Abstract
Using molecular dynamics simulations, we have explored the effect of asymmetric lipids—specifically those that contain one polyunsaturated (PUFA) and one saturated fatty acid chain—on phase separation in heterogeneous membranes. These lipids are prevalent in neuronal membranes, particularly in synaptic membranes, where the Parkinson's Disease protein α-Synuclein (αS) is found. We have therefore explored the relationship between asymmetric, PUFA-containing lipids, and αS. The simulations show that asymmetric lipids partition to the liquid disordered (Ld) phase of canonical raft mixtures because of the highly disordered PUFA chain. In the case of a membrane built to mimic the lipid composition of a synaptic vesicle, the PUFA-containing asymmetric lipids completely disrupt phase separation. Because αS is positively charged, we show that it partitions with negatively charged lipids, regardless of the saturation state of the chains. Additionally, αS preferentially associates with the polyunsaturated fatty acid tails of both charged and neutral lipids. This is a consequence of those chains' ability to accommodate the void beneath the amphipathic helix. This article is part of a Special Issue entitled: Lipid order/lipid defects and lipid-control of protein activity edited by Dirk Schneider.
Keywords: α-Synuclein, Lipid rafts, Polyunsaturated fatty acids, Molecular dynamics simulations, Phase separation, Amphipathic helices
1. Introduction
Amphipathic α-helical proteins bind to biological membranes and alter their physical characteristics, including shape [1–3], order [4], and phase [5]. Yet the biophysical details as to how these proteins affect the organization of lipids in complex biological membranes remain poorly understood. α-Synuclein (αS) is a physiologically important amphipathic helix because of its role in Parkinson's Disease [6–8]. Recently, solid-state NMR was used to show that αS homogenizes phase-separated, model membranes containing both liquid ordered (Ld, enriched in POPC) and liquid disordered (Lo, enriched in sphingomyelin and cholesterol) phases, reducing both the bilayer's overall order and thickness [5]. Synaptic vesicle (SV) membranes, a primary physiological location for αS [9], have profoundly different lipid compositions than these canonical lipid raft mixtures. In particular, >60% of the non-cholesterol lipids are asymmetric, meaning they contain one polyunsaturated fatty acid (PUFA) and one saturated fatty acid; cholesterol makes up 45% of the total lipid content; and sphingomyelin makes up just 4% of the composition [10,11]. Konyakhina & Feigenson recently solved a phase diagram for a mixture containing asymmetric PUFA-containing lipids, sphingomyelin, and cholesterol [12]. In the region of Lo/Ld phase coexistence, these PUFA-containing lipids form the Ld phase while sphingomyelin and cholesterol pack into the Lo phase, much like a canonical raft mixture.
The motivation for this current computational study is to begin to understand how αS relates to the complex lipid constituency of synaptic vesicles. Specifically, coarse-grained molecular dynamics (MD) simulations are used to predict how αS interacts with asymmetric PUFA-containing lipids in a variety of complex bilayers. MD simulations have previously been used to study PUFA-protein interactions and revealed that the transmembrane protein rhodopsin has a preference for the PUFA chain in asymmetric PUFA-containing lipids [13]. The extreme flexibility of PUFAs allows them to adopt irregular conformations that maximize contact with proteins, which is not possible for rigid saturated fatty acids. The details of the interactions between PUFA-chains and amphipathic helices, on the other hand, have not been investigated with MD simulations. Relatedly, we have recently used simulations to show that αS disorders lipid chains directly beneath the protein when embedded in simple, single-component bilayers composed of asymmetric lipids containing a monounsaturated chain and negatively charged headgroup (POPG) [4].
Because it is polybasic, αS binds with much greater affinity to bilayers composed of negatively charged lipids [14,15]. How this relates to the interaction between αS and synaptic vesicles remains unknown. In synaptic vesicles, the vast majority of charged lipids have a phosphatidylserine (PS) headgroup, and those make up 12% of the total phospholipid content [10]. Of these PS lipids, 98% are asymmetric with a PUFA-chain, underscoring the likely biological importance of αS/PUFA interactions. There are contrasting reports of αS's preference for Lo or Ld phases [16–18], both in model biophysical settings and in cells. Given Feigenson's recent phase diagram [12], this raises the question as to whether the preference of the PUFA chain for the Ld phase drives a synaptic vesicle's PS lipids into that phase and, if so, whether this brings αS along for the ride. We have designed a series of simulations aimed at addressing the possibility that αS localizes to whichever phase is enriched in PS lipids. In so doing, we address whether αS (and likely other amphipathic helices) has an intrinsic attraction to a given phase, or whether the electrostatics and/or chain disorder drive the protein's partitioning behavior.
We conducted four sets of simulations: to test 1) the behavior of asymmetric PUFA-containing lipids in phase-separated bilayers, we simulated lipid raft mixtures (4:3:3 DPPC:DLiPC:CHOL) with and without 12% PUFA-containing lipids (18:0–22:6 PS (SDPS)); 2) the phase behavior and lipid mixing of synaptic vesicle membranes, we simulated a synaptic vesicle mimic with 45% PUFA-containing lipids (88:12 SDPC:SDPS), 45% cholesterol, and 10% saturated lipids (DPPC); 3) how the fatty acid composition of anionic PS lipids influences the phase partitioning of the cationic αS helix, we simulated αS bound to lipid raft mixtures with and without 12% PS lipids of three different fatty acid compositions: mixed saturated and unsaturated (6% DPPS, 6% DLiPS), unsaturated (DLiPS), and asymmetric saturated polyunsaturated (SDPS); and 4) whether αS interacts with PUFA tails independent of lipid charge, we simulated αS bound to the synaptic mimic, which contains both charged and neutral PUFA-containing lipids.
Coarse-grained molecular dynamics (MD) simulations used the MARTINI force field [19], with the GROMACS v.4 simulation software [20,21]. To model αS, we used a single extended helix, MARTINI model of residues 1–100 (the amphipathic membrane-binding helix) [22,23], with a pre-defined secondary structure, a limitation currently imposed by the MARTINI force field. We have previously shown the equivalence of an extended helix and a broken helix in the protein's impact on membrane curvature. It is possible that increased protein dynamics (e.g. in a broken helix configuration) may influence partitioning in complex bilayers, though we have not investigated that possibility here. We have previously employed the MARTINI force-field to study Lo/Ld phase-behavior in lipid raft mixtures [24, 25], and also to study membrane-bound αS on bilayers [3,4]. We based our raft simulations on prior MARTINI studies of planar lipid bilayers which used a 4:3:3 ratio of di-16:0PC (DPPC), di-18:2PC (DLiPC) and cholesterol (CHOL) [26].
2. Methods
2.1. Simulation methods
Each simulation was carried out using coarse-grain (CG) MARTINI force field parameters in the GROMACS v4.5.3 molecular dynamics software package. We used the MARTINI lipid [19], MARTINI2.2 protein [27], and MARTINI2·P polarizable water parameters [28]. Lipids were constructed using previously published MARTINI parameters. Palmitic acid (P) and linoleic acid (Li) chains were modeled with four MARTINI beads [26,29], stearic acid (S) was modeled with five [30], and docosahexaenoic acid (D) was modeled with six (Table S1) [31]. Each system was run in an isothermal-isobaric ensemble (NPT) using the velocity-rescaling thermostat [32], and Berendsen barostat [33], with a temperature of 295 K and compressibility of 3e–4 bar-1 with a 10 fs time step. Pressure coupling was applied semi-isotropically with the xy- and z-dimensions coupled separately. For non-bonded interactions, the Lennard-Jones potential was shifted to zero between 0.9 and 1.2 nm and the Van-der-Waals between 0 and 1.2 nm.
