Abstract
The human body carries vast communities of microbes that provide many benefits. Our microbiome is complex and challenging to understand, but evolutionary theory provides a universal framework with which to analyse its biology and health impacts. Here we argue that to understand a given microbiome feature, such as colonization resistance, host nutrition or immune development, we must consider how hosts and symbionts evolve. Symbionts commonly evolve to compete within the host ecosystem, while hosts evolve to keep the ecosystem on a leash. We suggest that the health benefits of the microbiome should be understood, and studied, as an interplay between microbial competition and host control.
Humans carry massive and diverse communities of symbiotic microbes1,2. These cells colonize almost every surface of the body: the skin, teeth, airways and, particularly, the epithelial surfaces of the gastrointestinal tract. They impact nutrition3, tissue and immune development4, pathogen resistance5,6 and perhaps even our behaviour7. The importance of the human microbiome—defined here as the microbes (the microbiota) plus the host environment8—is matched by its complexity. At each body site, many different species and strains occur, each with the potential to interact with the host by modulating metabolism and the immune system that is itself a vastly complex biological system4. Moreover, each species has the potential to exert diverse effects on neighbouring microbes9. Some species kill others with dedicated toxins10–12, while others invest in enzymes that feed other species for mutual benefit13.
The microbiome is therefore a complex and dynamic ecosystem in which species are in continual flux. It is now clear that tools from theoretical ecology will have a central role in understanding and predicting the dynamics of the microbiota1,14–17. What has been less clear is the value in applying evolutionary thinking to understand the form and function of the whole microbiome (Box 1). The relationship between mammalian hosts and microbes is just one of a myriad of evolved symbioses that date back to the dawn of multicellular life17. Many of these have striking similarities to our own microbiome, suggesting that common evolutionary principles and processes are at play (Fig. 1).
BOX 1. Identifying biological function in the microbiome.
“The question inevitably arises as to how such an abundance of misinterpretation has arisen. I believe that the major factor is that biologists have no logically sound and generally accepted set of principles and procedures for answering the question: ‘What is its function?’” G. C. Williams94.
Darwin provided a unified framework to understand biological functions based on the question: why did a trait evolve? A peacock’s tail has evolved to attract mates, honeybees collect nectar to feed their colony and male polar bears fight over females. However, as G. C. Williams made clear, evolutionary theory has too often been neglected when discussing function94. Assigning function can also be challenging, particularly when multiple individuals or species influence traits50. This occurs in both the vertebrate immune system95 and the mammalian microbiome. Nevertheless, the study of function provides a universal logic to understand complex biological systems50 and how to manipulate them96. Here we discuss the function of several much-discussed benefits of our microbiota.
Colonization resistance
Microbial symbionts often protect against pathogens3,5,6,54. Is host protection an evolved function of symbionts? In diverse communities, symbionts are often strongly affected by competition from other microbes (Box 2). Host protection may, therefore, arise as a fortunate by-product of natural selection on microbes to avoid being replaced by other species28,97. Once in place, host mechanisms may evolve to promote the most protective microbes, which would make colonization resistance a composite of two functions, namely basic microbial competition and host control of the microbiome.
Polysaccharide digestion and butyrate production
Digestion of carbohydrates by bacteria such as Clostridia is thought to provide us with nutritional benefits through butyrate production47. Why evolve to use polysaccharides? Again, competition within microbiota appears to be central; cells are feeding themselves, their clonemates, or less commonly, another species that provides help in return13. Why make butyrate? Butyrate is a metabolic waste product that is also made by bacteria outside of a host98. Combined with the weak evolutionary incentive for microbiota species to help the host (Box 2), this suggests that butyrate production in the gut arose as a by-product of microbial metabolism, which was then reinforced by host evolution. Consistent with host adaptation, gut epithelial cells directly take up butyrate47 and a lack of butyrate is associated with immune dysregulation in the intestine99 (main text).
Development of the immune system and tolerance
The microbiota facilitates immune system development4,100. Have symbionts evolved to influence immune processes and limit negative impacts on the host (increasing host tolerance) because this improves host fitness90? Evolutionary theory suggests otherwise (Box 2). Natural selection may favour symbionts that influence host immunity but, again, a probable function is to improve competitiveness and niche occupation within the microbiome. From the host’s perspective, a long association with symbionts can drive the evolution of tolerance to symbionts. But it can also lead to ‘evolved dependence’18, whereby immune system development evolves to rely on certain microbial phenotypes, but without the bacteria needing to evolve dedicated functions for the host.
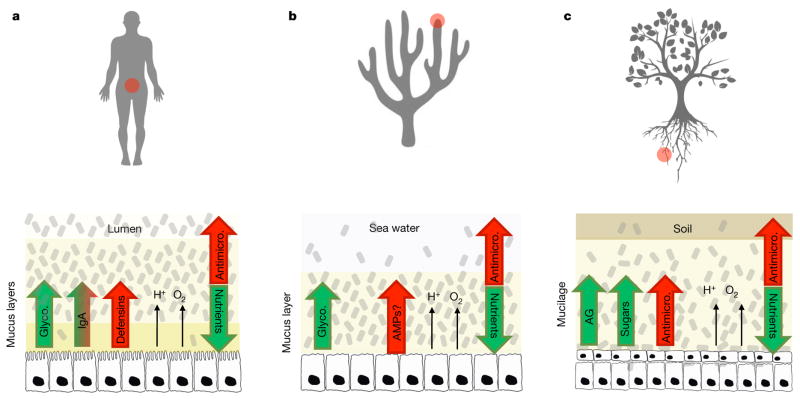
Figure 1. Convergent evolution of the host epithelial interface with the microbiota.
a–c, The study of the mammalian microbiota is most developed but it is becoming clear that diverse animals (a, b) and plants (c) possess epithelial surfaces where a complex microbiota can grow. In these systems, the host releases nutrients, antimicrobials, and a slimy matrix of mucus or mucilage, which are all thought to help control the microbiota (host control). In return, the symbionts may provide nutrients and protection from pathogens through antimicrobial (Antimicro.) release and other mechanisms3,6,36,47,92. Notably, the common ancestor of plants and animals is a single-cell organism93, which means that these adaptations have evolved convergently after multicellularity evolved in the two lineages. This convergence is an indicator of common evolutionary principles across diverse systems. a, Human large intestine. The host secretes glycoproteins (Glyco.), such as mucins, which certain microbes attach to and feed on. Large amounts of IgA are released4, which may both help and harm symbionts by affecting adhesion56. Defensins (antimicrobial peptides), acids and oxygen release also shape the symbiotic community. b, Coral epidermis. Corals have many of the same features as the mammalian intestine, including mucins that contain microbes54, acids and oxygen53. Whether antimicrobial peptides (AMPs) are released from the epidermis is not yet clear but the innate immune system shapes the epithelial microbiota in the coral-relative Hydra41. c, Plant-root epidermis. Plants release mucilage that contains arabinogalactan (AG) proteins55, which appear to be functionally similar to mucins, and sugars and other carbon sources in large quantities, which all provide nutrients for the root microbiota36,37 The release of oxygen, antimicrobials, and particularly organic acids, also shapes the microbiota of the rhizosphere3,36. Icons made by Freepick from http://www.flaticon.com/ (a) and https://www.vecteezy.com/ (b, c).
Here we use the predictions of evolutionary theory to interpret microbiome data, with a particular focus on the intensely studied mammalian microbiome and the benefits it provides to the host. Our goal throughout is to understand biological function: why did a particular feature of interest evolve? For example, why do the microbiota protect against pathogens or promote immune maturation (Box 1)? Such features can arise as a complex composite of both microbial and host evolution. To break down this complexity, we reduce the microbiome to three classes of effect, each of which has its own evolutionary characteristics: microbe-to-host, host-to-microbe and finally, microbe-to-microbe.
