Abstract
Breast cancer cells with stem cell properties are key contributors to metastatic disease, and there remains a need to better understand and target these cells in human cancers. Here, we identified rare stem-like cells in patients’ tumors characterized by low levels of the proapoptotic molecule p53-upregulated modulator of apoptosis (PUMA) and showed that these cells play a critical role in tumor progression that is independent of clinical subtype. A signaling axis consisting of the integrin αvβ3, Src kinase, and the transcription factor Slug suppresses PUMA in these cells, promoting tumor stemness. We showed that genetic or pharmacological disruption of αvβ3/Src signaling drives PUMA expression, specifically depleting these stem-like tumor cells; increases their sensitivity to apoptosis; and reduces pulmonary metastasis, with no effect on primary tumor growth. Taken together, these findings point to PUMA as a key vulnerability of stem-like cells and suggest that pharmacological upregulation of PUMA via Src inhibition may represent a strategy to selectively target these cells in a wide spectrum of aggressive breast cancers.
Keywords: Oncology, Stem cells
Keywords: Breast cancer
Introduction
Metastasis is the leading cause of breast cancer–related death (1). Despite progress in treating primary breast cancers with chemotherapy, radiation, and surgery, there remains a significant and stubbornly untreatable population of tumors that metastasize to distant organs. To distinguish patients with a high risk of progression, breast cancers are commonly divided into subtypes on the basis of immunohistochemical expression of certain receptors or analysis of gene expression profiles. In addition to tumors expressing the estrogen and progesterone hormone receptors (ER/PR) or human epidermal growth factor receptor 2 (HER2), or lacking all 3 (triple-negative), most patients’ tumors can also be classified according to gene expression profiles into 5 molecular subtypes (luminal A, luminal B, HER2, basal-like and claudin-low) (2–4). While these subtypes may be useful for predicting the course of the disease and response to therapy in some patients, they fail to adequately distinguish which tumors will metastasize, resulting in the overtreatment of many women diagnosed with “aggressive” subtypes. Additionally, while triple-negative or basal-like subtypes are often emphasized in metastasis research, metastases can form in patients diagnosed with most subtypes, although the latency periods may differ significantly (5).
In addition to these global differences between individual patient’s tumors, breast cancers also display significant intratumoral heterogeneity. A single tumor may contain several distinct types of cancer cells representing different mammary lineages (6). This is best exemplified by subpopulations of tumor-initiating cells present in aggressive breast cancers, some of which possess similarities with normal mammary stem cells (MaSCs) (2, 7–11). Cell-surface markers such as CD44+CD24lo (7) and CD49f+EpCAMlo (12) are commonly used to enrich for tumor-initiating cells, and these cells display similarities with the claudin-low molecular subtype, including expression of mesenchymal proteins such as Slug and vimentin (12–14). While these cell populations are routinely used to enrich for tumor cells with stem-like characteristics, they have very limited capacity in this respect, since they represent only general markers of cells that reside within the basal cell layer of the normal mammary gland (12, 15). This property makes these markers overly broad and limits their ability to identify tumors that are likely to progress (16). A better understanding of the mechanisms that regulate normal MaSC behavior will allow identification in patients’ tumors of the relevant stem-like cell types that are important for tumor progression.
Our previous studies show that the integrin αvβ3 plays a critical role in MaSCs important for mammary gland remodeling during pregnancy. Whereas both luminal progenitors and MaSCs in the normal mammary gland express αvβ3, we show that it is specifically required for MaSC activation and remodeling during pregnancy, with no effect on mammary development (17), which is consistent with a broad role for αvβ3 as a marker of activated cell types (18). Importantly, αvβ3 promotes stem cell expansion by enhancing expression of the transcription factor Slug, a master regulator of mammary stemness (19) and remodeling (20). Therefore, the presence of Slug distinguishes αvβ3-expressing MaSCs from luminal progenitors that also express this integrin. These results show that hormonal cues, such as those present during pregnancy, are important for mobilizing MaSCs from an otherwise less active state (21, 22) and highlight the coexpression of αvβ3 and Slug as markers of activated MaSCs. Our findings from the normal mammary gland suggest that tumor cells bearing an activated stem cell profile may represent particularly aggressive breast cancer cells that are distinct from those identified with basal cell markers alone.
In the present study, we use our unique insight from the normal mammary gland to examine the contribution of cells bearing an activated MaSC profile to breast cancer progression. Unexpectedly, in patients’ tumor samples, coexpression of αvβ3 and Slug identified rare stem-like cells that were associated with tumor progression, independent of hormone receptor status or molecular subtype. While these cells expressed traditional stem-like profiles including CD44+CD24lo, CD49f+EpCAMlo, epithelial-mesenchymal transformation (EMT), and the claudin-low molecular subtype, they are not synonymous with these classical markers but instead represent a distinct stem-like cell population. Importantly, we identify expression of the proapoptotic molecule p53-upregulated modulator of apoptosis (PUMA) as a key vulnerability of these cells. While PUMA is normally suppressed by an αvβ3/Src/Slug signaling axis, we found that disruption of this pathway could drive PUMA expression, specifically targeting stem-like cells and decreasing metastasis. Altogether, our findings define PUMA as a critical metastasis suppressor that may be pharmacologically induced by Src inhibition to target stem-like cells in a broad spectrum of aggressive human breast cancers.
Results
Rare αvβ3+ tumor cells identify aggressive breast cancers across clinical subtypes.
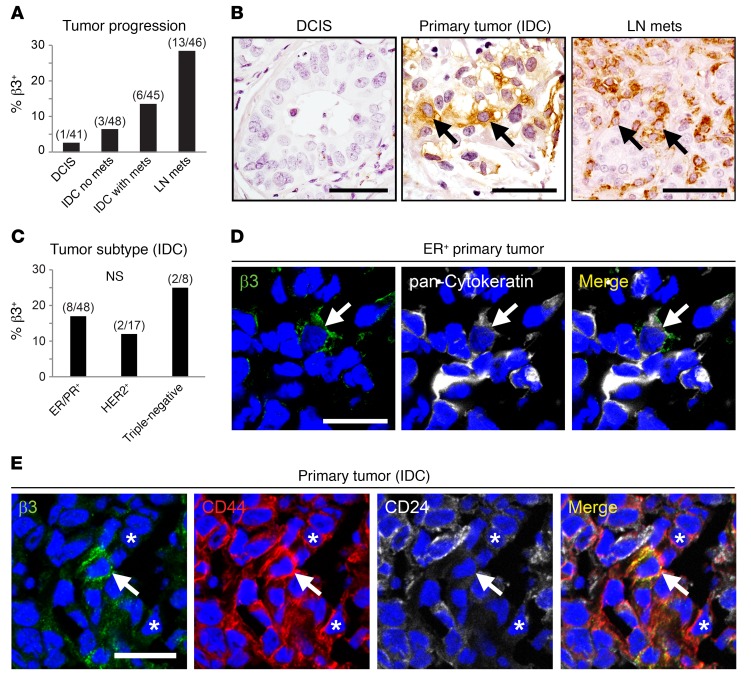
We previously identified the integrin αvβ3 as an important regulator of MaSC behavior during epithelial remodeling events associated with pregnancy (17). To explore whether this developmental function for αvβ3 in activated MaSCs may be related to its role in aggressive breast cancers, we first assessed αvβ3 expression in patients’ tumor samples. Immunohistochemical staining of a breast cancer microarray showed that tumor cell αvβ3 was associated with breast cancer progression (Figure 1, A and B). While αvβ3 was generally absent from premalignant ductal carcinoma in situ (DCIS), expression became more frequent in primary tumors from patients with invasive ductal carcinoma (IDC) and was most pronounced in lymph node metastases (Figure 1, A and B). Abundant αvβ3 in tumor blood vessels precluded the use of β3 mRNA levels for assessing tumor cell expression of this integrin in patients’ samples. Despite the association with more aggressive disease, analysis of tumors representing different clinical subtypes unexpectedly showed that αvβ3+ cells were not limited to any particular subtype (Figure 1C). Instead, we found these cells to reside in tumors representing all 3 major subtypes including ER+/PR+, HER2+, and triple-negative cancers (Figure 1C). This tumor cell expression of αvβ3 was confirmed by costaining with pan-cytokeratin (Figure 1D). Interestingly, our staining results showed that αvβ3 was limited to only a small fraction of tumor cells (Figure 1, B and D), consistent with the possibility that they may possess stem cell characteristics. Indeed, αvβ3+ tumor cells coexpressed the CD44+CD24lo stem profile (7) in frozen tumor samples from patients with breast cancer (Figure 1E and Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI93707DS1). Intriguingly, numerous CD44+CD24lo cells lacking αvβ3 could also be found in these cancers (Figure 1E and Supplemental Figure 1A), indicating that αvβ3+ cells may represent a distinct stem-like cell subset present in a wide range of aggressive tumors.
Figure 1. In breast cancer patients’ samples, αvβ3 identifies stem-like cells associated with tumor progression, independent of clinical subtype.
(A–C) Immunohistochemical staining for β3 in breast cancer tissue microarrays representing various stages of disease progression (A and B)and clinical breast cancer subtypes (C). (A) Histogram displaying the percentage of tumors containing β3+ cells at different stages of progression of breast cancer including DCIS, IDC, with or without metastases (mets), and lymph node metastases (LN mets). P = 0.0001, by χ2 test for trend. (B) Representative examples of β3-expressing tumor cells (arrows) are shown for the breast cancer patients’ samples in A. (C) Quantitation of the frequency of β3+ tumors in different clinical breast cancer subtypes, including tumors expressing ER+/PR+ or HER2+ receptors or lacking all 3 (Triple-negative). (A and C) Numbers above each bar indicate the β3+ tumors per total number of tumors for each category. (D and E) Representative examples of immunofluorescence staining for β3 and pan-cytokeratin (D) or CD44 and CD24 (E) in frozen human breast cancer tissue microarrays. Arrows indicate β3+ cells coexpressing cytokeratins (D) or the stem profile CD44+CD24lo (E). Nuclei are stained blue in all panels. Asterisks in E mark CD44+CD24lo cells that lack β3. n = 6/17 (β3+CD44+CD24lo tumors/total). Scale bars: 50 μm (B); 20 μm (D and E). See also Supplemental Figure 1.
