Abstract
Poly(ADP-ribose) polymerase (PARP)10 is a PARP family member that performs mono-ADP-ribosylation of target proteins. Recent studies have linked PARP10 to metabolic processes and metabolic regulators that prompted us to assess whether PARP10 influences mitochondrial oxidative metabolism. The depletion of PARP10 by specific shRNAs increased mitochondrial oxidative capacity in cellular models of breast, cervical, colorectal and exocrine pancreas cancer. Upon silencing of PARP10, mitochondrial superoxide production decreased in line with increased expression of antioxidant genes pointing out lower oxidative stress upon PARP10 silencing. Improved mitochondrial oxidative capacity coincided with increased AMPK activation. The silencing of PARP10 in MCF7 and CaCo2 cells decreased the proliferation rate that correlated with increased expression of anti-Warburg enzymes (Foxo1, PGC-1α, IDH2 and fumarase). By analyzing an online database we showed that lower PARP10 expression increases survival in gastric cancer. Furthermore, PARP10 expression decreased upon fasting, a condition that is characterized by increases in mitochondrial biogenesis. Finally, lower PARP10 expression is associated with increased fatty acid oxidation.
Introduction
Poly(ADP-ribosyl)ation (PARylation) is an evolutionarily conserved biochemical reaction that involves the catalytic cleavage of NAD+ to nicotinamide and ADP-ribose and the subsequent addition of the ADP-ribose (ADPR) moiet(ies) to acceptor proteins [1]. Poly(ADP-ribose) polymerase (PARPs/ARTDs) are the enzymes responsible for cellular PARylation. “Classical” PARP enzymes (e.g. PARP1, PARP2) cleave NAD+, then transfer ADPR to target proteins in an iterative process that results in the synthesis of protein-bound ADPR polymers (PAR). Not all PARPs are capable of PARylation. Family members that lack a catalytic glutamate are unable to perform this iterative transfer of ADPR units, hence, they use a mechanism called substrate assisted catalysis. These enzymes, including PARP10/ARTD10, only mono-ADP-ribosylate their substrates [2]. PARPs are involved in numerous cellular processes, among them metabolic regulation (for review see [3]). PARP1-evoked mitochondrial damage was described in 1998 [4], in light of recent investigations it seems that PAR metabolism has a rather complex regulatory role on mitochondrial oxidative capacity [5]. To date the expression and activity of PARP1, PARP2 showed characteristic inverse correlation with mitochondrial activity [5].
Mitochondria are intracellular organelles that are responsible for a wide array of biochemical processes, however, mitochondria are best known for harboring biological oxidation. Mitochondrial oxidative capacity is a very efficient way to generate ATP in cells. Therefore, in tissues with a high energy requirement mitochondrial oxidative capacity has a pivotal role in providing ATP, and as a logical continuation, mitochondrial damage is deleterious in these tissues [6]. Chronic decrease in mitochondrial oxidative capacity correlates with metabolic diseases (e.g. type II diabetes), with aging and with cancer development [6–8]. Acute changes in mitochondrial activity occur in response to nutrient availability, for example during fasting mitochondrial activity is enhanced [9].
PARP10 (ARTD10) is multidomain protein that performs mono-ADP-ribosylation of target proteins [2]. PARP10 activity seems to be regulated by posttranslational modifications (for comprehensive review see [10]). In quiescent, non-stressed cells the majority of PARP10 is cytosolic due to a strong nuclear export signal [11]. Both nuclear (e.g. c-MYC, RAN, and histones) and cytosolic (e.g. GSK3β, NEMO/IKKγ, and PCNA) substrates of PARP10 have been identified [11–16]. Importantly, PARP10 seems to be involved in metabolic regulation. PARP10 mono-ADP-ribosylates glycogen synthase kinase 3β (GSK3β) [12] and interacts with c-MYC [11]. Both GSK3β and c-MYC are involved in various metabolism-related signaling events [17]. Furthermore, genome wide association and proteome studies linked PARP10 to lipid metabolism [18, 19]. These preliminary data prompted us to assess a potential role of PARP10 in mitochondria-associated metabolic activities.
Methods
Chemicals
Chemicals were from Sigma-Aldrich (Budapest, Hungary) if not stated otherwise.
Cell culture
MCF-7, HeLa, CaCo2 and Capan3 cells were maintained in MEM (Sigma-Aldrich, Budapest, Hungary), 10% fetal bovine serum (Sigma-Aldrich), 1% Penicillin/Streptomycin (Invitrogen, Carlsbad, California, USA), 2 mM L-Glutamine in humidified atmosphere 95% air-5% CO2 at 37°C. Cells were serum or glucose-fasted for 12 hours where indicated.
The cell lines used in the study are available from European Collection of Authenticated Cell Cultures (ECACC).
Constructs, transfections
In PARP10 silencing experiments the pSUPER RNAi system was used [20]. Constructs expressing shRNAs specific for PARP10 have been described in [12]; in the experiments shown in the current manuscript a mixture of two PARP10 shRNA constructs were used. Transfections were performed on two consequtive days using Jet-PEI transfection reagent (Polyplus Transfection, New York, USA).
Assessment of cellular proliferation
For assessing cell proliferation we applied the sulphoradamine B (SRB) assay, or, as an alternative, BRDU-incorporation assay using the Cell Proliferation ELISA, Brdu (colorimetric) kit (Hoffmann-La Roche, Basel, Switzerland) according to the manufacturer’s instructions. In the SRB assay cells were seeded in 96-well plate (3000–5000 cells/well) and depleted of PARP10 using specific shRNA expressing constructs. At the end of the treatment cells were fixed in situ by 50% trichloroacetic acid (TCA) and subsequently stained with sulforhodamine B (SRB) solution (0.4% in 1% acetic acid). Unbound dye was removed by washing with 1% acetic acid. Bound stain was solubilized in 10 mM TRIS base. Absorbance was read on an automated plate reader (Thermo Labsystems Multiskan MS, Walthman, Massachusetts, USA) at 540 nm.
Measurement of superoxide production
Superoxide was measured using hydroethidine (HE) staining. Cells were seeded in 96-well plate (25 000 cells/well). PARP10 was depleted for 2 days, then cells were stained with 2 μM DioC6 for 30 min and subsequently harvested by trypsin/EDTA. Cells were then subjected to flow cytometric analysis (FACSCalibur, BD Biosciences, Mountain View, CA) with 20,000 events collected for each sample.
Oxygen consumption
Oxygen consumption was measured using an XF96 oxymeter (Seahorse Biosciences, North Billerica, MA, USA) similarly to a previously published protocol [21]. In brief, cells were seeded in 96-well assay plates (~2000 cell/well) and PARP10 was depleted by shRNA transfection. Oxygen consumption rate (OCR, reflecting mitochondrial oxidative capacity) and changes in pH, termed extracellular acidification rate (ECAR, reflecting glycolysis) were recorded. Cells were treated with etomoxir (50 μM), then oligomycin (10 μM) and finally antimycin (10 μM). Data were normalized to protein content and normalized readings were used for calculations. OCR values after antimycin treatment were subtracted from all other values. We named the readings for the untreated cells baseline OCR. OCR after etomoxir treatment represents the oxygen consumption (mostly) related to glucose and amino acid oxidation (Glc+AA), while the difference between baseline and Glc+AA OCR represents fatty acid oxidation (FAO). The oligomycin-resistant respiration gives information on the leakage through the inner membrane of the mitochondria (proton leak).
RNA isolation, reverse transcription-coupled QPCR (RT-qPCR)
Total RNA was prepared using the TRIzol reagent (Invitrogen, Carlsbad, California, USA) according to the manufacturer’s instructions. Two micrograms of RNA were used for reverse transcription (High Capacity cDNA Reverse Transcription Kit, Applied Biosystems, Foster City, CA, USA). Diluted cDNA was used for reverse transcription-coupled quantitative PCR (RT-qPCR). The qPCR reactions were performed with the qPCRBIO SyGreen Lo-ROX Supermix (PCR Biosystems, London, UK) using a Light-Cycler 480 system (Roche Applied Science, Basel, Switzerland). Gene expression was normalized to the geometric mean of human 36B4, 18S, and cyclophyllin values. Primers are listed in Table 1.