To explore the interactions between SDPS, αS, lipid rafts, and SV mimics we constructed the following bilayers: 1) 4:3:3 DPPC:DLiPC:CHOL, a standard model lipid raft system [26]; 2) 12% SDPS and 88% 4:3:3 DPPC:DLiPC:CHOL; 3) 45% CHOL, 40% SDPC, 10% DPPC, 5% SDPS, a synaptic vesicle mimic based on the makeup of rat synaptic vesicles [10]; 4) bilayer 1 with αS; 5) bilayer 2 with αS; 6) bilayer 3 with αS; 7) 6% DPPS and 6% DLiPS added to 88% 4:3:3 DPPC:DLiPC:CHOL with αS; 8) 12% DLiPS added to 88% 4:3:3 DPPC:DLiPC:CHOL with αS (Table S2). Each system contains ~3500 lipids randomly seeded in the bilayer plane and 56,000 polarizable water molecules with the appropriate counter ions to balance the system to a net neutral charge. Systems 1–3 and 7 were simulated for 5.5 µs of simulation time, and systems 4–6 and 8–9 were simulated for 10.5 µs.
For the systems with αS, lipids were seeded around a single protein with 23 lipids removed from the protein leaflet to accommodate the amphipathic helix [3]. The membrane-binding domain of αS (residues 1–100) was used. Proteins in MARTINI require secondary structure constraints [27]. Residues 1–93were modeled as α-helical and residues 94–100 as random coil [4].
Each system was energy minimized using the GROMACS steepest-descent energy minimization algorithm for 100,000 steps or until machine precision. Then solvent molecules were added and the system was simulated for 50–100 ps with the lipids fixed in the Z-dimension to allow the solvent to accommodate the membrane. An additional 1–3 ns were simulated without lipid constraints to allow the system to relax before beginning our production runs of 5.5 or 10.5 µs (actual simulation time).
2.2. Data analysis
Simulation results were analyzed with GROMACS (v.4.5.3) utilities (trjconv, trjcat, make_ndx) to manipulate the trajectory files and MATLAB (v. R2013) to perform analysis and generate figures.
2.3. Number density profiles
To show localization of the lipid components in each simulation, colored density maps were created from the trajectory file coordinate information for each lipid. The bilayer was first parsed into upper and lower leaflets by defining an undulating reference surface for each leaflet and averaging the two surfaces [34]. The systems with αS were oriented with the protein aligned on the x-axis. For the non-protein systems, the Cartesian reference orientation (default xyz-coordinates) was used. Each 30 × 30 nm leaflet was then binned into a 200 × 200 grid and the number of CG beads was counted in each bin for each lipid species. Colored density plots were then produced from a surface using the bin contents for each species and normalized using the number of bins and number of beads so the volume under the surface is 1. These plots are displayed in Fig. 1 and S1. A 500 ns time window was used to highlight domain formation and average out transient lipid diffusion (Fig. S2).
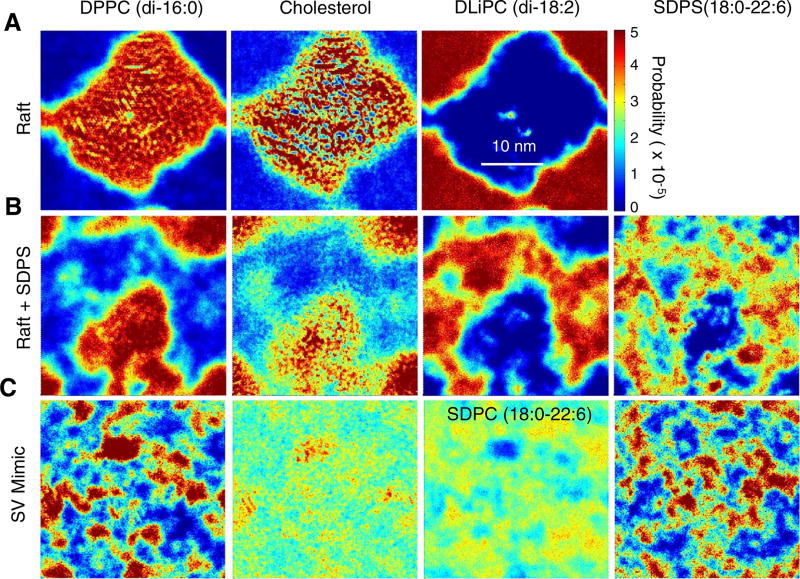
Fig. 1.
Local densities of lipid species in a (A) lipid raft mixture, (B) lipid raft mixture with 12% SDPS, and (C) synaptic vesicle mimic mixture (45:40:10:5 Chol:SDPC:DPPC:SDPS). Each plot is a 500 ns time average of a single periodic cell taken from 5 to 5.5 µs. Hot colors indicate high density and cold colors indicate low density.
To determine the density of a given acyl chain near αS, the protein-species distances were divided into 0.1 nm bins and the shortest distance between any protein bead and the center of mass of the species of interest was selected. Additionally, the shortest distance between any protein bead and any bead contained by the species of interest was selected for comparison as shown in Fig. S3. We plotted the number of each acyl chain near the protein for SDPS and SDPC in Fig. 5A.
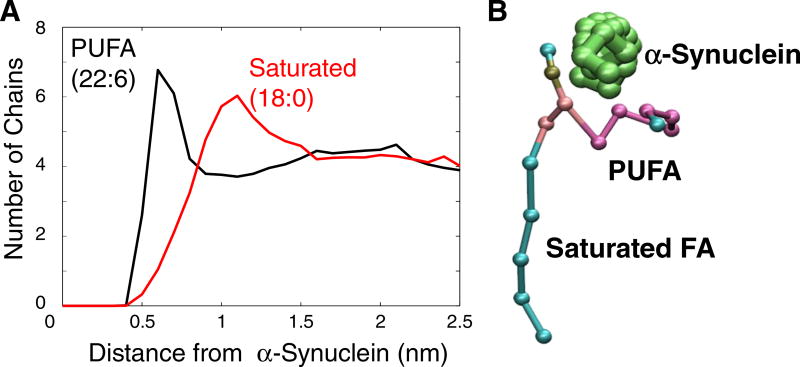
Fig. 5.
(A) Distribution of lipid-protein distances between PUFA-containing lipids (SDPC, SDPS) and AS. The black plot shows the distances between DHA (22:6) and the protein, and the red plot shows the distances between Steroyl (18:0) and the protein. This data was sampled using 0.1 nm radial bins and averaged over the last 500 ns of the 10.5 µs simulation. (B) Snapshot of a single asymmetric SDPC lipid near the αS helix highlighting the orientation shown in panel A.
2.4. Total lipid local order parameters
To show the localization of phases in each system, we calculated total lipid order parameters as described in our previous work [4]. Order parameters were interpolated onto a uniform 100 × 100 grid encompassing the 30 × 30 nm leaflet and the resulting surface was then plotted using the average order parameters at each point on the grid, as seen in Figs. 3A–C, 4, and S4.
