Abstract.
An important step in establishing the diagnostic potential for emerging optical imaging techniques is accurate registration between imaging data and the corresponding tissue histopathology typically used as gold standard in clinical diagnostics. We present a method to precisely register data acquired with a point-scanning spectroscopic imaging technique from fresh surgical tissue specimen blocks with corresponding histological sections. Using a visible aiming beam to augment point-scanning multispectral time-resolved fluorescence spectroscopy on video images, we evaluate two different markers for the registration with histology: fiducial markers using a 405-nm CW laser and the tissue block’s outer shape characteristics. We compare the registration performance with benchmark methods using either the fiducial markers or the outer shape characteristics alone to a hybrid method using both feature types. The hybrid method was found to perform best reaching an average error of . This method provides a profound framework to validate diagnostical abilities of optical fiber-based techniques and furthermore enables the application of supervised machine learning techniques to automate tissue characterization.
Keywords: registration, histology, point-scanning, fluorescence, shape matching, fiducials
1. Introduction
One of the major goals of cancer treatment is to excise the entire malignant tissue surrounded by a rim of normal tissue to rule out residual malignant cells. Studies have demonstrated that negative resection margins minimize the risk of local recurrence and maximize overall survival.1 Frozen section and retrospective histological analysis are currently the gold standard to evaluate negative margins and to grade tissue on a benign–malignancy scale. However, these methods are time consuming, subject to sampling error, require specialized personnel, and have intrinsic variability in subjective interpretation.
Optical methods have demonstrated considerable potential for intraoperatively localization of tumor regions in real time. Several studies have indicated the diagnostic relevance of optical techniques for tumor margin assessment. This includes Raman spectroscopy,2,3 elastic scattering spectroscopy,4,5 optical coherence tomography,6,7 infrared spectroscopy,8,9 photoacoustic tomography,10 and fluorescence techniques.11,12 Particularly, fiber-based fluorescence lifetime imaging is one of the most promising approaches, as it can rapidly characterize structural, biochemical, or functional properties of tissue.
A major challenge in demonstrating the diagnostic value of any of these optical technologies is the ability to validate the optical data against gold standard methods. For example, to evaluate the tumor margins in breast cancer lumpectomy specimens, measurements from optical imaging modalities must be correlated with pathologic findings. Histology is the gold standard for cancer diagnosis, grading, and localization. However, due to uneven tissue fixation and gland sectioning, histological preparation results in duct dilation, gland deformation, and tissue loss, making the registration with imaging data a highly challenging task.
Recent studies correlate fluorescence lifetime signatures with histological findings based on visual correspondence13 or ink markers14 being either imprecise or restricted to single points of interest. For other modalities, histological registration has been addressed by correlating different kinds of features across modalities or combinations thereof. The most common approach involves fiducial markers.15 Fiducial markers are nonanatomical and artificial landmarks that appear across different modalities. Specifically, these markers provide robustness to appearance variations. As fiducial marking destroys or hides histological information, the number of markers should be kept at a necessary minimum. Registration approaches based on anatomic image features16,17 could overcome this issue but demand the crossmodality presence of anatomical features. Other approaches compare and correlate anatomical surface profiles in different modalities.18,19 Because of possible tissue deformation and loss resulting from histological preparation this becomes particularly challenging for histological registration.
Using a previously described method enabling a registration between autofluorescence imaging data and video white-light images,20 we describe a method to register the white-light images with the associated crosssectional histology slide of specimen blocks, which can be used to correlate autofluorescence signatures with histology data. Moreover, all necessary steps such as marker placement, imaging, and registration can principally be fully automated using a mechanical stage. To the best of our knowledge, this paper presents the first report on the registration of autofluorescence imaging data with ex vivo histological images. To demonstrate this method, we (a) generated fiducial laser markers using a 405-nm CW laser diode on the tissue block, (b) analyzed the registration performance when the fiducial markers or the outer shape characteristics are used for registration purposes, and (c) analyzed the registration performance of a hybrid approach, which combines both feature types to achieve an optimal registration with a minimum number of fiducial markers. The method was demonstrated using optical data acquired using a point-scanning fluorescence lifetime imaging system from 12 distinct human breast lumpectomy or mastectomy specimen blocks.
