Abstract
Regulatory B cells, also well-known as IL-10-producing B cells, play a role in the suppression of inflammatory responses. However, the epigenetic modulation of regulatory B cells is largely unknown. Recent studies showed that the bromodomain and extra-terminal domain (BET) protein inhibitor JQ1 controls the expression of various genes involving cell proliferation and cell cycle. However, the role of BET proteins on development of regulatory B cells is not reported. In this study, JQ1 potently suppressed IL-10 expression and secretion in murine splenic and peritoneal B cells. While bromodomain-containing protein 4 (BRD4) was associated with NF-κB on IL-10 promoter region by LPS stimulation, JQ1 interfered the interaction of BRD4 with NF-κB on IL-10 promoter. In summary, BRD4 is essential for toll like receptor 4 (TLR4)-mediated IL-10 expression, suggesting JQ1 could be a potential candidate in regulating IL-10-producing regulatory B cells in cancer.
Keywords: BRD4, IL-10, JQ1, NF-κB, Regulatory B cells
INTRODUCTION
B cells have long been known to have antibody production and antigen presenting functions. Recent studies have also confirmed the existence of a regulatory B cell subset with the ability to regulate the inflammatory response just as regulatory T cells (Tregs) do (1). In addition, the immune regulation of IL-10-producing regulatory B cells (Breg or B10) has been reported in inflammatory immune diseases such as contact hypersensitivity, collagen-induced arthritis, experimental autoimmune encephalomyelitis, anaphylaxis, and food allergy (2–7). Recently, the distribution and partial functions of regulatory B cells have been reported in some cancers, and regulatory B cells are expected to have a mechanism to inhibit immune cell activity against cancer cells like Tregs (8–11).
IL-10-producing regulatory B cells have various phenotypes such as CD1dhiCD5+, CD21hiCD23+ (Transitional 2-marginal zone precursor (T2-MZP)), Tim-1+, CD9+, and are known to produce anti-inflammatory cytokines such as interleukin (IL)-10 or Transforming growth factor (TGF)-β (12–15). The deficiency of IL-10-producing regulatory B cells further exacerbates the development of a variety of inflammatory immune diseases but the mechanism of IL-10 production from regulatory B cells in anti-inflammatory responses remains unclear. While recent studies have uncovered the mechanism regulating the production of IL-10 in the cytoplasm of B cells such as the B-cell linker (BLNK) subtype signaling pathway and the phosphoinositide 3-kinase (PI3K), there is a lack of research on the signaling system such as transcription in the nucleus (16, 17).
The bromodomain and extra-terminal domain (BET) protein family consists of four proteins including BRD2, BRD3, BRD4, and tetris-specific BRDt, is known to read acetylated lysine on histones in the nucleus and change chromatin structure through their bromodomain (18, 19). As development of a couple of BET protein inhibitors, the role of BET proteins have been highlighted and they play critical functions in a variety of cellular processes such as cell growth, cell cycle, inflammation, and cancer development (20–22). Among BET protein inhibitors, JQ1 has been widely used as a potent, relative BRD4 selective and the first generation inhibitor (23).
In B cell immunology fields, BET bromodomain has been known to be involved in the development of germinal center B cells and the switching of immunoglobulins in B cells (24, 25), but it has not yet been studied how BET bromodomain functions in IL-10-producing regulatory B cells. In this study, we demonstrate for the first time that IL-10 production in regulatory B cells is reduced via interfering interaction of BRD4 at the promoter of IL-10 which NF-kB co-binds by JQ1, and verify that BRD4 plays an important role in transcriptional activation for the production of IL-10 in regulatory B cells. Our results suggest that JQ1 can be used as a novel therapeutic molecule for anti-cancer immunity targeting regulatory B cells.
RESULTS
LPS-stimulated IL-10 production is controlled by JQ1 in B cells
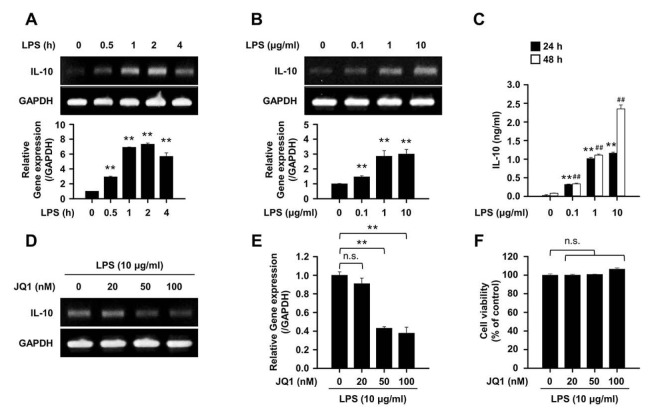
First, the expression of IL-10 gene in splenic B cells was confirmed by lipopolysaccharide (LPS) stimulation. IL-10 gene expression in splenic B cells was increased in response to LPS stimulation in a time- and dose-dependent manner (Fig. 1A and B). It was also found that IL-10 secretion by splenic B cells was also increased by LPS stimulation (Fig. 1C). Next, the expression of IL-10 in B cells by JQ1, a specific inhibitor of BRD4, was examined. IL-10 gene expression decreased in a dose-dependent manner when splenic B cells stimulated with LPS were treated with JQ1 (Fig. 1D and E). The concentration of JQ1 treatment in vitro were determined at the preliminary experiment (Supplementary Fig. 1), and the JQ1-mediated regulation of IL-10 gene expression and secretion by LPS was not due to cytotoxicity (Fig. 1F).