Many studies have focused on how the microbiota affects human health (microbe-to-host). However, here we argue that the other effects (host-to-microbe and microbe-to-microbe) explain why most functions evolve within the microbiome. Specifically, natural selection on the microbiota alone is not expected to make them beneficial to the host; rather, microbiota evolution is dominated by the need for each species to compete and persist within the host9. However, at the same time hosts are under strong natural selection to shape their microbiota to be beneficial. Thus, rather than being intrinsically helpful, the microbiome is a dynamic microbial ecosystem held on an ever-evolving leash by the host (Fig. 2). We discuss the implications of our perspective for microbiome health and disease, and lay out key next steps for microbiome research.
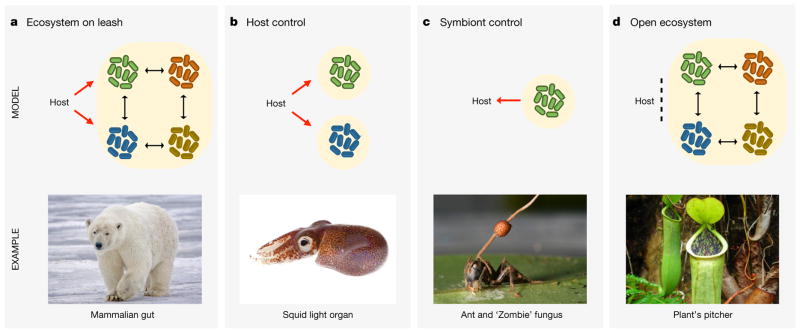
Figure 2. Models of host–microbiome interaction.
Black arrows represent ecological interactions within the microbiota, red arrows indicate mechanisms of control. a, Ecosystem on a leash. When host species interact with a diverse but beneficial microbiota, as occurs in mammals, evolutionary theory predicts that the microbial functions will centre on persistence in the microbiome ecosystem, while the host will attempt to control the microbiota, hence the ‘leash’ (Boxes 1 and 2). Image courtesy of A. D. Wilson. b, Host control. For interactions involving few microbial strains, ecological complexity is reduced and microbes are primarily shaped by the host environment. Natural selection on the host, therefore, can result in strong shaping and control of the phenotypes of beneficial microbes. The bobtail squid has a specialized light organ, which controls both the access and light production of the symbiotic bacteria that grow inside79,80. One hypothesis is that host enzymes generate bacteriocidal compounds from substrates that become available if the bacteria do not perform the light-producing reaction80. Photo of Euprymna scolopes, the Hawaiian bobtailed squid, by M. McFall-Ngai, PBRC, University of Hawaii-Manoa, published with permission. c, Symbiont control. Low microbial diversity also increases the potential for microbes to affect global host traits—including survival, reproduction and behaviour—and receive a fitness benefit from doing so (Box 2). This may select for adaptations that function to increase host fitness, such as enzymes that feed the host, but slow microbial growth. However, this can also enable symbiont manipulation of the host, such as for ‘zombie’ fungi, infection with which causes ants to move to a position ideal for fungal development84. Photo of Ophiocordyceps unilateralis and ant by D. Hughes, Penn. State, published with permission. d, Open ecosystem. A host carries a complex ecosystem without evolved control mechanisms beyond compartmentalization. This is most likely to occur if the microbiota are rarely either a threat or a benefit. Pitcher plants use pools of water to kill and digest prey. Although these plants regulate the pool by releasing enzymes and acids to promote digestion, there is currently little evidence that the plants have dedicated mechanisms to regulate the pool microbiota86. Image adapted from P. J. Ding used under Creative Commons Licence.
Microbe to host: the problem of a diverse microbiome
We first consider the evolutionary origins of the best-studied aspect of the microbiome, the beneficial effects of the microbes on the host. Interactions between hosts and their microbiota appear to include many examples of biological mutualism: interactions that provide fitness benefits to all the species involved18. However, a degree of caution is required; although benefits to the host have been well documented, it is difficult to show that microbes benefit, because we typically do not know if better alternatives exist for microbes outside of the host19. If microbes do not benefit, the prediction is that natural selection will favour strategies to escape from the microbiome, or adaptions that increase within-host fitness, even if this harms the host. Nevertheless, for many species in environments like the human gut, the combination of relatively stable conditions, nutrients and warmth is likely to improve microbial fitness relative to host-free environments.
For microbes that benefit from living inside a host, it might seem inevitable that natural selection would favour the evolution of microbe-to-host benefits. Evolutionary theory, however, shows otherwise. Consider a bacterial strain that generates a nutrient for the host, but must divide more slowly to do so. If this is the only strain within the host, then it can persist and continue to help its host, as this slow-growing strain will have no microbial competitors. However, in a diverse microbiota, the slow-growing strain runs the risk of being outcompeted by other faster-growing genotypes that do not make the nutrient (Box 2, Box 2 Figure). As a result, natural selection is predicted to favour microbes that invest in their own reproduction, rather than help the host20–22.
BOX 2. The problem of evolved cooperation from microbiota to host.
“If it could be proved that any part of the structure of any one species had been formed for the exclusive good of another species, it would annihilate my theory, for such could not have been produced through natural selection.” Charles Darwin26.
We now understand many examples of evolved cooperation between species20, including between members of the mammalian gut microbiota13. However, it is less clear why our symbionts help us. The problem is simple, the mammalian microbiota comprises many strains and species subject to continual turnover22,56. Consider a new mutation in a focal microbe that benefits the host but comes at a cost to the microbe. The mutant strain will divide slightly less rapidly than the parent and, even when the fitness cost is small, the strain is predicted to be rapidly lost (Box 2 Fig.). This effect is expected to greatly constrain the potential for a symbiotic microbe to evolve a cooperative trait that helps the host.
We consider three potential exceptions to this conclusion. First, if the advantage of increased competitiveness within the microbiota (within-group selection) is outweighed by the negative effects on the host (between-group selection)101. Taken to the extreme, if the lack of a cooperative trait in one strain has always led to immediate host death, this trait may be selected within the microbiota. However, the focal strain must have a very strong impact on host fitness amongst the hundreds to thousands of other microbial genotypes. Transmission from parents to offspring might increase the fitness of a helpful symbiont, as it can now also benefit by helping the host to have offspring32. However, with imperfect vertical transmission, it is difficult to see how these effects can protect against loss of a less competitive microbe23.
Second, when strains differ in their benefits to the host, but not in competitive ability, host-level selection has a higher potential to favour beneficial strains. Consider a hypothetical case where only some hosts carry a strain that synthesizes a vitamin in order to grow and compete within the microbiota; the vitamin-producing strain might increase in frequency during a famine that kills vitamin-deficient hosts. This would mean that the function of vitamin production is both for microbial competition and host benefit (Box 1). Such selection might even occur at the community level if traits of multiple species interact to create a host benefit102,103. However, the requirement for strains to differ strongly in host benefits but not in competitive ability appears stringent and may rarely be satisfied.
Finally, as discussed in the main text, the host appears to have evolved mechanisms that encourage beneficial traits in the microbiota, such as immune suppression of harmful microbes and preferential feeding of beneficial strains. When effective, a symbiont may thereby evolve traits that provide benefits to the host. The result can be that the two sides of the interaction—host and microbiota—both evolve to invest in one another, generating an evolutionarily stable mutualism18. We believe this to be the most important of the three exceptions for the mammalian microbiota.
Box 2 Figure. Simulation of bacterial growth on host epithelium.