2. Materials and Method
2.1. Specimen and Histological Preparation
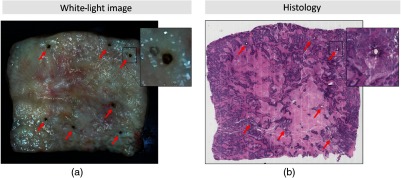
We obtained fresh specimens through approved consent from patients (three lumpectomy and seven mastectomy) at the University of California Davis Health System (UCDHS). The UCDHS Institutional Review Board approved this study. The tissue was assessed by a pathologist and a block with a diameter of 15 to 30 mm that was assumed to contain a tumor that was cut out for each specimen (one block per patient). Prior to imaging, a 405-nm CW laser diode was used to generate clearly observable landmarks (fiducial points) on the tissue block, as shown in Fig. 1. These landmarks can be easily detected in both modalities: white light and histology images. Another advantage of the thermal laser landmarks is that they can be integrated into a mechanical stage to automate the placement of the fiducial markers. The landmarks were distributed evenly and the number of laser markers ranged from 4 to 8 per specimen block depending on its size. Following imaging procedure, all histology sections were cut at thickness using a microtome, parallel to the imaging plane. The first few cuts from such specimens were discarded and the slice that is stained is the first continuous large section. The tissues were processed using standard histological processing protocols and the tissue sections were stained with hematoxylin and eosin (H&E). The histology slides were scanned with an Aperio Digital Pathology Slide Scanner (Leica Biosystems) to generate a digital image of the histology.
Fig. 1.
(a) White-light image of an excised breast specimen block and (b) histology slide with H&E staining. The laser marks can be identified in both modalities serving as fiducial markers for the developed registration method.
2.2. Instrumentation
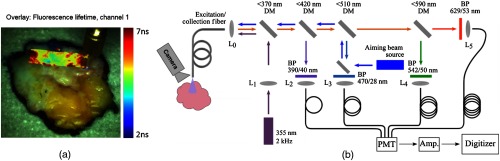
The registration method was demonstrated with experimental data from the time-domain multispectral time-resolved fluorescence spectroscopy (ms-TRFS) with an integrated aiming-beam system [Fig. 2(b)] described elsewhere.20,21 In brief, this system consists of a fluorescence excitation source, a wavelength-selection module, and a fast-response fluorescence detector and digitizer. The excitation source (Teem Photonics, France) emits pulses at 355 nm and 2-kHz repetition rate through a fiber. The same fiber collects and directs the autofluorescence to a wavelength-selection module consisting of dichroic mirrors and bandpass filters. Each channel outputs the autofluorescence to an optical delay fiber to merge the decay waveforms of the four channels into a single output, which is subsequently amplified (RF amplifier, AM-1607-3000, 3-GHz bandwidth, Miteq) and digitized (PXIe-5185, National Instruments, sampling rate).
Fig. 2.
(a) White-light image of an excised breast specimen block augmented with fluorescence lifetime from channel 1 centered at 390 nm and (b) schematic diagram of the instrumentation used for imaging purposes. A single fiber is used for excitation and autofluorescence collection. DM, dichroic mirror; BP, bandpass filter; , lens; PMT, photomultiplier; and amp, amplifier.
An external camera (point gray Chameleon3 1.3 MP Color USB3 vision with Fujinon HF9HA-1B 2/3″9 mm lens) captures the entire specimen block during the scanning procedure. To register fluorescence decay parameters with the video images, we use an aiming beam integrated into the second channel of the wavelength-selection module and emitted through the same optical fiber as the excitation source as described by Gorpas et al.20 The camera images are transferred to the HSV color space and thresholded in the Hue channel to localize the blue aiming-beam color and overlaid with fluorescence decay parameters. The scanning can be either performed motorized or free-hand. A motorized scan of an excised breast tissue specimen block augmented with fluorescence lifetime parameters is shown in Fig. 2(a).
2.3. Principle of the Registration Method
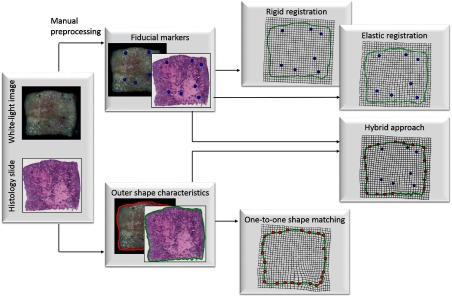
The dynamic aiming-beam augmentation provides registration of the fluorescence decay parameters with white-light video images. In this section, we will address the histological registration of white-light images obtained from the camera setup that was described in the previous section. We focus on two different features for registration, i.e., fiducial laser markers, as described in Sec. 2.1, and the outer shape characteristics of the specimen block. Here, we describe the rigid and elastic registration based on fiducial markers, a shape matching procedure that relies only on the outer shape characteristics, and finally introduce a hybrid method combining both feature types (Fig. 3).
Fig. 3.
Rigid and elastic registration based on fiducial markers, a shape matching procedure that relies only on outer shape characteristics, and finally introduce a hybrid method combining both feature types.
2.3.1. Registration based on fiducial markers
Given a pair consisting of histological image and white-light image , our goal is to derive an accurate alignment between and , which ideally can account for the potential local deformations. Recall that a number of fiducial markers are annotated on both and , which are denoted as and , respectively. For a given pair of corresponding markers and , let denote the correspondence assignment from the different modalities: , where and are the corresponding landmarks in histology and white-light image.
