Abstract
We previously reported the involvement of zinc-finger protein 143 (ZNF143) on cancer cell motility in colon cancer cells. Here, ZNF143 was further characterized in breast cancer. Immunohistochemistry was used to determine the expression of ZNF143 in normal tissues and in tissues from metastatic breast cancer at various stages. Notably, ZNF143 was selectively expressed in duct and gland epithelium of normal breast tissues, which decreased when the tissue became malignant. To determine the molecular mechanism how ZNF143 affects breast cancer progression, it was knocked down by infecting benign breast cancer cells with short-hairpin (sh) RNA-lentiviral particles against ZNF143 (MCF7 sh-ZNF143). MCF7 sh-ZNF143 cells showed different cell-cell contacts and actin filament (F-actin) structures when compared with MCF7 sh-Control cells. In migration and invasion assays, ZNF143 knockdown induced increased cellular motility in breast carcinoma cells. This was reduced by the recovery of ZNF143 expression. Taken together, these results suggest that ZNF143 expression contributes to breast cancer progression.
Keywords: Invasive ductal carcinoma, Metastasis, Motility, ZNF143
INTRODUCTION
Breast cancer is a heterogeneous disorder that is one of the most common cancers. Li and colleagues categorized 135,157 invasive breast cancer cases into nine histological subtypes based on surveillance, epidemiology and end result program data that showed several characteristics of histological subtypes (1). The most common histological types of invasive breast cancer are infiltrating ductal carcinoma (IDC) and infiltrating lobular carcinoma (ILC), which have been compared in many studies. IDC showed increased invasion of vessels, less differentiation, and decreased tumor size when compared with ILC (1, 2).
Since Perou and colleagues suggested four subtypes of human breast cancers including the oestrogen receptor α (ER)-positive/luminal-like, basal-like, ERBB2-positive, and a normal breast type based on gene expression patterns (3), a number of studies have reported specific molecular characteristics of breast cancer subtypes (4, 5). Recent studies have also reported relationships between molecular subtypes and histological phenotypes of breast cancers (6, 7). However, there is still limited knowledge of the relationships between histological subtypes and molecular subtypes, and little is known about how the breast cancer signature genes communicate during progression within the tumor.
During epithelial to mesenchymal transition (EMT) in tumor progression, transformed epithelial cells lose their tight interactions with neighboring cells, and become mobile by deconstructing cell junctions and polarities, and by changing cytoskeletal components after altered gene expression (8, 9). For example, the downregulation of E-cadherin, an important component of adherens junctions, is a marker for the EMT. The intermediate filament proteins, such as vimentin and α-smooth muscle actin, increased, while cytokeratins decreased in epithelial cells during the EMT (10, 11). In the early stages of the EMT, several transcription factors such as SNAI1, TWIST, and zinc finger E-box-binding (ZEB) transcription factors, the so-called “EMT drivers,” were activated and found to be involved in coordinating the repression of epithelial genes and the induction of the mesenchymal genes (8).
Since zinc-finger protein 143 (ZNF143) was identified as a human homolog of the Xenopus selenocysteine tRNA gene transcription activating factor (Staf) (12), accumulating evidence has suggested roles of ZNF143 in a variety of cellular and pathogenic processes (13–22). When injected with morpholino antisense oligonucleotides that target ZNF143 mRNA, developing zebrafish embryos showed phenotypic abnormalities, suggesting a transcriptional regulatory role of ZNF143 during development (14). ZNF143 has also been shown to contribute to chromatin interactions and gene expression at gene promoters, by acting as a chromatin-looping factor through sequence specificity (13), suggesting a role of ZNF143 beyond that of a general transcription factor. Carbon et al. reported that two ZNF143 binding sites, SBS1 and SBS2, were modulated by Notch1 and THAP11, as well as by ZNF143, in T-acute lymphoblastic leukemia (16), suggesting that ZNF143 is involved in tumor development and that ZNF143 targeting of genes is regulated by other transcription factors as well as by ZNF143 in cancer cells. Besides leukemia, ZNF143 has been studied in a variety of cancers such as lung adenocarcinoma (19), colon cancer cells (21), prostate cancer (23), gastric cancer (24) and breast cancers (25).
ZNF143 has reported to modulate cell survival by regulating glutathione peroxidase 1 activity through the transcriptional activation of selenocysteine transfer RNA (22, 26). Kohno and colleagues suggested that ZNF143 regulates DNA replication and cell-cycle-associated genes involved in cell growth and proliferation (23). We also reported that ZNF143 knockdown enhanced cellular motility by regulating ZEB1 and E-cadherin in colon cancer cells (21). Despite these results, there is still limited knowledge of the possible roles of ZNF143 in tumor malignancy. The possible relationships between the expression and function of ZNF143 in cancer cells and tissues should therefore be validated to establish definitively the role of ZNF143 in cancer biology. In the present study, we thus characterized the relationships between ZNF143 expression in tissues and the role of ZNF143 during tumor progression, especially in breast cancers.