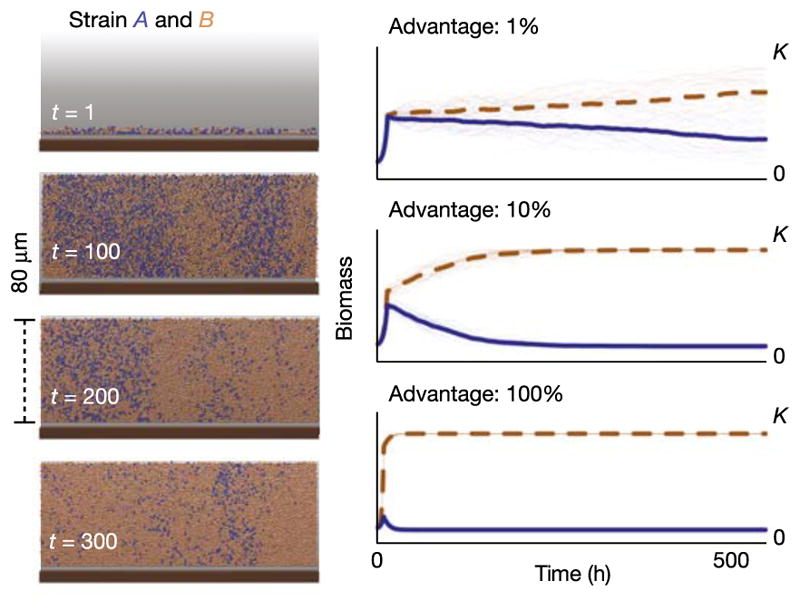
Left, brown bacterial cells (strain B) have a 1% growth rate advantage over blue bacterial cells (strain A). Even with a modest growth rate advantage, strain B succeeds and strain A is outcompeted in a few days. Right, plots of thirty independent simulations of bacterial competition. Development of biomass of strain B (brown dashed) and A (blue) with growth rate advantages for strain B of 1%, 10%, and 100% and environmental capacity K. The thick lines are mean values. From ref. 22.
Crucially then, we cannot assume that the host and microbiota are a single evolutionary unit acting with a common interest, as is sometimes done in applications of the ‘holobiont’ or ‘superorganism’ metaphors23 (although see ref. 24). Rather, the host and each individual microbial strain are distinct entities with potentially divergent selective pressures. The potential for divergent interests is made abundantly clear by the existence of pathogens within the microbiome, such as Clostridium difficile6, and more transient species such as Salmonella enterica25. However, these pathogens appear to be exceptions—the microbiota comprises mostly helpful, or at least neutral, microbial species. We are left again with the fundamental question: why do so many members of the microbiota benefit the host? To understand this, we next turn to the host.
Host to microbe: the importance of host control
An answer to why microbes help their hosts is suggested by general evolutionary theory. Here, the question of what drives between-species cooperation—a trait that evolves because of a beneficial effect on another species—has received a lot of attention18,20,21,26,27. A recurring theme throughout the evolutionary literature is the importance of control of one species by the other20. Applying this finding, we predict that hosts are under strong natural selection to control their microbiota22, an idea also raised by some applications of the holobiont metaphor24. Host control over the microbes (as opposed to microbial control of the host) can be predicted, because there is only one host in the interaction, in contrast to the myriad microbes. Thus, unlike individual microbes, a host can easily influence the entire microbiome, and benefit from doing so.
Evolutionary theory then predicts that host-to-microbe effects—rather than the much-studied impacts of microbe on host—are critical for microbiome form and function. Here we discuss evidence for four key aspects of host control: immigration, compartmentalization, monitoring and targeting.
Immigration and compartmentalization
Ecosystems are often shaped by which species happen to arrive from a global pool of species. The order of species arrivals is particularly important when there are priority effects, such that early arriving species resist invasion by later ones28. Microbial dispersal is thus expected to be an important shaper of host microbiomes. Nevertheless, a host can exert control by influencing which microbial strains and species it encounters, and which species make it to each epithelial surface. This can occur at the cognitive and behavioural level, such as by conditioned avoidance of rancid foods containing harmful microbes29. Once eaten, stomach acid can further reduce microbial diversity and abundance, and protect against pathogens30, although the primary biological function of acidity may be digestion.
Vertical transmission from parents to offspring will promote immigration of particular species, and active transmission has evolved in species including leafcutter ants31 and the beewolf32. The importance of parent–offspring transmission remains unclear for the mammalian microbiome33. However, the infant microbiome is similar to that of the mother’s vagina after natural birth, but not after Caesarean section, suggesting a role for parent–offspring transmission in early colonizers of infants. Similarly, breast milk contains large amounts of oligosaccharides that the baby cannot metabolize, but the microbiota can, which might cement transmission of specific symbionts34. Picking up a beneficial microbe from a parent, however, does not guarantee it will remain beneficial throughout a host’s life, because the microbiota can rapidly evolve (Box 2, Box 2 Figure). A host needs additional mechanisms of control.
Hosts also exert control over their microbiota by compartmentalization, which keeps some regions largely microbe free3,4. In many species, the epithelial barrier greatly limits tissue invasion by microbes (although less so in plants than animals3). In animals, mucus also plays a key part at this barrier by permitting access to host tissue for many diffusive molecules, while limiting both the access and colonization of microbes35. The secretion of mucilage in plants may have a similar role36,37. Finally, antimicrobial factors, including proteins such as RegIIIγ (ref. 38) and Lypd8 (ref. 39), also appear to be central in maintaining the epithelial barrier by discouraging microbial growth when it is too close to the epithelial surface (Fig. 1).
Monitoring and targeting
A host can benefit from monitoring the species in its microbiota, and targeting species to either promote or hinder their proliferation. Monitoring the location of microbes is one important strategy. In the mammalian intestine, microbe-specific features, for example lipopolysaccharides or flagellin, are detected by pattern-recognition receptors such as toll-like receptors. The host response to these structural features—often known as pathogen-associated molecular patterns (PAMPs)—is strongest in those instances that involve intestinal breach, and can include both tissue repair and secretion of anti-microbial factors38,40,41. Hosts can detect when microbes enter a cell using pattern-recognition receptors inside endosomes and other locations42. Hosts also monitor cell damage, such that the effects of bacterial toxins activate the inflammasome43, and they detect cellular changes induced by pathogens, such as actin polymerization44. Such surveillance is not restricted to mammals. Toll-like receptors are important for microbiome composition in more basal animals (Cnidarians)41, and innate immunity and pattern recognition have convergently evolved in plants45 (Fig. 1).
Another host strategy is to monitor incoming benefits. Legumes house and monitor bacteria, such as Bradyrhizobium japonicum, in root nodules. If B. japonicum bacteria do not fix nitrogen, the plant cuts off the supply of nutrients to the nodule21,46. In the vertebrate gut, short-chain fatty acids, such as butyrate and acetate, provide energy for colonic epithelial cells47. Strikingly, butyrate also has potent anti-inflammatory properties through the regulation of inflammatory gene expression and induction of regulatory T cells48. We speculate that butyrate monitoring, and specifically recognizing its absence, may drive host responses to restructure the microbiota in order to restore butyrate availability (Box 1). More generally, we predict that monitoring will occur for other nutrients, within both the mammalian microbiome and other systems.
Detecting harmful and beneficial traits (trait-based discrimination) is a robust way for a host to monitor microbiota that can rapidly evolve. Should a beneficial microbe evolve to be harmful, a host can detect this and respond. In this way, a host can link the fitness of a microbe to the benefits it provides, which will lead to natural selection for desirable microbial phenotypes. The evolution of such pleiotropic links is a general way to promote cooperation that may feature commonly in host–microbiome evolution50. The immune system can do this through localized responses—such as the inflammatory response—that target harmful microbes at the position where harm is detected or inferred43. However, this is fallible, as shown by the ability of strains of Salmonella enterica to prosper in the presence of an inflammatory response in the gut25.
An alternative to trait-based discrimination is to monitor or target microbial genotypes through unique chemical moieties (genotype-based discrimination). There is significant variability in antimicrobial peptides both within and between host species, suggesting that the secretion of these peptides from the host epithelium helps to determine which microbial genotypes prosper38,41. Genotype discrimination can be limited by an inability to detect a beneficial strain that evolves a pathogenic phenotype, or vice versa. The adaptive immune system can solve this problem by learning new associations during infection—combining trait and genotypic information—and adaptive immunity can impact the microbiota49,51. However, it is not yet clear whether this results in effective control of the microbiota at epithelial surfaces.