Table 1. List of primers used in the study.
| Cyclophilin A (human) | F: GTCTCCTTTGAGCTGTTTGCAGAC |
| Cyclophilin A (human) | R: CTTGCCACCAGTGCCATTAT |
| FOXO1 (human) | F: GTTCATTGAGCGCTTAGACTG |
| FOXO1 (human) | R: AAGTGTAACCTGCTCACTAACCC |
| Fumarase (human) | F: CTCGTTTTGGCCTCCGAACG |
| Fumarase (human) | R: TAACTGGGGTTGGCATGCGT |
| IDH2 (human) | F: GGTGGAGATGGATGGTGATGA |
| IDH2 (human) | R: GTGATGGTGGCACACTTGACT |
| PGC-1α (human) | F. AGAATTGGCTTATGGATGTACAGG |
| PGC-1α (human) | R: TTTGTTGATCATTTCCAGCAATAAT |
| 18s (human) | F: TCGAGGCCCTGTAATTGGAAT |
| 18s (human) | R: TCCCAAGATCCAACTACGAGCTT |
| 36B4 (human) | F: CCATTGAAATCCTGAGTGATGTG |
| 36B4 (human) | R: GTCGAACACCTGCTGGATGAC |
| PARP10 (human) | F: CTGTGGACCTGCTGTTGCTG |
| PARP10 (human) | R: GGATGTCGTAGTGGGGGACA |
| PARP1 (human) | F: CACTGGTACCACTTCTCCTGCTTC |
| PARP1 (human) | R: CTTTGCCTGTCACTCCTCCAG |
| PARP2 (human) | F: GCTAAATCAGACCAATCTCC |
| PARP2 (human) | R: CAGGCTGTGCTGTCCCATTT |
| PARP3 (human) | F: CTTCCTGGGCCTCATCCTCTG |
| PARP3 (human) | R: CAACCGCTTCTTCACCTGCTG |
| PARP4 (human) | F: AAAAGCGCACAGACTGCAAAG |
| PARP4 (human) | R: CACTGGCTAGGTCAGGGAGGT |
| PARP5a (human) | F: AACATCCTTCCTTCCAAAACCT |
| PARP5a (human) | R: GGCAAACGTAAATGCAAAGG |
| PARP5b (human) | F: AAGGTTACCCGGCAAAAGA |
| PARP5b (human) | R: TGGGTGTCCAGTTCACAAAG |
| PARP6 (human) | F: GTCTTGGGATCAGTGGGGTCC |
| PARP6 (human) | R: CCTTCTACACACTGGGCGTCA |
| PARP7 (human) | F: TGGACAGCCTTCGTAGTTGGT |
| PARP7 (human) | R: GGCAGATTTGAATGCCATGA |
| PARP8 (human) | F: ATGTGAACGGGAGCTGTGTGT |
| PARP8 (human) | R: TTGAAGGCCAACATCTGAGGA |
| PARP9 (human) | F: CGGATGTCCCTGGCAGAAGAA |
| PARP9 (human) | R: ACTCGACACCTTGCGATCCAA |
| PARP11 (human) | F: AAAGCTTCCTTGACCACCGAGA |
| PARP11 (human) | R: ACATGCGACCTCCTTCCAAAGA |
| PARP12 (human) | F: CAGAGGGCCAGGTTCTGTACTC |
| PARP12 (human) | R: GACCCAAATACGTGTCTCCCCA |
| PARP14 (human) | F: CACCTGGAAGATGATGGAGCCA |
| PARP14 (human) | R: GAGGTTCACTTTCTGCTGCACC |
| PARP15 (human) | F: CAAGAGGACAGTGGGGTAGGTG |
| PARP15 (human) | R: AGTCTGGGAGGGAATTGCACAA |
| PARP16 (human) | F: ACCACTGACAGCTTGGACACTT |
| PARP16 (human) | R: GCCTTTGATGATGTTCCCAGCC |
| NDUFA2 (human) | F: CGATATTAACAAGGATGGCGG |
| NDUFA2 (human) | R: TCTCAATGAAGTCCCTGACGC |
| NDUFA3 (human) | F: AGACAAAGATGGCTGCGAGAG |
| NDUFA3 (human) | R: CGTGGCCTTGTTGATCATGAC |
| NDUFB5 (human) | F: GTATTCATTGGTCAAGCTGAACTAG |
| NDUFB5 (human) | R: CAGCTCCTTTACCCGTAATTCAGC |
| PGC1-β (human) | F: GTGCTGACAAGAAATAGGAGAGG |
| PGC1-β (human) | R: CTCTTCTGAATTGGAATCGTAGTC |
| UCP2 (human) | F: CTACAAGACCATTGCCCGGAG |
| UCP2 (human) | R: ACAATGGCATTACGAGCAACA |
| ATP5G1 (human) | F: CTAAACAGCCTTCCTACAGCAACTT |
| ATP5G1 (human) | R: TGAACCAGCCACACCAACTGT |
| CYCS (human) | F: TAAGAACAAAGGCATCATCTGG |
| CYCS (human) | R: AGGCAGTGGCCAATTATTACTC |
| COX7A1 (human) | F: ATACGGAAACAGGCTCGGAGGT |
| COX7A1 (human) | R: ATCCGTTTCGGTCTCGGAATTT |
| COX17 (human) | F: GATGCGTGTATCATCGAGAAAG |
| COX17 (human) | R: CAGCAGACCACCATTTCATATTT |
| CPT1A (human) | F: CAGGCGAGAACACGATCTTC |
| CPT1A (human) | R: GCGGATGTGGTTTCCAAAG |
| CPT2 (human) | F: CCAGGCTGCCTATTCCCAAAC |
| CPT2 (human) | R: AGGGTCCCGAAATGTAGCTTG |
| MCAD (human) | F: AGAATTGGCTTATGGATGTACAGG |
| MCAD (human) | R: TTTGTTGATCATTTCCAGCAATAAT |
| ME2 (human) | F: TGTTAAGGCTGTTGTAGTGACTGA |
| ME2 (human) | R: TAAGAGTGCGATATTATCAGTTCCC |
| GAPDH (human) | F: CGTATTGGGCGCCTGGTC |
| GAPDH (human) | R: GGAATTTGCCATGGGTGGAA |
| MLYCD (human) | F: AACATCCAGGCAATCGTGAAG |
| MLYCD (human) | R: GAGGAAACTCTCTCTGCAACTCC |
| PKLR (human) | F: CACCAAGAACCCCAGAACTCC |
| PKLR (human) | R: GGAATAAGGGAAAGGCCCAAG |
| CAT (human) | F: TGAAAATTTGTGCATCCTTCA |
| CAT (human) | R: ATTCTGGAGAAGTGCGGAGA |
| SOD1 (human) | F: CCCACCGTGTTTTATGGATA |
| SOD1 (human) | R: AGGTGTGGGGAAGCATTAAA |
| SOD2 (human) | F: AATCAGGATCCACTGCAAGG |
| SOD2 (human) | R: TAAGCGTGCTCCCACACAT |
| SDHB (human) | F: CCTTCGGGTGCAAGCTAGAGT |
| SDHB (human) | R: CAAGGCTGGAGACAAACCTCA |
| PARP1 (mouse) | F: GGAGCTGCTCATCTTCACC |
| PARP1 (mouse) | R: GCAGTGACATCCCCAGTACA |
| PARP2 (mouse) | F: GGAAGGCGAGTGCTAAATGAA |
| PARP2 (mouse) | R: AAGGTCTTCACAGAGTCTCGATTG |
| PARP3 (mouse) | F: CCTGCTGATAATCGGGTCAT |
| PARP3 (mouse) | R: TTGTTGTTGTTGCCGATGTT |
| PARP4 (mouse) | F: GTTAAATTTTGCACTCCTGGAGA |
| PARP4 (mouse) | R: AATGTGAACACTGTCAAGAGGAAC |
| PARP5a (mouse) | F: TAGAGGCATCGAAAGCTGGT |
| PARP5a (mouse) | R: CAGGCATTGTGAAGGGG |
| PARP5b (mouse) | F: GGCCCTGCTTACACCATTG |
| PARP5b (mouse) | R: CGTGCTTGACCAGAAGTTCA |
| PARP6 (mouse) | F: TTTCCAGCCATCGAATAAGG |
| PARP6 (mouse) | R: ACCACTTGCCTTGAACCAAC |
| PARP7 (mouse) | F: AAAACCCCTGGAAATCAACC |
| PARP7 (mouse) | R: AGAAGGATGCGCTTCTGGTA |
| PARP8 (mouse) | F: TCCACCATTAAATCGCACAA |
| PARP8 (mouse) | R: GCTCCATTTTCGATGTCTTG |
| PARP9 (mouse) | F: ACCTGAAGAATGGCCTATTACATGG |
| PARP9 (mouse) | R: ACAGCTCAGGGTAGAGATGC |
| PARP10 (mouse) | F: CAAGATCCTGCAGATGCAAA |
| PARP10 (mouse) | R: TTGGAGAAGCACACGTTCTG |
| PARP11 (mouse) | F: CAATGAGCAGATGCTATTTCATG |
| PARP11 (mouse) | R: CACCAATTAGCACTCGAGCA |
| PARP12 (mouse) | F: CGGATCCAVAACATGGGC |
| PARP12 (mouse) | R: GGCATCTCTCGCAAAGTAGC |
| PARP14 (mouse) | F: GGCAAACGCAATGGAACTAT |
| PARP14 (mouse) | R: AGCACGTTCCTAAGCCTTGA |
| PARP16 (mouse) | F: CCGTGTGCCTTATGGAAACT |
| PARP16 (mouse) | R: TGGATTGTGTCTGGGCAC |
SDS-PAGE and Western blotting
Cells were lysed in RIPA buffer (50 mM Tris, 150 mM NaCl, 10% SDS, 1% Nonidet P-40, 1 mM Na3VO3, 1 mM NaF, 0.5% sodium deoxycholate, 1 mM phenylmethylsulfonyl fluoride, protease inhibitor mixture, pH 8.0). Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 8% acrylamide gels and blotted onto nitrocellulose membranes. After blocking in 5% (w/v) non-fat dry milk, the membranes were washed with 1x TW-TBS and incubated with primary antibodies at 4°C overnight. The primary antibodies are listed in Table 2. Blots were quantified by densitometry using the Image J software. Upon evaluation the values of the pSuper-transfected cells were considered 1, hence, the shPARP10 values are fold changes. Due to the lack of variance in the control values we did not perform statistical evaluation. The pAMPK/AMPK ratio was calculated using the average pAMPK and average AMPK values.
