ABSTRACT
Targeting post-translationally modified epitopes may provide a new strategy for generating tumor specific immune responses. Citrullination is the post-translational modification of arginine to citrulline catalyzed by peptidylarginine deaminase (PAD) enzymes. Presentation of citrullinated peptides on MHC-II has been associated with autophagy. Tumors upregulate autophagy and present citrullinated peptides in response to stresses including nutrient deprivation, oxygen deprivation, redox stress and DNA damage, making them good targets for immune attack. The ubiquitous glycolytic enzyme α-enolase (ENO1) is often citrullinated and degraded during autophagy. Immunization of mice with two citrullinated ENO1 peptides (ENO1 241–260cit253 or 11–25cit15) induced strong Th1 responses that recognize the post-translationally modified, but not the wild type unmodified epitope. ENO1 11–25cit15 induced tumor therapy of melanoma cells in C57Bl/6 (B16F1 50% survival p = 0.0026) and ENO1 241–260cit253 in HLA-DR4 transgenic mice (B16-DR4 50% survival p = 0.0048). In addition, ENO1 241–260cit253 induced therapy of pancreatic (Pan02-DR4 50% survival p = 0.0076) and lung (LLC/2-DR4 40% survival p = 0.0142) tumors in HLA-DR4 transgenic mice. The unmodified epitope induced no anti-tumor response. Minimal regression of class II negative B16 or LLC/2 tumor was seen, confirming direct recognition of MHC-II was required. Most tumors only express MHC-II in the presence of IFNγ; an IFNγ inducible model showed strong responses, with rejection of tumors in up to 90% of animals (p = 0.0001). In humans, a repertoire to ENO1 241–260cit253 was observed in healthy donors. This response was CD4 mediated and seen in people with a variety of HLA types suggesting a broad application for this vaccine in human cancer therapy.
KEYWORDS: Enolase, Citrullination, Cancer, Tumor immunotherapy, CD4 T cells, Autophagy, MHC-II
Introduction
The challenge for cancer vaccines is to induce potent immune responses which can break tolerance and overcome the immunosuppressive tumor environment. One way to overcome immune suppression is to induce pro-inflammatory CD4 T cells. CD4 T cells are the main orchestrators of immune responses generating inflammatory cytotoxic cytokines such as IFNγ and TNFα, and chemokine gradients to promote infiltration of other immune cells including antigen presenting cells (APCs), macrophages and CD8 T cells.1–4 CD4 T cells can also differentiate to potent killer T cells.5–7 The difficulty is raising CD4 responses to tumor antigens which overcome self-tolerance. Recent strategies have targeted specific tumor mutations.8,9 However, this strategy is patient specific and is hampered by the heterogeneous nature and ongoing adaptation of tumors.10 An alternative way to overcome self-tolerance is to target post-translational modifications. Citrullination is a post-translational modification which converts positively charged arginine to neutral citrulline. This process is catalyzed by a family of peptidylarginine deaminase (PAD) enzymes which are activated by high (mM) concentrations of calcium.11 Citrullination can occur in viable cells, either within the nucleus where PAD4 citrullinates regulatory proteins, or within autophagosomes where PAD2 citrullination plays a role in protein degradation. Autophagy has also been demonstrated to be an efficient mechanism to enable processing of endogenous antigens for presentation on MHC class II molecules in professional APCs as well as epithelial cells.11 More recently it has been shown that antigen processing cells have constitutively active autophagy pathways and present citrullinated peptides on MHC class II to stimulate CD4 T cell responses.12 If cellular stress is accompanied by inflammation then these CD4 T cells can orchestrate the removal of the stressed cell.13 Cells undergo autophagy experiencing stresses such as nutrient deprivation, oxygen deprivation, redox stress and DNA damage, to promote their survival. As such it is unsurprising that autophagy is upregulated in rapidly proliferating tumor cells.14 We have previously shown that vaccination with citrullinated epitopes from vimentin, a protein involved in epithelial to mesenchymal transition (EMT), generated cytotoxic CD4 T cells that rapidly eliminated tumors with no associated toxicity.15
α-enolase (ENO1) is a glycolytic enzyme catalyzing the penultimate step in glycolysis.16 Many tumors switch to generating their energy via glycolysis even under normoxic conditions in a process termed the “Warburg effect”. As such ENO1 is overexpressed in a wide range of tumors.17–20 Due to its ubiquitous expression and abundance in most cells, ENO1 is also degraded during autophagy. Previous studies have shown that ENO1 can also be citrullinated.21,22 Therefore, like vimentin, α-enolase may represent a good target for anti-tumor immunity.
In this study, we identified citrullinated ENO1 peptides which induce strong citrulline-specific Th1 responses in conventional and HLA transgenic mice, and these led to an anti-tumor effect in melanoma, pancreatic and lung cancer models. Further, a T cell repertoire to citrullinated ENO1 was detected in healthy human donors. Together our data shows that citrullinated ENO1 is a promising target for human cancer therapy.
Methods
Laboratory practice
These studies were conducted in a laboratory that operates under exploratory research principles. Standard operating procedures were used for all human and mouse T cell assays. These studies were performed using general research investigative assays. Procedures and raw data can be obtained from Scancell Limited through corresponding author and may be subject to non-disclosure agreements. Unless otherwise stated all reagents were obtained from Sigma-Aldridge.
Cell lines and culture
The murine melanoma B16F1 cell line (ATCC-CRL-6323), murine lung carcinoma line LLC/2 (ATCC-CRL-1642), murine prostate adenocarcinoma cell line TrampC1 (ATCC-CRL-2730) and HeLa cells (ATCC-CCL-2) were obtained from the ATCC. The murine pancreatic cancer cell line Pan02 (CVCL_D627) was obtained from the DCTD Tumor/Cell line repository at the National Cancer Institute. Murine ovarian cancer cell line ID8 was obtained from Prof. K Roby, University of Kansas medical Centre. RTLCL Lymphoblastoid cell line was kindly donated by Prof. Alan Rickinson, University of Birmingham. B16F1, Pan02 and RTLCL cell lines were cultured in RPMI medium 1640 with L-glutamine (2 mmol/L), and sodium bicarbonate buffered at pH 7 supplemented with 10% fetal calf serum (FCS). LLC/2 and ID8 cell lines were cultured in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% FCS, L-glutamine (2 mmol/L), and sodium bicarbonate buffered. Cell lines utilized were mycoplasma free, authenticated by suppliers (STR profiling), and used within ten passages.
Plasmids and transfections
Cell lines were transfected using the Lipofectamine Transfection Reagent (Invitrogen) utilizing the protocol previously described.15 B16F1, LLC/2 and Pan02 cells were transfected with constitutive HLA-DR4 using the pDC Vitro 2 chimeric HLA-DR401 plasmid. B16F1 cells were also transfected with the IFNγ-inducible HLA-DR4 using the pDC GAS chimeric HLA-DR401 plasmids where chimeric HLA-DR401 is under expression of the IFNγ-inducible promoter. Plasmid details have previously been described in full.15
Transfected cells were selected by growth in the presence of zeocin (300 μg/mL), or hygromycin B (300 μg/mL). Lines were cloned by limiting dilution and expression was confirmed by flow cytometry using the HLA-DR PE conjugated antibody (clone L243, eBioscience). Cells transfected with the IFNγ-inducible plasmid were incubated overnight in the absence or presence of murine IFNγ (30 ng/mL, Gibco Life Technologies) before staining with the antibody.
