Abstract
PKnox1 (also known as Prep1) belongs to the TALE family of homeodomain transcription factors that are critical for regulating growth and differentiation during embryonic and postnatal development in vertebrates. We demonstrate here that PKnox1 is required for adult spermatogenesis in a germ cell-intrinsic manner. Tamoxifen-mediated PKnox1 loss in the adult testes, as well as its germ cell-specific ablation, causes testis hypotrophy with germ cell apoptosis and, as a consequence, compromised spermatogenesis. In PKnox1-deficient testes, spermatogenesis was arrested at the c-Kit+ spermatogonia stage, with a complete loss of the meiotic spermatocytes, and was accompanied by compromised differentiation of the c-Kit+ spermatogonia. Taken together, these results indicate that PKnox1 is a critical regulator of maintenance and subsequent differentiation of the c-Kit+ stage of spermatogonia in the adult testes.
Introduction
Spermatogenesis is a complex and highly ordered cell differentiation process in which the germ cell lineage gives rise to functional gametes in the male. During adult spermatogenesis in mice, spermatogonia are localized closely attached onto the basement membrane of seminiferous tubules, and their descendants are arranged towards the lumen. Distinct spermatogonia differentiation stages have been defined based on morphological features: Asingle (As; isolated single cells), Apaired (Apr; chains of 2 cells), and Aaligned (Aal; chains of 4 or 8 cells) are referred to as early undifferentiated spermatogonia [1,2]. Subsequently, Aal cells give rise to the late undifferentiated spermatogonia (Aal16~32), and then to differentiating spermatogonia (A1 to A4), which are committed to meiosis[3,4].
The balance between maintenance of the undifferentiated state and differentiation is controlled by a complex interplay of germ cell-intrinsic mechanisms and -extrinsic factors secreted by Sertoli cells that support germ cells within the seminiferous tubules[5]. Several transcription factors expressed in the germ cells have been implicated in the regulation of spermatogenesis, including PLZF[6,7], Taf4b[8] and SOHLH1/2[9,10]. With regard to Sertoli cell-derived factors, glial cell line-derived neurotrophic factor (GDNF) supports self-renewal of undifferentiated spermatogonia through binding to its receptor consisting of GFRα1and RET[11], while signaling from c-Kit, when bound by its ligand stem cell factor expressed by Sertoli cells, plays crucial roles in regulating proliferation, survival and the entry of spermatogonia into meiosis [12]. Furthermore, retinoic acid, the biologically active form of vitamin A supplied by Sertoli cells, has also been shown to regulate spermatogonia differentiation, as vitamin A-deficient mice are infertile because of an arrest of spermatogonia differentiation at the Aal-A1 transition[13].
The three-amino-acid-loop-extension (TALE) class of homeodomain transcription factors are recognized as critical for regulating growth and differentiation during embryonic and postnatal development in vertebrates[14]. The TALE homeodomain transcription factors, including the Meis, PKnox and Pbx families, share a conserved atypical homeodomain through which they can bind to the target DNA as well as interact with Hox proteins[15]. In addition, PKnox and Meis family members have conserved protein interaction domains, MEIS-A and MEIS-B (also termed HM1 and HM2), in their N-terminal region that function as an interface for heterodimerization with Pbx family members, promoting their nuclear translocation and also affecting DNA-binding specificity[16–19]. PKnox1 (Pbx/Knotted homeobox 1), also known as Prep1, is expressed ubiquitously in embryonic and adult tissues but at distinct levels in different organs[20]. A PKnox1/Prep1 null mutation causes lethality shortly after implantation[21], while PKnox1/Prep1-hypomorphic mice display a leaky lethal phenotype characterized mostly by hematopoietic and angiogenic defects by embryonic day (E) 17.5[22]. In the adult mouse testes, another TALE family member, Pbx4, is preferentially expressed in the germ cell lineage[23], and has been reported to cooperate with PKnox/Meis family members to initiate testis-specific transcription of the Pgk2 gene[24,25], suggestive of a potential involvement of PKnox1 in adult spermatogenesis. While the testis is one of the tissues where PKnox1 is highly expressed[20], defects in spermatogenesis have not been reported in the PKnox1/Prep1-hypomorphic mice.
In the present study, we thus examined the function of PKnox1 in adult spermatogenesis by generating PKnox1 conditional knockout mice and demonstrated that PKnox1 is required for the differentiation of the c-Kit+ stage of spermatogonia in a cell-intrinsic manner.
Results
PKnox1 is expressed in germ cells of the adult testes
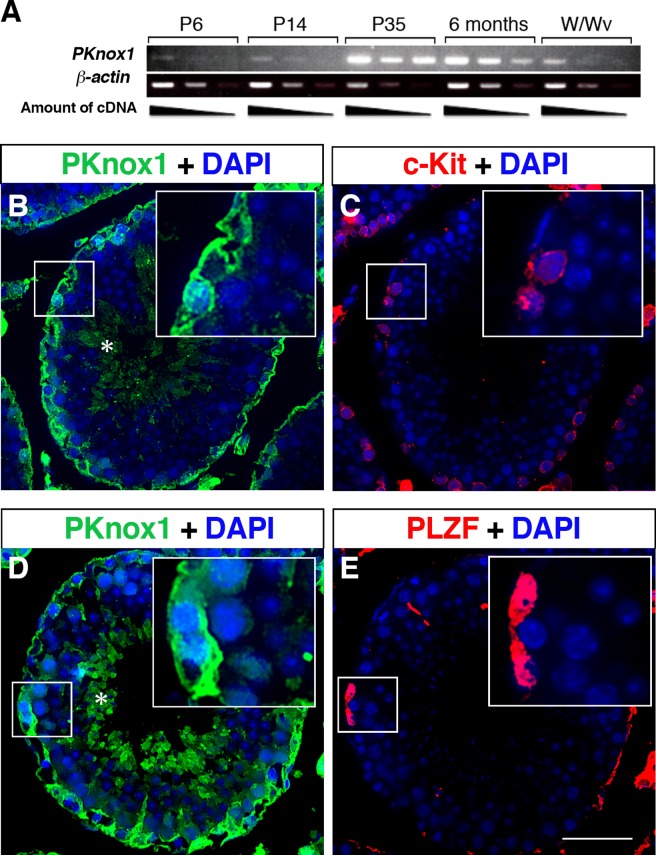
We examined the expression of PKnox1 in the testes at postnatal day (P) 6, 14, 35 and adult (6 months) by RT-PCR. PKnox1 expression was first detectable at P6, when gonocytes have been shown to complete their migration to the basement membrane of the testes[5]. The expression level of Prep1 then increased with age and reached a plateau at P35, a time point of termination of the first wave of spermatogenesis (Fig 1A). In contrast to wild-type adult testes, only weak PKnox1 expression was detected in the adult testes of W/Wv mice, which have an arrest in spermatogenesis at the c-Kit+ spermatogonia stage[26,27]. Immunohistochemical analysis revealed that a high level expression of PKnox1 was detected in most of the germ cells lining the basement membrane (Fig 1B and 1D), and some of it co-localized with cells expressing the late and early stage spermatogonia markers, c-Kit (insert in Fig 1B and 1C) and PLZF (insert in Fig 1D and 1E), respectively. In addition, expression of PKnox1 was also observed in haploid cells near the lumen of the seminiferous tubule (asterisks in Fig 1B and 1E). These findings suggest that PKnox1 is expressed in germ cells, from the early to the late stages of spermatogonia as well as at the haploid developmental stages in the adult testes.