Coexpression of αvβ3 and Slug defines a unique subset of stem-like cells but does not contribute to EMT.
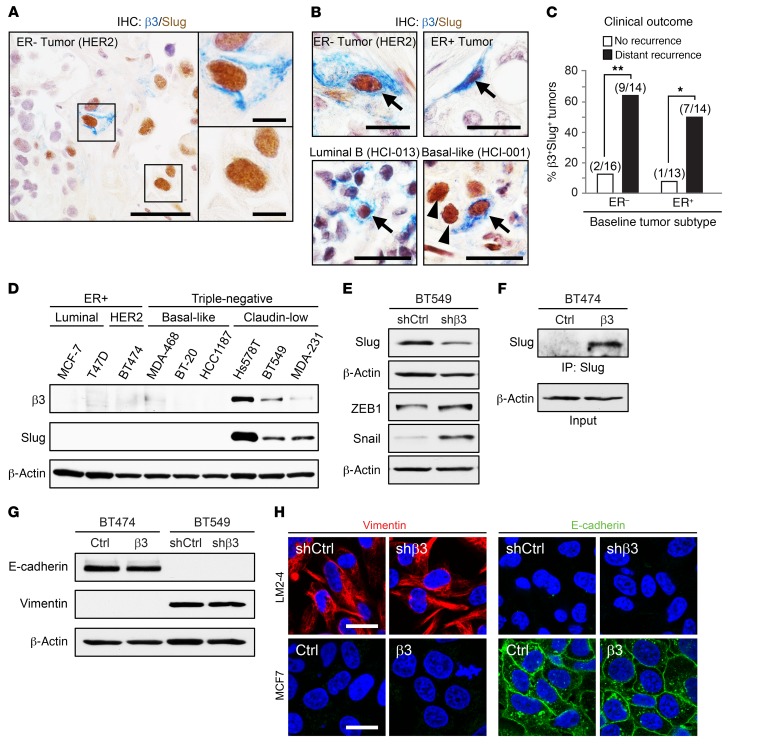
In the normal adult mammary gland, we previously found that αvβ3 was expressed by both luminal progenitor cells and MaSCs (17). We further showed in MaSCs that αvβ3 is critical for expression of the transcription factor Slug (17), a master regulator of mammary stemness (19). Therefore, Slug distinguished αvβ3-expressing MaSCs from luminal progenitors that also expressed this integrin. To examine which cell type was more similar to αvβ3-expressing cells in breast cancer, we used IHC to assess whether αvβ3 and Slug were coexpressed in patients’ tumor samples. Consistent with the stem-like nature of these cells, we identified a tumor cell subset coexpressing αvβ3 and Slug that occurred in approximately 15% to 20% of all primary breast cancers (Figure 2A), whereas cells expressing αvβ3 alone were decidedly rare. Notably, we also identified numerous Slug+ cancer cells lacking αvβ3 in these tumors (Figure 2A), indicating that αvβ3+Slug+ cells represent a unique subset of Slug-expressing cells in human breast cancers. In fact, both Slug+ and CD44+CD24lo cells were surprisingly frequent in breast cancers, sometimes comprising the majority of cells in a given tumor. Conversely, αvβ3+ cells were never abundant in any of the more than 400 primary tumor samples examined. Consistent with our staining data for αvβ3 alone, we found no association between αvβ3+Slug+ cells and any particular cell subtype (Figure 2B and Supplemental Figure 2A). Analysis of tissue microarrays showed that αvβ3+Slug+ cells were similarly represented in ER+/PR+, HER2+, and triple-negative disease, both in terms of frequency and cell numbers (Supplemental Figure 2, B and C). In addition to ER– and ER+ tumors, we also identified αvβ3+Slug+ cells in cancers representing the luminal B and basal-like molecular subtypes (Figure 2B). These surprising observations characterize our αvβ3+Slug+ cells as a distinct subset of stem-like tumor cells present in a diverse array of clinical breast cancer subtypes.
Figure 2. Coexpression of αvβ3 and Slug reveals unique stem-like cells in a broad-spectrum of clinical subtypes but does not impact EMT.
(A and B) Representative images of immunohistochemical staining for β3 (blue) and Slug (brown) in breast cancer samples from patient-derived xenografts (A and B, bottom) and tissue microarrays (B, top). (A) Shown is a tumor with heterogeneous staining for β3 and Slug. n = 5/30, β3+Slug+ tumors/total. Scale bars: 40 μm and 10 μm (enlarged insets). (B) β3+Slug+ cells (arrows) are shown for ER– and ER+ tumors (top) as well as tumors representing different intrinsic molecular subtypes (bottom). (B, top) n = 19/125, β3+Slug+ tumors/total: n = 10, ER+; n = 4, HER2+; n = 5, triple-negative (TN). (B, bottom) n = 4/12, β3+Slug+ tumors/total: n = 2, luminal B; n = 2, basal-like. Scale bars: 20 μm. (C) Frequency of β3+Slug+ cells in immunohistochemically stained “baseline” breast cancer samples from recurrence-free patients and patients who later progressed to form distant recurrences. Numbers above each bar indicate the number of β3+Slug+ tumors per total number of tumors. P = 0.0068 (ER–) and P = 0.0329 (ER+), for no recurrence versus distant recurrence. *P < 0.05 and **P < 0.01. Statistical analysis was performed by Fisher’s exact test. (D–G) Western blot analysis for the indicated proteins in a panel of breast cancer cell lines representing distinct subtypes (D), BT549 cells stably expressing β3 shRNA (shβ3) or a nonsilencing control shRNA (shCtrl) (E and G), or BT474 cells expressing ectopic β3 cDNA (β3) or a vector-only control (Ctrl) (F and G). For all immunoblots, data shown are representative of 3 independent experiments, and β-actin was used as a loading control. (H) Representative immunofluorescence images showing vimentin (red) and E-cadherin (green) in LM2-4 and MCF7 cells, with or without β3. Nuclei are stained blue in all panels. Scale bars: 20 μm. See also Supplemental Figures 2 and 3.
To examine a potential association between our αvβ3+Slug+ cells and clinical outcome, we examined patients’ samples collected as part of the Women’s Healthy Eating and Living (WHEL) clinical study (23). This large clinical study collected baseline tumor biopsies from patients, along with information on their associated ER status, and provided follow-up data on each patient over a period of approximately 10 years. Therefore, we were able to acquire tumor sections representing ER– and ER+ cancers from age-matched patients who either developed distant recurrences at some point during the study period or remained recurrence-free. Our staining of these tumor sections showed an association between αvβ3+Slug+ cells and distant recurrence in both ER– and ER+ breast cancers compared with tumors from recurrence-free patients (Figure 2C). We identified αvβ3+Slug+ cells in 64% of ER– tumors and 50% of ER+ tumors from patients who developed distant recurrences compared with only 12% and 8%, respectively, in tumors from patients with no recurrences. Thus, our findings show that αvβ3+Slug+ cells in patients’ tumor samples are associated with disease progression, independent of ER status.
To further characterize the tumor cell types that coexpress αvβ3 and Slug, we examined the expression of αvβ3 and Slug in cell lines representing different breast cancer subtypes based on hormone receptor status or gene expression profile (2, 13). Consistent with our results from patients’ tumors, both αvβ3 and Slug were only expressed in cell lines representing the claudin-low molecular subtype (Figure 2D and Supplemental Figure 2D), which are enriched for the CD44+CD24lo marker profile (14). Additionally, the claudin-low tumor subtype is the most similar to MaSCs in the normal mammary gland (2, 12), in agreement with the role that we characterized for αvβ3 and Slug in MaSCs during pregnancy (17). Further, coexpression of αvβ3 and Slug in claudin-low cells did not appear to depend on any particular genomic mutation, since each of the cell lines examined contained a different oncogenic driver (Supplemental Figure 2D). Thus, cell lines coexpressing αvβ3 and Slug share many properties with similar cells in patients’ tumors, supporting their use as in vitro models for exploring the contribution of αvβ3/Slug signaling to stem-like tumor cells and their role in breast cancer progression.
We previously showed that αvβ3 was required for Slug expression in MaSCs during early pregnancy (17), suggesting that a similar relationship may exist in stem-like breast cancer cells. Indeed, stable shRNA knockdown of endogenous β3 in BT549 cells reduced Slug protein expression (Figure 2E and Supplemental Figure 2E), whereas ectopic expression of β3 was sufficient to drive Slug expression in BT474 cells (Figure 2F and Supplemental Figure 2F). Together, these findings show that αvβ3 is necessary and sufficient to drive Slug expression in breast cancer cells, consistent with our prior results from other cell lines (17). Importantly, these effects were specific to Slug, as β3 knockdown not only failed to reduce other EMT transcription factors such as ZEB1 and Snail, but actually caused them to increase (Figure 2E). Despite this role in promoting Slug expression, we observed no effect of αvβ3 on EMT markers (Figure 2, G and H, and Supplemental Figure 3, A–D). Analysis of multiple stable breast cancer cell lines with β3 knockdown or ectopic β3 expression (Supplemental Figure 2, E and F) showed no changes in the EMT markers vimentin or E-cadherin by immunoblotting (Figure 2G and Supplemental Figure 3, A and B) or immunofluorescence staining (Figure 2H and Supplemental Figure 3D). Similarly, αvβ3 had little effect the on the mRNA expression of other EMT-related genes such as HDAC1, MUC1, and CLDN1, as assessed by quantitative PCR (qPCR) (Supplemental Figure 3C). Thus, our results show that αvβ3 has a limited effect on EMT markers in breast cancer cells, despite being necessary and sufficient for Slug expression. This is consistent with a recently published finding that Snail, and not Slug, is the primary EMT transcription factor in breast cancer (24). Given these observations, we propose that αvβ3 directs a non-EMT function for Slug in stem-like breast cancer cells.