Fig. 1.
LPS-induced IL-10 expression is regulated by JQ1 in B cells. (A) Representative images and relative gene expression of IL-10. Splenic B cells were stimulated with LPS (10 μg/ml) for indicated times or (B) indicated LPS concentration for 4 h. Representative band images and data are the mean ± SEM from three independent experiments. **P<0.01. (C) Quantitative analysis of secreted IL-10 from LPS-stimulated splenic B cells. Cells were stimulated with LPS for the indicated times and supernatant were harvested. IL-10 released into the medium were analyzed by enzyme-linked immunosorbent assay (ELISA). **P < 0.01 (24 h) or ##P < 0.01 (48 h) when compared with no LPS stimulation group. (D) Representative images and (E) relative gene expression of IL-10 gene expression in LPS stimulated splenic B cells with or without JQ1 (0–100 nM) for 4 h. (F) Splenic B cells were incubated in triplicate LPS with or without JQ1 (0–100 nM) for 24 h. Cell viability was determined using a cell counting kit-8. Representative band images (D) and data (E, F) are the mean ± SEM from three independent experiments. **P<0.01. n.s., not significant.
BRD4 pathway is associated with the development of IL-10-producing B cells
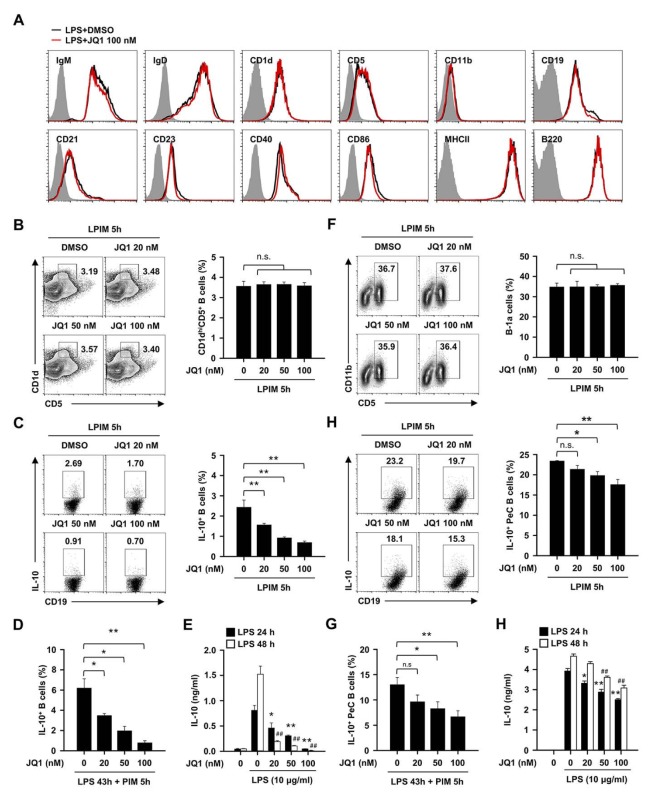
We observed that the treatment of JQ1 did not change the phenotype of B cells (Fig. 2A). It is well known that the frequency of IL-10-producing B cells is closely related to the splenic CD1dhiCD5+ phenotype (12). We next checked whether the decrease in IL-10 production function of B cells by JQ1 correlates with the decrease of CD1dhiCD5+ B cells. Unexpectedly, there was no frequency change of CD1dhiCD5+ B cells by a JQ1 treatment (Fig. 2B). Flow cytometry analysis showed that IL-10 production in LPS-induced B cells decreased in a dose-dependent manner (Fig. 2C). In previous study, Tedder and coworkers reported that precursor cells of regulatory B cells can differentiate into mature regulatory B cells by long-term stimulation of LPS (26). The effect of JQ1 on the differentiation of precursor cells by long-term LPS stimulation was also investigated. JQ1 reduced the differentiation of precursor cells to mature IL-10-producing regulatory B cells (Fig. 2D and E).
Fig. 2.
Effect of JQ1 on regulatory B subsets and IL-10 producing ability in splenic and peritoneal cavity B cells. (A) Expressions of the B cell surface markers from DMSO or JQ1 (100 nM) treated splenic B cells that were incubated with LPS for 4 h. Black histograms represent B cells treated with DMSO (control), red histograms represent JQ1-treated B cells, and gray filled histograms show cells with isotype control. Plots are representative images of at least three independent experiments. (B and C) Splenic B cells were incubated with 10 μg/ml of LPS + PIM for 5 h in presence or absence of the indicated dose of JQ1 (0–100 nM). (B) Representative plots and frequencies of CD1dhiCD5+ subset and (C) IL-10+ in splenic B cells. **P < 0.01. n.s., not significant. (D) Frequencies of IL-10+ splenic B cells that contained LPS (10 μg/ml) for 43 h + last PIM 5 h in the presence of the indicated dose of JQ1. *P < 0.05, **P < 0.01. (E) Secretion of IL-10 in LPS stimulated splenic B cells with or without JQ1 (0–100 nM) for 24 and 48h. *P < 0.05, **P < 0.01 (24 h) or ##P < 0.01 (48h) when compared with JQ1 untreated B cells under LPS stimulation group. (F and G) PeC B cells were incubated with 10 μg/ml of LPS + PIM for 5 h in presence or absence of the indicated dose of JQ1. (F) Representative plots and frequencies of CD5+CD11b+ B-1a cells and (G) IL-10+ in peritoneal cavity B cells. *P < 0.05, **P < 0.01. n.s., not significant. (H) Frequencies of IL-10+ peritoneal cavity B cells that contained LPS for 43 h + last PIM 5 h in the presence of the indicated dose of JQ1. *P<0.05, **P < 0.01. n.s., not significant. (I) Secretion of IL-10 in LPS stimulated peritoneal cavity B cells with or without JQ1 (0–100 nM) for 24 and 48 h. *P < 0.05, **P < 0.01 (24 h) or ##P < 0.01 (48 h) when compared with JQ1 untreated B cells under LPS stimulation group. All plots are representative images and data are the mean ± SEM from three independent experiments.