Fig 1. Expression of PKnox1 in postnatal testes.
(A) Expression of PKnox1 and β-actin (control) mRNA transcripts in testes at postnatal (P) days 6, 14, 35 and 6 months obtained from wild-type mice and 12-week-old W/Wv mice. (B-E) Immunohistochemical analysis to localize PKnox1-expressing cells in the adult testis. Serial tissue sections of the testes from 8-week-old wild-type mice were stained with a PKnox1-specific antibody, in combination with anti-c-Kit (B, C) or PLZF antibodies (D, E). Inserts indicate cells co-expressing both Prep1 and c-Kit or PLZF. Asterisks indicate haploid cells positive for PKnox1. Data are representative of 3 independent experiments. Scale bar, 50 μm.
Loss of PKnox1 causes defects in adult spermatogenesis
As the early embryonic lethality resulting from germline deletion of the PKnox1 gene precludes any study of spermatogenesis in the adult testes, we generated mice harboring conditional alleles of PKnox1, in which exon 3 of the PKnox1 gene encoding the N-terminal part of the Pbx-binding domain is floxed by loxP sites (S1A Fig). Removal of exon 3 is predicted to induce a frameshift mutation, leading to the incorporation of 11 unrelated amino acids before encountering a termination codon within exon 4. Even if a truncated polypeptide generated from this mutant transcript could exist as a stable protein, the mutant protein would lack almost all functional PKnox1 domains, including the two Pbx-binding domains and the homeodomain, thus rendering PKnox1 a null allele. To examine a potential involvement of PKnox1 in adult spermatogenesis, we chose to study the consequence of PKnox1 ablation in the testes by crossing the mice carrying the PKnox1fl allele with the Rosa26 gene-driven tamoxifen-responsive Rosa26-CreERT2 knock-in mouse line, which causes highly efficient excision of loxP-flanked DNA in a ubiquitous manner after induction by tamoxifen[28]. Three intragastric administrations of tamoxifen into Rosa26-CreERT2; PKnox1fl/fl mice were sufficient to induce efficient deletion of the floxed PKnox1 locus and the loss of PKnox1 protein in the adult testes (S1B and S1C Fig).
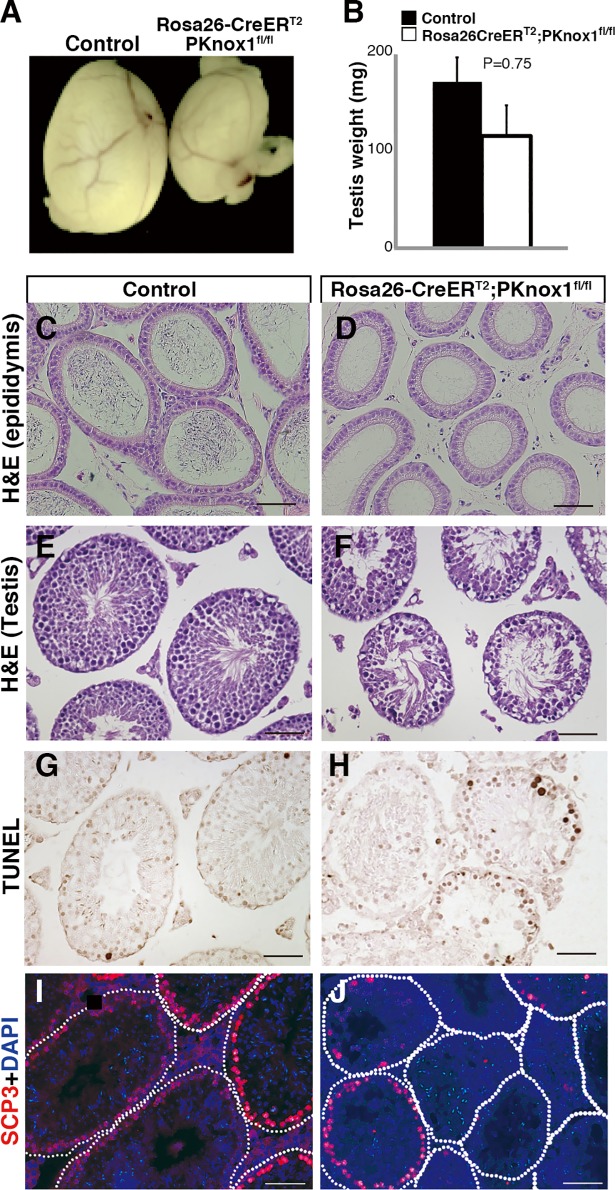
We first analyzed spermatogenesis in adult Rosa26-CreERT2; PKnox1fl/fl mice three weeks after tamoxifen treatment (hereafter referred to as PKnox1-CKO) and compared it to that of similarly treated Rosa26-CreERT2; PKnox1fl/+ control littermates. Three weeks after the induction of PKnox1 deletion, the size and weight of the testis was markedly, albeit not significantly, reduced in PKnox1-CKO mice compared to similarly treated controls (Fig 2A and 2B). Histological analysis of epididymides from PKnox1-CKO mice did not contain sperms (Fig 2C and 2D), and examination of sections of testes revealed atrophied seminiferous tubules containing very few spermatocytes (Fig 2E and 2F), while germ cells beneath the basement membrane, probably representing spermatogonia and haploid cells, appeared to exist (Fig 2F). Furthermore, we observed a profound increase in TUNEL+ apoptotic cells in PKnox1-CKO testes when compared with those in controls (Fig 2G and 2H). Immunohistochemical analyses further revealed that most seminiferous tubules in PKnox1-CKO testes lack SCP3+ meiotic cells (Fig 2I and 2J). These findings indicate that PKnox1 is indispensable for spermatogenesis in the adult testes.
