Emory University; and by the Winship Endowment Fund (to XD). We are grateful to Dr. Vera Gorbunova (University of Rochester) for kindly providing pGFP-Pem1-Ad2 and pDsRed2-N1, Dr. Tanya T. Paul (University of Texas at Austin) for kindly providing Flag-Mre11and His6-Rad50 constructs, Dr. Joseph Opferman (St. Jude Children’s Research Hospital) for kindly providing Mcl-1-/- MEFs, and Dr. Maria Jasin (Memorial Sloan-Kettering Cancer Center) for kindly providing U2OS DR-GFP cells. We thank Maohua Xie for technical support for protein purification and Ku-DNA binding. We thank Anthea Hammond for editing of the manuscript.
Version 1. 12/11/2017
Electronic publication
Version 2. 01/02/2018
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2018;128(1):500–516.https://doi.org/10.1172/JCI92742.
Contributor Information
Guo Chen, Email: guo.chen@emory.edu.
Ke Xu, Email: cola519@163.com.
Dongkyoo Park, Email: dpark33@emory.edu.
Xingming Deng, Email: xdeng4@emory.edu.
References
- 1.Aparicio T, Baer R, Gautier J. DNA double-strand break repair pathway choice and cancer. DNA Repair (Amst) 2014;19:169–175. doi: 10.1016/j.dnarep.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet. 2001;27(3):247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- 3.Fugger K, et al. FBH1 co-operates with MUS81 in inducing DNA double-strand breaks and cell death following replication stress. Nat Commun. 2013;4:1423. doi: 10.1038/ncomms2395. [DOI] [PubMed] [Google Scholar]
- 4.Santos-Pereira JM, Herrero AB, García-Rubio ML, Marín A, Moreno S, Aguilera A. The Npl3 hnRNP prevents R-loop-mediated transcription-replication conflicts and genome instability. Genes Dev. 2013;27(22):2445–2458. doi: 10.1101/gad.229880.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y, Park E, Kim CS, Paik JH. ZNF365 promotes stalled replication forks recovery to maintain genome stability. Cell Cycle. 2013;12(17):2817–2828. doi: 10.4161/cc.25882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chapman JR, Taylor MR, Boulton SJ. Playing the end game: DNA double-strand break repair pathway choice. Mol Cell. 2012;47(4):497–510. doi: 10.1016/j.molcel.2012.07.029. [DOI] [PubMed] [Google Scholar]
- 7.Dynan WS, Yoo S. Interaction of Ku protein and DNA-dependent protein kinase catalytic subunit with nucleic acids. Nucleic Acids Res. 1998;26(7):1551–1559. doi: 10.1093/nar/26.7.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li X, Heyer WD. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 2008;18(1):99–113. doi: 10.1038/cr.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peng G, et al. Genome-wide transcriptome profiling of homologous recombination DNA repair. Nat Commun. 2014;5:3361. doi: 10.1038/ncomms4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kakarougkas A, Jeggo PA. DNA DSB repair pathway choice: an orchestrated handover mechanism. Br J Radiol. 2014;87(1035):20130685. doi: 10.1259/bjr.20130685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annu Rev Genet. 2011;45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- 12.Yun MH, Hiom K. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature. 2009;459(7245):460–463. doi: 10.1038/nature07955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shibata A, et al. DNA double-strand break repair pathway choice is directed by distinct MRE11 nuclease activities. Mol Cell. 2014;53(1):7–18. doi: 10.1016/j.molcel.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tomimatsu N, et al. Phosphorylation of EXO1 by CDKs 1 and 2 regulates DNA end resection and repair pathway choice. Nat Commun. 2014;5:3561. doi: 10.1038/ncomms4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Opferman JT, et al. Obligate role of anti-apoptotic MCL-1 in the survival of hematopoietic stem cells. Science. 2005;307(5712):1101–1104. doi: 10.1126/science.1106114. [DOI] [PubMed] [Google Scholar]
- 17.Wang B, et al. Role of Ku70 in deubiquitination of Mcl-1 and suppression of apoptosis. Cell Death Differ. 2014;21(7):1160–1169. doi: 10.1038/cdd.2014.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mojsa B, Lassot I, Desagher S. Mcl-1 ubiquitination: unique regulation of an essential survival protein. Cells. 2014;3(2):418–437. doi: 10.3390/cells3020418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perciavalle RM, Opferman JT. Delving deeper: MCL-1’s contributions to normal and cancer biology. Trends Cell Biol. 2013;23(1):22–29. doi: 10.1016/j.tcb.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harley ME, Allan LA, Sanderson HS, Clarke PR. Phosphorylation of Mcl-1 by CDK1-cyclin B1 initiates its Cdc20-dependent destruction during mitotic arrest. EMBO J. 2010;29(14):2407–2420. doi: 10.1038/emboj.2010.