Abstract
Perforin-1 mutations result in a potentially fatal hemophagocytic lymphohistiocytosis (HLH) with heightened immune activation, hypercytokinemia, pancytopenia, and end-organ damage. At present, hematopoietic stem cell (HSC) transplantation is curative, but limited by donor availability and associated mortality, making gene therapy an attractive alternative approach for HLH. We reported that perforin expression driven by cellular promoters in lentiviral (LV) vectors resulted in significant, albeit partial, correction of the inflammatory features in a murine model of HLH. We hypothesized that the level of perforin expression achieved per cell from ectopic moderate-strength cellular promoters (phosphoglycerate kinase gene/perforin-1 gene) is inadequate and thus engineered an LV vector using a viral promoter (MND; a modified Moloney murine leukemia virus long terminal repeat with myeloproliferative sarcoma virus enhancer) containing microRNA126 target sequences to restrict perforin expression in HSCs. We show here that the MND-LV vector restored perforin expression to normal levels in a perforin-deficient human natural killer cell line and perforin gene-corrected Perforin1−/− transplant recipients, whereas cellular promoters drove only partial correction. On lymphocytic choriomeningitis virus challenge, the clinical scores and survival improved only with the MND-LV vector, but inflammatory markers and cytotoxicity were improved with all LV vectors. Our studies suggest that although moderate levels of expression can result in partial amelioration of the HLH phenotype, high levels of perforin expression per cell are required for complete correction of HLH.
Introduction
Hemophagocytic lymphohistiocytosis (HLH) is a potentially fatal immune regulatory disorder principally caused by impaired lymphocyte cytotoxicity. Patients experience episodes of extreme immune activation triggered by a variety of infectious organisms, but most commonly viruses. HLH is marked by an exaggerated release of cytokines, such as interferon (IFN)-γ and interleukin (IL)-6, IL-10, and IL-1; hemophagocytosis of hematopoietic cells; lymphoproliferation; and end-organ damage.1–4 Patients with HLH present with signs and symptoms that include high-grade fever, hepatitis, seizures, cytopenias, lymphadenopathy, and splenomegaly, and may ultimately progress to death from respiratory and/or cardiovascular collapse.
Familial HLH (FHLH) results from an underlying genetic defect in the cytotoxic or degranulation pathway. The known mutations causing FHLH are either in the degranulation/granule trafficking pathway genes, such as Munc13-4, Stx11, Rab27a, and StxBP2, or the cytolytic pathway gene, Perforin-1 (PRF1).5,6 The most common cause of FHLH (in 20–40% of cases) is perforin gene mutations, designated as FHLH2.7 Perforin is a critical component of lymphocyte cytotoxicity and is expressed in CD8+ T cells and natural killer (NK) cells.8–10
The short-term treatment of HLH is suppression of the overwhelming immune activation combined with identification and treatment of potential triggering infections. The long-term curative option for FHLH is allogeneic hematopoietic stem cell transplantation (HCT). HCT, however, is limited by the availability of donors, can have potential serious adverse effects such as graft-versus-host disease, and is associated with 20–50% mortality. HCT-related mortality has improved by using reduced intensity conditioning; however, mixed chimerism or graft rejection still poses challenges in 30–50% of transplanted patients.11,12
We have previously shown that 10–20% of wild-type chimerism corrects HLH symptoms in a Prf1−/− mouse model.13,14 We reported significant, but partial, correction of cytopenias and cytotoxicity in Prf1−/− mice, using lentiviral (LV) vectors expressing perforin from cellular promoters of the phosphoglycerate kinase gene (PGK) or the perforin-1 gene (PRF1).15 We theorized that this incomplete rescue may have been due to an insufficient level of perforin expression driven by these promoters. It has been previously shown that cellular promoters from ubiquitously expressed or tissue-restricted genes were inadequate for correcting the defect in leukocyte adhesion deficiency, where a strong viral promoter was required for phenotypic correction of this defect.16 Similar results have been reported in chronic granulomatous disease17,18 and Wiskott–Aldrich syndrome (WAS).19,20
We hypothesized that a high level of perforin expression from a viral promoter would be necessary for complete correction of the HLH phenotype. Perforin expression is restricted to cytotoxic T lymphocytes (CTLs) and NK cells.10 To permit high perforin expression in cytotoxic lymphocytes and to prevent potential toxicity due to perforin expression in HSCs, we placed microRNA-126 (miR126) target sequences in the 3′ untranslated region of the perforin cDNA in LV vectors driven by the MND promoter/enhancer. MND-driven perforin expression restored cytotoxicity in perforin-deficient human NK cells and murine CTLs. Perforin-deficient mice that received gene-corrected HSCs with the MND vector exhibited a dose-responsive increase in survival and viral clearance after lymphocytic choriomeningitis virus (LCMV) infection.
Materials and Methods
Lentiviral vectors
The sequence for hsa-miR126-3p was obtained from the miRbase directory. To create a vector with four target sites of the miR126-3p sequence, we designed an oligonucleotide to include four repeats of the miR126-3p sequence (5′-CTAGAATGCATCGCATTATTACTCACGGTACGACTAGCGCATTATTACTCACGGTACGAACCGGTCGCATTATTACTCACGGTACGATCACCGCATTATTACTCACGGTACGAGTTTAAACCCC-3′. The underlined region of the sequence is complementary to the hsa-miR126-3p sequence. This oligonucleotide was cloned, using XbaI and SmaI, into LV.NGFR.CMV.PGK.GFP.WPRE (kindly provided by Dr. Luigi Naldini, San Raffaele Institute, Milan, Italy) to derive LV.NGFR.CMV.PGK.GFP.4TmiR126 (GFP4T).
For the construction of LV.NGFR.CMV.PGK.Perforin1.WPRE.4TmiR126 (PGK4T), an EcoRI/XhoI PGK.Perforin1 fragment was isolated from LV.PGK.PRF.IRES.GFP (PGK), as we previously published.15 The EcoRI end was blunted and PGK.Perforin1 was ligated into the EcoRV/SalI-digested GFP4T. The same strategy was used to clone LV.NGFR.CMV.PRF.Perforin1.WPRE.4TmiR126 (PRF4T), with the fragment PRF.Perforin1 from the LV.PRF.Perforin1.IRES.GFP (PRF),15 into GFP4T. The miR126 target sites were then removed from PRF4T with XbaI and religated to create vector PRF0T.
LV.NGFR.CMV.MND.Perforin1.WPRE.4TmiR126 (MND4T) was generated by ligating the EcoRI/XbaI PRF.Perforin1 fragment into an intermediate pBluescript (pBS) vector from LV.PRF.Perforin.I.GFP.15 The EcoRI/AgeI MND promoter was isolated from the MND-eGFP-WPRE vector and ligated into XhoI/AgeI pBS-PRF.Perforin1 to create the intermediate plasmid pBS-MND-Perforin1, from which the SpeI (blunt)/XhoI fragment of MND.Perforin1 was ligated into the EcoRV/SalI sites of the GFP4T vector. LV vector was produced by transient transfection of 293T cells and viral titers were determined by transduction of mouse erythroleukemia cells after serial dilutions, followed by flow cytometry (all previously described).15
Mice
Perforin-1-deficient mice (C57BL/6-Prf1tm1Sdz/J; Prf1−/−) were obtained from the Jackson Laboratory (Bar Harbor, ME) and bred in the animal facility at Cincinnati Children's Hospital Medical Center (CCHMC, Cincinnati, OH). For HCT, 6- to 8-week-old recipient Prf1−/− mice were treated with busulfan (20 mg/kg from days −4 to −1) and cyclophosphamide (100 mg/kg on days −2 and −1) and transplanted on day 0 (24 hr after chemotherapy conditioning) with either Prf1−/− or C57/BL6 (wild-type) Lin−Sca-1+cKit+ (LSK) cells or with LSK cells transduced with one of the three LV vectors. Transplanted mice were challenged with 200 plaque-forming units of LCMV intraperitoneally 16 weeks post-HCT. Clinical scoring of the mice was done as previously described.21 Eye infections, dehydration, and ascites were graded on a 0–2 scale, and weight, coordination, and hunched posture were graded on a 0–3 scale. All experimental procedures in mice were performed in accordance with the protocol approved by the Institutional Animal Care and Use Committee at CCHMC.