Rigid registration:
Given a cost function , the image registration process can be formulated as a minimization problem
| (1) |
where is a parameterized transform22 aiming to spatially align images and , and is a vector containing the transformation parameters. For similarity transformations , includes scaling , rotation , and translation , so that reads as
| (2) |
For optimally adapting , we exploit the fiducial markers and to estimate a similarity transformation between and by minimizing the mean distance of corresponding markers
| (3) |
Elastic registration:
The rigid registration provides only a rough alignment of histological and white-light images. Uneven tissue fixation and sectioning causes local deformations such as shrinkage, expansion, tears, and folds demanding for elastic registration models. To account for these deformations, the transformation consists of a rigid transformation and an elastic component
| (4) |
The rigid component captures only translation, rotation, and scaling, whereas the elastic component models local deformations. Because it is difficult to describe these local deformations with a parameterized transformation model, we use a B-spline free-form deformation model allowing deformation of an image by manipulating its underlying mesh of control points.23,24 The idea is to form a spatial subdivision of the histological and white-light images and solve a local transformation for each corresponding local region. The method can be efficiently implemented and provides a smooth and continuous transformation.23 Given a rectangular domain in the space
| (5) |
and a mesh with the control points with uniform spacing , a superposition of locally defined tensor products of cubic B-splines is used to model the spatial deformation
| (6) |
where , , and refers to the ’th B-spline basis function
and .
Generally, is not uniquely defined so that desirable properties can be incorporated using additional constraints. One of the most common constraints incorporates smoothness criteria, which can implemented by minimizing a penalty term
| (7) |
to obtain continuity.
In this paper, we use an algorithm proposed by Lee et al.23 to interpolate the fiducial markers. Because of the compact support of the B-Spline functions, computational efficiency, and the -continuity of the approximation function, the method has found wide applications.
2.3.2. Registration based on shape matching
Shape matching refers to the problem of finding pointwise correspondences between two shapes. The shape context descriptor has been proposed by Belongie et al.25,26 In this approach, a log-polar histogram is built for each contour point characterizing local properties of the shape. The histogram
| (8) |
represents the relative distribution of any contour point to the remaining points and is referred to as shape context, where denotes the ’th bin of the log-polar space. The costs of matching two points are given by the test statistic
| (9) |
Let be a permutation of the point indices. Minimizing the overall matching cost
| (10) |
for two different shapes and , where and result in an optimal bipartite graph matching providing pointwise correspondences between and . The minimization problem can be solved using the Hungarian method.27 See Belongie et al.25 for a detailed description of the implementation.
The shape context matching minimizes a Euclidean metric, which does not take tissue shrinkage happening after histological preparation of the tissue are taken into account. A modified version of shape context matching was introduced by Ling and Jacobs.28 They replaced the Euclidean metric with the inner distance, which is defined as the shortest path between contour points within a shape silhouette. Additionally, the relative angle between two contour points is replaced by the angle between the tangent at the starting point and the initial direction of the shortest path, referred to as the inner angle.28 These modifications make the matching insensitive to shape articulations.
2.3.3. Hybrid registration approach
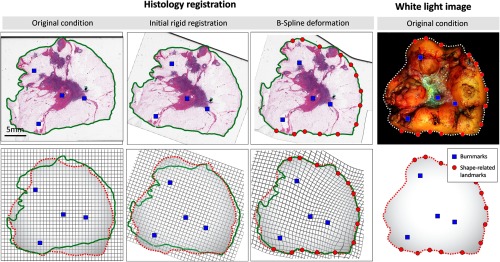
We present a hybrid approach that merges information obtained from fiducial markers and the outer shape of the specimen block to increase registration precision. The registration pipeline is basically built as a two-stage process and shown in Fig. 4. First, an initial rough registration is obtained from a rigid transformation based on the laser markers (see Sec. 2.3.1). Because tissue shrinkage and loss may affect the shape of the specimen block, shape information is often preserved only locally. In contrast to one-to-one boundary mappings as described in Sec. 2.3.2, we perform piecewise shape matching to increase the number of point correspondences and refine the initial rigid registration. We denote the enriched point sets for histology and white-light image as and . The final registration is obtained from a B-spline free-form deformation model using the fiducial markers and the point correspondences estimated by the piecewise shape matching are described in the following.
Fig. 4.
Registration pipeline: original conditions of histology image and white-light image are shown in the left-most and right-most columns. The boundaries obtained from thresholding are illustrated in green for histology and dashed white/red for the white-light image. The initial rigid alignment (second column) minimizes the distance between corresponding fiducial makers (blue points). The final (third column) registration is obtained from a B-spline free-form deformation involving the fiducial burnmarks as well as the shape-related landmarks obtained from the piecewise shape matching.
