Abstract
The embryo-specific Dc3 gene promoter driving the VvMybA1 anthocyanin regulatory gene was used to develop a visual selection system for the genetic transformation of citrus. Agrobacterium-mediated transformation of cell suspension cultures resulted in the production of purple transgenic somatic embryos that could be easily separated from the green non-transgenic embryos. The somatic embryos produced phenotypically normal plants devoid of any visual purple coloration. These results were also confirmed using protoplast transformation. There was minimal gene expression in unstressed one-year-old transgenic lines. Cold and drought stress did not have any effect on gene expression, while exogenous ABA and NaCl application resulted in a minor change in gene expression in several transgenic lines. When gas exchange was measured in intact leaves, the transgenic lines were similar to controls under the same environment. Our results provide conclusive evidence for the utilization of a plant-derived, embryo-specific visual reporter system for the genetic transformation of citrus. Such a system could aid in the development of an all-plant, consumer-friendly GM citrus tree.
Introduction
In plants, the 5’ flanking regions of most protein-coding genes contain DNA sequences that interact with the basic transcription initiation complexes and various transcription factors [1]. These DNA sequences regulating the function of the downstream gene are termed promoters. Most plant promoters are composed of three essential components: the transcription start site, the TATA box, and several hundred sequences that include upstream activating sequences (UASs), enhancers, upstream repressing sequences (URSs), and silencers [2]. Promoters are important elements used in genetic engineering, and they can either direct gene expression uniformly in most tissues and cells [3–5] or target expression to a specific tissue [6–8]. Targeted gene expression in a transgenic plant by utilizing a tissue specific promoter can result in enhanced transgene protein production in the organ of interest and does not cause a metabolic drain in the overall plant [9, 10]. This is because many tissue-specific promoters can provide tightly regulated gene expression in only the tissue of interest [11–13].
Seed-specific promoters target the gene expression in the seed tissues. Several seed-specific promoters and their cis-elements have been characterized [14–16], and a number of them have been utilized to target the transgene product to different parts of the seed. Metabolic engineering using these promoters have allowed scientists to increase the plant’s nutritional content by upregulating oil production and the manipulation of phenolic compounds for better grain quality and yield, resulting in a number of improved transgenic cultivars [17, 18]. Studies on the molecular and cellular events during the critical stages of early embryogenesis and seed development have resulted in the generation of a wealth of knowledge on the function of some seed-specific promoters [8, 19, 20].
Citrus transformation studies have mainly utilized constitutive promoters to express both the transgene of interest and the selectable marker gene (usually an antibiotic resistance marker) with or without a reporter gene in the genome of the plant [21–23]. Reporter genes code for proteins that can be easily detectable and are therefore particularly useful for selection systems. Green fluorescent protein (GFP), β-glucuronidase (GUS) and luciferase (LUC) are the most commonly used reporter genes in plant transformation [24]. Some studies have also exploited anthocyanin accumulation as a visual marker in cereal and fruit transformation [25–27]. Anthocyanins are endogenous pigments that are responsible for the red, purple and blue colors in flowering plants. The anthocyanin reporter system is nondestructive, does not require an exogeneous substrate, or produce a toxic compound and has no related environmental or health concerns [24].
Unlike the gene of interest, selectable marker/reporter gene(s) are important mainly in the early stages of genetic transformation. Consequently, the constitutive expression of these genes is unnecessary in the regenerated plants and can even be associated with substantial fitness penalties [28]. In addition, development of transgenic plants that have the selectable marker/reporter gene(s) either excised from the system or switched off can alleviate public concerns on the environmental safety of genetically modified plants [29]. Marker-free plants have been produced in citrus, mainly through site specific recombination [30–32] or PCR analyses of all regenerated plantlets [33]. These procedures are either very time consuming or result in a small population of successfully excised transgenic lines.
Accordingly, it is beneficial to develop a robust selection system for marker-free transformation of citrus. Using tissue-specific or inducible promoters in association with reporter genes may overcome the shortcomings of constitutive expression. In this report, we outline a novel method of selectable reporter gene silencing by utilizing the tightly regulated embryo-specific promoter of the carrot Dc3 gene, which is highly expressed during the initial phases of embryogenesis [34], coupled with a visual anthocyanin reporter gene (the Vitis vinifera-derived VvMybA1 [35]), following transformation of citrus using either cell suspension cultures or protoplasts to regenerate reporter gene expression-free citrus plants.
Materials and methods
Construction of plant transformation vectors
The 1.5 kb Dc3 promoter [34] in pBluescript KS and cloned between the PstI and HindIII sites was a kind gift from Dr. Terry Thomas (Dept. of Biology, Texas A&M University, College Station, TX). The promoter fragment was amplified utilizing primers DC3-F and DC3-R (Table 1) and the Phusion® Hot Start Flex 2X Master Mix (New England Biolabs, Ipswich, MA) to introduce a HindIII restriction site at the 5’ end and a BamHI site at the 3’ end. Two constructs were produced for this study: the first utilizing a modified pCAMBIA1300 binary plasmid for Agrobacterium-mediated transformation and the second utilizing a modified pUC18 Escherichia coli cloning vector.
Table 1. Primer sequences used in this study.
| Target gene | Primer | Purpose | Primer sequence 5’→ 3’ |
|---|---|---|---|
| Dc3 | DC3-F | Cloning | AAGCTTTGCTGTACCATATCTTTGTAGCC |
| DC3-R | GGATCCGGTGGCTTTCTTTGCAGATGT | ||
| VvmybA1 | VVM-F | qPCR | CCAGGAAGAAGGGAGAGATAAAC |
| VVM-R | CTAACAGGCTTTCCCACCATA | ||
| GAPC | GAPC-F | qPCR | GGAAGGTCAAGATCGGAATCAA |
| GAPC-R | CGTCCCTCTGCAAGATGACTCT |
The Sanger-sequence-verified Dc3 amplicon was cloned by replacing the 35S promoter at complementary sites of the pCAM1300-VvMyb to produce the plasmid pCAMDC3-VvMyb (S1 Fig). A pUC18-based intermediate cloning vector previously constructed to contain the VvMybA1 transgene (GenBank accession no. AB097923 [35]) under the control of a 35S promoter and a terminator sequence of the Pisum sativum-derived RuBisCO small subunit (rbcS) was modified to replace the 35S promoter with the DC3 promoter (pUDC3-VvMyb; S1 Fig).
Initiation of cell suspension cultures
Embryogenic callus from Citrus sinensis (L.) Osbeck cv. ‘Hamlin’ was initiated from unfertilized ovules as outlined earlier [22]. Cell suspension cultures were initiated from actively dividing one-year-old embryogenic callus (S2 Fig). Five grams of embryogenic callus was incubated in 25 ml of liquid H + H cell proliferation medium and sub-cultured on a 2-week transfer cycle [36]. Cells were harvested for transformation between 5 to 7 days after subculture.
Agrobacterium-mediated transformation of cell suspension cultures
Five milliliters of a vigorously growing Agrobacterium culture (EHA105; [37]) initiated the night before containing the pCAMDC3-VvMyb construct was seeded into 45 ml YEP medium containing appropriate antibiotics and allowed to grow for an additional 3 hours [38]. One gram of suspension cells were harvested and subsequently incubated in a 0.3-OD Agrobacterium suspension that had been re-suspended in DOG medium (EME sucrose supplemented with 5 mg L-1 kinetin) [39] for 20 minutes. Cells were subsequently blotted dry on sterile Whatman filter paper disks (GE Healthcare, Chicago, IL), plated on solid DOG medium supplemented with 100 μM acetosyringone, and incubated in the dark at 25°C for 5 days before transfer to EME medium supplemented with maltose (EME-M) for selection. The EME-M medium was supplemented with 400 mg L-1 timentin (Duchefa Biochemie B.V., Netherlands) and 25 mg L-1 hygromycin B. After a month of culture, the cells were overlaid with 2 ml of a 1:2 (v:v) mixture of 0.6 M BH3 and 0.15 M EME-M liquid media [39]. Anthocyanin-overexpressing globular-stage somatic embryos were individually cultured over 0.22-mm cellulose acetate membrane filters on embryo maturation EME-M medium [40] before transfer into B+ germination medium [36]. Well-developed PCR positive plantlets were transferred for further root development by transferring into RMAN rooting medium [36]. Plantlets that did not root were micrografted [38]. Well-acclimated plants after a year of transfer to the greenhouse were utilized for all subsequent molecular analyses.