Table 2. List of the primary antibodies used in the current study.
| Target | Company | Dilution |
|---|---|---|
| AMPK | Sigma-Alrdich | 1:500 |
| phospho-AMPK (pAMPK) | Cell Signaling, Danvers MA, USA | 1:1000 |
| PARP10 | rat anti ARTD10 Clone 5H11 [16] | 1:1000 |
| anti-beta-actin peroxidase-coupled antibody | Sigma-Alrdich | 1:20000 |
| PGC-1α | Abcam, Cambridge UK | 1:1000 |
The secondary antibody was IgG peroxidase HRP conjugate (Sigma-Alrdich, 1:2000), anti-rat-POD 1:1000. Bands were visualized by enhanced chemiluminescence reaction (West Pico ECL Kit, Thermo Scientific, Walthman, Massachusetts, USA).
Electron microscopy
Pellets of cells were processed for electron microscopic investigation. Briefly, cells were fixed in 3% glutaraldehyde dissolved in 0.1 M cacodylate buffer (pH: 7.4) containing 5% sucrose for 1 hour at RT. After washing several times in cacodylate buffer (pH: 7.4), cells were post-fixed in 2% osmium tetroxide for 2 hours at RT. Following several washes in cacodylate buffer (pH: 7.4), cells were dehydrated and embedded into Durcupan ACM resin. Ultrathin sections were cut, collected on Formvar-coated single-slot grids, and counterstained with uranyl acetate and lead citrate. Sections were investigated with a JEOL 1010 transmission electron microscope and photographed at a magnification of 6000-10000x with an Olympus Veleta CCD camera. Digitalized images were processed with Adobe Photoshop CS5 software.
Morphometric assessment was accomplished using the Image J software as follows. The EM pictures of at least 20 different cells of each group were analyzed. Twenty cells in each experimental group were analyzed in single EM micrographs. The numbers of mitochondria were counted. The cross-sectional area of mitochondria and the whole area of the investigated cells were measured. From the measured values the total and the average crossectional areas of mitochondria were calculated. Finally, the proportion of the surface area of the cell covered by the mitochondria was also calculated. From the measured and calculated data obtained from the 20 investigated cells average values and standard errors of means were calculated. Statistical differences among the data sets were calculated by using the Student’s paired t-test.
Annexin-PI double staining
To assess changes in apoptotic and necrotic cell death upon the silencing of PARP10 Annexin-PI double staining was performed similarly to [22] using the FITC Annexin V/Dead Cell Apoptosis Kit (Molecular Probes, Eugene, Oregon, USA) kit according to the manufacturer’s instructions.
ATP measurement
ATP content was measured using an ATP assay Kit (Sigma) following the manufacturer’s instructions.
Database screening
The effect of PARP10 expression on lung, breast, colon, and ovarian cancer survival was assessed through the Kaplan-Meier plotter database (http://kmplot.com/analysis/ [23, 24]). Overall survival rates were analyzed.
Animal experiments
All animal experiments were carried out according to the local national, EU and NIH ethical guidelines and were approved and authorized by the Institutional Animal Care and Use Committee (IACUC) at the University of Debrecen (7/2010 DE MÁB). C57/Bl6J male mice, purchased from Charles River Laboratories (Wilmington, MA, USA), had ad libitum access to water and food, and were kept under a 12/12 h dark-light cycle (light 7 a.m.– 7 p.m., night 7 p.m.– 7 a.m.). Mice were kept on chow diet (10 kcal% of fat) (SAFE, Augy, France). Mice were subjected to CLAMS experiment as described in [25, 26].
Statistical analysis
Significance between groups was analyzed by paired Student’s t-test.
Results
Silencing of PARP10 induces mitochondrial oxidation and AMPK activity
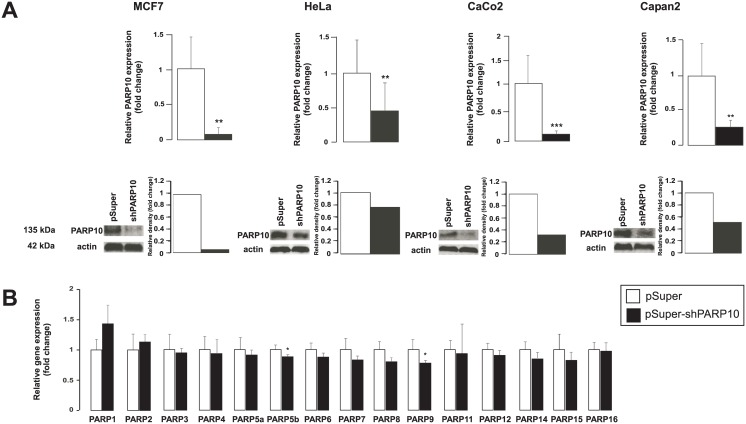
We used different, well-characterized cancer-derived cell lines in our experiments: MCF-7 as a model of breast cancer, HeLa as a model of cervix cancer, CaCo2 as a model of colorectal adenocarcinoma and Capan2 as a model for pancreas adenocarcinoma. In these models we silenced PARP10 using a mixture of two PARP10-specific pSuper constructs and using the empty pSuper construct as control. The mixture of the two PARP10 specific shRNA constructs [12] efficiently decreased the expression PARP10 in the four cell lines both at the mRNA and at the protein level (Fig 1A). The PARP10-specific shRNA proved to be specific for PARP10, none of the other members of the PARP family showed decreased expression, only slight (~10–20%) decrease was observed in the expression of PARP9 and PARP5b (TNK2) (Fig 1B).
Fig 1. The characteristics of PARP10 silencing in MCF7, HeLa, CaCo2 and Capan2 cell lines.
(A) PARP10 silencing in the indicated cell lines was assessed by RT-qPCR (n = 6/6) and Western blotting. A typical experiment is shown as average ± SD. ** and *** indicate statistically significant difference between control and transfected samples at p<0.01 and p<0.001, respectively. (B) In CaCo2 cells the specificity of the PARP10 shRNA constructs was assessed by measuring the expression of the PARP family members in RT-qPCR reactions (n = 4/4). A typical experiment is shown as average ± SD. * indicate statistically significant difference between control and transfected samples at p<0.05.
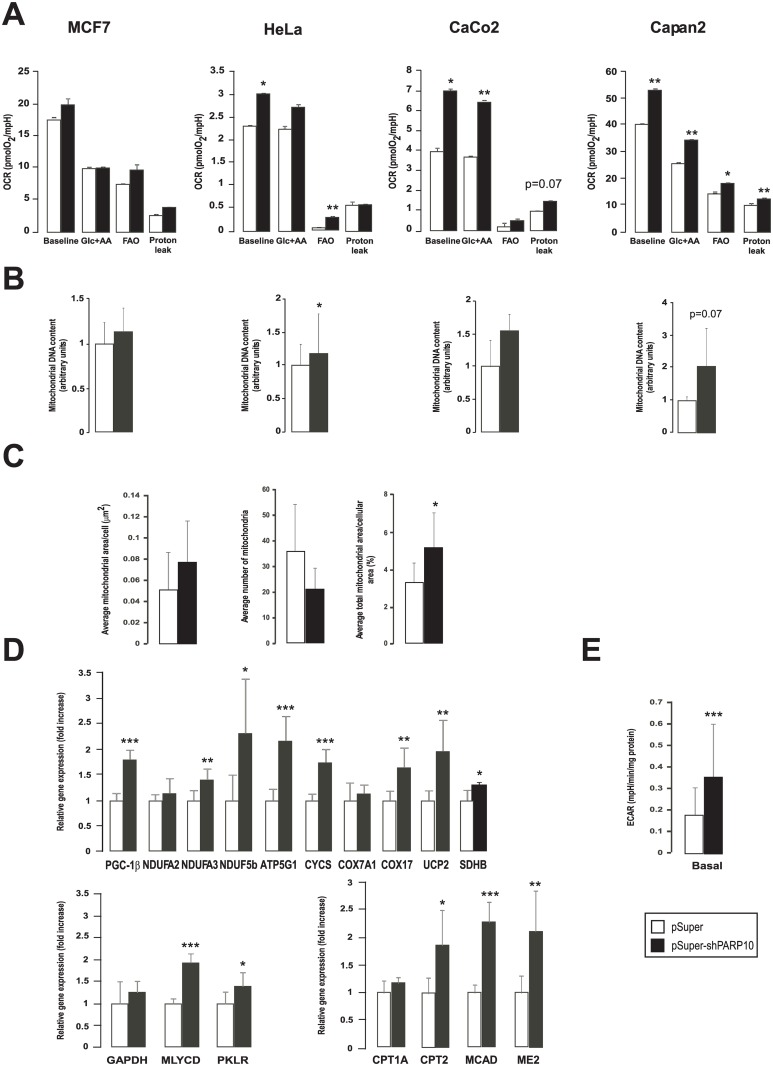
We assessed changes in mitochondrial oxidative metabolism through oximetry (Fig 2A). The silencing of PARP10 enhanced baseline OCR in Hela, CaCo2 and Capan2, with the largest effect seen in CaCo2 cells. In MCF7 cells a trend to enhanced OCR was seen but the increase was not significant. Next, we assessed changes in substrate preference for mitochondrial catabolism. Mitochondrial fatty acid oxidation was blocked by 50 μM of etomoxir, which inhibits carnitin palmitoyltransferase and substrate preference was judged as change in OCR. Fatty acid oxidation (oxygen consumption blocked by etomoxir) was enhanced in all cell lines upon PARP10 silencing, however it was statistically significant only in Hela and Capan2 cells (Fig 1B, FAO = baseline—Glc+AA). The etomoxir resistant respiration (mostly attributed to glucose and amino acid oxidation (Glc+AA)) was also enhanced by the ablation of PARP10 except for MCF7. PARP10 silencing enhanced oligomycin-resistant respiration, except in HeLa cells, that is an approximation of the proton leak through the mitochondrial inner membrane. In line with the increased mitochondrial oxidation rate, cellular mitochondrial DNA content increased in HeLa and Capan2 cells, while the increase in MCF7 and CaCo2 cells was not significant (Fig 2B).