RESULTS
ZNF143 expression decreases during tumor progression in breast tissues
Some studies have suggested a role for ZNF143 in tumor progression and malignancy in lung adenocarcinoma (19) and colon cancer cells (21). However, the expression levels of ZNF143 and the function of ZNF143 in cancer development are still unknown.
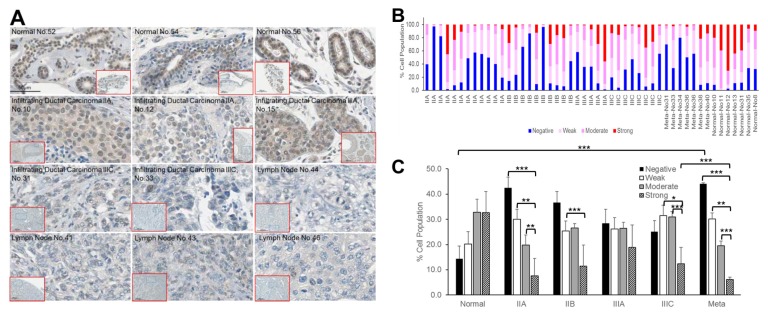
We therefore characterized the role of ZNF143 in breast cancer tissues and cancer cells, by determining its expression in breast tissues using immunohistochemistry (Fig. 1). Breast tissues on tissue arrays (SuperBioChips Laboratories, Slide CBA4; Fig. S1) were stained with hematoxylin and mouse anti-ZNF143 antibody, and manually delineated ROIs were extracted from the digitized slide data. A total of 90 tumor and normal images (from 42 tumors and seven normal tissues) were collected and used to extract features such as the intensities of ZNF143 and hematoxylin using HistoQuest® analysis software (TissueGnostics). This procedure quantitated the percentages of ZNF143-positive nuclei (weak, moderate, or strong intensities) relative to the total number of hematoxylin-positive nuclei per tumor. The results showed strong ZNF143 expression in normal breast tissues, especially in epithelial cells of ducts and glands (Fig. 1A). In advanced tumor tissues, ZNF143 intensity decreased compared with that in normal tissues. Although the intensities of ZNF143 in tumor tissues varied, the percentages of ZNF143-negative cells relative to whole hematoxylin-positive nuclei per tissue were higher in stage II, III, and metastasized tumors (25.1 ± 4.4–44.1 ± 0.4%) than in normal tissues (14.4 ± 5.1%) (Fig. 1B and C). Breast tissue arrays contained infiltrating ductal carcinoma (46 tissues), a sarcomatoid carcinoma (1 tissue), an atypical medullary carcinoma (1 tissue), a metastatic carcinoma (1 tissue), involving stage IIA, IIB, IIIA, IIIC and lymph nodes from metastatic carcinomas (10 tissues). As shown in Fig. 1A, ZNF143 was strongly detected in nuclei of ductal epithelial cells in normal breast tissues. In addition, most tissues from tissue arrays were from infiltrating ductal carcinoma. We therefore emphasized studies of invasive ductal carcinomas as a disease model as further work to define the possible relationships between ZNF143 and breast cancer malignancies. Low expression of ZNF143 significantly correlated with increased overall survival, as determined using an online meta-analysis tool (27), involving Kaplan-Meier analyses of breast cancer patients (Fig. S2; P < 0.05), suggesting a correlation between ZNF143 expression and breast cancer malignancy.
Fig. 1.
ZNF143 expression decreases tumor progression in invasive ductal carcinoma. (A) Breast tissues from a human breast tissue array (Super Bio Chips CBA4; human breast cancer, metastatic, or normal tissue) were stained with hematoxylin and mouse anti- ZNF143 antibody, then detected using DAB. Scale bar: 50 μm. (B) The percentages of ZNF143-positive nuclei (weak, moderate, or strong intensities) relative to the total number of hematoxylin- positive nuclei per tumor were quantitated using HistoQuest® software. (C) The graph depicts the mean percentage of (B) in each stage (IIA, IIB, IIIA, IIIC, metastasized, and normal). The data are expressed as means ± S. E. of at least three independent experiments. Statistical significance was assessed using the paired Student’s t-test (*P < 0.001; **P < 0.01; ***P < 0.05).