Protoplast-mediated transformation
Plasmid DNA containing the pUDC3-VvMyb construct was utilized for all protoplast transformation experiments. Cell suspension cultures were used for the isolation of protoplasts essentially as described earlier [41]. Enzymatically digested protoplasts obtained from 1 gram of cells were purified by centrifugation on a sucrose-mannitol gradient (S2 Fig) and suspended in 0.6 M BH3 protoplast culture medium to a final concentration of 2 × 106 cells ml-1 [36]. Then, 20 μg of plasmid DNA suspended in sterile water (2 mg ml-1 concentration) was added to a 15-ml polystyrene culture test tube (Thermo Fisher Scientific Inc, Waltham, MA) containing 0.25 ml of the protoplast suspension. The mixture was incubated for 10 minutes followed by the addition of 0.5 ml of a 40% polyethylene glycol (PEG) solution. After a 30-minute incubation, four drops of the PEG-protoplast mixture were pipetted into the center of a 60 mm × 15 mm Petri dish. PEG elution and protoplast washing were carried out as described earlier [36]. The washed protoplasts were cultured in a liquid medium containing a 1:1 mixture of 0.6 M BH3 protoplast culture medium with 0.6 M EME medium [36] at 25–27°C under diffused light for 4–6 weeks. Transgenic cells were visualized with the naked eye. Purple colored transgenic somatic embryos were separated from the non-transgenic green embryos and placed in embryo maturation and germination medium, essentially in a manner similar to that described for the cell suspension transformation process. In vitro plantlets were tested by PCR, and the positive plants acclimated in a similar manner as those obtained from the Agrobacterium-mediated transformation and were utilized for subsequent molecular analyses.
Molecular analysis of transformants
Initial verification of transgenic status was performed on in vitro leaves. Leaves were harvested from putative transgenic plants and non-transgenic control and verified by PCR utilizing the Extract-N-Amp™ Plant PCR Kit (Sigma-Aldrich, St. Louis, MO) and gene-specific primers [42]. Total RNA was isolated utilizing the RNeasy Mini Kit (Qiagen Inc. Valencia, CA) according to the manufacturer’s protocol. A 1-μg aliquot of RNA was reverse transcribed using RevertAid First-Strand cDNA Synthesis Kit (Thermo Fisher Scientific Inc). Real-time quantitative PCR (qPCR) was performed using the StepOne Plus system (Thermo Fisher Scientific Inc) utilizing SYBR Green reagents. The relative mRNA levels were compared to those of the Citrus sinensis glyceraldehyde-3-phosphate dehydrogenase (GAPC) gene [43] and calculated using the 2-ΔΔCT method [44]. Citrus genomic DNA from one-year-old transgenic trees and non-transgenic control was isolated using the PureLink™ Genomic Plant DNA Purification Kit (Thermo Fisher Scientific Inc). Transgene presence was re-confirmed by PCR. The transgene copy number in the selected lines was also validated using qPCR, essentially as previously described [45, 46]. All primer sequences used in this study are listed in Table 1.
Stress analyses
Two node cuttings from seven randomly selected year old transgenic lines (five from the Agrobacterium-mediated transformation and two from the protoplast transformation) were rooted in a mist bed [47]. Six-month-old well-rooted cuttings containing a minimum of 4–6 leaves and growing in 5 × 18 cm Deepot cells (Stuewe and Sons, Inc. Tangent, Oregon) were used for subsequent experiments. All stress treatments were conducted in a Percival Scientific (Percival Scientific Inc., Perry, Iowa) growth chamber under controlled conditions. The light intensity in the growth chamber as measured 15 cm from the lamps was 300 μmol m-2 s-1. For cold stress experiments, plants were kept at 8° C ± 2° C for 7 days before evaluation. Plants were drought stressed by withholding irrigation and maintained under these conditions until leaves had fully wilted (5 days). Exogenous ABA ((±)-cis,trans-abscisic acid, Sigma, St. Louis, MO) was applied by spraying the cuttings with 10 ml of 100 μM (±)ABA (Fluka) in 0.5% (v/v) Tween 20. Two drenches of 200 mM NaCl were applied at a weekly interval before leaf sampling.
Leaf gas exchange measurements
Seven selected transgenic lines along with a non-transgenic control were subjected to leaf gas exchange measurements. All plants were watered to soil field capacity one day prior to measurement. Measurements were taken on the same day in the morning (11 am-12:30 pm) with the quantum of external light averaging 329.35 μmol m-2 s-1. The leaf temperatures at the time of measurement were 40.46 ± 1.38°C, and air temperatures were 38.72 ± 0.63°C. Three distinct fully expanded leaves were utilized for the physiological responses of net assimilation of CO2 (A, μmol m-2 s-1), transpiration rate (E, mol m-2 s-1), stomatal conductance to water vapor (gsw, mol m-2 s-1) and intercellular CO2 (Ci, μmol mol-1). Measurements were conducted with a portable photosynthesis system (LI-6800; LICOR Inc., Lincoln, NE). Instantaneous water use efficiency (WUEinstantaneous; μmol CO2, mmol H2O-1) was calculated by the ratio of net assimilation and transpiration rate [48].
Data analysis and statistics
The data were analyzed by one-way analysis of variance (ANOVA, JMP® Pro, Version 12.2.0., SAS Institute Inc., Cary, NC) as a completely randomized design. Significant differences in mean values were separated by pair comparisons using Tukey-Kramer HSD with α = 0.05. Bar graphs represent the mean of each line for each parameter, and error bars represent the standard deviation from the mean.
Results
The Dc3 promoter is suitable for embryo-specific gene expression in citrus
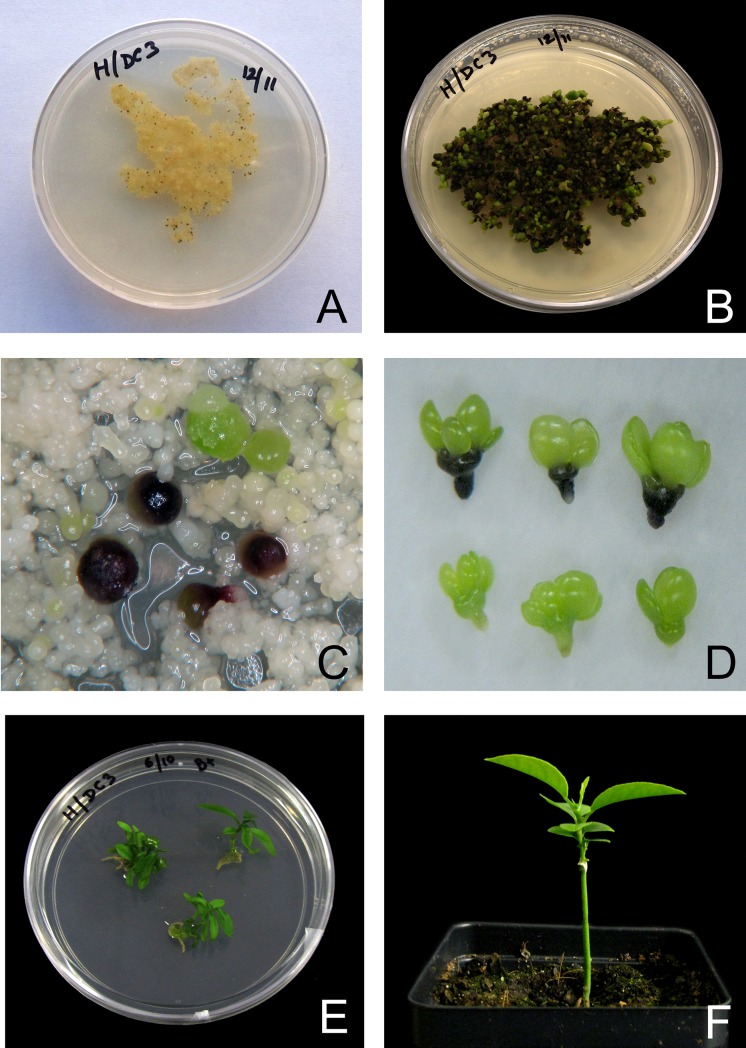
The present study was conducted to develop a simple selectable marker system for the genetic transformation of citrus, utilizing an embryo-specific, plant-derived visual reporter gene system. Initial studies utilizing an Agrobacterium mediated transformation protocol was conducted to test the suitability of the Dc3 promoter in citrus. Agrobacterium-mediated transformation of cell suspension cultures utilizing the pCAMDC3-VvMyb construct produced 18 ‘Hamlin’ sweet orange transgenic lines. Each line was confirmed for the presence of the transgene twice—first in vitro and subsequently after acclimatization in the greenhouse. Two lines tested negative in the second PCR confirmation step (S3 Fig). The cells did not exhibit any visual coloration following transformation. After two months in the selection medium, purple embryos were produced (Fig 1A and 1B). Globular stage transgenic embryos could be easily identified from the non-transgenic escapes based on their coloration (Fig 1C) and 56 individual embryos were manually selected and individually placed in maturation medium (Table 2). All germinating embryos gradually began to lose coloration and 18 phenotypically normal transgenic lines were regenerated. None of the developing cotyledons had the characteristic purple coloration derived from expression of the VvMybA1 transgene (Fig 1D). All lines that regenerated into normal plants were phenotypically normal and devoid of any visual purple coloration (Fig 1E and 1F). Transgenic lines remained visually free of reporter gene expression in the greenhouse.