The major population of regulatory B cells in the peritoneal cavity (PeC) has the phenotype of CD5+CD11b+ (B-1a). The changes of CD5+CD11b+ regulatory B cells by JQ1 were examined and it was found not to induce any phenotypic change as in splenic regulatory B cells (Fig. 2F). However, it was also observed that IL-10 production of LPS-induced B cells was decreased by JQ1 as in splenic regulatory B cells (Fig. 2G). Consistently, the differentiation of precursor cells of PeC regulatory B cells by the long-time treatment of LPS and the IL-10 secretion of PeC B cells were also suppressed by JQ1 (Fig. 2H and I). Of note, the production of IL-10 in splenic CD1dhiCD5+ and PeC CD5+CD11b+ B cell subsets, but not the other subsets, were suppressed by JQ1 (Supplementary Fig. 2). These results suggest that the BRD4 pathway affects LPS-induced IL-10 production in spleen and peritoneal cavity but not regulatory B subset frequency. Therefore, we hypothesized that the BRD4 pathway is associated with the signal transduction pathway for the production of IL-10 in regulatory B cells.
LPS treatment does not affect expression of BET proteins and NF-κB in B cells
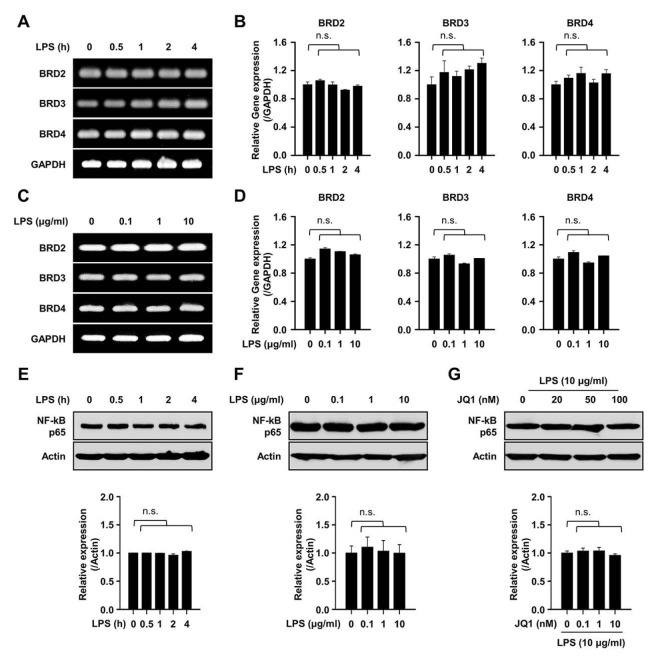
Next, we investigated whether LPS stimulation can change the expression of BET proteins themselves. As a result, any change of BET proteins was not observed in splenic B cells by stimulation of LPS (Fig. 3A to D). The expression of NF-κB p65 was also not changed by the treatment of LPS (Fig. 3E and F). Additionally, we hypothesized that the suppression of IL-10 production from B cells by JQ1 is associated with the expression of NF-kB p65. We then found that JQ1 did not alter the expression level of NF-κB p65 (Fig. 3G), suggesting that the negative mechanism of JQ1 is not directly associated with the levels of NF- kB p65 in the process of IL-10 production in regulatory B cells.
Fig. 3.
Expression of BET proteins and NF-κB in LPS stimulated B cells. (A and B) Splenic B cells were stimulated by 10 μg/ml of LPS for indicated times (0–4 h). (A) Representative image of BET (BRD2, BRD3, and BRD4) genes and (B) relative gene expression level of BET proteins in a time dependent manner. (C and D) Splenic B cells were stimulated by indicated doses (0–10 μg/ml) of LPS for 4 h. (C) Representative image of BET (BRD2, BRD3, and BRD4) genes and (D) relative gene expression level of BET proteins in a dose dependent manner. Representative images (A, C) and data (B, D) are the mean ± SEM from three independent experiments. n.s., not significant. (E and F) Cells were stimulated by indicated doses (0–10 μg/ml) of LPS for indicated times (0–4 h). Representative band image and relative protein expression level of NF-κB in LPS stimulated B cells. (G) Representative band image and relative protein expression level of NF-κB in LPS stimulated B cells in the presence of the indicated dose of JQ1 for 4 h. Representative band images and data are the mean ± SEM from three independent experiments. n.s., not significant.