Fig 2. Loss of PKnox1 in the adult testis causes defective spermatogenesis.
The size (A) and weight (B) of adult testes from 12-week-old PKnox1-CKO and control mice 3 weeks after induction of PKnox1 deletion. Bars are the mean and standard deviation of the weight of testes. (n = 4 for each genotype). Tissue sections from control and CKO epididymis stained with H&E (C, D) and testes stained with H&E (E, F), TUNEL (G, H) and anti-SCP3 antibody with DAPI (I, J). Data are representative of 4 independent experiments. Scale bars, 50 μm.
PKnox1 regulates adult spermatogenesis in a germ cell-intrinsic manner
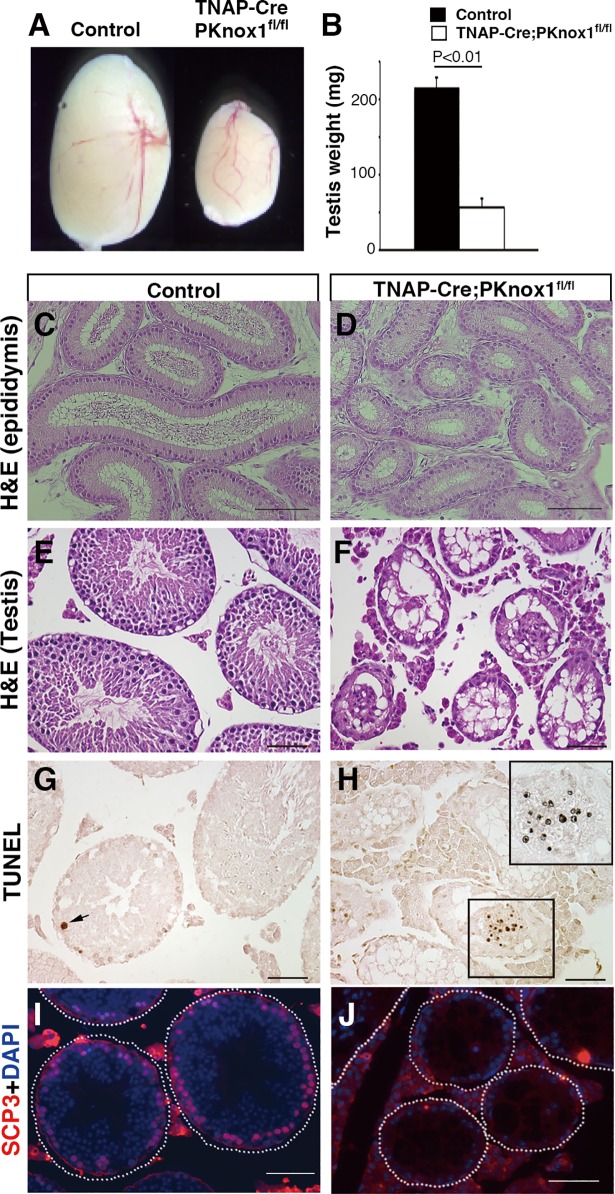
While the preceding data suggested that PKnox1 deficiency caused the differentiation arrest in spermatogenesis in the adult testes, Rosa26-CreERT2-mediated PKnox1 deletion is not limited to germ cells. Thus, to determine whether the PKnox1-deficient spermatogenesis phenotype reflects germ cell-intrinsic or -extrinsic effects of PKnox1 deletion, we crossed PKnox1fl mice with TNAP-Cre mice[29], with germ cell-specific Cre activity, to generate germ cell-specific PKnox1-deficient mice (TNAP-Cre; PKnox1fl/fl: referred to as GKO) and TNAP-Cre; PKnox1fl/+ control mice. The phenotype of the testes in GKO mice was similar but even more severe than that observed in the PKnox1-CKO mice, which is probably due to the residual PKnox1 activity in germ cells from CKO mice that retained the remaining floxed PKnox1 allele in the testes (S1B and S1C Fig). The size and weight of the testes from PKnox1-GKO mice (3-month-old) were significantly lower than those from littermate controls (Fig 3A and 3B). The epididymides from PKnox1-CKO mice lacked sperm (Fig 3C and 3D), and almost all the seminiferous tubules were atrophied with a loss of germ cells in the PKnox1-GKO testes (Fig 3E and 3F). An accumulation of TUNEL+ apoptotic cells inside the lumen was observed in a few tubules, probably the remaining apoptotic germ cells before their clearance from the tissue (Fig 3G and 3H). Furthermore, the PKnox1-GKO testes contained no SCP3+ meiotic cells (Fig 3I and 3J). Taken together, these findings indicate a germ cell-intrinsic requirement for PKnox1 in adult spermatogenesis.
Fig 3. Germ cell-specific PKnox1 loss causes defective spermatogenesis.
The size (A) and weight (B) of testes from 12-week-old PKnox1-GKO and control mice. Bars are the mean and standard deviation of the weight of testes. (n = 4 for each genotype). Tissue sections from control and GKO epididymis stained with H&E (C, D) and testes stained with H&E (E, F), TUNEL (G, H) and anti-SCP3 antibody with DAPI (I, J). Data are representative of 4 independent experiments. Scale bars, 50 μm.
Defects in the spermatogonia populations upon PKnox1 loss
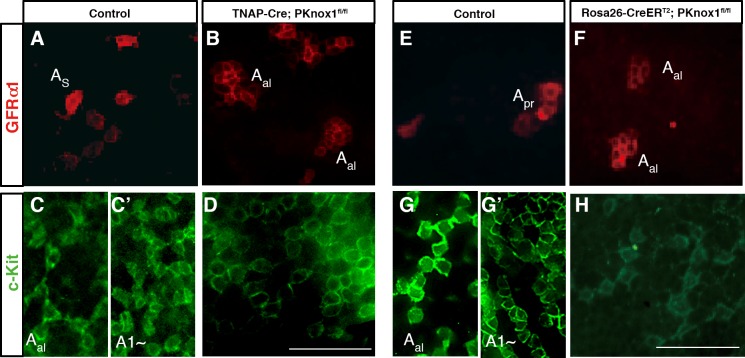
We further examined the differentiation of spermatogonia in the PKnox1-GKO and–CKO testis by whole-mount immunohistochemistry. This analysis reveled that GFRα1+ cells, corresponding to As~pr spermatogonia in the control testis (Fig 4A and 4E), form Aal-like morphologies in both PKnox1-GKO and -CKO testes (Fig 4B and 4F). Furthermore, various developmental stages of c-Kit+ cells, including late undifferentiated (Aal16-32) and differentiating spermatogonia (A1~), were observed in distinct areas of the seminiferous tubules from control testes, while the PKnox1-GKO testes contained relatively few c-Kit+ cells in which the level of c-Kit expression was weak, as compared to the control testes (S2 Fig). The detailed analysis revealed that c-Kit+ cells aligned more than 32 appear to be absent in the PKnox1-GKO and -CKO testes (Fig 4D and 4H), in contrast to the presence of Aal and A1-type spermatogonia in the control testes (Fig 4C, 4C’, 4G and 4G’).