The study of immune responses naturally leads to a focus on negative selection, but there is also the intriguing possibility that hosts target beneficial traits or genotypes. A recent theoretical study predicts that feeding of the microbiota by the host epithelium is a powerful mechanism of positive selection22. Fed strains will bloom and push other strains and species away from the surface. But how can a host target specific strains? One trait-based mechanism is to provide substrates that are the most easily used by microbes with desirable metabolic capabilities. Vertebrate hosts feed their microbiota with diverse glycans that must be removed from the mucin glycoproteins in mucus (Fig. 1), which is expected to favour bacteria, such as Bacteroidaceae22,52, with enzymes that are able to digest complex carbohydrates.
Mucins also have the potential to select particular symbionts in corals, where a remarkable 20–45% of photosynthate is released as mucus53, which houses a diverse microbiota54. Plants appear to have convergently evolved a comparable system to animals (Fig. 1). They can release 25% of photosynthate into the soil, much of it as root mucilage, which houses and feeds rhizosphere microbes36,37. Notably, plant mucilage contains large amounts of ‘arabinogalactan’ proteins55, which show intriguing structural and functional similarities to mucins (also glycoproteins). Symbiotic microbes not only feed on arabinogalactan proteins and mucins, but also attach to them55–57. Theoretical work suggests that a host can make use of this attachment by secreting specific glycoproteins, and molecules such as immunoglobulin A (IgA), that promote the colonization of beneficial strains56. This raises the possibility that adaptive immunity—via IgA—is used to hold onto beneficial symbionts (and not just inhibit harmful ones).
Microbe to microbe: surviving the microbiome jungle
While hosts may have evolved multiple means to regulate their microbiota, the control of all strains in a community is challenging and—in the face of vast microbial diversity—probably impossible. Thus, which microbial species persist will not only depend on host control, but also on their ability to compete in the microbiome jungle58. In order to understand the function of microbial traits, therefore, we must understand what is needed for a strain to succeed in the microbiome.
Evolution within the microbiota
A key determinant of biological function is resource competition (Box 1). Host diet has a major impact on the available resources within animals and, accordingly, which microbial species and types of metabolism can dominate59. Resources may also come from a host’s attempts to exert control, such as through mucin secretion22,52. Competition over resources in an evolving microbial population can drive rapid evolutionary radiations where different strains diverge into different niches to reduce competition (character displacement)60,61. Notably, this suggests that we each carry strains that are tuned to our specific set of niches, which may in turn promote colonization resistance by ensuring that invading strains are less evolutionarily adapted. Availability of a resource to exploit, however, is not sufficient for persistence. Microbial cells influence each other in many ways9, and these interactions can determine whether any given strain can persist6 and, more generally, which traits are needed to compete in a given community62.
Microbes use diverse mechanisms to compete with other members of the microbiota6,62. This includes resource acquisition as discussed above, but also physical properties such as adhesiveness63, production of antibiotics and bacteriocins11, and toxic effector proteins delivered by the spear-like type-VI secretion system10,12. These activities eliminate competitors and contribute to determining which strains persist in the gut10,11. More subtle competition occurs by monitoring and manipulating signalling molecules of competitors, which appears to occur for the quorum-sensing molecule autoinducer-2 (AI-2)64. Bacteriophages (or phages) are also abundant in the gut65 and can impact microbial competition and promote diversity; phages tend to spread easily through host bacteria that are plentiful, which can give rarer species an advantage14. Finally, phages drive horizontal gene transfer (HGT). The evolutionary impacts of HGT, by phage and other means, is an important area of microbiome research. Even when rare, HGT can have major effects66,67 and move a single function, such as antibiotic resistance, horizontally through competing strains and species66.
Many microbes also employ cooperative traits to remain competitive within communities. Cells secrete enzymes that degrade complex molecules, siderophores that scavenge iron, and quorum-sensing molecules that function as a communication system to report on cell density, diffusion conditions and genetic mixing9. In vitro and genomic studies indicate that host-associated microbes exhibit many of these phenotypes68,69, but inferring the function of extracellular enzymes can be challenging. Bacteroides thetaiotaomicron, an abundant species in the human gut microbiome, carries extracellular enzymes that degrade complex carbohydrates. However, the import of breakdown products is so effective that few products are actually shared with others, which renders carbohydrate breakdown by this species a largely private function13,70.
Indeed, the evolution of microbial cooperation is only expected under certain conditions. Genotypes that benefit from cooperative traits, but do not provide them (sometimes called ‘cheaters’) can invade cooperating populations and replace them, rendering cooperation unstable. This invasion can be prevented, if cells grow in single-genotype patches where one strain cannot use the resources of another. As a result, we expect that the spatiogenetic structure is fundamental to how microbes evolve within the microbiome9,71,72. We still understand little of this spatiogenetic structure, but recent work revealed elaborate structure in dental communities73 and (more modest) structuring in gut communities74. While cell–cell benefits often evolve to help clonemates9, cooperation can also evolve between species. For example, B. ovatus breaks down the carbohydrate inulin at a cost to itself; this species prefers to import undigested inulin. However, the breakdown feeds other species, including B. vulgatus, that provide benefits in return13. More simply, one species can benefit from the waste products of a second species. However, waste production is not formally a cooperative function; waste is a metabolic byproduct and did not evolve to benefit other cells (Box 1). This distinction is important for understanding the robustness of communities because cooperative functions are predicted to be more unstable than traits like waste production9,71,72.
Consequences of microbe–microbe interactions
How do the diverse microbial functions we have discussed combine at the level of the whole microbiome, and what is the effect on the host? Ecological theory provides a map between the properties of individual species and the properties of the whole community1,14. The mammalian microbiota often responds robustly to perturbations, allowing a host to keep key species for long periods75,76. Theory suggests that the key to this stability lies in how species interact with one another. Weak and competitive interactions are stabilizing14—they limit positive feedback loops and the possibility that, if one species goes down, it will take others with it. Another key property is productivity, that is the efficiency of converting resources into energy. In this case, it is cooperative interactions that can improve a community by preventing wasteful functions such as antibiotic competition9. A host may then face a tension between communities that are highly productive and those that are stable14.
Another community-level property is redundancy: the coexistence of species with a similar impact on the ecosystem. Redundancy may benefit a host, because other microbes can then compensate for the loss of a beneficial strain by providing the same benefit. There is evidence of considerable functional redundancy in microbiome systems, including the bovine rumen77 and the polysaccharide utilizers of the human intestine69. But this does not necessarily imply that hosts are regulating the diversity of the microbiota to promote redundancy. Theory predicts that redundancy will evolve in all ecosystems, whether host-associated or not, through a combination of competition and stochastic processes that allows highly similar species to persist together for long periods78. To fully apply ecological theory, however, we need more data on microbe-to-microbe interactions within the microbiome14.
An ecosystem on a leash
The mammalian microbiome comprises an ecosystem within which microbes must compete to survive and persist. What makes it so remarkable is that all of this complexity occurs inside a living host, which is itself evolved. Unlike a rainforest or river ecosystem, therefore, the microbiome is not only driven from the bottom up by species interactions, but the host is under strong natural selection to shape the microbiota from the top down and foster a community that is beneficial. We arrived at this characterization of the microbiome—an ecosystem held on a leash by the host—by applying evolutionary and ecological theory to microbiome data (above, Boxes 1 and 2). Next, we explore the plausible alternatives to our perspective and, looking forward, what predictions distinguish our model from these alternatives.
Alternative models of host–microbiota systems
We contrast the leash model to three alternatives: a host-control model, a symbiont-control model and an open-ecosystem model (Fig. 2). Although many permutations of the models are possible, these alternatives satisfy two key criteria. First, they are consistent with evolutionary theory—their evolution is predicted given certain conditions—and, second, each model is consistent with real-world examples.