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jamil S, Stoica C, Hackett TL, Duronio V. MCL-1 localizes to sites of DNA damage and regulates DNA damage response. Cell Cycle. 2010;9(14):2843–2855. doi: 10.4161/cc.9.14.12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pawlikowska P, et al. ATM-dependent expression of IEX-1 controls nuclear accumulation of Mcl-1 and the DNA damage response. Cell Death Differ. 2010;17(11):1739–1750. doi: 10.1038/cdd.2010.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jamil S, Mojtabavi S, Hojabrpour P, Cheah S, Duronio V. An essential role for MCL-1 in ATR-mediated CHK1 phosphorylation. Mol Biol Cell. 2008;19(8):3212–3220. doi: 10.1091/mbc.E07-11-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mattoo AR, et al. MCL-1 depletion impairs DNA double-strand break repair and reinitiation of stalled DNA replication forks. Mol Cell Biol. 2017;37(3):e00535-16. doi: 10.1128/MCB.00535-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beucher A, et al. ATM and Artemis promote homologous recombination of radiation-induced DNA double-strand breaks in G2. EMBO J. 2009;28(21):3413–3427. doi: 10.1038/emboj.2009.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quennet V, Beucher A, Barton O, Takeda S, Löbrich M. CtIP and MRN promote non-homologous end-joining of etoposide-induced DNA double-strand breaks in G1. Nucleic Acids Res. 2011;39(6):2144–2152. doi: 10.1093/nar/gkq1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yarbro JW. Mechanism of action of hydroxyurea. Semin Oncol. 1992;19(3 suppl 9):1–10. [PubMed] [Google Scholar]
- 28.Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol Cell. 2010;37(4):492–502. doi: 10.1016/j.molcel.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hanada K, et al. The structure-specific endonuclease Mus81 contributes to replication restart by generating double-strand DNA breaks. Nat Struct Mol Biol. 2007;14(11):1096–1104. doi: 10.1038/nsmb1313. [DOI] [PubMed] [Google Scholar]
- 30.Modesti M, Kanaar R. DNA repair: spot(light)s on chromatin. Curr Biol. 2001;11(6):R229–R232. doi: 10.1016/S0960-9822(01)00112-9. [DOI] [PubMed] [Google Scholar]
- 31.Wang Q, Gao F, May WS, Zhang Y, Flagg T, Deng X. Bcl2 negatively regulates DNA double-strand-break repair through a nonhomologous end-joining pathway. Mol Cell. 2008;29(4):488–498. doi: 10.1016/j.molcel.2007.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baumann P, West SC. Role of the human RAD51 protein in homologous recombination and double-stranded-break repair. Trends Biochem Sci. 1998;23(7):247–251. doi: 10.1016/S0968-0004(98)01232-8. [DOI] [PubMed] [Google Scholar]
- 33.Lundin C, Schultz N, Arnaudeau C, Mohindra A, Hansen LT, Helleday T. RAD51 is involved in repair of damage associated with DNA replication in mammalian cells. J Mol Biol. 2003;328(3):521–535. doi: 10.1016/S0022-2836(03)00313-9. [DOI] [PubMed] [Google Scholar]
- 34.Bianchi V, Pontis E, Reichard P. Changes of deoxyribonucleoside triphosphate pools induced by hydroxyurea and their relation to DNA synthesis. J Biol Chem. 1986;261(34):16037–16042. [PubMed] [Google Scholar]
- 35.Johnson N, et al. Compromised CDK1 activity sensitizes BRCA-proficient cancers to PARP inhibition. Nat Med. 2011;17(7):875–882. doi: 10.1038/nm.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pierce AJ, Hu P, Han M, Ellis N, Jasin M. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev. 2001;15(24):3237–3242. doi: 10.1101/gad.946401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13(20):2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu Y, Scully R, Sobhian B, Xie A, Shestakova E, Livingston DM. RAP80-directed tuning of BRCA1 homologous recombination function at ionizing radiation-induced nuclear foci. Genes Dev. 2011;25(7):685–700. doi: 10.1101/gad.2011011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seluanov A, Mittelman D, Pereira-Smith OM, Wilson JH, Gorbunova V. DNA end joining becomes less efficient and more error-prone during cellular senescence. Proc Natl Acad Sci U S A. 2004;101(20):7624–7629. doi: 10.1073/pnas.0400726101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bae J, Leo CP, Hsu SY, Hsueh AJ. MCL-1S, a splicing variant of the antiapoptotic BCL-2 family member MCL-1, encodes a proapoptotic protein possessing only the BH3 domain. J Biol Chem. 2000;275(33):25255–25261. doi: 10.1074/jbc.M909826199. [DOI] [PubMed] [Google Scholar]
- 41.Day CL, Chen L, Richardson SJ, Harrison PJ, Huang DC, Hinds MG. Solution structure of prosurvival Mcl-1 and characterization of its binding by proapoptotic BH3-only ligands. J Biol Chem. 2005;280(6):4738–4744. doi: 10.1074/jbc.M411434200. [DOI] [PubMed] [Google Scholar]
