One of the most difficult questions that clinicians face is “Why does my loved one have a mental illness?” The answer has changed dramatically over time. We have come a long way from blaming mental illness on poor parenting. Yet despite our increased discernment of potential causative factors in recent years, providing a satisfying answer is still a near insuperable task. Researchers are hopeful that continued breakthroughs in our understanding of the genetic basis of these illnesses may soon enable more complete answers.
One relatively recent breakthrough that radically shifted our understanding of genetics was the discovery of micro-RNA (miRNA). The first miRNA was discovered in 1993 while examining the development of the model organism Caenorhabditis elegans, a species of nematode. The investigators discovered that miRNAs were critical for proper development. Since then, functional miRNAs have been identified in many other animals, plants, and some viruses. Different miRNAs are found across tissues and cell types and many are expressed in specific temporal patterns, suggesting a diversity of roles and functions. For example, miR-21 is found in relatively high abundance in multiple tissues, suggestive of a universal role (1), while miR-9 is most abundantly expressed in the brain, suggestive of a unique role in the brain (2). To date, roughly 2500 mature miRNA sequences have been identified in humans, the majority of which are expressed in the brain.
So what exactly are miRNAs, what do they do, and why might they play a role in psychiatric illness? miRNAs are part of a larger class of molecules called noncoding RNA. They are refered to as “noncoding” because they do not ultimately code for protein products, but are instead fully functional as RNAs. miRNAs (and many other noncoding RNAs) largely function by influencing gene expression, the process by which genetic information encoded in DNA is transformed into functional protein products. Under ordinary circumstances, there are two major steps in this process (Figure 1): transcription refers to the creation of messenger RNA (mRNA) from the DNA sequence of a gene; translation refers to the synthesis of protein from this mRNA. Gene expression regulation refers to a range of processes that can up- or downregulate transcription and translation, allowing the cell to maintain tight control of protein levels. miRNAs largely function by inhibiting the second step of the gene expression process, the translation of mRNAs into protein (3). This occurs because each miRNA has a nucleotide sequence that is complementary to sequences within coding target RNAs. By binding the target RNAs at these sequences, the miRNAs diminish the availability of the target RNAs to the protein synthesis machinery. Of note, an individual miRNA can regulate tens to hundreds of different genes based on imperfect complimentary binding to multiple mRNAs.
Figure 1.
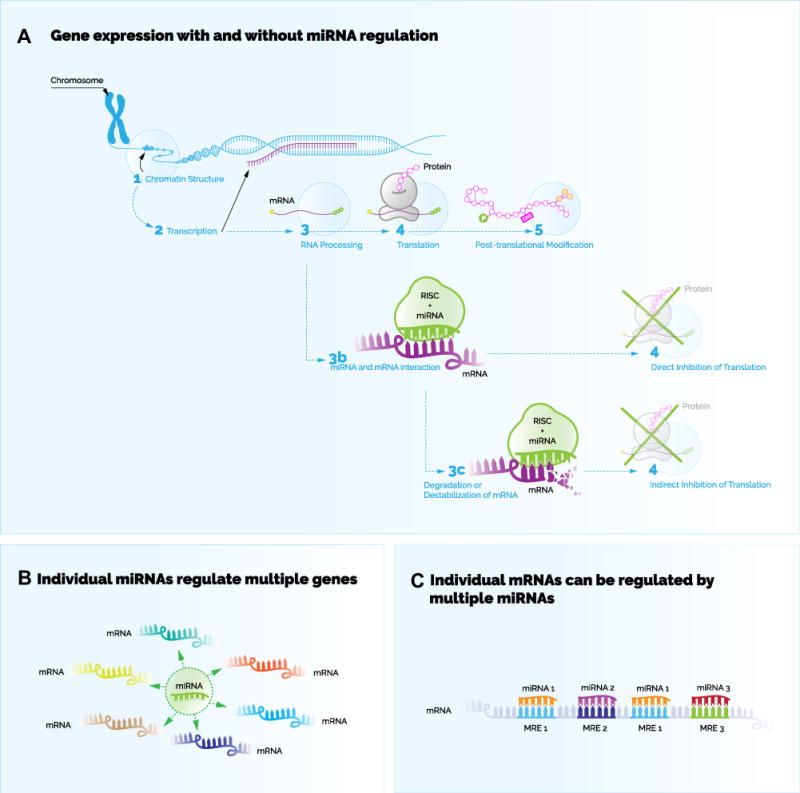