Western blotting
Cell lysates were prepared by washing cell pellets twice with cold phosphate buffered saline (PBS) and lysed with radioimmunoprecipitation buffer containing protease inhibitor cocktail (Roche) and endonuclease at 50 U/mL (Benzonase, Millipore). Cells were kept on ice for at least 30 mins. Equal volumes of lysates were resolved by 4–12% denaturation SDS-PAGE gel (Thermofisher), 140 V for 75 mins. Proteins were transferred to polyvinylidene fluoride transfer membrane (GE healthcare) (30 V, 90 min) and blocked in 5% bovine serum albumin (BSA) in 0.1% PBS Tween for 1 hr. The membrane was probed with antibodies to ENO1 (clone EPR10863(B), Abcam 1/1000) and β-actin (clone AC-15 1/2000). Proteins were visualized using fluorescent secondary antibodies at 1/15,000 against mouse (for ENO1) or rabbit (for β-actin) and a Licor detection system. Quantification was performed using Image Studio software and the intensity of respective bands was normalized to β-actin loading control.
Peptides
Peptides (see Table 1) were synthesized at >90% purity (Genscript), aliquoted to single use vials and stored lyophilized at −80°C. On the day of use they were reconstituted to the appropriate concentration in PBS.
Table 1.
Peptide sequences, aa coordinates and screening pools, and murine homology.
| Predicted binding (below 20) |
|||||
|---|---|---|---|---|---|
| aa coordinates | Sequence | Species | DR4 | DR1 | I-Ab |
| 1–20 | MSILKIHAREIFDSRGNPTV | Hu* | 14.35 | ||
| MSILRIHAREIFDSRGNPTV | Mo | ||||
| 6–25 | IHAREIFDSRGNPTVEVDLF | Hu* | |||
| IHAREIFDSRGNPTVEVDLY | Mo | ||||
| 11–25 | IFDSRGNPTVEVDLF | Hu* | |||
| IFDSRGNPTVEVDLY | Mo | ||||
| 21–40 | EVDLFTSKGLFRAAVPSGAS | Hu* | 15.51 | ||
| EVDLYTAKGLFRAAVPSGAS | Mo | 11.10 | |||
| 26–45 | TSKGLFRAAVPSGASTGIYE | Hu* | 2.16 | 3.02 | |
| TAKGLFRAAVPSGASTGIYE | Mo | 0.57 | |||
| 36–55 | PSGASTGIYEALELRDNDKT | Homo* | |||
| 46–65 | ALELRDNDKTRYMGKGVSKA | Hu* | 19.25 | ||
| ALELRDNDKTRFMGKGVSQA | Mo | 10.11 | |||
| 56–75 | RYMGKGVSKAVEHINKTIAP | Hu* | |||
| RFMGKGVSQAVEHINKTIAP | Mo | 17.15 | |||
| 121–140 | AGAVEKGVPLYRHIADLAGN | Homo* | |||
| 126–145 | KGVPLYRHIADLAGNSEVIL | Hu* | 2.76 | 12.77 | |
| KGVPLYRHIADLAGNPEVIL | Mo | ||||
| 171–190 | LPVGAANFREAMRIGAEVYH | Hu* | |||
| LPVGASSFREAMRIGAEVYH | Mo | ||||
| 176–195 | ANFREAMRIGAEVYHNLKNV | Hu* | |||
| SSFREAMRIGAEVYHNLKNV | Mo | ||||
| 241–260 | VIGMDVAASEFFRSGKYDLD | Hu* | 7.15 | ||
| VIGMDVAASEFYRSGKYDLD | Mo | ||||
| 246–265 | VAASEFFRSGKYDLDFKSPD | Hu* | |||
| VAASEFYRSGKYDLDFKSPD | Mo | ||||
| 256–275 | KYDLDFKSPDDPSRYISPDQ | Hu* | |||
| KYDLDFKSPDDPSRYITPDQ | Mo | ||||
| 261–280 | FKSPDDPSRYISPDQLADLY | Hu* | |||
| FKSPDDPSRYITPDQLADLY | Mo | ||||
| 316–335 | VGDDLTVTNPKRIAKAVNEK | Hu* | 14.07 | ||
| VGDDLTVTNPKRIAKAASEK | Mo | ||||
| 321–340 | TVTNPKRIAKAVNEKSCNCL | Hu* | 8.50 | ||
| TVTNPKRIAKAASEKSCNCL | Mo | ||||
| 326–345 | KRIAKAVNEKSCNCLLLKVN | Hu* | 4.16 | ||
| KRIAKAASEKSCNCLLLKVN | Mo | ||||
| 361–380 | QANGWGVMVSHRSGETEDTF | Hu* | 4.78 | 13.7 | |
| QSNGWGVMVSHRSGETEDTF | Mo | ||||
| 366–385 | GVMVSHRSGETEDTFIADLV | Homo* | |||
| 391–410 | GQIKTGAPCRSERLAKYNQL | Homo* | |||
| 396–415 | GAPCRSERLAKYNQLLRIEE | Hu* | |||
| GAPCRSERLAKYNQILRIEE | Mo | ||||
| 401–420 | SERLAKYNQLLRIEEELGSK | Hu* | 6.94 | ||
| SERLAKYNQILRIEEELGSK | Mo | ||||
| 406–425 | KYNQLLRIEEELGSKAKFAG | Hu* | 14.6 | ||
| KYNQILRIEEELGSKAKFAG | Mo | ||||
| 416–434 | ELGSKAKFAGRNFRNPLAK | Hu* | 19.53 | ||
| ELGSKAKFAGRSFRNPLAK | Mo | ||||
Arginine residues are shown in bold, non-homologous peptides are shown in italics Hu = human, Mo = Mouse, Homo = homologous.
= immunizing peptides for screen.
Immunization protocol
Animal experiments were carried out with ethical approval and under Home Office approved project licenses. HLA-DR4 mice (Model #4149, Taconic), HLA-A2/DR1 (HHDII/DR1, Pasteur Institute) or C57Bl/6 J mice (Charles River) aged 8–12 weeks were used. For all studies, mice were randomized into different groups and processed without blinding. Peptides were dissolved in PBS to 1 mg/mL and then emulsified with 6 μg/mouse each of CpG ODN 1826 and MPLA (Invivogen). Peptides (25 μg/mouse) were injected subcutaneously at the base of the tail. Mice were immunized on day 1, 7 and 14 they were then humanely euthanized and their spleens removed for analysis at day 21.
For tumor challenge experiments, mice were challenged with 2.5 × 104 B16-DR4 cells, 1 × 105 Pan02-DR4 cells or 1.5 × 106 LLC/2-DR4 cells subcutaneously on the right flank 3 days before primary immunization (unless stated otherwise) and subsequently immunized as above. Tumor growth was monitored at 3 to 4 day intervals and mice were humanely euthanized once tumor reached ≥10 mm in diameter.
Ex vivo ELISpot assay
ELISpot assays were performed using murine IFNγ or IL10 capture and detection reagents according to the manufacturer's instructions (Mabtech). In brief, the IFNγ or IL10-specific antibodies were coated onto wells of 96-well Immobilin-P plates. Synthetic peptides (10 μg/mL) and 5 × 105 per well splenocytes were added to the wells in quadruplicate. Plates were incubated for 40 hrs at 37 °C in an atmosphere of 5% CO2. After incubation, captured IFNγ or IL10 were detected by biotinylated specific IFNγ or IL-10 antibodies and developed with a streptavidin alkaline phosphatase and chromogenic substrate. Spots were analyzed and counted using an automated plate reader (Cellular Technologies Ltd). Lipopolysaccharide (LPS) at 5 μg/mL was used as a positive control. For MHC blocking studies 20 μg/mL of the CD8 (clone 2.43), and CD4 (clone GK1.5) antibodies purchased from BioXcell were added to ELISpot assays.
Granzyme B ELISA
Supernatant from ex vivo IFNγ ELISpot assays on splenocytes were removed after 40 hrs and assessed for Granzyme B by ELISA assay (R&D Systems) according to manufacturer's instructions.
