Abstract
Charcot-Marie-Tooth disease type 1A (CMT1A) is caused by duplication of peripheral myelin protein 22 (PMP22) and is the most common hereditary peripheral neuropathy. CMT1A is characterized by demyelination and axonal loss, which underlie slowed motor nerve conduction velocity (MNCV) and reduced compound muscle action potentials (CMAP) in patients. There is currently no known treatment for this disease. Here, we show that antisense oligonucleotides (ASOs) effectively suppress PMP22 mRNA in affected nerves in 2 murine CMT1A models. Notably, initiation of ASO treatment after disease onset restored myelination, MNCV, and CMAP almost to levels seen in WT animals. In addition to disease-associated gene expression networks that were restored with ASO treatment, we also identified potential disease biomarkers through transcriptomic profiling. Furthermore, we demonstrated that reduction of PMP22 mRNA in skin biopsies from ASO-treated rats is a suitable biomarker for evaluating target engagement in response to ASO therapy. These results support the use of ASOs as a potential treatment for CMT1A and elucidate potential disease and target engagement biomarkers for use in future clinical trials.
Keywords: Neuroscience, Therapeutics
Keywords: Drug therapy, Gene therapy, Monogenic diseases
Introduction
Inherited peripheral neuropathies, also known as Charcot-Marie-Tooth disease (CMT), are one of the most common heritable diseases of the nervous system, affecting approximately 1 in 2,500 individuals (1). Mutations in more than 90 distinct genes cause CMT, the most common of which is a 1.4-Mb duplication on human chromosome 17, classified as CMT1A (2, 3). The peripheral myelin protein 22 (PMP22) gene, which encodes the major myelin protein, peripheral myelin protein 22, resides within the 1.4-Mb duplicated interval (4–7). PMP22 is an intrinsic membrane protein of myelin that alters lipid organization/distribution (8, 9) and is developmentally induced within Schwann cells as they initiate myelination of peripheral nerves (10). Subsequent rodent studies demonstrated that overexpression of PMP22 is sufficient to cause a demyelinating neuropathy (11–13), and proof-of-concept conditional knockout studies demonstrated that reduction of PMP22 overexpression led to remyelination (14). Interestingly, the deletion of the same 1.4-Mb region results in the loss of a PMP22 allele and causes a distinct neuropathy known as hereditary neuropathy with liability to pressure palsies (15), further demonstrating that gene dosage is the critical determinant in these neuropathies. Finally, elevated levels of PMP22 protein have been demonstrated in dermal or sural nerves of CMT1A patients (16, 17). A logical therapeutic approach would be to decrease PMP22 expression in myelinating Schwann cells.
Several approaches to reduce PMP22 expression have been proposed, yet no therapy is currently available to patients. For instance, PMP22 overexpression in rodent models can be reduced by high-dose ascorbic acid (18); however, in clinical trials, ascorbic acid did not reduce the level of PMP22 mRNA in skin biopsies from treated CMT1A patients (19–21). Progesterone antagonists and GABAB agonists have also been shown to reduce PMP22 mRNA expression (22, 23), but their potential is hampered by diverse effects on the gene-regulation program of Schwann cells and possibly other cell types, which may complicate a chronic treatment of an inherited disease. Thus, we propose a direct therapeutic strategy targeting PMP22 RNA with antisense oligonucleotides (ASOs).
ASOs have recently emerged as viable therapeutics to treat multiple disorders, including neurological conditions, and can be readily translated into the clinic (24). ASOs are single-stranded synthetic nucleic acids that bind target mRNA via Watson-Crick base pairing, resulting in degradation of target mRNA by RNAse H, a ubiquitously expressed mammalian enzyme (25). Phosphothioate-modified DNA and 2′-sugar modifications enable ASOs to be water soluble, resistant to exonucleases, and diffusible and to exhibit dose-dependent activity in vitro and in vivo (24). A key advance is the demonstration that ASOs can target various cell types in the CNS following intrathecal delivery and Schwann cells in the peripheral nervous system (PNS) following systemic subcutaneous injections (26–28).
Here, we describe the development of ASOs that reduce PMP22 mRNA levels and result in long-term improvement in 2 well-established rodent models of CMT1A. In C22 mice, which overexpress human PMP22 and have a severe demyelinating neuropathy (13, 29–33), ASO-mediated reduction of PMP22 mRNA levels markedly improves and even reverses several neuropathy end points, such as motor, electrophysiology, pathology, and transcriptomic changes. Independent studies of the CMT1A rat model, a well-characterized model of CMT1A (12, 34), show that ASOs suppress Pmp22 mRNA levels in several affected nerves and restore myelination and electrophysiological properties of motor axons. We also identify disease biomarker genes that appear to be modulated by ASO treatment and demonstrate that PMP22 RNA levels in skin biopsies may be a suitable target engagement biomarker. These results demonstrate the utility of ASOs in treating CMT1A, providing a precedent that can be applied to other copy number variation (CNV) disorders as well as other peripheral neuropathies.
Results
Effects of ASO treatment in C22 mice.
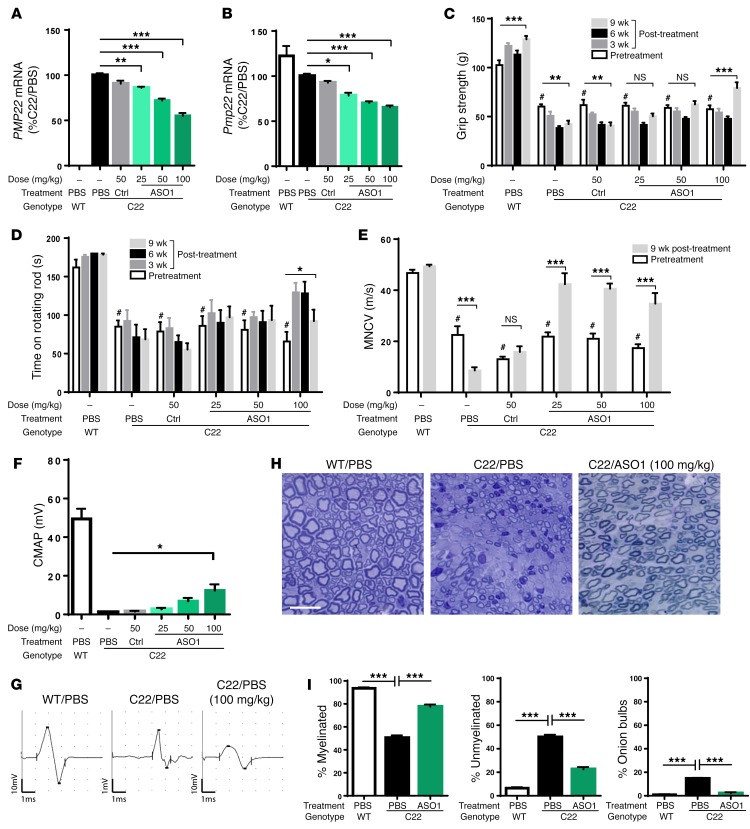
The C22 mouse model contains 7 copies of a human YAC encompassing the PMP22 gene, resulting in a severe, demyelinating neuropathy that is evident morphologically and behaviorally at early time points (29, 32, 33, 35). To determine whether ASO-mediated lowering of PMP22 mRNA would be beneficial in this model, we screened and identified the most potent ASO (ASO1), which targets the 3′ UTR of the human PMP22 gene (Supplemental Figure 1, A and B; supplemental material available online with this article; https://doi.org/10.1172/JCI96499DS1). Weekly subcutaneous injection of ASO1 at 50 mg/kg per week for 2 weeks in adult C22 mice resulted in approximately 25% suppression of PMP22 mRNA relative to PBS-treated mice (Figure 1C). Furthermore, weekly subcutaneous injection of ASO1 at 25, 50, or 100 mg/kg per week for 2 weeks in WT mice resulted in dose-dependent reduction in Pmp22 mRNA, as this site is homologous for the mouse Pmp22 gene (Supplemental Figure 1D). To determine whether ASO1 could ameliorate the neuropathy in C22 mice, mice were treated with weekly subcutaneous injections of saline, a control ASO (50 mg/kg), or ASO1 at 25, 50, or 100 mg/kg per week for 9 weeks. Injections were started in 5-week-old C22 mice (and their WT littermates), an age at which C22 mice exhibit CMT phenotypes including slowed motor nerve conduction velocity (MNCV), reduced compound muscle action potentials (CMAP), and