JQ1 inhibits the recruitment of BRD4 and NF-κB p65 at IL-10 promoter region upon LPS stimulation
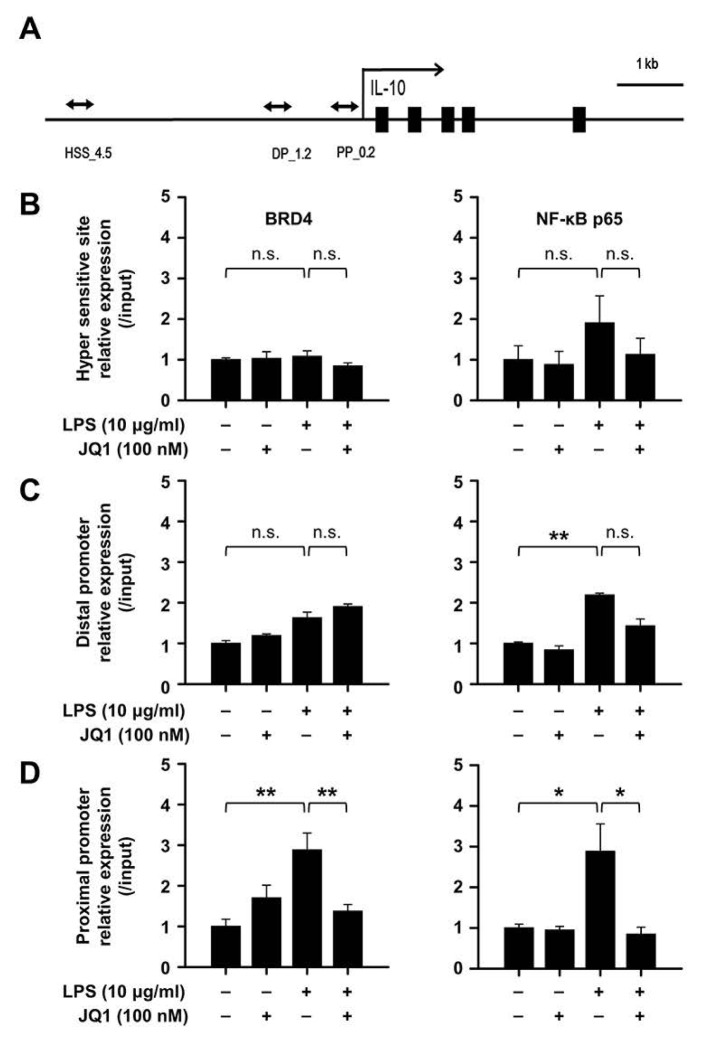
In the TLR signal transduction pathway by LPS stimulation, several reports demonstrated that the multiple chromatin complex with NF-kB p65 are formed at IL-10 DNA regulatory regions (27, 28). Besides, NF-kB was also known to interact with BRD4 in other cells (29). Based to these, we designed three sites for IL-10 DNA regulatory regions covering the hyper sensitive site (HSS, upstream 4.5 kb) (30), distal promoter (DP, upstream 1.2 kb), and proximal promoter (PP, upstream 0.2 kb) (31, 32) which are known as functional DNA elements for IL-10 transcription (Fig. 4A). We then performed chromatin immunoprecipitation (ChIP) assay with antibody against BRD4 and NF-κB p65 in splenic B cells. We found that NF-κB p65 binding, but not BRD4, was increased at two promoter sites by LPS (Fig. 4C and D). Interestingly, the ChIP analysis revealed significant increase in binding of BRD4 on IL-10 proximal promoter among three sites where the greater NF-κB p65 binding was observed concurrently and these recruitments suppressed by JQ1 (Fig. 4C). Therefore, we argue that BRD4 may be critical for NF-κB p65 binding on IL-10 proximal promoter for the production of IL-10 in regulatory B cells.
Fig. 4.
JQ1 negatively regulates the IL-10-NF-κB complex via suppression of BRD4 binding at the IL-10 promoters. (A) Schematic diagram of the IL-10 hyper sensitive site (HSS), distal promoter (DP), and proximal promoter (PP) are indicated. (B) Splenic B cells stimulated by 10 μg/ml of LPS with or without 100 nM of JQ1 for 2 h. Chromatin was immunoprecipitated with anti-BRD4 or anti-NF-κB p65 antibody and each proteins binding at the hyper sensitive site, (C) distal promoter, and (D) proximal promoter regions of IL-10 was analyzed by real time PCR. The relative expression graph data are the mean ± SEM from three independent experiments. *P < 0.05, **P < 0.01. n.s., not significant.
DISCUSSION
In the past decade, the suppressive effects, mainly through the secretion of IL-10, of regulatory B cells on inflammatory responses have been reported in a variety of immune disorders (33–36). Additionally, immune regulation through the interaction of immune cells with the intrinsic phenotype of regulatory B cells (e.g., CD1dhiCD5+, T2-MZP, Tim-1+, and CD9+) were demonstrated in various diseases, and it plays a critical role in autoimmune diseases (37). In recent studies, functional studies in cancer diseases are emerging (38–40). In particular, the change of the distribution of regulatory B cells in cancer tissue is considered to one of important indicators (8–10). Emerging evidence suggests that regulatory B cells suppress effector immune cells including IFN-γ-producing cytotoxicity cells in various cancer diseases through the secretion of IL-10 (11). Although regulatory B cells have to play the suppressive role on the effector function of T cells in autoimmune diseases to cure diseases (41), regulatory B cells need to be suppressed to induce anti-cancer immunity to cure cancer.