Peripheral blood mononuclear cell (PBMC) isolation
PBMC experiments were carried out with ethical approval. Demographics of healthy donors are given in Table 2. Peripheral blood sample (approx. 50 mL) was drawn into lithium heparin tubes (Becton Dickinson). Samples were maintained at room temperature and processed immediately following venepuncture. PBMCs were isolated by density gradient centrifugation using Ficoll-Hypaque. Proliferation and cultured ELISpot assay of PBMCs were performed immediately after PBMC isolation. The median number of PBMCs routinely derived from healthy donor samples was 1.04 × 106 PBMC/mL whole blood (range: 0.6 × 106 – 1.48 × 106 / mL). The median viability as assessed by trypan blue exclusion was 93% (range 90–95%).
Table 2.
Healthy donor details.
| Donor | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| Age (years) | 20–40 | 40–60 | 20–40 | 40–60 | 20–40 | 20–40 |
| Sex | Male | Male | Female | Male | Male | Female |
| Smoking Status | Ex-smoker | Non-smoker | Non-smoker | Ex-smoker | Non-smoker | Non-smoker |
| HLA type | A2 | A3, 32 | A3, 24 | A1, 2 | A2, 29 | A2, 29 |
| B7, 41 | B14, 35 | B15, 27 | B8, 44 | B44, 51 | B15, 44 | |
| C7, 17 | C4, 8 | C2, 3 | C5, 7 | DR7, 11 | C3, 16 | |
| DR7, 52 a, 13 | DR2, 11 | DR4, 52 a | DR3, 51 a, 15, 52 a | DQ2, 0301 | DR4, 7, 53 a | |
| DQ2, 3 | DQ1, 7 | DQ3 | DQ2, 6 | DP4, 5 | DQ2, 3 | |
| DP1, 4 | DP3, 4 | DP4, 9 | DP1, 4 | DP2 |
For CD25 depletion PBMCs were processed as above and then immediately enriched using anti-CD25 microbeads and MACS Cell Separation Columns (Miltenyi).
Thymidine proliferation assay
PBMCs (1.5 × 106 cells/well) were stimulated with individual peptides at a final concentration of 10 μg/mL in 2.0 mL of complete media (RPMI supplemented with L-glutamine and penicillin/streptomycin, final concentration of 0.1 mg/mL streptomycin and 100 units/mL penicillin, and 10% (v/v) autologous plasma). Stimulation with purified protein derivative (Statens Serum Institute, final concentration 20 µg/mL) and CEF, a mixture of peptides from cytomegalovirus, Epstein Barr and influenza viruses (Mabtech, final concentration of 4 µg/mL), served as positive controls. As a negative control, PBMCs were incubated with peptide vehicle alone. PBMCs were cultured at 37 °C in 5% CO2 and proliferative capacity assessed at day 4, 7 and 11 post-stimulation by overnight 3H-thymidine incorporation (Perkin Elmer). Data was expressed in terms of proliferative index (PI) defined as the mean peptide-specific counts per minute (cpm) divided by the mean negative control cpm at a given time point. A PI of >2 was defined as a positive response following predefined parameters.
Proliferation/phenotyping assessed by CSFE labelling
Freshly isolated PBMCs or CD25 depleted PBMCs were loaded with carboxyfluorescein succinimidyl ester (CFSE) (ThermoFisher). Briefly, a 50 µM stock solution in warm PBS was prepared from a master solution of 5 mM in DMSO. CFSE was rapidly added to PBMCs (5 × 106 cells/mL loading buffer (PBS with 5% v/v heat inactivated FCS)) to achieve a final concentration of 5 µM. PBMCs were incubated at room temperature in the dark for 5 mins after which non-cellular incorporated CFSE was removed by washing twice with excess (× 10 v/v volumes) of loading buffer (300 g × 10 mins). Cells were made up in complete media to 1.5 × 106/mL and plated and stimulated with vehicle (negative control), PHA (positive control, final concentration 10 µg/mL) or peptide (10 µg/mL) as described above.
On days 4, 7 and 10, 500 µL of cells were removed from culture, washed in PBS and stained with 1:50 dilution of anti-CD4 (PE-Cy5, clone RPA-T4, ThermoFisher) and anti-CD8 efluor 450, clone RPA-T8, ThermoFisher) and anti-CD134 (PE-Cy7, Clone REA621, Miltenyi). Cells were washed, fixed and permeabilized using intracellular fixation/permeablization buffers (both ThermoFisher) according to the manufactures instructions. Intracellular staining for cytokines was performed using a 1:50 dilution of anti-IFNγ (clone 4 S.B3, ThermoFisher) or anti-Granzyme B (PE, Clone GB11, Thermofisher). Stained samples were analyzed immediately on a MACSQuant 10 flow cytometer equipped with MACSQuant software version 2.8.168.16380 using stained vehicle stimulated controls to determine suitable gates.
Human interferon-gamma (IFNγ) ELISpot assay
Freshly isolated PBMCs (3 × 106 cells/well in 24-well flat-bottomed plates), were stimulated with individual peptides at a final concentration of 10 μg/mL in 4.0 mL of complete media supplemented with human recombinant IL-7 (Peprotech, final concentration 20 ng/mL) and IL-15 (Peprotech, final concentration 10 ng/mL). PBMCs stimulated by vehicle alone served as a negative control. PBMCs were cultured at 37 °C in an atmosphere of 5% CO2 and supplemented with 10 U/mL human recombinant interleukin-2 (Peprotech) after 3 days. Cytokine release by stimulated T cells was assessed after 13 days in an IFNγ ELISpot assay using capture and detection reagents according to the manufacturer's instructions (Mabtech). In brief, an anti-IFNγ antibody was coated onto the wells of a 96-well Immobilin-P plate Millipore). Cultured PBMCs were used at 5 × 104 cells/well (in quadruplicate) and stimulated with the relevant peptides at 10 µg/mL (200 µL final volume). Addition of vehicle or PHA (10 µg/mL) served as negative and positive controls respectively. Cells were incubated for 20 hrs at 37 °C in an atmosphere of 5% CO2, then captured IFNγ was detected by the addition of a biotinylated anti-IFNγ antibody and spots were identified with a streptavidin alkaline phosphatase and chromogenic substrate (BioRad). Spots were analyzed and counted using an automated plate reader (Cellular Technologies Ltd.) using Immunospot version 5.1.33 software. Analysis was performed using “autocount easy blue” with an optical sensitivity of 240, background 10, diffuse spot process 20 and min-max spot range of 0.002–8.77 mm.
Statistical analysis
Comparative analysis of the ELISpot results was performed by applying paired or unpaired ANOVA or Student t test as appropriate with values of P calculated accordingly. Comparison of tumor survival was assessed by log-rank test using the GraphPad Prism software version 7. P < 0.05 values were considered statistically significant and p < 0.01 values were considered highly significant. The error bars shown in the figures represent the mean and standard error of the mean (SEM).
Results
T cell responses to citrullinated ENO1 can be induced by peptide vaccination in mice
To determine if citrullinated ENO1 is a good target for anti-tumor immunity, all potential citrullinated epitopes were screened in transgenic HLA-DR4, HLA-DR1 and wild type C57Bl/6 mice for their ability to stimulate T cell responses. A panel of peptides was derived by dividing the human ENO1 sequence into 20 amino acid (aa) fragments which overlapped by 15aa. All peptides containing arginine were identified. Peptides were synthesized with the arginine residues replaced with citrulline and these peptides were then used to immunize mice (Table 1). Mice were immunized with pools of peptides covering non-overlapping regions of the human ENO1 sequence. Responses were then screened by IFNγ ELISpot using the human peptide sequences and where relevant, the corresponding murine peptide sequences.