Host-directed control that tightly regulates microbial phenotypes is most feasible when a host individually monitors one or a few strains20,21. Although all hosts will also interact with microbes they cannot control, fine-scale host control has evolved. One example is legumes, which hold nitrogen-fixing bacteria in root nodules46. Host control also dominates the mutualism between the bobtail squid and its luminescent bacteria, wherein the host uses multiple ‘winnowing’ mechanisms to ensure that each crypt of its light organ contains a single strain of light-producing Vibrio fischeri79,80 (Fig. 2).
Contrasting host control is the possibility of symbiont-directed control, where a microbe alters global host phenotypes—such as reproduction, survival or behaviour—in order to increase its own fitness. Like host control, symbiont control is predicted when there is low microbial diversity and, critically, limited competition between microbes81. This prediction arises because a strain must be able to invest resources into host manipulation without being outcompeted by other strains not investing the resources81 (as also predicted for cooperation with the host, Box 2). A master manipulator of insect hosts is the endosymbiotic bacteria Wolbachia, which, by living inside cells, avoids competition and ensures it can affect all host tissues. Wolbachia can benefit the host by providing metabolites82, but it also impairs host reproduction. For example, Wolbachia kills male offspring of hosts in order to promote its transmission, which occurs cytoplasmically only through females83. Another example is the fungal parasite Ophiocordyceps unilateralis, which induces its ant host to climb into the forest canopy and bite down on a plant, whereupon the fungus kills the ant and makes a fruiting body to disperse its spores84 (Fig. 2c).
Finally, we consider an open-ecosystem model where the host exerts little control over the microbiome. Taken to the extreme, this can be envisaged as a dead or dying host that is simply a resource for the microbiota. In healthy systems, limited host control is most likely to occur when microbiota have weak effects on host fitness. This appears consistent with bromeliads85, and perhaps some pitcher plants, whose leaves create a rainwater pool that contains microbes86. In pitchers, the host may affect the microbiota through its digestive enzymes and hydrogen ions that digest prey. However, beyond compartmentalizing the microbes in the pool, there is no evidence yet of evolved mechanisms of control86.
Predictions of the leash model
We next consider how the leash model can be distinguished from these alternatives. One approach is to study competition between two symbionts, for example bacteria, that are otherwise isogenic, except for the presence of a trait that benefits the host. There are potential complexities but, in general, the leash model predicts that the trait that benefits the host will also help bacteria to persist in the microbiome, such that a null-mutant without the trait would be outcompeted. The reason that the mutant loses might be either due to the leash (host control) or the ecosystem (the inability to compete within the microbiota), or both. By contrast, the expectation of symbiont control is that the trait is energetically costly to the bacteria so that the mutant will win, as long as it can reinvest the energy used to manipulate the host into growth81. The host control and open-ecosystem models, such as the leash model, predict mutant loss, but they are more restrictive on why. Host control predicts it is due to host manipulation, whereas the open-ecosystem model predicts an inability of the mutant to compete in the microbiome ecosystem that is independent of host control.
We can apply these predictions to data on B. ovatus, which break down complex carbohydrates in the mammalian intestine13 (Box 2). Here, symbiont control is unlikely to occur, because carbohydrate utilization gives the bacteria a competitive advantage within the microbiota, which occurs via cross-feeding another species13. Moreover, this advantage occurs both in a mouse model and ex vivo13, suggesting that host control is not central to the outcome. Although one cannot reject the open-ecosystem model, therefore, the data are consistent with the leash model. In another example, light-producing V. fischeri outcompete non-luminescent mutants in the light organ of the squid, which is also incompatible with symbiont control. Moreover, the advantage to the luminescent bacteria is hypothesized to be driven by host-produced enzymes, which argues against the open-ecosystem model (Fig. 2b). However, as no other bacterial species are found in the light organ, the leash model (where both ecosystem and species interactions are important) is also rejected. Host control, therefore, appears to be the best model for the squid symbiosis.
In contrast to the squid system, species interactions are known to affect the mammalian microbiota10,13. How is it possible to hold such a diverse, dynamic community on a leash? Unlike the host-control model, the leash model does not predict near-faultless control. Rather, hosts might focus on certain hub species87 that are important for community function. Hosts will also benefit from influencing the global properties of the microbiota. We, therefore, expect natural selection to favour hosts that act as ecosystem engineers that influence not only individual species but also community-level properties, such as stability and productivity14. Possible mechanisms of this control include the immune system49 and epithelial mucus secretion, which can weaken ecological interactions by regulating species density and increasing spatial structure14. Interrupting the immune system or mucus secretion22,52, therefore, may lead to a less stable, and thus less diverse, microbiota. Any mechanism of control also has to be protected against easy escape, which is why we predict that microbes who mutate to pull against the leash will typically sustain a fitness cost within the dynamic ecosystem (see ‘Monitoring and targeting’).
The importance of host control does not imply that community composition will remain static. Omnivorous hosts, in particular, may benefit from a flexible microbiota that can respond to changing metabolic demands. The fact that microbiome communities can shift strongly with host diet59, therefore, is not in itself evidence that a host is powerless to influence communities. Indeed, humans display a remarkable ability to keep major microbial lineages within our microbiota75,76, to the extent that several bacterial lineages appear to have co-speciated with us88. As for many other hosts, this suggests that humans have evolved to create an environment that selects for specific bacterial lineages. Strong perturbations, however, may force a host to deal with extinctions, followed by stochastic recolonization as new species arrive at random. This potential for recolonization is expected to promote trait-based discrimination in a host (see ‘Monitoring and targeting’), which applies general selection for microbes based on their benefits rather than targeting specific genotypes. As a corollary, hosts may be sometimes blind to an immigrating strain outcompeting a resident, so long as the new strain has the equivalent effect on a host.
Coevolution
Our conceptual model includes the possibility of coevolution, which is distinct from the potentially related process of co-speciation88. Coevolution describes reciprocal evolutionary adaptations in different species in response to one another89. A classic example is the bullhorn acacia that gets its name from the horns it evolved to house ants. The ants fiercely defend the plant, both from insects and other plants that contact the acacia27. This defence is so effective that the plant appears to have lost the normal defences against herbivory: without ants, the plant suffers severe defoliation and death. To our knowledge, there is no comparable evidence yet of such reciprocal adaptation between mammalian hosts and their beneficial microbiota. The hypothesis that there has been some degree of coevolution is a reasonable one. However, it remains possible that hosts control their microbiota and that the microbiota has evolved in response, but that these evolutionary responses across diverse microbial species are too weak to drive reciprocal adaptation in the host. Distinguishing this null model for more elaborate, and true, coevolutionary dynamics is an important empirical goal for the future. The evolution, and coevolution, of host–microbe interactions is also an interesting area for new theoretical work, especially given the complexity and rapid evolution of microbiota.
Treating and engineering the human microbiome
There is an ongoing effort to identify individual species within the human microbiome that have particular health benefits. While this pursuit has great value, our evolutionary approach suggests that too much focus on microbe-to-host effects can be misleading. If we are correct, the benefits that microbes provide are typically by-products of microbe species striving to persist in the microbiome; improving host fitness does not provide significant evolutionary benefits to most of the microbiota (Boxes 1 and 2). To better understand the form and function of the microbiome, therefore, we should focus more on the evolutionary and ecological challenges that the microbiota face. These challenges come both from competition with other microbes and from the selective influences of the host, including the effects of innate and adaptive immunity.
The list of benefits provided by the microbiota is increasing and includes improved nutrition3, colonization resistance5,6, immune system function4 and perhaps even mental health7. However, it is an open empirical question as to how many species actually provide benefits. Moreover, to improve health outcomes, we need to understand whether species provide unique benefits and, linked to this, what the functional basis of the benefit provided is. For example, certain microbial strains appear to have unique effects on immune system development, which might be interpreted as individual strains evolving specific benefits for a host90 (Box 1). However, this specificity might be better explained by host evolution and the complex actions of the immune system. Consistent with this, a virus, which acts through innate cytokine mediators, such as type I interferons, can drive immune maturation of the mouse, similar to the bacterial microbiota91. The effects on immune maturation, therefore, appear to be more because of the way the immune system functions than the identity of specific symbionts.