impaired motor performance (33). Similar to the results observed with short-term ASO1 exposure (Supplemental Figure 1), both mouse Pmp22 and human PMP22 mRNA exhibited a dose-dependent decrease relative to PBS following ASO1 treatment, with a 50% reduction of the human (transgenic) PMP22 mRNA at the end of 9 weeks at the highest dose tested (Figure 1, A and B). The grip strength and rotarod performance of these mice were measured before treatment began (pretreatment) and at 3, 6, and 9 weeks following ASO treatment. Consistent with previous reports (33), pretreatment/baseline grip strength (102.5 ± 4.9 g in WT treated with PBS vs. 60.1 ± 2.4 g in C22 treated with PBS, Figure 1C) and the rotarod performance (161.9 ± 10.3 s in WT treated with PBS vs. 85.1 ± 8.0 s in C22 treated with PBS, Figure 1D) of 5-week-old C22 mice were significantly impaired compared with those in WT controls. We observed a significant loss of grip strength over the 9-week testing period in C22 mice treated with PBS (60.1 ± 2.4 g at pretreatment/baseline vs. 42.2 ± 3.5 g at 9 weeks, P < 0.001). ASO1 treatment not only slowed progression at low doses, but also reversed the decline and improved performance over the pretreatment/baseline at the highest dose (100 mg/kg) after 9 weeks of treatment (Figure 1C). The highest dose of ASO1 also improved rotarod performance even at early time points (3 and 6 weeks after treatment; Figure 1D). Notably, improvement was observed compared not only with the saline treatment at equivalent time points, but also relative to pretreatment/baseline in the C22 model. Importantly, the control ASO had no effect on either human or mouse PMP22 mRNA or disease progression, as characterized by behavioral end points. Taken together, these data show that ASO1 resulted in a dose-dependent reduction in PMP22 mRNA and reversed motor deficits in symptomatic C22 mice.
Figure 1. Behavioral, electrophysiological, and pathological improvement after ASO treatment in C22 model.
Five-week-old C22 mice were treated with weekly subcutaneous injections of PB, control ASO (CTRL; 50 mg/kg), or ASO1 at 25, 50, or 100 mg/kg per week for 9 weeks. n = 8 per group. WT littermates treated with PBS were included as controls. n = 8. CTRL, control. (A and B) Human PMP22 and mouse Pmp22 mRNA levels in sciatic nerves of treated mice. One-way ANOVA with Dunnett’s post test. *P < 0.05; **P < 0.01; ***P < 0.001. (C) The hind limb grip strength (g) and (D) time remaining on rotarod (s) were measured before treatment began (pretreatment) and at 3, 6, and 9 weeks following treatment. Two-way ANOVA with Tukey’s post test was used to compare pretreated C22 vs. WT (#P < 0.05) or pretreated and posttreated disease groups. *P < 0.05; **P < 0.01; ***P < 0.001. (E) MNCV was measured at pretreatment and at 9 weeks following treatment. Two-way ANOVA with Tukey’s post test was used to compare pretreated C22 versus WT (#P < 0.05) or pretreated and posttreated disease groups. ***P < 0.001. (F) CMAP was measured at 9 weeks after treatment. ASO1-treated groups were compared with PBS group using 1-way ANOVA with Dunnett’s post test. *P < 0.05. (G) Representative electrophysiological trace and (H) representative histological images of cross-sectioned sciatic nerves of a WT treated with PBS, a C22 treated with PBS, or a C22 treated with 100 mg/kg of ASO1 for 9 weeks. Scale bar: 5 μm. (I) Quantification of percentage of myelinated, percentage of unmyelinated, and percentage of onion bulb axons. One-way ANOVA with Dunnett’s post test was used to compare between C22 treated with PBS or ASO1 and WT treated with PBS. ***P < 0.001
Abnormal myelination can be measured by electrophysiological analysis. MNCV is substantially slowed in CMT1A patients and in animal models of CMT1A, including C22 mice (35, 36), and can be done repeatedly in humans and rodents. Therefore, we measured MNCV of the sciatic nerve at pretreatment/baseline and at 9 weeks after treatment in the same cohorts of C22 and WT mice described above. MNCVs were substantially slower in C22 mice compared with their WT littermates at pretreatment time points (~20 m/s at baseline/pretreatment in C22 mice vs. ~45 m/s in WT mice; Figure 1E). While PBS and control ASO had no effect on MNCVs, all 3 doses of ASO1 resulted in substantial improvement in MNCVs that approached those in WT mice at 9 weeks after treatment (Figure 1E).
The amplitude of the CMAP correlates with the number of muscle fibers that can be activated from the site where their motor nerve is stimulated. The CMAP amplitude recorded from the tibialis anterior muscle following stimulation of the sciatic nerve at the hip was increased in a dose-dependent manner after 9 weeks of treatment with ASO1, but did not approach that of WT mice (Figure 1, F and G).
The presence of demyelinated axons and “onion bulbs” are pathological hallmarks of CMT1A. While C22 mouse nerves have some onion bulbs, the most conspicuous findings are unmyelinated axons with a diameter of more than 1 micron that are invariably myelinated in WT nerves. These become evident during early myelination and persist in adult C22 mice (32, 33). Histological examination of cross-sectioned sciatic nerves revealed significant reduction of unmyelinated axons of more than 1 micron in diameter and onion bulbs, together with a concomitant increase in myelinated axons in C22 mice following 9 weeks of ASO1 treatment (Figure 1, H and I). ASO1 resulted in improved myelination in C22 mice, as measured by motor behaviors, electrophysiology, and histology.
Transcriptional changes following ASO treatment in C22 mice.
We next investigated the transcriptional consequences of ASO-mediated suppression of PMP22 mRNA in C22 mice. We therefore performed 3′ mRNA poly-A+ sequencing (RNA-seq) on sciatic nerves of a separate cohort of naive 5-week-old C22 versus WT littermates (C22 5-week and WT 5-week) and a cohort of C22 and WT littermates that were treated with either PBS or ASO1 at 100 mg/kg weekly for 9 weeks (C22 PBS 15-week and C22 ASO1 15-week). Using variance-model–based differential expression analysis, we determined 2,080 differentially expressed genes (DEGs) with a fold change greater than 2 at a false discovery rate (FDR) of 0.00001 or less in 5-week-old C22 versus WT mice and 1,056 DEGs in 15-week-old C22 versus WT mice that had been treated with PBS for 9 weeks (Supplemental Figure 2A). Of note, 748 of 1,056 DEGs observed in 15-week-old mice were also altered in 5-week-old mice, suggesting C22 mice were already symptomatic at 5 weeks of age. To further confirm this observation, fold-change coherence analysis of these DEGs revealed similar direction and magnitude of changes of all DEGs observed in 5-week-old and 15-week-old mice (Supplemental Figure 2B). These findings were consistent with the motor behavior and electrophysiology characterizations of the C22 cohort described in Figure 1.
