Abstract
Viruses are completely dependent upon cellular machinery to support replication and have therefore developed strategies to co-opt cellular processes to optimize infection and counter host immune defenses. Many viruses, including human immunodeficiency virus type 1 (HIV-1), encode a relatively small number of genes. Viruses with limited genetic content often encode multifunctional proteins that function at multiple stages of the viral replication cycle. In this review, we discuss the functions of HIV-1 regulatory (Tat and Rev) and accessory (Vif, Vpr, Vpu, and Nef) proteins. Each of these proteins has a highly conserved primary activity; however, numerous additional activities have been attributed to these viral proteins. We explore the possibility that HIV-1 proteins leverage their multifunctional nature to alter host transcriptional networks to elicit a diverse set of cellular responses. Although these transcriptional effects appear to benefit the virus, it is not yet clear whether they are strongly selected for during viral evolution or are a ripple effect from the primary function. As our detailed knowledge of these viral proteins improves, we will undoubtedly uncover how the multifunctional nature of these HIV-1 regulatory and accessory proteins, and in particular their transcriptional functions, work to drive viral pathogenesis.
Keywords: human immunodeficiency virus, host-pathogen interactions, transcription, virus replication, multifunctional proteins
INTRODUCTION
A defining feature of retroviruses is the reverse transcription of an RNA genome into a DNA copy, which is subsequently integrated into the host cell genome. The family Retroviridae can be further subdivided at the genus level: Alpharetrovirus, Betaretrovirus, Gammaretrovirus, Deltaretrovirus, Lentivirus, and Spumavirus. Gammaretroviruses, such as Moloney murine leukemia virus (M-MLV), and alpharetroviruses, such as avian sarcoma leukosis virus (ASLV), encode only the gag, pol, and env structural and enzymatic genes to complete their life cycle. In contrast, other species of retroviruses, such as human immunodeficiency virus type 1 (HIV-1) of the Lentivirus, encode several additional accessory and regulatory proteins to modulate host cell immune responses or control the expression of the viral genome. The accessory proteins of HIV-1 are Vif, Vpr, Vpu, and Nef and the regulatory proteins are Tat and Rev. Since the discovery in the early 1980s that HIV-1 is the causative agent of acquired immune deficiency syndrome (AIDS), intensive research has been dedicated to understanding how these viral proteins subvert the host cell machinery to support active replication. Generally, each of these proteins has a highly conserved function that is considered its primary activity. However, numerous additional activities have been attributed to these viral proteins (secondary activities), leading to the conclusion that the accessory and regulatory factors are highly multifunctional. In this review we summarize the primary, most conserved function of the accessory and regulatory proteins but also describe secondary activities attributed to these relatively small proteins. These secondary activities have not been conclusively proven in all cases and we do not exhaustively cover all reports in the literature, nor is it clear whether these functions simply reflect pleiotropic effects indirectly stemming from their primary functions. However, it is intriguing that many of the secondary functions alter either viral or cellular transcription, prompting us to explore the possibility that HIV-1 utilizes multifunctional proteins to rewire host transcription networks. By emphasizing this one function common to all the accessory and regulatory proteins (transcription), we illustrate how perturbations in diverse cellular pathways (i.e., the primary function of the proteins) can be funneled into a single type of secondary output. Although these transcriptional effects may benefit the virus in various ways, it is not yet clear whether they are strongly selected for during viral evolution. It will be important to explore separation-of-function mutants in these viral proteins to establish whether they truly encode multiple essential functions or whether the secondary transcriptional effects are entirely indirect. If the transcriptional responses are indeed a ripple effect from the primary function, these viral factors may still have opportunities to evolve and better exploit these secondary functions for virus replication given that HIV-1 was transmitted to its human host only approximately one hundred years ago (1).
Tat
Primary Function
Following HIV-1 integration into the host genome, RNA polymerase II (RNAP II) assembles at the viral promoter located in the 5′ long terminal repeat (LTR) and begins the process of transcribing viral RNA. The promoter contains cis elements for host transcription factors, including three SP1 sites and two nuclear factor κB (NF-κB) sites, which provide an entirely cell-intrinsic mechanism to activate transcription, especially in activated T cells targeted by HIV-1 where the NF-κB pathway is induced (2). However, despite this appropriation of cellular mechanisms to increase gene expression, transcription complexes that assemble at the HIV-1 promoter generate predominantly short, incomplete viral transcripts (3).
To bypass this block to transcription elongation, HIV-1 encodes its own transcription factor, Tat, which increases the processivity of RNAP II to generate full-length viral mRNAs (Figure 1) (4). Tat activates transcription by binding to a nascent, 5′ stem-loop RNA structure termed the transactivation response element (TAR) (5). A major host cofactor for Tat is cyclin T1 (CCNT1) (6), which, together with cyclin-dependent kinase 9 (CDK9), constitutes positive transcription elongation factor b (P-TEFb). This heterodimeric host kinase phosphorylates and activates paused RNAP II and regulates elongation at most cellular genes (7). The earliest models of Tat activity proposed that Tat recruited the P-TEFb complex to the nascent TAR RNA, which positioned the kinase in proximity to the stalled polymerase for phosphorylation-dependent activation (8). Interestingly, CCNT1 also contributes to RNA binding as it contacts bases in the TAR loop to achieve a high-affinity interaction (9). The Tat-P-TEFb complex potently stimulates viral gene expression, initiating the postintegration steps of the life cycle, which eventually leads to viral budding and the infection of new cells.
Figure 1.
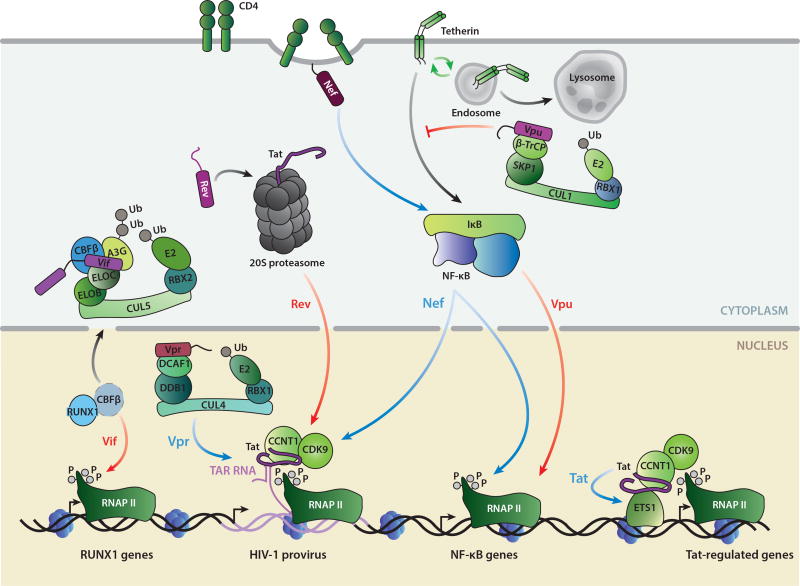
Primary effects of HIV-1 accessory and regulatory proteins. Tat recruits P-TEFb to a paused RNAP II at the HIV-1 promoter in the nucleus to activate viral transcription. Tat utilizes P-TEFb from both the 7SK snRNP and the superelongation complex to activate transcription (indicated by blue arrows). Rev exports partially and fully unspliced viral messages from the nucleus to the cytoplasm. Vif, Vpr, and Vpu all utilize Cullin-RING ubiquitin (Ub) ligases for their primary activities of A3G degradation, DNA damage response/ G2 arrest activation, and CD4/tetherin degradation, respectively. Nef induces the lysosomal degradation of CD4 at the plasma membrane. Gray arrows indicate the viral targeting of cellular substrates to the 26S proteasome or lysosome for degradation. Viral proteins are shaded in purple and hijacked host complexes in green and blue.
The majority of P-TEFb in the cell is sequestered in an RNA-based inactive complex termed the 7SK small nuclear ribonucleoprotein (snRNP) (10, 11) in which the HEXIM1 protein inhibits the kinase activity of CKD9 in an RNA-dependent manner (12). Consistent with the original model of Tat activity, HIV-1 replication (13, 14) or Tat expression alone (14) releases a significant fraction of P-TEFb from the inhibitory complex. This release is due to competitive binding between Tat and HEXIM1 for CCNT1, and the P-TEFb freed from the 7SK snRNP can then be delivered to a paused RNAP II at the viral promoter for activation.