Isolation and transduction of hematopoietic stem and progenitor cells and NK cell line
Bone marrow LSK cells and Lin−Sca-1−cKit+ (LK) cells from femurs and tibias of Prf1−/− or wild-type mice were isolated by a combination of magnetic labeling and fluorescence-activated cell sorting (FACS) as previously published.15 LSK and LK cells were both cultured in StemSpan serum-free expansion medium (SFEM) (STEMCELL Technologies, Vancouver, BC, Canada) supplemented with 2% fetal bovine serum (FBS), 0.5% lipoprotein (low density; Sigma-Aldrich, St. Louis, MO), 0.1% 10 mM deoxynucleotide (dNTP) solution mix (New England BioLabs, Ipswich, MA), 1% penicillin–streptomycin (Fisher Scientific, Hampton, NH), mouse IL-3 at 10 ng/ml and mouse stem cell factor (SCF) at 50 ng/ml (R&D Biosystems, Minneapolis, MN). LSK cells from Prf1−/− mice were transduced as described below; LK cells were irradiated with 3000 rads before coinjection into recipient mice.
LSK cells were prestimulated overnight and transduced twice at 8-hr intervals. The multiplicity of infection (MOI) of transduction was maintained at 25–30 per transduction. After 36 hr of culture, the cells were harvested and 4–5 × 104 LSK cells along with 6–8 × 104 irradiated LK cells were injected intravenously into recipient mice. A portion of the LSK cells was kept in culture for 6–7 days to measure initial transduction efficiency.
KHYG1 cells (from the Japanese Collection of Research Bioresources Cell Bank, JCRB0156) were transduced with retrovirus to overexpress miR30-based short hairpin RNAs (shRNAs) targeting the 3′ untranslated region (UTR) of PRF1 with mCherry expressed in cis22 (a kind gift from I. Voskoboinik, P. MacCallum Cancer Centre, Melbourne, Australia). Transduced KHYG1 cells were sorted for mCherry expression 7 days later to obtain the KHYG1 KD line. KHYG1 cells were maintained in complete RPMI with recombinant IL-2 (rIL-2) (100 U/ml; Roche, Indianapolis, IN). LV vector transduction was performed at an MOI of 5. Seventy-two hours posttransduction, the cells were sorted for truncated nerve growth factor receptor (tNGFR) after staining with Alexa 647 (anti-CD271 antibody, Cat. No. 560326; BD Biosciences, San Jose, CA) and then sorted for Alexa 647-positive, MicroBead-labeled cells by passage through a magnetic column (Cat. No. 130-091-395; Miltenyi Biotec, Bergisch Gladbach, Germany).
Human umbilical cord blood (CB) cells were used to isolate CD34+ cells, using the Miltenyi CD34 MicroBead kit (Cat. No. 130-046-702) as per the manufacturer's protocol. CD34+ cells were prestimulated overnight in 1 ml of Gibco α-MEM (Fisher Scientific) containing 10% human serum (Sigma-Aldrich) and 5% fetal calf serum (FCS; Sigma-Aldrich) supplemented with 10 mM Gibco HEPES (Fisher Scientific), 10 mM Gibco sodium pyruvate (Fisher Scientific), Gibco penicillin–streptomycin (Fisher Scientific), recombinant human SCF (rhSCF, 20 ng/ml; PeproTech, Rocky Hill, NJ), and rhIL-15 at 20 ng/ml (PeproTech) and transduced with LV vector at an MOI of 10 twice at 12-hr intervals. After 36 hr of culture, some of the cells were analyzed for initial transduction efficiency and the rest were transferred for NK cell differentiation, as previously described with minor modifications.23 Briefly, cells were seeded on a preestablished confluent layer of murine MS-5 stromal cells in the presence of rhSCF and rhIL-15 for 3 weeks. MS-5 cells were routinely expanded in α-MEM containing 20% FCS and passaged in 24-well plates at a concentration of 6 × 104 cells/ml per well 24 hr before initiation of the coculture. Cultures were maintained at 37°C, 5% CO2, in an air atmosphere saturated with humidity. Half of the medium was renewed twice per week and cells were tested for CD56+ cell percentage once per week. Eight days after the LCMV challenge, a subset of transplanted mice was sacrificed and spleens were harvested as single-cell suspensions for CTL assays. Splenocytes were analyzed for tNGFR expression and stained for CD8 biotin and sorted for RNA isolation.
Flow cytometry
Chimerism in the transplanted mice was measured 10–12 weeks posttransplantation by staining for BD Pharmingen tNGFR (CD271) PE/APC (BD Biosciences) in various cell lineages (BD Pharmingen CD8, CD4, and Nk1.1; BD Biosciences) in peripheral blood. KHYG1 cells were stained for tNGFR PE/Alexa 647 and perforin was stained with APC (BD Pharmingen). FACS analysis was done with a BD FACSCanto II (BD Biosciences).
Quantitative PCR and quantitative reverse transcription PCR
Genomic DNA was isolated with Qiagen cell lysis solution, according to the manufacturer's protocol, from bone marrow, spleen, and peripheral blood. The vector copy number (VCN) was determined by qPCR using woodchuck posttranscriptional regulatory element (WPRE) primers for vector sequences and Flk-1 primers for endogenous control. iTaq Superprobes (Bio-Rad, Hercules, CA) was used in accordance with the manufacturer's protocol, on a Bio-Rad CFX96 thermocycler. The sequences of the primers and probes used were as follows:
Flk-1: FP, AAGACCTTGAAGTTGGCAACGCAG; RP, AGGAAGAATGGGCAGATGGTCACA; probe, HEX/TGCCGTGAATTGCAGAGCTGTTGTGT
WPRE: FP, TGTATAAATCCTGGTTGCTGTC; RP, GCACACCACGCCACGTTG; probe, FAM/ATGAGGAGTTGTGGCCCGTTGT
RNA was isolated from KHYG1 cells, spleen CD8+ cells, and bone marrow cells with QIAzol lysis reagent (Qiagen, Hilden, Germany), using the manufacturer's protocol. Perforin mRNA expression was measured with primers that span exons 2–3 (Life Technologies 20× assay mix, Cat. No. 4331182; Thermo Fisher). The endogenous control used was human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) with the primers specific to exon 3 (Life Technologies 20× assay mix, Cat. No. 4333764; Thermo Fisher) or mouse β-actin (Life Technologies 20× assay mix, Cat. No. 4352341E; Thermo Fisher). For LCMV mRNA, iTaq universal SYBR green supermix was used to amplify the following primers:
LCMV FP: GAGTCCAGAAGCTTTCTGATGTCAT
LCMV RP: CAAGTATTCACACGGCATGGAT
iTaq Superprobes (Bio-Rad) were used in accordance with the manufacturer's protocol on an Applied Biosystems 7900HT fast real-time PCR system.