PUMA suppression by αvβ3 and Slug specifically promotes anchorage independence.
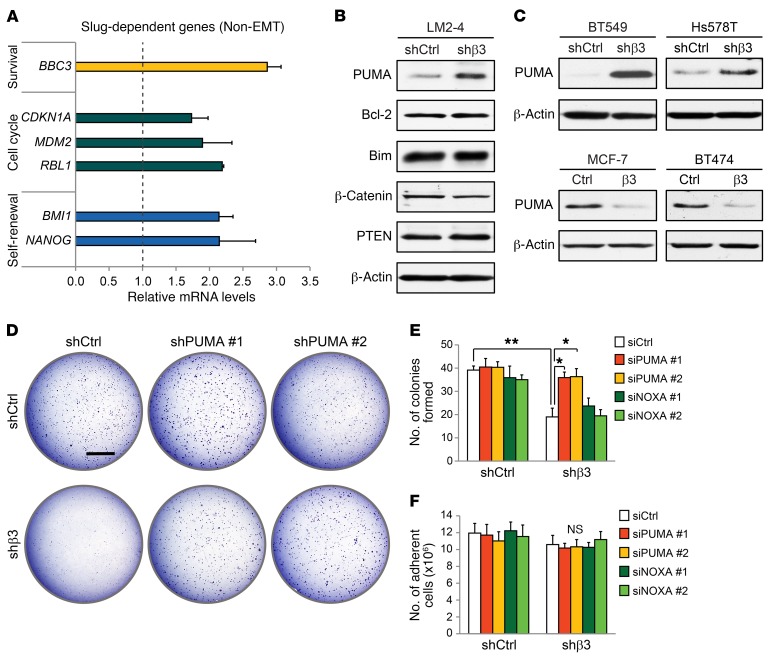
In addition to EMT, Slug is a transcriptional repressor of several genes important for cell survival, cell-cycle progression, and self-renewal (25). To explore the genes most impacted by αvβ3-driven Slug expression, we assessed mRNA levels in the highly metastatic LM2-4 breast cancer cell line (26) expressing control or β3 shRNA. Interestingly, BBC3, the gene for PUMA, showed the largest increase (≈3-fold) compared with the control (Figure 3A), with a similar increase in PUMA protein levels, confirmed by immunoblotting, relative to other Slug-dependent proteins (Figure 3B). A member of the proapoptotic BH3-only subset of the Bcl-2 protein family, PUMA is an important p53 effector that interacts with Bcl-2 to promote apoptosis (27). However, we observed no effect of αvβ3 on the protein expression of Bcl-2 in these cells (Figure 3B). As the sole BH3-only family member regulated by Slug (28), we show that PUMA is indeed suppressed by Slug in breast cancer cells (Supplemental Figure 4, A and B). In fact, others have shown that PUMA is specifically repressed by Slug and not other EMT transcription factors, such as Snail (29). This may account for the more pronounced increase in PUMA in our β3-knockdown cells, since Snail may compensate for the effects of reduced Slug expression on other genes. Our findings were not limited to LM2-4 cells, since αvβ3 was both necessary and sufficient to suppress PUMA mRNA and protein expression in several other breast cancer cell lines as well (Figure 3C and Supplemental Figure 4C). These findings show that αvβ3 directs a non-EMT role for Slug in suppressing PUMA expression, which may be important for controlling the behavior of stem-like cells.
Figure 3. PUMA is a critical Slug-dependent gene suppressed by αvβ3 to promote anchorage independence.
(A) qPCR analysis of non-EMT, Slug-dependent genes in β3-knockdown LM2-4 cells. Fold change (2–ΔΔCT) in β3-knockdown cells is relative to cells expressing a control nonsilencing shRNA. Cyclophilin A was used as a loading control. (B and C) Immunoblots showing PUMA protein levels in breast cancer cell lines expressing either control nonsilencing shRNA or β3 shRNA (B and C, top) or ectopic β3 cDNA (β3) or a vector-only control (C, bottom). β-Actin was used as a loading control. (D) Soft agar tumorsphere assay comparing PUMA shRNA (shPUMA) knockdown in control and β3-knockdown LM2-4 cells. Scale bar: 2 mm. (B–D) Data shown are representative of 3 independent experiments. (E and F) Quantitation of tumorsphere formation per field (E) and total number of adherent cells (F) after siRNA knockdown of PUMA (siPUMA) or NOXA (siNOXA) compared with control siRNA (siCtrl) in LM2-4 cells, with or without β3. (E) P = 0.0072 (siCtrl; shCtrl vs. shβ3); P = 0.0331 (shβ3; siCtrl vs. siPUMA no. 1); P = 0.0275 (shβ3; siCtrl vs. siPUMA no. 2); (E and F) *P < 0.05 and **P < 0.01. Statistical analysis was performed by 2-way ANOVA with Tukey’s multiple comparisons test. Data represent the mean ± SEM. n = 3 independent experiments (A, E, and F). Each sample was run in triplicate. See also Supplemental Figure 4.
Our prior studies found that αvβ3 is an important driver of anchorage independence, a hallmark of aggressive tumors and an important property of stem-like cells (17, 30, 31). We therefore examined whether PUMA suppression may be important for these effects. Specifically, we assessed whether PUMA knockdown could rescue defective anchorage-independent tumorsphere formation in β3-knockdown cells. As expected, β3-deficient cells formed fewer soft agar colonies than did control cells (Figure 3D and Supplemental Figure 4, D and E). We further showed that this was due to increased PUMA expression, since knockdown with either of 2 PUMA shRNAs specifically rescued colony numbers in our β3-deficient cells, with no impact on control cells (Figure 3D and Supplemental Figure 4, D and E), an effect replicated by transient PUMA knockdown with either of the 2 siRNAs (Figure 3E and Supplemental Figure 4F). Importantly, these effects were specific to PUMA, since reduced expression of NOXA, a related Bcl-2 family member, failed to rescue αvβ3-dependent colony formation in these experiments (Figure 3E and Supplemental Figure 4F). Overall, these findings point to a role for αvβ3 in aiding the survival of stem-like tumor cells by suppressing PUMA expression. Consistent with this, blocking apoptosis with a pan-caspase inhibitor rescued colony formation specifically in β3-knockdown cells (Supplemental Figure 4G), supporting the idea that αvβ3 plays a role in prosurvival signaling. Interestingly, PUMA knockdown failed to affect adherent cell survival (Figure 3F). Thus, the increased PUMA expression we observed as a result of β3 knockdown was only sufficient to promote cell death in anchorage-independent conditions and did not appear to affect adherent cells. This is consistent with our prior work characterizing an adhesion-independent role for αvβ3 (30). These new findings identify an unexpected function for αvβ3 in suppressing PUMA-mediated cell death, which contributes to tumorsphere formation, a hallmark of stem-like cells and aggressive disease.
PUMA is a metastasis suppressor.
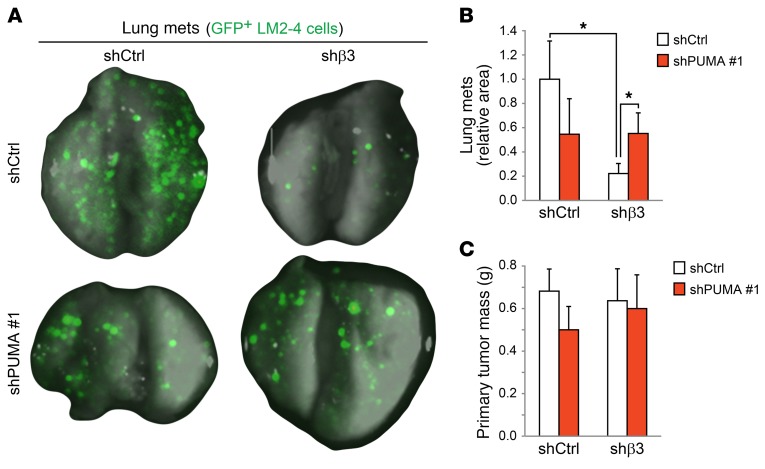
Stem-like cells play an important role in tumor progression and metastasis (32). Since our findings show that αvβ3 deficiency drives PUMA expression and reduces tumorsphere formation in vitro, this suggests that PUMA may similarly be responsible for reducing the tumor growth and metastasis of stem-like cells in vivo. To assess this possibility, we examined whether the increased PUMA expression associated with β3 knockdown is sufficient to suppress tumor growth and metastasis in the LM2-4 cell line and whether this effect could be rescued by stable PUMA shRNA knockdown (Figure 4, A–C and Supplemental Figure 4D). For these experiments, we injected GFP-labeled LM2-4 cells orthotopically into the mammary fat pads of adult female NOD/SCID/IL-2 receptor γ chain–KO mice (NSG) mice and assessed primary tumor growth and metastasis after 6 weeks. Pulmonary metastasis was quantified by calculating the area of the lungs covered by GFP+ cells. We observed that β3 knockdown caused a nearly 5-fold reduction in metastasis relative to controls (Figure 4, A and B). Importantly, increased PUMA levels contributed directly to this effect, as PUMA shRNA knockdown rescued lung metastasis specifically in β3-deficient cells (Figure 4, A and B). These findings reveal the role of PUMA as an important metastasis suppressor. Interestingly, the increased PUMA levels associated with β3 knockdown were not sufficient to affect primary tumor mass (Figure 4C), suggesting that disseminated tumor cells may be more vulnerable to PUMA than are primary tumor cells. Similarly, PUMA shRNA alone failed to affect primary tumor size, consistent with the low endogenous expression levels of PUMA in these cells (Figure 4C). These in vivo findings are in agreement with the specific role we observed for PUMA in anchorage-independent tumorsphere assays in vitro (Figure 3, E and F), indicating that these assays more closely resemble the stresses present during metastasis compared with adherent cell growth. Taken together, our results suggest that PUMA upregulation may be an important vulnerability of αvβ3+Slug+ stem-like cells that serves to reduce tumor progression.