However, 7SK snRNP–inhibited P-TEFb can be recruited to the HIV-1 promoter and associates with RNAP II (15), which provides a simple explanation for why RNAP II transcription is nonprocessive in its basal state. When Tat is synthesized, it can also bind the HIV-1 promoter together with the 7SK snRNP even in the absence of TAR, which demonstrates that the viral RNA is not necessary for recruitment of Tat or P-TEFb. Once the TAR RNA is transcribed by RNAP II, the proteins of the inhibitory 7SK snRNP are displaced from DNA-bound transcription complexes, which activate the kinase activity of P-TEFb to phosphorylate RNAP II. In addition, Tat forms a soluble Tat–7SK snRNA–P-TEFb complex upon ejection of HEXIM1 (16), which itself might be recruited to the viral promoter. TAR expression would then hand off Tat from the cellular RNA to the viral RNA, completing the displacement of the inhibitory complex. In addition to the 7SK snRNP, Tat binds a larger P-TEFb complex termed the superelongation complex (SEC) (16, 17). The 7SK snRNP and SEC are distinct complexes, although recent work has demonstrated that the AFF1 scaffold protein of the SEC is a ubiquitous P-TEFb partner and therefore a component of the 7SK snRNP (18). The SEC is required for full Tat activation and the AFF4 scaffold increases the affinity of Tat-P-TEFb for TAR by 30-fold, likely by limiting the flexibility of the Tat-TAR recognition motif (TRM) of CCNT1 (19). The extent of cross talk between the 7SK snRNP and the SEC is still uncertain; however, Tat clearly utilizes multiple host complexes to achieve potent transcriptional stimulation of the integrated provirus.
Secondary Function
In addition to activating the viral promoter, Tat alters the expression of cellular genes to facilitate viral replication. For example, HIV-1 infection or Tat expression increased the transcription and secretion of four chemokines (IP-10, HuMIG, MCP-2, and MCP-3) in immature dendritic cells (iDCs) (20), which then induced the chemotaxis of T cells and monocytes. The migration of these HIV-1 target cells was proposed as a means for the virus to amplify the infection. Despite the induction of chemokines, Tat did not cause generic iDC maturation, arguing for a specific cellular response. As Tat is localized to the nucleus, cytoplasm, and cellular membrane (21, 22), its effect on cellular transcription could be direct or indirect.
Several groups have demonstrated a direct effect of Tat on host transcription by localizing the viral transcription factor to cellular promoters. Chromatin immunoprecipitation (ChIP)-on-chip with a microarray chip containing human promoter sequences illustrated that Tat could bind approximately 450 promoters in Jurkat cells, although changes in the expression of these genes were not globally determined (23). However, Tat specifically bound the promoters and increased the expression of two regulatory proteins of the PP2A phosphatase, PPP2R1B and PPP2R5E. PP2A dephosphorylates FOXO3a, which then translocates to the nucleus to activate a proapoptotic pathway. HIV-1 infection can cause apoptosis in CD4+ T cells (23), and specific knockdown of the regulatory phosphatase subunits prevented the observed Tat-dependent apoptosis in Jurkat cells. Therefore, Tat induction of PPP2R1B and PPP2R5E expression is a plausible mechanism for HIV-1-triggered apoptosis of infected cells. More recent Tat ChIP-sequencing and RNA-sequencing experiments in Jurkat cells have also uncovered 456 cellular genes that are bound by Tat and experience changes in gene expression; however, the overlap with the previously identified Tat gene targets was not evaluated (24). Some genes are stimulated, whereas others are downregulated, and regulation may occur via transcription initiation or elongation. TAR-like RNA structures were not identified on the nascent transcripts that might provide binding sites for Tat, but instead, ETS1, a T cell master transcription factor, was enriched near most Tat peaks (Figure 2). Indeed, Tat bound ETS1 and knockdown of ETS1 reduced Tat recruitment at target genes, suggesting that Tat affects host gene transcription through the ETS1 interaction. It is not yet clear how hijacking cellular transcription networks may benefit the virus or whether the interaction with ETS1 is under selective pressure.
Figure 2.
Transcriptional effects of HIV-1 accessory and regulatory proteins. Tat directly deregulates multiple cellular genes through the ETS1 transcription factor. Rev-induced degradation of Tat through the 20S proteasome decreases HIV-1 transcription. Vif sequesters CBFβ from RUNX1 to decrease RUNX1-dependent transcription, including the A3G gene. Vpr DNA damage response activation and G2 arrest activate HIV-1 transcription. Membrane-bound Nef modulates signaling pathways through NF-κB to activate both viral and cellular transcription. Vpu inhibits tetherin signaling through NF-κB to decrease transcription at NF-κB target genes. Blue arrows indicate positive effects on transcription, whereas red arrows indicate inhibitory effects.
Tat may also alter cellular transcription indirectly, as many genes that show changes in expression do not show corresponding physical enrichment of Tat (24). This may occur by altering the activity of other transcription factors, which consequently affect downstream gene expression. For example, Tat increases IRF7 and STAT1 expression (20), which regulates interferon-inducible genes. Tat may indirectly affect transcription of other genes through competition with protein-protein interactions. For example, in forming the Tat–P-TEFb complex for viral transcription, Tat buries 3,500 Å2 of surface area on P-TEFb, the majority on the CCNT1 subunit (25). This interaction surface is competitive with several host proteins that control P-TEFb activity, including HEXIM1 (14), BRD4 (26), and CIITA (27), and indeed, Tat expression, even at low physiological levels during infection, releases a substantial amount of P-TEFb from the 7SK snRNP (13, 14). The pool of freed P-TEFb released from the inhibited 7SK complexes would then be available to activate cellular gene expression, either with or without Tat. At the CD69 promoter, for example, Tat binding increases P-TEFb recruitment and transcription and may require ETS1 (24). In another example, competition between Tat and CIITA for CCNT1 binding decreases the expression of major histocompatibility complex (MHC) class II genes and inhibits antigen presentation in macrophages, potentially reflecting another strategy for Tat to establish optimal replication conditions (27). Finally, BRD4 inhibitors activate Tat-dependent HIV-1 transcription by increasing the pool of P-TEFb available for Tat, consistent with a competition model between Tat and BRD4 (28). These results suggest that Tat may alter many host P-TEFb regulatory pathways by titrating the elongation factor away from other binding partners.
Tat protein produced during infection can be secreted (22) and enter neighboring cells by endocytosis (29). Although the biological importance of extracellular Tat has not yet been conclusively established, the secreted Tat pool can activate T cells independently of antigen but not increase T cell proliferation. These activated T cells are then prone to infection, further amplifying viral replication (30, 31). This activity appears to be largely a transcriptional effect, as exogenous Tat deregulates 94 genes in primary T cells, leading to the secretion of interleukin 17 (IL-17) to generate a proinflammatory state suited to viral infection (30), although it is unclear whether this transcriptional effect is direct or indirect.
It will be interesting to determine whether the functions of Tat in viral and cellular transcription entirely overlap at the genetic level or whether certain amino acids in Tat specifically affect cellular transcription. In this regard, several amino acids in Tat that do not overlap with other protein reading frames in the virus are highly conserved in patients (Glu9, Pro10, Trp11, Gln17, Thr20, and Ala21) yet display neutral selection during replication competition experiments in Sup-T1 cells (32). Future work will reveal whether some of these residues have undergone positive selection for host transcription responses or other functions that are not recapitulated in tissue culture, or whether these residues function in a cell-type-specific manner.
Rev
Primary Function
After transcriptional activation by Tat, expression of the full-length viral transcripts is regulated by splicing and export. Complete cellular processing yields spliced RNAs that are exported from the nucleus through the canonical TAP pathway and results in translation of only three viral proteins: Tat, Rev, and Nef. Rev is then reimported into the nucleus through its nuclear localization sequence (NLS), where it directs the export of partially and fully unspliced messages that are translated into the remaining viral proteins and provides genomic RNA for packaging into budding virions (33). Rev, an RNA-binding protein similar to Tat, exports the intron-containing RNAs by binding specifically to the ~350-nucleotide, highly structured Rev response element (RRE) contained in the Env sequence (Figure 1) (34). The Rev NLS, which is an arginine-rich motif (ARM), doubles as its RNA-binding domain. Rev also encodes two hydrophobic oligomerization domains (ODs) and a leucine-containing nuclear export sequence (NES), which are critical for its export activity. Rev-mediated nuclear export is a multistep process that is initiated when the ARM contacts a high-affinity site in stem IIB of the RRE RNA (33). Rev then likely dimerizes on the RNA through the OD when a second Rev monomer contacts the adjacent stem IIABC in the RRE, followed by further Rev oligomerization on the RNA to yield an export-competent RNP containing 6–10 Rev subunits (35–39).
The major host cofactor for Rev export is chromosome maintenance factor 1 (CRM1) (40). CRM1 normally exports cellular proteins that contain a leucine-rich NES and is not typically involved in mRNA export. Rev hijacks CRM1 for viral RNA export by functioning as an adaptor, wherein the Rev NES engages the host export factor and the ARM engages RRE-containing messages. Interestingly, recent electron microscopy reconstructions demonstrate that the export complex contains a dimer of CRM1 (41). Given that CRM1 had been shown to function only as a monomer for known cellular cargos, Rev-dependent CRM1 dimerization is likely a way to increase recognition of the low-affinity Rev NES sites. Assembly of this virus-host export complex results in robust cytoplasmic trafficking of intron-containing HIV-1 messages to complete the later stages of the virus life cycle. It is currently unclear whether the dimerization of CRM1 is unique to the Rev-RRE RNP or reflects a mode of binding of other host cargos. It also is not known whether Rev-RRE binding significantly alters the endogenous pool of CRM1 and thereby affects host RNA export or gene expression as a secondary consequence, or whether Rev at physiological concentrations binds and exports cellular messages that contain RNA structural elements similar to the RRE or other RNA features.