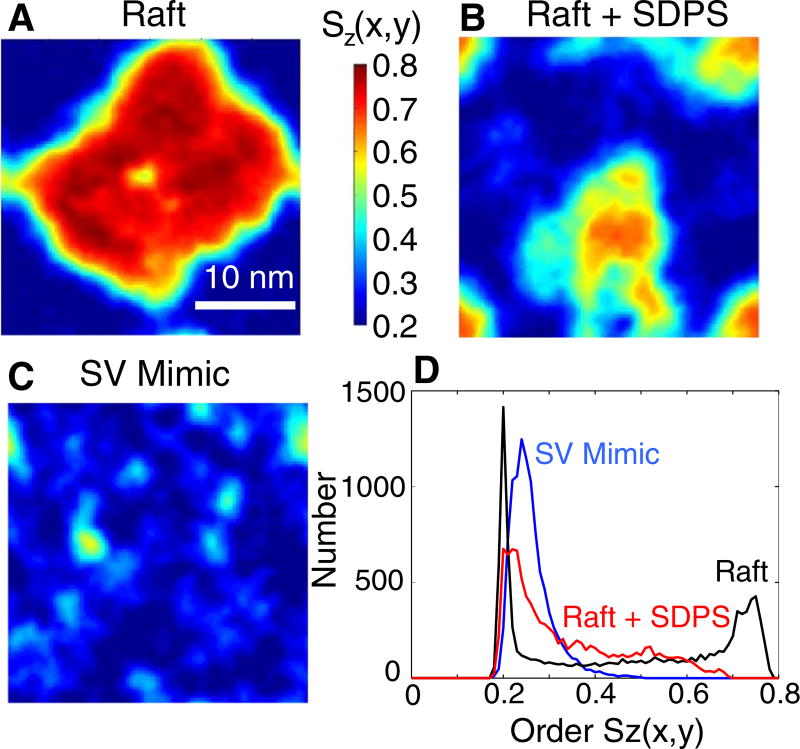
Fig. 3.
Local order parameter distributions. (A) Local order parameters in the raft mixture. (B) Order parameters in the raft + SDPS mixture. (C) SV mimic order parameters. (D) Order parameter histograms of the data shown in A, B, and C. Each plot is the average from 5 to 5.5 µs of simulation time and shows a single periodic cell.
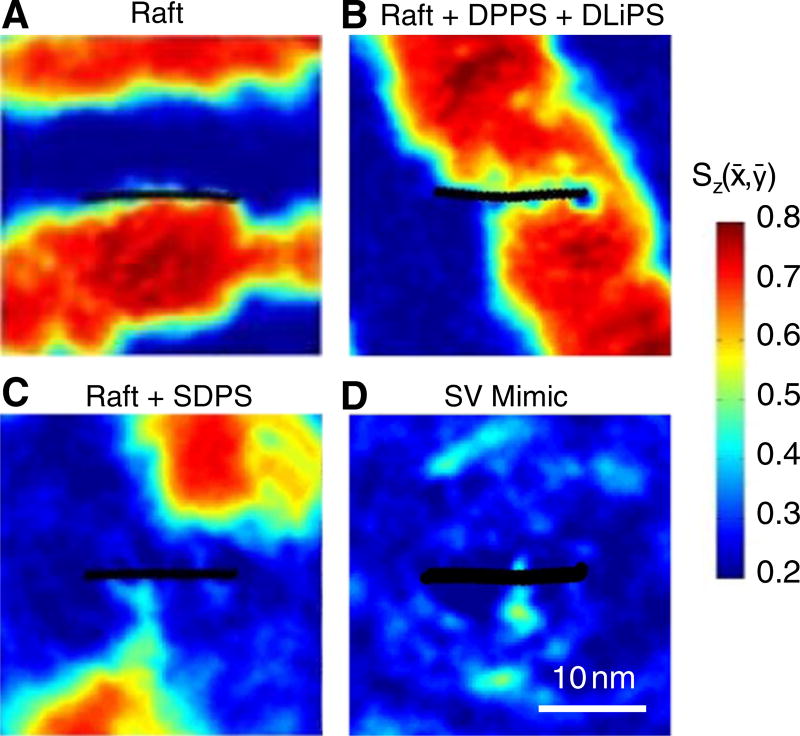
Fig. 4.
Time-averaged local order parameter Sz (x̄,ȳ) for each system containing αS. Hot colors are more ordered than cool colors. αS is indicated with black circles and is oriented with the N-terminus on the left. Each image is a time-average of 500 ns taken from 10 to 10.5 µs centered on the protein and represents a single periodic box of 30 × 30 nm.
To generate one-dimensional order histograms, the order parameter values for each pixel were divided into 0.01 (order parameter units) bins. Fig. 3D was generated in this manner.
2.5. Radial distribution functions
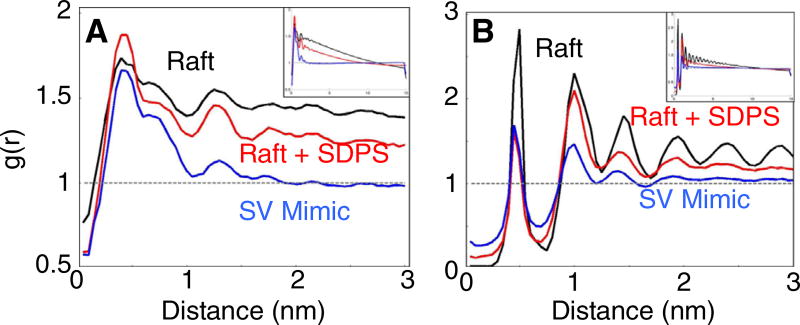
To calculate the radial distribution between the species in each of our systems, the distances between all species in the system for each leaflet were binned according to length using only x- and y-coordinates. Each system was then normalized to its bulk density. The radial distributions between each species in each system are found in Fig. S5. These distribution plots are truncated to 3 nm to show enrichment and depletion at short distances.
3. Results
3.1. Asymmetric PUFA-containing lipids disrupt phase separation
In order to investigate the impact and localization of the asymmetric lipid SDPS in Lo/Ld separated bilayers, we first simulated the canonical raft mixture (4:3:3 DPPC:DUPC:CHOL) established by Marrink absent SDPS [26]. Fig. 1A shows the two-dimensional lipid distribution as calculated over a 500 ns window, and recapitulates the original findings of this mixture, showing strong phase separation. The Lo and Ld are single, well-defined and contiguous regions separated by clear boundaries that reflect the nearly complete isolation of cholesterol within the saturated lipid phase. It is important to note that the exact size and boundaries of the phase-separated domains portrayed in our analysis are not characteristic of a single snapshot from the system. The temporal averaging across the 500 ns window smoothes the phase boundaries and results in a subtle loss of refined structural detail. Nevertheless, the overarching results are consistent across all systems and are converged over separate 500 ns windows.
Fig. 1B shows that when SDPS is added to this mixture at 12 mol% it partitions to the Ld phase. Unlike in the case of an asymmetric, monounsaturated lipid (POPC), which was shown to partition to the boundary between the Lo and Ld phases [35], the polyunsaturated-containing SDPS is peppered throughout the Ld domains. The consequences of this on the separation of the phases are profound. Qualitatively, it is apparent that the phases are less well defined. Most notably, the exclusion of cholesterol from the Ld phase appears to be less complete. While cholesterol is still excluded from the centers of the Ld domains, it is more likely to encroach upon the boundaries of the Ld domains when SDPS is present.
In the SV mimic system, in which 45% of the lipids contain a PUFA, large cohesive domains are no longer visible (Fig. 1C). Nevertheless, close inspection reveals co-localization of DPPC and cholesterol into small nanodomains; for example there is a cluster near the top center edge of the simulation box. Additionally, in this location there is a depletion of PUFA-containing lipids, most noticeable as a blue spot in the third panel, which shows the density of SDPC.