This was, to our knowledge, the first RNA-seq profiling of a CMT1A model, and the analysis showed some categories of genes in common with microarray profiles of other CMT1A models (34, 37, 38). For example, there was a significant decrease in cholesterol and lipid biosynthetic genes as well as myelin-related genes in 5-week-old C22 compared with WT littermates (Supplemental Figure 2, C and F). The increased gene categories include a number involved in cell signaling, as well as those involved in axon guidance mechanisms (RhoA, Plxna1, and Rac1) and integrin signaling (Itgb8) (Supplemental Figure 2D). Expression of myelin inhibitors such as Id2, Id4, and Ednrb was also increased, suggesting an unfavorable environment for remyelination (Supplemental Figure 2E). Dysregulation of several transcription factors regulating Schwann cell differentiation was also observed in the C22 mice (Supplemental Figure 2F). Of note were the high levels of Pou3f1 (also known as Oct6), a transcription factor that is expressed by promyelinating Schwann cells (39) and was previously reported to be upregulated in other models of CMT1A (34, 37–40). Furthermore, the CMT1A phenotype has previously been thought to recapitulate certain aspects of nerve injury, in which Schwann cells are reprogrammed to undergo demyelination and initiate a program involving macrophage recruitment, downregulation of myelin genes, and secretion of factors that ultimately support nerve regeneration. One of the critical regulators of Schwann cell responses to nerve injury is the c-Jun transcription factor (41), which is also induced at the protein level in both rodent models of CMT1A and also in Schwann cells of skin biopsies from human patients (40, 42, 43). While c-Jun was induced in C22 mice, the data obtained here allowed us to determine whether the broader profile of deregulated genes in C22 overlap with those found in nerve injury. Somewhat surprisingly, of the 2,080 DEGs that are induced in the C22 model, only 43 were found to be induced in expression analyses of different time points after nerve injury (41, 44, 45), including the Pou3f1 transcription factor expressed in promyelinating Schwann cells as well as other transcription factors, such as Sox4 and Id2 (Supplemental Figure 2, E and F). Indeed, none of the previously described c-Jun target genes (41) were induced in the C22 model, indicating that other transcriptional programs were enacted by PMP22 overexpression in this model. Relatively few cytokines/chemokines were induced in the C22 model, with the exception of Cxcl14 (46), and immune cell markers were not an enriched gene ontology category with this data set (Supplemental Figure 2G).
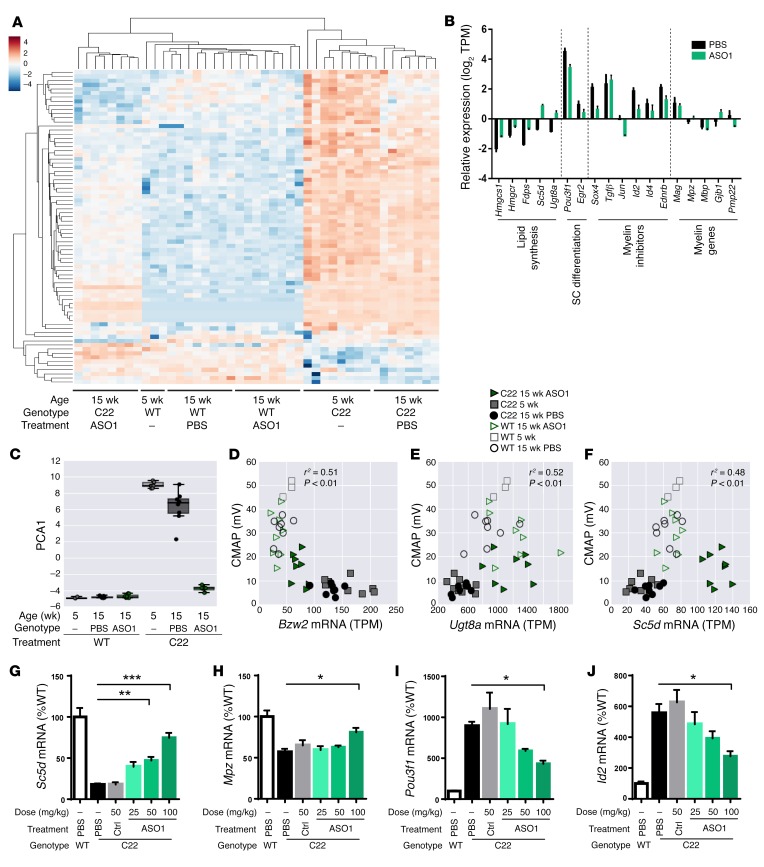
We next determined whether ASO-mediated suppression could restore gene-expression levels toward WT levels and found 97 DEGs that behaved in this manner. Among this set, 76 overlapped with the 746 DEGs that significantly changed in the comparison of C22 and WT cohorts at 5 or 15 weeks (Supplemental Table 1). Hierarchical clustering of the expression profiles of these DEGs showed that ASO treatment improved and in some instances reversed the transcriptional changes in C22 mice to almost WT levels (Figure 2A). Specifically, ASO-mediated suppression of PMP22 mRNA increased cholesterol/lipid biosynthetic and myelin-related genes, while decreasing levels of myelin inhibitors and dedifferentiated Schwann cell markers, supporting the interpretation that there was ongoing remyelination and restoration of normal Schwann cell function (Figure 2B). Principal component analysis (PCA) of these 76 DEGs generated a first principal component (PCA1) that accounted for 80.1% variance. PCA1 further demonstrated a transcriptional profile of C22 mice treated with ASO1 for 9 weeks similar to that of WT mice (Figure 2C). Genes with the largest eigenvalues or weights in PCA1, such as Bzw2, Cuedc2, Mpzl1, Padi2, and Snx16, were previously found to be regulated by Sox10 (47), a crucial regulator of several phases of Schwann cell development, suggesting reprogramming of dysregulated Schwann cells following ASO treatment. Our studies did not reveal statistically significant changes in Sox10 mRNA. It is likely that a Sox10-interacting factor is responsible for these changes, since Sox10 interacts with a wide range of transcription factors (47).
Figure 2. Transcriptional changes in C22 mice following ASO treatment.
(A) Hierarchical clustering of the expression profiles of 76 DEGs. (B) Subset of lipid biosynthetic genes, Schwann cell differentiation markers, myelin inhibitors, and myelin-related genes altered by 9 weeks after ASO1 treatment. (C) PCA of 76 DEGs. Correlation analyses of CMAP amplitude and mRNA expression of (D) Bzw2 (Pearson r2 = 0.51, P < 0.01), (E) Ugt8a (Pearson r2 = 0.52, P < 0.01), and (F) Sc5d (Pearson r2 = 0.48, P < 0.01). (G–J) qRT-PCR confirmation of mRNA expression of Sc5d, Mpz, Pou3f1, and Id2 in ASO1-treated sciatic nerves. ASO1-treated groups were compared with PBS group using 1-way ANOVA with Dunnett’s post test. *P < 0.05; **P < 0.01; ***P < 0.001.
CMAP, MNCV, and distal motor latency were also measured in these mice. Thus, Pearson correlation was computed between expression of each of the strongly weighted PCA genes and each of these electrophysiological measures to determine whether novel disease biomarkers could be identified. Expression of Bzw2, a repressor of translation as a nonfunctional mimic of eIF2α, was negatively correlated with electrophysiological measurements, while higher expression levels of lipid synthetic genes, Ugt8a and Scd5, were found to be positively correlated with these measures (Figure 2, D–F, and Supplemental Figure 3), suggesting they could serve as additional disease biomarkers.