HLA-DR4 transgenic mice show a dominant response to a peptide spanning aa241-260 (Fig. 1A) this peptide includes a citrulline at position 253 (ENO1 241-260cit253). Subdominant responses to peptides spanning aa21-40, aa316-335 and aa321-340 were also observed. HHDII/DR1 mice show responses to aa126-145, aa316-335 and a low response to aa1-20 (Fig. 1B). C57Bl/6 mice show responses to aa21-40, aa261-280 and aa316-335 (Fig. 1C).
Figure 1.

Screening IFNγ responses to peptide pools. Transgenic mouse strains with human DR4 (A) or DR1/HHDII (B) and parental C57Bl/6 (C and D) mice were used to screen IFNγ responses to peptides. Mice were immunized with pools of 4–6 non-overlapping human citrullinated ENO1 peptides. Ex vivo responses to stimulation with human and mouse (Mo) equivalent citrullinated peptides were assessed by IFNγ ELISpot. Media only responses were used as a negative control. For each pool n = 3. Statistical significance of peptide compared to media responses for each pool was determined by ANOVA with Dunnett's post-hoc test * p<0.05, ** p<0.01, *** p<0.001.
The screening panel used the human (Hu) ENO1 sequence, but these sequences are not homologous with the mouse (Mo) sequence. Therefore, cross reactivity with the murine epitopes was also tested. HLA-DR4 mice showed cross reactivity to murine ENO1 241-260cit253. In HLA-DR4 mice, ENO1 21-40cit32 peptide cross reacts with the murine peptide but ENO1 316-335cit327 does not cross react with mouse ENO1 316-335cit327. Likewise, analysis of responses in C57Bl/6 and HHDII/DR1 mice show that in C57Bl/6 only the ENO1 21-40cit32 response cross reacts with the murine homologue and none of the responses in the HHDII/DR1 mice cross react (Fig. 1). Peptides which did not cross react were disregarded as these responses are probably due to the human peptide inducing foreign responses in mice which may not be citrulline specific and are likely to be deleted in humans. Rescreening of ENO1 peptides 21–40cit32, 126–145cit132 and 316–335cit327 was performed but these peptides gave low frequency IFNγ responses (Supplementary Fig. 1). The responses to ENO1 241-260cit253 were also rescreened in the C57Bl/6 and HHDI/DR1 mice. No significant responses were observed in either of these strains (Supplementary Fig. 1D).
In C57Bl/6 mice, immunization with the peptide ENO1 6-25cit9,15 induced no responses in the initial screen. However, in rheumatoid arthritis (RA) a shorter version of this peptide has been linked to increased binding to a number of HLA types.22 C57Bl/6 mice were therefore immunized with a shorter ENO1 11–25cit15 epitope with CpG/MPLA as an adjuvant. IFNγ responses to human ENO1 11–25cit15 peptide were shown and cross reacted with the mouse peptide (Fig. 1D).
The results of this screen demonstrate that citrullinated ENO1 peptides can induce IFNγ responses in several mouse models. Of the peptides tested Hu ENO1 241–260cit253 showed the highest frequency response and cross reacted with the murine peptide. Therefore, further studies in HLA-DR4 transgenic mice concentrated on this peptide. Studies in C57Bl/6 mice concentrate on the mouse ENO1 11-25cit15 peptide.
Vaccination with ENO1 241–260cit253 or ENO1 11–25cit15 peptides induce citrulline specific Th1-type responses in HLA-DR4 transgenic and C57Bl/6 mice
To further characterize the citrullinated ENO1 specific responses, mice were immunized with citrullinated human ENO1 peptides with CpG/MPLA as an adjuvant. IFNγ responses were shown to be citrulline specific and cross react specifically with the mouse citrullinated peptides (Fig. 2A). No IL-10 responses were observed (Supplementary Fig. 1E).
Figure 2.
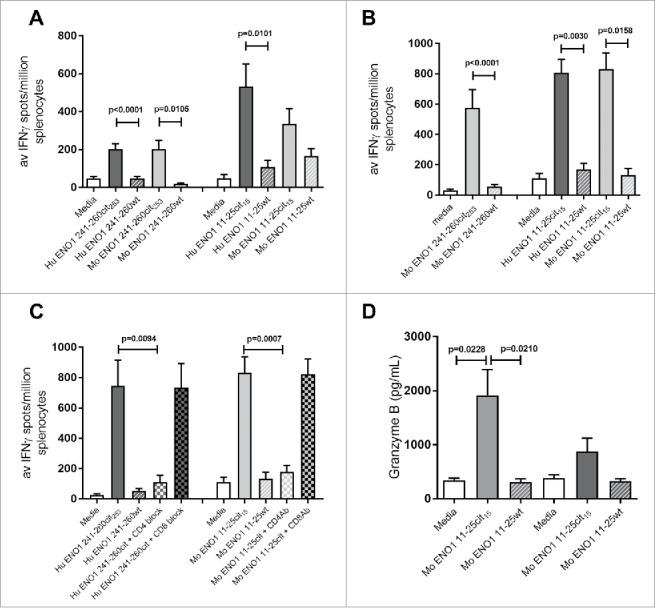
Human ENO1 241–260cit253 and 11–25cit15 peptide induces strong IFNγ responses. Transgenic HLA-DR4 or parental C57Bl/6 mice were immunized with human (Hu) ENO1 241–260cit253 or Hu ENO1 11–25cit15 peptide, respectively. Ex vivo ELISpot was used to determine the IFNγ responses to citrullinated (cit) peptide and wild type (wt) peptides (A). Responses were also determined after immunization with Mouse (Mo) equivalent peptides (B). Further, IFNγ responses after ex vivo addition of anti-CD4 or anti-CD8 blocking antibodies were determined (C) and ex vivo Granzyme B production in response to peptides determined by ELISA (D).
The human ENO1 peptides are one amino acid different to the mouse peptides (Table 1) and may be inducing foreign responses in the murine system and thus not subject to thymic deletion. To prove that mice had a repertoire to the self-antigen they were also immunized with the murine peptides and responses were analyzed. Murine ENO1 241–260cit253 and ENO1 11–25cit15 peptides induced citrulline specific IFNγ immune responses (Fig. 2B).
To determine whether the citrullinated ENO1 peptide specific responses are CD4 mediated, responses were assessed in the presence of CD4 or CD8 blocking antibodies. The ENO1 241–260cit253 response was blocked by addition of an anti-CD4 antibody (p = 0.0417) but was not blocked by an anti-CD8 antibody (Fig. 2C). This suggests that the ENO1 241–260cit253 response is mediated by CD4 T cells. The same was true for the ENO1 11–25cit15 specific response in C57Bl/6 mice (Fig. 2C).
These results show that the citrullinated ENO1 peptides induce proinflammatory CD4-mediated immune response in the transgenic HLA-DR4 and C57Bl/6 mouse strains. In addition to IFNγ we also show that the ENO1 11–25cit15 peptide induced Granzyme B to the citrullinated peptide but not the native sequence (Fig. 2D).
Vaccination against citrullinated ENO1 provides efficient tumor therapy
To determine which tumor cell lines would provide potential targets for this vaccine, Western blots were performed for the expression of ENO1. All tumor cell lines tested showed high expression of ENO1 including the melanoma cell line B16, the pancreatic tumor cell line Pan02 and Lewis lung carcinoma cell line LLC/2 (Fig. 3A). These cells are therefore good targets for tumor therapy studies.
Figure 3.