Fig 4. Loss of PKnox1 causes accumulation of GFRα1+ cells and differentiation arrest of c-Kit+ spermatogonia.
Whole-mount immunodetection of cells expressing GFRα1 (red) and c-Kit (green) in seminiferous tubules of 12-week-old littermate controls (A, C, C’, E, G and G’), PKnox1-GKO (B, D), and -CKO mice (F, H). As; single cell, Apr; paired cells, Aal; aligned cells. Data are representative of 3 independent experiments. Scale bars, 50μm.
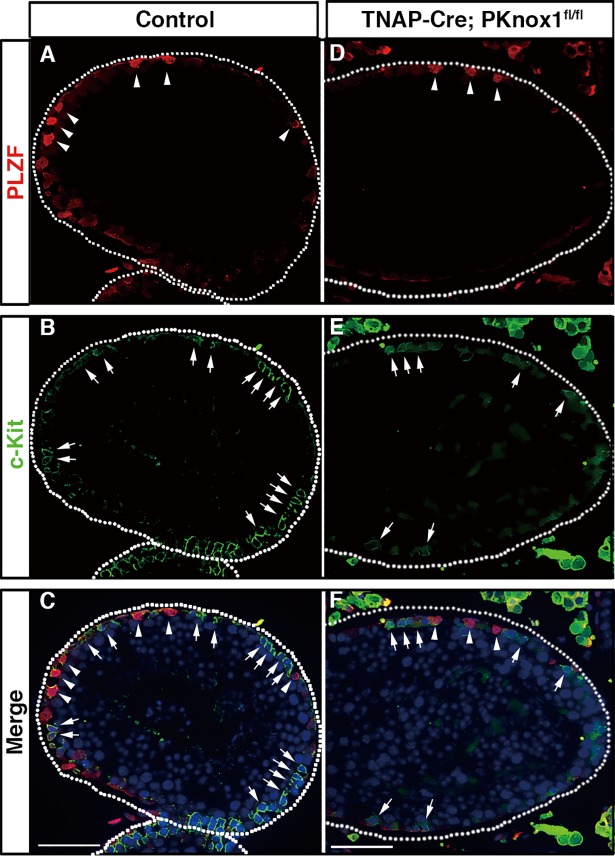
To further understand the defects in spermatogonia differentiation in the PKnox1-deficient testes, we examined whether sequential differentiation of spermatogonia occurs in PKnox1-GKO testes by simultaneous identification of markers differentially expressed during early to late spermatogonia differentiation, PLZF and c-Kit. PLZF is a transcription factor expressed in the early stage of spermatogonia differentiation, and is required not only for self-renewal[6,7] but also for the maintenance of spermatogonial stem cells in an undifferentiated state by repressing transcription of the c-Kit gene[30]. Thus, the expression of c-Kit begins after the cessation of PLZF expression at the later stage of spermatogonial cell differentiation. As similar to the non-overlapping pattern of PLZF and c-Kit expression in the control testes (Fig 5A–5C), distinct populations of germ cells expressed PLZF and c-Kit in the PKnox1-GKO testes (Fig 5D–5F), suggesting that PKnox1 is dispensable for initial differentiation of undifferentiated spermatogonia to the c-Kit+ stage.
Fig 5. PLZF and c-Kit are expressed in a distinct subset of spermatogonia in PKnox1-deficient testes.
Immunohistochemical analysis of PLZF (A, D) and c-Kit (B, E) expression in the testis from 12-week-old PKnox1-GKO and control mice with merged images (C, F). Tissue sections were double stained with the indicated combinations of antibodies and counterstained with DAPI. Arrowheads and arrows indicate PLZF+ and c-Kit+ cells, respectively. Broken lines indicate the basement membrane of seminiferous tubules. Data are representative of 3 independent experiments. Scale bars, 50 μm.
PKnox1 is required for differentiation of the c-Kit+ spermatogonia
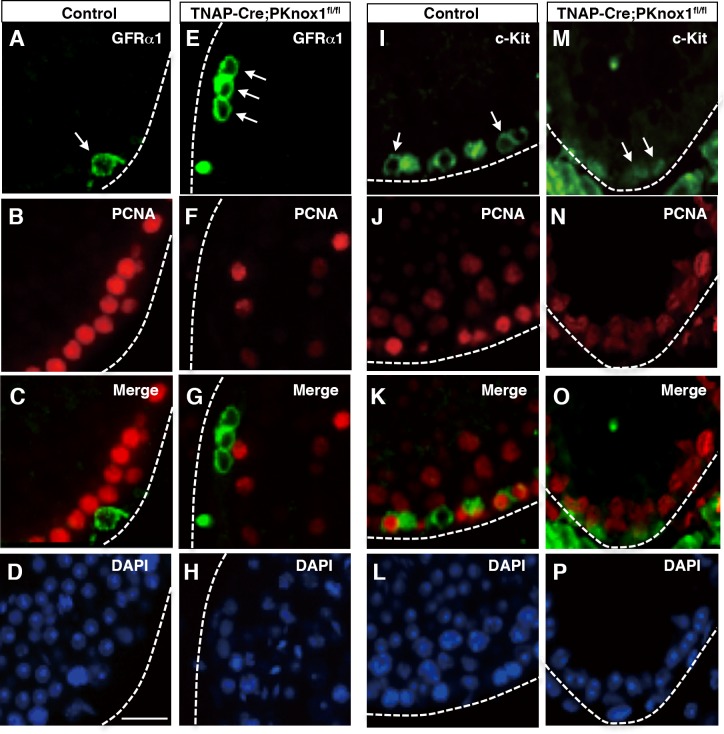
Given that spermatogenesis proceeded at least to the c-Kit+ spermatogonial differentiation stage but subsequent stages including meiosis were absent in the PKnox1-GKO testes, we finally focused our attention on alterations in the c-Kit+ stage of spermatogonia differentiation, when rapid cell proliferation occurs through signals from c-Kit[31]. In the control testes, GFRα1+ cells, representing the most immature spermatogonial cells, were negative for PCNA, one of markers of proliferating cells (Fig 6A–6D), whereas c-Kit+ spermatogonial cells expressed a high level of PCNA(Fig 6I–6L). Although GFRα1+ cells in in the PKnox1-GKO testes were PCNA-negative (Fig 6E–6H), as similar to the control testes, c-Kit+ spermatogonia in the PKnox1-GKO testes appeared to be negative for PCNA (Fig 6M–6P), suggestive of a possibility that proliferation defects in PKnox1-defficient c-Kit+ spermatogonia might be associated with the subsequent differentiation arrest of spermatogenesis.