As development of BET protein inhibitors, there are tremendous effort to apply these drug to various fields such as cancer and immune disorders. In cancer, they are well known as pivotal regulators for the expression of several oncogenes, such as c-Myc and Bcl-2 (42, 43). Moreover, several BET protein inhibitors have been under clinical research (44). However, there is no study on the relationship between BET proteins and regulatory B cells, especially for IL-10 production. Furthermore, the epigenetic mechanism the production of IL-10 in the nucleus of regulatory B cells is largely unknown. This study proposes BRD4, a chromatin reader is a critical modulator of regulatory B cells. We found that the gene expression and secretion of IL-10 by LPS stimulation were reduced in regulatory B cells by the treatment of JQ1 in a dose-dependent manner (Fig. 1). The effect of JQ1 is not due to the induction of changes of regulatory B cell phenotype but modulation of the IL-10 production in regulatory B cells (Fig. 1G and Fig. 2).
It is generally accepted that LPS-mediated TLR4 signal pathway is critical to increase the frequency of IL-10 producing regulatory B cells. However, the epigenetic regulation on the production of IL-10 in regulatory B cells by LPS is not well unknown. It has reported that BRD4 is interacted with various proteins including several transcription factors. Among them, NF-kB is critical for the production of IL-10 (27–29). In this study, we examined whether the relationship between BET proteins and NF-κB (a major transcription factor for IL-10 production by LPS) were altered by LPS stimulation. The expression of total BRD4 (also BRD2 and BRD3) and NF-κB p65 proteins was not affected by LPS stimulation or JQ1 treatment in B cells (Fig. 3A to G). These results suggest that BRD4 may be directly involved in transcriptional activation IL-10 via the TLR4 signal by LPS. Therefore, we further assessed the role of BRD4 in LPS-mediated IL-10-producing B cells by using ChIP assay. Three major sites such as hyper sensitive site (HSS), distal promoter (DP), and proximal promoter (PP) were investigated by ChIP assay. LPS stimulation caused recruitment of BRD4 and NF-kB to the IL-10 proximal promoter region of B cells, and this process was inhibited by JQ1 (Fig. 4D), suggesting that BRD4 play critical role for production of IL-10 of regulatory B cells. Although it was reported that BRD4 directly binds to acetylated NF-kB p65 in LPS stimulation (45), whether both proteins are directly interacted in regulatory B cells remained to be determined.
In summary, this study demonstrates that BRD4 as a novel epigenetic regulator directly participates in the transcriptional process for IL-10 production via altering chromatin structure in regulatory B cells upon LPS simulation and presumably this mechanism could contribute anti-cancer effects of JQ1 in various cancer diseases.
MATERIALS AND METHODS
Mice
C57BL/6 (5–6-week-old male) mice were purchased from Orient Bio Inc. (Gyeonggi, Korea). Mice were housed under specific pathogen free facility at Koukuk University (Seoul, Korea). The animal study was done in accordance with the institutional guidelines. The protocol was approved by the Institutional Animal Care and Use Committee (IACUC) at Konkuk University.
Reverse transcriptase-polymerase chain reaction (RT-PCR)
Splenic B cells were purified with CD19 mAb-conjugated microbeads (Miltenyi Biotech, Auburn, CA) according to the manufacturer’s instructions, to > 95% purity. Total RNA was isolated from sorted Splenic CD19+ B cells (1 × 107 cells) by using easy-BLUE (iNtRON Biotechnology, Gyeonggi, Korea) and reverse-transcribed with the ImProm-II reverse transcription first-strand synthesis system (Promega, Madison, WI) according to the manufacturer’s protocol. PCR was performed at 95°C for 2 min, 30 cycles of 95°C for 20 sec, 58°C for 40 sec, 72°C for 30 sec, and 72°C for 5 min. Primers used as follow: mouse BRD2 (forward 5′-CCACGAAAAGACTTGCC TGA-3′, reverse 5′-CAGCGTGCTTCTTTGAGAGC-3′); mouse BRD3 (forward 5′-CTATGCGTGGCCCTTTTACA-3′, reverse 5′-CTTCCTTTTGACCGTGCTGA-3′); mouse BRD4 (forward 5′-CAAAAGGAAGAGGACGAGGG-3′, reverse 5′-ACAGGTG GAGGAGGGTTCTG-3′); mouse IL-10 (forward 5′-GGCCCAG AAATCAAGGAGCA-3′, reverse 5′-GGGGGATGACAGTAGGG GAA-3′); mouse GAPDH forward 5′-TGACGTGCCGCCTGGA GAAA-3′, reverse 5′-AGTGTAGCCCAAGATGCCCTTCAG-3′.
Measurement of Interleukin-10 release by ELISA
Isolated splenic CD19+ B cells (3 × 106 cells/well) and Peritoneal cavity fluid (PeC)-derived CD19+ B cells (1 × 106 cells/well) were stimulated with LPS (0, 0.1, 1, and 10 μg/ml) for 24 or 48 h, the level of IL-10 by using a mouse BD OptEIA IL-10 ELISA kit according to the manufacturer’s instructions (BD Biosciences, San Jose, CA).