Importantly, while ASO1 treatment in WT mice reduced Pmp22 expression (Supplemental Figure 2H), no significant transcriptional changes were observed between WT mice treated with PBS or ASO1 for 9 weeks (Figure 2, A, C–F). These data suggested that approximately 35% suppression of endogenous Pmp22 mRNA is tolerated, although further dose titration studies will be needed.
Quantitative reverse-transcriptase PCR (qRT-PCR) analyses using the sciatic lysates from the dose-response cohort in Figure 1 further confirmed the RNA-seq findings. Specifically, there were dose-dependent increases in the mRNA levels of several lipid biosynthetic and myelin genes (Figure 2, G and H, and Supplemental Figure 4, A–C) as well as dose-dependent decreases of dedifferentiation markers and myelin inhibitor (Figure 2, I and J, and Supplemental Figure 4, D–F) following 9 weeks of ASO1 treatment. ASO1 treatment caused a profound shift in the transcriptional profile of C22 sciatic nerve toward that of WT mice.
ASO treatment of a rat model of CMT1A.
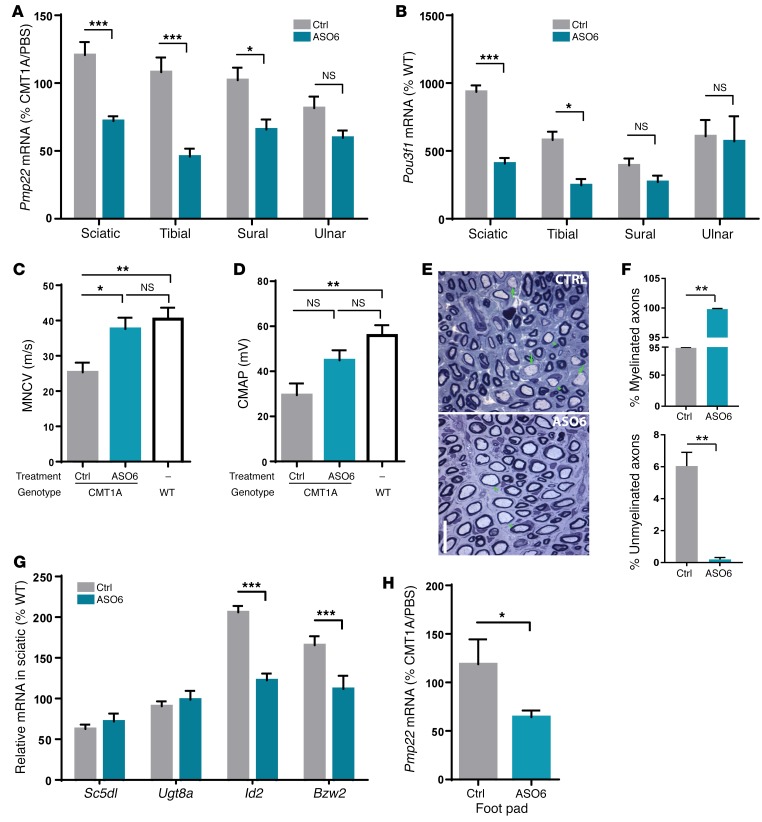
The results obtained from treating C22 mice were highly encouraging. Thus, we also investigated ASO treatment in a second model, the heterozygous CMT1A rat (12, 34, 48), which contains 3 copies of a cosmid containing mouse Pmp22 and exhibits a milder demyelinating neuropathy than C22 mice. CMT1A rats exhibit grip-strength deficit starting at 5 weeks of age and significantly reduced CMAP and demyelination by 9 weeks of age (48). We performed in vitro screening in RT4-D6P2T cells and in vivo screening of ASOs in Sprague-Dawley rats and identified an active ASO (ASO6) that effectively suppressed rat Pmp22 mRNA in a dose-dependent manner, but had no effect on mouse Pmp22 mRNA (Supplemental Figure 5, A–D). To test the efficacy of ASO6 in reducing rat Pmp22 mRNA, we treated 6-week-old CMT1A rats weekly for 12 weeks with PBS, control ASO (12.5 mg/kg), or ASO6 (12.5 mg/kg) and sacrificed the rats following electrophysiological testing performed 1 week after the last dose. To characterize the activity of ASO6 in different nerves, we dissected the sciatic, tibial, sural, and ulnar nerves and observed a robust decrease of rat Pmp22 mRNA in all of these nerves (Figure 3A). Consistent with published reports (34, 48) and our data in C22 mice, reducing Pmp22 mRNA was associated with reduced Pou3f1 expression in these nerves (Figure 3B).
Figure 3. Myelination and electrophysiological improvement after ASO treatment in a rat model of CMT1A.
CMT1A rats were treated weekly for 12 weeks with PBS, control ASO (12.5 mg/kg), or ASO6 (12.5 mg/kg). n = 10 per group. WT littermates treated with PBS were included as control. n = 10 per group. (A) Rat Pmp22 mRNA in various nerves of control and ASO6-treated rats. ASO6-treated group was compared with control group in each nerve using Student’s t test. *P < 0.05; ***P < 0.001. (B) Pou3f1 mRNA in various nerves of control and ASO6-treated rats. PMP22 ASO–treated group was compared with control group in each nerve using Student’s t test. *P < 0.05; ***P < 0.001. (C and D) MNCV and CMAP were measured at 12 weeks following treatment. (E) Representative histological image of cross-sectioned femoral motor nerves of CMT1A rats treated with control or ASO6. Arrows and arrowheads indicate unmyelinated and myelinated axon of greater than 1 micron in diameter, respectively. Scale bar: 5 μm. (F) Quantification of percentage of myelinated and percentage of unmyelinated axons. ASO6-treated group was compared with control group using Student’s t test. **P < 0.01. (G) qRT-PCR confirmation of mRNA expression of lipid biosynthetic (Sc5dl and Ugt8a), myelin inhibitor (Id2), and translational repressor (Bzw2) genes in the sciatic nerve of control vs. ASO6-treated rats. ASO6-treated group was compared with control group in each nerve using Student’s t test. ***P < 0.001. (H) Rat Pmp22 mRNA in footpad following control or ASO6 treatment. ASO6-treated group was compared with control group using Student’s t test. *P < 0.05.
Electrophysiological analysis revealed that ASO6 treatment completely reversed the slow MNCVs of CMT1A rats to velocities that were comparable to those of their WT littermates (Figure 3C). ASO6 also increased the CMAP amplitude compared with that in control-treated CMT1A rats, suggesting more motor axons made functional connections to the corresponding tibialis anterior muscle (Figure 3D). CMT1A rat nerves had unmyelinated axons greater than 1 micron in diameter even at 5 weeks of age (12) prior to onset of the treatment at 6 weeks. Histological analysis of the femoral motor nerve demonstrated CMT1A rats treated with ASO6 had fewer unmyelinated axons of more than 1 micron in diameter than did the animals that were treated with the control, whereas the total number of myelinated and unmyelinated axons was not significantly different (Figure 3, E and F).
Next, we asked whether the disease biomarkers identified in the C22 transcriptomic study can be confirmed in the CMT1A rat model. Consistent with findings in C22 mice, the mRNA levels of lipid biosynthetic genes such as Sc5d and Ugt8a were decreased in CMT1A rats, although to a lesser extent than in C22 mouse nerves. ASO6 treatment trended toward increased mRNA levels of these genes (Figure 3G). Furthermore, the mRNA levels of Id2 and Bzw2 were significantly increased in CMT1A rats compared with WT rats and were substantially decreased following ASO6 treatment (Figure 3G).