Figure 4. PUMA suppression rescues metastasis in β3-knockdown cells.
(A–C) In vivo orthotopic breast cancer experiments comparing the effect of PUMA knockdown on lung metastasis or primary tumor growth in LM2-4 cells, with or without β3. (A) Representative images of whole mouse lungs with GFP-expressing metastases for each of the indicated LM2-4 stable cell types. (B) Bar graph shows the relative area of each lung covered by GFP-expressing metastases for each cell type. P = 0.0416 (shCtrl; shCtrl vs. shβ3); P = 0.0142 (shβ3; shCtrl vs. shPUMA no. 1); *P < 0.05. Statistical analysis was performed by 1-way ANOVA with Tukey’s multiple comparisons test. (C) Primary tumor mass for each cell type. Data in B and C represent the mean ± SEM. n = 6, shCtrl; n = 5, shPUMA no. 1; n = 7, shβ3 and shCtrl; n = 7, shPUMA no. 1. Data are from 2 independent experiments.
PUMA is upregulated by blocking αvβ3/Src signaling.
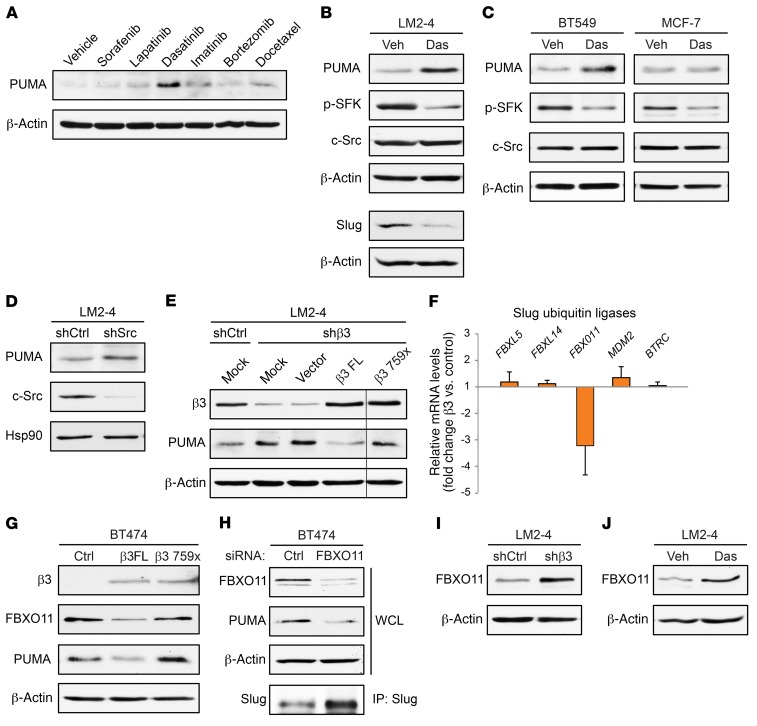
The importance of PUMA suppression for the aggressive behavior of αvβ3+Slug+ cells led us to consider whether there were any therapies available that could induce PUMA expression and therefore target these stem-like cells. Since there are no therapies that directly inhibit Slug, we screened existing clinically approved inhibitors with diverse targets (Supplemental Figure 5A) for their ability to induce PUMA expression in the LM2-4 breast cancer cell line. Surprisingly, only the Src family kinase inhibitor dasatinib was able to significantly increase PUMA expression in these cells at any of the doses examined (Figure 5A and Supplemental Figure 5B). The related inhibitor imatinib failed to increase PUMA (Figure 5A and Supplemental Figure 5B), despite having many of the same targets as dasatinib, with the exception of the Src family kinases (Supplemental Figure 5A). This effect was not limited to dasatinib, as another Src family kinase inhibitor, saracatinib, also induced PUMA expression (Supplemental Figure 5C). Importantly, dasatinib also reduced Slug expression (Figure 5B), consistent with the role of Slug in suppressing PUMA and offering a potential mechanism to account for the ability of dasatinib to enhance PUMA expression. We observed similar results in the BT549 stem-like cells, but not in the non-stem MCF7 cells (Figure 5C), suggesting that the ability of dasatinib to increase PUMA is specific to αvβ3+Slug+ cells. In addition to pharmacological inhibition of Src kinase activity, shRNA knockdown of c-Src also induced PUMA expression (Figure 5D). Furthermore, αvβ3 activation of Src is required for its ability to suppress PUMA, as a β3 Src-binding domain mutant was unable to rescue PUMA expression compared with the full-length β3 subunit (Figure 5E). These findings highlight an important role for αvβ3/Src/Slug signaling in suppressing PUMA expression in stem-like cells.
Figure 5. Disrupting αvβ3/Src signaling specifically drives PUMA expression in stem-like cells.
(A) PUMA immunoblot of LM2-4 cells treated with the indicated drugs for 24 hours. All drugs were dosed at 100 nM, except for bortezomib and docetaxel, which were added at a sublethal concentration of 5 nM. (B and C) Immunoblots of the indicated cell types after treatment with DMSO vehicle control (Veh) or 100 nM dasatinib (Das) for 24 hours. (D and E) Western blot analysis of LM2-4 cells stably expressing c-Src shRNA (shSrc) or a nonsilencing shRNA control (shCtrl) (D) or transiently transfected with mock, vector alone, full-length β3 cDNA (β3 FL), or a β3 Src–binding domain mutant (β3 759x) (E). (E) All lanes were run on the same gel, but the β3 759x lane was noncontiguous (black line). (F) qPCR analysis of ubiquitin ligases that regulate Slug expression. Fold change (2–ΔΔCT) in BT474 cells stably expressing β3 cDNA is shown relative to vector control cells. β-Actin was used as a loading control. n = 3 independent experiments. Each sample was run in triplicate. Data represent the mean ± SEM. (G–J) Western blot analysis for the indicated proteins in BT474 cells stably expressing vector control, full-length β3 cDNA, or the β3 759x mutant (G); BT474 cells with transient siRNA knockdown of FBXO11 (H); LM2-4 β3–knockdown cells (I); and LM2-4 cells treated with 100 nM dasatinib (J). (A–E and G–J) β-Actin or Hsp90 was used as a loading control. Data shown are representative of 3 independent experiments. See also Supplemental Figure 5.
We further investigated a potential mechanism to explain this unanticipated effect of αvβ3/Src signaling on Slug and PUMA. We previously showed that αvβ3 promoted Slug protein stability in normal mammary cells, with no effect on Slug mRNA (17). Consistent with these results, ectopic β3 expression in BT474 cells significantly upregulated Slug protein levels (Figure 2F), but not SLUG mRNA (Supplemental Figure 5D). Taken together, these findings suggest that genes involved in regulating Slug protein stability may play a role in our pathway. Slug is a highly labile protein that is rapidly degraded by the proteasome following ubiquitination, and several ubiquitin ligases that decrease Slug protein expression in various cell types have been well described (33–37). We therefore examined whether any of these ubiquitin ligases previously characterized to decrease Slug expression may be downregulated by αvβ3/Src signaling. Surprisingly, we found that only 1 of these genes, FBXO11, was consistently decreased in BT474 cells expressing ectopic β3 by over 3-fold relative to vector control cells (Figure 5F). Ectopic β3 also reduced FBXO11 protein expression, and this required Src activation by αvβ3 (Figure 5G). Importantly, these decreased levels of FBXO11 due to ectopic β3 corresponded with reduced levels of PUMA in the same cells (Figure 5G). This is consistent with a previous study (35) showing that low levels of FBXO11 cause Slug to increase, resulting in PUMA suppression. Indeed, we now show that siRNA knockdown of FBXO11 not only increases Slug, but causes a reciprocal decrease in PUMA as well (Figure 5H). Additionally, reduced levels of either αvβ3 (Figure 5I) or Src activation (Figure 5J) increase FBXO11 expression in LM2-4 cells. Thus, we have uncovered what we believe to be a unique role for αvβ3/Src signaling in reducing the expression of a ubiquitin ligase known to suppress Slug, offering a potential explanation as to how Src inhibition may result in increased PUMA expression. Together, our findings highlight a role for Src in suppressing PUMA and reveal Src inhibition as a therapeutic approach to drive PUMA expression in stem-like breast cancer cells.
PUMA upregulation selectively targets stem-like cells in distinct tumor subtypes.