Fig 6. Loss of PKnox1 affects PCNA expression in c-Kit+ spermatogonia.
Tissue sections from the testis from 12-week-old PKnox1-GKO and control mice were double stained with the indicated antibody combinations and counterstained with DAPI. Arrows indicate GFRα1+ and c-Kit+ cells with or without PCNA expression. Broken lines indicate the basement membrane of seminiferous tubules. Data are representative of 3 independent experiments. Scale bars, 25 μm.
Discussion
Here we demonstrate that PKnox1 is critical for the maintenance of spermatogenesis in the post pubertal testis and is prerequisite for sustained spermatogenesis in adult male mice. Tamoxifen-mediated deletion of PKnox1 in the adult testes as well as its germ cell-selective ablation causes testis hypotrophy with germ cell apoptosis and, as a consequence, compromised spermatogenesis resulting in the loss of germ cells at the stage of meiosis.
Upon PKnox1 inactivation, the undifferentiated spermatogonia expressing GFRα1 appeared to form clusters. There might be two potential explanations for this phenotype after PKnox1 loss, abnormal differentiation and/or proliferation of spermatogonia. In this regard, while c-Kit+, but not GFRα1+, spermatogonia in the control testes expressed the cell proliferation marker, PCNA, neither of these spermatogonia populations in PKnox1-GKO testes expressed PCNA. Thus, the appearance of Aal-like GFRα1+ spermatogonia is not likely to be a consequence of proliferation of GFRα1+ spermatogonia themselves. Furthermore, as PLZF represses c-Kit transcription in the As and Apr undifferentiated spermatogonia to maintain these cells in the G1/G0 phase of cell cycle[30], PLZF expression was not detected in the more differentiated c-Kit+ stage of spermatogonia in the PKnox1-GKO testes, indicating that differentiation of GFRα1+spermatogonia to the c-Kit+ stage occurs in the absence of PKnox1. Therefore, it seems reasonable to propose that the accumulation of undifferentiated GFRα1+ spermatogonia observed in the absence of PKnox1 results from an indirect consequence of a failure in maintenance and/or subsequent differentiation of the more differentiated c-Kit+ stage of spermatogonia[32]. Combining the previous findings that spermatogonia enter the S phase of cell cycle after differentiation into the A1 stage[33,34] and our present data that PKnox1-deficient c-Kit+ spermatogonia were PCNA-negative, the prime spermatogenic defect in PKnox1 deficiency appears to be a failure of the c-Kit+ Aal spermatogonia to differentiate into A1 spermatogonia. As a result, most seminiferous tubules of PKnox1-deficient testes only contain As, Apr and Aal spermatogonia, with a loss of subsequent differentiation stages of germ cells.
The importance of c-Kit-mediated signaling, particularly that mediated by phosphatidylinositol 3’-kinase (PI3K), in spermatogenesis is well established[12]. Mice bearing a point mutation in c-Kit that specifically disrupts a binding site for the p85 subunit of PI3-kinase fail to produce sperm due to decreased proliferation by the cKit+ spermatogonia and increased apoptosis, probably caused by the arrest of subsequent spermatogenic differentiation[35]. In the PKnox1-deficient testes, Aal spermatogonia expressed c-Kit, albeit its expression level was weaker than that of controls, and appeared to fail to proliferate, suggesting possible defects in c-Kit-mediated signaling. Although there have been no studies indicating a possible function of PKnox1 in c-Kit-mediated signaling, Meis3, another member of the PKnox/Meis family of transcription factors, was reported to transcriptionally regulate expression of PDK1, a serine/threonine kinase functioning downstream of PI3K[36]. The potential involvement of PKnox1 in c-Kit-mediated signaling warrants further investigation.
Considering the binding partners and potential upstream regulators of PKnox1 expressed in the spermatogenic process, Pbx4 is preferentially expressed in pachytene spermatocytes corresponding to meiotic spermatocytes, but not in spermatogonia[23]. In addition, a potential upstream regulator of TALE family transcription factors, MLL5, is thought to participate only in the late stage of spermatogenesis, as its deficiency causes defects predominantly in the post meiotic stages of spermatogenesis that lead to spermatozoa[37]. Since PKnox1 is expressed not only in spermatogonia but also in haploid germ cells, PKnox1 might have additional roles in the post meiotic stage of spermatogenesis in conjunction with Pbx4 and MLL5, such as activation of Pgk2 gene expression, as previously suggested[24,25].
Overall, our findings strongly indicate that PKnox1 represents a new intrinsic regulator of maintenance and subsequent differentiation of the c-Kit+ stage of spermatogonia, adding important insight into our understanding of the molecular mechanisms regulating spermatogenesis, a process that is essential for sustaining the germline in adulthood and is therefore required for male fertility.
Materials and methods
Ethics statement
All animal experiments were carried out under the ethical guidance of Tokyo University of Science, and protocols were reviewed and approved by the Tokyo University of Science Animal Care and Use Committee.