Flow cytometry analysis
Spleen and PeC CD19+ B cells (3 × 106 cells/well) stimulated for 5 or 48 h with LPS (10 μg/ml, Sigma-Aldrich, St. Louis, MO). The B cells were incubated with LPS alone or with JQ1 (0, 20, 50, and 100 nM) for 5 h or indicated times in figure legends, and phorbol 12-myristate 13-acetate (PMA, 50 ng/ml, Sigma-Aldrich), ionomycin (500 ng/ml, Sigma-Aldrich), and Monensin (2 μM, eBioscience, San Diego, CA) were added during last 5 h. Prior to surface staining, Fcγ receptors were blocked with anti-CD16/CD32 monoclonal antibodies (2.4G2, BD Biosciences). Cells were fixed and permeabilized with a Cytofix/Cytoperm kit (eBioscience) and then were stained with anti-IL-10 (JES5-16E3, eBioscience) (46). The antibodies against surface proteins were as follows: CD1d (1B1), CD5 (53-7.3), CD11b (M1/70), CD19 (eBio1D3), CD21/CD35 (eBioBD9), CD23 (B3B4), CD40 (HM40-3), CD86 (GL1), B220 (RA3-6B2), IgM (eB121-15F9), IgD (11–26), MHCII (M5/114.15.2), all purchased from eBioscience. Cells were analyzed with FACSCanto II (BD Biosciences, San Jose, CA) and FlowJo V10 software (TreeStar, Ashland, OR). The gate strategy for IL-10-producing B cells was illustrated in Supplementary Fig. 3.
Measurement of cell viability
Mouse splenic CD19+ B cells (3 × 106 cells/well) were plated on 24-well plates in LPS contained media with or without JQ1 for 24h. Then the cell viability was determined by using a cell counting kit-8 (CCK-8, Dojindo Labpratpries, Kumamoto, Japan), according to the manufacturer’s protocol.
Immunoblotting
Splenic CD19+ B cells (1 × 107 cells/well) were lysed in RIPA buffer containing protease inhibitor on ice for 10 min. Lysate centrifuged at 13,000 ×g for 10 min at 4°C. Protein lysates of each sample were analyzed by western blotting using specific antibodies (47). Antibodies against NF-κB p65 and actin were purchased from Cell Signaling Technology (Danvers, MA, USA). Immunoreactive proteins were detected with HRP-coupled secondary antibodies and enhanced chemiluminescence according to the manufacturer’s protocol (Amersham Biosciences, Piscataway, NJ).
Chromatin immunoprecipitation (ChIP)
ChIP was performed with the splenic B cells following to instructions from Upstate Biotechnology (Lake Placid, NY, USA). Cells (2 × 107 cells/well) were harvested from the culture and fixed with 37% formaldehyde for 10 min. For each assay, sheared by a sonication (the DNA fragment size was 200 to 400 bp), was precleared with protein A magnetic beads and then 50 μg DNA was precipitated by BRD4 (Bethyl Laboratories, Montgomery, TX) or NF-κB p65 (Cell Signaling Technology). After immunoprecipitation, recovered chromatin fragments were subjected to real-time PCR. IgG control experiments were performed for all ChIPs and incorporated into the IP/Input (1%) by presenting the results as (IPIgG)/(Input-IgG). The primers used as follow: ChIP_IL-10 hyper sensitive site (forward 5′-GCCCGAAATATCACCTATTGC-3′, reverse 5′-CCGGATTGAATGTCCTGAGA-3′); ChIP_IL-10 Distal Promoter (forward 5′-CCCTGGTGTGGTAACCCTCT-3′, reverse 5′-ACCCTGGGCAAGCAACTACT-3′); ChIP_IL-10 Proximal Promoter (forward 5′-GCAGAAGTTCATTCCGACCA-3′, reverse 5′-GCCTTGTGGCTTTGGTAGTG-3′).
Statistical analysis
The data are expressed as the mean ± SEM. Statistical analysis was determined using Student’s t-test or one-way ANOVA. All statistical significance (*P < 0.05 and **P < 0.01) was performed using the software SigmaStat (Systat Software, Inc., San Jose, CA, USA).
Supplementary Materials
ACKNOWLEDGEMENTS
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIP, NRF-2013R1A4A1069575 and NRF-2016R1A2B3015840) and in part by NRF-2017R1D1B03028379.
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting interests.