Fig 2. The shRNA silencing of PARP10 induces mitochondrial oxidative activity.
(A) The mitochondrial oxygen consumption rate (OCR) were assayed by Seahorse XF analyzer after control shRNA or shPARP10 transfection as described in Materials and Methods (n = 23/23/23/23). A typical experiment is shown as average ± SD. * and ** indicate statistically significant difference between control and transfected cells at p<0.05 and p<0.01, respectively. (B) Mitochondrial DNA content was analyzed using qPCR in the indicated cell lines two days post transfection with control shRNA or shPARP10 (n = 3/3/3/3) as described in the Materials and Methods. Bars represent fold changes relative to control samples. All gene abbreviations are listed in the text. A typical experiment is shown as average ± SD. *, ** indicate statistically significant difference between control and transfected cells at p<0.05 and p<0.01, respectively. (C) To assess mitochondrial content and morphology electron microscopy was performed on control and PARP10-depleted CaCo2 cells followed by morphometry (n = 10/10). A typical experiment is shown as average ± SD. Representative images are provided, bar equals 2.5 μm. * indicate statistically significant difference between control and transfected cells at p<0.05. (D) The expression of genes involved in mitochondrial oxidative capacity, fatty acid oxidation and glycolysis was assessed in RT-qPCR reactions in control and PARP10-depleted CaCo2 (n = 4/4). Abbreviations are in the text. A typical experiment is shown as average ± SD. *, ** or *** indicate statistically significant difference between control and transfected cells at p<0.05 p<0.01 or p<0.001 respectively. Bars represent fold changes relative to control samples. (E) ECAR was assayed by Seahorse XF analyzer after two days transfection as described in Materials and Methods (n = 23/23/23/23). A typical experiment is shown as average ± SD. *** indicate statistically significant difference between control and transfected cells at p<0.001.
We assessed changes in mitochondrial content by performing morphometry on electron microscopy sections. The shRNA silencing of PARP10 resulted in a decrease in the numbers mitochondria. However, the average surface areas of mitochondria and the total surface areas of mitochondria in proportion to the surface areas of the cytoplasm of the investigated cells were much higher in the PARP10-silenced cells (Fig 2C). These readouts may suggest mitochondrial biogenesis and mitochondrial fusion, both linked to enhanced mitochondrial oxidative capacity [25–30]. Improved mitochondrial oxidative capacity was also evidenced by the enhanced expression of mitochondrial genes (Nduf2a, Nduf3a, Nduf5b, peroxisome proliferator activated receptor cofactor (PGC)-1β, uncoupling protein-2 (UCP2), ATP5g1, cytochrome c (Cytc), Cox7a1, COX17, succinate dehydrogenase (SDH)), fatty acid oxidation genes (carnitin palmitoyl transferase (CPT)1, CPT2, medium-chain acyl-CoA dehydrogenase (MCAD) and malic enzyme 2 (ME2)) and genes involved in glycolysis (glyceraldehyde-3-phosphate dehydrogenase (GAPDH), malonyl-CoA decarboxylase (MYLCD), pyruvate kinase liver isoform (PKLR)) (Fig 2D). The induction of glycolytic genes was translated into enhanced glycolytic flux (Fig 2E).
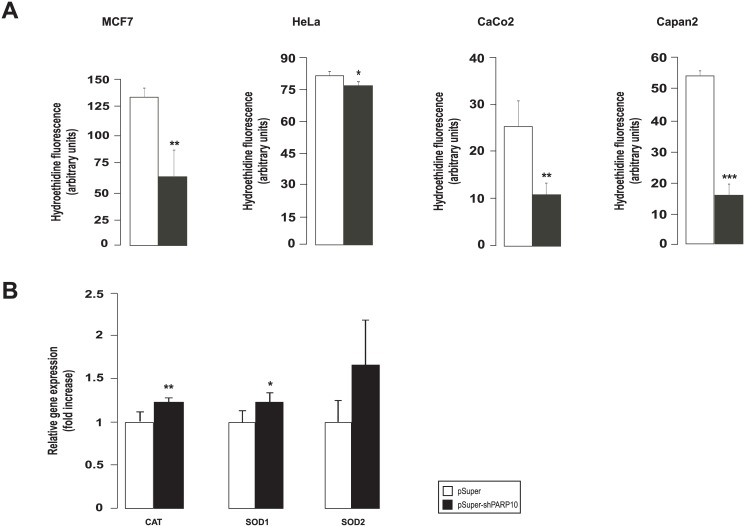
We also assessed cellular reactive species production and found that cellular hydroethidine fluorescence decreased (Fig 3A) suggesting lower superoxide production in PARP10-silenced cells. These were in line with the increased the mRNA expression of catalase (CAT), superoxide dismutase (SOD)1 and SOD2 increased (Fig 3B).
Fig 3. The shRNA silencing of PARP10 reduces free radical production.
(A) Superoxide production was measured in the indicated cell lines using hydroethidine (HE) staining after transfecting cell twice with control shRNA or shPARP10 as described in Materials and Methods. 20,000 events collected for each sample, experiments were performed three times, results are the average of these experiments ± SEM. *, ** and *** indicate statistically significant difference between control and transfected cells at p<0.05, p<0.01 and p<0.001, respectively. (B) The expression of antioxidant genes was assessed in RT-qPCR reactions in control and PARP10-depleted CaCo2 cells (n = 4/4). A typical experiment is shown as average ± SD. *, ** indicate statistically significant difference between control and transfected cells at p<0.05 and p<0.01, respectively. Bars represent fold changes relative to control samples. Abbreviations are in the text.
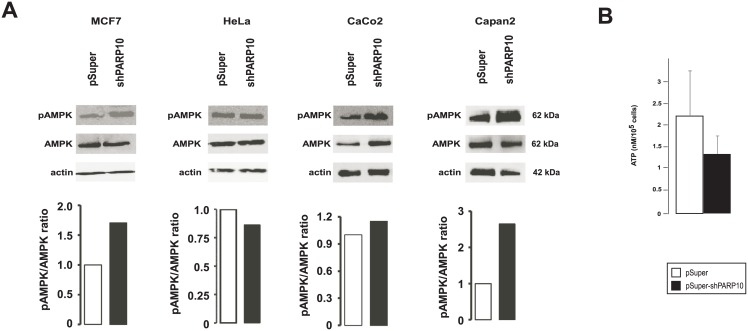
We aimed to identify the molecular background of the enhancement of mitochondrial oxidative capacity. As PARP10 was described to interfere with energy/metabolite sensor pathways (e.g. GSK3β [12] or c-MYC [11]) we also assessed a typical energy sensor pathway that converges on the mitochondria, AMP-activated kinase (AMPK) [31]. AMPK acts as a heterotrimer (α, β and γ subunits) that is activated by cellular energy stress in response to increased in AMP/ATP ratio. Upon activation AMPK inhibits anabolism and stimulates catabolism by phosphorylating a plethora of target proteins [31]. Upon silencing of PARP10 the phosphorylation of the AMPKα subunit increased except for Hela cells (Fig 4A), suggesting enhanced AMPK activation in MCF7, CaCo2 and Capan cell lines, although the change in phosphorylation varied between cell lines. Furthermore, silencing of PARP10 reduced ATP levels that is a likely trigger for AMPK (Fig 4B).
Fig 4. The shRNA silencing of PARP10 induces AMPK.
(A) The indicated cells were transfected with pSuper or the shPARP10 constructs for 2 days then cells were harvested and Western blot was performed. AMPK activity was determined by Western blot analysis of AMPK phosphorylation. The number of biological replicates for the AMPK blots in MCF7 is 3, Hela is 4, For CaCo2 is 3 and for Capan is 3; for the pAMPK blots in MCF7 is 4, Hela is 2, For CaCo2 is 3 and for Capan is 2. Actin was used as a loading control. Western blots were subjected to densitometry using the ImageJ software, a typical results is shown. On the pAMPK blots for HeLa and MCF7 brightness and contrast was adjusted. Abbreviations are in the text. (B) In control and PARP10-depleted CaCo2 (n = 4/4) ATP levels were determined. A typical experiment is shown as average ± SD.
Silencing of PARP10 exerts anti-Warburg features in tumors of the gastrointestinal tract
As next step, we sought (patho)physiological processes where PARP10-mediated in oxidative metabolism may play role. Tumor cells are characterized by a complex rearrangement of metabolic processes that is referred to as the Warburg effect [8, 32]. Although, the biochemical changes that underlie this are diverse, a commonly occurring feature is the suppression of mitochondrial oxidative capacity [8]. It is also of note that the Warburg effect can be reverted by enhancing the mitochondrial oxidative metabolism, which leads to a slowdown of cell proliferation in susceptible tumors [8] suggesting that the silencing of PARP10 through inducing mitochondrial catabolism may have anti-proliferative effects.