Fig 1. Steps in the Agrobacterium mediated transformation of citrus suspension cells with a construct containing the VvmybA1 gene driven by the Dc3 embryo-specific promoter.
A) Citrus cell suspension cultures on EME-M medium at the beginning of embryonic development; B) Anthocyanin producing transgenic somatic embryos after 60 days in EME-M medium; C) Anthocyanin expression in transgenic (purple) embryos with non-transgenic (green) embryos; D) Anthocyanin free cotyledon development in transgenic embryos (top) and non-transgenic embryos at similar stage of development (bottom); E) Transgenic embryos germinating in the plant regeneration medium (B+ medium); F) A normal regenerated transgenic plant without any visual purple coloration (no VvmybA1 gene expression) micro grafted on Carrizo rootstock.
Table 2. Transformation efficiency of ‘Hamlin’ sweet orange from 1 gram of suspension cell derived cultures.
| Transformation system |
VvmybA1-negative embryos a | VvmybA1-positive embryos a |
VvmybA1-positive shoots (Plant recovery efficiency (%))b |
|---|---|---|---|
| Suspension Cells | 74 | 56 | 18 (32) |
| Protoplast | 194 | 21 | 11 (52) |
a Data from three independent experiments. Results indicate the total number of embryos produced (VvmybA1 positive and negative)
b Plant recovery efficiency was calculated as the total number of VvmybA1-positive shoots/total number of VvmybA1-positive embryos produced X 100.
The Dc3 promoter can be utilized to produce transgenic plants using protoplast transformation
In contrast to Agrobacterium-mediated transformation where transient gene expression could not be detected, transient VvMybA1-expressing cells following protoplast transformation utilizing the plasmid DNA containing the pUDC3-VvMyb construct could be identified within 3 days following transformation (Fig 2). Primary selection was based on the purple coloration produced as a result of VvMybA1 gene expression. Eleven phenotypically normal plants were regenerated using this system from a total of 21 purple embryos that were placed on maturation medium (Table 2). Similar to that observed in the cell suspension transformation system, we were able to produce transgenic plants that had the marker gene switched off in the developing plantlets.
Fig 2. Steps in the protoplast transformation process with a construct containing the VvmybA1 gene driven by the Dc3 embryo-specific promoter.
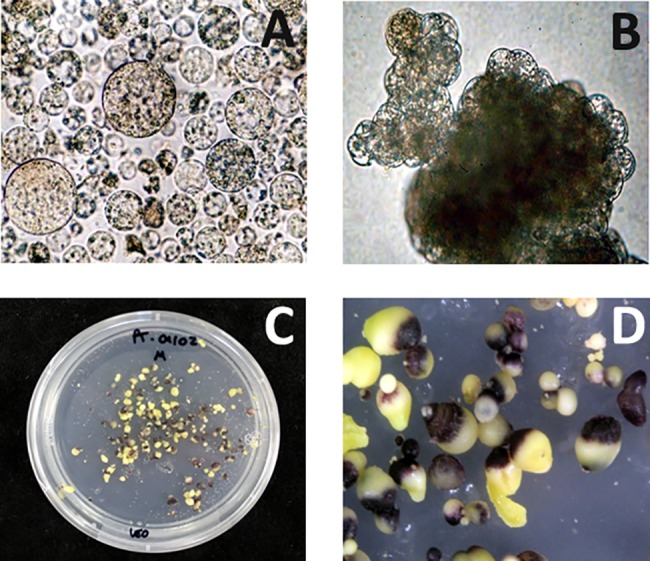
A) Transformed protoplast derived cells producing anthocyanins; B) Micro-calli derived from protoplasts; C) Anthocyanin producing somatic embryos; D) Closeup of the regenerating anthocyanin producing somatic embryos.
Expression level of VvMybA1 in one-year-old transgenic lines
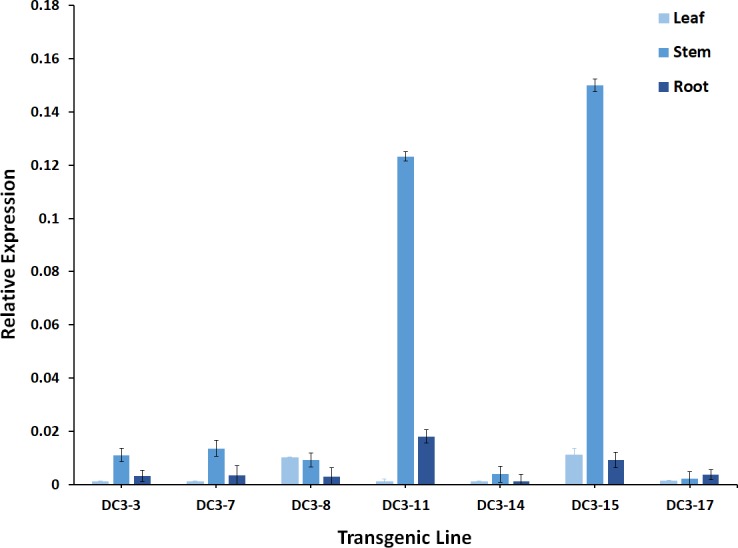
PCR confirmed the transgenic ‘Hamlin’ sweet orange lines regenerated following either the Agrobacterium-mediated transformation protocol or the protoplast protocol had the VvMybA1 transgene stably incorporated into the genome (S3 Fig). Transgene copy number validated using qPCR ranged from 1 to 4 copies in all the PCR positive lines (data not presented), and there was no significant variation in plants regenerated from either method. The copy numbers of the 7 selected lines evaluated in this study (5 lines obtained through the Agrobacterium mediated transformation method and 2 through the protoplast transformation method) are presented in Table 3 and ranged from 1–3. VvMybA1 transgene levels were analyzed in each of these lines. Transgene expression could not be detected in the leaves of most transgenic lines apart from DC3-8 and DC3-15 (0.01-fold). Transgene expression could however be detected in some transgenic stems. However, the results were variable and depended on the transgenic line evaluated. The relative gene expression was minimal and ranged from 0.02-fold (DC3-17) to 0.15-fold (DC3-15). VvMybA1 expression was negligible in the roots and could not be detected in most lines (Fig 3). DC3-11 had a 0.18-fold change in the expression level when compared to the non-transgenic control.