To quantify the association between cholesterol and DPPC and understand the extent of ordered domain formation, the radial distribution function between DPPC and cholesterol for each of the three systems was calculated. Fig. 2A shows that the likelihood of finding cholesterol in the first shell surrounding a DPPC lipid is quite similar in each system. At increasing distances, the probability of DPPC-cholesterol interactions drops off in the Raft + SDPS system and even more so in the SV mimic mixture, indicating the formation of smaller ordered domains than in the raft mixture. The addition of PUFA-containing lipids causes the size of DPPC-cholesterol rafts to shrink. In the SV mimic, the RDF reaches the bulk value (g(r) = 1) at ~2 nm, whereas the Raft systems do not reach bulk until ~15 nm (half the box dimension).
Fig. 2.
Cholesterol radial distributions. (A) Radial distribution plots between the cholesterol OH group and the phosphate bead of DPPC are displayed for the raft mixture (black) the raft + SDPS mixture (red), and the SV mimic mixture (blue). (B) Radial distribution plots between cholesterol molecules in each system. Each RDF is a time average from 5 to 5.5 µs of simulation time. The full-length (15 nm) radial distribution plots are shown as insets.
Additionally, cholesterol-cholesterol interactions are ameliorated when PUFA-containing lipids are present (Fig. 2B), reflecting the migration of cholesterol out of the Lo phase. In both the Raft + SDPS and the SV mimic simulations, the probability of finding a cholesterol-cholesterol interaction in the first shell is roughly half as likely as in the Raft system. Interestingly, although the first-shell values for these two PUFA-containing systems are the same, the g(r) values are much lower in the SV mimic at greater distances. This provides evidence that cholesterol cluster size is inversely related to PUFA concentration. Furthermore, there is a greater probability of an SDPS/CHOL interaction than a DLiPC/CHOL interaction (Sup. Fig. S5), presumably reflecting a preferential interaction between the cholesterol and the saturated chains that are in the Ld phase. Likewise, there are increased DPPC/DLiPC interactions in the SDPS system.
The changes in lipid distributions induced by PUFA-containing lipids described in Figs. 1 and 2 manifest in the calculated lipid order parameter profile, which further highlights the impact of these lipids on the phase behavior of the membrane. Fig. 3 shows the local order parameters for the raft system with and without SDPS and from the SV mimic mixture. As is clear from the data, SDPS severely diminishes the order of the Lo phase. Fig. 3D shows a histogram of the two-dimensional order parameter data, quantifying the extent to which SDPS reduces phase separation. The figure shows that the signature of the Lo phase (peak at Sz ~ 0.75) is completely eliminated, giving way to a broad, mixed state in the raft mixture with 12% SDPS. Furthermore, with 45% PUFA-containing lipid, the region with mixed order parameter (Sz = 0.4 to 0.6) is also eliminated. What remains in this SV mimic mixture is a single wider Ld peak (Sz ~ 0.25). In this representation of the data, an ordering effect of SDPS on the Ld phase can also be seen, though the change in the distribution is less striking than the impact on the Lo phase. That this effect on the Ld phase is relatively small is somewhat surprising, given that the Ld phase contains both additional cholesterol and the saturated chains of SDPS. Thus, this must reflect the severity to which the polyunsaturated chain disorders the surrounding lipids, even at low mol%, mostly overcoming the presence of the cholesterol and stearoyl chains. In the SV mimic system, this increase in order is more pronounced, most likely resulting from the significantly increased cholesterol content.
3.2. SDPS draws α-Synuclein into the Ld phase
Having established that the polyunsaturated chain on the asymmetric SDPS lipid disrupts phase separation by preferential association with the Ld phase, we asked how this might impact the behavior of αS. As stated earlier, it is well known that αS has a strong preference for charged lipids and does not bind strongly to neutral membranes in the fluid phase [14,18,36–38]. Previously, atomistic simulations showed that the lysine residues in αS form salt bridges with charged headgroup [39]. Thus, we hypothesized that the phase-distribution of the charged lipid should dictate the partitioning of the protein. This was tested with multiple simulations of αS bound to the 4:3:3 membrane mixture doped with either: i) 12% SDPS; ii) 12% DLiPS; iii) 6% DPPS + 6% DLiPS; or iv) no additional charged lipids and the vi) SV mimic mixture. DLiPS and DPPS contain the same fatty acids as the raft components DLiPC and DPPC, but have anionic phosphatidylserine headgroups rather than zwitterionic phosphatidylcholine.
Fig. 4A shows that in the raft mixture, αS exhibits no obvious preference for either the Lo or the Ld phase, instead partitioning to the phase interface. In contrast, Fig. 4C shows that in the raft + 12% SDPS bilayer, αS partitions to the Ld phase, matching the behavior of the lipid (Fig. 1). As in the protein-free simulations, the Lo region is small and weakly formed (details below). Tracking the protein's position over the full length of the simulated trajectory shows that as the ordered regions coalesced they never encroached on the protein (that is, the protein never sampled the Lo phase). A control simulation of the 4:3:3 mixture + 12% DLiPS was used as a first test of whether the localization of αS to the Ld domain is due to the PS headgroup localization, as opposed to something unique about the 18:0–22:6 chains. Here too, αS partitions to the Ld phase. Of the four αS simulations these were the only two where the protein remained squarely embedded in a single phase throughout the entire trajectory.
Next, we tested whether the partitioning of αS into the Ld phase in the 12% SDPS bilayer is in fact driven by the location of the PS headgroup, or whether the protein itself imposes a separate (non-electrostatic) driving force into the disordered region. To do so, the 4:3:3 mixture + 6% DPPS + 6% DLiPS was simulated, controlling against the headgroup charge localization to a particular phase. Fig. 4B shows that αS exhibits dramatically different partitioning behaviors in this case. As in the SDPS simulation, over the first 2.5 µs of the trajectory αS is excluded from the coalescing Lo phase (Fig. S4). At this time point, the Lo phase is almost fully formed, though its shape remains somewhat diffuse. After this time point, the Lo phase becomes more regular (a single stripe) and engulfs the protein. Thus, the result shown in Fig. 4B for the equimolar DPPS/DLiPS system should be taken carefully. It is very possible that the protein became stuck in the Lo phase due to the slow exchange of saturated PS lipids, rather than having a true energetic preference for that phase. Given infinite sampling, one might expect that the protein would continue to sample both phases, with the Lo phase weighted heavily by the damped lipid diffusion and exchange. It is noted that there are two small patches of unsaturated lipids in the Lo phase at the site of the protein, and that the order parameter profile immediately under the protein reflects a slightly less-ordered lipid milieu (Fig. 4B).
In a further attempt to understand whether the protein has an intrinsic preference for the Lo over the Ld phase (absent electrostatics), we next simulated the protein in the PS-free raft mixture. In this case, the protein was never fully engulfed in the Lo phase. It was again excluded from the Lo phase in the initial portion of the trajectory, however over the second half of the trajectory it partitioned to the interface between the Lo and Ld domains. This result is consistent with the idea that the Lo/Ld interface may act as a defect (due to a thickness mismatch) that increases αS binding affinity to gel-phase, charge-free membranes [5,15,40]. Regardless, the failure of the protein to sample the Lo phase in this mixture further suggests that it is not the palmitoyl chains that drive the protein into the Lo phase in the 6% DLiPS/6% DPPS system. Collectively, these results suggest that the protein has very little intrinsic drive towards either the Lo or the Ld phase. Rather, the protein's localization to the Ld phase in the 12% SDPS bilayer (Fig. 4A) is due almost entirely to the protein's attraction to the PS headgroup on the SDPS lipid, which has its own intrinsic preference for the Ld phase.