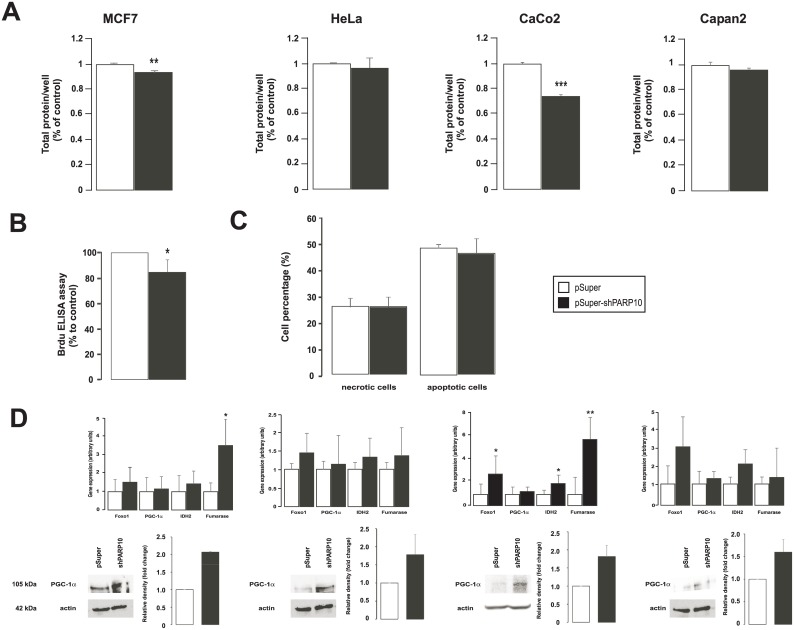
We assessed cellular proliferation upon the ablation of PARP10 in the four tumor cell lines. The silencing of PARP10 in MCF7 and in CaCo2 resulted in a significant reduction of cellular proliferation (Fig 5A) as shown in sulphorhodamine B assays. The biggest change was observed in the case of CaCo2 cells. We verified reduced proliferation in BRDU-incorporation assays in CaCo2 cells that showed a similar pattern as the SRB assay (Fig 5B). Importantly, we have not detected changes in the rate of necrosis and apoptosis (Fig 5C) that underlined the cytostatic, but not cytotoxic property of PARP10 silencing. Next, we assessed the expression of genes involved in Warburg-type metabolism in different tumors. These genes are often mutated or underexpressed in tumors [33–38]. Although, in all cell lines the silencing of PARP10 increased the mRNA and protein expression of these genes to a certain extent, increases were statistically significant only in CaCo2 cells (Fig 5D). This coincided well with the observed stimulation of mitochondrial oxidative capacity and reduction of cell proliferation.
Fig 5. The shRNA silencing of PARP10 reduces the proliferation of MCF7 and CaCo2 cells.
(A) Cell proliferation was determined by sulphorhodamine B assay upon ablation of PARP10 in the cell lines indicated as described in Materials and Methods (n = 3). Experiments were performed three times, results are the average of these experiments ± SEM. (B) Cell proliferation was assessed in CaCo2 cells (n = 8/8) by measuring BRDU incorporation after two days of control or PARP10 shRNA transfection. A typical experiment is shown as acverage ± SD. (C) The ratio of apoptotic and necrotic cells were determined in CaCo2 cells (n = 3/3) transfected with control of PARP10 shRNA for 2 days. A typical experiment is shown. *, ** and *** indicate statistically significant difference between control and transfected cells at p<0.05, p<0.01 and p<0.001, respectively. (D) The expression of the genes indicated was determined in RT-qPCR assays (n = 3, average ± SEM). In the same cells PGC-1α protein levels were determined by Western blotting. The number of the biological replicates for MCF7 is 2, for Hela is 2, for CaCo is 2 and for Capan is 3. On the bar charts the average of the results and SEM is shown.
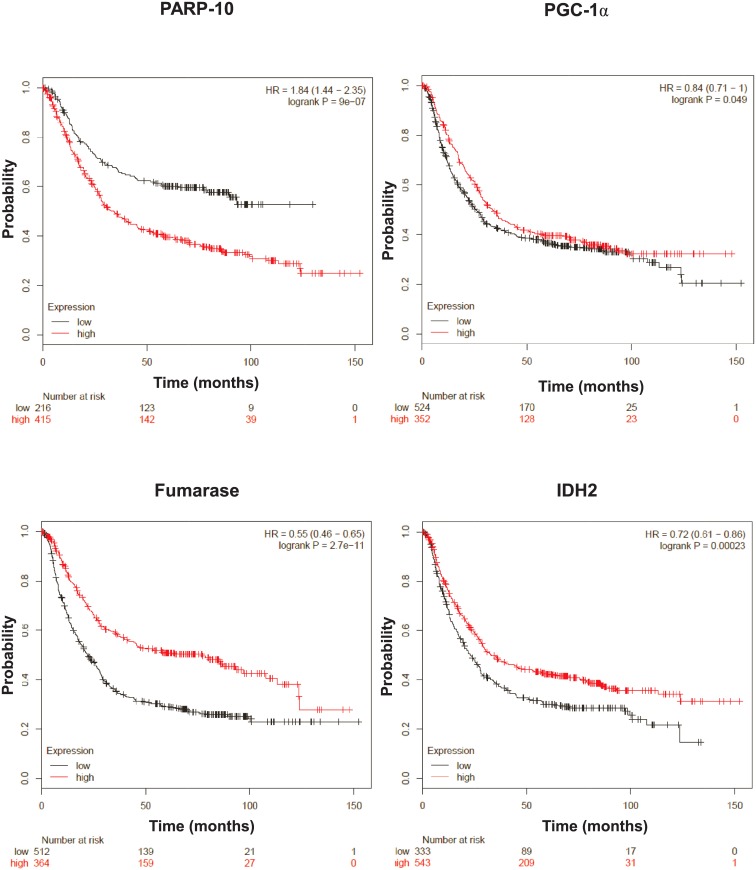
Subsequently, we assessed whether these findings may have human implications. We assessed an online database (kmplot.com) that collects gene expression profiles human tumors and survival the corresponding cancer patients. Using this database we found that low PARP10 expression, as well as, higher expression of the previously described marker genes (PGC-1α, fumarase and isocitrate dehydrogenase-2 (IDH2)) provides longer survival in gastric cancer (Fig 6).
Fig 6. Lower expression of PARP10 and higher expression of enzymes involved in oxidative phosphorylation confer protection against gastric cancer.
kmplot.com, a freely accessible database was screened for the genes indicated in patients suffering from gastric cancer. Overall survival rates were analyzed and all patients are depicted.
PARP10 expression in metabolic tissues correlates with fasting
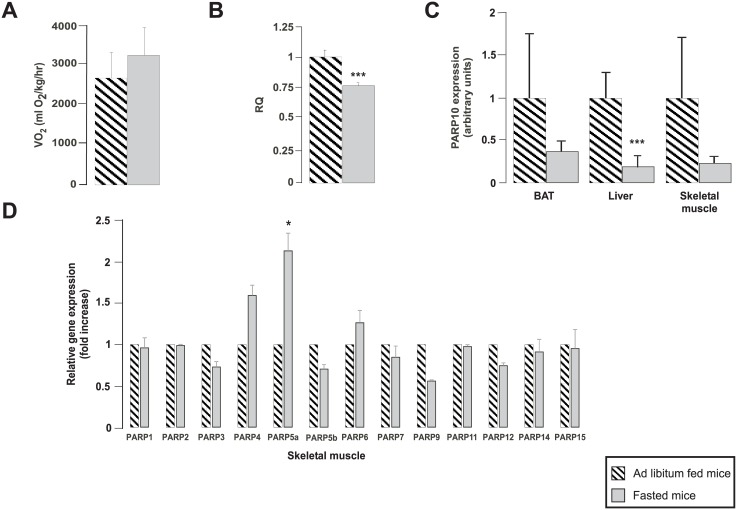
Besides Warburg metabolism another physiological stress that induces mitochondrial catabolism is fasting [9]. Under fasting, anabolic processes are shut down, while catabolism is enhanced to support survival. A key factor for adaptation to starving is the activation of mitochondrial oxidative capacity [9] that prompted us to assess changes in PARP10 expression in response to fasting in metabolic tissues. Fasting and ad libitum (AL)-fed mice were subjected to an indirect calorimetry experiment. Fasted mice were deprived of food for 16 hours that led to an increase in oxygen consumption (Fig 7A), indicating mitochondrial activation and reduction in respiratory quotient (RQ) suggesting enhanced fatty acid oxidation (a process that frequently involves mitochondrial activation) (Fig 7B). In line with enhanced mitochondrial oxidative capacity PARP10 expression was reduced significantly in the liver and PARP10 expression in the brown adipose tissue and skeletal muscle showed a similar trend too (Fig 7C). We assessed whether the expression of other PARPs also changes upon fasting. Interestingly, the expression of PARP4, PARP5a and PARP6 were induced, while the mRNA expression of PARP3, PARP5b, PARP9 and PARP12 were reduced upon fasting (Fig 7D).
Fig 7. PARP10 expression changes inversely as mitochondrial oxidative capacity in mice.