IFN-γ ELISA
An IFN-γ ELISA was performed on plasma, as previously described.15,21
LCMV quantification
LCMV quantification was performed on lysates from bone marrow as previously described.24
Chromium release assay
A chromium-51 (51Cr) release assay was performed as previously described.15 Briefly, EL4 target cells were pulsed with LCMV gp33-41 (100 ng/ml) for 1 hr before 51Cr labeling. Effector and EL4 target cells (5000 cells per well) were coincubated for 4 hr at 37°C. 51Cr release into the supernatant was counted on a 96-well LumaPlate (PerkinElmer, Wellesley, MA) with a gamma counter (TopCount NXT; PerkinElmer). Cytotoxicity was reported as the percentage of 51Cr released into the supernatant according to the following calculation: (test − spontaneous)/(max lysis − spontaneous) × 100. Maximum lysis was determined by addition of 1% Triton X-100 (Sigma-Aldrich) to 5000 EL4 target cells.
Western blot analysis
Cell were lysed with lysis buffer (2% Nonidet P-40 [NP-40]; 150 mM NaCl; 50 mM Tris-HCl; 1 mM MgCl2, pH 8; 2.5 mM EDTA; leupeptin at 2 mg/ml; aprotinin at 2 mg/ml; phenylmethylsulfonyl fluoride at 50 mg/ml; pepstatin at 1 mg/ml). Lysate (15 μg) was prepared with Laemmli sample buffer without 2-mercaptoethanol and run on a 10% Bis-Tris gel. Protein was transferred to a nitrocellulose membrane, blocked with Odyssey blocking buffer, probed by perforin antibody clone P1-8 (diluted 1:500; Kamiya Biomedical, Seattle, WA), and visualized with a LICOR Odyssey CLx scanner. Equal loading was monitored by the detection of actin clone AC-15 (diluted 1:5000; Sigma-Aldrich).
Statistical analysis
Statistical analysis was done with GraphPad Prism software (GraphPad, San Diego, CA).
Results
Restriction of expression in hematopoietic stem and progenitor cells but not in cytotoxic cell progeny
miR126 target sequences placed 3′ to transgenes expressed from LV vectors have been reported to restrict transgene expression in HSCs, but to allow robust expression in its myeloid progeny.25,26 However, reports on miR126 detection in NK cells are conflicting.26–28 We evaluated whether miR126 target sequences would permit transgene expression in CD8+ and NK cell progeny. We thus engineered LV vectors with a PGK promoter, GFP transgene, and either four (GFP-4T) or no (GFP-0T) repeats of the miR126 target sequences downstream of the GFP cDNA. Both LV vectors also included tNGFR driven by a minimal cytomegalovirus (CMV) promoter in the opposite orientation, so that tNGFR expression would be unaffected by miR126 target sequences and permit determination of gene-modified chimerism (Fig. 1a).
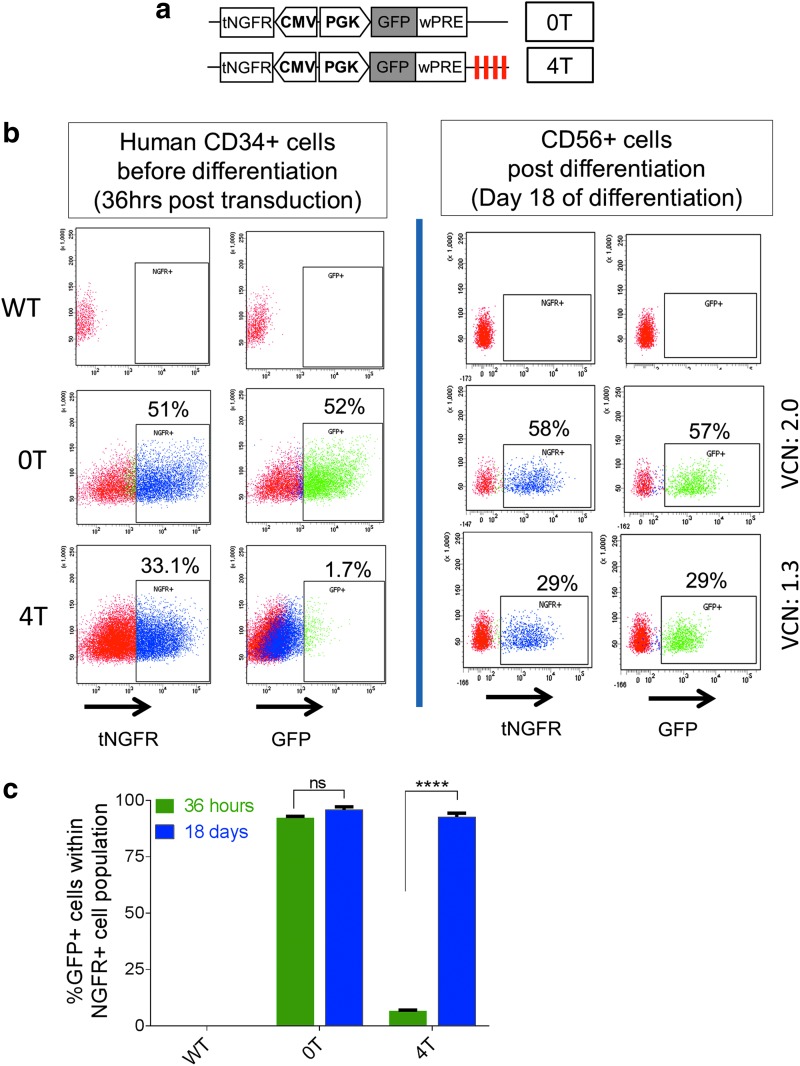
Figure 1.
MicroRNA-126 (miRNA126) target elements in lentiviral (LV) vectors restrict transgene expression in human CD34+ hematopoietic stem and progenitor cells but allow robust transgene expression in natural killer (NK) cells. (a) Bidirectional lentiviral vector design with a mini-cytomegalovirus (CMV) promoter driving a truncated nerve growth factor receptor (tNGFR) cDNA in one direction and phosphoglycerate kinase (PGK) promoter driving enhanced green fluorescent protein (eGFP) cDNA with (4T) or without (0T) four repeats of the miR126 target sequences in the opposite direction. (b) Human CD34+ cells were transduced with the two LV vectors. Analysis of tNGFR and eGFP expression by flow cytometry 36 hr posttransduction (left); and after 18 days of NK (CD56+) cell differentiation (right) is shown. The cells expressing tNGFR or eGFP are gated on the basis of appropriate negative controls, and their percentage is indicated in each dot plot. (c) Cumulative data on the proportion of transduced CD34+ cells (tNGFR+; blue columns) that are expressing GFP (green columns) 36 hr after transduction or after 18 days of NK cell differentiation with the 0T and 4T LV vectors. Error bars represent the SEM. Statistics were done by two-way analysis of variance with Sidak correction. ****p < 0.0001.
We transduced human umbilical cord blood CD34+ hematopoietic stem and progenitor cells (HSPCs) with the GFP-4T and GFP-0T LV vectors within the first 24 hr of isolation, and at 36 hr subjected a portion of the HSPCs to flow cytometry and differentiated the rest to NK cells (Fig. 1b and c). Nearly 50 and 30% of HSPCs showed transduction (tNGFR expression) with the GFP-0T and GFP-4T vectors, which corresponded with a VCN per cell of 1.3 and 2, respectively. GFP expression was minimal in the CD34+ cells transduced with the GFP-4T vector, but robust in those transduced with the GFP-0T vector, confirming the previously reported restriction in expression in HSPCs.25,26 However, on differentiation into NK cells, GFP was robustly expressed from the GFP-4T vector in the same proportion of NK cells that expressed tNGFR. Collectively, we found that miR126 target sequences, while effectively restricting transgene expression in HSPCs, allowed expression in their differentiated NK cell progeny at levels comparable to the vector without the miR126 target sequences.