3.3. α-Synuclein shows a preference for PUFA chains
We showed recently that the lipids in the solvating shell of αS in a simple bilayer (comprised of asymmetric monounsaturated lipids) are disordered by the protein, and that this has consequences for the protein's curvature-inducing properties [4]. Thus, we were interested in exploring the lipid organization in the solvating shell of αS. Interestingly, we find that there is a rotational preference of the PUFA chains for the protein (Fig. 5A), which is mindful of previous studies that have shown the propensity of PUFA chains to interact with transmembrane proteins [13,41,42]. In this case, we presume that the preference is a consequence of the ease with which the more disordered chain accommodates the void beneath the protein. In the SV mimic mixture, this effect was observed for both SDPS and SDPC (Fig. 5). This result is particularly surprising given that SDPC lacks the electrostatic attraction to αS possessed by SDPS and suggests that the acyl-chain disordering is a driving force for lipid partitioning/orienting near αS. Fig. 5B shows a representative snapshot of a single lipid near a segment of theαS monomer, clearly revealing the extent to which the PUFA curls underneath the helix. In addition to the rotational preference, there is an overall enrichment of PUFA-containing lipids near the protein, which is highlighted in Sup. Fig. S3, which compares the average number of each type of fatty acid and cholesterol near the protein.
In addition to the electrostatic interactions favoring αS association with SDPS and the disordering process favoring partitioning near αS for asymmetric PUFA lipids, we caution that the PUFA chain parameterization in the MARTINI CG forcefield may potentially bias PUFA-protein interactions. PUFA chains are slightly more hydrophilic than a typical saturated fatty acid chain, and as such are more attracted to typical CG protein beads. However, both CG and all-atom simulation studies have identified a preference for unsaturated chains to associate with proteins [13,41,42]. Although we cannot rule out a potential bias for PUFA-protein interaction, we are confident that the two driving forces (electrostatics and disordering) dominate the observation.
4. Discussion
Because of its amphipathic nature, αS wedges into the headgroup region of a bilayer. This creates a void beneath the protein that forces the lipids to undergo a dynamic rearrangement that reduces their order [4,40,43]. In a recent NMR study of a raft-like mixture containing sphingomyelin (SM) but lacking PUFA chains, the protein was shown to disorder the lipids in the bilayer and cause a dramatic thinning [5]. Our previous combined X-ray scattering and simulation study used a simple monounsaturated lipid mixture, and showed a much smaller thinning effect [44], despite a substantial decrease in the order of the lipids closest to the protein [4]. Here, too, we have seen no impact of the protein on the total bilayer order (Sup. Table S3). This is not surprising in PUFA-containing bilayers where we have shown that the overall order of the protein-free bilayer is already substantially reduced (Fig. 3D).
More interesting is thatαS also has no effect on order or thickness in the simulated 4:3:3 DPPC/DLiPC/Chol bilayer. This simulated mixture more closely mimics the composition used in the NMR study (equimolar mix of EYSM/POPC/Chol). The authors proposed that the protein acts by disrupting the interactions between lipid chains in defect zones, increasing diffusion and therefore mixing. In our simulation, interestingly, the protein spent considerable time either in the Ld (t < 6 µs) or in the interface of the two domains (Sup. Fig. S4). A major difference exists between the two systems, of course, namely the high-melting lipid: DPPC (our simulations) and sphingomyelin (the NMR experiments). It was speculated that the source of the homogenization in the SM mixture was due, at least in part, to specific interactions between αS and the sphingosine backbone of SM that do not exist with glycerolipids (hydrogen bonding and electrostatics). Indeed, a recent MD simulation showed a more complex electrostatic potential in an SM membrane in the backbone region suggestive of this difference [45]. Thus, there appear to be at least two molecular driving forces for domain mixing in the SM mixture, only one of which exists in our DPPC mixture: 1) in both cases, the more dynamic, unsaturated lipid chain is drawn to the void under the protein: we saw this in the orientation of the PUFA chain (Fig. 5A), and also in the 6% DLiPS/6% DPPS bilayer, where even in the Lo domain there are small hot-spots of DLiPS and DLiPC near the protein; and 2) in the SM mixture only, sphingosine backbone-driven electrostatics and h-bonding to the protein.
While the available information regarding phase behavior of PUFA-containing bilayers is limited [42,46,47], there has been considerable progress made in understanding how chain asymmetry in monounsaturated lipids affects the separation of phases. Recently, by using a combination of fluorescence microscopy, FRET and ESR, nanodomains that are smaller than the large micron-sized domains and that are invisible to fluorescence microscopy, have been detected [48,49]. The core finding was that chain asymmetry in the unsaturated lipid (e.g. POPC or SOPC) results in much smaller domains (as small as 5–10 nm in diameter, or approximately 50–150 lipids) than in bilayers containing symmetric chained lipids, such as DOPC. Additionally, in Feigenson's ternary PUFA mixture, the substitution of small amounts of POPC for SDPC resulted in a transition from macroscopic to nanoscopic phase separation. Significant questions remain regarding the nature of these nanodomains: why should small domains be stable if the penalty of a high line tension would tend to push the system towards larger domains? One possibility is that in the asymmetric lipids, the monounsaturated oleoyl chains face inwards, interacting with the liquid disordered (Ld) domain. This chain-rotational bias may decrease the energetic mismatch at the domain boundary—and hence the line tension—and therefore significantly decreases the driving force for large domain size [35]. Indeed, in the simulated bilayers with asymmetric PUFA-containing lipids, there is a clear blurring of the boundaries between domains (Figs. 1 and 2). In this case, however, the PUFA chain drives the lipids into the interiors of the Ld domains, dramatically reducing the size of the Lo domain and slightly increasing the order of the Ld domain. In fact, the phase boundaries are so poorly defined in these simulations that it was impossible to determine the extent to which the asymmetric chains might be oriented (although, interestingly, they are oriented at the binding site for αS). The partitioning of PUFA-containing lipids into the Ld domain in this study is consistent with the phase diagram of Feigenson [12], and is consistent with the observations of Wassall [42].
While our simulations of asymmetric lipids containing PUFA chains are revealing in their own right, they take only one small step in the direction of building physically accurate models of synaptic vesicles. In addition to PUFAs, there are other characteristics of the SV lipid composition that will need to be addressed in future studies. For example, one important characteristic of SVs missing from our current model is the 2:1 protein:phospholipid w/w ratio, and that an estimated 20% of the SV surface area is covered by protein [10]. It has been suggested that this high protein/lipid ratio depicts SVs as rigid constructs composed of large protein patches surrounded by thin shells of lipid, rather than phase-separated fluid membranes [10,50,51]. These thin shells of lipid may look something like the nanodomains described above.