REFERENCES
- 1.DiLillo DJ, Matsushita T, Tedder TF. B10 cells and regulatory B cells balance immune responses during inflammation, autoimmunity, and cancer. Ann N Y Acad Sci. 2010;1183:38–57. doi: 10.1111/j.1749-6632.2009.05137.x. [DOI] [PubMed] [Google Scholar]
- 2.Watanabe R, Fujimoto M, Ishiura N, et al. CD19 expression in B cells is important for suppression of contact hypersensitivity. Am J Pathol. 2007;171:560–570. doi: 10.2353/ajpath.2007.061279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang M, Deng J, Liu Y, et al. IL-10-producing regulatory B10 cells ameliorate collagen-induced arthritis via suppressing Th17 cell generation. Am J Pathol. 2012;180:2375–2385. doi: 10.1016/j.ajpath.2012.03.010. [DOI] [PubMed] [Google Scholar]
- 4.Matsushita T, Yanaba K, Bouaziz JD, Fujimoto M, Tedder TF. Regulatory B cells inhibit EAE initiation in mice while other B cells promote disease progression. J Clin Invest. 2008;118:3420–3430. doi: 10.1172/JCI36030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim HS, Kim AR, Kim DK, et al. Interleukin-10-producing CD5+ B cells inhibit mast cells during immunoglobulin E-mediated allergic responses. Sci Signal. 2015;8:ra28. doi: 10.1126/scisignal.2005861. [DOI] [PubMed] [Google Scholar]
- 6.Mangan NE, Fallon RE, Smith P, van Rooijen N, McKenzie AN, Fallon PG. Helminth infection protects mice from anaphylaxis via IL-10-producing B cells. J Immunol. 2004;173:6346–6356. doi: 10.4049/jimmunol.173.10.6346. [DOI] [PubMed] [Google Scholar]
- 7.Kim AR, Kim HS, Kim DK, et al. Mesenteric IL-10-producing CD5+ regulatory B cells suppress cow’s milk casein-induced allergic responses in mice. Sci Rep. 2016;6:19685. doi: 10.1038/srep19685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou J, Min Z, Zhang D, Wang W, Marincola F, Wang X. Enhanced frequency and potential mechanism of B regulatory cells in patients with lung cancer. J Transl Med. 2014;12:304. doi: 10.1186/s12967-014-0304-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang H, Yue R, Zhao P, et al. Proinflammatory follicular helper T cells promote immunoglobulin G secretion, suppress regulatory B cell development, and correlate with worse clinical outcomes in gastric cancer. Tumour Biol. 2017;39:1010428317705747. doi: 10.1177/1010428317705747. [DOI] [PubMed] [Google Scholar]
- 10.Lu Y, Meng F, Yang Y, et al. Significance of B10 cell in patients with thymoma complicated with myasthenia gravis. Oncotarget. 2017;8:73774–73786. doi: 10.18632/oncotarget.17908. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Sarvaria A, Madrigal JA, Saudemont A. B cell regulation in cancer and anti-tumor immunity. Cell Mol Immunol. 2017;14:662–674. doi: 10.1038/cmi.2017.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yanaba K, Bouaziz JD, Haas KM, Poe JC, Fujimoto M, Tedder TF. A regulatory B cell subset with a unique CD1dhiCD5+ phenotype controls T cell-dependent inflammatory responses. Immunity. 2008;28:639–650. doi: 10.1016/j.immuni.2008.03.017. [DOI] [PubMed] [Google Scholar]
- 13.Evans JG, Chavez-Rueda KA, Eddaoudi A, et al. Novel suppressive function of transitional 2 B cells in experimental arthritis. J Immunol. 2007;178:7868–7878. doi: 10.4049/jimmunol.178.12.7868. [DOI] [PubMed] [Google Scholar]
- 14.Xiao S, Brooks CR, Sobel RA, Kuchroo VK. Tim-1 is essential for induction and maintenance of IL-10 in regulatory B cells and their regulation of tissue inflammation. J Immunol. 2015;194:1602–1608. doi: 10.4049/jimmunol.1402632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun J, Wang J, Pefanis E, et al. Transcriptomics Identify CD9 as a Marker of Murine IL-10-Competent Regulatory B Cells. Cell Rep. 2015;13:1110–1117. doi: 10.1016/j.celrep.2015.09.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin G, Hamaguchi Y, Matsushita T, et al. B-cell linker protein expression contributes to controlling allergic and autoimmune diseases by mediating IL-10 production in regulatory B cells. J Allergy Clin Immunol. 2013;131:1674–1682. doi: 10.1016/j.jaci.2013.01.044. [DOI] [PubMed] [Google Scholar]
- 17.Matsushita T, Le Huu D, Kobayashi T, et al. A novel splenic B1 regulatory cell subset suppresses allergic disease through phosphatidylinositol 3-kinase-Akt pathway activation. J Allergy Clin Immunol. 2016;138:1170–1182. doi: 10.1016/j.jaci.2015.12.1319. [DOI] [PubMed] [Google Scholar]
- 18.Owen DJ, Ornaghi P, Yang JC, et al. The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase gcn5p. EMBO J. 2000;19:6141–6149. doi: 10.1093/emboj/19.22.6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeng L, Zhou MM. Bromodomain: an acetyl-lysine binding domain. FEBS Lett. 2002;513:124–128. doi: 10.1016/S0014-5793(01)03309-9. [DOI] [PubMed] [Google Scholar]
- 20.Mele DA, Salmeron A, Ghosh S, Huang HR, Bryant BM, Lora JM. BET bromodomain inhibition suppresses TH17-mediated pathology. J Exp Med. 2013;210:2181–2190. doi: 10.1084/jem.20130376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Asangani IA, Dommeti VL, Wang X, et al. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature. 2014;510:278–282. doi: 10.1038/nature13229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia PL, Miller AL, Kreitzburg KM, et al. The BET bromodomain inhibitor JQ1 suppresses growth of pancreatic ductal adenocarcinoma in patient-derived xenograft models. Oncogene. 2016;35:833–845. doi: 10.1038/onc.2015.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Filippakopoulos P, Qi J, Picaud S, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao F, Yang Y, Wang Z, Gao X, Zheng B. BRAD4 plays a critical role in germinal center response by regulating Bcl-6 and NF-κB activation. Cell Immunol. 2015;294:1–8. doi: 10.1016/j.cellimm.2015.01.010. [DOI] [PubMed] [Google Scholar]