ZNF143 knockdown reduces cell-cell interactions and increases motility in breast cancer cells
To investigate further the molecular mechanism involving possible correlations between ZNF143 expression and breast cancer characteristics, we selected breast cancer cells originating from invasive ductal carcinomas, such as MCF7 and T47D cells (28). ZNF143 expression was knocked down by lentiviral particles against ZNF143 or control lentiviral particles in MCF7 cells (Fig. 2E and F), as described previously (29).
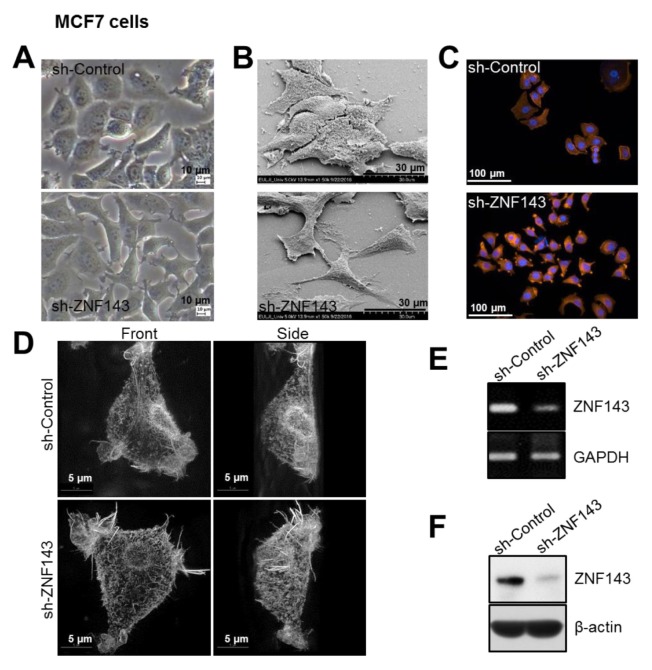
Fig. 2.
ZNF143 knockdown reduces cell-cell adhesion and induces morphological changes in MCF7 breast cancer cells. (A) Images of growing cells were captured at 20 × magnification using an inverted light microscope. (B) Subconfluent cells were fixed and characterized using SEM to show differences in cell-cell interactions in MCF sh-Control cells compared with sh-ZNF143 cells. The images were obtained at 1,500 × magnification. (C, D) Growing cells were fixed and stained with rhodamine-phalloidin and DAPI. Digital images were obtained at 20 × magnification by using a Cytation 3 cell imaging multi-mode reader (C) or at 60 × magnification using a DeltaVision OMXTM SR (D). (E, F) Subconfluent MCF7 sh-Control and MCF7 sh-ZNF143 cells were harvested for RT-PCR (E) and immunoblotting (F) to confirm the expression of ZNF143. All results shown are representative of at least three independent experiments.
Differences in cell morphology involved differences in cell shape and cell-cell interactions (Fig. 2A and B). Cell-cell interactions in MCF7 sh-Control cells were tight and close, in a manner similar to that in normal epithelial cells (Fig. 2A and B). In contrast, MCF7 sh-ZNF143 cells interacted with neighboring cells at certain loci and overall contacts between cells were decreased (Fig. 2A and B). Notably, MCF7 sh-ZNF143 cells showed a mesenchymal morphology. Among the many factors that could affect cellular morphology, we investigated cellular structural proteins such as F-actin (30, 31). Although the amount of F-actin was not significantly altered in MCF7 sh-Control and MCF7 sh-ZNF143 cells (data not shown), the cellular distribution of F-actin in MCF7 sh-ZNF143 cells differed from that in MCF7 sh-Control cells (Fig. 2C and D). These results suggested a possible role of ZNF143 in the EMT and/or cellular motility, as previously reported in colon cancer cells (21).