(A) Gene expression with and without microRNA (miRNA) regulation. In situations where the expression of a gene is not regulated by a miRNA, then the general process of gene expression occurs to produce a protein product from a sequence of DNA. First, if the chromatin structure is conducive for transcription, then transcription of the DNA into messenger RNA (mRNA) occurs, followed by processing of the mRNA, and finally followed by translation of the mRNA into protein. The final stages include posttranslational modifications of the protein product. The general process of gene expression is generally halted when miRNA regulation occurs. In this case, miRNA-containing sequences called seed sequences bind to complimentary miRNA response elements within mRNAs. A molecular complex called RNA-induced silencing complex (RISC) is guided by a miRNA to inhibit translation of bound mRNAs. This occurs by directly inhibiting translation of bound mRNAs or indirectly by mechanisms of destabilization or degradation of the bound mRNA. (B) Individual miRNAs regulate multiple genes. A single miRNA can regulate many different mRNAs corresponding to different protein coding genes. (C) Individual mRNAs can be regulated by multiple miRNAs. A single mRNA may contain multiple miRNA response elements (MREs) for the same miRNA or for different miRNAs. [Adapted with permission from Elsevier (10).]
miRNAs are now known to mediate gene expression involved in nearly every type of biological process, including many brain-specific processes, such as synaptic activity and neurogenesis. It is estimated that 60% of all mammalian protein coding genes are regulated by at least one miRNA (4). Some genes are regulated by multiple miRNAs, enabling tight and multilayered control of the protein expression of these genes. This points to the compelling possibility that miRNA may be quite influential in complex polygenic diseases, including psychiatric illnesses.
Our knowledge of miRNAs is beginning to help answer difficult clinical questions for a variety of complex diseases (5). Improper expression or function of miRNAs has been linked to nearly every category of common disease, including cancer, cardiovascular disease, autoimmune disease, infectious disease, neurodegenerative disease, intellectual disabilities, and, more recently, psychiatric disorders. The most advanced translational miRNA research to date has been in oncology. Many miRNAs have been identified to be tumor suppressors and enhancers, as well as promoters and inhibitors of metastasis. miRNA expression profiling—both in tumor tissue and in circulating blood—is being used as a biomarker strategy to aid in diagnosis, tumor classification, and making treatment decisions. New therapies are also being designed to attempt to either mimic miRNAs or inhibit the regulatory activity of miRNAs. For example, in vitro studies have used small molecules to block transcription of miR-21, an miRNA that promotes cancer by inhibiting the expression of apoptosis-related genes. This miRNA has been associated with a variety of cancers, including breast, ovarian, and glioblastoma (1). Clinical trials have begun for several other miRNA therapies. One example, called MRX34, mimics miR-34a, which represses the expression of over 30 oncogenes and is underexpressed in many cancers. This miRNA mimic has shown promising efficacy and safety in a phase I clinical trial when administered intravenously to patients with solid tumors (6). These new therapies are in the early stages of development, and more are expected to come to clinical trials in the next few years.