- 25.Stanlie A, Yousif AS, Akiyama H, Honjo T, Begum NA. Chromatin reader Brd4 functions in Ig class switching as a repair complex adaptor of nonhomologous end-joining. Mol Cell. 2014;55:97–110. doi: 10.1016/j.molcel.2014.05.018. [DOI] [PubMed] [Google Scholar]
- 26.Yoshizaki A, Miyagaki T, DiLillo DJ, et al. Regulatory B cells control T-cell autoimmunity through IL-21-dependent cognate interactions. Nature. 2012;491:264–268. doi: 10.1038/nature11501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geijtenbeek TB, Gringhuis SI. Signalling through C-type lectin receptors: shaping immune responses. Nat Rev Immunol. 2009;9:465–479. doi: 10.1038/nri2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Z, Wang P, Chen H, et al. Drug Discovery Targeting Bromodomain-Containing Protein 4. J Med Chem. 2017;60:4533–4558. doi: 10.1021/acs.jmedchem.6b01761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brown JD, Lin CY, Duan Q, et al. NF-κB directs dynamic super enhancer formation in inflammation and atherogenesis. Mol Cell. 2014;56:219–231. doi: 10.1016/j.molcel.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saraiva M, Cheistensen JR, Tsytsykova AV, et al. Identification of a macrophage-specific chromatin signature in the IL-10 locus. J Immunol. 2005;175:1041–1046. doi: 10.4049/jimmunol.175.2.1041. [DOI] [PubMed] [Google Scholar]
- 31.Hedrich CM, Rauen T, Apostolidis SA, et al. Stat3 promotes IL-10 expression in lupus T cells through trans-activation and chromatin remodeling. Proc Natl Acad Sci U S A. 2014;111:13457–13462. doi: 10.1073/pnas.1408023111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choi SY, Lee HH, Lee JH, et al. TonEBP suppresses IL-10-mediated immunomodulation. Sci Rep. 2016;6:25726. doi: 10.1038/srep25726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mauri C, Bosma A. Immune regulatory function of B cells. Annu Rev Immunol. 2012;30:221–241. doi: 10.1146/annurev-immunol-020711-074934. [DOI] [PubMed] [Google Scholar]
- 34.Mizoguchi A, Mizoguchi E, Takedatsu H, Blumberg RS, Bhan AK. Chronic intestinal inflammatory condition generates IL-10-producing regulatory B cell subset characterized by CD1d upregulation. Immunity. 2002;16:219–230. doi: 10.1016/S1074-7613(02)00274-1. [DOI] [PubMed] [Google Scholar]
- 35.Fillatreau S, Sweenie CH, McGeachy MJ, Gray D, Anderton SM. B cells regulate autoimmunity by provision of IL-10. Nat Immunol. 2002;3:944–950. doi: 10.1038/ni833. [DOI] [PubMed] [Google Scholar]
- 36.Mauri C, Gray D, Mushtaq N, Londei M. Prevention of arthritis by interleukin 10-producing B cells. J Exp Med. 2003;197:489–501. doi: 10.1084/jem.20021293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosser EC, Mauri C. Regulatory B cells: origin, phenotype, and function. Immunity. 2015;42:607–612. doi: 10.1016/j.immuni.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 38.Balkwill F, Montfort A, Capasso M. B regulatory cells in cancer. Trends Immunol. 2013;34:169–173. doi: 10.1016/j.it.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 39.He Y, Qian H, Liu Y, Duan L, Li Y, Shi G. The roles of regulatory B cells in cancer. J Immunol Res. 2014;2014;215471 doi: 10.1155/2014/215471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y, Gallastegui N, Rosenblatt JD. Regulatory B cells in anti-tumor immunity. Int Immunol. 2015;27:521–530. doi: 10.1093/intimm/dxv034. [DOI] [PubMed] [Google Scholar]
- 41.Carter NA, Rosser EC, Mauri C. Interleukin-10 produced by B cells is crucial for the suppression of Th17/Th1 responses, induction of T regulatory type 1 cells and reduction of collagen-induced arthritis. Arthritis Res Ther. 2012;14:R32. doi: 10.1186/ar3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nicodeme E, Jeffrey KL, Schaefer U, et al. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468:1119–1123. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fu LL, Tian M, Li X, et al. Inhibition of BET bromodomains as a therapeutic strategy for cancer drug discovery. Oncotarget. 2015;6:5501–5516. doi: 10.18632/oncotarget.3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chaidos A, Caputo V, Karadimitris A. Inhibition of bromodomain and extra-terminal proteins (BET) as a potential therapeutic approach in haematological malignancies: emerging preclinical and clinical evidence. Ther Adv Hematol. 2015;6:128–141. doi: 10.1177/2040620715576662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barrett E, Brothers S, Wahlestedt C, Beurel E. I-BET151 selectively regulates IL-6 production. Biochim Biophys Acta. 20141842:1549–1555. doi: 10.1016/j.bbadis.2014.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim HS, Jang JH, Lee MB, et al. A novel IL-10-producing innate lymphoid cells (ILC10) in a contact hypersensitivity mouse model. BMB Rep. 2016;49:293–296. doi: 10.5483/BMBRep.2016.49.5.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim HS, Lee JH, Han HD, et al. Autocrine stimulation of IL-10 is critical to the enrichment of IL-10-producing CD40(hi)CD5(+) regulatory B cells in vitro and in vivo. BMB Rep. 2015;48:54–59. doi: 10.5483/BMBRep.2015.48.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.