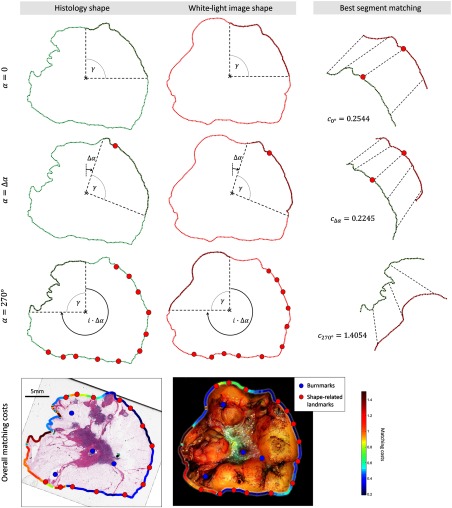
Figure 5 shows the difficulties arising from finding point correspondences on the outer shape of histology and white-light images. A comparison of both shapes points out a problem often occurring in the context of histology registration: while the outer shape on the right side of the specimen block remains intact after histologic preparation, it is significantly altered for the left side. To counteract these difficulties, we perform piecewise shape matching on the outer contours using a modified version of shape context matching with the inner-distance metric where local shape environments are matched.
Fig. 5.
The coregistration algorithm extracts segments from the outer shape and performs piecewise shape matching. Each pair of segments from white light and histological images are matched and the points with the lowest matching costs are added to and . Three different iterations are shown in the first three rows for , , 270 deg, respectively. The matching costs are encoded in the color of the outer boundary in the last row. The right side of the specimen block has considerably lower matching costs than the left side, resulting in additional landmarks on the right side.
Because shape information is preserved only locally, we identify matches in local shape environments. Here, the presented method is as follows: first, the outer contour is extracted from and using a simple threshold criterion. The specimen block is imaged on a black background facilitating an easy separation of fore- and background. Starting from the centroid of each contour
| (11) |
where is the number of contour points, the segment spanned by the vertical line passing the centroid, and the angle is extracted from and , respectively. Since the images have been previously aligned, the segments roughly correspond to the same part of the block. The inner-distance matching provides an optimal matching of the segments and the corresponding overall matching cost . The goodness of fit was obtained by dividing the overall matching cost by the number of matched points in the segment .
In this context, the choice of the angle is a crucial input as it represents a compromise between the precision and number of landmarks. If the angle is chosen too large, finding an additional landmark on the outer shape requires that large parts of the outer shape stay intact after histological preparation. On the other hand, if the angle is chosen too small, the correspondence of landmarks in both modalities might be inaccurate, leading to a less precise image registration. To filter out inaccurate shape matchings, a threshold criterion is used allowing only reasonable matches to produce additional landmarks. If the matching costs are below the given threshold , we add the correspondence with the lowest matching costs in the mid half of the current segment to the point sets and . We consider only points in the mid section of the segment to avoid matchings with a one-sided environment that might imply imprecise matching. Subsequently, the angular range is rotated clockwise by a constant offset , and the whole matching process is performed again until the entire contour is covered (Fig. 5). The matching cost , given for the entire shape of the specimen block, is presented in the last row of Fig. 5. On the left side of the specimen block, high matching costs indicate considerably altered shape characteristics making it more difficult to find corresponding landmarks on the shape. In this regard, no landmarks are found for the left side. Thus, changing the threshold will directly affect the number of landmarks on the outer shape. The registration will involve the fiducial markers only if chosen too low.
There are three basic parameters influencing the outcome of the piecewise shape matching: , , and . To optimize the parameter setting, we perform an iterative grid search for the three parameters (, , and ) on 11 of the 12 specimen blocks and test it on the one that has been left out.
3. Experimental Results
3.1. Registration Precision
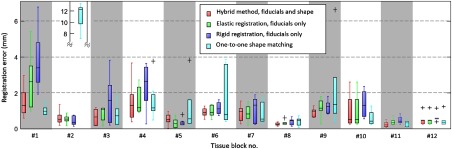
The fiducial markers can be seen as ground truth for the registration. An ideal registration minimizes the displacements between corresponding fiducial markers. In this regard, a leave-one-out strategy is pursued to evaluate the performance of different methods. For fiducial markers on each modality, we perform the registration with markers and evaluate the registration error occurring for the fiducial marker that has been left out within the histology image. Because the scale of histology slides is known, the errors are computed in the histology domain. The registration error for each specimen block is shown in Fig. 6. Evidently, none of the evaluated methods consistently outperforms all the other methods. However, the hybrid method obtains the minimum per-block average error of in contrast to , , and for B-spline approximation using the fiducials only, rigid registration, and shape matching. A nonparametric two-sided Wilcoxon signed rank test corroborated a significant difference between the hybrid and the B-spline approximation using the fiducials only (). For the hybrid method, the optimal parameter setting , , and was obtained from the leave-one-out optimization.
Fig. 6.
Registration error evaluated in the histology domain. The error is obtained from a leave-one-out validation scheme.