Certainly, evolutionary theory does not predict that each symbiont strain will provide a benefit, unique or otherwise, but it does predict that all strains will be effective competitors (Boxes 1 and 2). For decades, there have been attempts to introduce particular cellulolytic bacteria into the bovine rumen in order to improve energy yield, but despite large doses, these introduced strains are often outcompeted and lost77. In order to design probiotics, therefore, we need to better understand how bacteria compete, be it through metabolism62, adhesion56,63 or compounds that inhibit other strains in a niche6,10,11. Understanding symbiont competitiveness, however, faces further challenges. We predict that the rapid diversification of microbes60,61 will personalize some symbionts to our individual biology and microbiota. This threatens any one-size-fits-all probiotic, because if a strain is not competitive in all ecosystems, it will only sporadically perform its desired function. One potential solution comes from exploiting the same evolutionary processes that are the barriers to colonization. Repeated large introductions of a strain should increase the chances that the probiotic will evolve and adapt to the host in a way that allows it to persist (see ‘Evolution within the microbiota’). However, even when a strain is well-adapted to a niche, there is no guarantee it will invade if there is a competing strain in place to exclude it.
A key question, therefore, is when and how do similar species avoid competitive exclusion? This question is much discussed in theoretical ecology78, and may help to explain much of the functional redundancy in the microbiome. Ecological theory can also be used to identify species that are important for microbiome function87 or stability14. Theory shows that, even if a species provides no direct benefit to the host, it may contribute to overall community stability, but we need more data on species interactions to develop and apply the theory14. Indeed, while ecological stability is typically seen as a desirable trait, it can be detrimental when attempting to introduce a probiotic, or when trying to reconstitute a healthy microbiota during domination of a dysbiotic yet stable community. In addition to understanding what makes a strain competitive, therefore, we need to understand how to make a community temporarily susceptible to invasion. Faecal microbiota transplantation, which pits large sets of strains against each other, represents a major ecological manipulation that may provide valuable insights into these questions.
In seeking to control the microbiome, we are presented with its daunting complexity. However, there is solace in the fact that diverse host species have faced, and largely overcome, the same challenge over evolutionary time. It may therefore be informative to spend a little less time investigating how our symbionts affect us, and more on how our biology affects them. What is the role of adaptive immunity? Does a host typically employ positive or negative selection? What types of traits and species are targeted? Improving our understanding of the way that a host exerts control over its microbiome may offer the potential to engineer communities with traits, species or communities that are most easily controlled by a host. The long evolutionary history of host control also raises the possibility of harnessing this control for therapeutic means. If a host already has ways to regulate the microbiota, augmenting these mechanisms offers an alternative to targeting the microbes themselves.
Acknowledgments
We thank J. Boomsma, J. Thompson, S. Knowles and N. Ruby for discussions and comments on the manuscript, and D. Hughes, A. Wilson, M. McFall-Ngai for providing images for Fig. 2. K.R.F. is funded by European Research Council grant 242670 and a Calleva Research Centre for Evolution and Human Science (Magdalen College, Oxford) grant. K.Z.C. is funded by a Sir Henry Wellcome Postdoctoral Research Fellowship (grant 201341/Z/16/Z). J.S. is funded by an NIH grant to E. Pamer and J. Xavier (project number 1U01AI124275-01). S.R.-N. is funded by NIH grant 1K08AI130392-01 and a Career Award for Medical Scientists from the Burroughs Wellcome Fund.
Footnotes
Author Contributions All authors contributed to the planning and writing of the manuscript.
The authors declare no competing financial interests.
Reviewer Information Nature thanks S. Brown, D. A. Relman and the other anonymous reviewer(s) for their contribution to the peer review of this work.
References
- 1.Costello EK, Stagaman K, Dethlefsen L, Bohannan BJ, Relman DA. The application of ecological theory toward an understanding of the human microbiome. Science. 2012;336:1255–1262. doi: 10.1126/science.1224203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hacquard S, et al. Microbiota and host nutrition across plant and animal kingdoms. Cell Host Microbe. 2015;17:603–616. doi: 10.1016/j.chom.2015.04.009. [DOI] [PubMed] [Google Scholar]
- 4.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van der Waaij D, Berghuis-de Vries JM, Lekkerkerk-van der Wees JEC. Colonization resistance of the digestive tract in conventional and antibiotic-treated mice. J Hyg (Lond) 1971;69:405–411. doi: 10.1017/s0022172400021653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buffie CG, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 2015;517:205–208. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cryan JF, Dinan TG. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci. 2012;13:701–712. doi: 10.1038/nrn3346. [DOI] [PubMed] [Google Scholar]
- 8.Marchesi JR, Ravel J. The vocabulary of microbiome research: a proposal. Microbiome. 2015;3:31. doi: 10.1186/s40168-015-0094-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nadell CD, Drescher K, Foster KR. Spatial structure, cooperation and competition in biofilms. Nat Rev Microbiol. 2016;14:589–600. doi: 10.1038/nrmicro.2016.84. [DOI] [PubMed] [Google Scholar]
- 10.Chatzidaki-Livanis M, Geva-Zatorsky N, Comstock LE. Bacteroides fragilis type VI secretion systems use novel effector and immunity proteins to antagonize human gut Bacteroidales species. Proc Natl Acad Sci USA. 2016;113:3627–3632. doi: 10.1073/pnas.1522510113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kommineni S, et al. Bacteriocin production augments niche competition by enterococci in the mammalian gastrointestinal tract. Nature. 2015;526:719–722. doi: 10.1038/nature15524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wexler AG, et al. Human symbionts inject and neutralize antibacterial toxins to persist in the gut. Proc Natl Acad Sci USA. 2016;113:3639–3644. doi: 10.1073/pnas.1525637113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rakoff-Nahoum S, Foster KR, Comstock LE. The evolution of cooperation within the gut microbiota. Nature. 2016;533:255–259. doi: 10.1038/nature17626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coyte KZ, Schluter J, Foster KR. The ecology of the microbiome: networks, competition, and stability. Science. 2015;350:663–666. doi: 10.1126/science.aad2602. [DOI] [PubMed] [Google Scholar]
- 15.Stein RR, et al. Ecological modeling from time-series inference: insight into dynamics and stability of intestinal microbiota. PLoS Comput Biol. 2013;9:e1003388. doi: 10.1371/journal.pcbi.1003388. Using ecological models fitted to data, this landmark study estimates ecological interactions within the microbiota and concludes there is a lot of competition and exploitation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat Rev Microbiol. 2008;6:776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Mazancourt C, Loreau M, Dieckmann U. Understanding mutualism when there is adaptation to the partner. J Ecol. 2005;93:305–314. [Google Scholar]
- 19.Mushegian AA, Ebert D. Rethinking “mutualism” in diverse host–symbiont communities. BioEssays. 2016;38:100–108. doi: 10.1002/bies.201500074. [DOI] [PubMed] [Google Scholar]
- 20.Foster KR, Wenseleers T. A general model for the evolution of mutualisms. J Evol Biol. 2006;19:1283–1293. doi: 10.1111/j.1420-9101.2005.01073.x. [DOI] [PubMed] [Google Scholar]