Mice and gene targeting
All animal experiments were carried out in accordance with the ethical guidance of Tokyo University of Science, and protocols were reviewed and approved by the Tokyo University of Science Animal Care and Use Committee. All of the genomic fragments used in constructing the targeting vector for the PKnox1 conditional allele, including a 0.9-kb genomic fragment immediately upstream of the loxP-flanked 1.2-kb fragment containing exon 3 of the PKnox1 gene and a 5.5-kb fragment immediately downstream of exon 3, were obtained from a C57BL/6 bacterial artificial chromosome (BAC) clone (RP23-99E16, Invitrogen) by using the RED/ET system (Gene Bridge GmbH, Heidelberg, Germany). These fragments were assembled in a modified pBluescript II SK vector containing a PGK promoter-driven FRT-flanked neomycin-resistant gene and an MC1 promoter-driven diphtheria toxin gene. The linearized targeting vector was electroporated into Bruce-4 ES cells and drug-resistant colonies were screened for homologous recombination. Targeted clones were injected into BALB/c blastocysts and the resultant chimeric mice were bred to produce progeny with germ line transmission of the mutated allele. F1 progeny harboring a targeted PKnox1 allele were then crossed with ubiquitous CAG promoter-driven FLPe mice on a C57BL/6 background[38] (obtained from RIKEN Bioresource Center) to remove the FRT-flanked neomycin-resistant gene cassette. Rosa26-CreERT2[28] and TNAP-Cre mice[29] on a C57BL/6 background were provided from Drs. T. Ludwig and A. Nagy, respectively. C57BL/6, BALB/c, and W/Wv mice were purchased from Japan SLC Inc. (Hamamatsu Japan). Mice were euthanized under general anesthesia with an isoflurane overdose (5%). The PCR primers for genotyping are listed in S1 Table.
Tamoxifen treatment
Tamoxifen (final concentration: 10 mg/ml, Sigma-Aldrich, St Louis, MO) was prepared as described previously[39]. In brief, 50 mg of tamoxifen was dissolved into 500 μl of ethanol at 55°C followed by addition of 4.5 ml of warmed sunflower oil and mixed thoroughly by vortexing. The solution was then filtered and stored at -20°C. For in vivo administration, 5 mg tamoxifen/30 g per body weight was delivered by intragastric gavage for three consecutive days to 3-month-old male mice.
RT-PCR
Total RNA was isolated using Trizol reagent (Invitrogen) and first-strand cDNA was synthesized using oligo (dT) primers and the Superscript RT-PCR kit (Invitrogen). Each cDNA sample was then subjected to PCR. Amplified signals were confirmed to be single bands by gel electrophoresis and were normalized to β-actin. Primer sequences and PCR conditions are listed in S1 Table.
Immunoblotting
Testes lysed in RIPA buffer were separated by SDS-PAGE and transferred to PVDF membranes. The blots were then probed with anti-PKnox1/Prep1 (Abcam, Cambridge, MA, USA) and anti-GAPDH (Santa Cruz, CA) antibody, followed by HRP-conjugated secondary antibodies and then visualized by enhanced chemiluminescence (ECL plus). Blots were scanned and analyzed using a Luminescent image analyzer (LAS-3000, FUJIFILM, Japan).
Immunohistochemistry
The testis samples were fixed with 4% paraformaldehyde (PFA) in PBS at 4°C for 16 hours, dehydrated in ethanol, cleared in xylene, and then routinely embedded in paraffin. The deparaffinized tissue sections were blocked with PBS containing 3% bovine serum albumin (Sigma) then incubated with primary antibodies diluted in Can Get Signal Immunostain buffer (Toyobo) at 4°C for 12 hours followed by a one-hour incubation at room temperature with the appropriate secondary antibodies. The following primary antibodies were used: rabbit anti-SCP3 (1:100 dilution; Santa Cruz), goat anti-mouse c-Kit (1:100 dilution; R&D systems), goat anti-mouse GFRα1 (1:100 dilution; R&D systems), mouse anti-PCNA (1:100 dilution; Santa Cruz), mouse anti-PLZF (1:100 dilution; Santa Cruz) and mouse anti-PKnox1/Prep1 (1:100 dilution; Abcam). Secondary antibodies were anti-rabbit IgG antibody conjugated with Alexa Fluor 488 or 647 (Cell Signaling), anti-mouse IgG antibody conjugated with Alexa Fluor 633 or 488 (Invitrogen), or anti-goat IgG antibody conjugated with Alexa Fluor 633. Nuclei were counterstained with Prolong Gold antifade reagent with DAPI (Invitrogen). For whole-mount immunohistochemistry, testes were removed from the tunica albuginea, fixed in 4% PFA for 16 hours at 4°C and washed with cold PBS. The seminiferous tubule fragments were incubated with goat anti-mouse GFRα1 (1:100 dilution; R&D systems), goat anti-mouse c-Kit (1:100 dilution; R&D systems) antibodies at 4°C for 12 hours. After washing with PBS, the samples were incubated with Alexa-488/555 conjugated secondary antibodies at room temperature for two hours. After counter-staining with DAPI, the samples were analyzed using a BIOREVO BZ-9000 microscope (KEYENCE). Whole mount samples of the seminiferous tubule were photographed (×400).
Apoptosis analysis
Paraffin sections were deparaffinized and subjected to the terminal deoxynucleotydyl transferase-mediated dUTP nick-end labeling (TUNEL) assay using the In Situ Cell Death Detection Kit (Roche, USA). The TUNEL working procedure was carried out following the manufacturer's protocols.
Statistical analysis
Statistical significance was calculated with the unpaired two-tailed Student’s t-test. Data were considered statistically significant when p values were less than 0.05.
Supporting information
(A) Diagram depicting exon 3 of the Prep1 locus and the targeting strategy used to generate mutant PKnox1 alleles (floxed and deleted alleles). LoxP sites (arrowheads) were inserted into intronic sites flanking exon 3 of the PKnox1 gene. The FRT-flanked neomycin gene (PGK-neo) selection cassette was removed by crossing to CAG-FLPe mice. PCR primers for verifying the Cre-mediated deletion of the loxP-flanked fragment are indicated by arrows. DT-A, a diphtheria toxin negative selection cassette; B, BamHI: E, EcoRI. (B) Confirmation of PKnox1 deletion. Genomic DNAs from testes of Rosa26-CreERT2; PKnox1fl/fl and Rosa26-CreERT2; PKnox1fl/+ mice that were treated with tamoxifen 3 weeks previously were subjected to PCR analysis using the indicated primer pairs. (C) Western blot analysis to confirm loss of PKnox1 protein expression. Whole-cell lysates from testes of Rosa26-CreERT2; PKnox1fl/fl and Rosa26-CreERT2; PKnox1fl/+ mice that were treated with tamoxifen 3 weeks previously were blotted with an anti-PKnox1 antibody. After stripping, the filter was reprobed with an anti-GAPDH antibody.