Breast cancers are heterogeneous, with cells representing several different mammary lineages often found in the same tumor, including populations of aggressive stem-like cells. We show in patients’ tumors that cells with the αvβ3+Slug+ stem profile represent a rare tumor cell subset associated with disease progression in a diverse array of breast cancer subtypes (Figure 1, A–E and Figure 2, A–C). Our findings suggest that PUMA expression induced by Src inhibition may specifically target these cells, even in tumors of different subtypes. To investigate this possibility in a situation that more closely resembles patients’ tumors, we chose the heterogeneous HCC38 and HCC1143 breast cancer cell lines, which represent the claudin-low and basal-like molecular subtypes, respectively (13). Previous studies have shown that both cell lines can be separated into 2 populations on the basis of surface expression of CD49f and epithelial cell adhesion molecule (EpCAM) (13). The CD49f+EpCAMhi cells represent a luminal-like cell type, whereas a CD49f+EpCAMlo profile exhibits MaSC properties (13). Thus, the HCC38 and HCC1143 breast cancer cell lines represent well-characterized heterogeneous models of distinct subtype that we could explore for the presence of αvβ3+Slug+ cells and their ability to upregulate PUMA in response to Src inhibition.
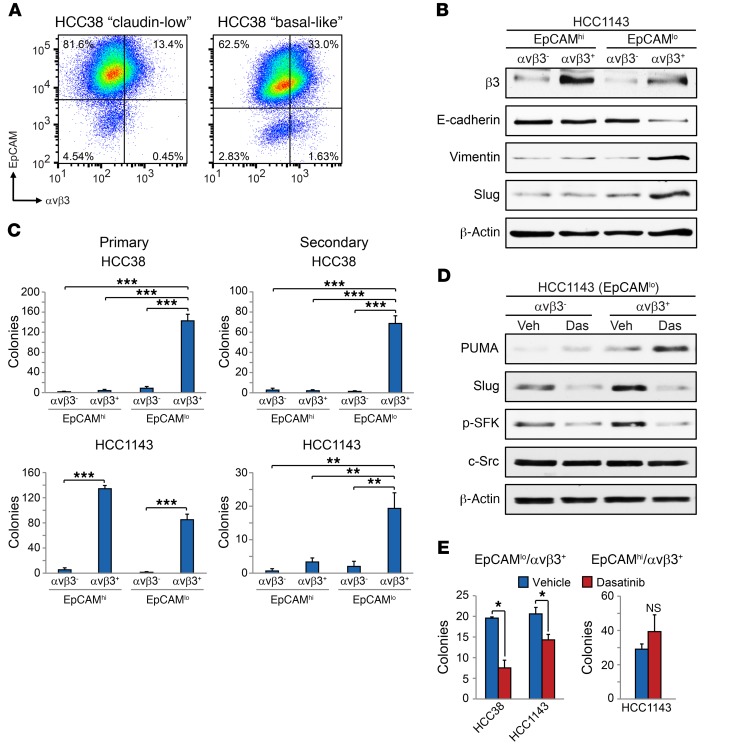
Given our previous results, we hypothesized that αvβ3 would be limited to the stem-like CD49f+EpCAMlo population in each cell type. Indeed we observed αvβ3 expression in a small fraction of EpCAMlo cells in both cell lines, representing approximately 0.5% of HCC38 and less than 2% of HCC1143 cells compared with controls (Figure 6A and Supplemental Figure 6A). These findings indicate that αvβ3 expression is not synonymous with either the CD49f+EpCAMlo profile or the claudin-low gene signature. Further analysis of these cell lines with the CD44+CD24–EpCAM+ (ESA) cancer stem cell marker profile (38) also showed minimal overlap with αvβ3 expression (Supplemental Figure 6B). Importantly, these rare EpCAMloαvβ3+ cells expressed a stem marker profile including high expression of Slug and vimentin and low levels of E-cadherin (Figure 6B). In contrast EpCAMhi cells expressing αvβ3 (Figure 6A) did not enrich for stem markers (Figure 6B). As further evidence of their stem-like properties, EpCAMloαvβ3+ cells were enriched for primary tumorsphere formation in both methylcellulose (Figure 6C) and soft agar (Supplemental Figure 6, C and D) and uniquely possessed the ability to self-renew, as assessed by secondary tumorsphere formation (Figure 6C). While the EpCAMhiαvβ3+ HCC1143 cells formed primary tumorspheres, they failed to self-renew (Figure 6C). These properties are reminiscent of luminal progenitor cells that also express αvβ3 in the normal mammary gland (39) and suggest that EpCAMhiαvβ3+ cells represent a distinct tumor cell population compared with EpCAMloαvβ3+ cells. Thus, as with patients’ tumors, we show that the HCC38 and HCC1143 cell lines contain a rare subset of αvβ3+ cells with stem-like properties, in addition to non–stem cell types.
Figure 6. Src inhibition upregulates PUMA and depletes stem-like cells in distinct breast cancer subtypes.
(A) Representative FACS density plots of the HCC38 and HCC1143 breast cancer cell lines showing the live, CD49f+ cells according to their cell-surface EpCAM and αvβ3 expression. (B) Immunoblot of the indicated FACS-sorted cell populations from HCC1143 cells. β-Actin was used as a loading control. (C) Primary and secondary tumorsphere assays in methylcellulose for the indicated sorted cell populations. Shown are the total number of colonies per well. Primary tumorspheres, HCC38; P < 0.0001 for EpCAMloαvβ3+ versus all other cell types, HCC1143; P < 0.0001 for EpCAMhiαvβ3– versus EpCAMhiαvβ3+ and EpCAMloαvβ3– versus EpCAMloαvβ3+. Secondary tumorspheres, HCC38; P < 0.0001 for EpCAMloαvβ3+ versus all other cell types, HCC1143 (EpCAMloαvβ3+); P = 0.0037 (vs. EpCAMhiαvβ3–), P = 0.0094 (vs. EpCAMhiαvβ3+), P = 0.0059 (vs. EpCAMloαvβ3–). Statistical analysis was performed by 1-way ANOVA with Tukey’s multiple comparisons test. n = 3 independent experiments. (D) Western blot analysis of the indicated HCC1143 sorted cells treated with DMSO vehicle or 100 nM dasatinib for 24 hours. β-Actin was used as a loading control. Data shown in A, B, and D are representative of 3 independent experiments. (E) Bar graphs show the number of tumorspheres per field formed in the indicated HCC38 and HCC1143 cell populations after treatment with vehicle (DMSO) or 500 nM dasatinib. The P values for vehicle versus dasatinib are as follows: P = 0.0208 (HCC38; EpCAMloαvβ3+); P = 0.0384 (HCC1143; EpCAMloαvβ3+); P = 0.4060 (HCC1143; EpCAMhiαvβ3+). Statistical analysis was performed by Student’s t test (E). n = 3 independent experiments performed in triplicate. Data in C and E represent the mean ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001. See also Supplemental Figure 6.
To compare the ability of Src inhibition to upregulate PUMA in our different sorted cell types, we treated EpCAMloαvβ3+ and EpCAMloαvβ3– sorted HCC1143 cells with dasatinib and analyzed them for PUMA protein expression. Interestingly, dasatinib treatment induced PUMA and reduced Slug expression specifically in EpCAMloαvβ3+ cells compared with EpCAMloαvβ3– cells (Figure 6D). We further showed that this increased PUMA expression was associated with fewer EpCAMloαvβ3+ tumorspheres in both cell lines after dasatinib treatment (Figure 6E). Importantly, the HCC1143 EpCAMhiαvβ3+ cells, which also represented a highly clonogenic cell type (Figure 6C and Supplemental Figure 6, D and E), were unaffected by dasatinib treatment (Figure 6E), further distinguishing these cells as a distinct tumor cell population. Our findings show that, even in heterogeneous tumors representing different subtypes, Src inhibition specifically drives PUMA expression in stem-like cells and reduces tumorsphere formation, in support of our data from more homogeneous cell types. Thus, PUMA upregulation may be an effective strategy for targeting stem-like cells to prevent tumor progression.
PUMA upregulation is a key vulnerability of stem-like cells.
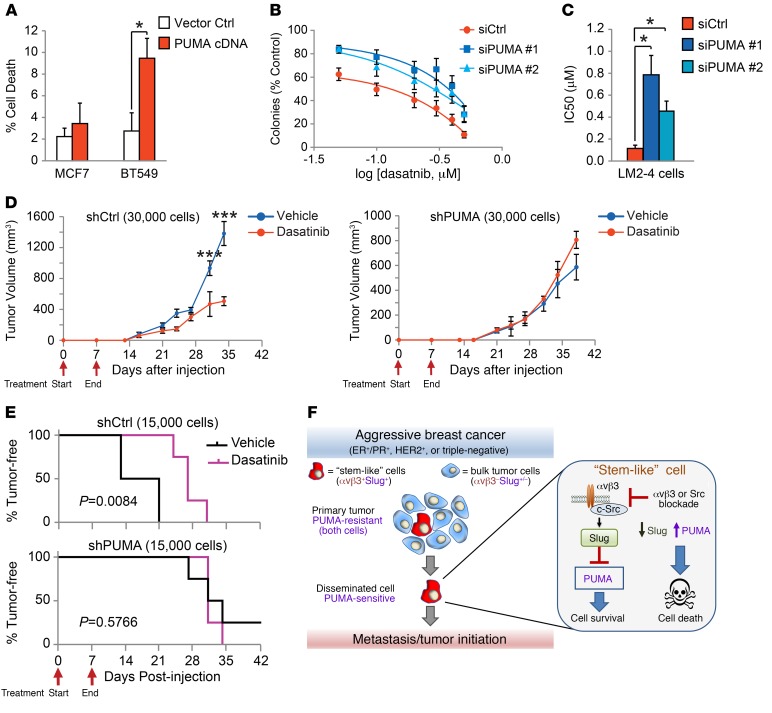
The increased PUMA expression driven by genetic or pharmacological blockade of αvβ3 or Src suggests that this may represent a critical vulnerability of our stem-like cells. To examine this possibility, we transiently expressed PUMA cDNA (27) or an empty vector control in MCF7 or BT549 cells and measured cell death by trypan blue staining (Figure 7A). While PUMA was sufficient to induce cell death in the BT549 cells, MCF7 cells were relatively unaffected (Figure 7A), despite high levels of ectopic PUMA expression (Supplemental Figure 7A). This suggests that PUMA was sufficient to induce cell death in our αvβ3+Slug+ cells, in contrast to other breast cancer cell types. Additionally, we used dasatinib to determine whether PUMA expression is required to block tumorsphere formation. For these experiments, we performed a dasatinib dose-response experiment with LM2-4 cells transfected with control siRNA or either of 2 different PUMA siRNAs and compared tumorsphere formation (Figure 7B and Supplemental Figure 7B). While control cells were highly sensitive to dasatinib, we observed that PUMA knockdown caused cells to become resistant to treatment (Figure 7B). Calculating the IC50, we show that PUMA knockdown decreased sensitivity to dasatinib by 8-fold and 4-fold, respectively, compared with the control (Figure 7C). In contrast, we found that dasatinib failed to inhibit cell viability in adherent cells at doses as high as 1 μM (Supplemental Figure 7C), similar to our findings with β3-knockdown cells (Figure 3F). Thus, dasatinib-induced PUMA expression appears to selectively inhibit the colony-forming ability of stem-like cells.