Finally, we sought to identify a target engagement biomarker in the CMT1A rats. Toward this end, we collected blood and footpad skin biopsies from treated CMT1A rats. Although there was detectable rat Pmp22 mRNA in the blood, there was no change in Pmp22 levels following ASO6 treatment (Supplemental Figure 5E). The level of rat Pmp22 mRNA in the footpad (containing dermal nerves and Schwann cells) was significantly reduced in ASO6- compared with control-treated rats, suggesting this could serve as a biomarker of target engagement (Figure 3H). In summary, ASO-mediated rat Pmp22 suppression in the physiological rat CMT1A model effectively reduced rat Pmp22 mRNA levels in several nerves, with concomitant transcriptional changes that indicate improved myelination, and marked improvement and even reversal of CMT1A electrophysiological and pathological features.
Discussion
Duplication of the PMP22 gene has been unequivocally determined to be the genetic cause of peripheral neuropathy in people with CMT1A. Thus, reducing the level of PMP22 mRNA in myelinating Schwann cells is an attractive and direct therapeutic approach. Here, we show that ASOs can penetrate the peripheral nerve barriers (perineurium and/or blood nerve barrier) and affect gene expression in Schwann cells. Further, we developed potent ASOs that decrease PMP22 mRNA levels and improve behavioral, electrophysiological, and pathological features of neuropathy in 2 animal models of CMT1A. Finally, ASO treatment alters the transcriptional profile of Schwann cells in favor of myelination. Together, our data suggest that ASOs result in the myelination of demyelinated axons, contributing to faster conduction and even more conducting myelinated motor axons. Remarkably, ASO treatment initiated after disease onset in C22 mice and CMT1A rats reversed the severity of the neuropathy.
In the C22 mouse model, where the human PMP22 gene is highly expressed, neuropathy end points could not only be arrested, but also reversed at the highest dose of the ASO. This raises the possibility that therapy may be effective not only in preventing disease progression, but also in causing some functional improvement, given that the affected Schwann cells in CMT1A remain viable during progression of the neuropathy. The results were not specific to one model, but were also demonstrated in the rat CMT1A model, in which overexpression of mouse Pmp22 is more modest. Even in this model, there was substantial improvement in the symptoms. Importantly, in both models, the appearance of unmyelinated axons of more than 1 micron in diameter that are normally myelinated was drastically improved. This also implies reversibility of the symptoms, since unmyelinated axons are apparent in the CMT1A rat model even at 5 weeks of age (12). Therefore, dramatic improvement is observed with cellular, electrophysiological, and functional measures of peripheral neuropathy. Despite the promising results, there remain several important questions, including whether sustained treatment will lead to continued improvement, whether it will prevent axonal loss, and whether it will work in older animals, in which axonal loss contributes to clinical manifestation (49). Future work will also be required to determine optimal dosing route and frequency as well as detailed characterization of ASO pharmacokinetic and pharmacodynamic properties in peripheral nerves.
In addition to the improvements induced by ASOs, these studies also afforded the opportunity to perform an RNA-seq profile of the C22 model, which identified several genes that were previously shown to be upregulated in CMT1A models (c-Jun, Pou3f1, and Cxcl14) and also those that are downregulated (lipid biosynthetic genes and other myelin-related genes). Several of the deregulated genes (c-Jun, Pou3f1, Id2, Ednrb) have been shown to inhibit myelination and/or myelin gene expression (41, 50–52). The profiling of ASO-treated animals provided further insight into those gene expression changes, such as those observed for Id2 and Bzw2, that are restored upon ASO treatment. Since this group is relatively small (<100), this provides important information on treatment-responsive gene expression changes that help normalize Schwann cell function. Future measurement of these genes along with PMP22 mRNA in skin biopsies (as done in the foot pad analysis) could provide important biomarkers that can be used in clinical trials. Since overreduction of PMP22 levels could theoretically cause the HNPP neuropathy caused by loss of a PMP22 allele, evaluation of PMP22 mRNA and other biomarkers can be used to monitor for this clinically. Overall, these studies demonstrate the potential therapeutic utility of ASOs in treating CMT1A and elucidate the gene expression networks that respond to successful restoration of normalized PMP22 levels.
Methods
Experimental design.
To determine effects of ASO treatment on C22 mice, 5-week-old C22 or WT littermates of mixed sex were randomized into treatment groups that received subcutaneous injections of saline, control ASO at 50 mg/kg, or ASO1 at 25, 50, and 100 mg/kg weekly for 9 weeks. Mice were sacrificed at 2 days after the last dose (n = 8 per genotype/treatment group). Mice were also subjected to hind limb grip strength and rotarod tasks at pretreatment/baseline, 3 weeks, 6 weeks, and 9 weeks following treatment and to electrophysiological recordings at pretreatment/baseline and 9 weeks after treatment in a blinded manner. For the transcriptomic study, 5-week-old C22 or WT littermates of mixed sex were subcutaneously dosed with saline or 100 mg/kg of ASO1 weekly for 9 weeks and were sacrificed at 2 days after the last dose for collection of sciatic nerves (n = 8 per genotype/treatment group). To determine efficacy of ASO treatment in CMT1A rats, male 6-week-old heterozygous CMT1A rats and WT littermates were subcutaneously dosed with saline (Gibco 14190, Thermo Fisher Scientific), control ASO, or ASO6 (25 mg/ml) at 12.5 mg/kg to the lower right flank once weekly for 12 weeks and were sacrificed following electrophysiology studies performed in the week following the last dose in a blinded manner (n = 10 per genotype/treatment group).
Animals.
Adult C57BL/6J females of 8 to 10 weeks of age were purchased from the Jackson Laboratory. C22 mice, which harbor 7 copies of a human PMP22 transgene (13), were obtained and generated by Taconic via a rapid expansion method. CMT1A rats, which harbor 3 copies of a mouse Pmp22 cosmid (12), were bred and maintained on the Sprague-Dawley background. Adult Sprague-Dawley rats of approximately 200 g were purchased from Envigo.
Oligonucleotides.
Synthesis and purification of all chemically purified ASOs were performed as previously described (53), and 16-mer chimeric (gapmer) phosphothioate oligonucleotides containing cEt groups at positions 1–3 and 14–16 were targeted to human/mouse PMP22 (ASO1), rat Pmp22 (ASO6), or a control. The sequences evaluated are listed in Supplemental Table 2.
Lead ASO identification.
Approximately 500 ASOs were designed against the full human PMP22 or rat Pmp22 genes, respectively. Electroporation of ASOs was carried out using the HT-200 BTX Electroporator with ElectroSquare Porator (ECM830) voltage source at 140 V in 96-well electroporation plates (BTX, 2 mm; Harvard Apparatus). Human PMP22 ASOs were screened in K-562 cells at 10 μM, while rat Pmp22 ASOs were screened in RT4-D6P2T cells at 7 μM. Cells were harvested at 24 hours after treatment for RNA extraction, and human PMP22 or rat Pmp22 mRNA was quantified by qRT-PCR. The most potent ASOs were taken into a 4-point dose response in K-562 or RT4-D6P2T cells, respectively. Five of the most potent human PMP22 ASOs that targeted identical sequences in human PMP22 and mouse Pmp22 were screened for tolerability by subcutaneous injections in adult C57BL/6J female mice (n = 4) at 50 mg/kg per week for 6 weeks. The most potent ASO (ASO1) was identified by screening in adult C22 mice by weekly subcutaneous injection at 25 mg/kg for 2 weeks. Three of the most potent rat Pmp22 ASOs were injected subcutaneously into adult Sprague-Dawley rats (n = 4) at 50 mg/kg per week for 6 weeks for the activity and tolerability screen, leading to the identification of the most potent ASO (ASO6).
qRT-PCR.