Recovery of ZNF143 expression increases cellular motility in breast cancer cells
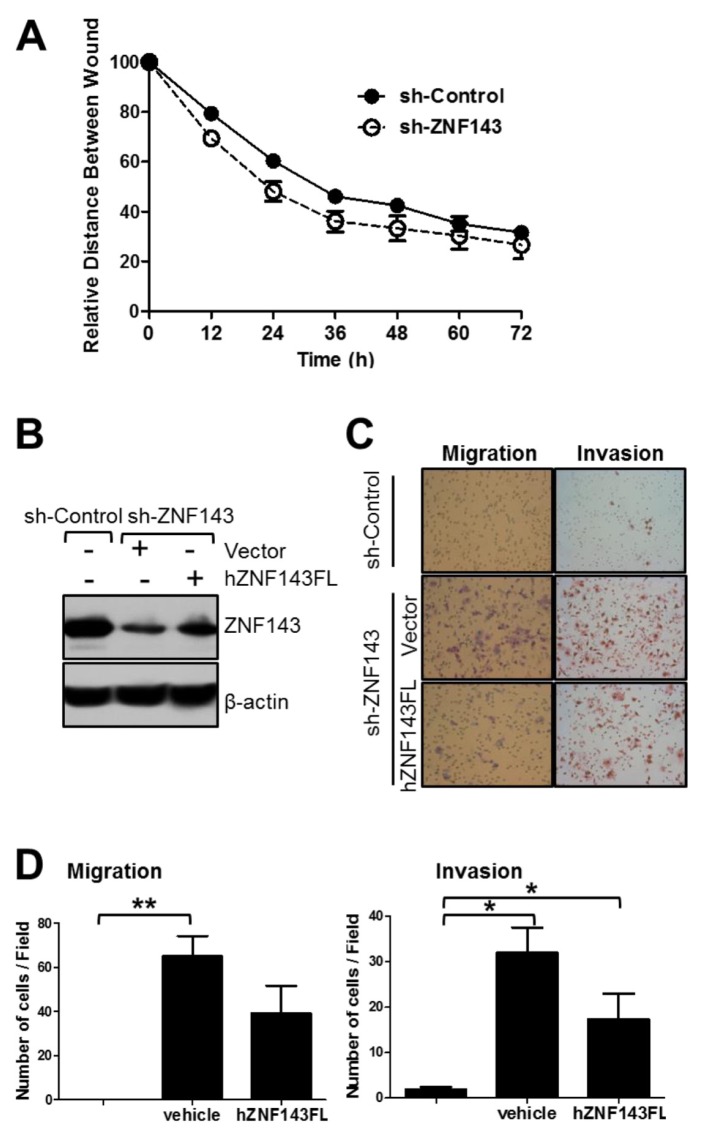
Three experimental approaches were performed to identify whether ZNF143 played a role in cellular motility in breast cancer cells. As expected, MCF7 sh-ZNF143 cells migrated and closed the wound faster than the MCF sh-Control cells (Fig. 3A), suggesting that ZNF143 knockdown contributed to epithelial cell characteristics by affecting cellular motility. To define the role of ZNF143 in cellular motility further, migration and invasion assays were performed (Fig. 3B–D). When cells were plated on the upper compartment of an 8 μm Transwell®, MCF7 sh-ZNF143 cells migrated more than MCF7 sh-Control cells, regardless of the presence of Matrigel®, which was consistent with the results from the wound healing assay. To clarify the involvement of ZNF143 in cellular motility, we induced the recovery of ZNF143 expression (Fig. 3B–D) by transfecting plasmids encoding human ZNF143 (21). As expected, the number of migrating cells passing through the transwell system was significantly reduced by expressing full-length human ZNF143 in MCF7 sh-ZNF143 cells, suggesting that the expression of ZNF143 at the protein level played a role in breast tumor cell motility.
Fig. 3.
ZNF143 knockdown increases the motility of MCF7 breast cancer cells. (A) Cells were grown until cells were fully confluent, and a uniform scratch was made down the center of the well, followed by washing with PBS to remove nonadherent cells. Digital images of the wound were obtained every 12 h until 72 h. The horizontal distance between the two wound sides was measured and statistically analyzed. Data are expressed as means ± S. E. of at least three independent experiments. Statistical significance was assessed using the paired Student’s t-test (***P < 0.0001). (B-D) MCF7 sh-Control and MCF7 sh-ZNF143 cells were transfected with pFLAG-CMV2-hZNF143FL or an empty vector using Lipofectamine® 2000, grown for 48 h, and harvested for migration (C, left panel, and d, left panel) and invasion (C, right panel, and D, right panel) assays, followed by immunoblotting (B). Cells were induced to move or invade through uncoated (migration) or Matrigel®-coated (invasion) Transwell® membranes. After 24 h, the migrated cells were fixed and stained. Digital images (C) were taken for cell counting (D). All results shown are representative of at least three independent experiments. Data are expressed as means ± S. E. of at least three independent experiments. Statistical significance was assessed using the paired Student’s t-test (*P < 0.0001; **P < 0.001).