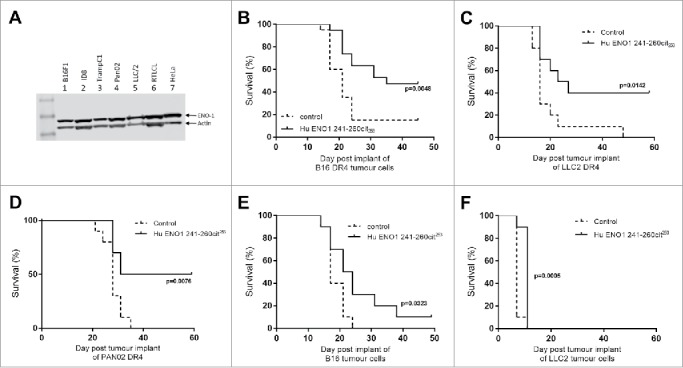
Immunization with citrullinated ENO1 peptides provides an in vivo survival advantage in anti-tumor studies. ENO1 Westerns blots show ENO1 protein expression in cancer cell lines, B16F1 (melanoma), ID8 (Ovarian cancer), TrampC1 (Prostate cancer), Pan02 (Pancreatic ductal adenocarcinoma), LLC/2 (Lung Carcinoma), RTLCL (Lymphoblastoid cell line) and in HeLa cells (A). HLA-DR4 mice were challenged with subcutaneous implant on HLA transgenic B16-DR4 (B), LLC2-DR4 (C), PAN02-DR4 (D) tumor cell lines on day 1. Survival is shown for unimmunized control animals and animals immunized with ENO1 241–260cit253 peptide on day 4, 11 and 18. Mice were also challenged with parental B16F1 (E) or LLC2 (F) cell lines before immunization as above.
To determine whether immunization with citrullinated ENO1 peptides provide an anti-tumor effect mice were implanted subcutaneously with tumor cells at day 1 and then immunized with peptides in combination with CpG/MPLA on days 4, 11 and 18. Responses were initially tested against tumor cell lines engineered to constitutively express human HLA-DR4 (Fig. 3B–D). Mice immunized with the human ENO1 241–260cit253 peptide showed a significant survival advantage when compared to unimmunized mice after challenge with B16-DR4 (p = 0.0048), LLC/2-DR4 (p = 0.0142) and Pan02-DR4 (p = 0.0076). To determine whether these responses were dependent on the tumor cells expressing HLA-DR4, responses were also tested in parental cell lines B16 (Fig. 3E) and LLC2 (Fig. 3F) with an HLA mismatch. In these cell lines the anti-tumor effect of immunization with ENO1 241-260cit253 was reduced with 10% survival for B16 and no survival for LLC/2 challenged mice. This suggests that the CD4 T cells were partially mediating their effects by indirect tumor recognition but that direct recognition of the citrullinated epitopes on MHC class II on the tumor was more potent.
Most tumors do not constitutively express MHC class II therefore the ENO1 241–260cit253 peptide was tested in a model where HLA-DR4 was expressed under an IFNγ inducible promoter. We have previously shown that the levels of HLA-DR4 are similar in our inducible and constitutive models.15 In this model ENO1 241–260cit253 immunization was associated with 90% survival compared to no survival in the control group (Fig. 4A, p<0.0001). The surviving mice were rechallenged with the same tumor on day 42, with no further immunization, to assess the formation of a memory response. Mice rejected a tumor rechallenge with 67% survival compared to no survival in a new control group (p = 0.0112, Fig. 4B). The anti-tumor effect of ENO1 11–25cit15 was also assessed (Fig. 4C) in the unmodified B16 model that does not constitutively express MHC class II but can be induced by IFNγ. For this experiment C57Bl/6 mice were implanted with parental B16F1 on day 1 and then immunized with human ENO1 11–25cit15 peptide on day 4, 11 and 18. Mice immunized with the citrullinated peptide showed a significant survival advantage (p = 0.0026).
Figure 4.
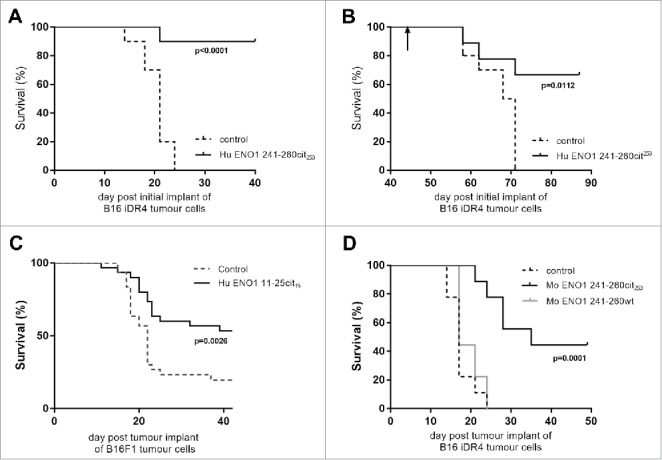
Anti-tumor advantage of immunized mice is seen in models with IFNγ-inducible MHC-II. Transgenic HLA-DR4 mice were challenged with B16 tumor with IFNγ inducible DR4 (B16-iDR4). Survival was determined after immunization with Hu ENO1 241–260cit253 on day 4, 11 and 18 (A). Surviving mice from the immunized group were rechallenged with the same tumor cell line at day 42 post initial tumor implant (indicated by arrow). Survival data for rechallenged mice and a previous unchallenged control group are shown (B). C57bl/6 mice were also challenged with parental B16F1 implanted subcutaneously on day 1 and survival was assessed in unimmunized control mice or mice immunized with ENO1 11–25cit15 peptide (C). In a separate experiment, mice implanted with the B16-iDR4 cell line were immunized with the mouse ENO1 241–260cit253 or 241–260 wt peptides (D). For tumor studies, each group had n = 10, significant p values are shown.
To confirm that the anti-tumor effect is not dependent on using the foreign human ENO1 peptide, mice challenged with the B16 melanoma expressing HLA-DR4 under the IFNγ inducible promoter were immunized with mouse ENO1 241-260cit253 or mouse ENO1 241–260 wild type peptide with arginine at position 253 (Fig. 4D). Mouse ENO1 241-260cit241 peptide immunization was associated with a significant survival advantage when compared to the mouse ENO1 241–260 wild type immunized mice (p = 0.0003) or control unimmunized mice (p = 0.0001).
These results show that the citrullinated ENO1 peptides induce anti-tumor effects against a number of different tumor cell lines derived from different cell types. This increase in survival was observed for both the human and mouse peptides and was not observed after immunization with the wild type peptide.
A T cell repertoire to citrullinated ENO1 can be identified in humans
To investigate whether a repertoire to citrullinated ENO1 241–260cit253 exists in humans, PBMCs were isolated from 6 healthy donors (Table 2) and cultured in the presence of human ENO1 241–260cit253 peptide. Thymidine proliferation assays were performed on the cells after 4, 7 and 11 days and the proliferation index for each was calculated (Fig. 5A). 5/6 of the donors showed proliferation to ENO1 241–260cit253 peptide on at least one of the samples days. For example, Donor 1 showed a proliferative response to ENO1 241-260cit253 at day 11 (mean 20.4) and day 7 (mean 28.6) but not at day 4 (mean 0.8). Responses to ENO1 241-260 wild type peptides with arginine at position 253, were consistently low at day 11 (mean 0.9), 7 (mean 1.2) and 4 (mean 0.3). In contrast, Donor 2 showed only a low-level response at day 11 (mean 2.7) and Donor 6 was a non-responder. For each donor, HLA types were determined and are shown in table 2.
Figure 5.