Figure 7. PUMA expression is a critical vulnerability of stem-like cells.
(A) Quantification of cell death by trypan blue staining after transient transfection of MCF7 and BT549 breast cancer cell lines with PUMA cDNA or empty control vector relative to mock-transfected controls. P = 0.0262 (BT549; PUMA cDNA vs. vector control), by Student’s t test. n = 3 independent experiments run in duplicate. (B and C) Dasatinib dose-response experiments with LM2-4 cells transfected with control siRNA or either of 2 different PUMA siRNAs. n = 5 independent experiments performed in duplicate. (B) Curves were fitted by nonlinear regression. (C) IC50 was calculated from the nonlinear regression curves. P = 0.0331 (siCtrl vs. siPUMA no. 1); P = 0.0141 (siCtrl vs. siPUMA no. 2), by 1-way ANOVA with Dunnett’s multiple comparisons test. (D and E) In vivo tumor studies comparing different numbers of shCtrl and shPUMA LM2-4 cells injected orthotopically into adult female mice and treated with 30 mg/kg dasatinib or vehicle (1% citric acid) once daily by oral gavage for the first 7 days after injection. Red arrows indicate the start and end of daily drug treatment. n = 4 mice per group. (D) Graphs display the tumor volume after injection of 30,000 cells. P < 0.0001 (days 31 and 34), by 2-way, repeated-measures ANOVA with Sidak’s multiple comparisons test. (A–D) Data represent the mean ± SEM. *P < 0.05 and ***P < 0.001. (E) Tumor latency after injection of 15,000 cells. P = 0.0084 (shCtrl; vehicle vs. dasatinib); P = 0.5766 (shPUMA; vehicle vs. dasatinib), by log-rank (Mantel-Cox) test. (F) Schematic depicting how PUMA expression driven by αvβ3/Src blockade can target highly vulnerable metastatic and tumor-initiating stem-like breast cancer cells compared with cells in the primary tumor. See also Supplemental Figure 7.
Our in vitro findings suggest that driving PUMA expression with Src inhibitors may be particularly effective at suppressing tumor progression in vivo. To test this possibility, we injected limiting numbers of control or PUMA-knockdown LM2-4 cells orthotopically into the mammary fat pads of adult female NSG mice and compared their response to once-daily treatment with 30 mg/kg dasatinib or vehicle control (1% citric acid). Since we expected dasatinib treatment to reduce cell viability by increasing PUMA expression, we reasoned that a short-term dosing regimen should be capable of producing a long-term effect. Thus, we began treatment immediately after tumor cell implantation and stopped treatment after only 7 days, approximately 1 to 2 weeks before most tumors formed. While this short-term treatment did not affect tumor incidence (Supplemental Figure 7D), it was sufficient to decrease tumor volume when 30,000 cells were injected (Figure 7D) and even to delay tumor initiation by over a week when 15,000 cells were used (Figure 7E). Importantly, PUMA-knockdown cells were resistant to these effects, as dasatinib-treated tumors showed latency and growth characteristics similar to those of vehicle controls (Figure 7, D and E). Our data show that Src inhibition can reduce the rate of tumor initiation and growth in a manner that requires PUMA expression (Figure 7, D and E). Unexpectedly, these findings also indicate that short-term administration of Src inhibitors prior to tumor formation can have long-term, lasting effects leading to reduced tumor growth (Figure 7D). Taken together, our findings present a model whereby enhanced PUMA expression driven by αvβ3 or Src blockade may be an effective means of targeting αvβ3+Slug+ stem-like breast cancer cells to reduce tumor progression (Figure 7F).
Discussion
There are approximately 40,000 deaths from breast cancer every year in the United States alone, and essentially all of these deaths are caused by metastatic disease (40). The prevailing theory suggests that a rare subset of stem-like cells may play an important role in contributing to metastatic progression, highlighting the need to identify critical molecular pathways that can be exploited therapeutically to target these cells. Here, we identify the proapoptotic molecule PUMA as a key vulnerability of a population of stem-like breast cancer cells characterized by coexpression of αvβ3 and Slug. These effects were specific to PUMA, as the related family member NOXA had no effect on these cells, which is consistent with its role in targeting basal-like breast cancer cells (41). Importantly, driving PUMA expression was sufficient to specifically deplete our stem-like cells and reduce metastasis in vivo, revealing its role as an important metastasis suppressor. Our findings suggest that pharmacologically upregulating PUMA expression with targeted therapies such as Src inhibition may represent a potential strategy to target stem-like cells and reduce deaths from breast cancer metastasis.
An emerging consensus supports the existence of a rare population of stem-like cells in aggressive breast cancers important for tumor progression. However, efforts to identify these cells in human tumors have been hampered by that fact that the cell-surface marker profiles used to date are overly broad and and have enrichment of cells similar to those in the basal cell layer of the normal mammary gland (12, 15). While gene expression profiles have proven useful in predicting the existence of stem-like cells and their possible contribution to recurrence (14) and metastasis (32), they cannot identify these cells in patients’ tumors in situ and preclude the prospective isolation and evaluation of their role in tumor progression or response to therapy. Our prior findings from the normal mammary gland highlighted a critical role for αvβ3 in regulating Slug expression in activated MaSCs important for remodeling during pregnancy (17), suggesting that a similar relationship may exist in human breast cancers. In this study, we now show that αvβ3+Slug+ cells represent a distinct stem-like tumor cell subset associated with aggressive disease in breast cancer patient samples. While these cells share properties with previously characterized stem-like cell markers including CD44+CD24lo, CD49f+EpCAMlo, EMT-associated proteins, or claudin-low profiles, in all cases, these cells represent a more refined subset of these relatively abundant cell types. Surprisingly, these cells could be found in tumors representing most clinical subtypes, suggesting that they may identify a broad spectrum of aggressive disease.
Despite this relationship between αvβ3 and Slug, our findings show that αvβ3 is dispensable for EMT and instead directs the role of Slug in suppressing expression of the proapoptotic molecule PUMA and promoting tumor stemness. This suggests that the well-established role of Slug in breast cancer metastasis (19, 42, 43) might be independent of EMT, consistent with a study showing that the related protein Snail is the primary EMT transcription factor in breast cancer cells (24). In addition to EMT-related genes, Slug is also capable of transcriptionally repressing many genes involved in non-EMT cellular functions as well, such as cell survival, cell-cycle progression, and self-renewal. In the present study, we unexpectedly discovered a critical role for PUMA as a Slug-dependent gene suppressed by αvβ3/Src signaling. The importance of this relationship is evidenced by the fact that increased PUMA expression alone accounted for a considerable portion of the effect of β3 knockdown on metastasis (Figure 4, A and B). The relatively specific regulation of PUMA by αvβ3/Src signaling compared with regulation of other Slug-dependent genes may be due to compensation for the effects of Slug on other gene targets by the related EMT transcription factors Snail and ZEB1, both of which are increased by β3 shRNA knockdown (Figure 2E). In fact, both Snail and ZEB1 share many of the same gene targets as Slug (44), with the exception of PUMA, which is uniquely repressed by Slug (29). Taken together, our findings characterize PUMA suppression by αvβ3/Src/Slug signaling as a critical property of stem-like breast cancer cells that promotes their aggressive behavior.
Since stem-like tumor cells exist as a small subpopulation in primary breast cancers but play a disproportionate role in initiating recurrent and metastatic disease, current studies are investigating whether these cells are best targeted in the adjuvant setting, or after surgery, to prevent the formation of local or distant recurrences (45). Therapies targeting stem-like breast cancer cells may best be administered in the adjuvant setting to obtain maximum clinical benefit (46), especially since residual tumors after conventional treatment show enrichment for tumor-initiating cells (14). We now show that driving PUMA expression with therapies such as Src inhibitors selectively targets αvβ3+Slug+ stem-like tumor cells. Importantly, these cells were most sensitive to PUMA expression while disseminating to distant sites compared with cells present within the primary tumor, consistent with the idea of a potential effectiveness of these drugs when used in the adjuvant setting. While prior studies suggested that Src inhibition may selectively affect mesenchymal triple-negative breast cancer cells (47–49), clinical trials with Src inhibitors have generally failed to improve disease outcome in these breast cancer patients, showing no effect in reversing established metastases (50–52). In contrast to these prior reports, we now show that Src inhibition is highly effective at driving PUMA expression by reducing Slug, thereby increasing the sensitivity of stem-like cells to apoptosis. Thus, our findings suggest that repurposing Src inhibitors for use in the adjuvant setting may be a more effective use of these drugs and may represent a potential therapeutic approach to prevent the emergence of new metastatic disease.