Regulated perforin LV vectors and their relative expression and CTL activity
We designed a new LV vector using a ubiquitously expressing MNDU3 promoter/enhancer derived from a modified murine myeloproliferative sarcoma retrovirus long terminal repeat (LTR),29,30 for comparison with the LV vector driven by the PGK promoter. Four repeats of the miR126 target sequences were placed 3′ to the perforin cDNA to restrict expression from both of these ubiquitously expressing promoters in HSCs (termed MND4T and PGK4T vectors hereafter).25,26 A third LV vector had perforin expression driven by the previously described PRF promoter (a −1430 bp 5′ UTR of the perforin-1 gene) to achieve CTL/NK-specific expression of perforin15; no miR126 target sequences were placed in this LV vector because expression from the PRF promoter is naturally restricted in HSCs (termed PRF0T). All LV vectors also contained the mini-CMV:tNGFR placed in the opposite orientation for unrestricted monitoring of gene transfer in HSCs (Fig. 2a).
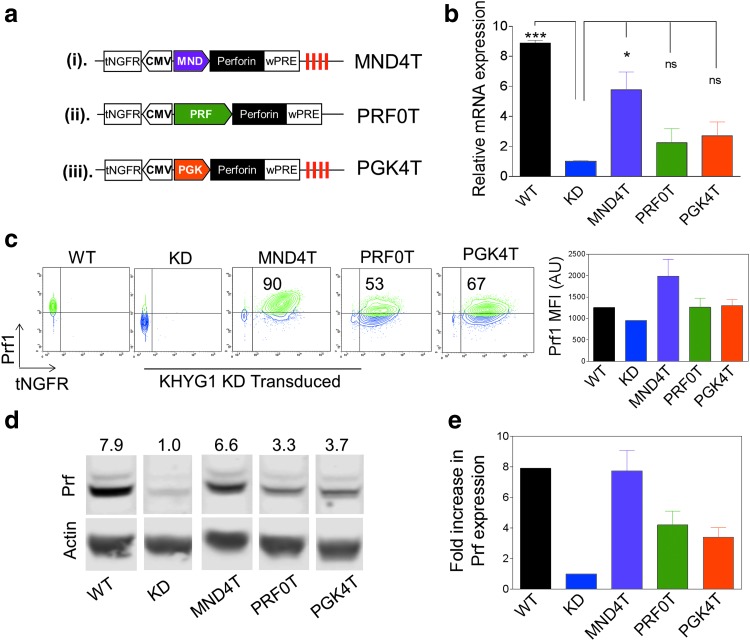
Figure 2.
Perforin expression in the human KHYG1 NK cell line from LV vectors carrying the MND, PGK, and PRF promoters. (a) Bidirectional LV vector design with a mini-CMV promoter driving the human tNGFR cDNA in one direction, and (i) MNDU3 promoter (MND4T), (ii) tissue-specific perforin gene promoter (PRF0T), or (iii) phosphoglycerate kinase (PGK4T) promoter driving expression of perforin in the opposite direction. miR126 target sequences (red vertical lines) were placed downstream of the woodchuck posttranscriptional regulatory element (WPRE) in the MND and PGK vectors. (b) Relative Prf1 mRNA expression in wild-type KHYG1 cells (WT), KHYG1 permanently transduced with prf1 shRNA to the 3′ UTR of the prf1 gene (KHYG1 KD), and KD cells transduced (and selected for tNGFR+) with the three LV vectors. Expression is normalized to KHYG1 KD mRNA expression (n = 3 experiments). (c) Representative fluorescence-activated cell-sorting (FACS) plots showing perforin versus tNGFR expression in wild-type and KD KHYG1 cells, and KHYG1 KD cells transduced with MND4T, PGK4T, or PRF0T LV vector; bar diagram shows the cumulative results of Prf1 mean fluorescence intensity (MFI) in these cells (n = 3 experiments). (d) Representative Western blot analysis showing relative perforin protein in KHYG1 WT, KD, and KD cells transduced with the three LV vectors. Numbers represent fold increase in expression over KD cells. (e) Cumulative data from Western blots showing fold increase in Prf1 protein expression compared with KHYG1 KD cells. Error bars represent the SEM. *p < 0.05.
We determined the relative perforin expression from the PGK4T, MND4T, and PRF0T LV vectors in the KHYG1 cell line, a human NK cell line where we have stably suppressed perforin expression by approximately 90% with shRNA specific to the 3′ UTR of perforin (KHYG1-KD).22,31 The perforin cDNA in LV vectors lacks the 3′ UTR and is hence unaffected by the shRNA. KHYG1-KD cells were transduced with all three LV vectors at a low MOI and sorted for tNGFR-expressing cells to enrich for transduced cells with single or low VCN and tested for perforin expression in three independent transductions. The mean VCNs were comparable: 1.22, 1.25, and 1.48 VCN in MND4T, PGK4T, and PRF0T KHYG1-KD cells, respectively.
The relative perforin mRNA expression in MND4T KHYG1-KD cells was about 2-fold higher than that in PGK4T and PRF0T cells and nearly comparable to wild-type perforin expression in KHYG1 cells (Fig. 2b). Flow cytometric analysis showed that both the percentage of cells and the mean fluorescence intensity of perforin were 2-fold higher in MND4T KYHG1-KD cells as compared with PGK4T and PRF0T KYHG1-KD cells (Fig. 2c). Results from Western blotting revealed a similar trend in perforin protein levels from the MND4T LV vector compared with PGK4T and PRF0T LV vectors, and that was comparable to wild-type perforin expression (Fig. 2d and e; original Western blot is shown in Supplementary Fig. S1; supplementary data are available online at www.liebertpub.com/hum). Taken together, expression of perforin from the MND4T vector in a human NK cell line was the highest compared with other vectors and was equivalent to near normal levels.
Comparison of HLH phenotype between transplanted and untransplanted Prf1−/− mice after LCMV challenge
The murine transplantation model we have previously used included lethal radiation to ablate the marrow in the recipient mice.15 In the current study, we administered busulfan and cyclophosphamide for myeloablation in order to better approximate clinical strategies used in human HCT. After stable engraftment at 16 weeks, perforin-deficient (Prf1−/−) mice were infected with LCMV to induce the HLH phenotype (experimental schema in Supplementary Fig. S2). Eight days after LCMV challenge, evaluation of CTL function in splenocytes from a subgroup of mice showed similar strong CTL activity from wild-type and transplanted wild-type (TxWT) mice (Fig. 3a), and similarly poor CTL function in the Prf1−/− or the transplanted (TxPrf1−/−) mice. A careful analysis showed distinct immunopathology in the transplanted mice compared with nontransplanted animals. We have previously shown that serum IFN-γ increases transiently after LCMV infection in wild-type mice, whereas in Prf1−/− mice, the increase in IFN-γ is highly exaggerated and persistent.14 The IFN-γ response was substantially blunted in the TxPrf1−/− compared with Prf1−/− mice, although still significantly higher than in TxWT mice (Fig. 3b). Cytopenias from the hemophagocytosis and inflammation, another characteristic feature of HLH, develop about 2 weeks after LCMV challenge in Prf1−/− mice, and these are normally minimal in wild-type mice. These differences were also blunted in TxPrf1−/− versus TxWT mice (Supplementary Fig. S3a and b).