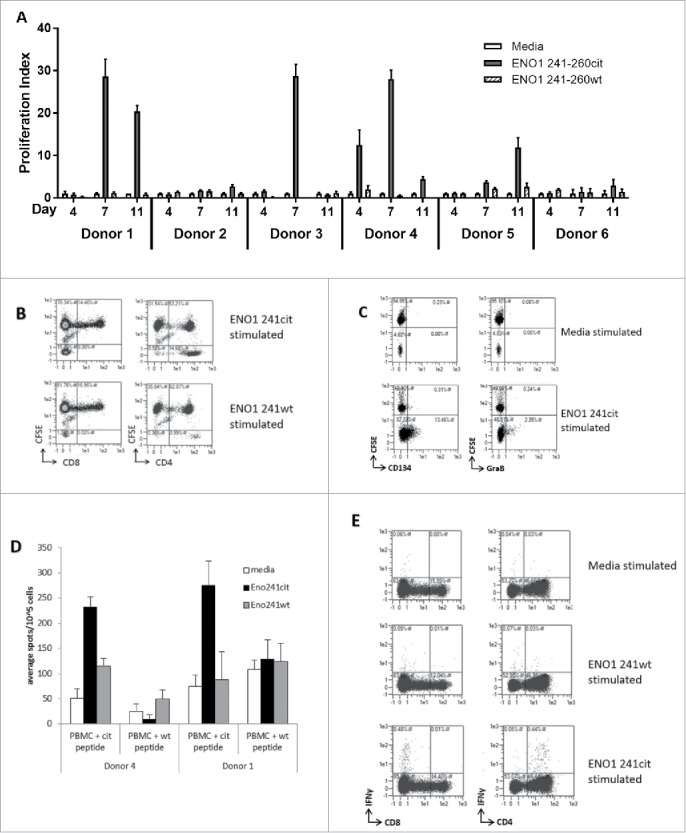
ENO1 241–260cit253 peptide induces responses in human PBMCs. PBMCs were isolated from 6 healthy donors and cultured with media, human ENO1 241–260cit253 or ENO1 241–260 wt peptide. Thymidine assays were performed to determine proliferation after 4, 7 and 11 days (A). HLA typing was performed on each donor (Table 2). PBMCs from donor 4 were labelled with CFSE prior to stimulation with peptides. The CD4 and CD8 populations within the CFSE labelled cell population was assessed by flow cytometry at day 10 (B). CD25-depleted CFSE labelled PBMCs were stimulated with media or ENO1 241–260cit253 peptide and analyzed by Flow cytometry on day 10. CD4+ gated samples are shown with staining for activation marker CD134 and cytokine Granzyme B (C). PBMCs from donor 1 and 4 were cultured for 13 days in the presence of ENO1 241–260cit253 or ENO1 241–260 wt peptide. PBMCs were then restimulated overnight and IFNγ responses were assessed by ELISpot (D). Flow cytometry staining at day 13 showed IFNγ staining in combination with CD4 and CD8 populations after restimulation with media, ENO1 241–260cit253 or ENO1 241–260 wt (E).
Donor 4 showed a high proliferation index at day 4 (mean 12.5) and day 7 (mean 28) and day 11 (4.4). This donor was chosen for further analysis. In order to determine the phenotype of proliferation responses, PBMCs from donor 4 were labelled with CFSE prior to ex vivo culture in the presence of peptides. Cells were assessed at day 10 for the phenotype of the proliferating population (Fig. 5B). Of the proliferating CFSE low population responding to the citrullinated ENO1 peptide, between 73-96% of cells were CD4+ and 0–2% were CD8+. ENO1 241–260cit253 peptide showed increased proportions of CFSElowCD4+ cells compared to ENO1 241–260 wild type peptide.
Although proliferation was observed in these samples the overall number of proliferating cells was relatively low and therefore difficult to reliably phenotype. Therefore, CFSE proliferation assays were also carried out on CD25 depleted PBMCs (Fig. 5C). In these samples the number of CD4+ cells which proliferated was significantly higher. The ENO1 241–260cit253 stimulated sample showed an increase in the proportions of CFSElow CD4+ cells expressing the activation marker CD134 and the cytokine Granzyme B.
To determine whether these responses were a Th1 phenotype PBMCs were cultured for 13 days with ENO1 241–260cit253 or wild type peptide, restimulated with citrullinated or wild type ENO1 peptide and cytokine release measured by IFNγ ELISpot assay. Fig. 5D shows results of the IFNγ ELISpot assay on donors 1 and 4. Cells from both donors show responses to the citrullinated peptide but not the wild type peptide. Further analysis of these responses by intracellular cytokine staining confirms IFNγ responses to be CD4 mediated (Fig. 5E).
These results suggest that healthy humans have a repertoire of CD4 T cells capable of responding to the ENO1 241–260cit253 peptide. This repertoire shows a citrulline specific proliferative response to ENO1 and the production of Th1 cytokines. Interestingly analysis of the HLA types of donors tested showed only one responding donor to be HLA-DR4 positive. This implies that citrullinated ENO1 specific responses are restricted through other HLA alleles. This has implications for vaccine design as it suggests the vaccine would be applicable to a broader population.
Discussion
The challenge for cancer immunotherapy is to generate immune responses that can recognize and destroy diverse cancer cells with minimal cross reactivity to healthy tissue. Targeting epitopes that are post-translationally modified under stress conditions may provide efficient targets for cancer vaccines.13 This study focused on identifying citrullinated epitopes from ENO1 that can be used in vaccines to induce potent CD4 T cell responses resulting in an anti-tumor effect. As ENO1 is over-expressed by a wide range of tumors this would allow us to identify a vaccine with a broad application to multiple tumor types.
Our results show that citrullinated ENO1 epitopes can be used to target cancer. We identified one ENO1 epitope corresponding to position 241–260 with a citrulline at position 253 which induces responses restricted via DR4 MHC-II allele. This study utilized transgenic mice which express human HLA-DR4 to demonstrate that immunization with ENO1 241–260cit253 can induce CD4-mediated IFNγ responses. In vivo HLA-transgenic tumor models were used to demonstrate that this epitope induces rejection of a number of tumor types including melanoma, lung and pancreas. RA pathogenesis has been related to enhanced binding of citrullinated epitopes to HLA-DR4.23 By also showing that a citrullinated ENO1 epitope can stimulate Th1 responses and induce tumor survival in C57Bl/6 mice, we suggest that this phenomenon is not specific to the transgenic mouse models or the HLA-DR4 haplotype and is more broadly applicable. This is supported by our human studies, in which PBMCs from donors with several HLA types were shown to respond to stimulation with the ENO1 241–260cit253 peptide. This suggests that humans have a repertoire to the same epitope that induced responses in the transgenic mouse model. It also implies that citrullinated peptide specific responses are not restricted to HLA-DR4 positive individuals since only one of five of the responding donors was HLA-DR4 positive. We therefore suggest that this response is not specific to the transgenic mouse models or HLA-DR4 positive individuals but implies that citrullinated peptides can bind to a range of HLA alleles. In concordance with this there is evidence emerging in the literature to suggest that citrullinated peptide specific responses can indeed be detected through other HLA alleles in humans.24–26 James et al. (2010), have previously shown RA patients could see T cell responses in vitro to the ENO1 11-25cit15 and in this study we also show it is a T cell epitope in C57Bl/6 mice. However, this peptide did not come up in our screen in HLA-DR4 transgenic mice.27 This could reflect the fact that the HLA-DR4 mice have a different repertoire or perhaps is a consequence of this peptide also being a B cell epitope and therefore possibly presented by B cells in the context of autoimmunity. In contrast ENO1 241-260cit253 was a good epitope in both the HLA-DR4 transgenic mice and normal donors. Gersner et al. (2016), have shown that both the wild type and the citrullinated versions of ENO1 241–255 epitope bind equally well to HLA-DR4.22 This suggests that citrulline is not always an MHC-II binding residue but can interact with the T cell receptor creating a neo-epitope.