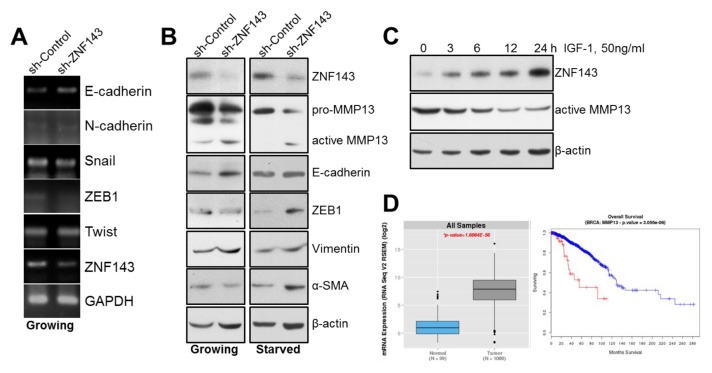
Some transcription factors that had been reported to induce tumor cell plasticity or the EMT were not changed by ZNF143 knockdown in MCF7 cells (Fig. 4A), implying the existence of an alternative mechanism for breast cancer cell motility or the existence of an alternative mechanism regulating the EMT. In a similar manner to the RT-PCR results, there was no significant difference in E-cadherin expression in MCF7 sh-ZNF143 cells compared with that in MCF7 sh-Control cells (Fig. 4B, left) at proteins. To determine the possible role of ZNF143 during the EMT in breast cancer cells, cells were starved for 15 h and the levels of specific proteins were determined (32). The levels of ZEB1, α-SMA, vimentin and active MMP13 increased in MCF sh-ZNF143 cells, suggesting a role for ZNF143 during the EMT (Fig. 4B, right and Fig. S3). Next, we investigated whether the altered proteins in ZNF143 knockdown cells correlated with tumorigenicity or survival in breast cancer patients. As shown in Fig. 4D, mRNA expression of the MMP13 gene in breast cancer patients was remarkably altered compared with that in normal breast tissues in TCGA datasets. In addition, survival analyses showed that higher than average expression levels of MMP13 correlated with poorer overall survival. The detailed mechanism how ZNF143 might be involved in MMP13 regulation is under being investigated.
Fig. 4.
ZNF143 is involved in vimentin and MMP13 expression in MCF7 breast cancer cells. (A) Subconfluent MCF7 sh-Control and MCF7 sh-ZNF143 cells were harvested, lysed, and analyzed for Snai1, ZEB1, and Twist by RT-PCR. (B) Growing or starved MCF7 sh-Control and MCF7 sh-ZNF143 cells were harvested, lysed, and analyzed for ZEB1, E-cadherin, and vimentin by immunoblotting. (C) Starved cells were incubated with 50 ng/ml IGF-1 for the indicated time periods and harvested for immunoblotting. All results shown are representative of at least three independent experiments. (D) Bioinformatics analyses of MMP13 in breast cancer patients. (left) The distribution of MMP13 gene expression in normal samples and breast cancer patients. The P-value = 1.6064 × 10−56. (right) In a similar manner as the clinical information dataset from the TCGA, higher levels of MMP13 expression were associated with reduced overall survival in the TCGA cohort. The log-rank value was 3.056 × 10−6. Patients with higher levels of MMP13 gene expression are shown in red and patients without higher levels of MMP13 gene expression are shown in blue.
DISCUSSION
The major findings of this study are as follows: (1) ZNF143 expression decreased with increasing stages in human breast cancer tissues; (2) ZNF143 knockdown altered cellular characteristics, including cell-cell interactions and motilities, which were recovered by ZNF143 expression in breast cancer cells; and (3) MMP13, an altered protein in breast cancer cells after ZNF143 knockdown, correlated with overall survival of breast cancer patients. Taken together, our results suggest that ZNF143 plays a role as a regulator of breast cancer metastasis.
MMP13, a member of the collagenase family, was first cloned from a breast tumor-derived cDNA library (33), and has been thought to play a role in breast cancer metastasis (34). In the present study, ZNF143 knockdown induced activation of MMP13 in breast cancer cells, suggesting that ZNF143 plays a role in tumor malignancy, especially during tumor invasion. When MCF7 cells were treated with insulin-like growth factor-1(IGF-1) to induce ZNF143 expression (29), MMP13 decreased (Fig. 4C), suggesting that ZNF143 activates MMP13 expression. However, the underlying mechanism by which ZNF143 is involved in MMP13 activation in breast cancer cells is still under investigation.