There is growing interest in deciphering the role of these gene expression regulators in a variety of psychiatric illnesses (7,8). Recent large genome-wide association studies of schizophrenia revealed that the second most statistically significantly associated region in the genome includes a sequence for a miRNA, miR-137, that has a known role in regulating neuronal maturation and differentiation (9). Several studies found that miR-137 may influence cognitive function and brain physiology. Another example is miR-16, an miRNA involved in inhibiting neurogenesis that appears to be decreased by fluoxetine, a treatment for depression. Several lines of evidence suggest that the miRNA plays a critical role in the effectiveness of the drug (4) and that the molecular effect of decreasing miR-16 expression may underlie the observation that antidepressants increase hippocampal neurogenesis. Furthermore, altered expression of various miRNAs has been identified for several psychiatric disorders based on studies of blood, postmortem brain, or cerebral spinal fluid (7). Among these, miR-132 was shown to be altered in brain tissue from subjects with schizophrenia and autism, as well as in serum of subjects with major depressive disorder and in animal models of depression and addiction (7,8). It has been proposed that this may be at least in part related to the role of this miRNA in regulating the circadian rhythm or brain-derived neurotrophic factor, a gene with a well-defined role in neurogenesis, cognition, and mood regulation.
Some argue that the widespread influence of miRNAs on gene expression regulation, though challenging for therapeutic development in the traditional sense, may actually offer a unique opportunity (8). Thus a single target—in this case, an miRNA—might be a therapeutic entry point to influence a complex network of relevant genes. In terms of biomarkers, reproducibly altered miRNAs have potential as an approach to differentiating patient subgroups for treatment decisions. Importantly, increased knowledge of miRNA and all causative factors involved in mental illness will help reduce the stigma associated with mental illness and help clinicians provide a more satisfying answer to patients when they ask, “Why do I have a mental illness?”
Acknowledgments
Clinical Commentaries are produced in collaboration with the National Neuroscience Curriculum Initiative (NNCI). David Ross, in his dual roles as co-chair of the NNCI and Education Editor of Biological Psychiatry, manages the development of these commentaries but plays no role in the decision to publish each commentary. The NNCI is supported by National Institutes of Health Grant Nos. R25 MH10107602S1 and R25 MH08646607S1.
We thank Amanda Wang for her assistance in developing the figure.
Footnotes
The authors report no biomedical financial interests or potential conflicts of interest.
References
- 1.Gumireddy K, Young DD, Xiong X, Hogenesch JB, Huang Q, Deiters A. Small-molecule inhibitors of microRNA miR-21 function. Angew Chem Int Ed. 2008;47:7482–7484. doi: 10.1002/anie.200801555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ludwig N, Leidinger P, Becker K, Backes C, Fehlmann T, Pallasch C, et al. Distribution of miRNA expression across human tissues. Nucleic Acids Res. 2016;44:3865–3877. doi: 10.1093/nar/gkw116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iwakawa H, Tomari Y. The functions of MicroRNAs: mRNA decay and translational repression. Trends Cell Biol. 2015;25:651–665. doi: 10.1016/j.tcb.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 4.O’Connor RM, Gururajan A, Dinan TG, Kenny PJ, Cryan JF. All roads lead to the miRNome: miRNAs have a central role in the molecular pathophysiology of psychiatric disorders. Trends Pharmacol Sci. 2016;37:1029–1044. doi: 10.1016/j.tips.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 5.Ardekani AM, Naeini MM. The role of microRNAs in human diseases. Avicenna J Med Biotechnol. 2010;2:161–179. [PMC free article] [PubMed] [Google Scholar]
- 6.Beg MS, Brenner AJ, Sachdev J, Borad M, Kang Y-K, Stoudemire J, et al. Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Invest New Drugs. 2017;35:180–188. doi: 10.1007/s10637-016-0407-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geaghan M, Cairns MJ. MicroRNA and posttranscriptional dysregulation in psychiatry. Biol Psychiatry. 2015;78:231–239. doi: 10.1016/j.biopsych.2014.12.009. [DOI] [PubMed] [Google Scholar]
- 8.Chan AW, Kocerha J. The path to microRNA therapeutics in psychiatric and neurodegenerative disorders. Front Genet. 2012;3:82. doi: 10.3389/fgene.2012.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wright C, Turner JA, Calhoun VD, Perrone-Bizzozero N. Potential impact of miR-137 and its targets in schizophrenia. Front Genet. 2013;4:58. doi: 10.3389/fgene.2013.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olfson E, Ross DA. Genes orchestrating brain function. Biol Psychiatry. 2017;82:e17–e19. doi: 10.1016/j.biopsych.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]