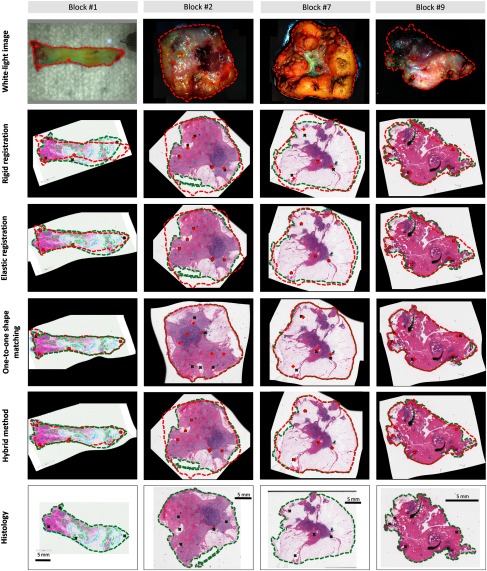
Figure 7 shows the results of the different registration methods for exemplarily chosen challenging cases. The fiducial markers are colored in red for the video domain and in green for histology. Note that for the elastic and hybrid registration all markers are perfectly matched, as they are used as control points for the B-spline interpolation. For the first block, the outer shape is considerably deformed but tissue is mostly preserved after histological preparation. In this case, the one-to-one shape matching performs best (according to Fig. 6) allowing for an adequate one-to-one matching due to the high degree of tissue preservation. Rigid registration does not account for tissue deformation and consequently provides poor alignment. Elastic registration follows the deformations to a certain degree but exhibits a contradicting larger histology domain. In contrast to block #1, parts of tissue are lost during histological preparation for block #2. In this case, the outer shape of the specimen block is considerably altered. Consequently, both modalities do not share the same outer shape and exploiting shape contour information alone results in severe misalignment. On the other hand, rigid and elastic registrations lead to a low error rate exploiting the fiducial markers only. For blocks 1 and 2, the hybrid registration provides the lowest overall error rate. For blocks 7 and 9, the favorable characteristics of the hybrid registration procedure can be observed because, it benefits from the partially well-preserved shape contours to enrich the set of corresponding landmarks to improve registration accuracy.
Fig. 7.
Examples of registration results from four different specimen blocks. Block 1 is considerably deformed but the tissue is mostly preserved after histological preparation, block 2 exhibits a large degree of tissue loss, block 7 has a well-preserved shape on the right side of the block whereas the opposing side is considerably deformed, and block 7 has a well preserved shape with only minor deformation.
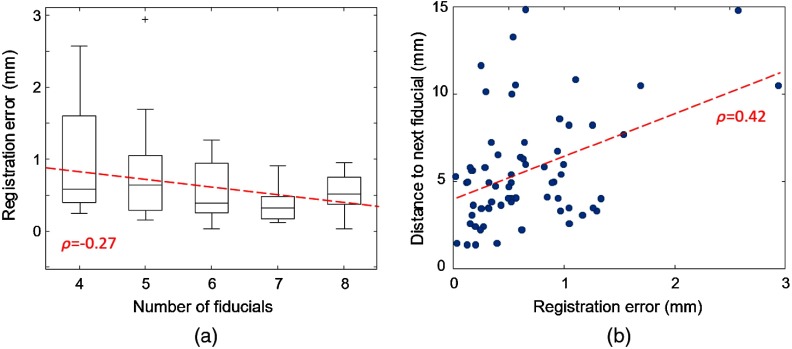
Figure 8 shows the correlation between the registration error and number of fiducials per specimen block (a) as well as the density of fiducial markers (b) using the hybrid registration method. A weak correlation between the number of fiducial markers and the registration precision can be observed. The maximum error drops monotonously with a higher number of fiducials. Moreover, a higher density of markers enhances registration precision.
Fig. 8.
Registration error for a varying (a) absolute number and (b) density of markers. Note that due to the leave-one-out validation strategy of fiducials are used for registration.
3.2. Histological Mapping of Fluorescence Decay Parameters
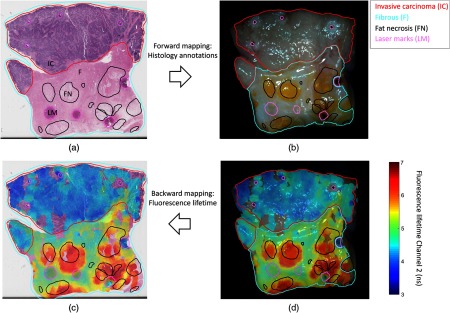
Figure 9 shows an application to map histological findings onto the white-light video image. (a) Regions of fibrous tissue, fat, and invasive cancer were traced by the pathologist and (b) can be placed on the video image. Inversely, the fluorescence decay parameters that have been coregistered with white-light image (d) can be mapped back to the histology slide (c) to directly correlate them with histological findings. Note that the overlay has been obtained from a free-hand scan and does not perfectly cover the whole specimen block.