Another critical perspective on understanding SV phase behavior is the impact of the high curvature imposed on SV membranes (SVs are small, with a diameter ~ 40 nm). αS is known to preferentially bind to vesicles of high curvature [15], and to induce curvature in large bilayer sheets [15,52–54]. Recent studies have shown that the Ld phase is recruited to tubules (curvature similar to that of an SV) pulled from the Lo phase in biphasic vesicles [55]. The high curvature stress induced in a tubule or a small vesicle does not favor the rigid lipid packing of ordered domains [56]. This perhaps explains the importance of such a high percentage of PUFAs in SVs relative to typical cell membranes [57], especially given our finding that PUFA-containing lipids reduce the extent of Lo phase formation. Additionally, PE phospholipids, which are curvature inducing and therefore may reduce the curvature-strain on SVs, make up 23 mol% of SV phospholipids. Perhaps more striking is that plasmalogen phosphatidylethanolamine (Pl-PE) makes up an astounding 19% of the SV lipid content. Pl-PE is a terribly understudied lipid that is likely to strongly impact the physical characteristic of SVs. In particular, Pl-PE has a vinyl ether linkage that promotes formation of the inverse hexagonal phase [58], and promotes membrane fusion [59].
Changes in a membrane's phase can influence its propensity to deform and fuse [56,60,61]. In complex membranes (e.g. those containing Lo/Ld phase coexistence), it is likely that any perturbations (e.g. protein binding) that might drive the system towards a homogenized, Ld state will ameliorate the driving force for fusion. Understanding the relationship between curvature strain, phase separation and αS binding will remain a critical focal point in the study of αS biology and pathology.
Supplementary Material
Acknowledgments
Simulations and data analyses were conducted at the Minnesota Supercomputing Institute. We also thank Drs. Stephen Wassall, Gerald Feigenson, and Ana West for valuable discussion.
Funding sources
This study was funded by the National Institutes of Health R01 NS084998-01 to J.N.S.
Abbreviations
- DPPC
1,2-dipalmitoyl-sn-glycero-3-phosphocholine
- DPPS
1,2-dipalmitoyl-sn-glycero-3-phospho-l-serine
- DLiPC
1,2-dilinoleoyl-sn-glycero-3-phosphocholine
- DLiPS
1,2-dilinoleoyl-sn-glycero-3-phospho-l-serine
- CHOL
cholesterol
- SDPS
1-stearoyl-2-docosahexaenoyl-sn-glycero-3-phospho-l-serine
- POPC
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- DOPC
1,2-dioleoyl-sn-glycero-3-phosphocholine
- SOPC
1-stearoyl-2-oleoyl-sn-glycero-3-phosphocholine
- SM
sphingomyelin
- EYSM
egg-yolk sphingomyelin
- SV
synaptic vesicle
- αS
α-Synuclein
- PUFA
polyunsaturated fatty acid
- PS
phosphatidylserine
- Pl-PE
plasmalogen phosphatidylethanolamine
- Lo
liquid ordered
- Ld
liquid disordered
- DHA
docosahexanoic acid
- FRET
Förster resonance energy transfer
- ESR
electron spin resonance
Footnotes
This article is part of a Special Issue entitled: Lipid order/lipid defects and lipid-control of protein activity edited by Dirk Schneider.
Transparency document
The Transparency document associated with this article can be found, in online version.
Appendix A. Supplementary data
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.bbamem.2016.10.006.
References
- 1.Campelo F, McMahon HT, Kozlov MM. The hydrophobic insertion mechanism of membrane curvature generation by proteins. Biophys. J. 2008;95:2325–2339. doi: 10.1529/biophysj.108.133173. http://dx.doi.org/10.1529/biophysj.108.133173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mim C, Unger VM. Membrane curvature and its generation by BAR proteins. Trends Biochem. Sci. 2012;37:526–533. doi: 10.1016/j.tibs.2012.09.001. http://dx.doi.org/10.1016/j.tibs.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braun AR, Sevcsik E, Chin P, Rhoades E, Tristram-Nagle S, Sachs JN. Alpha-synuclein induces both positive mean curvature and negative Gaussian curvature in membranes. J. Am. Chem. Soc. 2012;134:2613–2620. doi: 10.1021/ja208316h. http://dx.doi.org/10.1021/ja208316h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braun AR, Lacy MM, Ducas VC, Rhoades E, Sachs JN. α-Synuclein-induced membrane remodeling is driven by binding affinity, partition depth, and interleaflet order asymmetry. J. Am. Chem. Soc. 2014;136:9962–9972. doi: 10.1021/ja5016958. http://dx.doi.org/10.1021/ja5016958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leftin A, Job C, Beyer K, Brown MF. Solid-state 13C NMR reveals annealing of raft-like membranes containing cholesterol by the intrinsically disordered protein α-synuclein. J. Mol. Biol. 2013;425:2973–2987. doi: 10.1016/j.jmb.2013.04.002. http://dx.doi.org/10.1016/j.jmb.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, Pike B, Root H, Rubenstein J, Boyer R, Stenroos ES, Chandrasekharappa S, Athanassiadou A, Papapetropoulos T, Johnson WG, Lazzarini AM, Duvoisin RC, DiIorio G, Golbe LI, Nussbaum RL. Mutation in the alpha synuclein gene identified in families with Parkinson's disease. Science. 1997;80(276):2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 7.Spillantini MG, Schmidt ML, Lee VMY, Trojanowski JQ. Alpha-synuclein in Lewy bodies. Nature. 1997;388:839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 8.Singleton AB, Farrer M, Johnson J, Singleton A, Hague S, Kachergus J, Hulihan M, Peuralinna T, Dutra A, Nussbaum RL, Lincoln S, Crawley A, Hanson M, Maraganore D, Adler C, Cookson MR, Muenter M, Baptista M, Miller D, Blancato J, Hardy J, Gwinn-Hardy K. Alpha-synuclein locus triplication causes Parkinson's disease. Science 80- 2003;(302):841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 9.Jakes R, Spillantini MG, Goedert M. Identification of two distinct synucleins from human brain. FEBS Lett. 1994;345:27–32. doi: 10.1016/0014-5793(94)00395-5. http://dx.doi.org/10.1016/0014-5793(94)00395-5. [DOI] [PubMed] [Google Scholar]
- 10.Takamori S, Holt M, Stenius K, Lemke Ea, Grønborg M, Riedel D, Urlaub H, Schenck S, Brügger B, Ringler P, Müller Sa, Rammner B, Gräter F, Hub JS, De Groot BL, Mieskes G, Moriyama Y, Klingauf J, Grubmüller H, Heuser J, Wieland F, Jahn R. Molecular anatomy of a trafficking organelle. Cell. 2006;127:831–846. doi: 10.1016/j.cell.2006.10.030. http://dx.doi.org/10.1016/j.cell.2006.10.030. [DOI] [PubMed] [Google Scholar]
- 11.Breckenrldge WC, Gombos G, Morgan IG. The lipid composition of adult rat brain synaptosomal plasma membranes. Biochim. Biophys. Acta. 1972;266:695–707. doi: 10.1016/0006-3002(72)90012-1. http://dx.doi.org/10.1016/0005–2736(72)90365-3. [DOI] [PubMed] [Google Scholar]