Secondary Function
Rev indirectly regulatesHIV-1 transcription in at least two ways. First, the fully spliced transcripts encoding Tat, Rev, and Nef lack the RRE; therefore, Rev export of RRE-containing messages decreases the accumulation of fully spliced messages. This posttranscriptional negative feedback loop decreases Tat protein levels (42). Second, Rev decreases Tat protein at a posttranslational step. In this mechanism, physiological expression of Rev during infection decreases the levels of the NQO1 protein, which is an inhibitor of the 20S core proteasome. Tat, an inherently unstructured protein, can be degraded by the 20S complex, and the Rev-dependent decrease in the inhibitory NQO1 protein activates the 20S proteasome to degrade Tat and decrease viral transcription (Figure 2) (43). Therefore, Rev inhibits Tat activity at posttranscriptional and posttranslational levels. Interestingly, several transcription factors, including p53 (44), are intrinsically disordered and are regulated by 20S proteasomal degradation. Future work may uncover that Rev also indirectly deregulates cellular transcription by inducing the degradation of transcription factors through NQO1 and the 20S proteasome.
Vif
Primary Function
Among the known lentiviral accessory proteins, Vif is found in all lentiviruses except equine infectious anemia virus (EIAV). The primary function of Vif is to counteract the antiviral effects of host APOBEC3 (A3) innate immune proteins, restriction factors that inhibit replication by inducing hypermutation of the viral genome (45–47). Vif antagonizes A3 by hijacking a cellular Cullin-RING ubiquitin ligase (CRL), resulting in the ubiquitination and subsequent targeting of A3 for proteasomal degradation (48–51). The HIV-1 Vif E3 ligase complex comprises CRL5, which includesCullin-5 (CUL5), elongin B (ELOB), elongin C (ELOC), and RING-box protein 2 (RBX2), as well as a noncanonical cofactor, core-binding factor beta subunit (CBFβ) (50–52). The recruitment of CBFβ to the Vif E3 ligase is surprising given that CBFβ is a transcription cofactor and not a component of a known cellular E3 ligase. Recruitment of CBFβ serves to stabilize HIV-1 Vif and is required for HIV-1 Vif-mediated A3 degradation activity in vivo (Figure 1) (50, 51, 53–55). More recent work has shown that CBFβ is required only for primate lentiviral infection, as it is dispensable for nonprimate Vif function (53, 56). These data suggest that there was an evolutionary pressure that promoted the acquisition of CBFβ to the primate Vif complex. A number of reviews cover the primary function of Vif to ubiquitinate APOBEC3 proteins (57–60); here, we discuss the role Vif has in altering the host transcriptome.
Secondary Function
CBFβ forms a heterodimer with members of the RUNX family of transcription factors, serving to both stabilize RUNX steady-state levels and enhance DNA-binding affinity to regulate the expression of a diverse set of genes (61, 62). Initial in vitro biochemical work established that Vif can outcompete RUNX for CBFβ binding, suggesting that the virus may utilize the Vif-CBFβ interaction to alter gene expression in infected T cells (54). Indeed, overexpression of HIV-1 Vif in permissive Jurkat T cells altered the expression patterns of a large number of genes that exhibit enriched RUNX1 binding sites (Figure 2) (54). Moreover, a pharmacological approach to inhibit RUNX1 demonstrated that RUNX1 and CBFβ play a role in reducing HIV-1 replication (63). Together, these data establish that the presence of Vif alters endogenous RUNX activity, potentially to the benefit of the virus.
More recent work by Anderson & Harris (64) presents an additional facet to the role the Vif-CBFβ interaction has in viral infectivity. In their efforts to investigate CBFβ function, they employed a separation-of-function mutant that would allow CBFβ to bind to either RUNX or Vif (64, 65). Using this separation-of-function CBFβ construct, they discovered that the CBFβ- RUNX interaction is required for APOBEC3 transcription. Reduction or ablation of CBFβ mRNA by RNA interference (RNAi) or CRISPRs reduced the expression of APOBEC3 (C, D, F, G, and H) mRNA, as detected by quantitative reverse transcription PCR in either CD4+ T cell lines or primary CD4+ T cells. Furthermore, an RNAi-resistant CBFβ complemented the CBFβ knockdown by increasing A3G protein expression levels, and this effect required interaction with the RUNX proteins. Importantly, ablation of CBFβ rendered nonpermissive H9 cells permissive to infection with a Vif-deficient HIV-1 virus, as the restrictive potential of APOBEC3 in these cells was almost completely suppressed (64). Additionally, these data might explain why primate Vif acquired the CBFβ interaction. It is tempting to speculate that the Vif-CBFβ interaction developed in an effort to allow Vif to disrupt the RUNX-mediated transcription of APOBEC3 proteins and thus counteract the APOBEC3 repertoire at the transcriptional and posttranslational levels. Although it is uncertain what driving force promoted the primate Vif-CBFβ interaction, together these findings highlight the large transcriptional changes that arose from Vif hijacking CBFβ and the significant implications this has on both the virus and the infected cell.
In addition to the transcriptional consequences of hijacking CBFβ, Vif remodels the cellular phosphoproteome during HIV-1 infection. A whole-cell proteomics study demonstrated that Vif was necessary and sufficient for the proteasomal degradation of the B56 family of regulatory subunits of the cellular phosphatase PP2A, and quantitative phosphoproteomics revealed Vif-dependent hyperphosphorylation of over 200 cellular proteins (66). Intriguingly, the ability of Vif to target PPP2R5 subunits is found in primate and nonprimate lentiviral lineages, suggesting that remodeling the cellular phosphoproteome is a conserved function of Vif. Although there is currently no direct link to transcription through PP2A degradation, the general importance of posttranslational modifications, especially phosphorylation, in regulating transcription is well-established. Therefore, future work will likely demonstrate that Vif-mediated remodeling of host phosphorylation will have major effects on cellular transcription.
Vpu
Primary Function
The vpu gene is found exclusively in HIV-1 and precursor simian immunodeficiency virus (SIV) strains and produces a small, transmembrane protein, Vpu, that is expressed late in the viral replication cycle (67–70). Initially, Vpu was observed to play a critical role in facilitating viral egress from the plasma membrane; however, these observations were cell type specific, suggesting the presence of a host restriction factor (67, 71). Almost 20 years after the discovery that Vpu promotes viral release, the host factor BST-2/Tetherin was identified as a target of Vpu (72, 73). The Vpu-mediated inhibition of BST-2/Tetherin is the most active area of current Vpu-related research; however, additional functions are attributed to this protein. These include primarily the downregulation of CD4 and MHC1 molecules, the inhibition of NF-κB activation, and the formation of a viroporin ion channel in the Golgi apparatus to alter membrane potential and possibly enhance virion release (74–78).
By removing membrane-bound host proteins that inhibit viral replication, particularly CD4 and BST-2/Tetherin (79), Vpu remodels the cell surface to carry out its functions. To downregulate CD4, Vpu targets newly synthesized CD4 while it is in the endoplasmic reticulum (ER), thus preventing it from trafficking to the plasma membrane (Figure 1). Mechanistically, Vpu does this by recruiting the CUL1–β-TrCP–Skp1–RBX1 E3 ligase complex to the ER, where it ubiquitinates newly synthesized CD4 molecules, leading to their retention in the ER (80). As a result, the ubiquitinated CD4 molecules are processed through the ER-associated degradation (ERAD) pathway and ultimately degraded by the proteasome (81, 82). In addition to functioning in the ER, Vpu acts at the plasma membrane to counteract the inhibitory role of BST-2. BST-2/Tetherin is a type II transmembrane protein that is thought to directly tether Vpu-deficient virions to the surface of infected cells (83, 84). Vpu decreases BST-2/Tetherin found on the cell surface by directly binding to it and inhibiting the recycling of internalized BST2 back to the plasma membrane (Figure 1) (73, 85, 86). Vpu can also ubiquitinate BST-2/Tetherin via the β-TrCP–Skp1–RBX1 E3 ligase complex, which destines BST-2/Tetherin for lysosomal degradation (87, 88). Lentiviruses neutralize host BST-2/Tetherin through multiple mechanisms; select SIV strains utilize Nef, HIV-2 utilizes its Env glycoprotein, and HIV-1 has evolved to utilize Vpu (89–96). That lentiviruses have evolved multiple ways to antagonize BST-2 emphasizes the importance of neutralizing this restriction factor for viral pathogenesis.