Cultured cells were lysed in 300 μl of RLT buffer (QIAGEN) containing 1% (v/v) 2-mercaptoethanol (BME, Sigma Aldrich). For RNA extraction, the left and sciatic nerves were dissected in mice, while the left sciatic, ulnar, femoral motor, and femoral sensory nerves were dissected in rats. Tissues were homogenized in 500 μl of RLT buffer containing 1% (v/v) BME. RNA was isolated from 20 μl of lysate with an RNeasy 96 Kit (QIAGEN) that included in-column DNA digestion with 50 U of DNase I (Invitrogen). RT-PCR was done using the StepOne Realtime PCR system (Applied Biosystems), as described previously (28). The sequences of primers and probes used are listed in Supplemental Table 3. PCR results were normalized by housekeeping genes, cyclophilin A/Ppia or Gapdh, or Gldn for foot pad skin biopsy and further normalized to the levels in PBS-treated mice or untreated cells.
Histological analyses.
Semi-thin sections of mouse sciatic nerve (2 mm piece cut from sciatic notch on the right side) were prepared as previously described (54). Briefly, nerves were fixed in 2% formaldehyde and 2% glutaraldehyde in 0.1 M phosphate buffer overnight at 4°C, osmicated, and dehydrated in increasing concentrations of ethanol, infiltrated, and then embedded in Embed 812. Sections (1 μm thick ) were cut from the distal site of sciatic notch using an Ultracut UTC ultramicrotome (Leica) with a glass knife, stained with 1% toluidine blue in 2% borate buffer, and imaged with an eVOS microscope (×40 lens). Approximately 400 axons were counted per animal, and the numbers of myelinated, unmyelinated axons (axons greater than 1 μm in diameter without a myelin sheath), and onion bulbs were expressed as percentage of total axons counted.
Anesthetized rats were transcardially perfused with 0.9% NaCl, followed by 4% paraformaldehyde and 2.5% glutaraldehyde in 0.1 M PB (pH 7.4). The femoral motor and sensory nerves were dissected together after fixation at 4°C, then osmicated, dehydrated, and infiltrated in Embed 812. Semi-thin sections (1 μm thick ) were stained with alkaline toluidine blue and imaged on a Leica DFC 295 camera with a ×100 objective. For counting axons, ×100 images of each femoral motor branch were photomerged by using Adobe Photoshop CC 2015.5. The myelinated axons and unmyelinated axons were identified, manually labeled, and counted in a blinded manner. We selected the femoral motor nerve because it was more affected than the femoral sensory nerve or the sciatic nerve and its branches.
Behavioral assessments and electrophysiology.
Behavioral assessments and electrophysiology were performed by an investigator who was blinded to treatment and genotypes. Tests were performed in the following order: rotarod, grip strength, then electrophysiology. There was 1 test per day. Mice were dosed with ASOs after all assessments were completed to avoid stress.
The rotarod test was performed as described (55). Hind limb grip strength was assessed using the Grip Strength Meter (Columbus Instruments). Electrophysiology was done as described with slight modifications: stimulating cathodes were placed at sciatic notch (proximal) and 10 mm distal to the sciatic notch, while recording electrodes were placed subdermally on the muscle belly of the tibialis anterior muscle (33). Detailed descriptions can be found in Supplemental Methods.
Digital gene expression.
Sciatic nerves from both legs were dissected, and total mRNA was extracted (TRIzol, Applied Biosciences) and quality controlled (Bioanalyzer, Agilent). Digital gene expression (DGE) was performed by sequencing fragment libraries from purified RNA. First-strand synthesis was performed using custom barcoded primers with oligo-dT priming. Libraries were then generated with a Lexogen 3′ QuantSeq multiplexing kit using random hexamers and barcoded amplification. Samples were sequenced (60 base pair reads) on an Illumina NextSeq500 to an average depth of 4.5 million uniquely mapped reads per sample. Demultiplexed reads were mapped using STAR alignment tool version 2.5.1b with the following run parameters: outFilterMultimapNmax 10, outFilterMatchNmin 23, seedSearchStartLmax 50. Aligned reads showed an average mapping rate of 61%. Transcript quantification was performed using Salmon version 0.7.1 using quasimapping-based mode with automated libtype detection (56, 57). TPM was computed for each gene by aggregating transcript counts and normalizing by total number of mapped reads. For alignment and expression, gene model indexes were built upon Ensembl Mus musculus build-81 cDNA sequences. Genes were identified as differentially expressed assuming a model of gene-expression variance derived from a negative binomial distribution and based upon gene-expression levels in the reference cohorts. For each comparison, the variance model was applied symmetrically, alternating both case and control groups as a reference for gene variation, and the 2 lists were subsequently aggregated. Gene-specific P values were computed for each biological replicate and median aggregated. Genes having a minimum P value of less than or equal to 0.1 in all replicates within a group were considered significant and used in downstream analysis. All original microarray data were deposited in the NCBI’s Gene Expression Omnibus database (GEO GSE103799).
Statistics.
For pairwise comparisons, Student’s t test was used. For comparisons of more than 2 treatment groups, 1-way ANOVA with Dunnett’s post hoc test was used. For comparisons of more than 2 treatment groups over time, 2-way ANOVA with Tukey’s post hoc test was used. Statistical significance was set at P < 0.05. Data are presented as mean ± SEM unless otherwise noted.
Study approval.
All experimental procedures involving animals were approved by the Institutional Animal Care and Use Committee at Ionis Pharmaceuticals, Psychogenics, and University of Philadelphia (Philadelphia, Pennsylvania, USA).
Author contributions
HTZ, GH, EES, MAS, and HBK conceived and coordinated the studies. HTZ, SD, KIL, SK, JL, AM, AK, MAS, SSS, and JS designed and performed experiments and analyzed results. HTZ, SSS, JS, and HBK wrote the paper with input from all authors. All authors approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank Toby Ferguson and Bob Baloh for thoughtful inputs in the initial experimental design, Christine Cho for critical review of the manuscript, and Donna Sipe, Johnnatan Tamayo, and Gemma Ebling for vivarium assistance, Psychogenics for breeding and dosing of CMT1A rats. This work was partially funded by the Charcot-Marie-Tooth Association (to JS) and the Judy Seltzer Levenson Memorial Fund for CMT Research (to JL and SSS).
Version 1. 12/04/2017
Electronic publication
Version 2. 01/02/2018
Print issue publication
Funding Statement
Charcot-Marie-Tooth Association has provided funding for John Svaren
Judy Seltzer Levenson Memorial Fund for CMT Research provides funding for Joel Li and Steve S. Scherer.
Footnotes
Conflict of interest: H.T. Zhao, S. Damle, K. Ikeda-Lee, S. Kuntz, A. Mohan, A. Kim, G. Hung, E.E. Swayze, and H.B. Kordasiewicz are full-time employees and stockholders of Ionis Pharmaceuticals Inc.
Reference information: J Clin Invest. 2018;128(1):359–368.https://doi.org/10.1172/JCI96499.
See the related Commentary at Antisense oligonucleotides offer hope to patients with Charcot-Marie-Tooth disease type 1A.
Contributor Information
Sagar Damle, Email: sdamle@ionisph.com.
Karli Ikeda-Lee, Email: kikeda-lee@ionisph.com.