- 12.Konyakhina TM, Feigenson GW. Phase diagram of a polyunsaturated lipid mixture: brain sphingomyelin/1-stearoyl-2-docosahexaenoyl-sn-glycero-3-phosphocholine/cholesterol. Biochim. Biophys. Acta Biomembr. 2015;1858:153–161. doi: 10.1016/j.bbamem.2015.10.016. http://dx.doi.org/10.1016/j.bbamem.2015.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feller SE, Gawrisch K, Woolf TB. Rhodopsin exhibits a preference for solvation by polyunsaturated docosohexaenoic acid. J. Am. Chem. Soc. 2003;125:4434–4435. doi: 10.1021/ja0345874. http://dx.doi.org/10.1021/ja0345874. [DOI] [PubMed] [Google Scholar]
- 14.Davidson WS, Jonas A, Clayton DF, George JM. Stabilization of alpha-synuclein secondary structure upon binding to synthetic membranes. J. Biol. Chem. 1998;273:9443–9449. doi: 10.1074/jbc.273.16.9443. http://dx.doi.org/10.1074/jbc.273.16.9443. [DOI] [PubMed] [Google Scholar]
- 15.Middleton ER, Rhoades E. Effects of curvature and composition on α-Synuclein binding to lipid vesicles. Biophys. J. 2010;99:2279–2288. doi: 10.1016/j.bpj.2010.07.056. http://dx.doi.org/10.1016/j.bpj.2010.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fortin DL, Troyer MD, Nakamura K, Kubo S, Anthony MD, Edwards RH. Lipid rafts mediate the synaptic localization of alpha-synuclein. J. Neurosci. 2004;24:6715–6723. doi: 10.1523/JNEUROSCI.1594-04.2004. http://dx.doi.org/10.1523/JNEUROSCI.1594-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kubo S, Nemani VM, Chalkley RJ, Anthony MD, Hattori N, Mizuno Y, Edwards RH, Fortin DL. A combinatorial code for the interaction of alpha-synuclein with membranes. J. Biol. Chem. 2005;280:31664–31672. doi: 10.1074/jbc.M504894200. http://dx.doi.org/10.1074/jbc.M504894200. [DOI] [PubMed] [Google Scholar]
- 18.Stöckl M, Fischer P, Wanker E, Herrmann A. Alpha-synuclein selectively binds to anionic phospholipids embedded in liquid-disordered domains. J. Mol. Biol. 2008;375:1394–1404. doi: 10.1016/j.jmb.2007.11.051. http://dx.doi.org/10.1016/j.jmb.2007.11.051. [DOI] [PubMed] [Google Scholar]
- 19.Marrink SJ, Risselada HJ, Yefimov S, Tieleman DP, de Vries AH. The MARTINI force field: coarse grained model for biomolecular simulations. J. Phys. Chem. B. 2007;111:7812–7824. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- 20.Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4:algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 21.Van der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 22.Georgieva ER, Ramlall TF, Borbat PP, Freed JH, Eliezer D. Membrane-bound alpha-synuclein forms an extended helix: long-distance pulsed ESR measurements using vesicles, bicelles, and rodlike micelles. J. Am. Chem. Soc. 2008;130:12856–12857. doi: 10.1021/ja804517m. http://dx.doi.org/10.1021/ja804517m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trexler AJ, Rhoades E. Alpha-synuclein binds large unilamellar vesicles as an extended helix. Biochemistry. 2009;48:2304–2306. doi: 10.1021/bi900114z. http://dx.doi.org/10.1021/bi900114z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perlmutter JD, Sachs JN. Interleaflet interaction and asymmetry in phase separated lipid bilayers: molecular dynamics simulations. J. Am. Chem. Soc. 2011;133:6563–6577. doi: 10.1021/ja106626r. http://dx.doi.org/10.1021/ja106626r. [DOI] [PubMed] [Google Scholar]
- 25.Perlmutter JD, Sachs JN. Inhibiting lateral domain formation in lipid bilayers: simulations of alternative steroid headgroup chemistries. J. Am. Chem. Soc. 2009;131:16362–16363. doi: 10.1021/ja9079258. http://dx.doi.org/10.1021/ja9079258. [DOI] [PubMed] [Google Scholar]
- 26.Risselada HJ, Marrink SJ. The molecular face of lipid rafts in model membranes. Proc. Natl. Acad. Sci. U. S. A. 2008;105:17367–17372. doi: 10.1073/pnas.0807527105. http://dx.doi.org/10.1073/pnas.0807527105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monticelli L, Kandasamv SK, Periole X, Larson RG, Tieleman DP, Marrink SJ. The MARTINI coarse-grained force field: extension to proteins. J. Chem. Theory Comput. 2008;4 doi: 10.1021/ct700324x. [DOI] [PubMed] [Google Scholar]
- 28.Yesylevskyy SO, Schäfer LV, Sengupta D, Marrink SJ. Polarizable water model for the coarse-grained MARTINI force field. PLoS Comput. Biol. 2010;6:e1000810. doi: 10.1371/journal.pcbi.1000810. http://dx.doi.org/10.1371/journal.pcbi.1000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khandelia H, Loubet B, Olżyńska A, Jurkiewicz P, Hof M. Pairing of cholesterol with oxidized phospholipid species in lipid bilayers. Soft Matter. 2014;10:639. doi: 10.1039/c3sm52310a. http://dx.doi.org/10.1039/c3sm52310a. [DOI] [PubMed] [Google Scholar]
- 30.Marrink SJ, De Vries AH, Harroun TA, Katsaras J, Wassall SR. Cholesterol shows preference for the interior of polyunsaturated lipid membranes. J. Am. Chem. Soc. 2008;130:10–11. doi: 10.1021/ja076641c. http://dx.doi.org/10.1021/ja076641c. [DOI] [PubMed] [Google Scholar]
- 31.Ingólfsson HI, Melo MN, Van Eerden FJ, Arnarez C, López CA, Wassenaar TA, Periole X, De Vries AH, Tieleman DP, Marrink SJ. Lipid organization of the plasma membrane lipid organization of the plasma membrane. J. Am. Chem. Soc. 2014 doi: 10.1021/ja507832e. [DOI] [PubMed] [Google Scholar]
- 32.Bussi G, Donadio D, Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126:14101. doi: 10.1063/1.2408420. http://dx.doi.org/10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 33.Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola a, Haak JR. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. http://dx.doi.org/10.1063/1.448118. [Google Scholar]
- 34.Braun AR, Brandt EG, Edholm O, Nagle JF, Sachs JN. Determination of electron density profiles and area from simulations of undulating membranes. Biophys. J. 2011;100:2112–2120. doi: 10.1016/j.bpj.2011.03.009. http://dx.doi.org/10.1016/j.bpj.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schäfer LV, Marrink SJ. Partitioning of lipids at domain boundaries in model membranes. Biophys. J. 2010;99:L91–L93. doi: 10.1016/j.bpj.2010.08.072. http://dx.doi.org/10.1016/j.bpj.2010.08.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hellstrand E, Grey M, Ainalem ML, Ankner J, Forsyth VT, Fragneto G, Haertlein M, Dauvergne MT, Nilsson H, Brundin P, Linse S, Nylander T, Sparr E. Adsorption of alpha-synuclein to supported lipid bilayers: positioning and role of electrostatics. ACS Chem. Neurosci. 2013;4:1339–1351. doi: 10.1021/cn400066t. http://dx.doi.org/10.1021/cn400066t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramakrishnan M, Jensen PH, Marsh D. Alpha-synuclein association with phosphatidylglycerol probed by lipid spin labels. Biochemistry. 2003;42:12919–12926. doi: 10.1021/bi035048e. http://dx.doi.org/10.1021/bi035048e. [DOI] [PubMed] [Google Scholar]