Secondary Function
Vpu also influences immune signaling, particularly through deregulation of the NF-κB pathway. This is thought to occur through two distinct mechanisms. First, Vpu restricts NF-κB signaling by downregulating BST-2/Tetherin. In addition to its role in preventing viral budding, BST-2/Tetherin activates the NF-κB pathway (97–99). Therefore, Vpu-induced reduction of BST-2/Tetherin at the plasma membrane also dampens NF-κB signaling (Figure 2). Second, Vpu sequesters the F-box protein β-TrCP, which it uses to ubiquitinate BST-2/Tetherin. During the normal activation of the NF-κB pathway, the SCFβ-TrCP ligase degrades the inhibitor of NF-κB (IκB), which allows the nuclear translocation of NF-κB. However, as noted above, Vpu, which contains a canonical DpSGxxpS phosphodegron bound by β-TrCP, also uses SCFβ-TrCP to ubiquitinate BST-2/Tetherin. In this model, Vpu binding of SCFβ-TrCP prevents the degradation of IκB, which further inhibits signaling through the NF-κB pathway (100, 101). These effects were also observed during infection, arguing against an overexpression artifact (100). A Vpu phosphodegron mutant that is deficient in binding to SCFβ-TrCP (S52/56N) is unable to inhibit the NF-κB pathway (100, 102, 103), which supports a role for the ubiquitin ligase in Vpu antitranscriptional activity and is consistent with both models. In support of the sequestration model, Vpu expression also stabilizes a number of SCFβ-TrCP substrates, including β-catenin. Importantly, this stabilization is lost with the Vpu S52/56N mutant.
Changes to NF-κB signaling likely has significant transcriptional consequences for the cell, because NF-κB is an important transcription factor that facilitates biological processes such as cell proliferation, cytokine production, and induction of apoptosis (104). The two-pronged deregulation of NF-κB signaling by Vpu also has similarities to the downregulation of A3G by Vif. In each case, a strong primary interaction (Vpu–β-TrCP or Vif–CBFβ) modulates two nodes of a pathway, with an ultimate effect on transcription. A simple model, then, for the pleiotropic effects of the HIV-1 regulatory and accessory proteins is that they target critical host proteins involved in many pathways, such that their deregulation leads to a multitude of effects in the cell.
Vpr
Primary Function
The Vpr protein is conserved across human and primate lentiviruses and is specifically incorporated into the viral particle by interactions with the p6 domain of Gag (105). Vpr has many reported functions, including LTR transactivation, nuclear import of the preintegration complex, cellular apoptosis, cell cycle arrest, and activation of the DNA damage response (106–111). The activation of the DNA damage response by Vpr is conserved across primate lentiviruses and is considered its primary function (112). The interaction of Vpr with CRL4DCAF1 is required for many of these phenotypes (113–115). For example, knockdown of DCAF1 prevents Vpr-induced G2 arrest, highlighting that the CRL4DCAF1 ubiquitin ligase complex is essential for Vpr’s primary function (Figure 1) (113, 114).
Extensive effort has been directed at identifying the cellular substrate of the Vpr-CRL4DCAF1-Vpr complex whose ubiquitination results in the DNA damage response and G2 arrest. Laguette et al. (116) recently proposed that aberrant activation of the SLX4 DNA damage response complex by Vpr induces cell cycle arrest. In this model, Vpr recruitment of DCAF1 and the kinase PLK1 to the MUS81-EME1 endonuclease of the SLX4 complex prematurely activates its nucleolytic activity, increasing FANCD2 foci and causing G2 arrest. However, other work has shown that Vpr can induce cell cycle arrest even if SLX4 is knocked out by CRISPR (117). Another model of Vpr-induced cell cycle arrest proposes that Vpr binds the CRL4DCAF1 ligase and sequesters it away from its normal substrates whose degradation is required for proper cell cycling. This model is supported by the fact that DDB1 knockdown alone causes cells to arrest in G2 (115, 118) and DCAF1 knockdown causes G1 and G2 arrests (113). A recent crystal structure of UNG2-Vpr-DCAF1-DDB1 complex supports this model, highlighting that Vpr binds to DCAF1 on the typical DCAF substrate interaction surface, potentially occluding the recruitment of normal substrates (119). UNG2 appears to be a neo-substrate for DCAF1, as Vpr adapts UNG2 to the ligase complex, providing the entirety of the interaction surface. Chen et al. (120) have proposed that UNG2 contributes to the Vpr-mediated decrease in error rate of reverse transcription. One issue with the sequestration model is that a specific point mutant in Vpr (R80A) is unable to arrest cells in G2 but maintains the interaction with DCAF1 (114, 116), suggesting that DCAF1 binding alone is not entirely sufficient to induce G2 arrest.
Secondary Function
Despite several proposed models of activity, Vpr clearly induces the DNA damage response with subsequent G2 arrest. Vpr can also modestly increase transcription from the HIV-1 LTR. It was originally assumed that this transcriptional activation was direct because Vpr interacts with cellular transcription factors, including SP1 (121), which provided a recruitment mechanism for LTR stimulation. Further, Vpr and Tat can interact with each other and with distinct regions of CycT1, resulting in synergistic activation of theHIV-1LTRthrough P-TEFb (122). However, other work has demonstrated that the transcriptional effect by Vpr appears to be largely indirect, as G2 arrest alone induces expression from the LTR (Figure 2) (123, 124). Moreover, Vpr mutants that are unable to arrest cells in G2, including R80A, are also unable to activate the LTR, whereas there is no correlation between Vpr nuclear import mutants and transactivation (124–126). Therefore, based on the mutant phenotypes, Vpr-mediated activation of the viral promoter may be solely an indirect effect. Given that Vpr increases the expression of numerous cellular genes (127), it will be interesting to determine whether this transcriptional effect is similarly due to activation of the DNA damage response and G2 arrest. In addition, many human genes are cell cycle regulated (128) and could be affected by Vpr expression.
It is intriguing that Vif, Vpu, and Vpr all target host ubiquitin ligase complexes. Integration into the ubiquitin-proteasome pathway may generally result in pleiotropic effects, including changes in transcription, as these ligases are components of signaling pathways and generally have multiple substrates. As degrons for ubiquitin ligases are often short, unstructured peptides, it might be relatively easy for the accessory factors to rapidly evolve degron-mimics to hijack these complexes.
Nef
Primary Function
Nef is a small viral accessory protein that is produced early in HIV-1 infection (129). Although not essential for viral replication in permissive cells, long-term infection with Nef-defective HIV-1 progresses to AIDS very slowly, if at all (130). Many functions have been credited to Nef, including modulating signaling by protein tyrosine kinases and downregulating CD4, MHC-I, BST2/Tetherin, and other cell surface receptors (92, 94, 96, 131"–135). Nef modulates host trafficking by binding to AP-1 and AP-2 clathrin adaptor complexes involved in coated vesicle budding, and the ESCRT machinery involved in degradative lysosomal sorting (136). MHC-I is rerouted from the trans-Golgi network to the endo-lysosomal system for degradation by Nef and AP-1 (137). Additionally, Nef and AP-2 bind CD4 at the plasma membrane, which triggers the recruitment of clathrin, budding, and eventual lysosomal destruction (Figure 1) (138). Membranous CD4 downregulation prevents superinfection and is thought to aid viral egress by preventing Env-CD4 interactions during budding (139). On the basis of recent findings, the mechanism by which Nef promotes HIV-1 infectivity may be mediated by the downregulation of the integral membrane protein, SERINC5 (140, 141). Little biochemical literature exists on SERINC5 and it is currently unclear whether SERINC5 binds Nef directly or via one or more additional factors, as AP-2 is essential for SERINC5 downregulation (140, 141). Nef-dependent downregulation of SERINC5 is proposed to be one of the most important contributions of Nef to infectivity by HIV-1 and is now a major focus of Nef-related research.
Secondary Function
Nef is the most abundantly expressed viral protein early in infection and can activate transcription through NF-κB or nuclear factor of activated T cells (NFAT) pathways (102, 142, 143). In contrast, Vpu is expressed late in viral infection and inhibits the NF-κB pathway. It is assumed that early stimulation of NF-κB by Nef helps initiate the potent positive transcriptional feedback loop of Tat. Using luciferase reporter assays to investigate the role Nef has in NFAT and NF-κB signaling, Wang et al. (142) found that Nef activation of an NFAT-luciferase reporter required myristoylation, suggesting an indirect effect on transcription through signaling events at the plasma membrane (Figure 2). Similarly, Nef expression alone did not increase NF-κB transcription but first required pathway stimulation, indicating that Nef regulates the response to stimulation (signal transduction) rather than directly activates NF-κB (144). Importantly, activation of the NF-κB pathway promotes transcription of viral genes, and in the case of HIV-1, binding of NF-κB p50–p65 heterodimers to the HIV-1 LTR is necessary for viral replication (145). Indeed, HIV-1 Nef enhances both LTR promoter activity and the transcription of HIV-1 provirus (102). Although it would appear that HIV-1 Nef exploits cellular signaling cascades to directly promote viral replication, it is possible that remodeling of host membrane proteins indirectly triggers cellular transcription. Mutational analyses are needed to better define how Nef is directly involved in deregulating cellular gene expression.