Methods
IHC.
Breast cancer tissue microarrays were purchased from US Biomax. For immunohistochemical staining of formalin-fixed, paraffin-embedded sections from breast cancer tissues, antigen retrieval was performed in citrate buffer, followed by incubation with primary and secondary antibodies, and staining was detected using DAB or Vector Blue substrate (Vector Laboratories). Stained tumor sections were scored by a blinded observer. See the Supplemental Methods for additional details.
Immunofluorescence staining.
Immunofluorescence staining was performed on frozen sections fixed in ice-cold acetone or on cultured cells fixed briefly in 2% paraformaldehyde and PBS at room temperature and permeabilized. All samples were blocked with normal goat serum in PBS before incubation in primary antibody overnight at 4°C, followed by incubation with Alexa Fluor–conjugated secondary antibodies. Additional details are provided in the Supplemental Methods.
Cell transfection and lentiviral transduction.
Plasmids containing the full-length β3 cDNA or the β3 759x Src binding mutant in the pcDNA3.1 vector were described previously (30). HA-tagged PUMA cDNA in the pCEP4 vector (pHA-PUMA) (Addgene; plasmid 16588) was a gift of Bert Vogelstein (Johns Hopkins University, Baltimore, Maryland, USA) (27). Transient transfections for all cDNAs were performed with Lipofectamine 3000 (Invitrogen, Thermo Fisher Scientific), while HiPerFect Transfection Reagent (QIAGEN) was used for siRNAs. All transfections were performed according to the manufacturers’ instructions. FlexiTube siRNAs (QIAGEN) included AllStars negative control, PUMA, NOXA, and FBXO11. Human-specific shRNAs targeting β3, PUMA, Slug, c-Src, or a nonsilencing control in the pGIPZ vector (Open BioSystems) were used for generating stable knockdowns with lentivirus and selected using puromycin. Additional stable cell lines expressing ectopic β3 were made using lentivirus from FG12 plasmid alone (empty vector) or lentivirus containing β3 cDNA (gift of the Shattil laboratory, UCSD), or by transient transfection with β3 or β3 759x cDNA and selection with G418. See the Supplemental Methods for additional details and cell line information.
Immunoblotting and immunoprecipitations.
Whole-cell lysates were prepared from cell lines with RIPA lysis buffer combined with scraping, and the lysates were cleared by centrifugation. Additionally, Slug was immunoprecipitated from cell lysates with protein A/G beads (Pierce, Thermo Fisher Scientific) and anti-Slug antibody overnight at 4°C. The beads were then washed prior to elution of proteins with sample buffer and analyzed by immunoblotting. For inhibitor studies with LM2-4 cells, treatments with drug and DMSO alone (vehicle) were performed 24 hours prior to harvesting the lysates. Standard Western blotting procedures were performed. For further details, see the Supplemental Methods.
Real-time qPCR.
qPCR experiments were performed by collecting total RNA from cultured cells using the RNeasy Mini Kit (QIAGEN) and reverse transcribing with the High Capacity cDNA Reverse Transcription Kit (Life Technologies, Thermo Fisher Scientific). Real-time qPCR was performed using LightCycler 480 SYBR Green 1 Master (Roche) and run on a LightCycler 480 qPCR System (Roche). See the Supplemental Methods for the complete list of qPCR primer sequences.
Tumorsphere assays.
Primary tumorsphere formation was assessed in cells grown under anchorage-independent conditions in either soft agar or methylcellulose. For soft agar, cells were suspended in 0.3% agar and complete DMEM medium and cultured on top of a bottom layer of 1% agar in 48-well dishes. Cells were cultured for 14 to 21 days prior to staining with crystal violet, and colonies consisting of at least 6 cells from 4 fields per well were counted. Alternatively, sorted HCC38 and HCC1143 cells were cultured in 1% methylcellulose in complete DMEM in poly-HEMA–coated 48-well dishes and the number of primary tumorspheres per well quantified after 14 days. Self-renewal was measured by collecting primary tumorspheres in PBS, dissociating with trypsin, and reseeding in 1% methylcellulose before evaluating secondary colonies after an additional 14 days. For the zVAD-fmk and dasatinib treatment studies, the concentrations of drug were added only once, immediately after embedding the cells.
Orthotopic breast cancer.
Tumors were generated by injection of the number of GFP-labeled LM2-4 stable cells (10,000 cells for metastasis assays) in 50 μl sterile PBS into the inguinal fat pads of 10- to 12-week-old adult female NSG mice (purchased from The Jackson Laboratory). For tumor initiation experiments, mice were treated with 30 mg/kg dasatinib or vehicle control (1% citric acid) delivered once daily by oral gavage for the first 7 days after injection. All mice were monitored weekly for tumor formation by gentle palpation. Tumor volume was measured with calipers twice weekly by a blinded observer. The experiment was concluded, and mice were sacrificed just prior to the vehicle control tumors reaching the maximum allowable size of 2 cm3. For metastasis experiments, tumors were harvested at 6 weeks, and GFP expression in the lungs was measured with an OV100 or IVIS imaging system. Primary tumor mass was determined by assessing the wet weight of the resected tumors.
Flow cytometry.
Single-cell suspensions were prepared from cultured cells, blocked in 0.5% BSA/PBS, and stained with the following antibodies prior to sorting: CD49-PE (GoH3; BD Biosciences); EpCAM–Alexa 647 (9C4; BioLegend); and αvβ3–Alexa 488 (LM609; MilliporeSigma). Alternatively, the following antibodies were used to evaluate CD44 and CD24 expression: CD44-PE (G44-26; BD Biosciences); CD24–Alexa 647 (ML5; BioLegend); EpCAM-FITC (VU-1D9; STEMCELL Technologies); αvβ3-biotin (LM609; MilliporeSigma); and Streptavidin–Brilliant Violet 421 (BioLegend). Propidium iodide (0.5 μg/ml) was used to detect dead cells. Viable cells were collected by sorting with a FACSDiva or FACSAria machine (BD Biosciences).
Statistics.
Statistical analysis was performed using GraphPad Prism 5 (GraphPad Software). A 2-tailed Student’s t test was used for comparison of 2 means, while ANOVA was performed for 3 or more data sets. Post-hoc analysis was performed using Tukey’s (for comparison of all groups), Dunnett’s (for comparison with controls), or Sidak’s (repeated-measures) multiple comparisons test. For all analyses, a P value of less than 0.05 was considered statistically significant.
Study approval.
All animal experiments were performed under protocols approved by the IACUC of UCSD and in accordance with the guidelines set forth in the NIH’s Guide for the Care and Use of Laboratory Animals (National Academies Press, 2011).
Author contributions
QS, JL, EL, and HW helped design and conduct experiments and analyze data. YSD and AW generated and provided sections from patient-derived xenograft (PDX) samples. DAC provided resources and guidance to support the project. JSD designed the project, performed experiments, analyzed the data, supervised the overall project, and wrote the manuscript.
Supplementary Material
Acknowledgments
We would like to thank Michael Karin (UCSD) for providing the human breast cancer progression tissue microarray and Sanford Shattil and Hisashi Kato (UCSD) for the lentiviral plasmids containing the β3 cDNA. We would also like to acknowledge the histology and flow cytometry shared resource facilities at the Moores Cancer Center of UCSD. This research was supported by funding from an American Cancer Society Institutional Research grant provided through the Moores Cancer Center of UCSD (70-002 , to JSD) and a Susan G. Komen Career Catalyst Research grant (CCR15330839, to JSD).
Version 1. 12/11/2017
Electronic publication
Version 2. 01/02/2018
Print issue publication
Funding Statement
Career Catalyst Research Grant
American Cancer Society Institutional Research Grant provided through the Moores Cancer Center
Footnotes
QS’s present address is: Department of General Surgery, The First Affiliated Hospital of Xi’an Jiaotong University, Xi’an, China.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2018;128(1):531–544.https://doi.org/10.1172/JCI93707.
Contributor Information
Qi Sun, Email: sunqi4875@163.com.
Jacqueline Lesperance, Email: jlesperance@ucsd.edu.
Hiromi Wettersten, Email: hwettersten@ucsd.edu.
Elaine Luterstein, Email: elaine.luterstein@gmail.com.