- 38.Pandey AP, Haque F, Rochet JC, Hovis JS. Clustering of alpha-synuclein on supported lipid bilayers: role of anionic lipid, protein, and divalent ion concentration. Biophys. J. 2009;96:540–551. doi: 10.1016/j.bpj.2008.10.011. http://dx.doi.org/10.1016/j.bpj.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perlmutter JD, Braun AR, Sachs JN. Curvature dynamics of alpha-synuclein familial Parkinson disease mutants. Molecular simulations of the micelle-and bilayer-bound forms. J. Biol. Chem. 2009;284:7177–7189. doi: 10.1074/jbc.M808895200. http://dx.doi.org/10.1074/jbc.M808895200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nuscher B, Kamp F, Mehnert T, Odoy S, Haass C, Kahle PJ, Beyer K. Alpha-synuclein has a high affinity for packing defects in a bilayer membrane: a thermodynamics study. J. Biol. Chem. 2004;279:21966–21975. doi: 10.1074/jbc.M401076200. http://dx.doi.org/10.1074/jbc.M401076200. [DOI] [PubMed] [Google Scholar]
- 41.Feller SE. Acyl chain conformations in phospholipid bilayers: a comparative study of docosahexaenoic acid and saturated fatty acids. Chem. Phys. Lipids. 2008;153:76–80. doi: 10.1016/j.chemphyslip.2008.02.013. http://dx.doi.org/10.1016/j.chemphyslip.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 42.Stillwell W, Wassall SR. Docosahexaenoic acid: membrane properties of a unique fatty acid. Chem. Phys. Lipids. 2003;126:1–27. doi: 10.1016/s0009-3084(03)00101-4. http://dx.doi.org/10.1016/S0009-3084(03)00101-4. [DOI] [PubMed] [Google Scholar]
- 43.Kamp F, Beyer K. Binding of alpha-synuclein affects the lipid packing in bilayers of small vesicles. J. Biol. Chem. 2006;281:9251–9259. doi: 10.1074/jbc.M512292200. http://dx.doi.org/10.1074/jbc.M512292200. [DOI] [PubMed] [Google Scholar]
- 44.Braun AR, Sevcsik E, Rhoades E, Tristram-Nagle S, Sachs JN. The nature of membrane curvature-induction by amphipathic alpha-helices relies upon protein length: simulations of alpha-synuclein and H0. Biophys. J. 2012;102:237 A–237 A. <Go to ISI>://000321561201495. [Google Scholar]
- 45.Niemelä P, Hyvönen MT, Vattulainen I. Structure and dynamics of sphingomyelin bilayer: insight gained through systematic comparison to phosphatidylcholine. Biophys. J. 2004;87:2976–2989. doi: 10.1529/biophysj.104.048702. http://dx.doi.org/10.1529/biophysj.104.048702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wassall SR, Stillwell W. Docosahexaenoic acid domains: the ultimate non-raft membrane domain. Chem. Phys. Lipids. 2008;153:57–63. doi: 10.1016/j.chemphyslip.2008.02.010. http://dx.doi.org/10.1016/j.chemphyslip.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 47.Stillwell W, Shaikh S, Zerouga M, Siddiqui R, Wassall S. Docosahexaenoic acid affects cell signaling by altering lipid rafts. Reprod. Nutr. Dev. 2005;45:559–579. doi: 10.1051/rnd:2005046. http://dx.doi.org/10.1051/rnd. [DOI] [PubMed] [Google Scholar]
- 48.Zhao J, Wu J, Heberle Fa, Mills TT, Klawitter P, Huang G, Costanza G, Feigenson GW. Phase studies of model biomembranes: complex behavior of DSPC/DOPC/cholesterol. Biochim. Biophys. Acta. 2007;1768:2764–2776. doi: 10.1016/j.bbamem.2007.07.008. http://dx.doi.org/10.1016/j.bbamem.2007.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heberle Fa, Wu J, Goh SL, Petruzielo RS, Feigenson GW. Comparison of three ternary lipid bilayer mixtures: FRET and ESR reveal nanodomains. Biophys. J. 2010;99:3309–3318. doi: 10.1016/j.bpj.2010.09.064. http://dx.doi.org/10.1016/j.bpj.2010.09.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Engelman DM. Membranes are more mosaic than fluid. Nature. 2005;438:578–580. doi: 10.1038/nature04394. http://dx.doi.org/10.1038/nature04394. [DOI] [PubMed] [Google Scholar]
- 51.Dupuy AD, Engelman DM. Protein area occupancy at the center of the red blood cell membrane. Proc. Natl. Acad. Sci. 2008;105:2848–2852. doi: 10.1073/pnas.0712379105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Braun AR, Sevcsik E, Chin P, Rhoades E, Tristram-Nagle S, Sachs JN. α-Synuclein induces both positive mean curvature and negative Gaussian curvature in membranes. J. Am. Chem. Soc. 2012;134:2613–2620. doi: 10.1021/ja208316h. http://dx.doi.org/10.1021/ja208316h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jensen MB, Bhatia VK, Jao CC, Rasmussen JE, Pedersen SL, Jensen KJ, Langen R, Stamou D. Membrane curvature sensing by amphipathic helices: a single liposome study using alpha-synuclein and annexin B12. J. Biol. Chem. 2011;286:42603–42614. doi: 10.1074/jbc.M111.271130. http://dx.doi.org/10.1074/jbc.M111.271130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jiang Z, de Messieres M, Lee JC. Membrane remodeling by alpha-synuclein and effects on amyloid formation. J. Am. Chem. Soc. 2013 doi: 10.1021/ja405993r. http://dx.doi.org/10.1021/ja405993r. [DOI] [PMC free article] [PubMed]
- 55.Heinrich M, Tian A, Esposito C, Baumgart T. Dynamic sorting of lipids and proteins in membrane tubes with a moving phase boundary. Proc. Natl. Acad. Sci. U. S. A. 2010;107:7208–7213. doi: 10.1073/pnas.0913997107. http://dx.doi.org/10.1073/pnas.0913997107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jiang H, Powers T. Curvature-driven lipid sorting in a membrane tubule. Phys. Rev. Lett. 2008;101:018103. doi: 10.1103/PhysRevLett.101.018103. http://dx.doi.org/10.1103/PhysRevLett.101.018103. [DOI] [PubMed] [Google Scholar]
- 57.Spector AA, Yorek MA. Membrane lipid composition and cellular function. J. Lipid Res. 1985;26:1015–1035. (doi:3906008) [PubMed] [Google Scholar]
- 58.Lohner K. Is the high propensity of ethanolamine plasmalogens to form nonlamellar lipid structures manifested in the properties of biomembranes? Chem. Phys. Lipids. 1996;81:167–184. doi: 10.1016/0009-3084(96)02580-7. [DOI] [PubMed] [Google Scholar]
- 59.Glaser PE, Gross RW. Plasmenylethanolamine Facilitates Rapid Membrane Fusion: A Stopped-flow Kinetic Investigation Correlating the Propensity of a Major Plasma Membrane Constituent to Adopt an HII Phase With Its Ability to Promote Membrane. 1994:5805–5812. doi: 10.1021/bi00185a019. [DOI] [PubMed] [Google Scholar]
- 60.Parthasarathy R, Yu C, Groves JT. Curvature-modulated phase separation in lipid bilayer membranes. Langmuir. 2006;22:5095–5099. doi: 10.1021/la060390o. http://dx.doi.org/10.1021/la060390o. [DOI] [PubMed] [Google Scholar]
- 61.van Meer G. Membrane curvature sorts lipids. EMBO J. 2005;24:1537–1545. doi: 10.1038/sj.embor.7400410. http://dx.doi.org/10.1038/sj.emboj.7600631. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.