A more global assessment of Nef-induced changes to cellular transcription revealed that Nef induces a gene expression program that is highly similar to anti-CD3 T cell activation (146). Much of this transcriptional induction required signaling through NF-κB or NFAT, supporting the earlier work on Nef function. Interestingly, several transcription factors that upregulate the LTR, including ETS1 and CDK9, were induced. CDK9 protein levels also increase upon Nef expression, and the Nef-dependent increase in LTR activation was sensitive to CDK9 inhibition. Furthermore, as highlighted above, ETS1 recruits Tat to many cellular genes for altered transcription (24). It is intriguing that Nef increases the expression of critical Tat host machinery and, in this way, acts to both increase viral transcription and further deregulate host transcription. Analogous to the Rev-stimulated degradation of Tat, the secondary effects of Nef provide a clear example of cross talk between the HIV-1 accessory and regulatory proteins to rewire the host for optimal infection.
CONCLUSION
Altogether, these examples highlight the multifunctional nature of HIV-1 regulatory and accessory proteins and emphasize a common ability to modulate cellular or viral transcription. Many viruses encode a relatively small number of genes and likely express multifunctional proteins that can alter host transcriptional networks. For example, adenovirus and human papillomavirus encode proteins that degrade p53 and alter p53-dependent transcription (147, 148); the Marburg virus VP24 protein disrupts the Keap1-Nrf2 interaction, which upregulates the transcription of cytoprotective genes (149); and HPV E7 and adenovirus E1A facilitate the expression of cell cycle–promoting genes to induce the transition from G1 to S phase (150–152).
Intriguingly, theHIV-1 transcriptional effects that we describe here appear to benefit the virus, yet it is unclear whether these effects are selected for during viral evolution. As our understanding of HIV-1 evolution continues to improve, we now recognize that many factors influence viral fitness. These factors include (a) a condensed genome and overlapping open reading frames, which influence the adaptability of neighboring genes in a manner that helps purge unfit viruses (32); (b) the host-pathogen arms race of accessory protein adaptation to maintain restriction factor counteraction; and (c) the acquisition of new binding partners such as CBFβ to potentially aid Vif in antagonizing APOBECs at the transcriptional and posttranslational levels. By improving our knowledge of viral proteins and using technologies that will enable us to study virus-host systems, we will undoubtedly uncover how the complex, multifunctional nature of accessory and regulatory HIV-1 proteins, particularly their transcriptional functions, work in concert to drive viral pathogenesis. It will be interesting to uncover how much cross talk exists between the viral proteins and the extent to which they cooperate to rewire the cell. Similar to the Rev-mediated degradation of Tat or the Nef-induced expression of Tat cofactors, the viral accessory and regulatory factors might have unexpected overlapping functional circuitry, so it will be important to evaluate both virus-host and virus-virus interactions while teasing apart the discrete activities of these multifunctional proteins.
Acknowledgments
We would like to thank Dr. Michael Emerman and Dr. Jason Fernandes for critically reading the review prior to publication. This work was supported by National Institutes of Health grants to A.D.F. (P50GM082250) and J.D.G. (P50GM082250), and a National Institutes of Health/National Institute of Allergy and Infectious Diseases postdoctoral fellowship F32-AI120867 to J.M.B.
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Worobey M, Gemmel M, Teuwen DE, Haselkorn T, Kunstman K, et al. Direct evidence of extensive diversity of HIV-1 in Kinshasa by 1960. Nature. 2008;455:661–64. doi: 10.1038/nature07390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen BK, Feinberg MB, Baltimore D. The κB sites in the human immunodeficiency virus type 1 long terminal repeat enhance virus replication yet are not absolutely required for viral growth. J. Virol. 1997;71:5495–504. doi: 10.1128/jvi.71.7.5495-5504.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lassen KG, Bailey JR, Siliciano RF. Analysis of human immunodeficiency virus type 1 transcriptional elongation in resting CD4+ T cells in vivo. J. Virol. 2004;78:9105–14. doi: 10.1128/JVI.78.17.9105-9114.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feinberg MB, Baltimore D, Frankel AD. The role of Tat in the human immunodeficiency virus life cycle indicates a primary effect on transcriptional elongation. PNAS. 1991;88:4045–49. doi: 10.1073/pnas.88.9.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berkhout B, Silverman RH, Jeang KT. Tat trans-activates the human immunodeficiency virus through a nascent RNA target. Cell. 1989;59:273–82. doi: 10.1016/0092-8674(89)90289-4. [DOI] [PubMed] [Google Scholar]
- 6.Wei P, Garber ME, Fang SM, Fischer WH, Jones KA. A novel CDK9-associated c-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–62. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- 7.Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, et al. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–45. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 9.Garber ME, Wei P, KewalRamani VN, Mayall TP, Herrmann CH, et al. The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev. 1998;12:3512–27. doi: 10.1101/gad.12.22.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nguyen VT, Kiss T, Michels AA, Bensaude O. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature. 2001;414:322–25. doi: 10.1038/35104581. [DOI] [PubMed] [Google Scholar]
- 11.Yang Z, Zhu Q, Luo K, Zhou Q. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature. 2001;414:317–22. doi: 10.1038/35104575. [DOI] [PubMed] [Google Scholar]
- 12.Michels AA, Fraldi A, Li Q, Adamson TE, Bonnet F, et al. Binding of the 7SK snRNA turns the HEXIM1 protein into a P-TEFb (CDK9/cyclin T) inhibitor. EMBO J. 2004;23:2608–19. doi: 10.1038/sj.emboj.7600275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sedore SC, Byers SA, Biglione S, Price JP, Maury WJ, Price DH. Manipulation of P-TEFb control machinery by HIV: recruitment of P-TEFb from the large form by Tat and binding of HEXIM1 to TAR. Nucleic Acids Res. 2007;35:4347–58. doi: 10.1093/nar/gkm443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barboric M, Yik JHN, Czudnochowski N, Yang Z, Chen R, et al. Tat competes with HEXIM1 to increase the active pool of P-TEFb for HIV-1 transcription. Nucleic Acids Res. 2007;35:2003–12. doi: 10.1093/nar/gkm063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D’Orso I, Frankel AD. RNA-mediated displacement of an inhibitory snRNP complex activates transcription elongation. Nat. Struct. Mol. Biol. 2010;17:815–21. doi: 10.1038/nsmb.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sobhian B, Laguette N, Yatim A, Nakamura M, Levy Y, et al. HIV-1 Tat assembles a multifunctional transcription elongation complex and stably associates with the 7SK snRNP. Mol. Cell. 2010;38:439–51. doi: 10.1016/j.molcel.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He N, Liu M, Hsu J, Xue Y, Chou S, et al. HIV-1 Tat and host AFF4 recruit two transcription elongation factors into a bifunctional complex for coordinated activation of HIV-1 transcription. Mol. Cell. 2010;38:428–38. doi: 10.1016/j.molcel.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu H, Li Z, Xue Y, Schulze-Gahmen U, Johnson JR, et al. AFF1 is a ubiquitous P-TEFb partner to enable Tat extraction of P-TEFb from 7SK snRNP and formation of SECs for HIV transactivation. PNAS. 2014;111:E15–24. doi: 10.1073/pnas.1318503111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schulze-Gahmen U, Lu H, Zhou Q, Alber T. AFF4 binding to Tat-P-TEFb indirectly stimulates TAR recognition of super elongation complexes at the HIV promoter. eLife. 2014;2014:1–13. doi: 10.7554/eLife.02375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Izmailova E, Bertley FM, Huang Q, Makori N, Miller CJ, et al. HIV-1 Tat reprograms immature dendritic cells to express chemoattractants for activated T cells and macrophages. Nat. Med. 2003;9:191–97. doi: 10.1038/nm822. [DOI] [PubMed] [Google Scholar]
- 21.Frankel AD, Pabo CO. Cellular uptake of the Tat protein from human immunodeficiency virus. Cell. 1988;55:1189–93. doi: 10.1016/0092-8674(88)90263-2. [DOI] [PubMed] [Google Scholar]