Table 3. Transgene copy number determination using quantitative real-time PCR by comparison of transgenic lines with external plasmid controls.
| Transgenic Line | Obtained using | Mean Cp A | STD Cp B | Copy number C |
|---|---|---|---|---|
| DC3-3 | Agrobacterium | 28.051 | 0.196 | 2 |
| DC3-7 | Agrobacterium | 27.834 | 0.368 | 2 |
| DC3-8 | Agrobacterium | 29.263 | 0.157 | 1 |
| DC3-11 | Agrobacterium | 28.930 | 0.0489 | 1 |
| DC3-14 | Agrobacterium | 28.441 | 0.0810 | 2 |
| DC3-15 | Protoplast | 29.047 | 0.263 | 1 |
| DC3-17 | Protoplast | 27.514 | 0.104 | 3 |
| Plasmid-1CD | - | 29.433 | 0.128 | 1 |
| Plasmid-2C | - | 28.506 | 0.141 | 2 |
| Plasmid-3C | - | 27.704 | 0.111 | 3 |
| Plasmid-4C | - | 27.276 | 0.213 | 4 |
| Plasmid-5C | - | 26.927 | 0.157 | 5 |
A Average values of crossing point from three sample replicates.
B Standard deviations.
C Approximate copy number derived from the average values of extrapolated concentration relative to a single transgene copy.
D Plasmid DNA used for copy number calculations
Fig 3. Relative VvMybA1 gene expression in leaves, stem and roots in selected transgenic lines.
Dc3 promoter in the transgenic plants respond variably to different abiotic stresses
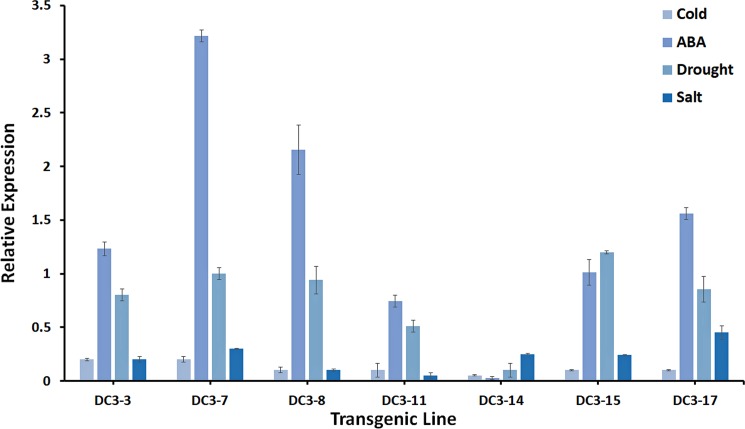
The Dc3 promoter responds to several external stress stimuli [49]. The regenerated Dc3 transgenic citrus plants were subjected to several stresses, and the relative gene expression of the VvMybA1 transgene was evaluated in 6 independent transgenic lines. Cold stress treatment did not result in any significant effect on transgene expression in all lines evaluated (Fig 4). Relative gene expression following water stress treatments increased and ranged from a 1.2-fold change in DC3-15 to 0.1-fold change in DC3-14 (Fig 4). ABA application resulted in the greatest change in expression levels in all transgenic lines, except the line DC3-14, which did not respond to exogenous ABA application. A 3.2-fold change in gene expression level was observed in the line DC3-7, while the other lines had a 2.1- to 0.7-fold change in expression levels. Two drenches of 200 mM NaCl applied weekly induced VvMybA1expression, again in all lines except DC3-14. The VvMybA1 expression levels ranged from 0.45-fold expression from line DC3-17 to 0.1-fold in DC3-8.
Fig 4. Relative VvMybA1 gene expression in response to various abiotic stress imposed on the selected transgenic lines.
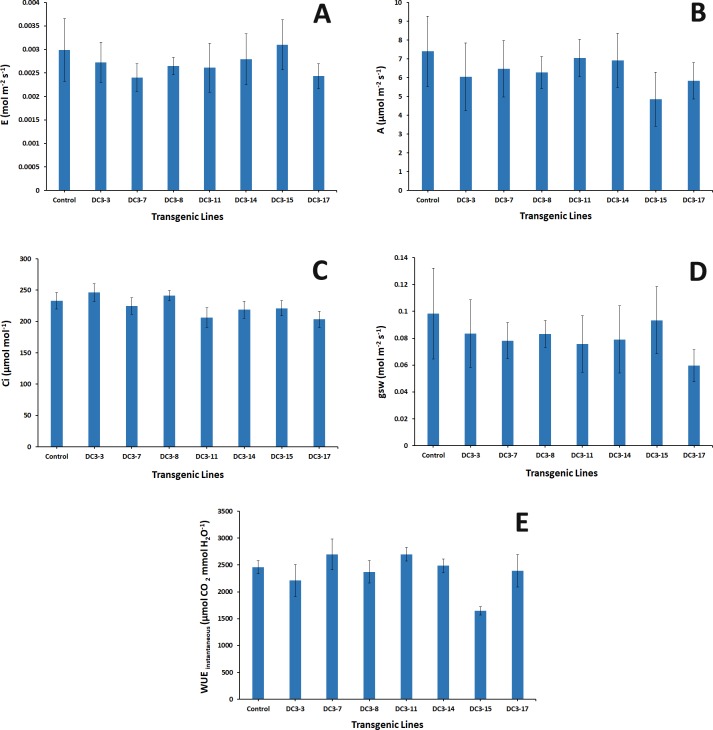
Transgenic lines show similar levels of physiological responses compared to non-transgenic control
Several physiological parameters were evaluated in the regenerated transgenic lines. None of the transgenic lines differed from the non-transgenic control for net assimilation of CO2, transpiration rate, intercellular CO2 content and stomatal conductance to water vapor responses. The WUEinstantaneous (a molar ratio of assimilation to transpiration) in transgenic line DC3-15 significantly differed from the control. All other transgenic lines were not significant (Fig 5)
Fig 5. Comparison of several physiological parameters between selected transgenic lines and control.
A) Transpiration rate; B) Net assimilation rate; C) Intercellular CO2 concentration; D) Stomatal conductance and E) Instantaneous physiological water use efficiency.
Discussion
The Dc3 gene isolated from carrot (Daucus carota L.) is a group III lea-class gene [50] that is highly expressed during the initial phases of embryogenesis [51]. Its promoter is expressed in developing seeds [52] and can therefore be utilized to target seed specific transgenes or, as in our study, to drive a visual reporter gene to select transgenic cells following genetic transformation. Citrus can be efficiently transformed using cell suspension cultures and regenerated through somatic embryogenesis [53]. In addition, cell suspension cultures allow for the genetic transformation of any polyembryonic citrus cultivar that can be established as an embryogenic cell suspension culture, including the otherwise hard-to-transform mandarins and seedless cultivars that cannot be transformed through conventional epicotyl mediated transformation techniques [38].
The genetic transformation process relies heavily on the ability to select transformed cells from most non-transformed cells. This is because, in most plant species, DNA integration into the genome following transformation usually occurs at a low frequency [54]. Transgenic plants are commonly selected following the expression of a selectable marker gene linked to the gene of interest [55]. Thus, selection of transformed cells for their ability to proliferate in the presence of the selective agent allows isolation of transgenic cells containing only the gene of interest [56]. Once a desired transgenic plant is produced from transgenic cells, the marker gene becomes useless [57]. However, recent studies have focused on excision systems for the removal of the marker gene [58–60].
In our current study, we tested the VvMybA1 anthocyanin regulatory gene (isolated from the grapevine) driven by the embryo-specific Dc3 promoter as a regulated visual selectable marker for the selection of transgenic citrus lines. Anthocyanin regulatory genes have been used for the selection of transgenic cells in other plant species [61–63]. In most cases, these studies have relied on the utilization of a strong constitutive promoter to drive the anthocyanin regulatory gene. This invariably resulted in high anthocyanin accumulation in the cells and the regenerated plants exhibited growth retardation when compared to the non-transgenic plants [62]. The Dc3 promoter is a weak promoter compared to the commonly used virus derived constitutive promoters and produces a much weaker gene expression pattern (our observations). Transgene expression under the control of the Dc3 promoter did not produce high levels of anthocyanins in the embryo and we did not observe any developmental differences between transgenic and non-transgenic embryos. In apple, transgenic plants were obtained after visual selection with a construct containing the Myb10 gene driven by its own promoter. This resulted in much lower gene expression and the production of phenotypically normal plants [63].
This system also enabled the selection of transgenic embryos that could be easily identified by their purple coloration from the non-transformed green embryos (Fig 1C). The construct was initially tested using conventional Agrobacterium-mediated transformation–a process that requires an antibiotic selection step. Transgenic embryos identified visually based on color could be regenerated into normal plants without any visual coloration (Fig 1). Our results demonstrated the specific activation of the Dc3 promoter in citrus embryo.