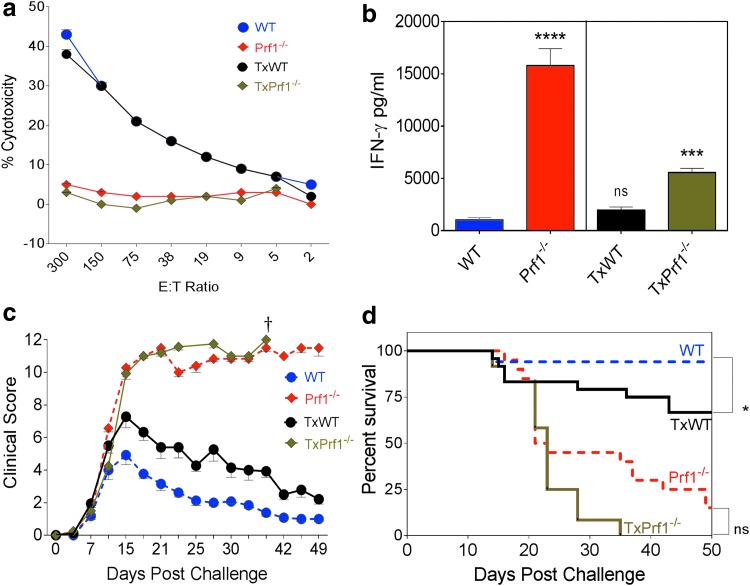
Figure 3.
Lymphocytic choriomeningitis virus (LCMV)-induced hemophagocytic lymphohistiocytosis (HLH) model comparing nontransplanted wild-type and Prf1−/− mice versus those transplanted with wild-type and Prf1−/− LSK cells. (a) Cytotoxic function analysis of splenocytes 8 days after LCMV challenge, using a 51Cr release assay with GP-33-loaded EL4 cell targets. Symbols represent means and error bars represent the SEM from four mice per group (three mice in the PGK4T group). (b) IFN-γ levels were measured in peripheral blood 8 days after LCMV infection. Statistics were done by one-way Dunnett's analysis of variance, comparing all groups with the wild-type mice. (c) Mice challenged with LCMV were clinically scored two or three times per week. Clinical scoring is represented for groups until they had ≥10% survival rate (n = 6 experiments; wild-type, n = 16; TxWT, n = 22; Prf1−/−, n = 16; TxPrf1−/−, n = 12 mice). (d) Survival of LCMV-challenged mice (n = 6 experiments; wild-type, n = 16; TxWT, n = 22; Prf1−/−, n = 20; TxPrf1−/−, n = 12 mice). Statistics were done by Mantel–Cox test. Error bars represent the SEM. †Death of all mice in that group. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
The clinical features of HLH were also compared. For clinical assessments, mice were graded for weight loss, activity, eye infections, skin turgor, and features of distress two or three times per week for 50 days after LCMV infection, after which the surviving mice were sacrificed. Parameters were graded on a 0–2/0–3 scale, with higher grades given for more severe signs or symptoms. In wild-type mice, clinical scores peaked 15 days after LCMV infection and then came down and stayed low for the remainder of the experiment. In TxWT mice, peak scores were higher than for untransplanted wild-type mice, and there was a delay before they returned to baseline (Fig. 3c). In the Prf1−/− and TxPrf1−/− mice, the clinical scores peaked and did not subside and mice succumbed to HLH, with the TxPrf1−/− mice dying earlier than the Prf1−/− mice. Finally, survival in TxWT mice was lower than among the wild-type mice (Fig. 3d). Hence, differences in clinical scores and survival reflected transplant-associated mortality, with earlier death in TxPrf1−/− mice and some mortality even in TxWT mice, compared with untransplanted counterparts.
These findings suggested that the HLH phenotype in LSK cell-transplanted mice with busulfan/cytoxan conditioning was distinct. Because of the transplant-related mortality noted in wild-type mice, we reasoned that large numbers of animals and multiple experiments would be necessary to distinguish between HCT- and HLH-related mortality in experiments evaluating gene correction of Prf1−/− animals.
High levels of stable transgene chimerism correct cytotoxicity and serum IFN-γ levels
We next evaluated the therapeutic efficacy of the MND4T, PGK4T, and PRF0T LV vectors in the Prf1−/− mouse model of HLH, using the same myeloablation protocol. Prf1−/− LSK cells transduced with the three LV vectors were transplanted into Prf1−/− mice (Supplementary Fig. S2). Expression of tNGFR from the mini-CMV promoter (Fig. 2a) provided a measure of transduction efficiency in PGK4T and MND4T LV vector-transduced HSCs (Supplementary Fig. S4a). Unexpectedly, however, HSCs transduced with the PRF0T vector had reduced expression of tNGFR in HSCs, but not in NK cells in the peripheral blood 12 weeks posttransplantation (Supplementary Fig. S4a). This suggested that the repressive elements that bind the PRF promoter in cells not meant to naturally express perforin,32 such as HSPCs, also repressed the CMV minimal promoter immediately adjacent to the perforin promoter in the PRF0T vector (Fig. 2a). Hence, tNGFR expression was also corepressed along with the perforin promoter in those cells. Just like the natural regulation of perforin, where NK cells express perforin constitutively,32 repression of the mini-CMV promoter-driven tNGFR was not observed in NK cells (Fig. 4a). Naive CD8+ cells do not constitutively express perforin, expressing it only when activated32; here, the repression of tNGFR expression was seen in naive CD8+ cells (Supplementary Fig. S4b), which was removed in activated CD8+ cells after LCMV infection (Fig. 4d, inset). Hence, the perforin promoter coregulated the adjacent mini-CMV promoter and conferred it the same lineage specificity, because tNGFR expression paralleled the natural expression of perforin. The chimerism of genetically modified cells was therefore determined by tNGFR expression in resting NK cells, which express perforin constitutively.
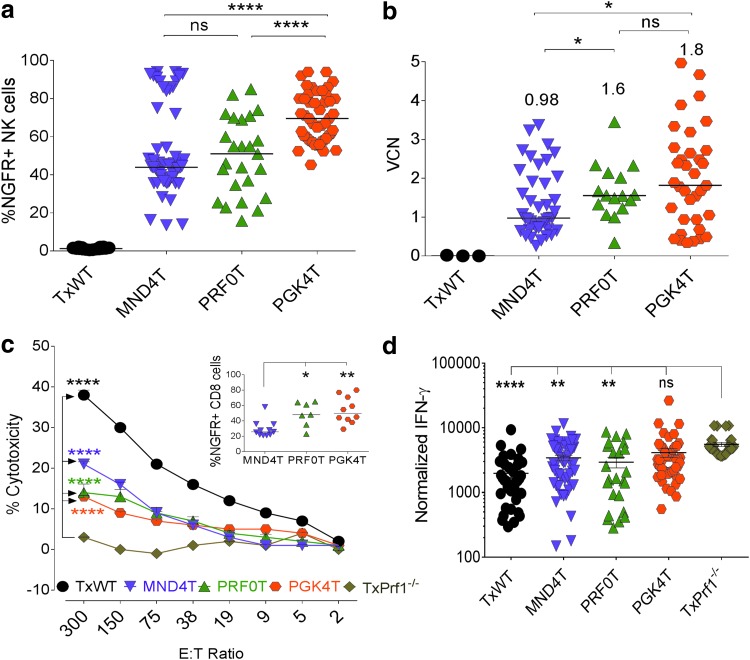
Figure 4.