- 22.Rayne F, Debaisieux S, Yezid H, Lin Y-L, Mettling C, et al. Phosphatidylinositol-(4,5)-bisphosphate enables efficient secretion of HIV-1 Tat by infected T-cells. EMBO J. 2010;29:1348–62. doi: 10.1038/emboj.2010.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim N, Kukkonen S, Gupta S, Aldovini A. Association of Tat with promoters of PTEN and PP2A subunits is key to transcriptional activation of apoptotic pathways in HIV-infected CD4+ T cells. PLOS Pathog. 2010;6:e1001103. doi: 10.1371/journal.ppat.1001103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reeder JE, Kwak YT, McNamara RP, Forst CV, D’Orso I. HIV Tat controls RNA polymerase II and the epigenetic landscape to transcriptionally reprogram target immune cells. eLife. 2015;4:1–44. doi: 10.7554/eLife.08955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tahirov TH, Babayeva ND, Varzavand K, Cooper JJ, Sedore SC, Price DH. Crystal structure of HIV-1 Tat complexed with human P-TEFb. Nature. 2010;465:747–51. doi: 10.1038/nature09131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang Z, Yik JHN, Chen R, He N, Moon KJ, et al. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell. 2005;19:535–45. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 27.Kanazawa S, Okamoto T, Peterlin BM. Tat competes with CIITA for the binding to P-TEFb and blocks the expression of MHC class II genes in HIV infection. Immunity. 2000;12:61–70. doi: 10.1016/s1074-7613(00)80159-4. [DOI] [PubMed] [Google Scholar]
- 28.Li Z, Guo J, Wu Y, Zhou Q. The BET bromodomain inhibitor JQ1 activates HIV latency through antagonizing Brd4 inhibition of Tat-transactivation. Nucleic Acids Res. 2013;41:277–87. doi: 10.1093/nar/gks976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mann DA, Frankel AD. Endocytosis and targeting of exogenous HIV-1 Tat protein. EMBO J. 1991;10:1733–39. doi: 10.1002/j.1460-2075.1991.tb07697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson TP, Patel K, Johnson KR, Maric D, Calabresi PA, et al. Induction of IL-17 and nonclassical T-cell activation by HIV-Tat protein. PNAS. 2013;110:13588–93. doi: 10.1073/pnas.1308673110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li CJ, Ueda Y, Shi B, Borodyansky L, Huang L, et al. Tat protein induces self-perpetuating permissivity for productive HIV-1 infection. PNAS. 1997;94:8116–20. doi: 10.1073/pnas.94.15.8116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fernandes J, Faust TB, Frankel AD. Functional segregation of overlapping genes in HIV. Cell. 2016;167:1762–66. e12. doi: 10.1016/j.cell.2016.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pollard VW, Malim MH. The HIV-1 Rev protein. Annu. Rev. Microbiol. 1998;52:491–532. doi: 10.1146/annurev.micro.52.1.491. [DOI] [PubMed] [Google Scholar]
- 34.Malim MH, Hauber J, Le SY, Maizel JV, Cullen BR. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature. 1989;338:254–57. doi: 10.1038/338254a0. [DOI] [PubMed] [Google Scholar]
- 35.Bai Y, Tambe A, Zhou K, Doudna JA. RNA-guided assembly of Rev-RRE nuclear export complexes. eLife. 2014;3:e03656. doi: 10.7554/eLife.03656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Daugherty MD, Booth DS, Jayaraman B, Cheng Y, Frankel AD. HIV Rev response element (RRE) directs assembly of the Rev homooligomer into discrete asymmetric complexes. PNAS. 2010;107:12481–86. doi: 10.1073/pnas.1007022107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jayaraman B, Crosby DC, Homer C, Ribeiro I, Mavor D, Frankel AD. RNA-directed remodeling of theHIV-1 protein Rev orchestrates assembly of the Rev-Rev response element complex. eLife. 2014;3:e04120. doi: 10.7554/eLife.04120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pond SJK, Ridgeway WK, Robertson R, Wang J, Millar DP. HIV-1 Rev protein assembles on viral RNA one molecule at a time. PNAS. 2009;106:1404–8. doi: 10.1073/pnas.0807388106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rausch JW, le Grice SFJ. HIV Rev assembly on the Rev response element (RRE): a structural perspective. Viruses. 2015;7:3053–75. doi: 10.3390/v7062760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–60. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 41.Booth DS, Cheng Y, Frankel AD. The export receptor CRM1 forms a dimer to promote nuclear export of HIV RNA. eLife. 2014;3:e04121. doi: 10.7554/eLife.04121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Felber BK, Drysdale CM, Pavlakis GN. Feedback regulation of human immunodeficiency virus type 1 expression by the Rev protein. J. Virol. 1990;64:3734–41. doi: 10.1128/jvi.64.8.3734-3741.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lata S, Ali A, Sood V, Raja R, Banerjea AC. HIV-1 Rev downregulates Tat expression and viral replication via modulation of NAD(P)H:quinine oxidoreductase 1 (nqo1) Nat. Commun. 2015;6:7244. doi: 10.1038/ncomms8244. [DOI] [PubMed] [Google Scholar]
- 44.Asher G, Tsvetkov P, Kahana C. A mechanism of ubiquitin-independent proteasomal degradation of the tumor suppressors p53 and p73. Genes Dev. 2005;19:316–21. doi: 10.1101/gad.319905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Larue RS, Lengyel J, Jónsson SR, Andrésdóttir V, Harris RS. Lentiviral Vif degrades the APOBEC3Z3/APOBEC3H protein of its mammalian host and is capable of cross-species activity. J. Virol. 2010;84:8193–201. doi: 10.1128/JVI.00685-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–50. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 47.Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, et al. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113:803–9. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- 48.Yu X, Yu Y, Liu B, Luo K, Kong W, et al. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science. 2003;302:1056–60. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- 49.Sheehy AM, Gaddis NC, Malim MH. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 2003;9:1404–7. doi: 10.1038/nm945. [DOI] [PubMed] [Google Scholar]
- 50.Jäger S, Kim DY, Hultquist JF, Shindo K, LaRue RS, et al. Vif hijacks CBF-β to degrade APOBEC3G and promote HIV-1 infection. Nature. 2011;481:371–75. doi: 10.1038/nature10693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang W, Du J, Evans SL, Yu Y, Yu X-F. T-cell differentiation factor CBF-β regulates HIV-1 Vif-mediated evasion of host restriction. Nature. 2012;481:376–69. doi: 10.1038/nature10718. [DOI] [PubMed] [Google Scholar]
- 52.Guo Y, Dong L, Qiu X, Wang Y, Zhang B, et al. Structural basis for hijacking CBF-β and CUL5 E3 ligase complex by HIV-1 Vif. Nature. 2014;505:229–33. doi: 10.1038/nature12884. [DOI] [PubMed] [Google Scholar]
- 53.Hultquist JF, Binka M, LaRue RS, Simon V, Harris RS. Vif proteins of human and simian immunodeficiency viruses require cellular CBFB to degrade APOBEC3 restriction factors. J. Virol. 2012;86:2874–77. doi: 10.1128/JVI.06950-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim DY, Kwon E, Hartley PD, Crosby DC, Mann S, et al. CBFβ stabilizes HIV Vif to counteract APOBEC3 at the expense of RUNX1 target gene expression. Mol. Cell. 2013;49:632–44. doi: 10.1016/j.molcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miyagi E, Kao S, Yedavalli V, Strebel K. CBFβ enhances de novo protein biosynthesis of its binding partners HIV-1 Vif and RUNX1 and potentiates the Vif-induced degradation of APOBEC3G. J. Virol. 2014;88:4839–52. doi: 10.1128/JVI.03359-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kane JR, Stanley DJ, Hultquist JF, Johnson JR, Mietrach N, et al. Lineage-specific viral hijacking of non-canonical E3 ubiquitin ligase cofactors in the evolution of Vif anti-APOBEC3 activity. Cell Rep. 2015;11:1236–50. doi: 10.1016/j.celrep.2015.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Salter JD, Morales GA, Smith HC. Structural insights for HIV-1 therapeutic strategies targeting Vif. Trends Biochem. Sci. 2014;39:373–80. doi: 10.1016/j.tibs.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rose KM, Marin M, Kozak SL, Kabat D. The viral infectivity factor (Vif) of HIV-1 unveiled. Trends Mol. Med. 2004;10:291–97. doi: 10.1016/j.molmed.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 59.Harris RS, Liddament MT. Retroviral restriction by APOBEC proteins. Nat. Rev. Immunol. 2004;4:868–77. doi: 10.1038/nri1489. [DOI] [PubMed] [Google Scholar]
- 60.Hultquist JF, Harris RS. Leveraging APOBEC3 proteins to alter the HIV mutation rate and combat AIDS. Future Virol. 2009;4:605. doi: 10.2217/fvl.09.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang G, Shigesada K, Ito K, Wee HJ, Yokomizo T, et al. Dimerization with PEBP2β protects RUNX1/AML1 from ubiquitin-proteasome-mediated degradation. EMBO J. 2001;20:723–33. doi: 10.1093/emboj/20.4.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tahirov TH, Inoue-Bungo T, Morii H, Fujikawa A, Sasaki M, et al. Structural analyses of DNA recognition by the AML1/Runx-1 Runt domain and its allosteric control by CBFβ. Cell. 2001;104:755–67. doi: 10.1016/s0092-8674(01)00271-9. [DOI] [PubMed] [Google Scholar]