We showed that ZNF143 knockdown affected both cell motility and cell morphology (Figs. 2 and 3). The SEM images showed marked differences in the morphologies of MCF7 sh-Control and MCF7 sh-ZNF143 cells. Furthermore, the cell contacts of MCF7 sh-ZNF143 cells were reduced compared with those of MCF7 sh-Control cells, and the cell surfaces of MCF7 sh-ZNF143 were much smoother than those of MCF7 sh-Control cells, suggesting a role of ZNF143 in the cellular integrity of epithelial cells. Increased long and thick F-actin fibrous structures in MCF7 sh-ZNF143 cells (Fig. 2D) were additional evidence for a role of ZNF143 in cellular morphology and motility. When F-actin, which is involved in cell motility and invasiveness (35, 36), was stained with rhodamine-phalloidin, long and thick F-actin structures were predominantly found in MCF7 sh-ZNF143 cells, which differed from the short F-actin structures and microvilli on the cell surface found in MCF7 sh-Control cells. The composition between the microvilli and the long and thick F-actin structures showed distinct differences in the two cell types, suggesting a role for ZNF143 in the maintenance of cellular morphology. However, it is still unclear how ZNF143 might contribute to cellular structures by regulating the cytoskeleton. The correlation between molecular classification and histological classification in breast cancers has been investigated in numerous studies. A previous study differentiated invasive lobular carcinoma from invasive ductal carcinoma based on a multidimensional molecular atlas, including genetic features (6). ZNF143 was specifically expressed in the duct epithelium of normal breast tissues, which was reduced in invasive ductal carcinoma tissues, suggesting a role for ZNF143 expression during invasive ductal carcinoma progression, as well as the use of ZNF143 as a marker for the prognosis of invasive ductal carcinoma.
MATERIALS AND METHODS
Detailed experimental procedures are described in Supplementary Information.
The short-hairpin (sh)RNA-mediated silencing of human ZNF143 in breast cancer cells
To achieve stable lentivirus-mediated expression of shRNA specific for the gene encoding ZNF143 in MCF7 cells, cells were grown for 24 h, incubated with 5 μg/ml polybrene for 30 minutes, and then infected as previously described (21). To avoid clonal variations, the individual clones for each stable cell line produced by infection were pooled. Established cell lines were propagated and their images were captured using an inverted light microscope (CKX53; Olympus, Tokyo, Japan).
Wound healing assay
Cells (5 × 105) were grown in 12-well plates for 24 h until they were fully confluent. A uniform scratch was then made and the plate was washed with PBS. Digital images of the wound were obtained every 12 h at 10 × magnification using the NCC cell observer system (Axiovert 200M; Carl Zeiss, Oberkochen, Germany). The horizontal distance between the two sides of the wound was then measured (21).
Transmigration and invasion assays
Cells were suspended in RPMI 1640 medium and placed in the upper compartment of an 8 μm Transwell® (3.2 mm diameter; Neuro Probe, Gaithersburg, MD, USA). The lower compartment was filled with RPMI 1640 medium supplemented with FBS. After 24 h, the filter was washed with PBS and the migrated cells on the filter membrane were stained using a Diff-Quik Stain Kit (Sysmex, Tokyo, Japan). For invasion assays, the upper compartment of an 8 μm Transwell® (6.5 mm diameter; Costar, Cambridge, MA, USA) was coated with Matrigel® (1 mg/ml) before starting the assay. Each assay was conducted at least three times, and three random fields using 20 × magnification were analyzed for each filter membrane (21).
Isolation of RNA and RT-PCR
Total cellular RNA was extracted using an RNeasy Kit (Qiagen, Valencia, CA, USA) and dissolved in diethyl pyrocarbonate-treated water (37). The RNA was quantified by UV scanning, and samples (5 μg) were reverse-transcribed in the presence of oligo(dT) primers as described previously. The identities of the primers for each gene are available upon request (21, 29). The PCR products were subjected to electrophoresis and the resulting bands were visualized with ethidium bromide and photographed using Gel Doc software (Bio-Rad, Hercules, CA, USA).
Immunoblotting
Protein samples were resolved by SDS-PAGE, followed by protein transfer to polyvinylidene-difluoride membranes for 1 h using a Bio-Rad transfer unit (Bio-Rad). The membranes were then blocked for 30 minutes in Tris-buffered saline containing 0.01% Tween 20 (TBST) and 5% nonfat dried milk, followed by incubation for 2 h with primary antibody in TBST containing 2% bovine serum albumin, and then 1 h of incubation with horseradish peroxidase-conjugated anti-mouse or rabbit antibody. The blots were developed with WEST-ZOL® plus western blot detection system (Intron Biotechnology, Daejeon, Republic of Korea). Quantitation of band intensities on the XAR-5 film (Eastman Kodak, NY, USA) (21, 38) was performed using Quantity One software (Bio-Rad).
TCGA provisional analyses
For the analyses of matrix metallopeptidase 13 (MMP13) from breast cancer patients, we first gathered gene expression data from RNA-Seq data at the Cancer Genome Atlas (TCGA) data portal. The gene expression data were normalized by RNA-Seq expression estimation using expectation-maximization (RSEM) (39). To determine the differential expression between normal and tumor tissues, boxplots were drawn and the significance was determined using the Student’s t-test and R software (https://www.r-project.org/about.html). To perform the survival analyses, clinical information of breast cancer patients was recorded and annotated according to gene expression levels by our own scripts, and then analyzed by the Kaplan-Meier method using the R software.