C57/Bl6 male mice were subjected to 16 hours of fasting or received ad libitum food (n = 4/4, 3 months of age, average ± SEM). (A) The oxygen consumption and (B) the RQ were determined in these animals in indirect calorymetry experiments. (C) After dissection, the expression of PARP10 mRNA was determined by RT-qPCR in the brown adipose tissue (BAT), liver and skeletal muscle. (D) The expression of the members of the PARP family was determined in skeletal muscle using RT-qPCR reactions. Data is represented as average ± SEM. *, *** indicate statistically significant difference between control and transfected cells at p<0.05 or p<0.001, respectively.
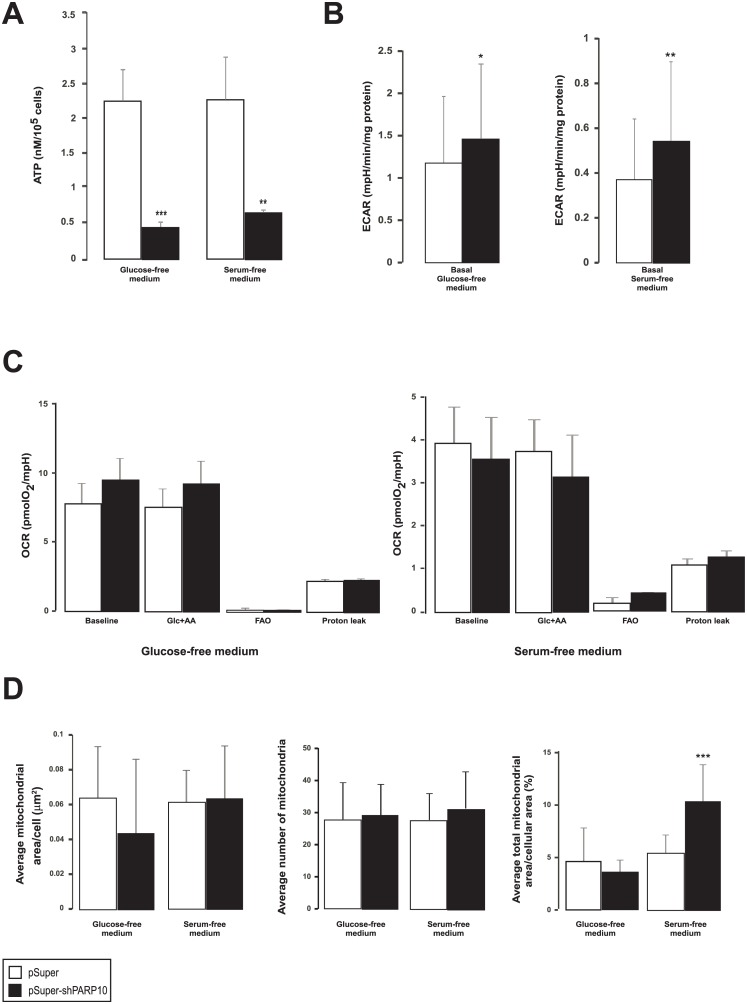
We performed additional assays to gain better insight into the PARP10-driven metabolic processes upon serum or glucose fasting. ATP levels were further reduced upon glucose or serum fasting as compared to controls (Figs 4C vs. 8A) suggesting aggravated energy stress. In serum and glucose-fasted cells ECAR vales were higher in the PARP10-silenced cells (Fig 8B), however, to a way lower extent than in non-fasted cells (Fig 2E). In a similar fashion to these, increases in OCR values upon PARP10 silencing were lower upon glucose fasting, while in serum-fasted cells OCR values were even lower in shPARP10 cells than in control cells (Fig 8C). The morphological changes suggesting mitochondrial biogenesis in non-fasted cells (Fig 2C) were also abolished by glucose or serum-fasting (Figs 2C vs. 8D).
Fig 8. PARP10-induced mitochondrial biogenesis is abolished upon glucose or serum fasting.
(A) CaCo2 cells (n = 4/4) were transfected with control or PARP10 shRNA for two days then ATP levels were determined. A typical experiment is shown as average ± SD. (B) ECAR was assayed by Seahorse XF analyzer in CaCo2 cells after two days transfection as described in Materials and Methods (n = 23/23/23/23). A typical experiment is shown as average ± SD. (C) The mitochondrial oxygen consumption rate (OCR) was assayed by Seahorse XF analyzer after two days transfection as described in Materials and Methods (n = 23/23/23/23). A typical experiment is shown as average ± SD. (D) To assess mitochondrial content and morphology electron microscopy was performed on control and PARP10-depleted CaCo2 cells (n = n = 8/13/8/18). A typical experiment is shown as average ± SD. **, *** indicate statistically significant difference between control and transfected cells at p<0.011 and p<0.001, respectively.
Discussion
In the present study we identify PARP10 as a metabolic regulator. The ablation of PARP10 enhanced mitochondrial oxidative capacity and glycolysis, furthermore, induced fatty acid oxidation in our experiments. As a possible molecular explanation to these observations we showed that changes in PARP10 expression inversely correlated with the activity of AMPK and with the expression of key mitochondrial genes, among them, PGC-1α.
What could be the link between PARP10, AMPK and PGC-1α? AMPK is activated upon cellular energetic stress marked by decreases in ATP and/or increases in AMP levels [39]. Our study revealed that ATP levels drop upon the silencing of PARP10 and therefore it is a possible explanation that the depletion of PARP10 declutches cellular energy stress that in turn activate AMPK. Continuing along this hypothesis, AMPK activation, provoked by energy stress, is responsible for the upregulation of catabolism, nevertheless, the actual steps how PARP10 achieves that are unknown warranting further investigations in this direction. Interestingly, when cells are further stressed by glucose or serum deprivation we were able to further depress ATP levels, but the signs of PARP10-induced mitochondrial biogenesis were generally reduced pointing out that fasting-induced and PARP10-induced energy stress are not additive. It is also of note that another ADP-ribosyltransferase, PARP-1, was reported to physically interact with AMPK [40] and it seems that both enzymes can cross activate each other. AMPK phosphorylates PARP-1 [40], while PAR degradation yields AMP [41] that stimulates AMPK [5].
We have also observed increases in the expression of PGC-1α in cells where PARP10 was silenced. PGC-1α is also a key metabolic regulator, increases in PGC-1α expression or activity supports mitochondrial biogenesis through facilitating the de novo expression of mitochondrial genes [30, 42, 43], furthermore, mutations or decrease in the expression of PGC-1α blunt mitochondrial oxidative metabolism and hence at the level of physiology may predispose to type II diabetes [44, 45]. The activation of other PARP enzymes, such as PARP1, PARP2 and TiPARP, were already shown to inhibit PGC-1α activity [25, 26, 46–48]. How PARP10 can influence PGC-1α expression is unknown, however, it should be mentioned that AMPK activation through the consequent induction of SIRT1 can positively regulate PGC-1α [29, 49] that may contribute to PGC-1α activation upon the silencing of PARP10.
PARP10-mediated AMPK activation can modulate metabolic adaptation in cancer cells and in different metabolic tissues upon fasting. AMPK activation in those cancers that rely on Warburg metabolism, like breast or colorectal cancer, counteracts cancer growth [38, 50]. Although, we detected AMPK activation in all cell lines we investigated, only two responded to PARP10 ablation by reduced proliferation, the breast cancer cell line MCF-7 and the colorectal carcinoma cell line CaCo2, however, the other two cell lines also displayed a tendency towards reduced proliferation. It seems therefore that other, cell/tissue-specific signaling pathways also contribute to reduced proliferation capacity upon the silencing of PARP10. It is also noteworthy that PARP10 is an interacting partner of c-Myc and through that, PARP10 protects against the transformation of embryonic fibroblasts [16]. Our data support that PARP10 is necessary for cellular proliferation, however add that the effects of PARP10 appears to be tissue/cancer dependent.
Changes in mitochondrial oxidative function plays a key role in fasting. Fasting induces mitochondrial oxidative activity that frequently correlates with fatty acid oxidation in metabolic tissues [9]. PARP10 expression correlated inversely with mitochondrial oxidative capacity (marked by increases in oxygen consumption) in the liver with similar tendency in the brown adipose tissue and skeletal muscle, all key tissues of energy expenditure, fatty acid and glucose homeostasis [9]. Taken together in cancer and metabolic models PARP10 expression was regulated inversely as mitochondrial oxidative capacity. On broader perspective, these data also suggest that lower PARP10 expression may bring about a negative energy balance through enhancing mitochondrial biogenesis and energy expenditure in vivo.
Another important observation in our studies was that low PARP10 expression associated with increases in fatty acid oxidation. Namely, increases in the expression of fatty acid oxidation genes or etomoxir-sensitive respiration and low RQ values in fasting that is associated with low PARP10 expression. In fact, AMPK activation has been shown to enhance fatty acid oxidation [31, 51, 52] that makes it tempting to speculate that PARP10-induced AMPK activation may stand behind enhanced fatty acid oxidation. A genome wide association study associated PARP10 to apolipoprotein B expression [18] further pointing towards a role for PARP10 in lipid homeostasis. It is of note that PARP1 and PARP2 are both associated with lipid metabolism [3]. PARP1 and PARP2 are involved in the regulation of organismal and tissular triglyceride and fatty acid homeostasis, furthermore cholesterol and lipoprotein metabolism [25, 26, 53–62]. Our study nominate PARP10 as a PARP that is involved in the regulation of fatty acid degradation.