High levels of stable transgene chimerism in transplanted Prf1−/− mice and significant correction of HLH phenotype. Twelve weeks posttransplantation (before LCMV challenge), peripheral blood from mice was analyzed for (a) gene-modified NK cell chimerism in peripheral blood by flow cytometry for tNGFR expression. Statistics were done by one-way Dunnett's analysis of variance (ANOVA) and (b) vector copy number (VCN) on leukocyte DNA by qPCR. Median tNGFR expression of VCN is indicated by the horizontal bars. Statistics were done by Mann-Whitney U test. At 16 weeks transplanted mice were challenged with LCMV. (c) Splenocytes were tested for cytotoxic function by a 51Cr release assay with GP-33-loaded EL4 target cells. Error bars represent the SEM from 8 to 12 mice per group or three mice in the TxPrf1−/− group. Statistics were done by two-way Dunnett's ANOVA. Inset: Percent tNGFR expression in CD8+ cells in the splenocytes 8 days after LCMV challenge (n = 10 experiments). Horizontal bars represent median values in each group. For the inset, statistics were done by one-way Dunnett's ANOVA. (d) Serum IFN-γ levels were measured in peripheral blood 8 days after LCMV infection and normalized to VCN in peripheral blood preinfection. Statistics were done by one-way Dunnett's ANOVA. Error bars represent the SEM. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Median (and mean ± SEM) gene-modified NK cell chimerism was 44% (52 ± 3%), 51% (50 ± 4%), and 70% (70 ± 2%) in MND4T, PRF0T and PGK4T mice, respectively, 12 weeks after HCT, with significantly higher chimerism in the PGK4T mice (Fig. 4a). Notably, the median VCN in blood leukocytes were comparable between the PGK4T and PRF0T groups of mice (1.8 and 1.6 VCN, respectively), but was significantly lower in MND4T mice at 0.98 (Fig. 4b). Wild-type and Prf1−/− HSPCs were also transplanted into Prf1−/− mice (TxWT and TxPrf1−/−) as controls for the vector-modified Prf1−/− groups. We then induced HLH with LCMV infection 4 months after HCT and studied restoration of cytotoxicity.
Eight days after LCMV infection, a subset of mice in each of these groups was sacrificed to determine CTL activity of splenocytes against target EL4 cells ex vivo; tNGFR expression was also determined on CD8+ T splenocytes. In this subset, median (mean ± SEM) gene-modified CD8+ T cells were lowest in the MND4T splenocytes (25% [29 ± 3%]) versus those in the PRF0T and PGK4T splenocytes (48% [49 ± 6%] and 50% [54 ± 6%], respectively [inset, Fig. 4c]). Despite half the chimerism, the CTL activity tended to be higher in the MND4T mice, closely followed by the PRF0T and PGK4T mice, albeit lower than that seen in TxWT mice, where all CTLs expressed perforin (Fig. 4c). IFN-γ levels in the MND4T and PRF0T groups were significantly lower than in the TxPrf1−/− mice, although IFN-γ levels remained high in the PGK4T mice (Fig. 4d). However, there were minimal differences in peripheral blood cytopenias within the three groups compared with TxPrf1−/− mice (Supplementary Fig. S3a and b).
Improved clinical score and survival are observed with the MND4T LV vector
The remaining mice were monitored for clinical symptoms of HLH for up to 50 days after LCMV challenge. The median (and mean ± SEM) gene-modified chimerism in these mice was 44% (51 ± 3%), 55% (54 ± 5%), and 67% (70 + 2%); and the VCN was 1 (1.4 ± 0.2), 1.6 (1.8 ± 0.2), and 1.9 (2 ± 0.2) in the MND4T, PRF0T, and PGK4T groups of animals, respectively (Supplementary Fig. S5a and b), similar to the chimerism and VCN in all of the mice in the three groups (Fig. 4a and b).
As expected, all animals became clinically sick within 2 weeks of LCMV infection, with the TxPrf1−/− and PGK4T animals having worse (higher) clinical scores than the PRF0T, MND4T, and TxWT animals. The TxWT and MND4T mice started improving thereafter, but the PRF0T mice continued to worsen clinically until their clinical scores were similar to those of the PGK4T and TxPrf1−/− animals. Overall, the MND4T group of mice had the lowest clinical scores, which correlated with the highest survival of the three vector groups (Fig. 5a and b). The PGK4T and PRF0T animals maintained similar high clinical scores, with almost all of the mice progressively succumbing to the challenge, even though the PRF0T mice had significantly lower IFN-γ, were alive much longer, and had lower gene-modified cell chimerism than the PGK4T mice.
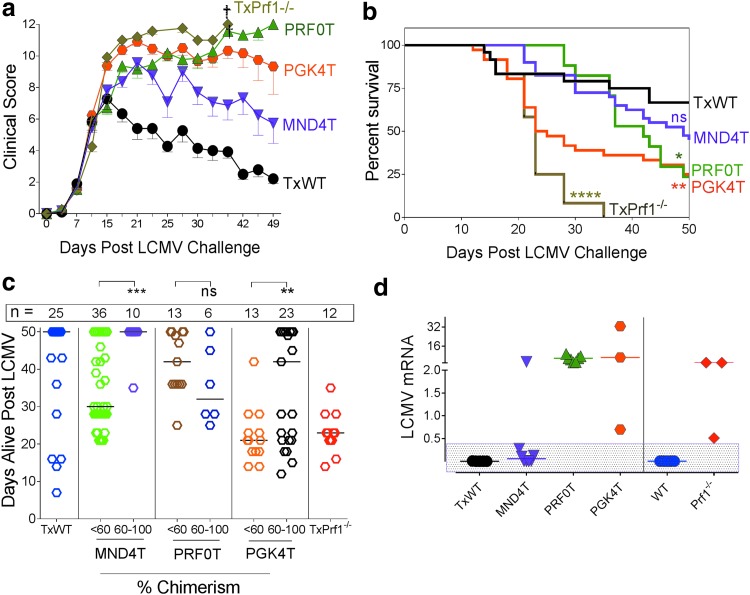
Figure 5.
Improved clinical score, viral clearance, and survival are observed with lineage-restricted MND4T vector expressing perforin. (a) Mice challenged with LCMV 16 weeks after transplantation were clinically scored three times weekly for 50 days. Clinical scoring is represented for groups until they had ≥10% survival per group (n = 10 experiments; TxWT, n = 20; MND4T, n = 35; PRF0T, n = 11; PGK4T, n = 29; TxPrf1−/−, n = 12 mice). Error bars represent the SEM. (b) Kaplan–Meier survival curves for the same group of mice as in (a), measured up to 50 days. Statistics were done by Mantel–Cox test. (c) Length of survival (in days) among the mice with low chimerism (<60%) versus those with high chimerism (60–100%) within each group after LCMV challenge. Each symbol represents one mouse; the number of mice per group is stated above the group. Percent chimerism is derived from tNGFR-expressing NK cells at 12 weeks posttransplantation. Medians are denoted by a horizontal line. Statistics: Unpaired t test with Welch correction within each vector group. (d) LCMV mRNA in the bone marrow of mice that were either alive on day 50 (mice represented in the rectangular shaded box) or were sacrificed when moribund. mRNA expression was normalized to expression in Prf1−/− mice sacrificed 8 days after LCMV challenge (n = 3 experiments). Medians are denoted by a horizontal line. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. †Death of all mice in that group.