Steven Kuntz, Email: skuntz@ionisph.com.
Apoorva Mohan, Email: amohan@ionisph.com.
Aneeza Kim, Email: AKim@ionisph.com.
Gene Hung, Email: ghung62@gmail.com.
John Svaren, Email: jpsvaren@wisc.edu.
References
- 1.Skre H. Genetic and clinical aspects of Charcot-Marie-Tooth’s disease. Clin Genet. 1974;6(2):98–118. doi: 10.1111/j.1399-0004.1974.tb00638.x. [DOI] [PubMed] [Google Scholar]
- 2.Lupski JR, et al. DNA duplication associated with Charcot-Marie-Tooth disease type 1A. Cell. 1991;66(2):219–232. doi: 10.1016/0092-8674(91)90613-4. [DOI] [PubMed] [Google Scholar]
- 3.Raeymaekers P, et al. Duplication in chromosome 17p11.2 in Charcot-Marie-Tooth neuropathy type 1a (CMT 1a). The HMSN Collaborative Research Group. Neuromuscul Disord. 1991;1(2):93–97. doi: 10.1016/0960-8966(91)90055-W. [DOI] [PubMed] [Google Scholar]
- 4.Patel PI, et al. The gene for the peripheral myelin protein PMP-22 is a candidate for Charcot-Marie-Tooth disease type 1A. Nat Genet. 1992;1(3):159–165. doi: 10.1038/ng0692-159. [DOI] [PubMed] [Google Scholar]
- 5.Timmerman V, et al. The peripheral myelin protein gene PMP-22 is contained within the Charcot-Marie-Tooth disease type 1A duplication. Nat Genet. 1992;1(3):171–175. doi: 10.1038/ng0692-171. [DOI] [PubMed] [Google Scholar]
- 6.Valentijn LJ, et al. The peripheral myelin gene PMP-22/GAS-3 is duplicated in Charcot-Marie-Tooth disease type 1A. Nat Genet. 1992;1(3):166–170. doi: 10.1038/ng0692-166. [DOI] [PubMed] [Google Scholar]
- 7.Matsunami N, et al. Peripheral myelin protein-22 gene maps in the duplication in chromosome 17p11.2 associated with Charcot-Marie-Tooth 1A. Nat Genet. 1992;1(3):176–179. doi: 10.1038/ng0692-176. [DOI] [PubMed] [Google Scholar]
- 8.Lee S, et al. PMP22 is critical for actin-mediated cellular functions and for establishing lipid rafts. J Neurosci. 2014;34(48):16140–16152. doi: 10.1523/JNEUROSCI.1908-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mittendorf KF, et al. Peripheral myelin protein 22 alters membrane architecture. Sci Adv. 2017;3(7):e1700220. doi: 10.1126/sciadv.1700220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Snipes GJ, Suter U, Welcher AA, Shooter EM. Characterization of a novel peripheral nervous system myelin protein (PMP-22/SR13) J Cell Biol. 1992;117(1):225–238. doi: 10.1083/jcb.117.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Magyar JP, et al. Impaired differentiation of Schwann cells in transgenic mice with increased PMP22 gene dosage. J Neurosci. 1996;16(17):5351–5360. doi: 10.1523/JNEUROSCI.16-17-05351.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sereda M, et al. A transgenic rat model of Charcot-Marie-Tooth disease. Neuron. 1996;16(5):1049–1060. doi: 10.1016/S0896-6273(00)80128-2. [DOI] [PubMed] [Google Scholar]
- 13.Huxley C, et al. Construction of a mouse model of Charcot-Marie-Tooth disease type 1A by pronuclear injection of human YAC DNA. Hum Mol Genet. 1996;5(5):563–569. doi: 10.1093/hmg/5.5.563. [DOI] [PubMed] [Google Scholar]
- 14.Perea J, et al. Induced myelination and demyelination in a conditional mouse model of Charcot-Marie-Tooth disease type 1A. Hum Mol Genet. 2001;10(10):1007–1018. doi: 10.1093/hmg/10.10.1007. [DOI] [PubMed] [Google Scholar]
- 15.Chance PF, et al. DNA deletion associated with hereditary neuropathy with liability to pressure palsies. Cell. 1993;72(1):143–151. doi: 10.1016/0092-8674(93)90058-X. [DOI] [PubMed] [Google Scholar]
- 16.Vallat JM, et al. Ultrastructural PMP22 expression in inherited demyelinating neuropathies. Ann Neurol. 1996;39(6):813–817. doi: 10.1002/ana.410390621. [DOI] [PubMed] [Google Scholar]
- 17.Katona I, et al. PMP22 expression in dermal nerve myelin from patients with CMT1A. Brain. 2009;132(Pt 7):1734–1740. doi: 10.1093/brain/awp113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Passage E, et al. Ascorbic acid treatment corrects the phenotype of a mouse model of Charcot-Marie-Tooth disease. Nat Med. 2004;10(4):396–401. doi: 10.1038/nm1023. [DOI] [PubMed] [Google Scholar]
- 19.Lewis RA, et al. High-dosage ascorbic acid treatment in Charcot-Marie-Tooth disease type 1A: results of a randomized, double-masked, controlled trial. JAMA Neurol. 2013;70(8):981–987. doi: 10.1001/jamaneurol.2013.3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nobbio L, et al. PMP22 messenger RNA levels in skin biopsies: testing the effectiveness of a Charcot-Marie-Tooth 1A biomarker. Brain. 2014;137(Pt 6):1614–1620. doi: 10.1093/brain/awu071. [DOI] [PubMed] [Google Scholar]
- 21.Pareyson D, et al. Ascorbic acid in Charcot-Marie-Tooth disease type 1A (CMT-TRIAAL and CMT-TRAUK): a double-blind randomised trial. Lancet Neurol. 2011;10(4):320–328. doi: 10.1016/S1474-4422(11)70025-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meyer zu Horste G, Prukop T, Liebetanz D, Mobius W, Nave KA, Sereda MW. Antiprogesterone therapy uncouples axonal loss from demyelination in a transgenic rat model of CMT1A neuropathy. Ann Neurol. 2007;61(1):61–72. doi: 10.1002/ana.21026. [DOI] [PubMed] [Google Scholar]
- 23.Sereda MW, Meyer zu Hörste G, Suter U, Uzma N, Nave KA. Therapeutic administration of progesterone antagonist in a model of Charcot-Marie-Tooth disease (CMT-1A) Nat Med. 2003;9(12):1533–1537. doi: 10.1038/nm957. [DOI] [PubMed] [Google Scholar]
- 24.Bennett CF, Baker BF, Pham N, Swayze E, Geary RS. Pharmacology of antisense drugs. Annu Rev Pharmacol Toxicol. 2017;57:81–105. doi: 10.1146/annurev-pharmtox-010716-104846. [DOI] [PubMed] [Google Scholar]
- 25.Cerritelli SM, Crouch RJ. Ribonuclease H: the enzymes in eukaryotes. FEBS J. 2009;276(6):1494–1505. doi: 10.1111/j.1742-4658.2009.06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kordasiewicz HB, et al. Sustained therapeutic reversal of Huntington’s disease by transient repression of huntingtin synthesis. Neuron. 2012;74(6):1031–1044. doi: 10.1016/j.neuron.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hung G, et al. Characterization of target mRNA reduction through in situ RNA hybridization in multiple organ systems following systemic antisense treatment in animals. Nucleic Acid Ther. 2013;23(6):369–378. doi: 10.1089/nat.2013.0443. [DOI] [PubMed] [Google Scholar]
- 28.Rigo F, et al. Pharmacology of a central nervous system delivered 2’-O-methoxyethyl-modified survival of motor neuron splicing oligonucleotide in mice and nonhuman primates. J Pharmacol Exp Ther. 2014;350(1):46–55. doi: 10.1124/jpet.113.212407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huxley C, et al. Correlation between varying levels of PMP22 expression and the degree of demyelination and reduction in nerve conduction velocity in transgenic mice. Hum Mol Genet. 1998;7(3):449–458. doi: 10.1093/hmg/7.3.449. [DOI] [PubMed] [Google Scholar]