On a broader perspective, other cell types and other processes can be under the influence of the interaction between PARP10 and AMPK/mitochondria (e.g. beige cells [63–65], macropahges [66]) warranting further studies towards the understanding of the role of PARP10 in metabolic regulation.
Data Availability
All raw data is available from figshare (https://figshare.com/s/05b649ebc545b73c6d35).
Funding Statement
Our work was funded by grants from NKFIH (K108308, K123975, KKP126710, GINOP-2.3.3-15-2016-00021, GINOP-2.3.2-15-2016-00006), the Momentum fellowship of the Hungarian Academy of Sciences and a Campus France fellowship to PB, and the German Science Foundation (DFG LU 466/16-1) to BL. PB was also financed by the University of Debrecen. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hottiger MO, Hassa PO, Luscher B, Schuler H, Koch-Nolte F. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem Sci. 2010;35(4):208–19 doi: 10.1016/j.tibs.2009.12.003 [DOI] [PubMed] [Google Scholar]
- 2.Kleine H, Poreba E, Lesniewicz K, Hassa PO, Hottiger MO, Litchfield DW, et al. Substrate-assisted catalysis by PARP10 limits its activity to mono-ADP-ribosylation. Mol Cell. 2008;32(1):57–69 doi: 10.1016/j.molcel.2008.08.009 [DOI] [PubMed] [Google Scholar]
- 3.Bai P. Biology of Poly(ADP-Ribose) Polymerases: The Factotums of Cell Maintenance. Mol Cell. 2015;58(6):947–58 doi: 10.1016/j.molcel.2015.01.034 [DOI] [PubMed] [Google Scholar]
- 4.Virag L, Salzman AL, Szabo C. Poly(ADP-ribose) synthetase activation mediates mitochondrial injury during oxidant-induced cell death. JImmunol. 1998;161(7):3753–9 [PubMed] [Google Scholar]
- 5.Bai P, Nagy L, Fodor T, Liaudet L, Pacher P. Poly(ADP-ribose) polymerases as modulators of mitochondrial activity. Trends Endocrinol Metab. 2015;26(2):75–83 doi: 10.1016/j.tem.2014.11.003 [DOI] [PubMed] [Google Scholar]
- 6.Nunnari J, Suomalainen A. Mitochondria: in sickness and in health. Cell. 2012;148(6):1145–59 doi: 10.1016/j.cell.2012.02.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Houtkooper RH, Mouchiroud L, Ryu D, Moullan N, Katsyuba E, Knott G, et al. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature. 2013;497(7450):451–7 doi: 10.1038/nature12188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74 doi: 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 9.Lopez-Lluch G, Hunt N, Jones B, Zhu M, Jamieson H, Hilmer S, et al. Calorie restriction induces mitochondrial biogenesis and bioenergetic efficiency. Proc Natl Acad Sci U S A. 2006;103(6):1768–73. doi: 10.1073/pnas.0510452103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaufmann M, Feijs KL, Luscher B. Function and regulation of the mono-ADP-ribosyltransferase ARTD10. Curr Top Microbiol Immunol. 2015;384:167–88 doi: 10.1007/82_2014_379 [DOI] [PubMed] [Google Scholar]
- 11.Kleine H, Herrmann A, Lamark T, Forst AH, Verheugd P, Luscher-Firzlaff J, et al. Dynamic subcellular localization of the mono-ADP-ribosyltransferase ARTD10 and interaction with the ubiquitin receptor p62. Cell Commun Signal. 2012;10(1):28 doi: 10.1186/1478-811X-10-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feijs KL, Kleine H, Braczynski A, Forst AH, Herzog N, Verheugd P, et al. ARTD10 substrate identification on protein microarrays: regulation of GSK3beta by mono-ADP-ribosylation. Cell Commun Signal. 2013;11(1):5 doi: 10.1186/1478-811X-11-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forst AH, Karlberg T, Herzog N, Thorsell AG, Gross A, Feijs KL, et al. Recognition of Mono-ADP-Ribosylated ARTD10 Substrates by ARTD8 Macrodomains. Structure. 2013;21(3):462–75 doi: 10.1016/j.str.2012.12.019 [DOI] [PubMed] [Google Scholar]
- 14.Nicolae CM, Aho ER, Vlahos AH, Choe KN, De S, Karras GI, et al. The ADP-ribosyltransferase PARP10/ARTD10 interacts with proliferating cell nuclear antigen (PCNA) and is required for DNA damage tolerance. J Biol Chem. 2014;289(19):13627–37. doi: 10.1074/jbc.M114.556340 Epub 2014 Apr 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Verheugd P, Forst AH, Milke L, Herzog N, Feijs KL, Kremmer E, et al. Regulation of NF-kappaB signalling by the mono-ADP-ribosyltransferase ARTD10. Nat Commun. 2013: doi: 10.1038/ncomms2672 [DOI] [PubMed] [Google Scholar]
- 16.Yu M, Schreek S, Cerni C, Schamberger C, Lesniewicz K, Poreba E, et al. PARP-10, a novel Myc-interacting protein with poly(ADP-ribose) polymerase activity, inhibits transformation. Oncogene. 2005;24(12):1982–93 doi: 10.1038/sj.onc.1208410 [DOI] [PubMed] [Google Scholar]
- 17.Cohen P, Goedert M. GSK3 inhibitors: development and therapeutic potential. Nat Rev Drug Discov. 2004;3(6):479–87 doi: 10.1038/nrd1415 [DOI] [PubMed] [Google Scholar]
- 18.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466(7307):707–13. doi: 10.1038/nature09270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kang JH, Ryu HS, Kim HT, Lee SJ, Choi UK, Park YB, et al. Proteomic analysis of human macrophages exposed to hypochlorite-oxidized low-density lipoprotein. Biochim Biophys Acta. 2009;1794(3):446–58. doi: 10.1016/j.bbapap.2008.11.015 [DOI] [PubMed] [Google Scholar]
- 20.Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296(5567):550–3 doi: 10.1126/science.1068999 [DOI] [PubMed] [Google Scholar]
- 21.Nagy L, Docsa T, Szántó M, Brunyánszki A, Hegedus C, Márton J, et al. Glycogen Phosphorylase Inhibitor N-(3,5-Dimethyl-Benzoyl)-N’-(β-D-Glucopyranosyl)Urea Improves Glucose Tolerance under Normoglycemic and Diabetic Conditions and Rearranges Hepatic Metabolism. PLoS ONE. 2013;8(7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bai P, Bakondi E, Szabo E, Gergely P, Szabo C, Virag L. Partial protection by poly(ADP-ribose) polymerase inhibitors from nitroxyl-induced cytotoxity in thymocytes. Free RadicBiolMed. 2001;31(12):1616–23 [DOI] [PubMed] [Google Scholar]
- 23.Gyorffy B, Surowiak P, Budczies J, Lanczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8(12):e82241 doi: 10.1371/journal.pone.0082241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szasz AM, Lanczky A, Nagy A, Forster S, Hark K, Green JE, et al. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016;30(10):10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bai P, Canto C, Oudart H, Brunyanszki A, Cen Y, Thomas C, et al. PARP-1 Inhibition Increases Mitochondrial Metabolism through SIRT1 Activation. Cell Metab. 2011;13(4):461–8 doi: 10.1016/j.cmet.2011.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bai P, Canto C, Brunyanszki A, Huber A, Szanto M, Cen Y, et al. PARP-2 Regulates SIRT1 Expression and Whole-Body Energy Expenditure. Cell Metab. 2011;13(4):450–60 doi: 10.1016/j.cmet.2011.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Silva Ramos E, Larsson NG, Mourier A. Bioenergetic roles of mitochondrial fusion. Biochim Biophys Acta. 2016;7(16):30085–8 [DOI] [PubMed] [Google Scholar]
- 28.Schrepfer E, Scorrano L. Mitofusins, from Mitochondria to Metabolism. Mol Cell. 2016;61(5):683–94. doi: 10.1016/j.molcel.2016.02.022 [DOI] [PubMed] [Google Scholar]
- 29.Canto C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, et al. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458(7241):1056–60. doi: 10.1038/nature07813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, et al. Resveratrol Improves Mitochondrial Function and Protects against Metabolic Disease by Activating SIRT1 and PGC-1alpha. Cell. 2006;127(6):1109–22 doi: 10.1016/j.cell.2006.11.013 [DOI] [PubMed] [Google Scholar]
- 31.Inoki K, Kim J, Guan KL. AMPK and mTOR in cellular energy homeostasis and drug targets. Annu Rev Pharmacol Toxicol. 2012;52:381–400 doi: 10.1146/annurev-pharmtox-010611-134537 [DOI] [PubMed] [Google Scholar]
- 32.Warburg O, Wind F, Negelein E. The Metabolism of Tumors in the Body. J Gen Physiol. 1927;8(6):519–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fathi AT, Wander SA, Faramand R, Emadi A. Biochemical, Epigenetic, and Metabolic Approaches to Target IDH Mutations in Acute Myeloid Leukemia. Semin Hematol. 2015;52(3):165–71. doi: 10.1053/j.seminhematol.2015.03.002 [DOI] [PubMed] [Google Scholar]
- 34.Liu X, Ling ZQ. Role of isocitrate dehydrogenase 1/2 (IDH 1/2) gene mutations in human tumors. Histol Histopathol. 2015;30(10):1155–60. doi: 10.14670/HH-11-643 [DOI] [PubMed] [Google Scholar]