Because the MND4T group had some mice with high chimerism (>60%) and those with moderate to low chimerism (Fig. 4a), we analyzed whether higher gene-modified cytotoxic cells conferred longer survival. Mice with 60% or greater chimerism in NK cells in the MND4T and PGK4T groups were alive for significantly more days than were mice with lower chimerism (below 60%; Fig. 5c). However, in the PRF0T group, there was no correlation of mortality with chimerism (or IFN-γ levels); and although they were alive longer than PGK4T mice, they had similar eventual mortality.
We then determined LCMV viral RNA in bone marrow from the limited number of surviving mice and from those that were sacrificed when moribund (Fig. 5d). LCMV mRNA was high in the PRF0T and PGK4T groups, and was at low levels in the MND4T mice. All surviving mice after LCMV challenge (shown in the rectangular shaded box, Fig. 5d) had extremely low levels of LCMV mRNA, consistent with viral clearance or low-grade chronic infection. These data were also confirmed with the LCMV plaque-forming assay (Supplementary Fig. S5c). Hence, our data suggest that low perforin expression improves some of the inflammatory parameters, but there is little to no LCMV clearance, which is associated with mortality from HLH in this model.
Discussion
Autologous transplantation with gene therapy provides a potentially attractive treatment option that is not limited by donor availability and the adverse immunological consequences associated with allogeneic HCT. Herein, we show that FHLH2 requires high (normal) levels of perforin expression per cell for complete correction.
Strong enhancers in the gammaretroviral LTR were shown to cause oncogenesis by activating proto-oncogenes surrounding insertion sites. Since then, LV vectors with self-inactivating (SIN) LTR design and internal tissue-specific/cellular promoters were developed to reduce genotoxicity. However, with the perforin and PGK promoter-driven LV vectors, despite high gene-modified chimerism, there was partial correction of HLH. We observed that the MND promoter/enhancer was necessary for normal levels of perforin expression from LV vectors in a human NK cell line, whereas the PGK/PRF promoters had significantly lower expression.
Similar phenomena are now being discovered in other diseases: In leukocyte adhesion deficiency (LAD), CD18 expression from PGK promoter-driven vector was insufficient to correct the LAD phenotype in dogs, and necessitated a strong promoter/enhancer from the MSCV-LTR U3 region for disease correction16; and in gene therapy for Wiskott–Aldrich syndrome (WAS), where the WAS cellular promoter significantly corrected the phenotype, but only partially corrected the thrombocytopenia.19,20 The MND(U3) region of a modified gammaretroviral LTR is a strong promoter/enhancer, and has been previously successfully used internally in an LV vector in a clinical trial of gene therapy for adrenoleukodystrophy, without evidence of genotoxicity after 10 years of follow-up; it has also been shown to adequately correct the WAS phenotype in preclinical studies.33–35
To avoid high expression of a pore-forming protein in HSPCs, we restricted expression in HSPCs by using miR126 target elements.25,26 Transgene expression in HSPCs in globoid leukodystrophy was found to be toxic to HSPCs, which was overcome by placing miR126 target sites in the vector.22 Although this may cause some reduction in the HSPC miR126 pool, studies in normal HSCs show that miR126 attenuation does not adversely affect HSPC function, and may even expand them.36
Our studies on functional correction of FHLH2 in the Prf1−/− mouse model were performed with LSK transplantation and myeloablation to simulate a human clinical gene therapy trial. Unexpectedly, with this modified protocol, we noted blunted cytopenias and IFN-γ responses in TxPrf1−/− mice compared with previous methodologies.14 Moreover, TxWT mice also experienced some illness and mortality after LCMV infection. We suspect these differences relate to relatively slower immune reconstitution with the revised protocol. Although there are limitations to the murine model, with a narrow margin for successful correction of HLH pathology, this approach is favored from a translational perspective. Patients will receive transduced CD34+ HSPCs, after a similar myeloablation. It may be beneficial to consider transplantation of genetically modified T cells along with genetically modified HSCs, to prevent adverse consequences from peritransplant infectious complications while a new immune system emerges from the gene-corrected HSCs.
Despite these caveats, transfer of the perforin gene into HSCs led to a significant correction of the IFN-γ levels and ex vivo cytotoxicity with the MND4T and PRF0T LV vectors, and partial correction with the PGK4T vector, as compared with TxPrf1−/− mice. Notably, the MND4T LV vector outperformed in all aspects, with nearly half the VCN and lower gene-modified cell chimerism. Also, despite similar VCN and perforin expression from the PGK4T and PRF0T mice, there was much better reduction in IFN-γ levels and longer survival in the PRF0T mice, suggesting a regulated promoter may be beneficial; although the eventual dismal outcome in both groups appears to be from subnormal perforin expression and inability to clear/suppress LCMV; TxWT and MND4T mice had LCMV mRNA levels consistent with low-grade chronic LCMV infection and/or clearance. It is to be noted that the PRF promoter comprises a small fragment (1.4 kb) of an otherwise 150-kb upstream cis-regulatory region that regulates perforin expression.32 Hence identification and inclusion of important cis-regulatory elements in the perforin promoter that can boost the level of perforin expression per cell may improve this cellular promoter.
The importance of perforin in the feedback control of immune activation has been studied in the context of HLH, and there exists a perforin-dependent reciprocal relationship between dendritic cells and CD8+ T cells.37,38 In the absence of perforin and in the face of a viral infection, Prf1−/−CD8+ T cells are conceivably incapable of providing feedback control of immune activation, besides being incapable of reducing the viral load, which rises and/or stabilizes in the liver and spleen of infected mice.31 Our data suggest that moderate, but subnormal levels of perforin in CD8+ T and NK cells are sufficient to provide the feedback to decrease the exaggerated inflammation and immune response; however, complete correction and recovery from HLH, including viral clearance, requires a much higher level of perforin expression (i.e., near normal levels), and was achievable by the MNDU3 promoter/enhancer. It will be of interest to study the expression of this promoter in primary CD8+ T cells and NK cells of perforin-deficient patients with HLH for robust validation.
In conclusion, our data suggest that for gene therapy of FHLH2, cellular promoters are capable of providing only partial phenotypic correction, and complete correction of the phenotype requires robust perforin expression that can be facilitated by a strong MND enhancer in an LV vector.
Supplementary Material
Acknowledgments
This work has been funded by a Translational Research Pilot Grant Award from the University of Cincinnati CCTST (M.B.J. and P.M.) and by a grant from Liam's Lighthouse Foundation (M.B.J. and K.A.R.). The authors acknowledge the support of Dr. H.B. Gaspar (Great Ormond Street Hospital Children's Charity). S.T., A.H., C.T., M.J., K.A.R., and P.M. designed the experiments and wrote the manuscript. S.T. performed the cloning and production of the lentiviral vectors. S.T. and C.T. performed the murine experiments. A.H. was responsible for the NK cell differentiation and the chromium release assays. P.A. and M.C. provided input and preliminary data for the design of the experiments. All authors have contributed to their respective portions of the manuscript and reviewed the manuscript. A special thanks to Anastacia Loberg, Katie Burke, Sarah Figuera, and Michael Wourms for their contributions to the murine experiments.
Author Disclosure
No competing financial interests exist.