- 63.Klase Z, Yedavalli VS, Houzet L, Perkins M, Maldarelli F, et al. Activation of HIV-1 from latent infection via synergy of RUNX1 inhibitor Ro5-3335 and SAHA. PLOS Pathog. 2014;10:e1003997. doi: 10.1371/journal.ppat.1003997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Anderson BD, Harris RS. Transcriptional regulation of APOBEC3 antiviral immunity through the CBF-β/RUNX axis. Sci. Adv. 2015;1:e1500296. doi: 10.1126/sciadv.1500296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hultquist JF, McDougle RM, Anderson BD, Harris RS. HIV type 1 viral infectivity factor and the RUNX transcription factors interact with core binding factor β on genetically distinct surfaces. AIDS Res. Hum. Retrovir. 2012;28:1543–51. doi: 10.1089/aid.2012.0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Greenwood EJ, Matheson NJ, Wals K, van den Boomen DJ, Antrobus R, et al. Temporal proteomic analysis of HIV infection reveals remodelling of the host phosphoproteome by lentiviral Vif variants. eLife. 2016;p5:e18296. doi: 10.7554/eLife.18296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Strebel K, Klimkait T, Martin M. A novel gene of HIV-1, Vpu, and its 16-kilodalton product. Science. 1988;241:1221–23. doi: 10.1126/science.3261888. [DOI] [PubMed] [Google Scholar]
- 68.Cohen EA, Terwilliger EF, Sodroski JG, Haseltine WA. Identification of a protein encoded by the vpu gene of HIV-1. Nature. 1988;334:532–34. doi: 10.1038/334532a0. [DOI] [PubMed] [Google Scholar]
- 69.Malim MH, Emerman M, An P, Duggal P, Wang LH, et al. HIV-1 accessory proteins–ensuring viral survival in a hostile environment. Cell Host Microbe. 2008;3:388–98. doi: 10.1016/j.chom.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 70.Matsuda Z, Chou M-J, Matsuda M, Huang J-H, Chen Y-M, et al. Human immunodeficiency virus type 1 has an additional coding sequence in the central region of the genome. PNAS. 1988;85:6968–72. doi: 10.1073/pnas.85.18.6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Terwilliger EF, Cohen EA, Lu Y, Sodroski JG, Haseltine WA. Functional role of human immunodeficiency virus type 1 vpu. Microbiology. 1989;86:5163–67. doi: 10.1073/pnas.86.13.5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Neil SJ, Zang T, Bieniasz PD. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451:425–30. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- 73.Van Damme N, Goff D, Katsura C, Jorgenson RL, Mitchell R, et al. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe. 2008;3:245–52. doi: 10.1016/j.chom.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Willey RL, Maldarelli F, Martin MA, Strebel K. Human immunodeficiency virus type 1 Vpu protein induces rapid degradation of CD4. J. Virol. 1992;66:7193–200. doi: 10.1128/jvi.66.12.7193-7200.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ewart GD, Sutherland T, Gage PW, Cox GB, Curtin J. The Vpu protein of human immunodeficiency virus type 1 forms cation-selective ion channels. J. Virol. 1996;70:7108–15. doi: 10.1128/jvi.70.10.7108-7115.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schubert U, Ferrer-Montiel AV, Oblatt-Montal M, Henklein P, Strebel K, Montal M. Identification of an ion channel activity of the Vpu transmembrane domain and its involvement in the regulation of virus release from HIV-1-infected cells. FEBS Lett. 1996;398:12–18. doi: 10.1016/s0014-5793(96)01146-5. [DOI] [PubMed] [Google Scholar]
- 77.Kerkau T, Bacik I, Bennink JR, Yewdell JW, Húnig T, et al. The human immunodeficiency virus type 1 (HIV-1) Vpu protein interferes with an early step in the biosynthesis of major histocompatibility complex (MHC) class I molecules. J. Exp. Med. 1997;185:1295–305. doi: 10.1084/jem.185.7.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Akari H, Bour S, Kao S, Adachi A, Strebel K. The human immunodeficiency virus type 1 accessory protein Vpu induces apoptosis by suppressing the nuclear factor κB-dependent expression of antiapoptotic factors. J. Exp. Med. 2001;194:1299–312. doi: 10.1084/jem.194.9.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Roy N, Pacini G, Berlioz-Torrent C, Janvier K. Mechanisms underlying HIV-1 Vpu-mediated viral egress. Front. Microbiol. 2014;5:177. doi: 10.3389/fmicb.2014.00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Margottin F, Bour SP, Durand H, Selig L, Benichou S, et al. A novel human WD protein, h-βTrCP, that interacts with HIV-1 Vpu connects CD4 to the ER degradation pathway through an F-box motif. Mol. Cell. 1998;1:565–74. doi: 10.1016/s1097-2765(00)80056-8. [DOI] [PubMed] [Google Scholar]
- 81.Magadán JG, Pérez-Victoria FJ, Sougrat R, Ye Y, Strebel K, et al. Multilayered mechanism of CD4 downregulation by HIV-1 Vpu involving distinct ER retention and ERAD targeting steps. PLOS Pathog. 2010;6:e1000869. doi: 10.1371/journal.ppat.1000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Magadán JG, Bonifacino JS. Transmembrane domain determinants of CD4 downregulation by HIV-1 Vpu. J. Virol. 2012;86:757–72. doi: 10.1128/JVI.05933-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kupzig S, Korolchuk V, Rollason R, Sugden A, Wilde A, Banting G. Bst-2/HM1.24 is a RAFT-associated apical membrane protein with an unusual topology. Traffic. 2003;4:694–709. doi: 10.1034/j.1600-0854.2003.00129.x. [DOI] [PubMed] [Google Scholar]
- 84.Perez-Caballero D, Zang T, Ebrahimi A, McNatt MW, Gregory DA, et al. Tetherin inhibits HIV-1 release by directly tethering virions to cells. Cell. 2009;139:499–511. doi: 10.1016/j.cell.2009.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Goffinet C, Allespach I, Homann S, Tervo H-M, Habermann A, et al. HIV-1 antagonism of CD317 is species specific and involves Vpu-mediated proteasomal degradation of the restriction factor. Cell Host Microbe. 2009;5:285–97. doi: 10.1016/j.chom.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 86.Iwabu Y, Fujita H, Kinomoto M, Kaneko K, Ishizaka Y, et al. HIV-1 accessory protein Vpu internalizes cell-surface Bst-2/tetherin through transmembrane interactions leading to lysosomes. J. Biol. Chem. 2009;284:35060–72. doi: 10.1074/jbc.M109.058305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mangeat B, Gers-Huber G, Lehmann M, Zufferey M, Luban J, et al. HIV-1 Vpu neutralizes the antiviral factor tetherin/Bst-2 by binding it and directing its β-TrCP2-dependent degradation. PLOS Pathog. 2009;5:e1000574. doi: 10.1371/journal.ppat.1000574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Douglas JL, Viswanathan K, McCarroll MN, Gustin JK, Früh K, Moses AV. Vpu directs the degradation of the human immunodeficiency virus restriction factor Bst-2/tetherin via a βTrCP-dependent mechanism. J. Virol. 2009;83:7931–47. doi: 10.1128/JVI.00242-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Abada P, Noble B, Cannon PM. Functional domains within the human immunodeficiency virus type 2 envelope protein required to enhance virus production. J. Virol. 2005;79:3627–38. doi: 10.1128/JVI.79.6.3627-3638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bour S, Strebel K. The human immunodeficiency virus (HIV) type 2 envelope protein is a functional complement to HIV type 1 Vpu that enhances particle release of heterologous retroviruses. J. Virol. 1996;70:8285–300. doi: 10.1128/jvi.70.12.8285-8300.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hauser H, Lopez LA, Yang SJ, Oldenburg JE, Exline CM, et al. HIV-1 Vpu and HIV-2 Env counteract BST-2/tetherin by sequestration in a perinuclear compartment. Retrovirology. 2010;7:51. doi: 10.1186/1742-4690-7-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jia B, Serra-Moreno R, Neidermyer W, Rahmberg A, Mackey J, et al. Species-specific activity of SIV Nef and HIV-1 Vpu in overcoming restriction by tetherin/BST2. PLOS Pathog. 2009;5:e1000429. doi: 10.1371/journal.ppat.1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Le Tortorec A, Neil SJD. Antagonism to and intracellular sequestration of human tetherin by the human immunodeficiency virus type 2 envelope glycoprotein. J. Virol. 2009;83:11966–78. doi: 10.1128/JVI.01515-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sauter D, Schindler M, Specht A, Landford WN, Münch J, et al. Tetherin-driven adaptation of Vpu and Nef function and the evolution of pandemic and nonpandemic HIV-1 strains. Cell Host Microbe. 2009;6:409–21. doi: 10.1016/j.chom.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yang SJ, Lopez LA, Hauser H, Exline CM, Haworth KG, Cannon PM. Anti-tetherin activities in Vpu-expressing primate lentiviruses. Retrovirology. 2010;7:13. doi: 10.1186/1742-4690-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang F, Wilson SJ, Landford WC, Virgen B, Gregory D, et al. Nef proteins from simian immunodeficiency viruses are tetherin antagonists. Cell Host Microbe. 2009;6:54–67. doi: 10.1016/j.chom.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Matsuda A, Suzuki Y, Honda G, Muramatsu S, Matsuzaki O, et al. Large-scale identification and characterization of human genes that activate NF-κB and MAPK signaling pathways. Oncogene. 2003;22:3307–18. doi: 10.1038/sj.onc.1206406. [DOI] [PubMed] [Google Scholar]