- 21.West SA, Kiers ET, Simms EL, Denison RF. Sanctions and mutualism stability: why do rhizobia fix nitrogen? Proc R Soc B. 2002;269:685–694. doi: 10.1098/rspb.2001.1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schluter J, Foster KR. The evolution of mutualism in gut microbiota via host epithelial selection. PLoS Biol. 2012;10:e1001424. doi: 10.1371/journal.pbio.1001424. Theoretical study predicting the importance of host control in the microbiome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Douglas AE, Werren JH. Holes in the hologenome: why host–microbe symbioses are not holobionts. MBio. 2016;7:e02099–15. doi: 10.1128/mBio.02099-15. Critical analysis of the holobiont concept. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bordenstein SR, Theis KR. Host biology in light of the microbiome: ten principles of holobionts and hologenomes. PLoS Biol. 2015;13:e1002226. doi: 10.1371/journal.pbio.1002226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rivera-Chávez F, Bäumler AJ. The pyromaniac inside you: Salmonella metabolism in the host gut. Annu Rev Microbiol. 2015;69:31–48. doi: 10.1146/annurev-micro-091014-104108. [DOI] [PubMed] [Google Scholar]
- 26.Darwin CR. On the Origin of Species by Means of Natural Selection or the Preservation of Favoured Races in the Struggle for Life. (Murray, 1859) [PMC free article] [PubMed] [Google Scholar]
- 27.Janzen DH. Coevolution of mutualism between ants and acacias in Central America. Evolution. 1966;20:249–275. doi: 10.1111/j.1558-5646.1966.tb03364.x. [DOI] [PubMed] [Google Scholar]
- 28.Scheuring I, Yu DW. How to assemble a beneficial microbiome in three easy steps. Ecol Lett. 2012;15:1300–1307. doi: 10.1111/j.1461-0248.2012.01853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Welzl H, D’Adamo P, Lipp HP. Conditioned taste aversion as a learning and memory paradigm. Behav Brain Res. 2001;125:205–213. doi: 10.1016/s0166-4328(01)00302-3. [DOI] [PubMed] [Google Scholar]
- 30.Imhann F, et al. Proton pump inhibitors affect the gut microbiome. Gut. 2016;65:740–748. doi: 10.1136/gutjnl-2015-310376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poulsen M, Boomsma JJ. Mutualistic fungi control crop diversity in fungus-growing ants. Science. 2005;307:741–744. doi: 10.1126/science.1106688. [DOI] [PubMed] [Google Scholar]
- 32.Kaltenpoth M, et al. Partner choice and fidelity stabilize coevolution in a Cretaceous-age defensive symbiosis. Proc Natl Acad Sci USA. 2014;111:6359–6364. doi: 10.1073/pnas.1400457111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bokulich NA, et al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci Transl Med. 2016;8:343ra382. doi: 10.1126/scitranslmed.aad7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.LoCascio RG, et al. Glycoprofiling of bifidobacterial consumption of human milk oligosaccharides demonstrates strain specific, preferential consumption of small chain glycans secreted in early human lactation. J Agric Food Chem. 2007;55:8914–8919. doi: 10.1021/jf0710480. [DOI] [PubMed] [Google Scholar]
- 35.Caldara M, et al. Mucin biopolymers prevent bacterial aggregation by retaining cells in the free-swimming state. Curr Biol. 2012;22:2325–2330. doi: 10.1016/j.cub.2012.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Philippot L, Raaijmakers JM, Lemanceau P, van der Putten WH. Going back to the roots: the microbial ecology of the rhizosphere. Nat Rev Microbiol. 2013;11:789–799. doi: 10.1038/nrmicro3109. [DOI] [PubMed] [Google Scholar]
- 37.Jones DL, Nguyen C, Finlay RD. Carbon flow in the rhizosphere: carbon trading at the soil–root interface. Plant Soil. 2009;321:5–33. [Google Scholar]
- 38.Vaishnava S, et al. The antibacterial lectin RegIIIγ promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okumura R, et al. Lypd8 promotes the segregation of flagellated microbiota and colonic epithelia. Nature. 2016;532:117–121. doi: 10.1038/nature17406. [DOI] [PubMed] [Google Scholar]
- 40.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 41.Franzenburg S, et al. MyD88-deficient Hydra reveal an ancient function of TLR signaling in sensing bacterial colonizers. Proc Natl Acad Sci USA. 2012;109:19374–19379. doi: 10.1073/pnas.1213110109. Evidence of an ancient role for the immune system in regulation of the microbiota. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barton GM, Kagan JC. A cell biological view of Toll-like receptor function: regulation through compartmentalization. Nat Rev Immunol. 2009;9:535–542. doi: 10.1038/nri2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lamkanfi M, Dixit VM. Mechanisms and functions of inflammasomes. Cell. 2014;157:1013–1022. doi: 10.1016/j.cell.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 44.Xu H, et al. Innate immune sensing of bacterial modifications of Rho GTPases by the Pyrin inflammasome. Nature. 2014;513:237–241. doi: 10.1038/nature13449. [DOI] [PubMed] [Google Scholar]
- 45.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 46.Kiers ET, Rousseau RA, West SA, Denison RF. Host sanctions and the legume–rhizobium mutualism. Nature. 2003;425:78–81. doi: 10.1038/nature01931. Evidence of powerful host control from plants. [DOI] [PubMed] [Google Scholar]
- 47.Roediger WE. Role of anaerobic bacteria in the metabolic welfare of the colonic mucosa in man. Gut. 1980;21:793–798. doi: 10.1136/gut.21.9.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arpaia N, Rudensky AY. Microbial metabolites control gut inflammatory responses. Proc Natl Acad Sci USA. 2014;111:2058–2059. doi: 10.1073/pnas.1323183111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barroso-Batista J, Demengeot J, Gordo I. Adaptive immunity increases the pace and predictability of evolutionary change in commensal gut bacteria. Nat Commun. 2015;6:8945. doi: 10.1038/ncomms9945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Foster KR. The sociobiology of molecular systems. Nat Rev Genet. 2011;12:193–203. doi: 10.1038/nrg2903. [DOI] [PubMed] [Google Scholar]
- 51.Kato LM, Kawamoto S, Maruya M, Fagarasan S. The role of the adaptive immune system in regulation of gut microbiota. Immunol Rev. 2014;260:67–75. doi: 10.1111/imr.12185. [DOI] [PubMed] [Google Scholar]
- 52.Sonnenburg JL, et al. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- 53.Brown BE, Bythell JC. Perspectives on mucus secretion in reef corals. Mar Ecol Prog Ser. 2005;296:291–309. [Google Scholar]
- 54.Glasl B, Herndl GJ, Frade PR. The microbiome of coral surface mucus has a key role in mediating holobiont health and survival upon disturbance. ISME J. 2016;10:2280–2292. doi: 10.1038/ismej.2016.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nguema-Ona E, Vicré-Gibouin M, Cannesan MA, Driouich A. Arabinogalactan proteins in root–microbe interactions. Trends Plant Sci. 2013;18:440–449. doi: 10.1016/j.tplants.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 56.McLoughlin K, Schluter J, Rakoff-Nahoum S, Smith AL, Foster KR. Host selection of microbiota via differential adhesion. Cell Host Microbe. 2016;19:550–559. doi: 10.1016/j.chom.2016.02.021. [DOI] [PubMed] [Google Scholar]
- 57.Huang JY, Lee SM, Mazmanian SK. The human commensal Bacteroides fragilis binds intestinal mucin. Anaerobe. 2011;17:137–141. doi: 10.1016/j.anaerobe.2011.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hibbing ME, Fuqua C, Parsek MR, Peterson SB. Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol. 2010;8:15–25. doi: 10.1038/nrmicro2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Turnbaugh PJ, et al. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rainey PB, Travisano M. Adaptive radiation in a heterogeneous environment. Nature. 1998;394:69–72. doi: 10.1038/27900. [DOI] [PubMed] [Google Scholar]
- 61.Barroso-Batista J, et al. The first steps of adaptation of Escherichia coli to the gut are dominated by soft sweeps. PLoS Genet. 2014;10:e1004182. doi: 10.1371/journal.pgen.1004182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goodman AL, et al. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe. 2009;6:279–289. doi: 10.1016/j.chom.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schluter J, Nadell CD, Bassler BL, Foster KR. Adhesion as a weapon in microbial competition. ISME J. 2015;9:139–149. doi: 10.1038/ismej.2014.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thompson JA, Oliveira RA, Djukovic A, Ubeda C, Xavier KB. Manipulation of the quorum sensing signal AI-2 affects the antibiotic-treated gut microbiota. Cell Rep. 2015;10:1861–1871. doi: 10.1016/j.celrep.2015.02.049. [DOI] [PubMed] [Google Scholar]