References
- 1.Henter JI, Horne A, Arico M, et al. HLH-2004: diagnostic and therapeutic guidelines for hemophagocytic lymphohistiocytosis. Pediatr Blood Cancer 2007;48:124–131 [DOI] [PubMed] [Google Scholar]
- 2.Risma K, Jordan MB. Hemophagocytic lymphohistiocytosis: updates and evolving concepts. Curr Opin Pediatr 2012;24:9–15 [DOI] [PubMed] [Google Scholar]
- 3.Rosado FG, Kim AS. Hemophagocytic lymphohistiocytosis: an update on diagnosis and pathogenesis. Am J Clin Pathol 2013;139:713–727 [DOI] [PubMed] [Google Scholar]
- 4.Jordan MB, Allen CE, Weitzman S, et al. How I treat hemophagocytic lymphohistiocytosis. Blood 2011;118:4041–4052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang K, Jordan MB, Marsh RA, et al. Hypomorphic mutations in PRF1, MUNC13-4, and STXBP2 are associated with adult-onset familial HLH. Blood 2011;118:5794–5798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gholam C, Grigoriadou S, Gilmour KC, et al. Familial haemophagocytic lymphohistiocytosis: advances in the genetic basis, diagnosis and management. Clin Exp Immunol 2011;163:271–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stepp SE, Dufourcq-Lagelouse R, Le Deist F, et al. Perforin gene defects in familial hemophagocytic lymphohistiocytosis. Science 1999;286:1957–1959 [PubMed] [Google Scholar]
- 8.Voskoboinik I, Smyth MJ, Trapani JA. Perforin-mediated target-cell death and immune homeostasis. Nat Rev Immunol 2006;6:940–952 [DOI] [PubMed] [Google Scholar]
- 9.Lopez JA, Susanto O, Jenkins MR, et al. Perforin forms transient pores on the target cell plasma membrane to facilitate rapid access of granzymes during killer cell attack. Blood 2013;121:2659–2668 [DOI] [PubMed] [Google Scholar]
- 10.Katano H, Cohen JI. Perforin and lymphohistiocytic proliferative disorders. Br J Haematol 2005;128:739–750 [DOI] [PubMed] [Google Scholar]
- 11.Marsh RA, Kim MO, Liu C, et al. An intermediate alemtuzumab schedule reduces the incidence of mixed chimerism following reduced-intensity conditioning hematopoietic cell transplantation for hemophagocytic lymphohistiocytosis. Biol Blood Marrow Transplant 2013;19:1625–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chandrakasan S, Filipovich AH. Hemophagocytic lymphohistiocytosis: advances in pathophysiology, diagnosis, and treatment. J Pediatr 2013;163:1253–1259 [DOI] [PubMed] [Google Scholar]
- 13.Terrell CE, Jordan MB. Mixed hematopoietic or T-cell chimerism above a minimal threshold restores perforin-dependent immune regulation in perforin-deficient mice. Blood 2013;122:2618–2621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jordan MB, Hildeman D, Kappler J, et al. An animal model of hemophagocytic lymphohistiocytosis (HLH): CD8+ T cells and interferon gamma are essential for the disorder. Blood 2004;104:735–743 [DOI] [PubMed] [Google Scholar]
- 15.Carmo M, Risma KA, Arumugam P, et al. Perforin gene transfer into hematopoietic stem cells improves immune dysregulation in murine models of perforin deficiency. Mol Ther 2015;23:737–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hunter MJ, Zhao H, Tuschong LM, et al. Gene therapy for canine leukocyte adhesion deficiency with lentiviral vectors using the murine stem cell virus and human phosphoglycerate kinase promoters. Hum Gene Ther 2011;22:689–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barde I, Laurenti E, Verp S, et al. Lineage- and stage-restricted lentiviral vectors for the gene therapy of chronic granulomatous disease. Gene Ther 2011;18:1087–1097 [DOI] [PubMed] [Google Scholar]
- 18.Chiriaco M, Farinelli G, Capo V, et al. Dual-regulated lentiviral vector for gene therapy of X-linked chronic granulomatosis. Mol Ther 2014;22:1472–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aiuti A, Biasco L, Scaramuzza S, et al. Lentiviral hematopoietic stem cell gene therapy in patients with Wiskott–Aldrich syndrome. Science 2013;341:1233151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hacein-Bey Abina S, Gaspar HB, Blondeau J, et al. Outcomes following gene therapy in patients with severe Wiskott–Aldrich syndrome. JAMA 2015;313:1550–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson TS, Terrell CE, Millen SH, et al. Etoposide selectively ablates activated T cells to control the immunoregulatory disorder hemophagocytic lymphohistiocytosis. J Immunol 2014;192:84–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brennan AJ, Chia J, Browne KA, et al. Protection from endogenous perforin: glycans and the C terminus regulate exocytic trafficking in cytotoxic lymphocytes. Immunity 2011;34:879–892 [DOI] [PubMed] [Google Scholar]
- 23.Carayol G, Robin C, Bourhis JH, et al. NK cells differentiated from bone marrow, cord blood and peripheral blood stem cells exhibit similar phenotype and functions. Eur J Immunol 1998;28:1991–2002 [DOI] [PubMed] [Google Scholar]
- 24.Hildeman D, Salvato M, Whitton JL, et al. Vaccination protects β2 microglobulin deficient mice from immune mediated mortality but not from persisting viral infection. Vaccine 1996;14:1223–1229 [DOI] [PubMed] [Google Scholar]
- 25.Gentner B, Visigalli I, Hiramatsu H, et al. Identification of hematopoietic stem cell-specific miRNAs enables gene therapy of globoid cell leukodystrophy. Sci Transl Med 2010;2:58ra84. [DOI] [PubMed] [Google Scholar]
- 26.Petriv OI, Kuchenbauer F, Delaney AD, et al. Comprehensive microRNA expression profiling of the hematopoietic hierarchy. Proc Natl Acad Sci U S A 2010;107:15443–15448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fehniger TA, Wylie T, Germino E, et al. Next-generation sequencing identifies the natural killer cell microRNA transcriptome. Genome Res 2010;20:1590–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Landgraf P, Rusu M, Sheridan R, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 2007;129:1401–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Challita PM, Skelton D, el-Khoueiry A, et al. Multiple modifications in cis elements of the long terminal repeat of retroviral vectors lead to increased expression and decreased DNA methylation in embryonic carcinoma cells. J Virol 1995;69:748–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robbins PB, Yu XJ, Skelton DM, et al. Increased probability of expression from modified retroviral vectors in embryonal stem cells and embryonal carcinoma cells. J Virol 1997;71:9466–9474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vrazo AC, Hontz AE, Figueira SK, et al. Live cell evaluation of granzyme delivery and death receptor signaling in tumor cells targeted by human natural killer cells. Blood 2015;126:e1–e10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pipkin ME, Rao A, Lichtenheld MG. The transcriptional control of the perforin locus. Immunol Rev 2010;235:55–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koldej RM, Carney G, Wielgosz MM, et al. Comparison of insulators and promoters for expression of the Wiskott–Aldrich syndrome protein using lentiviral vectors. Hum Gene Ther Clin Dev 2013;24:77–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cartier N, Hacein-Bey-Abina S, Bartholomae CC, et al. Lentiviral hematopoietic cell gene therapy for X-linked adrenoleukodystrophy. Methods Enzymol 2012;507:187–198 [DOI] [PubMed] [Google Scholar]
- 35.Cartier N, Hacein-Bey-Abina S, Bartholomae CC, et al. Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science 2009;326:818–823 [DOI] [PubMed] [Google Scholar]
- 36.Lechman ER, Gentner B, van Galen P, et al. Attenuation of miR-126 activity expands HSC in vivo without exhaustion. Cell Stem Cell 2012;11:799–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Terrell CE, Jordan MB. Perforin deficiency impairs a critical immunoregulatory loop involving murine CD8+ T cells and dendritic cells. Blood 2013;121:5184–5191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lykens JE, Terrell CE, Zoller EE, et al. Perforin is a critical physiologic regulator of T-cell activation. Blood 2011;118:618–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.