References
- 1.Weigelt B, Peterse JL, van ‘t Veer LJ. Breast cancer metastasis: markers and models. Nat Rev Cancer. 2005;5(8):591–602. doi: 10.1038/nrc1670. [DOI] [PubMed] [Google Scholar]
- 2.Prat A, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12(5):R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perou CM, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 4.Herschkowitz JI, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8(5):R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kennecke H, et al. Metastatic behavior of breast cancer subtypes. J Clin Oncol. 2010;28(20):3271–3277. doi: 10.1200/JCO.2009.25.9820. [DOI] [PubMed] [Google Scholar]
- 6.Zhang M, Rosen JM. Developmental Insights into Breast Cancer Intratumoral Heterogeneity. Trends Cancer. 2015;1(4):242–251. doi: 10.1016/j.trecan.2015.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100(7):3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lim E, et al. Transcriptome analyses of mouse and human mammary cell subpopulations reveal multiple conserved genes and pathways. Breast Cancer Res. 2010;12(2):R21. doi: 10.1186/bcr2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prat A, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12(5):R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pece S, et al. Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content. Cell. 2010;140(1):62–73. doi: 10.1016/j.cell.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 11.Spike BT, Engle DD, Lin JC, Cheung SK, La J, Wahl GM. A mammary stem cell population identified and characterized in late embryogenesis reveals similarities to human breast cancer. Cell Stem Cell. 2012;10(2):183–197. doi: 10.1016/j.stem.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lim E, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15(8):907–913. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- 13.Prat A, et al. Characterization of cell lines derived from breast cancers and normal mammary tissues for the study of the intrinsic molecular subtypes. Breast Cancer Res Treat. 2013;142(2):237–255. doi: 10.1007/s10549-013-2743-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Creighton CJ, et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci U S A. 2009;106(33):13820–13825. doi: 10.1073/pnas.0905718106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shipitsin M, et al. Molecular definition of breast tumor heterogeneity. Cancer Cell. 2007;11(3):259–273. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 16.Abraham BK, Fritz P, McClellan M, Hauptvogel P, Athelogou M, Brauch H. Prevalence of CD44+/CD24-/low cells in breast cancer may not be associated with clinical outcome but may favor distant metastasis. Clin Cancer Res. 2005;11(3):1154–1159. [PubMed] [Google Scholar]
- 17.Desgrosellier JS, et al. Integrin αvβ3 drives slug activation and stemness in the pregnant and neoplastic mammary gland. Dev Cell. 2014;30(3):295–308. doi: 10.1016/j.devcel.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Desgrosellier JS, Cheresh DA. Integrins in cancer: biological implications and therapeutic opportunities. Nat Rev Cancer. 2010;10(1):9–22. doi: 10.1038/nrc2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo W, et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell. 2012;148(5):1015–1028. doi: 10.1016/j.cell.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Phillips S, et al. Cell-state transitions regulated by SLUG are critical for tissue regeneration and tumor initiation. Stem Cell Reports. 2014;2(5):633–647. doi: 10.1016/j.stemcr.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Asselin-Labat ML, et al. Control of mammary stem cell function by steroid hormone signalling. Nature. 2010;465(7299):798–802. doi: 10.1038/nature09027. [DOI] [PubMed] [Google Scholar]
- 22.Joshi PA, et al. Progesterone induces adult mammary stem cell expansion. Nature. 2010;465(7299):803–807. doi: 10.1038/nature09091. [DOI] [PubMed] [Google Scholar]
- 23.Pierce JP, et al. Influence of a diet very high in vegetables, fruit, and fiber and low in fat on prognosis following treatment for breast cancer: the Women’s Healthy Eating and Living (WHEL) randomized trial. JAMA. 2007;298(3):289–298. doi: 10.1001/jama.298.3.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ye X, et al. Distinct EMT programs control normal mammary stem cells and tumour-initiating cells. Nature. 2015;525(7568):256–260. doi: 10.1038/nature14897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bermejo-Rodríguez C, Pérez-Caro M, Pérez-Mancera PA, Sánchez-Beato M, Piris MA, Sánchez-García I. Mouse cDNA microarray analysis uncovers Slug targets in mouse embryonic fibroblasts. Genomics. 2006;87(1):113–118. doi: 10.1016/j.ygeno.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 26.Munoz R, et al. Highly efficacious nontoxic preclinical treatment for advanced metastatic breast cancer using combination oral UFT-cyclophosphamide metronomic chemotherapy. Cancer Res. 2006;66(7):3386–3391. doi: 10.1158/0008-5472.CAN-05-4411. [DOI] [PubMed] [Google Scholar]
- 27.Yu J, Zhang L, Hwang PM, Kinzler KW, Vogelstein B. PUMA induces the rapid apoptosis of colorectal cancer cells. Mol Cell. 2001;7(3):673–682. doi: 10.1016/S1097-2765(01)00213-1. [DOI] [PubMed] [Google Scholar]
- 28.Wu WS, et al. Slug antagonizes p53-mediated apoptosis of hematopoietic progenitors by repressing puma. Cell. 2005;123(4):641–653. doi: 10.1016/j.cell.2005.09.029. [DOI] [PubMed] [Google Scholar]
- 29.Escrivà M, et al. Repression of PTEN phosphatase by Snail1 transcriptional factor during gamma radiation-induced apoptosis. Mol Cell Biol. 2008;28(5):1528–1540. doi: 10.1128/MCB.02061-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Desgrosellier JS, et al. An integrin alpha(v)beta(3)-c-Src oncogenic unit promotes anchorage-independence and tumor progression. Nat Med. 2009;15(10):1163–1169. doi: 10.1038/nm.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seguin L, et al. An integrin β3-KRAS-RalB complex drives tumour stemness and resistance to EGFR inhibition. Nat Cell Biol. 2014;16(5):457–468. doi: 10.1038/ncb2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lawson DA, et al. Single-cell analysis reveals a stem-cell program in human metastatic breast cancer cells. Nature. 2015;526(7571):131–135. doi: 10.1038/nature15260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu ZQ, Li XY, Hu CY, Ford M, Kleer CG, Weiss SJ. Canonical Wnt signaling regulates Slug activity and links epithelial-mesenchymal transition with epigenetic Breast Cancer 1, Early Onset (BRCA1) repression. Proc Natl Acad Sci U S A. 2012;109(41):16654–16659. doi: 10.1073/pnas.1205822109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang SP, et al. p53 controls cancer cell invasion by inducing the MDM2-mediated degradation of Slug. Nat Cell Biol. 2009;11(6):694–704. doi: 10.1038/ncb1875. [DOI] [PubMed] [Google Scholar]
- 35.Jin Y, et al. FBXO11 promotes ubiquitination of the Snail family of transcription factors in cancer progression and epidermal development. Cancer Lett. 2015;362(1):70–82. doi: 10.1016/j.canlet.2015.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vernon AE, LaBonne C. Slug stability is dynamically regulated during neural crest development by the F-box protein Ppa. Development. 2006;133(17):3359–3370. doi: 10.1242/dev.02504. [DOI] [PubMed] [Google Scholar]
- 37.Díaz VM, Viñas-Castells R, García de Herreros A. Regulation of the protein stability of EMT transcription factors. Cell Adh Migr. 2014;8(4):418–428. doi: 10.4161/19336918.2014.969998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fillmore CM, Kuperwasser C. Human breast cancer cell lines contain stem-like cells that self-renew, give rise to phenotypically diverse progeny and survive chemotherapy. Breast Cancer Res. 2008;10(2):R25. doi: 10.1186/bcr1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Asselin-Labat ML, et al. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat Cell Biol. 2007;9(2):201–209. doi: 10.1038/ncb1530. [DOI] [PubMed] [Google Scholar]
- 40. Breast Cancer Facts & Figures 2015-2016. American Cancer Society. https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-statistics/breast-cancer-facts-and-figures/breast-cancer-facts-and-figures-2015-2016.pdf Accessed November 13, 2017.
- 41.Petrocca F, et al. A genome-wide siRNA screen identifies proteasome addiction as a vulnerability of basal-like triple-negative breast cancer cells. Cancer Cell. 2013;24(2):182–196. doi: 10.1016/j.ccr.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chakrabarti R, et al. Elf5 inhibits the epithelial-mesenchymal transition in mammary gland development and breast cancer metastasis by transcriptionally repressing Snail2. Nat Cell Biol. 2012;14(11):1212–1222. doi: 10.1038/ncb2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim S, et al. Slug promotes survival during metastasis through suppression of Puma-mediated apoptosis. Cancer Res. 2014;74(14):3695–3706. doi: 10.1158/0008-5472.CAN-13-2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kurrey NK, et al. Snail and slug mediate radioresistance and chemoresistance by antagonizing p53-mediated apoptosis and acquiring a stem-like phenotype in ovarian cancer cells. Stem Cells. 2009;27(9):2059–2068. doi: 10.1002/stem.154. [DOI] [PubMed] [Google Scholar]
- 45.Brooks MD, Burness ML, Wicha MS. Therapeutic Implications of Cellular Heterogeneity and Plasticity in Breast Cancer. Cell Stem Cell. 2015;17(3):260–271. doi: 10.1016/j.stem.2015.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ithimakin S, et al. HER2 drives luminal breast cancer stem cells in the absence of HER2 amplification: implications for efficacy of adjuvant trastuzumab. Cancer Res. 2013;73(5):1635–1646. doi: 10.1158/0008-5472.CAN-12-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Finn RS, et al. Dasatinib, an orally active small molecule inhibitor of both the src and abl kinases, selectively inhibits growth of basal-type/”triple-negative” breast cancer cell lines growing in vitro. Breast Cancer Res Treat. 2007;105(3):319–326. doi: 10.1007/s10549-006-9463-x. [DOI] [PubMed] [Google Scholar]
- 48.Huang F, et al. Identification of candidate molecular markers predicting sensitivity in solid tumors to dasatinib: rationale for patient selection. Cancer Res. 2007;67(5):2226–2238. doi: 10.1158/0008-5472.CAN-06-3633. [DOI] [PubMed] [Google Scholar]
- 49.Lehmann BD, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121(7):2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Finn RS, et al. Dasatinib as a single agent in triple-negative breast cancer: results of an open-label phase 2 study. Clin Cancer Res. 2011;17(21):6905–6913. doi: 10.1158/1078-0432.CCR-11-0288. [DOI] [PubMed] [Google Scholar]
- 51.Gucalp A, et al. Phase II trial of saracatinib (AZD0530), an oral SRC-inhibitor for the treatment of patients with hormone receptor-negative metastatic breast cancer. Clin Breast Cancer. 2011;11(5):306–311. doi: 10.1016/j.clbc.2011.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Herold CI, et al. Phase II trial of dasatinib in patients with metastatic breast cancer using real-time pharmacodynamic tissue biomarkers of Src inhibition to escalate dosing. Clin Cancer Res. 2011;17(18):6061–6070. doi: 10.1158/1078-0432.CCR-11-1071. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.