- 65.Waller AS, et al. Classification and quantification of bacteriophage taxa in human gut metagenomes. ISME J. 2014;8:1391–1402. doi: 10.1038/ismej.2014.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Niehus R, Mitri S, Fletcher AG, Foster KR. Migration and horizontal gene transfer divide microbial genomes into multiple niches. Nat Commun. 2015;6:8924. doi: 10.1038/ncomms9924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McInerney JO, McNally A, O’Connell MJ. Why prokaryotes have pangenomes. Nat Microbiol. 2017;2:17040. doi: 10.1038/nmicrobiol.2017.40. [DOI] [PubMed] [Google Scholar]
- 68.Ji B, Nielsen J. From next-generation sequencing to systematic modeling of the gut microbiome. Front Genet. 2015;6:219. doi: 10.3389/fgene.2015.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rakoff-Nahoum S, Coyne MJ, Comstock LE. An ecological network of polysaccharide utilization among human intestinal symbionts. Curr Biol. 2014;24:40–49. doi: 10.1016/j.cub.2013.10.077. Experimental evidence of complex ecological interactions between Bacteroides species of the human gut via polysaccharide utilization. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cuskin F, et al. Human gut Bacteroidetes can utilize yeast mannan through a selfish mechanism. Nature. 2015;517:165–169. doi: 10.1038/nature13995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nadell CD, Foster KR, Xavier JB. Emergence of spatial structure in cell groups and the evolution of cooperation. PLoS Comput Biol. 2010;6:e1000716. doi: 10.1371/journal.pcbi.1000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schluter J, Schoech AP, Foster KR, Mitri S. The evolution of quorum sensing as a mechanism to infer kinship. PLoS Comput Biol. 2016;12:e1004848. doi: 10.1371/journal.pcbi.1004848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mark Welch JL, Rossetti BJ, Rieken CW, Dewhirst FE, Borisy GG. Biogeography of a human oral microbiome at the micron scale. Proc Natl Acad Sci USA. 2016;113:E791–E800. doi: 10.1073/pnas.1522149113. Striking evidence of the spatial structure of different bacterial species in the oral microbiome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Earle KA, et al. Quantitative imaging of gut microbiota spatial organization. Cell Host Microbe. 2015;18:478–488. doi: 10.1016/j.chom.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Faith JJ, et al. The long-term stability of the human gut microbiota. Science. 2013;341:1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci USA. 2011;108:4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Weimer PJ. Redundancy, resilience, and host specificity of the ruminal microbiota: implications for engineering improved ruminal fermentations. Front Microbiol. 2015;6:296. doi: 10.3389/fmicb.2015.00296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Scheffer M, van Nes EH. Self-organized similarity, the evolutionary emergence of groups of similar species. Proc Natl Acad Sci USA. 2006;103:6230–6235. doi: 10.1073/pnas.0508024103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nyholm SV, McFall-Ngai MJ. The winnowing: establishing the squid–vibrio symbiosis. Nat Rev Microbiol. 2004;2:632–642. doi: 10.1038/nrmicro957. [DOI] [PubMed] [Google Scholar]
- 80.Schwartzman JA, Ruby EG. A conserved chemical dialog of mutualism: lessons from squid and vibrio. Microbes Infect. 2016;18:1–10. doi: 10.1016/j.micinf.2015.08.016. A review of the remarkable squid and Vibrio fischeri mutualism that shows strong host control. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Brown SP. Cooperation and conflict in host-manipulating parasites. Proc R Soc B. 1999;266:1899–1904. [Google Scholar]
- 82.Brownlie JC, et al. Evidence for metabolic provisioning by a common invertebrate endosymbiont, Wolbachia pipientis, during periods of nutritional stress. PLoS Pathog. 2009;5:e1000368. doi: 10.1371/journal.ppat.1000368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Correa CC, Ballard J. Wolbachia associations with insects: winning or losing against a master manipulator. Front Ecol Evol. 2016;3:153. [Google Scholar]
- 84.Andersen SB, et al. The life of a dead ant: the expression of an adaptive extended phenotype. Am Nat. 2009;174:424–433. doi: 10.1086/603640. [DOI] [PubMed] [Google Scholar]
- 85.Louca S, et al. High taxonomic variability despite stable functional structure across microbial communities. Nat Ecol Evol. 2016;1:0015. doi: 10.1038/s41559-016-0015. [DOI] [PubMed] [Google Scholar]
- 86.Adlassnig W, Peroutka M, Lendl T. Traps of carnivorous pitcher plants as a habitat: composition of the fluid, biodiversity and mutualistic activities. Ann Bot (Lond) 2010;107:181–194. doi: 10.1093/aob/mcq238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Agler MT, et al. Microbial hub taxa link host and abiotic factors to plant microbiome variation. PLoS Biol. 2016;14:e1002352. doi: 10.1371/journal.pbio.1002352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Moeller AH, et al. Cospeciation of gut microbiota with hominids. Science. 2016;353:380–382. doi: 10.1126/science.aaf3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Thompson JN. Interaction and Coevolution. Chicago Univ. Press; 2014. [Google Scholar]
- 90.Ayres JS. Cooperative microbial tolerance behaviors in host–microbiota mutualism. Cell. 2016;165:1323–1331. doi: 10.1016/j.cell.2016.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kernbauer E, Ding Y, Cadwell K. An enteric virus can replace the beneficial function of commensal bacteria. Nature. 2014;516:94–98. doi: 10.1038/nature13960. Challenges the notion that commensal bacteria provide specific benefits to the host by showing that some of these benefits are also provided by a virus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rypien KL, Ward JR, Azam F. Antagonistic interactions among coral-associated bacteria. Environ Microbiol. 2010;12:28–39. doi: 10.1111/j.1462-2920.2009.02027.x. [DOI] [PubMed] [Google Scholar]
- 93.Knoll AH. The multiple origins of complex multicellularity. Annu Rev Earth Planet Sci. 2011;39:217–239. [Google Scholar]
- 94.Williams GC. Adaptation and Natural Selection: A Critique of Some Current Evolutionary Thought. Princeton Univ. Press; 1966. Seminal text on evolutionary adaptation and the importance of understanding biological function [Google Scholar]
- 95.Medzhitov R. Toll-like receptors and innate immunity. Nat Rev Immunol. 2001;1:135–145. doi: 10.1038/35100529. [DOI] [PubMed] [Google Scholar]
- 96.Allen RC, Popat R, Diggle SP, Brown SP. Targeting virulence: can we make evolution-proof drugs? Nat Rev Microbiol. 2014;12:300–308. doi: 10.1038/nrmicro3232. [DOI] [PubMed] [Google Scholar]
- 97.King KC, et al. Rapid evolution of microbe-mediated protection against pathogens in a worm host. ISME J. 2016;10:1915–1924. doi: 10.1038/ismej.2015.259. Experimental evolution shows that colonization resistance can evolve as a byproduct of microbial competition. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nölling J, et al. Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J Bacteriol. 2001;183:4823–4838. doi: 10.1128/JB.183.16.4823-4838.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Smith PM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 101.Wilson DS. A theory of group selection. Proc Natl Acad Sci USA. 1975;72:143–146. doi: 10.1073/pnas.72.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wilson DS. Biological communities as functionally organized units. Ecology. 1997;78:2018–2024. [Google Scholar]
- 103.Williams HT, Lenton TM. Environmental regulation in a network of simulated microbial ecosystems. Proc Natl Acad Sci USA. 2008;105:10432–10437. doi: 10.1073/pnas.0800244105. [DOI] [PMC free article] [PubMed] [Google Scholar]