(TIF)
Distribution of c-Kit+ cells in the seminiferous tubules of 12-week-old littermate controls (upper panels) and PKnox1-GKO mice (lower panels). Arrows indicate c-Kit+ cells in PKnox1-GKO testes. Asterisks indicate the area of the seminiferous tubules lacking c-Kit+ cells in PKnox1-GKO testes.
(TIF)
(DOC)
Acknowledgments
The authors would like to thank Drs. Thomas Ludwig and Andras Nagy for providing ROSA26-CreERT2 mice and TNAP-Cre mice, respectively, and Dr. Peter D. Burrows for critical reading of the manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the Grant-in-Aid for Scientific Research (KAKENHI 22780264 and 24780290 to YK and 25292194 and 25111513 to RG) of the Japan Society for the Promotion of Science. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hara K, Nakagawa T, Enomoto H, Suzuki M, Yamamoto M, et al. (2014) Mouse spermatogenic stem cells continually interconvert between equipotent singly isolated and syncytial states. Cell Stem Cell 14: 658–672. doi: 10.1016/j.stem.2014.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shirakawa T, Yaman-Deveci R, Tomizawa S, Kamizato Y, Nakajima K, et al. (2013) An epigenetic switch is crucial for spermatogonia to exit the undifferentiated state toward a Kit-positive identity. Development 140: 3565–3576. doi: 10.1242/dev.094045 [DOI] [PubMed] [Google Scholar]
- 3.Nakagawa T, Sharma M, Nabeshima Y, Braun RE, Yoshida S (2010) Functional hierarchy and reversibility within the murine spermatogenic stem cell compartment. Science 328: 62–67. doi: 10.1126/science.1182868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Rooij DG, Russell LD (2000) All you wanted to know about spermatogonia but were afraid to ask. J Androl 21: 776–798. [PubMed] [Google Scholar]
- 5.Yang QE, Oatley JM (2014) Spermatogonial stem cell functions in physiological and pathological conditions. Curr Top Dev Biol 107: 235–267. doi: 10.1016/B978-0-12-416022-4.00009-3 [DOI] [PubMed] [Google Scholar]
- 6.Buaas FW, Kirsh AL, Sharma M, McLean DJ, Morris JL, et al. (2004) Plzf is required in adult male germ cells for stem cell self-renewal. Nat Genet 36: 647–652. doi: 10.1038/ng1366 [DOI] [PubMed] [Google Scholar]
- 7.Costoya JA, Hobbs RM, Barna M, Cattoretti G, Manova K, et al. (2004) Essential role of Plzf in maintenance of spermatogonial stem cells. Nat Genet 36: 653–659. doi: 10.1038/ng1367 [DOI] [PubMed] [Google Scholar]
- 8.Falender AE, Freiman RN, Geles KG, Lo KC, Hwang K, et al. (2005) Maintenance of spermatogenesis requires TAF4b, a gonad-specific subunit of TFIID. Genes Dev 19: 794–803. doi: 10.1101/gad.1290105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ballow D, Meistrich ML, Matzuk M, Rajkovic A (2006) Sohlh1 is essential for spermatogonial differentiation. Dev Biol 294: 161–167. doi: 10.1016/j.ydbio.2006.02.027 [DOI] [PubMed] [Google Scholar]
- 10.Toyoda S, Miyazaki T, Miyazaki S, Yoshimura T, Yamamoto M, et al. (2009) Sohlh2 affects differentiation of KIT positive oocytes and spermatogonia. Dev Biol 325: 238–248. doi: 10.1016/j.ydbio.2008.10.019 [DOI] [PubMed] [Google Scholar]
- 11.Meng X, Lindahl M, Hyvonen ME, Parvinen M, de Rooij DG, et al. (2000) Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science 287: 1489–1493. [DOI] [PubMed] [Google Scholar]
- 12.Zhang L, Tang J, Haines CJ, Feng HL, Lai L, et al. (2011) c-kit and its related genes in spermatogonial differentiation. Spermatogenesis 1: 186–194. doi: 10.4161/spmg.1.3.17760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hogarth CA, Griswold MD (2010) The key role of vitamin A in spermatogenesis. J Clin Invest 120: 956–962. doi: 10.1172/JCI41303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Longobardi E, Penkov D, Mateos D, De Florian G, Torres M, et al. (2014) Biochemistry of the tale transcription factors PREP, MEIS, and PBX in vertebrates. Dev Dyn 243: 59–75. doi: 10.1002/dvdy.24016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Piper DE, Batchelor AH, Chang CP, Cleary ML, Wolberger C (1999) Structure of a HoxB1-Pbx1 heterodimer bound to DNA: role of the hexapeptide and a fourth homeodomain helix in complex formation. Cell 96: 587–597. [DOI] [PubMed] [Google Scholar]
- 16.Knoepfler PS, Calvo KR, Chen H, Antonarakis SE, Kamps MP (1997) Meis1 and pKnox1 bind DNA cooperatively with Pbx1 utilizing an interaction surface disrupted in oncoprotein E2a-Pbx1. Proc Natl Acad Sci U S A 94: 14553–14558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rieckhof GE, Casares F, Ryoo HD, Abu-Shaar M, Mann RS (1997) Nuclear translocation of extradenticle requires homothorax, which encodes an extradenticle-related homeodomain protein. Cell 91: 171–183. [DOI] [PubMed] [Google Scholar]
- 18.Berthelsen J, Zappavigna V, Mavilio F, Blasi F (1998) Prep1, a novel functional partner of Pbx proteins. EMBO J 17: 1423–1433. doi: 10.1093/emboj/17.5.1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ryoo HD, Marty T, Casares F, Affolter M, Mann RS (1999) Regulation of Hox target genes by a DNA bound Homothorax/Hox/Extradenticle complex. Development 126: 5137–5148. [DOI] [PubMed] [Google Scholar]
- 20.Ferretti E, Schulz H, Talarico D, Blasi F, Berthelsen J (1999) The PBX-regulating protein PREP1 is present in different PBX-complexed forms in mouse. Mech Dev 83: 53–64. [DOI] [PubMed] [Google Scholar]