- 30.Norreel JC, Jamon M, Riviere G, Passage E, Fontes M, Clarac F. Behavioural profiling of a murine Charcot-Marie-Tooth disease type 1A model. Eur J Neurosci. 2001;13(8):1625–1634. doi: 10.1046/j.0953-816x.2001.01535.x. [DOI] [PubMed] [Google Scholar]
- 31.Robaglia-Schlupp A, et al. PMP22 overexpression causes dysmyelination in mice. Brain. 2002;125(Pt 10):2213–2221. doi: 10.1093/brain/awf230. [DOI] [PubMed] [Google Scholar]
- 32.Robertson AM, Huxley C, King RH, Thomas PK. Development of early postnatal peripheral nerve abnormalities in Trembler-J and PMP22 transgenic mice. J Anat. 1999;195(Pt 3):331–339. doi: 10.1046/j.1469-7580.1999.19530331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verhamme C, et al. Myelin and axon pathology in a long-term study of PMP22-overexpressing mice. J Neuropathol Exp Neurol. 2011;70(5):386–398. doi: 10.1097/NEN.0b013e318217eba0. [DOI] [PubMed] [Google Scholar]
- 34.Vigo T, et al. Experimental Charcot-Marie-Tooth type 1A: a cDNA microarrays analysis. Mol Cell Neurosci. 2005;28(4):703–714. doi: 10.1016/j.mcn.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 35.Huxley C, et al. Construction of a mouse model of Charcot-Marie-Tooth disease type 1A by pronuclear injection of human YAC DNA. Hum Mol Genet. 1996;5(5):563–569. doi: 10.1093/hmg/5.5.563. [DOI] [PubMed] [Google Scholar]
- 36.Huxley C, et al. Correlation between varying levels of PMP22 expression and the degree of demyelination and reduction in nerve conduction velocity in transgenic mice. Hum Mol Genet. 1998;7(3):449–458. doi: 10.1093/hmg/7.3.449. [DOI] [PubMed] [Google Scholar]
- 37.Giambonini-Brugnoli G, Buchstaller J, Sommer L, Suter U, Mantei N. Distinct disease mechanisms in peripheral neuropathies due to altered peripheral myelin protein 22 gene dosage or a Pmp22 point mutation. Neurobiol Dis. 2005;18(3):656–668. doi: 10.1016/j.nbd.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 38.Fledrich R, et al. A rat model of Charcot-Marie-Tooth disease 1A recapitulates disease variability and supplies biomarkers of axonal loss in patients. Brain. 2012;135(Pt 1):72–87. doi: 10.1093/brain/awr322. [DOI] [PubMed] [Google Scholar]
- 39.Arroyo EJ, Bermingham JR, Rosenfeld MG, Scherer SS. Promyelinating Schwann cells express Tst-1/SCIP/Oct-6. J Neurosci. 1998;18(19):7891–7902. doi: 10.1523/JNEUROSCI.18-19-07891.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fledrich R, et al. Soluble neuregulin-1 modulates disease pathogenesis in rodent models of Charcot-Marie-Tooth disease 1A. Nat Med. 2014;20(9):1055–1061. doi: 10.1038/nm.3664. [DOI] [PubMed] [Google Scholar]
- 41.Arthur-Farraj PJ, et al. c-Jun reprograms Schwann cells of injured nerves to generate a repair cell essential for regeneration. Neuron. 2012;75(4):633–647. doi: 10.1016/j.neuron.2012.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hutton EJ, et al. c-Jun expression in human neuropathies: a pilot study. J Peripher Nerv Syst. 2011;16(4):295–303. doi: 10.1111/j.1529-8027.2011.00360.x. [DOI] [PubMed] [Google Scholar]
- 43.Hantke J, et al. c-Jun activation in Schwann cells protects against loss of sensory axons in inherited neuropathy. Brain. 2014;137(Pt 11):2922–2937. doi: 10.1093/brain/awu257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim Y, et al. The MMP-9/TIMP-1 axis controls the status of differentiation and function of myelin-forming Schwann cells in nerve regeneration. PLoS One. 2012;7(3):e33664. doi: 10.1371/journal.pone.0033664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barrette B, Calvo E, Vallières N, Lacroix S. Transcriptional profiling of the injured sciatic nerve of mice carrying the Wld(S) mutant gene: identification of genes involved in neuroprotection, neuroinflammation, and nerve regeneration. Brain Behav Immun. 2010;24(8):1254–1267. doi: 10.1016/j.bbi.2010.07.249. [DOI] [PubMed] [Google Scholar]
- 46.Barbaria EM, et al. The alpha-chemokine CXCL14 is up-regulated in the sciatic nerve of a mouse model of Charcot-Marie-Tooth disease type 1A and alters myelin gene expression in cultured Schwann cells. Neurobiol Dis. 2009;33(3):448–458. doi: 10.1016/j.nbd.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 47.Srinivasan R, et al. Genome-wide analysis of EGR2/SOX10 binding in myelinating peripheral nerve. Nucleic Acids Res. 2012;40(14):6449–6460. doi: 10.1093/nar/gks313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fledrich R, et al. A rat model of Charcot-Marie-Tooth disease 1A recapitulates disease variability and supplies biomarkers of axonal loss in patients. Brain. 2012;135(Pt 1):72–87. doi: 10.1093/brain/awr322. [DOI] [PubMed] [Google Scholar]
- 49.Krajewski KM, et al. Neurological dysfunction and axonal degeneration in Charcot-Marie-Tooth disease type 1A. Brain. 2000;123(Pt 7):1516–1527. doi: 10.1093/brain/123.7.1516. [DOI] [PubMed] [Google Scholar]
- 50.Mager GM, Ward RM, Srinivasan R, Jang SW, Wrabetz L, Svaren J. Active gene repression by the Egr2.NAB complex during peripheral nerve myelination. J Biol Chem. 2008;283(26):18187–18197. doi: 10.1074/jbc.M803330200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Quintes S, et al. Zeb2 is essential for Schwann cell differentiation, myelination and nerve repair. Nat Neurosci. 2016;19(8):1050–1059. doi: 10.1038/nn.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ryu EJ, et al. Misexpression of Pou3f1 results in peripheral nerve hypomyelination and axonal loss. J Neurosci. 2007;27(43):11552–11559. doi: 10.1523/JNEUROSCI.5497-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Swayze EE, et al. Antisense oligonucleotides containing locked nucleic acid improve potency but cause significant hepatotoxicity in animals. Nucleic Acids Res. 2007;35(2):687–700. doi: 10.1093/nar/gkl1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bauder AR, Ferguson TA. Reproducible mouse sciatic nerve crush and subsequent assessment of regeneration by whole mount muscle analysis. J Vis Exp. 2012;(60):3606. doi: 10.3791/3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tran HT, et al. Α-synuclein immunotherapy blocks uptake and templated propagation of misfolded α-synuclein and neurodegeneration. Cell Rep. 2014;7(6):2054–2065. doi: 10.1016/j.celrep.2014.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14(4):417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.