Anti-tumor CD4 T cells can induce anti-tumor immunity either indirectly or directly. If they work indirectly the anti-tumor effect would not require MHC-II expression on the tumor cells and as most tumor cells do not constitutively express MHC-II this could be important. CD4 T cells can be activated at the tumor site by APCs which have processed and presented tumor antigens on MHC-II. The CD4 T cells can then release cytotoxic cytokines which can have a cytostatic or cytotoxic effect on the tumor cells and/or can recruit other immune effectors cell such as CD8 T cells.1–4 This response would work equally well on MHC mismatched tumors. In this study, although a small anti-tumor response was seen in the HLA-DR4 mice against the HLA mismatched tumor lines, the response against HLA-DR4 expressing lines was far superior. Although most tumors do not express MHC-II, it can be induced by IFNγ, it was therefore important to create a model in which MHC-II was under the control of the IFNγ promoter. As similar anti-tumor responses were generated in both the constitutive and inducible HLA-DR4 models it suggests that the potent Th1 CD4 cells can secrete sufficient IFNγ to induce class II expression. It also implies that the anti-tumor response was primarily mediated by direct CD4 tumor recognition. In this context, it is of interest that in both the mouse model and the human in vitro assays the CD4 T cells were expressing Granzyme B suggesting that they were cytotoxic CD4 T cells. Cytotoxic CD4 cells have been difficult to induce by vaccination but adoptively transferred naïve CD4 cells recognizing self-antigens have been shown to differentiate to cytotoxic CTL in lymphopenic hosts and this can be enhanced with OX40 engagement or following CD137/CD134 costimulation.5–7 It is also of interest that in our study these cells do not appear to be terminally differentiated as rechallenge of animals that had rejected their initial tumor showed a memory response that efficiently rejected the new tumors.
As well as working as a glycolytic enzyme, ENO1 can also be expressed on the cell surface where it acts as a plasminogen binding receptor.16 Several groups have either used monoclonal antibodies or induced antibody responses with a DNA vaccine encoding unmodified ENO1, to induce anti-tumor responses.20,28 The benefit of targeting citrullinated ENO1 has the potential to increase the specificity of the immune response generated and to minimize the normal tissue toxicity. In this study, we showed efficient anti-tumor responses against a range of tumor models and the ability to induce a memory response that rejects a new challenge with fresh tumor with no associated toxicity in either transgenic or normal mice.
Citrullination can occur during cell death when osmotic control is lost and high levels of cytoplasmic calcium activate PAD enzymes. Citrullinated proteins are precipitated and if the cell death is accompanied by inflammation, they can be recognized by activated B cells. The B cells process and present the citrullinated proteins on MHC-II which stimulates a CD4 response, allowing antibody subclass switching and affinity maturation. In autoimmune diseases such as RA, collagen II-induced arthritis, sarcoidosis, celiac disease and psoriasis, inflammatory cell death results in antibody responses to a wide range of citrullinated proteins and is known to be involved in the pathogenesis of these diseases.29–32 This study suggests that inducing CD4 responses in the absence of an antibody response, does not have pathogenic consequences. This needs to be carefully monitored in clinical trials and it may be wise to initially exclude cancer patients with autoimmunity to prevent exacerbating this condition.
Our results identify a citrullinated ENO1 epitope which can be used to generate strong Th1 responses that can preferentially target tumor cells. The intention is to combine this epitope with the previously described citrullinated vimentin epitopes15 to create a novel vaccine for rapid translation into the clinic for the treatment of solid cancers.
Supplementary Material
Disclosure statement
V.A. Brentville, R.L Metheringham and L.G Durrant have ownership interest in patent WO 2017013425 A1. L.G. Durrant is the joint CEO of Scancell Ltd has ownership interest (including patents) in Scancell Ltd, is a consultant/advisory board member of Scancell Ltd, and has provided expert testimony for Scancell Ltd. All authors are employees of Scancell Ltd.
Acknowledgments
The authors would like to thank Barbara Gunn for her technical support and Prof. Alan Rickinson (University of Birmingham) for contributing the RTLCL cell line. The authors would also like to thank Dr Tina Parsons for help in proofreading the manuscript.
Funding details
This work was funded by Scancell Ltd.
References
- 1.Pardoll DM, Topalian SL. The role of CD4+ T cell responses in antitumor immunity. Curr Opin Immunol. 1998;10:588–94. doi: 10.1016/S0952-7915(98)80228-8. PMID:9794842. [DOI] [PubMed] [Google Scholar]
- 2.Quezada SA, Peggs KS. Tumor-reactive CD4+ T cells: plasticity beyond helper and regulatory activities. Immunotherapy. 2011;3:915–7. doi: 10.2217/imt.11.83. PMID:21843076. [DOI] [PubMed] [Google Scholar]
- 3.Hung K, Hayashi R, Lafond-Walker A, Lowenstein C, Pardoll D, Levitsky H. The central role of CD4(+) T cells in the antitumor immune response. J Exp Med. 1998;188:2357–68. doi: 10.1084/jem.188.12.2357. PMID:9858522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ossendorp F, Mengede E, Camps M, Filius R, Melief CJ. Specific T helper cell requirement for optimal induction of cytotoxic T lymphocytes against major histocompatibility complex class II negative tumors. J Exp Med. 1998;187:693–702. doi: 10.1084/jem.187.5.693. PMID:9480979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xie Y, Akpinarli A, Maris C, Hipkiss EL, Lane M, Kwon EK, Muranski P, Restifo NP, Antony PA. Naive tumor-specific CD4(+) T cells differentiated in vivo eradicate established melanoma. J Exp Med. 2010;207:651–67. doi: 10.1084/jem.20091921. PMID:20156973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hirschhorn-Cymerman D, Budhu S, Kitano S, Liu C, Zhao F, Zhong H, Lesokhin AM, Avogadri-Connors F, Yuan J, Li Y, et al.. Induction of tumoricidal function in CD4+ T cells is associated with concomitant memory and terminally differentiated phenotype. J Exp Med. 2012;209:2113–26. doi: 10.1084/jem.20120532. PMID:23008334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qui HZ, Hagymasi AT, Bandyopadhyay S, St Rose MC, Ramanarasimhaiah R, Menoret A, Mittler RS, Gordon SM, Reiner SL, Vella AT, et al.. CD134 plus CD137 dual costimulation induces Eomesodermin in CD4 T cells to program cytotoxic Th1 differentiation. J Immunol. 2011;187:3555–64. doi: 10.4049/jimmunol.1101244. PMID:21880986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robbins PF, Lu YC, El-Gamil M, Li YF, Gross C, Gartner J, Lin JC, Teer JK, Cliften P, Tycksen E, et al.. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat Med. 2013;19:747–52. doi: 10.1038/nm.3161. PMID:23644516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Linnemann C, van Buuren MM, Bies L, Verdegaal EM, Schotte R, Calis JJ, Behjati S, Velds A, Hilkmann H, Atmioui DE, et al.. High-throughput epitope discovery reveals frequent recognition of neo-antigens by CD4+ T cells in human melanoma. Nat Med. 2015;21:81–5. doi: 10.1038/nm.3773. PMID:25531942. [DOI] [PubMed] [Google Scholar]