- 42.Xie M, et al. Bcl2 inhibits recruitment of Mre11 complex to DNA double-strand breaks in response to high-linear energy transfer radiation. Nucleic Acids Res. 2015;43(2):960–972. doi: 10.1093/nar/gku1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun J, Lee KJ, Davis AJ, Chen DJ. Human Ku70/80 protein blocks exonuclease 1-mediated DNA resection in the presence of human Mre11 or Mre11/Rad50 protein complex. J Biol Chem. 2012;287(7):4936–4945. doi: 10.1074/jbc.M111.306167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaidi A, Weinert BT, Choudhary C, Jackson SP. Human SIRT6 promotes DNA end resection through CtIP deacetylation. Science. 2010;329(5997):1348–1353. doi: 10.1126/science.1192049. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 45.Sartori AA, et al. Human CtIP promotes DNA end resection. Nature. 2007;450(7169):509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Unno J, et al. FANCD2 binds CtIP and regulates DNA-end resection during DNA interstrand crosslink repair. Cell Rep. 2014;7(4):1039–1047. doi: 10.1016/j.celrep.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 47.Huertas P. DNA resection in eukaryotes: deciding how to fix the break. Nat Struct Mol Biol. 2010;17(1):11–16. doi: 10.1038/nsmb.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Han B, et al. Small-molecule Bcl2 BH4 antagonist for lung cancer therapy. Cancer Cell. 2015;27(6):852–863. doi: 10.1016/j.ccell.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jin L, et al. Glutamate dehydrogenase 1 signals through antioxidant glutathione peroxidase 1 to regulate redox homeostasis and tumor growth. Cancer Cell. 2015;27(2):257–270. doi: 10.1016/j.ccell.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krell T, et al. Characterization of molecular interactions using isothermal titration calorimetry. Methods Mol Biol. 2014;1149:193–203. doi: 10.1007/978-1-4939-0473-0_16. [DOI] [PubMed] [Google Scholar]
- 51.Linkuvienė V, Krainer G, Chen WY, Matulis D. Isothermal titration calorimetry for drug design: precision of the enthalpy and binding constant measurements and comparison of the instruments. Anal Biochem. 2016;515:61–64. doi: 10.1016/j.ab.2016.10.005. [DOI] [PubMed] [Google Scholar]
- 52.Gu L, et al. Discovery of dual inhibitors of MDM2 and XIAP for cancer treatment. Cancer Cell. 2016;30(4):623–636. doi: 10.1016/j.ccell.2016.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mei Y, Du W, Yang Y, Wu M. Puma(*)Mcl-1 interaction is not sufficient to prevent rapid degradation of Mcl-1. Oncogene. 2005;24(48):7224–7237. doi: 10.1038/sj.onc.1208873. [DOI] [PubMed] [Google Scholar]
- 54.Liu T, Huang J. DNA end resection: facts and mechanisms. Genomics Proteomics Bioinformatics. 2016;14(3):126–130. doi: 10.1016/j.gpb.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hustedt N, Durocher D. The control of DNA repair by the cell cycle. Nat Cell Biol. 2016;19(1):1–9. doi: 10.1038/ncb3452. [DOI] [PubMed] [Google Scholar]
- 56.Escribano-Díaz C, et al. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol Cell. 2013;49(5):872–883. doi: 10.1016/j.molcel.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 57.Ismail IH, et al. The RNF138 E3 ligase displaces Ku to promote DNA end resection and regulate DNA repair pathway choice. Nat Cell Biol. 2015;17(11):1446–1457. doi: 10.1038/ncb3259. [DOI] [PubMed] [Google Scholar]
- 58.Bernstein KA, Rothstein R. At loose ends: resecting a double-strand break. Cell. 2009;137(5):807–810. doi: 10.1016/j.cell.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jimeno S, Fernández-Ávila MJ, Cruz-García A, Cepeda-García C, Gómez-Cabello D, Huertas P. Neddylation inhibits CtIP-mediated resection and regulates DNA double strand break repair pathway choice. Nucleic Acids Res. 2015;43(2):987–999. doi: 10.1093/nar/gku1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hefferin ML, Tomkinson AE. Mechanism of DNA double-strand break repair by non-homologous end joining. DNA Repair (Amst) 2005;4(6):639–648. doi: 10.1016/j.dnarep.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 61.Burma S, Chen BP, Chen DJ. Role of non-homologous end joining (NHEJ) in maintaining genomic integrity. DNA Repair (Amst) 2006;5(9–10):1042–1048. doi: 10.1016/j.dnarep.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 62.Siddiqui WA, Ahad A, Ahsan H. The mystery of BCL2 family: Bcl-2 proteins and apoptosis: an update. Arch Toxicol. 2015;89(3):289–317. doi: 10.1007/s00204-014-1448-7. [DOI] [PubMed] [Google Scholar]
- 63.Thomas LW, Lam C, Edwards SW. Mcl-1; the molecular regulation of protein function. FEBS Lett. 2010;584(14):2981–2989. doi: 10.1016/j.febslet.2010.05.061. [DOI] [PubMed] [Google Scholar]
- 64.Kolacsek O, et al. Reliable transgene-independent method for determining Sleeping Beauty transposon copy numbers. Mobile DNA. 2011;2(1):5. doi: 10.1186/1759-8753-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.