The protoplast expression vector was constructed to test an all-plant DNA sequence as a reporter. Protoplast transformation utilizing naked DNA offers the simplest technique to create an all-plant DNA-containing transgenic or even a cisgenic/intragenic plant. An added advantage of utilizing a visual marker with protoplast transformation is that it does not depend on a plant antibiotic-selectable marker for plant regeneration [64]. Although we utilized a plasmid DNA containing an E. coli antibiotic selection backbone in this study, our results indicated that protoplast transformation can efficiently incorporate the construct and allow the regeneration of a phenotypically normal plant similar to that obtained using Agrobacterium-mediated transformation (Fig 2). Protocols can subsequently be developed to incorporate a linear all-plant DNA construct or eventually an all-citrus DNA construct (cisgenic or intragenic) into the genome through protoplast-mediated transformation.
Chimeric transgenic plant production is a problem during citrus transformation and has been frequently observed with epicotyl-mediated transformation [65]. Chimeric plants arise due to multiple cells producing the shoot apical meristem–some cells are transformed and others are non-transgenic [66]. Somatic embryogenesis utilizes the ability of single cells to produce plants. A single-cell origin of somatic embryos has been reported in both monocots and dicots [67–70]. We did not regenerate any chimeric plants from the Agrobacterium mediated transformation system.
Protoplast transformation relies on the ability of PEG to agglutinate neighboring protoplasts and DNA [71]. During protoplast transformation, the cells are brought into close contact with the exogenous DNA, and a small amount of DNA is incorporated by endocytosis [72]. This process, however, also predisposes transient transformed cells in contact with neighboring untransformed cells to fuse and produce chimeric cell masses. This occurs when two adjacent cells (one transformed and the other wild type) fuse to produce a chimeric plant. In rare instances, two transformed cells (each containing a different copy number) can fuse to regenerate a visually non-chimeric plant. We observed the formation of a few chimeric embryos during protoplast transformation but they were not regenerated and subsequently discarded.
VvMybA1 gene expression was low to negligible in the transgenic plants (Fig 3). Acid methanol extracts also did not produce a discernable change in coloration in these tissue samples (results not shown). Anthocyanins are a major component of many fruits [73] and vegetables [74]. There have been detailed studies on the safety of anthocyanins [75], and anthocyanins present in grape juice, wine and fruits (mostly produced as a result of the VvMybA1 gene) have been consumed by humans for several millennia [76]. The anthocyanin selection system is comparable to the GUS and the GFP selection systems in grapes [77]. While GUS is used as a destructive marker [78], GFP is considered non-destructive [79]. Even though marker genes have not been implicated in any safety issues [80–82], it is our understanding that an anthocyanin-based marker would be safer and more consumer friendly than either GUS or GFP. Anthocyanin regulatory genes have been utilized as visual markers in other crop systems with variable success [83–85]. However, these studies relied on the use of a constitutive promoter that made practical application meaningless.
The Dc3 promoter is influenced by external abiotic stimuli [49, 86], and we observed a variable response to different external stimuli. Exogenous ABA application produced the greatest change; this was expected since the promoter is ABA inducible. Since citrus trees are grafted, we did not test gene expression in the roots following treatment. The Dc3 promoter is highly inducible in transgenic tobacco seedlings by salt and water stress [86]. The lower expression levels observed in our transgenic lines could be crop specific, with citrus being a woody perennial in comparison to published results from herbaceous seedlings [49, 52, 86]. It is possible to affirm that even though VvMybA1 gene expression under the control of the Dc3 promoter was variable in response to different abiotic stresses, it was not highly induced in response to any stress. Thus, VvMybA1 expression is virtually suppressed, and the induced levels are low to negligible under the normal stress that plants usually suffer. This is important to maintain a lack of marker gene expression in transgenic plants. GAPC was used as the reference gene since it has been thoroughly evaluated in citrus and deemed to a superior housekeeping gene for qPCR studies [43].
To understand if the VvMybA1 gene had any physiological influence on the transgenic plants, we used an open gas exchange system, the LI-6800, which rapidly measures changes in the gas concentration using infrared gas analyzers (IRGAs) [87]. Measuring gas exchange in intact leaves is a precise and reliable method to evaluate plant response to environment, disease pressure and input rate limitations [88–90]. The transgenic lines behaved similarly when compared to controls under the same environment, and these results agree with citrus physiology characteristics and response to changes in environment [91–93]. The addition of the VvMybA1 transgene did not change the transpiration rate, photosynthesis assimilation, CO2 assimilation rate or intercellular CO2 levels when compared to those of control non-transgenic trees (Fig 5). However, there was a significant change in the WUEinstantaneous in one of the transgenic lines which can be attributed to the lower photosynthesis rate reading in that line [94].
Conclusion
Our research provides conclusive evidence supporting the utilization of a plant-derived, embryo-specific visual reporter system for the genetic transformation of citrus. It is possible that there could be transgene expression in the seeds, but due to the long juvenile phase in citrus, it was not possible to study gene expression in the fruit in the current study. However, there is an increasing demand for the cultivation of seedless citrus [95]. Most of the commercially available fresh-market citrus, such as clementines [96], Tango mandarin [97], and navel oranges [98], are already seedless. In addition, juice oranges are processed in specialized facilities where it would be possible to discard seeds before they reach the consumers. This coupled with the utilization of an anthocyanin regulatory gene as a marker may alleviate safety concerns among the producers, processors and consumers. In addition, identification of similar tightly regulated, embryo-specific promoters from the citrus genome coupled with citrus-derived anthocyanin regulatory genes would result in the creation of an intragenic component for the genetic transformation of citrus.
Supporting information
(PDF)
Cells used in the transformation experiment. A) Embryogenic citrus callus B) Citrus suspension cultures C) Protoplast ring in a sucrose-mannitol gradient following enzymatic digestion of suspension derived cells.
(TIF)
PCR results from transgenic lines regenerated from either cell suspension transformation (A) or protoplast transformation (B) and successfully acclimated to a greenhouse.