Fig. 9.
Example of the forward and backward mapping between white light and histology: the forward mapping (a → b) allows to precisely see the pathologist’s annotations directly on the video frame. Inversely (d → c), the fluorescence lifetime parameter can be overlaid onto the histology slide using the backward mapping.
Following the mapping of pathologist delineations to the white-light images [Fig. 9(b)], fluorescence lifetime around 470 nm provides contrast between adipose, fibrous, and tumor tissue and follows similar trends as previously reported.13,14 Specifically, the average lifetime values for adipose tissue are higher compared with fibrous tissue and invasive carcinoma demonstrates lower values.
4. Discussion
Optical tissue diagnostics methods have demonstrated potential for tumor margin delineation as they can probe tissue nondestructively and provide information about structural, biochemical, or functional properties of the interrogated area. To demonstrate the diagnostic value e.g., for intraoperative diagnosis, it is essential to validate the imaging data against gold standard methods such as histopathology. In particular, the combination of optical imaging with supervised machine learning techniques requires a comprehensive database of specimens or specimen blocks correlated with histology slides.
In this paper, we present a histological registration of a point-scanning imaging technique implemented using an ms-TRFS system. To the best of our knowledge, this paper presents the first report on the registration of autofluorescence imaging data with ex vivo histological images. As histology is the gold standard for cancer diagnosis, grading, and localization, a precise histological registration will help to validate the ability of imaging techniques to delineate tumor margins. Using a previously described aiming-beam optical marker, point measurements can be localized on the video image. To further register white-light images with histology, we assess the value of fiducial laser markers and local shape characteristics of excised breast specimen blocks. Additionally, we present a hybrid method that uses the fiducial markers to prealign both modalities and use a B-spline free-form deformation to refine the registration involving data from the outer shape of the specimen block. The hybrid approach using both type of features exhibited overall smaller mean and standard deviation of the registration error and overall smaller maximum error compared with methods relying on the fiducials or shape information only.
As expected and shown in Fig. 8, a higher number and density of fiducial markers can enhance the registration precision but the laser marks also destroy histological information. Thus, a more optimized and automatically chosen number and positions of the fiducial markers could considerably improve registration results. The number of markers has been chosen between 4 and 8 but can be generally increased. Note that the leave-one-out validation uses only of the fiducial markers for validation. A considerable decrease in the maximum error was observed for a higher number of fiducial laser landmarks. The density should be generally higher for narrow stripes that are prone to deformations. A likewise high impact on the registration precision is related to strong deformations after histological preparation. In these cases, the hybrid method provided a smaller error and thus, a considerably smaller maximum error compared with rigid alignment or one-to-one shape matching. To further optimize the preparation of the specimen, the specimen block should ideally have a constant thickness toward the edges to minimize the risk of tissue loss, the shape should not be longitudinal to minimize deformation, and the markers should be evenly distributed. Furthermore, there are additional factors that impact the precision. All histology sections are cut at thickness, parallel to the imaging plane. However, surgical specimen topography is often nonuniform. In current pathology practice, the first few cuts from such specimens are discarded and the slice that is stained is the first continuous large section. Therefore, the stained section might be further away from the imaged surface in a surgical specimen with topographical undulations. This process complicates the registration by the fact that the stained slice is different from the surface of the specimen block that is observed with the camera. Specifically, this causes a lack or shift of significant cross-modality information that is seen as reasonable assumption for many registration methods for clinical applications.29
Compared with other approaches that rely on a dense distribution of anatomical landmarks, the reported error rates are rather high as the presented approach is premised on only a few artificial landmarks combined with shape information that is only partially preserved. In this respect, the entire information that is available is used for the registration process. Strong deformations combined with a low local density of landmarks cause high error rates. Other approaches dealing with histology to ex vivo registration with similar error metrics reported 1.25 to 3.45 mm for histology and ultrasound fusion30 and 15 for magnetic resonance imaging. Recently, Prabhu et al.31 reported error rates of for registering volumetric intravascular optical coherence tomography with cryo-image volumes. Using cryo imaging overcomes limitations such as spatial distortion caused by histological slicing and staining as it has been reported to adequately characterize vascular features. Both modalities are able to accurately segment the lumen providing a dense set of registration landmarks. However, this dense set of cross-modal landmarks is not available for the presented registration problem.
One possible solution to enlarge the set of cross-modal landmarks is to take a picture of the tissue embedded in the paraffin block prior to sectioning. In doing so, the tissue that gets lost during the slicing and staining process could be identified precisely. If tissue is missing after staining, it could be obtained from deeper sections to make sure that tissue loss is kept at a minimum level. This would make the shape of the stained section a better approximation of the processed tissue and thus, probably improve the overall registration accuracy.