- 21.Iotti G, Longobardi E, Masella S, Dardaei L, De Santis F, et al. (2011) Homeodomain transcription factor and tumor suppressor Prep1 is required to maintain genomic stability. Proc Natl Acad Sci U S A 108: E314–322. doi: 10.1073/pnas.1105216108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Penkov D, Di Rosa P, Fernandez Diaz L, Basso V, Ferretti E, et al. (2005) Involvement of Prep1 in the alphabeta T-cell receptor T-lymphocytic potential of hematopoietic precursors. Mol Cell Biol 25: 10768–10781. doi: 10.1128/MCB.25.24.10768-10781.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wagner K, Mincheva A, Korn B, Lichter P, Popperl H (2001) Pbx4, a new Pbx family member on mouse chromosome 8, is expressed during spermatogenesis. Mech Dev 103: 127–131. [DOI] [PubMed] [Google Scholar]
- 24.Yoshioka H, Geyer CB, Hornecker JL, Patel KT, McCarrey JR (2007) In vivo analysis of developmentally and evolutionarily dynamic protein-DNA interactions regulating transcription of the Pgk2 gene during mammalian spermatogenesis. Mol Cell Biol 27: 7871–7885. doi: 10.1128/MCB.00990-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Z, Yoshioka H, McCarrey JR (2013) Sequence-specific promoter elements regulate temporal-specific changes in chromatin required for testis-specific activation of the Pgk2 gene. Reproduction 146: 501–516. doi: 10.1530/REP-13-0311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng LX, Ravindranath N, Dym M (2000) Stem cell factor/c-kit up-regulates cyclin D3 and promotes cell cycle progression via the phosphoinositide 3-kinase/p70 S6 kinase pathway in spermatogonia. J Biol Chem 275: 25572–25576. doi: 10.1074/jbc.M002218200 [DOI] [PubMed] [Google Scholar]
- 27.Dolci S, Pellegrini M, Di Agostino S, Geremia R, Rossi P (2001) Signaling through extracellular signal-regulated kinase is required for spermatogonial proliferative response to stem cell factor. J Biol Chem 276: 40225–40233. doi: 10.1074/jbc.M105143200 [DOI] [PubMed] [Google Scholar]
- 28.Nie Y, Han YC, Zou YR (2008) CXCR4 is required for the quiescence of primitive hematopoietic cells. J Exp Med 205: 777–783. doi: 10.1084/jem.20072513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lomeli H, Ramos-Mejia V, Gertsenstein M, Lobe CG, Nagy A (2000) Targeted insertion of Cre recombinase into the TNAP gene: excision in primordial germ cells. Genesis 26: 116–117. [PubMed] [Google Scholar]
- 30.Filipponi D, Hobbs RM, Ottolenghi S, Rossi P, Jannini EA, et al. (2007) Repression of kit expression by Plzf in germ cells. Mol Cell Biol 27: 6770–6781. doi: 10.1128/MCB.00479-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rossi P, Sette C, Dolci S, Geremia R (2000) Role of c-kit in mammalian spermatogenesis. J Endocrinol Invest 23: 609–615. doi: 10.1007/BF03343784 [DOI] [PubMed] [Google Scholar]
- 32.Nagai R, Shinomura M, Kishi K, Aiyama Y, Harikae K, et al. (2012) Dynamics of GFRalpha1-positive spermatogonia at the early stages of colonization in the recipient testes of W/Wnu male mice. Dev Dyn 241: 1374–1384. doi: 10.1002/dvdy.23824 [DOI] [PubMed] [Google Scholar]
- 33.Huckins C (1971) Cell cycle properties of differentiating spermatogonia in adult Sprague-Dawley rats. Cell Tissue Kinet 4: 139–154. [DOI] [PubMed] [Google Scholar]
- 34.Oakberg EF (1971) Spermatogonial stem-cell renewal in the mouse. Anat Rec 169: 515–531. doi: 10.1002/ar.1091690305 [DOI] [PubMed] [Google Scholar]
- 35.Kissel H, Timokhina I, Hardy MP, Rothschild G, Tajima Y, et al. (2000) Point mutation in kit receptor tyrosine kinase reveals essential roles for kit signaling in spermatogenesis and oogenesis without affecting other kit responses. EMBO J 19: 1312–1326. doi: 10.1093/emboj/19.6.1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu J, Wang Y, Birnbaum MJ, Stoffers DA (2010) Three-amino-acid-loop-extension homeodomain factor Meis3 regulates cell survival via PDK1. Proc Natl Acad Sci U S A 107: 20494–20499. doi: 10.1073/pnas.1007001107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yap DB, Walker DC, Prentice LM, McKinney S, Turashvili G, et al. (2011) Mll5 is required for normal spermatogenesis. PLoS One 6: e27127 doi: 10.1371/journal.pone.0027127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kanki H, Suzuki H, Itohara S (2006) High-efficiency CAG-FLPe deleter mice in C57BL/6J background. Exp Anim 55: 137–141. [DOI] [PubMed] [Google Scholar]
- 39.Hirayama T, Asano Y, Iida H, Watanabe T, Nakamura T, et al. (2014) Meis1 is required for the maintenance of postnatal thymic epithelial cells. PLoS One 9: e89885 doi: 10.1371/journal.pone.0089885 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Diagram depicting exon 3 of the Prep1 locus and the targeting strategy used to generate mutant PKnox1 alleles (floxed and deleted alleles). LoxP sites (arrowheads) were inserted into intronic sites flanking exon 3 of the PKnox1 gene. The FRT-flanked neomycin gene (PGK-neo) selection cassette was removed by crossing to CAG-FLPe mice. PCR primers for verifying the Cre-mediated deletion of the loxP-flanked fragment are indicated by arrows. DT-A, a diphtheria toxin negative selection cassette; B, BamHI: E, EcoRI. (B) Confirmation of PKnox1 deletion. Genomic DNAs from testes of Rosa26-CreERT2; PKnox1fl/fl and Rosa26-CreERT2; PKnox1fl/+ mice that were treated with tamoxifen 3 weeks previously were subjected to PCR analysis using the indicated primer pairs. (C) Western blot analysis to confirm loss of PKnox1 protein expression. Whole-cell lysates from testes of Rosa26-CreERT2; PKnox1fl/fl and Rosa26-CreERT2; PKnox1fl/+ mice that were treated with tamoxifen 3 weeks previously were blotted with an anti-PKnox1 antibody. After stripping, the filter was reprobed with an anti-GAPDH antibody.
(TIF)
Distribution of c-Kit+ cells in the seminiferous tubules of 12-week-old littermate controls (upper panels) and PKnox1-GKO mice (lower panels). Arrows indicate c-Kit+ cells in PKnox1-GKO testes. Asterisks indicate the area of the seminiferous tubules lacking c-Kit+ cells in PKnox1-GKO testes.
(TIF)
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.