(TIF)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the Citrus Research & Development Foundation (www.citrusrdf.org/). The Citrus Research & Development Foundation provided support in the form of salaries for author MD, but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Dutt M, Dhekney SA, Soriano L, Kandel R, Grosser JW. Temporal and spatial control of gene expression in horticultural crops. Hortic Res. 2014;1: 14047 doi: 10.1038/hortres.2014.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee TI, Young RA. Transcription of eukaryotic protein-coding genes. Annu Rev Genet. 2000;34: 77–137. doi: 10.1146/annurev.genet.34.1.77 [DOI] [PubMed] [Google Scholar]
- 3.Christensen AH, Quail PH. Ubiquitin promoter-based vectors for high-level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Res. 1996;5: 213–218. [DOI] [PubMed] [Google Scholar]
- 4.McElroy D, Zhang W, Cao J, Wu R. Isolation of an efficient actin promoter for use in rice transformation. Plant Cell. 1990;2: 163–171. doi: 10.1105/tpc.2.2.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiao K, Zhang C, Harrison M, Wang Z-Y. Isolation and characterization of a novel plant promoter that directs strong constitutive expression of transgenes in plants. Mol Breeding. 2005;15: 221–231. [Google Scholar]
- 6.Bäumlein H, Nagy I, Villarroel R, Inzé D, Wobus U. Cis-analysis of a seed protein gene promoter: the conservative RY repeat CATGCATG within the legumin box is essential for tissue-specific expression of a legumin gene. Plant J. 1992;2: 233–239. [PubMed] [Google Scholar]
- 7.Twell D, Yamaguchi J, McCormick S. Pollen-specific gene expression in transgenic plants: coordinate regulation of two different tomato gene promoters during microsporogenesis. Development. 1990;109: 705–713. [DOI] [PubMed] [Google Scholar]
- 8.Thomas TL. Gene expression during plant embryogenesis and germination: an overview. Plant Cell. 1993;5: 1401 doi: 10.1105/tpc.5.10.1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Streatfield SJ. Approaches to achieve high-level heterologous protein production in plants. Plant Biotechnol J. 2007;5: 2–15. doi: 10.1111/j.1467-7652.2006.00216.x [DOI] [PubMed] [Google Scholar]
- 10.Yang J, Barr LA, Fahnestock SR, Liu Z-B. High yield recombinant silk-like protein production in transgenic plants through protein targeting. Transgenic Res. 2005;14: 313–324. [DOI] [PubMed] [Google Scholar]
- 11.Chakraborty S, Chakraborty N, Datta A. Increased nutritive value of transgenic potato by expressing a nonallergenic seed albumin gene from Amaranthus hypochondriacus. Proc Natl Acad Sci U S A. 2000;97: 3724–3729. doi: 10.1073/pnas.050012697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dutt M, Ananthakrishnan G, Jaromin M, Brlansky R, Grosser J. Evaluation of four phloem-specific promoters in vegetative tissues of transgenic citrus plants. Tree Physiol. 2012;32: 83–93. doi: 10.1093/treephys/tpr130 [DOI] [PubMed] [Google Scholar]
- 13.Urwin PE, Atkinson HJ, Waller DA, McPherson MJ. Engineered oryzacystatin-I expressed in transgenic hairy roots confers resistance to Globodera pallida. Plant J. 1995;8: 121–131. [DOI] [PubMed] [Google Scholar]
- 14.Falco S, Guida T, Locke M, Mauvais J, Sanders C, Ward R, Webber P. Transgenic canola and soybean seeds with increased lysine. Biotechnology (N Y). 1995;13: 577–582. [DOI] [PubMed] [Google Scholar]
- 15.Seo M, Kanno Y, Frey A, North HM, Marion-Poll A. Dissection of Arabidopsis NCED9 promoter regulatory regions reveals a role for ABA synthesized in embryos in the regulation of GA-dependent seed germination. Plant Sci. 2016;246: 91–97. doi: 10.1016/j.plantsci.2016.02.013 [DOI] [PubMed] [Google Scholar]
- 16.Urriola J, Rathore KS. Temporal and spatial activities of a rice glutelin promoter in transgenic sorghum. Plant Cell Tissue Organ Cult. 2014;116: 227–234. [Google Scholar]
- 17.Potenza C, Aleman L, Sengupta-Gopalan C. Invited review: Targeting transgene expression in research, agricultural, and environmental applications: Promoters used in plant transformation. In Vitro Cell Dev Biol Plant. 2004;40: 1–22. [Google Scholar]
- 18.Hood EE, Love R, Lane J, Bray J, Clough R, Pappu K, Drees C, Hood KR, Yoon S, Ahmad A, Howard JA. Subcellular targeting is a key condition for high-level accumulation of cellulase protein in transgenic maize seed. Plant Biotechnol J. 2007;5: 709–719. doi: 10.1111/j.1467-7652.2007.00275.x [DOI] [PubMed] [Google Scholar]
- 19.Fehér A. Somatic embryogenesis—stress-induced remodeling of plant cell fate. Biochim Biophys Acta. 2015;1849: 385–402. doi: 10.1016/j.bbagrm.2014.07.005 [DOI] [PubMed] [Google Scholar]
- 20.Tian Q, Lin Y, Lai Z, Wang T. Cloning and bioinformatic analysis of the promoters of DlRan3A and DlRan3B from embryogenic callus in Dimocarpus longan. Chin J Trop Crops. 2014;35: 82–89. [Google Scholar]
- 21.Bunnag S, Tangpong D. Genetic Transformation of Citrus sinensis L. with an antisense ACC oxidase Gene. Am J Plant Sci. 2012;3: 1336. [Google Scholar]
- 22.Grosser JW, Gmitter FG. Protoplast fusion for production of tetraploids and triploids: applications for scion and rootstock breeding in citrus. Plant Cell Tissue Organ Cult. 2011;104:343–357. [Google Scholar]
- 23.Hao G, Pitino M, Duan Y, Stover E. Reduced susceptibility to Xanthomonas citri in transgenic citrus expressing the FLS2 receptor from Nicotiana benthamiana. Mol Plant Microbe Interact. 2016;29: 132–142. doi: 10.1094/MPMI-09-15-0211-R [DOI] [PubMed] [Google Scholar]
- 24.Anami S, Njuguna E, Coussens G, Aesaert S, Van Lijsebettens M. Higher plant transformation: principles and molecular tools. Int J Dev Biol. 2013;57: 483–494. doi: 10.1387/ijdb.130232mv [DOI] [PubMed] [Google Scholar]
- 25.Li ZT, Kim K-H, Jasinski JR, Creech MR, Gray DJ. Large-scale characterization of promoters from grapevine (Vitis spp.) using quantitative anthocyanin and GUS assay systems. Plant Sci. 2012;196: 132–142. doi: 10.1016/j.plantsci.2012.08.009 [DOI] [PubMed] [Google Scholar]
- 26.Kawahigashi H, Hirose S, Iwai T, Ohashi Y, Sakamoto W, Maekawa M, Ohkawa Y. Chemically induced expression of rice OSB2 under the control of the OsPR1. 1 promoter confers increased anthocyanin accumulation in transgenic rice. J Agric Food Chem. 2007;55: 1241–1247. doi: 10.1021/jf062339w [DOI] [PubMed] [Google Scholar]
- 27.Gao X, Zhang L, Zhou S, Wang C, Deng X, Zhang H, Yang G, Javeed H, He G. AtMYB12 gene: a novel visible marker for wheat transformation. Mol Biol Rep. 2011;38: 183–190. doi: 10.1007/s11033-010-0093-3 [DOI] [PubMed] [Google Scholar]
- 28.Gurr SJ, Rushton PJ. Engineering plants with increased disease resistance: what are we going to express? Trends Biotechnol. 2005;23: 275–282. doi: 10.1016/j.tibtech.2005.04.007 [DOI] [PubMed] [Google Scholar]
- 29.Puchta H. Marker-free transgenic plants. Plant Cell Tissue Organ Cult. 2003;74: 123–134. [Google Scholar]
- 30.Ballester A, Cervera M, Peña L. Efficient production of transgenic citrus plants using isopentenyl transferase positive selection and removal of the marker gene by site-specific recombination. Plant Cell Rep. 2007;26: 39–45. doi: 10.1007/s00299-006-0197-3 [DOI] [PubMed] [Google Scholar]
- 31.Zou X, Peng A, Xu L, Liu X, Lei T, Yao L, He Y, Chen S. Efficient auto-excision of a selectable marker gene from transgenic citrus by combining the Cre/loxP system and ipt selection. Plant Cell Rep. 2013;32: 1601–1613. doi: 10.1007/s00299-013-1470-x [DOI] [PubMed] [Google Scholar]
- 32.Peng A, Xu L, He Y, Lei T, Yao L, Chen S, et al. Efficient production of marker-free transgenic ‘Tarocco’ blood orange (Citrus sinensis Osbeck) with enhanced resistance to citrus canker using a Cre/loxP site-recombination system. Plant Cell Tissue Organ Cult. 2015;123: 1–13. [Google Scholar]