Future work will focus on a full automation of the image acquisition and registration process. This involves the placement of fiducial markers, motorized scanning, and registration. The laser marker placement and imaging procedure can be easily performed with a mechanical stage. The benefit of automation is twofold: first, it does not require trained personnel, and it will considerably speed up data acquisition process thus leading to higher data throughput. In a second step, we aim to build up a large-scale database of imaged ex-vivo breast specimen blocks and corresponding histology slides to evaluate the suitability of fluorescence lifetime imaging for intraoperative tumor margin delineation. The registration will provide a reliable dataset for supervised machine learning to classifiers with the aim to delineate tumor margins in real time.
5. Conclusion
Optical tissue diagnostic technologies have shown potential for intraoperative and real-time delineation or assessment of tumor margins. Correlation of optical imaging data with tissue histology is a crucial step in demonstrating the clinical diagnostic value of these technologies. Specifically, accurate coregistration of the optically interrogated tissue and the corresponding histology slides is an essential prerequisite to demonstrate their sensitivity and specificity in tissue diagnostics. Current results show the performance of the hybrid method introduced here for histological registration of optical imaging data from ex-vivo breast cancer specimens. The hybrid approach outperformed methods relying on either fiducials or shape information alone. Moreover, it showed high accuracy even for specimen blocks with a high degree of tissue loss or deformation. Current findings pave the way for future evaluation of the ability of ms-TRFS as well as other point-scanning techniques to delineate tumor margins in excised breast specimen.
Acknowledgments
This study was supported in part by the Comprehensive Cancer Center at UC Davis and NIH Grant 5R01CA187427.
Biography
Biographies for the authors are not available.
Disclosures
No potential conflicts of interest relevant to this article were reported.
References
- 1.Sandison A., “The surgical resection margin,” Head Neck Oncol. 1(Suppl. 1), I1 (2009). 10.1186/1758-3284-1-S1-I1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lau D., et al. , “Raman spectroscopy for optical diagnosis in normal and cancerous tissue of the nasopharynx-preliminary findings,” Lasers Surg. Med. 32(3), 210–214 (2003). 10.1002/(ISSN)1096-9101 [DOI] [PubMed] [Google Scholar]
- 3.Stone N., et al. , “Raman spectroscopy for optical diagnosis in normal and cancerous tissue of the nasopharynx-preliminary findings,” Laryngoscope 110, 1756–1763 (2000). 10.1097/00005537-200010000-00037 [DOI] [PubMed] [Google Scholar]
- 4.Bigio I. J., et al. , “Diagnosis of breast cancer using elastic-scattering spectroscopy: preliminary clinical results,” J. Biomed. Opt. 5(2), 221–228 (2000). 10.1117/1.429990 [DOI] [PubMed] [Google Scholar]
- 5.Mueller M. G., et al. , “Spectroscopic detection and evaluation of morphologic and biochemical changes in early human oral carcinoma,” Cancer 97(7), 1681–1692 (2003). 10.1002/cncr.v97:7 [DOI] [PubMed] [Google Scholar]
- 6.Vakoc B. J., et al. , “Cancer imaging by optical coherence tomography: preclinical progress and clinical potential,” Nat. Rev. Cancer 12, 363–368 (2012). 10.1038/nrc3235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haka A. S., et al. , “Diagnosing breast cancer by using Raman spectroscopy,” PNAS 102(35), 12371–12376 (2005). 10.1073/pnas.0501390102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kondepati V. R., Heise H. M., Backhaus J., “Recent applications of near-infrared spectroscopy in cancer diagnosis and therapy,” Anal. Bioanal. Chem. 390(1), 125–139 (2008). 10.1007/s00216-007-1651-y [DOI] [PubMed] [Google Scholar]
- 9.Hirosawa N., et al. , “In vivo investigation of progressive alterations in rat mammary gland tumors by near-infrared spectroscopy,” Anal. Biochem. 305(2), 156–165 (2002). 10.1006/abio.2002.5649 [DOI] [PubMed] [Google Scholar]
- 10.Li R., et al. , “Assessing breast tumor margin by multispectral photoacoustic tomography,” Biomed. Opt. Express 6(4), 1273–1281 (2015). 10.1364/BOE.6.001273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dhingra J. K., et al. , “Early diagnosis of upper aerodigestive tract cancer by autofluorescence,” Arch. Otolaryngol.–Head Neck Surg. 122(11), 1181–1186 (1996). 10.1001/archotol.1996.01890230029007 [DOI] [PubMed] [Google Scholar]