- 35.Rodriguez FJ, Vizcaino MA, Lin MT. Recent Advances on the Molecular Pathology of Glial Neoplasms in Children and Adults. J Mol Diagn. 2016;18(16):005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Desideri E, Vegliante R, Ciriolo MR. Mitochondrial dysfunctions in cancer: genetic defects and oncogenic signaling impinging on TCA cycle activity. Cancer Lett. 2015;356(2 Pt A):217–23. doi: 10.1016/j.canlet.2014.02.023 [DOI] [PubMed] [Google Scholar]
- 37.Massari F, Ciccarese C, Santoni M, Brunelli M, Piva F, Modena A, et al. Metabolic alterations in renal cell carcinoma. Cancer Treat Rev. 2015;41(9):767–76. doi: 10.1016/j.ctrv.2015.07.002 [DOI] [PubMed] [Google Scholar]
- 38.Fodor T, Szanto M, Abdul-Rahman O, Nagy L, Der A, Kiss B, et al. Combined Treatment of MCF-7 Cells with AICAR and Methotrexate, Arrests Cell Cycle and Reverses Warburg Metabolism through AMP-Activated Protein Kinase (AMPK) and FOXO1. PLoS One. 2016;11(2):e0150232 doi: 10.1371/journal.pone.0150232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mihaylova MM, Shaw RJ. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nature Cell Biology. 2011;13(9):1016–23 doi: 10.1038/ncb2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walker JW, Jijon HB, Madsen KL. AMP-activated protein kinase is a positive regulator of poly(ADP-ribose) polymerase. Biochem Biophys Res Commun. 2006;342(1):336–41 doi: 10.1016/j.bbrc.2006.01.145 [DOI] [PubMed] [Google Scholar]
- 41.Formentini L, Macchiarulo A, Cipriani G, Camaioni E, Rapizzi E, Pellicciari R, et al. Poly(ADP-ribose) catabolism triggers AMP-dependent mitochondrial energy failure. J Biol Chem. 2009;284(26):17668–76 doi: 10.1074/jbc.M109.002931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin J, Wu PH, Tarr PT, Lindenberg KS, St-Pierre J, Zhang CY, et al. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell. 2004;119(1):121–35 doi: 10.1016/j.cell.2004.09.013 [DOI] [PubMed] [Google Scholar]
- 43.Lin J, Wu H, Tarr PT, Zhang CY, Wu Z, Boss O, et al. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418(6899):797–801. doi: 10.1038/nature00904 [DOI] [PubMed] [Google Scholar]
- 44.Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434(7029):113–8 doi: 10.1038/nature03354 [DOI] [PubMed] [Google Scholar]
- 45.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–73. doi: 10.1038/ng1180 [DOI] [PubMed] [Google Scholar]
- 46.Mohamed JS, Hajira A, Pardo PS, Boriek AM. MicroRNA-149 Inhibits PARP-2 and Promotes Mitochondrial Biogenesis via SIRT-1/PGC-1alpha Network in Skeletal Muscle. Diabetes. 2014;63(5):1546–59 doi: 10.2337/db13-1364 [DOI] [PubMed] [Google Scholar]
- 47.Fang EF, Scheibye-Knudsen M, Brace LE, Kassahun H, Sengupta T, Nilsen H, et al. Defective Mitophagy in XPA via PARP-1 Hyperactivation and NAD(+)/SIRT1 Reduction. Cell. 2014;157(4):882–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Diani-Moore S, Zhang S, Ram P, Rifkind AB. Aryl hydrocarbon receptor activation by dioxin targets phosphoenolpyruvate carboxykinase (PEPCK) for ADP-ribosylation via TCDD-inducible poly(ADP-ribose) polymerase (TiPARP). J Biol Chem. 2013:14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Canto C, Houtkooper RH, Pirinen E, Youn DY, Oosterveer MH, Cen Y, et al. The NAD(+) Precursor Nicotinamide Riboside Enhances Oxidative Metabolism and Protects against High-Fat Diet-Induced Obesity. Cell Metab. 2012;15(6):838–47 doi: 10.1016/j.cmet.2012.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rutter GA, Leclerc I. The AMP-regulated kinase family: enigmatic targets for diabetes therapy. Mol Cell Endocrinol. 2009;297(1–2):41–9 [DOI] [PubMed] [Google Scholar]
- 51.Ahmadian M, Abbott MJ, Tang T, Hudak CS, Kim Y, Bruss M, et al. Desnutrin/ATGL is regulated by AMPK and is required for a brown adipose phenotype. Cell Metab. 2011;13(6):739–48 doi: 10.1016/j.cmet.2011.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xin C, Liu J, Zhang J, Zhu D, Wang H, Xiong L, et al. Irisin improves fatty acid oxidation and glucose utilization in type 2 diabetes by regulating the AMPK signaling pathway. Int J Obes (Lond). 2015;40(3):443–51 [DOI] [PubMed] [Google Scholar]
- 53.Lehmann M, Pirinen E, Mirsaidi A, Kunze FA, Richards PJ, Auwerx J, et al. ARTD1-induced poly-ADP-ribose formation enhances PPARgamma ligand binding and co-factor exchange. Nucleic Acids Res. 2015;43(1):129–42 doi: 10.1093/nar/gku1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cerutti R, Pirinen E, Lamperti C, Marchet S, Sauve AA, Li W, et al. NAD(+)-dependent activation of Sirt1 corrects the phenotype in a mouse model of mitochondrial disease. Cell metabolism. 2014;19(6):1042–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Erener S, Hesse M, Kostadinova R, Hottiger MO. Poly(ADP-Ribose)Polymerase-1 (PARP1) Controls Adipogenic Gene Expression and Adipocyte Function. Mol Endocrinol. 2012;26(1):79–86 doi: 10.1210/me.2011-1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Erener S, Mirsaidi A, Hesse M, Tiaden AN, Ellingsgaard H, Kostadinova R, et al. ARTD1 deletion causes increased hepatic lipid accumulation in mice fed a high-fat diet and impairs adipocyte function and differentiation. Faseb J. 2012;26(6):2631–8 doi: 10.1096/fj.11-200212 [DOI] [PubMed] [Google Scholar]
- 57.Bai P, Houten SM, Huber A, Schreiber V, Watanabe M, Kiss B, et al. Poly(ADP-ribose) polymerase-2 controls adipocyte differentiation and adipose tissue function through the regulation of the activity of the retinoid X receptor/peroxisome proliferator-activated receptor-gamma heterodimer. JBiolChem. 2007;282(52):37738–46 [DOI] [PubMed] [Google Scholar]
- 58.Peng H, Zhu QS, Zhong S, Levy D. Transcription of the Human Microsomal Epoxide Hydrolase Gene (EPHX1) Is Regulated by PARP-1 and Histone H1.2. Association with Sodium-Dependent Bile Acid Transport. PLoS One. 2015;10(5):e0125318 doi: 10.1371/journal.pone.0125318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shrestha E, Hussein MA, Savas JN, Ouimet M, Barrett TJ, Leone S, et al. Poly(ADP-ribose) Polymerase 1 Represses Liver X Receptor-mediated ABCA1 Expression and Cholesterol Efflux in Macrophages. J Biol Chem. 2016;291(21):11172–84. doi: 10.1074/jbc.M116.726729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Szántó M, Brunyánszki A, Márton J, Vámosi G, Nagy L, Fodor T, et al. Deletion of PARP-2 induces hepatic cholesterol accumulation and decrease in HDL levels. Biochimica et Biophysica Acta—Molecular Basis of Disease. 2014;1842(4):594–602 [DOI] [PubMed] [Google Scholar]
- 61.Kiss B, Szanto M, Szklenar M, Brunyanszki A, Marosvolgyi T, Sarosi E, et al. Poly(ADP) ribose polymerase-1 ablation alters eicosanoid and docosanoid signaling and metabolism in a murine model of contact hypersensitivity. Mol Med Rep. 2015;11(4):2861–7 doi: 10.3892/mmr.2014.3044 [DOI] [PubMed] [Google Scholar]
- 62.Mukhopadhyay P, Horvath B, Rajesh M, Varga ZV, Gariani K, Ryu D, et al. PARP inhibition protects against alcoholic and non-alcoholic steatohepatitis. J Hepatol. 2017;66(3):589–600. doi: 10.1016/j.jhep.2016.10.023 [DOI] [PubMed] [Google Scholar]
- 63.Claussnitzer M, Dankel SN, Kim KH, Quon G, Meuleman W, Haugen C, et al. FTO Obesity Variant Circuitry and Adipocyte Browning in Humans. N Engl J Med. 2015;373(10):895–907 doi: 10.1056/NEJMoa1502214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Abdul-Rahman O, Kristóf E, Doan-Xuan QM, Vida A, Horváth A, Simon J, et al. AMP-activated kinase (AMPK) activation by AICAR in human white adipocytes derived from pericardial white adipose tissue stem cells induces a partial beige-like phenotype. PLoS One. 2016;11(6):e0157644 doi: 10.1371/journal.pone.0157644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu J, Bostrom P, Sparks LM, Ye L, Choi JH, Giang AH, et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell. 2012;150(2):366–76 doi: 10.1016/j.cell.2012.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van der Burgh R, Boes M. Mitochondria in autoinflammation: cause, mediator or bystander? Trends Endocrinol Metab. 2015;26(5):263–71. doi: 10.1016/j.tem.2015.03.004 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All raw data is available from figshare (https://figshare.com/s/05b649ebc545b73c6d35).