- 33.Ballester A, Cervera M, Peña L. Selectable marker-free transgenic orange plants recovered under non-selective conditions and through PCR analysis of all regenerants. Plant Cell Tissue Organ Cult. 2010;102: 329–336. [Google Scholar]
- 34.Seffens WS, Almoguera C, Wilde HD, Vonder Haar RA, Thomas TL. Molecular analysis of a phylogenetically conserved carrot gene: developmental and environmental regulation. Dev Genet. 1990;11: 65–76. doi: 10.1002/dvg.1020110108 [DOI] [PubMed] [Google Scholar]
- 35.Kobayashi S, Goto-Yamamoto N, Hirochika H. Retrotransposon-induced mutations in grape skin color. Science. 2004;304: 982 doi: 10.1126/science.1095011 [DOI] [PubMed] [Google Scholar]
- 36.Grosser JW, Gmitter FG. Protoplast Fusion and Citrus Improvement In: Janick J, editor. Plant Breeding Reviews. John Wiley & Sons, Inc.; 1990. pp. 339–374. [Google Scholar]
- 37.Hood EE, Gelvin SB, Melchers LS, Hoekema A. New Agrobacterium helper plasmids for gene transfer to plants. Transgenic Res. 1993;2: 208–218. [Google Scholar]
- 38.Dutt M, Grosser J. Evaluation of parameters affecting Agrobacterium-mediated transformation of citrus. Plant Cell Tissue Organ Cult. 2009;98: 331–340. [Google Scholar]
- 39.Dutt M, Vasconcellos M, Song K, Gmitter F, Grosser J. In vitro production of autotetraploid Ponkan mandarin (Citrus reticulata Blanco) using cell suspension cultures. Euphytica. 2010;173: 235–242. [Google Scholar]
- 40.Niedz RP, Hyndman SE, Wynn ET, Bausher MG. Normalizing sweet orange (C. sinensis (L.) Osbeck) somatic embryogenesis with semi-permeable membranes. In Vitro Cell Dev Biol-Plant. 2002;38: 552–557. [Google Scholar]
- 41.Fleming GH, Olivares-Fuster O, Del-Bosco SF, Grosser JW. An alternative method for the genetic transformation of sweet organce. In Vitro Cell Dev Biol-Plant. 2000;36:450. [Google Scholar]
- 42.Dutt M, Stanton D, Grosser JW. Ornacitrus: Development of Genetically Modified Anthocyanin-expressing Citrus with Both Ornamental and Fresh Fruit Potential. J Am Soc Hortic Sci. 2016;141: 54–61. [Google Scholar]
- 43.Mafra V, Kubo KS, Alves-Ferreira M, Ribeiro-Alves M, Stuart RM, Boava LP, Rodrigues CM, Machado MA. Reference Genes for Accurate Transcript Normalization in Citrus Genotypes under Different Experimental Conditions. Plos One. 2012;7: e31263 doi: 10.1371/journal.pone.0031263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Methods. 2001;25: 402–408. [DOI] [PubMed] [Google Scholar]
- 45.Dutt M, Li ZT, Dhekney SA, Gray DJ. A co-transformation system to produce transgenic grapevines free of marker genes. Plant Sci. 2008;175(3):423–30. [Google Scholar]
- 46.Dutt M, Erpen L, Ananthakrishnan G, Barthe G, Brlansky R, Maiti I, Grosser JW. Comparative expression analysis of five caulimovirus promoters in citrus. Plant Cell Tissue Organ Cult. 2016;126: 229–238. [Google Scholar]
- 47.Grosser J, Chandler J. Somatic hybridization of high yield, cold-hardy and disease resistant parents for citrus rootstock improvement. J Hortic Sci Biotechnol. 2000;75: 641–644. [Google Scholar]
- 48.Polley HW. Implications of atmospheric and climatic change for crop yield and water use efficiency. Crop Sci. 2002;42: 131–140. [PubMed] [Google Scholar]
- 49.Vivekananda J, Drew MC, Thomas TL. Hormonal and Environmental Regulation of the Carrot lea-Class Gene Dc3. Plant Physiol. 1992;100: 576–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dure L 3rd, Crouch M, Harada J, Ho TH, Mundy J, Quatrano R, Thomas T, Sung ZR. Common amino acid sequence domains among the LEA proteins of higher plants. Plant Mol Biol. 1989;12: 475–486. doi: 10.1007/BF00036962 [DOI] [PubMed] [Google Scholar]
- 51.Wilde HD, Nelson WS, Booij H, Vries SC, Thomas TL. Gene-expression programs in embryogenic and non-embryogenic carrot cultures. Planta. 1988;176: 205–211. doi: 10.1007/BF00392446 [DOI] [PubMed] [Google Scholar]
- 52.Chak RKF, Thomas TL, Quatrano RS, Rock CD. The genes ABI1 and ABI2 are involved in abscisic acid-and drought-inducible expression of the Daucus carota L. Dc3 promoter in guard cells of transgenic Arabidopsis thaliana (L.) Heynh. Planta. 2000;210: 875–883. doi: 10.1007/s004250050692 [DOI] [PubMed] [Google Scholar]
- 53.Dutt M, Grosser JW. An embryogenic suspension cell culture system for Agrobacterium-mediated transformation of citrus. Plant Cell Rep. 2010;29: 1251–1260. doi: 10.1007/s00299-010-0910-0 [DOI] [PubMed] [Google Scholar]
- 54.Narusaka Y, Narusaka M, Yamasaki S, Iwabuchi M. Methods to transfer foreign genes to plants In: Çiftçi OY, editor. Transgenic Plants-Advances and Limitations. Çiftçi Yelda Ozden (Ed.), InTech, 2012. doi: 10.5772/32773 [Google Scholar]
- 55.Brasileiro A, Dusi DMA. Transformação genética de plantas In: Torres AC, Caldas LS, Buso JA, editors. Cultura de tecidos e transformação genética de plantas. Brasília: Embrapa; 1999. pp. 679–735. [Google Scholar]
- 56.Gleave A. Elimination of selectable marker genes from transgenic crops. Molecular Methods of Plant Analysis. 2002;22: 73–94. [Google Scholar]
- 57.Park J, Lee YK, Kang BK, Chung WI. Co-transformation using a negative selectable marker gene for the production of selectable marker gene-free transgenic plants. Theor Appl Genet. 2004;109: 1562–1567. doi: 10.1007/s00122-004-1790-x [DOI] [PubMed] [Google Scholar]
- 58.Hare PD, Chua NH. Excision of selectable marker genes from transgenic plants. Nat Biotechnol. 2002; 20:575–580. doi: 10.1038/nbt0602-575 [DOI] [PubMed] [Google Scholar]
- 59.García-Almodóvar R, Petri C, Padilla I, Burgos L. Combination of site-specific recombination and a conditional selective marker gene allows for the production of marker-free tobacco plants. Plant Cell Tissue Organ Cult. 2014;116: 205–215. [Google Scholar]
- 60.Mészáros K, Éva C, Kiss T, Bányai J, Kiss E, Téglás F, Láng L, Karsai I, Tamás L. Generating marker-free transgenic wheat using minimal gene cassette and cold-inducible Cre/Lox System. Plant Mol Biol Rep. 2015;33: 1221–1231. [Google Scholar]
- 61.Kandel R, Dutt M, Grosser JW, Gray DJ, Li ZT, Sitther V, Bergey DR, Dhekney SA. Evaluation of plant-based reporter systems for improvement of cold-hardy grape cultivars. Acta Hortic. 2016; 115: 57–62. [Google Scholar]
- 62.Lim SH, Sohn SH, Kim DH, Kim JK, Lee JY, Kim YM, Ha SH. Use of an anthocyanin production phenotype as a visible selection marker system in transgenic tobacco plant. Plant Biotechnol Rep. 2012;6: 203–211. [Google Scholar]
- 63.Kortstee A, Khan S, Helderman C, Trindade L, Wu Y, Visser R, Brendolise C, Allan A, Schouten HJ, Jacobsen E. Anthocyanin production as a potential visual selection marker during plant transformation. Transgenic Res. 2011;20: 1253–1264. doi: 10.1007/s11248-011-9490-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guo W, Duan Y, Olivares-Fuster O, Wu Z, Arias CR, Burns JK, Grosser JW. Protoplast transformation and regeneration of transgenic Valencia sweet orange plants containing a juice quality-related pectin methylesterase gene. Plant Cell Rep. 2005;24: 482–486. doi: 10.1007/s00299-005-0952-x [DOI] [PubMed] [Google Scholar]
- 65.Domínguez A, Cervera M, Pérez RM, Romero J, Fagoaga C, Cubero J, López MM, Juárez JA, Navarro L, Peña L. Characterisation of regenerants obtained under selective conditions after Agrobacterium-mediated transformation of citrus explants reveals production of silenced and chimeric plants at unexpected high frequencies. Mol Breeding. 2004;14: 171–183. [Google Scholar]