- 12.Shin D., et al. , “Advances in fluorescence imaging techniques to detect oral cancer and its precursors,” Future Oncol. 6(7), 1143–1154 (2010). 10.2217/fon.10.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorpas D., et al. , “Fluorescence lifetime spectroscopy for breast cancer margins assessment,” Proc. SPIE 9318, 93180B (2015). 10.1117/12.2079622 [DOI] [Google Scholar]
- 14.Sharma V., et al. , “Auto-fluorescence lifetime and light reflectance spectroscopy for breast cancer diagnosis: potential tools for intraoperative margin detection,” Biomed. Opt. Express 3(8), 1825–1840 (2012). 10.1364/BOE.3.001825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gibson E., et al. , “Registration of prostate histology images to ex vivo MR images via strand-shaped fiducials,” J. Magn. Reson. Imaging 36(6), 1402–1412 (2012). 10.1002/jmri.v36.6 [DOI] [PubMed] [Google Scholar]
- 16.Chappelow J., et al. , “Elastic registration of multimodal prostate MRI and histology via multiattribute combined mutual information,” Med. Phys. 38(4), 2005–2018 (2011). 10.1118/1.3560879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bechara F. G., et al. , “Histomorphologic correlation with routine histology and optical coherence tomography,” Skin Res. Technol. 10(3), 169–173 (2004). 10.1111/j.1600-0846.2004.00038.x [DOI] [PubMed] [Google Scholar]
- 18.Jacobs M. A., et al. , “Registration and warping of magnetic resonance images to histological sections,” Med. Phys. 26(8), 1568–1578 (1999). 10.1118/1.598671 [DOI] [PubMed] [Google Scholar]
- 19.Pelizzari C. A., “Accurate three-dimensional registration of CT, PET, and/or MR images of the brain,” J. Comput. Assisted Tomogr. 13(1), 20–26 (1989). 10.1097/00004728-198901000-00004 [DOI] [PubMed] [Google Scholar]
- 20.Gorpas D., et al. , “Real-time visualization of tissue surface biochemical features derived from fluorescence lifetime measurements,” IEEE Trans. Med. Imaging 35(8), 1802–1811 (2016). 10.1109/TMI.2016.2530621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yankelevich D. R., et al. , “Design and evaluation of a device for fast multispectral time-resolved fluorescence spectroscopy and imaging,” Rev. Sci. Instrum. 85(3), 034303 (2014). 10.1063/1.4869037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klein S., et al. , “Elastix: a toolbox for intensity-based medical image registration,” IEEE Trans. Med. Imaging 29(1), 196–205 (2010). 10.1109/TMI.2009.2035616 [DOI] [PubMed] [Google Scholar]
- 23.Lee S., Wolberg G., Shin S. Y., “Scattered data interpolation with multilevel B-splines,” IEEE Trans. Visual Comput. Graphics 3(3), 228–244 (1997). 10.1109/2945.620490 [DOI] [Google Scholar]
- 24.Lee S., et al. , “Image metamorphosis with scattered feature constraints,” IEEE Trans. Visual Comput. Graphics 2(4), 337–354 (1996). 10.1109/2945.556502 [DOI] [Google Scholar]
- 25.Belongie S., Malik J., Puzicha J., “Shape matching and object recognition using shape contexts,” IEEE Trans. Pattern Anal. Mach. Intell. 24(9), 509–522 (2002). 10.1109/34.993558 [DOI] [Google Scholar]
- 26.Belongie S., Malik J., Puzicha J., “Shape context: a new descriptor for shape matching and object recognition,” in Advances in Neural Information Processing Systems 13: Proc. of the 2000 Conf., Leen T. K., Dietterich T. G., Tresp V., Eds., pp. 831–837 (2000). [Google Scholar]
- 27.Papadimitriou C., Stieglitz K., Combinatorial Optimization: Algorithms and Complexity, Prentice Hall, Upper Saddle River, New Jersey: (1982). [Google Scholar]
- 28.Ling H., Jacobs D. W., “Shape classification using the inner-distance,” IEEE Trans. Pattern Anal. Mach. Intell. 29(2), 286–299 (2007). 10.1109/TPAMI.2007.41 [DOI] [PubMed] [Google Scholar]
- 29.Toth D., et al. , “Registration with adjacent anatomical structures for cardiac resynchronization therapy guidance,” in Statistical Atlases and Computational Models of the Heart. Imaging and Modelling Chalenges: 7th Int. Workshop, STACOM 2016, pp. 127–134, Springer International Publishing; (2017). [Google Scholar]
- 30.Porter B. C., et al. , “Histology and ultrasound fusion of excised prostate tissue using surface registration,” in Proc. IEEE Ultrasonics Symp., Vol. 2, pp. 1473–1476 (2001). 10.1109/ULTSYM.2001.991998 [DOI] [Google Scholar]
- 31.Prabhu D., et al. , “Three-dimensional registration of intravascular optical coherence tomography and cryo-image volumes for microscopic-resolution validation,” J. Med. Imaging 3(2), 026004 (2016). 10.1117/1.JMI.3.2.026004 [DOI] [PMC free article] [PubMed] [Google Scholar]