- 10.Lipinski KA, Barber LJ, Davies MN, Ashenden M, Sottoriva A, Gerlinger M. Cancer evolution and the limits of predictability in precision cancer medicine. Trends Cancer. 2016;2:49–63. doi: 10.1016/j.trecan.2015.11.003. PMID:26949746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Witalison EE, Thompson PR, Hofseth LJ. Protein arginine deiminases and associated citrullination: physiological functions and diseases associated with dysregulation. Curr Drug Targets. 2015;16:700–10. doi: 10.2174/1389450116666150202160954. PMID:25642720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ireland JM, Unanue ER. Autophagy in antigen-presenting cells results in presentation of citrullinated peptides to CD4 T cells. J Exp Med. 2011;208:2625–32. doi: 10.1084/jem.20110640. PMID:22162830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Durrant LG, Metheringham RL, Brentville VA. Autophagy, citrullination and cancer. Autophagy. 2016;12:1055–6. doi: 10.1080/15548627.2016.1166326. PMID:27145231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marino ML, Pellegrini P, Di Lernia G, Djavaheri-Mergny M, Brnjic S, Zhang X, Hägg M, Linder S, Fais S, Codogno P, et al.. Autophagy is a protective mechanism for human melanoma cells under acidic stress. J Biol Chem. 2012;287:30664–76. doi: 10.1074/jbc.M112.339127. PMID:22761435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brentville VA, Metheringham RL, Gunn B, Symonds P, Daniels I, Gijon M, Cook K, Xue W, Durrant LG. Citrullinated vimentin presented on MHC-II in tumor cells is a target for CD4+ T-cell-mediated antitumor immunity. Cancer Res. 2016;76:548–60. doi: 10.1158/0008-5472.CAN-15-1085. PMID:26719533. [DOI] [PubMed] [Google Scholar]
- 16.Miles LA, Dahlberg CM, Plescia J, Felez J, Kato K, Plow EF. Role of cell-surface lysines in plasminogen binding to cells: identification of alpha-enolase as a candidate plasminogen receptor. Biochemistry. 1991;30:1682–91. doi: 10.1021/bi00220a034. PMID:1847072. [DOI] [PubMed] [Google Scholar]
- 17.Zhao M, Fang W, Wang Y, Guo S, Shu L, Wang L, Chen Y, Fu Q, Liu Y, Hua S, et al.. Enolase-1 is a therapeutic target in endometrial carcinoma. Oncotarget. 2015;6:15610–27. doi: 10.18632/oncotarget.3639. PMID:25951350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cappello P, Tomaino B, Chiarle R, Ceruti P, Novarino A, Castagnoli C, Migliorini P, Perconti G, Giallongo A, Milella M, et al.. An integrated humoral and cellular response is elicited in pancreatic cancer by alpha-enolase, a novel pancreatic ductal adenocarcinoma-associated antigen. Int J Cancer. 2009;125:639–48. doi: 10.1002/ijc.24355. PMID:19425054. [DOI] [PubMed] [Google Scholar]
- 19.Fu QF, Liu Y, Fan Y, Hua SN, Qu HY, Dong SW, Li RL, Zhao MY, Zhen Y, Yu XL, et al.. Alpha-enolase promotes cell glycolysis, growth, migration, and invasion in non-small cell lung cancer through FAK-mediated PI3 K/AKT pathway. J Hematol Oncol. 2015;8:22. doi: 10.1186/s13045-015-0117-5. PMID:25887760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Principe M, Ceruti P, Shih NY, Chattaragada MS, Rolla S, Conti L, Bestagno M, Zentilin L, Yang SH, Migliorini P, et al.. Targeting of surface alpha-enolase inhibits the invasiveness of pancreatic cancer cells. Oncotarget. 2015;6:11098–113. doi: 10.18632/oncotarget.3572. PMID:25860938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lundberg K, Kinloch A, Fisher BA, Wegner N, Wait R, Charles P, Mikuls TR, Venables PJ. Antibodies to citrullinated alpha-enolase peptide 1 are specific for rheumatoid arthritis and cross-react with bacterial enolase. Arthritis Rheum. 2008;58:3009–19. doi: 10.1002/art.23936. PMID:18821669. [DOI] [PubMed] [Google Scholar]
- 22.Gerstner C, Dubnovitsky A, Sandin C, Kozhukh G, Uchtenhagen H, James EA, Rönnelid J, Ytterberg AJ, Pieper J, Reed E, et al.. Functional and Structural Characterization of a Novel HLA-DRB1*04:01-Restricted alpha-Enolase T Cell Epitope in Rheumatoid Arthritis. Front Immunol. 2016;7:494. doi: 10.3389/fimmu.2016.00494. PMID:27895642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hill JA, Southwood S, Sette A, Jevnikar AM, Bell DA, Cairns E. Cutting edge: the conversion of arginine to citrulline allows for a high-affinity peptide interaction with the rheumatoid arthritis-associated HLA-DRB1*0401 MHC class II molecule. J Immunol. 2003;171:538–41. doi: 10.4049/jimmunol.171.2.538. PMID:12847215. [DOI] [PubMed] [Google Scholar]
- 24.James EA, Moustakas AK, Bui J, Papadopoulos GK, Bondinas G, Buckner JH, Kwok WW. HLA-DR1001 presents “altered-self” peptides derived from joint-associated proteins by accepting citrulline in three of its binding pockets. Arthritis Rheum. 2010;62:2909–18. doi: 10.1002/art.27594. PMID:20533291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Catalan D, Aravena O, Zuniga R, Silva N, Escobar A, Sabugo F, Wurmann P, Soto L, González R, Alfaro J, et al.. Weak CD4+ T-cell responses to citrullinated vimentin in rheumatoid arthritis patients carrying HLA-DR9 alleles. Rheumatol Int. 2012;32:1819–25. doi: 10.1007/s00296-011-2039-z. PMID:21769486. [DOI] [PubMed] [Google Scholar]
- 26.Kampstra AS, van Heemst J, Moustakas AK, Papadopoulos GK, Huizinga TW, Toes RE. The increased ability to present citrullinated peptides is not unique to HLA-SE molecules: arginine-to-citrulline conversion also enhances peptide affinity for HLA-DQ molecules. Arthritis Res Ther. 2016;18:254. doi: 10.1186/s13075-016-1153-4. PMID:27809896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.James EA, Rieck M, Pieper J, Gebe JA, Yue BB, Tatum M, Peda M, Sandin C, Klareskog L, Malmström V, et al.. Citrulline-specific Th1 cells are increased in rheumatoid arthritis and their frequency is influenced by disease duration and therapy. Arthritis Rheumatol. 2014;66:1712–22. doi: 10.1002/art.38637. PMID:24665079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cappello P, Rolla S, Chiarle R, Principe M, Cavallo F, Perconti G, Feo S, Giovarelli M, Novelli F. Vaccination with ENO1 DNA prolongs survival of genetically engineered mice with pancreatic cancer. Gastroenterology. 2013;144:1098–106. doi: 10.1053/j.gastro.2013.01.020. PMID:23333712. [DOI] [PubMed] [Google Scholar]
- 29.Choy E. Understanding the dynamics: pathways involved in the pathogenesis of rheumatoid arthritis. Rheumatology (Oxford). 2012;51(Suppl 5):v3–11. doi: 10.1093/rheumatology/kes113. PMID:22718924. [DOI] [PubMed] [Google Scholar]
- 30.Grunewald J, Eklund A. Role of CD4+ T cells in sarcoidosis. Proc Am Thorac Soc. 2007;4:461–4. doi: 10.1513/pats.200606-130MS. PMID:17684290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coimbra S, Figueiredo A, Castro E, Rocha-Pereira P, Santos-Silva A. The roles of cells and cytokines in the pathogenesis of psoriasis. Int J Dermatol. 2012;51:389–95; quiz 95-8. doi: 10.1111/j.1365-4632.2011.05154.x. PMID:22435425. [DOI] [PubMed] [Google Scholar]
- 32.Holmdahl R, Klareskog L, Rubin K, Larsson E, Wigzell H. T lymphocytes in collagen II-induced arthritis in mice. Characterization of arthritogenic collagen II-specific T-cell lines and clones. Scand J Immunol. 1985;22:295–306. doi: 10.1111/j.1365-3083.1985.tb01884.x. PMID:2413528. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