Supplementary Information
ACKNOWLEDGEMENTS
We thank Mi Ae Kim of the Microscopy Core (National Cancer Center), Tae Sik Kim of the FACS core (National Cancer Center) and Mi Sun Park of Animal Laboratory (National Cancer Center) for their expert assistance and helpful suggestions. This work was supported in part by National Cancer Center Grant NCC-1710180 (to HJ YOU) and National Research Foundation of Korea Grant funded by the Korea Government (MSIP, South Korea) (No.2015R1A2A2A01003829).
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting interests.
REFERENCES
- 1.Li CI, Uribe DJ, Daling JR. Clinical characteristics of different histologic types of breast cancer. Br J Cancer. 2005;93:1046–1052. doi: 10.1038/sj.bjc.6602787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pestalozzi BC, Zahrieh D, Mallon E, et al. Distinct clinical and prognostic features of infiltrating lobular carcinoma of the breast: combined results of 15 International Breast Cancer Study Group clinical trials. J Clin Oncol. 2008;26:3006–3014. doi: 10.1200/JCO.2007.14.9336. [DOI] [PubMed] [Google Scholar]
- 3.Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 4.Gatza ML, Silva GO, Parker JS, Fan C, Perou CM. An integrated genomics approach identifies drivers of proliferation in luminal-subtype human breast cancer. Nat Genet. 2014;46:1051–1059. doi: 10.1038/ng.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ciriello G, Gatza ML, Beck AH, et al. Comprehensive Molecular Portraits of Invasive Lobular Breast Cancer. Cell. 2015;163:506–519. doi: 10.1016/j.cell.2015.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heng YJ, Lester SC, Tse GM, et al. The molecular basis of breast cancer pathological phenotypes. J Pathol. 2017;241:375–391. doi: 10.1002/path.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nieto MA, Cano A. The epithelial-mesenchymal transition under control: global programs to regulate epithelial plasticity. Semin Cancer Biol. 2012;22:361–368. doi: 10.1016/j.semcancer.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 11.Huang RY, Guilford P, Thiery JP. Early events in cell adhesion and polarity during epithelial-mesenchymal transition. J Cell Sci. 2012;125:4417–4422. doi: 10.1242/jcs.099697. [DOI] [PubMed] [Google Scholar]
- 12.Myslinski E, Krol A, Carbon P. ZNF76 and ZNF143 are two human homologs of the transcriptional activator Staf. J Biol Chem. 1998;273:21998–22006. doi: 10.1074/jbc.273.34.21998. [DOI] [PubMed] [Google Scholar]
- 13.Bailey SD, Zhang X, Desai K, et al. ZNF143 provides sequence specificity to secure chromatin interactions at gene promoters. Nat Commun. 2015;2:6186. doi: 10.1038/ncomms7186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Halbig KM, Lekven AC, Kunkel GR. The transcriptional activator ZNF143 is essential for normal development in zebrafish. BMC Mol Biol. 2012;13:3. doi: 10.1186/1471-2199-13-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Myslinski E, Gerard MA, Krol A, Carbon P. A genome scale location analysis of human Staf/ZNF143-binding sites suggests a widespread role for human Staf/ZNF143 in mammalian promoters. J Biol Chem. 2006;281:39953–39962. doi: 10.1074/jbc.M608507200. [DOI] [PubMed] [Google Scholar]
- 16.Ngondo-Mbongo RP, Myslinski E, Aster JC, Carbon P. Modulation of gene expression via overlapping binding sites exerted by ZNF143, Notch1 and THAP11. Nucleic Acids Res. 2013;41:4000–4014. doi: 10.1093/nar/gkt088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vinckevicius A, Parker JB, Chakravarti D. Genomic Determinants of THAP11/ZNF143/HCFC1 Complex Recruitment to Chromatin. Mol Cell Biol. 2015;35:4135–4146. doi: 10.1128/MCB.00477-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishiguchi H, Izumi H, Torigoe T, et al. ZNF143 activates gene expression in response to DNA damage and binds to cisplatin-modified DNA. Int J Cancer. 2004;111:900–909. doi: 10.1002/ijc.20358. [DOI] [PubMed] [Google Scholar]