- 66.Peña L, Cervera M, Juárez J, Ortega C, Pina J, Durán-Vila N, Navarro L. High efficiency Agrobacterium-mediated transformation and regeneration of citrus. Plant Sci. 1995;104: 183–191. [Google Scholar]
- 67.Nagmani R, Becwar MR, Wann SR. Single-cell origin and development of somatic embryos in Picea abies (L.) Karst. (Norway spruce) and P. glauca (Moench) Voss (white spruce). Plant Cell Rep. 1987;6: 157–159. doi: 10.1007/BF00276677 [DOI] [PubMed] [Google Scholar]
- 68.Toonen MAJ, Hendriks T, Schmidt EDL, Verhoeven HA, van Kammen A, de Vries SC. Description of somatic-embryo-forming single cells in carrot suspension cultures employing video cell tracking. Planta. 1994;194: 565–572. [Google Scholar]
- 69.Dudits D, Bgre L, Gyrgyev J. Molecular and cellular approaches to the analysis of plant embryo development from somatic cells in vitro. J. Cell Sci. 1975;99: 473–482. [Google Scholar]
- 70.Shoemaker RC, Couche LJ, Galbraith DW. Characterization of somatic embryogenesis and plant regeneration in cotton (Gossypium hirsutum L.). Plant Cell Rep. 1986;5: 178–181. doi: 10.1007/BF00269112 [DOI] [PubMed] [Google Scholar]
- 71.Constabel F, Kao K. Agglutination and fusion of plant protoplasts by polyethylene glycol. Can J Bot. 1974;52:1603–1606. [Google Scholar]
- 72.Davey MR, Anthony P, Power JB, Lowe KC. Plant protoplasts: status and biotechnological perspectives. Biotechnol Adv. 2005;23:131–171. doi: 10.1016/j.biotechadv.2004.09.008 [DOI] [PubMed] [Google Scholar]
- 73.Moyer RA, Hummer KE, Finn CE, Frei B, Wrolstad RE. Anthocyanins, phenolics, and antioxidant capacity in diverse small fruits: Vaccinium, Rubus, and Ribes. J Agric Food Chem. 2002;50: 519–525. [DOI] [PubMed] [Google Scholar]
- 74.Böhm H, Mazza G, Miniati E. Anthocyanins in Fruits, Vegetables and Grains 362 Seiten, zahlr. Abb. und Tab. CRC Press; 1993. pp. 343.–343. [Google Scholar]
- 75.Zafra-Stone S, Yasmin T, Bagchi M, Chatterjee A, Vinson JA, Bagchi D. Berry anthocyanins as novel antioxidants in human health and disease prevention. Mol Nutr Food Res. 2007;51: 675–683. doi: 10.1002/mnfr.200700002 [DOI] [PubMed] [Google Scholar]
- 76.Greene JA. The beginnings of grape cultivation and wine production in Phoenician/Punic north Africa In: McGovern PE, Fleming SF, Katz SH, editors. Origins and Ancient History of Wine Food and Nutrition in History and Anthropology Series. London: Routledge; 1996. pp. 311–337. [Google Scholar]
- 77.Kandel R, Bergey D, Dutt M, Sitther V, Li Z, Gray D, Dhekney SA. Evaluation of a grapevine-derived reporter gene system for precision breeding of Vitis. Plant Cell Tissue Organ Cult. 2016;124: 599–609. [Google Scholar]
- 78.Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6: 3901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Joshi M, Niu C, Fleming G, Hazra S, Chu Y, Nairn CJ, Yang H, Ozias-Akins P. Use of green fluorescent protein as a non-destructive marker for peanut genetic transformation. In Vitro Cell Dev Biol-Plant. 2005;41: 437–445. [Google Scholar]
- 80.Yeh SD, Gonsalves D. Practices and perspective of control of papaya ringspot virus by cross protection In: Harris KF, editor. Advances in Disease Vector Research. New York: Springer; 1994. pp. 237–257. [Google Scholar]
- 81.Fuchs M, Gonsalves D. Safety of virus-resistant transgenic plants two decades after their introduction: lessons from realistic field risk assessment studies. Annu Rev Phytopathol. 2007;45: 173–202. doi: 10.1146/annurev.phyto.45.062806.094434 [DOI] [PubMed] [Google Scholar]
- 82.Shelton AM, Zhao JZ, Roush RT. Economic, ecological, food safety, and social consequences of the deployment of Bt transgenic plants. Annu Rev Entomol. 2002;47: 845–881. doi: 10.1146/annurev.ento.47.091201.145309 [DOI] [PubMed] [Google Scholar]
- 83.Ludwig SR, Bowen B, Beach L, Wessler SR. A regulatory gene as a novel visible marker for maize transformation. Science. 1990;247: 449–451. doi: 10.1126/science.247.4941.449 [DOI] [PubMed] [Google Scholar]
- 84.Naing AH, Lim KB, Kim CK. The usage of snapdragon Delila (Del) gene as a visible selection marker for the antibiotic-free transformation system. J Plant Biol. 2015;58: 110–116. [Google Scholar]
- 85.Mentewab A, Letellier V, Marque C, Sarrafi A. Use of anthocyanin biosynthesis stimulatory genes as markers for the genetic transformation of haploid embryos and isolated microspores in wheat. Cereal Res Commun. 1999; 27: 17–24. [Google Scholar]
- 86.Siddiqui NU, Chung H-J, Thomas TL, Drew MC. Abscisic acid-dependent and-independent expression of the carrot late-embryogenesis-abundant-class gene Dc3 in transgenic tobacco seedlings. Plant Physiol. 1998;118: 1181–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stinziano JR, Morgan PB, Lynch DJ, Saathoff AJ, McDermitt DK, Hanson DT. The rapid A–Ci response: photosynthesis in the phenomic era. Plant Cell Environ. 2017;40: 1256–1262. doi: 10.1111/pce.12911 [DOI] [PubMed] [Google Scholar]
- 88.Melgar J, Dunlop J, Syvertsen J. Growth and physiological responses of the citrus rootstock Swingle citrumelo seedlings to partial rootzone drying and deficit irrigation. J Agric Sci. 2010;148: 593–602. [Google Scholar]
- 89.Melgar J, Syvertsen J, Martínez V, García-Sánchez F. Leaf gas exchange, water relations, nutrient content and growth in citrus and olive seedlings under salinity. Biol Plant. 2008;52: 385–390. [Google Scholar]
- 90.Ribeiro R, Machado E, Santos M, Oliveira R. Photosynthesis and water relations of well-watered orange plants as affected by winter and summer conditions. Photosynthetica. 2009;47: 215–222. [Google Scholar]
- 91.César Bachiega Zambrosi F, Mattos D, Syvertsen JP. Plant growth, leaf photosynthesis, and nutrient-use efficiency of citrus rootstocks decrease with phosphite supply. J. Plant Nutr. Soil Sci. 2011;174: 487–495. [Google Scholar]
- 92.Pons E, Peris JE, Peña L. Field performance of transgenic citrus trees: assessment of the long-term expression of uidA and nptII transgenes and its impact on relevant agronomic and phenotypic characteristics. BMC Biotechnol. 2012;12: 41 doi: 10.1186/1472-6750-12-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Syvertsen J, Garcia-Sanchez F. Multiple abiotic stresses occurring with salinity stress in citrus. Environ Exp Bot. 2014;103: 128–137. [Google Scholar]
- 94.Field C, Merino J, Mooney HA. Compromises between water-use efficiency and nitrogen-use efficiency in five species of California evergreens. Oecologia. 1983;60: 384–389. doi: 10.1007/BF00376856 [DOI] [PubMed] [Google Scholar]
- 95.Stover E, Castle W, Chao C-CT. Trends in US sweet orange, grapefruit, and mandarin-type cultivars. Horttechnology. 2005;15: 501–506. [Google Scholar]
- 96.Garcia-Martinez J, Garcia-Papi M. Influence of gibberellic acid on early fruit development, diffusible growth substances and content of macronutrients in seedless Clementine mandarin. Sci Hortic. 1979;11: 337–347. [Google Scholar]
- 97.Roose M, Williams T. Tango Mandarin: A new seedless mid-late season irradiated selection of W. Murcott (Afourer) mandarin developed by the University of California Citrus Breeding Program. 2006.
- 98.Shamel A. Why navel oranges are seedless. J. Hered. 1918;9: 246–249. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
Cells used in the transformation experiment. A) Embryogenic citrus callus B) Citrus suspension cultures C) Protoplast ring in a sucrose-mannitol gradient following enzymatic digestion of suspension derived cells.
(TIF)
PCR results from transgenic lines regenerated from either cell suspension transformation (A) or protoplast transformation (B) and successfully acclimated to a greenhouse.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.