- 19.Kawatsu Y, Kitada S, Uramoto H, et al. The combination of strong expression of ZNF143 and high MIB-1 labelling index independently predicts shorter disease-specific survival in lung adenocarcinoma. Br J Cancer. 2014;110:2583–2592. doi: 10.1038/bjc.2014.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Myslinski E, Gerard MA, Krol A, Carbon P. Transcription of the human cell cycle regulated BUB1B gene requires hStaf/ZNF143. Nucleic Acids Res. 2007;35:3453–3464. doi: 10.1093/nar/gkm239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paek AR, Lee CH, You HJ. A role of zinc-finger protein 143 for cancer cell migration and invasion through ZEB1 and E-cadherin in colon cancer cells. Mol Carcinog. 2014;53(Suppl 1):E161–168. doi: 10.1002/mc.22083. [DOI] [PubMed] [Google Scholar]
- 22.Lu W, Chen Z, Zhang H, Wang Y, Luo Y, Huang P. ZNF143 transcription factor mediates cell survival through upregulation of the GPX1 activity in the mitochondrial respiratory dysfunction. Cell Death Dis. 2012;3:e422. doi: 10.1038/cddis.2012.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Izumi H, Yasuniwa Y, Akiyama M, et al. Forced Expression of ZNF143 Restrains Cancer Cell Growth. Cancers (Basel) 2011;3:3909–3920. doi: 10.3390/cancers3043909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei S, Wang L, Zhang L, et al. ZNF143 enhances metastasis of gastric cancer by promoting the process of EMT through PI3K/AKT signaling pathway. Tumour Biol. 2016;77:12813–12821. doi: 10.1007/s13277-016-5239-z. [DOI] [PubMed] [Google Scholar]
- 25.Ngondo RP, Carbon P. Transcription factor abundance controlled by an auto-regulatory mechanism involving a transcription start site switch. Nucleic Acids Res. 2014;42:2171–2184. doi: 10.1093/nar/gkt1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chambers I, Frampton J, Goldfarb P, Affara N, McBain W, Harrison PR. The structure of the mouse glutathione peroxidase gene: the selenocysteine in the active site is encoded by the ‘termination’ codon, TGA. EMBO J. 1986;5:1221–1227. doi: 10.1002/j.1460-2075.1986.tb04350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gyorffy B, Lanczky A, Eklund AC, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 28.Neve RM, Chin K, Fridlyand J, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paek AR, Kim SH, Kim SS, Kim KT, You HJ. IGF-1 induces expression of zinc-finger protein 143 in colon cancer cells through phosphatidylinositide 3-kinase and reactive oxygen species. Exp Mol Med. 2010;42:696–702. doi: 10.3858/emm.2010.42.10.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choi JA, Jung YS, Kim JY, Kim HM, Lim IK. Inhibition of breast cancer invasion by TIS21/BTG2/Pc3-Akt1-Sp1-Nox4 pathway targeting actin nucleators, mDia genes. Oncogene. 2016;35:83–93. doi: 10.1038/onc.2015.64. [DOI] [PubMed] [Google Scholar]
- 31.Blanchoin L, Boujemaa-Paterski R, Sykes C, Plastino J. Actin dynamics, architecture, and mechanics in cell motility. Physiol Rev. 2014;94:235–263. doi: 10.1152/physrev.00018.2013. [DOI] [PubMed] [Google Scholar]
- 32.Li J, Yang B, Zhou Q, et al. Autophagy promotes hepatocellular carcinoma cell invasion through activation of epithelial-mesenchymal transition. Carcinogenesis. 2013;34:1343–1351. doi: 10.1093/carcin/bgt063. [DOI] [PubMed] [Google Scholar]
- 33.Freije JM, Diez-Itza I, Balbin M, et al. Molecular cloning and expression of collagenase-3, a novel human matrix metalloproteinase produced by breast carcinomas. J Biol Chem. 1994;269:16766–16773. [PubMed] [Google Scholar]
- 34.Pivetta E, Scapolan M, Pecolo M, et al. MMP-13 stimulates osteoclast differentiation and activation in tumour breast bone metastases. Breast Cancer Res. 2011;13:R105. doi: 10.1186/bcr3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alexandrova AY. Plasticity of tumor cell migration: acquisition of new properties or return to the past? Biochemistry (Mosc) 2014;79:947–963. doi: 10.1134/S0006297914090107. [DOI] [PubMed] [Google Scholar]
- 36.Chhabra ES, Higgs HN. The many faces of actin: matching assembly factors with cellular structures. Nat Cell Biol. 2007;9:1110–1121. doi: 10.1038/ncb1007-1110. [DOI] [PubMed] [Google Scholar]
- 37.Lee JJ, Kim M, Kim HP. Epigenetic regulation of long noncoding RNA UCA1 by SATB1 in breast cancer. BMB Rep. 2016;49:578–583. doi: 10.5483/BMBRep.2016.49.10.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee CS, Ghim J, Song P, Suh P-G, Ryu SH. Loss of phospholipase D2 impairs VEGF-induced angiogenesis. BMB Rep. 2016;49:191–196. doi: 10.5483/BMBRep.2016.49.3.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.