- 98.Tokarev A, Suarez M, Kwan W, Fitzpatrick K, Singh R, Guatelli J. Stimulation of NF-κB activity by the HIV restriction factor BST2. J. Virol. 2013;87:2046–57. doi: 10.1128/JVI.02272-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Galão RP, Le Tortorec A, Pickering S, Kueck T, Neil SJD. Innate sensing of HIV-1 assembly by tetherin induces NF-κB-dependent proinflammatory responses. Cell Host Microbe. 2012;12:633–44. doi: 10.1016/j.chom.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bour S, Perrin C, Akari H, Strebel K. The human immunodeficiency virus type 1 Vpu protein inhibits NF-κB activation by interfering with βTrCP-mediated degradation of IκB. J. Biol. Chem. 2001;276:15920–28. doi: 10.1074/jbc.M010533200. [DOI] [PubMed] [Google Scholar]
- 101.Besnard-Guerin C, Belaïdouni N, Lassot I, Segeral E, Jobart A, et al. HIV-1 Vpu sequesters β-transducin repeat-containing protein (βTrCP) in the cytoplasm and provokes the accumulation of β-catenin and other SCFβTrCP substrates. J. Biol. Chem. 2004;279:788–95. doi: 10.1074/jbc.M308068200. [DOI] [PubMed] [Google Scholar]
- 102.Sauter D, Hotter D, Van Driessche B, Stürzel CM, Kluge SF, et al. Differential regulation of NF-kB-mediated proviral and antiviral host gene expression by primate lentiviral Nef and Vpu proteins. Cell Rep. 2015;10:586–600. doi: 10.1016/j.celrep.2014.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Manganaro L, de Castro E, Maestre AM, Olivieri K, García-Sastre A, et al. HIV Vpu interferes with NF-κB activity but not with interferon regulatory factor 3. J. Virol. 2015;89:9781–90. doi: 10.1128/JVI.01596-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hayden MS, Ghosh S, Hayden MS, Ghosh S. NF-kB, the first quarter-century: remarkable progress and outstanding questions. Genes Dev. 2012;26:203–34. doi: 10.1101/gad.183434.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kondo E, Mammano F, Cohen EA, Göttlinger HG. The p6gag domain of human immunodeficiency virus type 1 is sufficient for the incorporation of Vpr into heterologous viral particles. J. Virol. 1995;69:2759–64. doi: 10.1128/jvi.69.5.2759-2764.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Andersen JL, Planelles V. The role of Vpr in HIV-1 pathogenesis. Curr. HIV Res. 2005;3:43–51. doi: 10.2174/1570162052772988. [DOI] [PubMed] [Google Scholar]
- 107.Lai M, Zimmerman ES, Planelles V, Chen J. Activation of the ATR pathway by human immunodeficiency virus type 1 Vpr involves its direct binding to chromatin in vivo. J. Virol. 2005;79:15443–51. doi: 10.1128/JVI.79.24.15443-15451.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Roshal M, Kim B, Zhu Y, Nghiem P, Planelles V. Activation of the ATR-mediated DNA damage response by the HIV-1 viral protein R. J. Biol. Chem. 2003;278:25879–86. doi: 10.1074/jbc.M303948200. [DOI] [PubMed] [Google Scholar]
- 109.Zimmerman ES, Sherman MP, Blackett JL, Neidleman JA, Kreis C, et al. Human immunodeficiency virus type 1 Vpr induces DNA replication stress in vitro and in vivo. J. Virol. 2006;80:10407–18. doi: 10.1128/JVI.01212-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Belzile JP, Abrahamyan LG, Gérard FCA, Rougeau N, Cohen ÉA. Formation of mobile chromatin-associated nuclear foci containing HIV-1 Vpr and VPRBP is critical for the induction of G2 cell cycle arrest. PLOS Pathog. 2010;6:e1001080. doi: 10.1371/journal.ppat.1001080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zimmerman ES, Chen J, Andersen JL, Ardon O, Dehart JL, et al. Human immunodeficiency virus type 1 Vpr-mediated G2 arrest requires Rad17 andHus1 and induces nuclear BRCA1 andγ-H2AX focus formation. Mol. Cell. Biol. 2004;24:9286–94. doi: 10.1128/MCB.24.21.9286-9294.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Planelles V, Jowett JBM, Li Q-X, Xie Y, Hahn B, Chen ISY. Vpr-induced cell cycle arrest is conserved among primate lentiviruses. J. Virol. 1996;70:2516–24. doi: 10.1128/jvi.70.4.2516-2524.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hrecka K, Gierszewska M, Srivastava S, Kozaczkiewicz L, Swanson SK, et al. Lentiviral Vpr usurps Cul4-DDB1[VPRBP] E3 ubiquitin ligase to modulate cell cycle. PNAS. 2007;104:11778–83. doi: 10.1073/pnas.0702102104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Belzile JP, Duisit G, Rougeau N, Mercier J, Finzi A, Cohen ÉA. HIV-1 Vpr-mediated G2 arrest involves the DDB1-CUL4AVPRBP E3 ubiquitin ligase. PLOS Pathog. 2007;3:0882–93. doi: 10.1371/journal.ppat.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Schröfelbauer B, Hakata Y, Landau NR. HIV-1 Vpr function is mediated by interaction with the damage-specific DNA-binding protein DDB1. PNAS. 2007;104:4130–35. doi: 10.1073/pnas.0610167104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Laguette N, Brégnard C, Hue P, Basbous J, Yatim A, et al. Premature activation of the SLX4 complex by Vpr promotes G2/M arrest and escape from innate immune sensing. Cell. 2014;156:134–45. doi: 10.1016/j.cell.2013.12.011. [DOI] [PubMed] [Google Scholar]
- 117.Fregoso OI, Emerman M. Activation of the DNA damage response is a conserved function of HIV-1 and HIV-2 Vpr that is independent of SLX4 recruitment. mBio. 2016;7:1–10. doi: 10.1128/mBio.01433-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lovejoy CA, Lock K, Yenamandra A, Cortez D. DDB1 maintains genome integrity through regulation of Cdt1. Mol. Cell. Biol. 2006;26:7977–90. doi: 10.1128/MCB.00819-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wu Y, Zhou X, Barnes CO, DeLucia M, Cohen AE, et al. The DDB1-DCAF1-Vpr-UNG2 crystal structure reveals how HIV-1 Vpr steers human UNG2 toward destruction. Nat. Struct. Mol. Biol. 2016;23:933–40. doi: 10.1038/nsmb.3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chen R, Le Rouzic E, Kearney JA, Mansky LM, Benichou S. Vpr-mediated incorporation of UNG2 into HIV-1 particles is required to modulate the virus mutation rate and for replication in macrophages. J. Biol. Chem. 2004;279:28419–25. doi: 10.1074/jbc.M403875200. [DOI] [PubMed] [Google Scholar]
- 121.Wang L, Mukherjee S, Jia F, Narayan O, Zhao LJ. Interaction of virion protein Vpr of human immunodeficiency virus type 1 with cellular transcription factor Sp1 and trans-activation of viral long terminal repeat. J. Biol. Chem. 1995;270:25564–69. doi: 10.1074/jbc.270.43.25564. [DOI] [PubMed] [Google Scholar]
- 122.Sawaya BE, Khalili K, Gordon J, Taube R, Amini S. Cooperative interaction between HIV-1 regulatory proteins Tat and Vpr modulates transcription of the viral genome. J. Biol. Chem. 2000;275:35209–14. doi: 10.1074/jbc.M005197200. [DOI] [PubMed] [Google Scholar]
- 123.Gummuluru S, Emerman M. Cell cycle- and Vpr-mediated regulation of human immunodeficiency virus type 1 expression in primary and transformed T-cell lines. J. Virol. 1999;73:5422–30. doi: 10.1128/jvi.73.7.5422-5430.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Goh WC, Rogel ME, Kinsey CM, Michael SF, Fultz PN, et al. HIV-1Vpr increases viral expression by manipulation of the cell cycle: a mechanism for selection of Vpr in vivo. Nat. Med. 1998;4:65–71. doi: 10.1038/nm0198-065. [DOI] [PubMed] [Google Scholar]
- 125.Forget J, Yao XJ, Mercier J, Cohen EA. Human immunodeficiency virus type 1 Vpr protein transactivation function: mechanism and identification of domains involved. J. Mol. Biol. 1998;284:915–23. doi: 10.1006/jmbi.1998.2206. [DOI] [PubMed] [Google Scholar]
- 126.Felzien LK, Woffendin C, Hottiger MO, Subbramanian RA, Cohen EA, Nabel GJ. HIV transcriptional activation by the accessory protein, Vpr, is mediated by the p300 co-activator. PNAS. 1998;95:5281–86. doi: 10.1073/pnas.95.9.5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zahoor MA, Xue G, Sato H, Murakami T, Takeshima SN, Aida Y. HIV-1 Vpr induces interferon-stimulated genes in human monocyte-derived macrophages. PLOS ONE. 2014;9:e106418. doi: 10.1371/journal.pone.0106418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Campbell MJ, Cho RJ, Huang M, Dong H, Steinmetz L, et al. Transcriptional regulation and function during the human cell cycle. Nat. Genet. 2001;27:48–54. doi: 10.1038/83751. [DOI] [PubMed] [Google Scholar]
- 129.Allan J, Coligan J, Lee T, McLane M, Kanki P, et al. A new HTLV-III/LAV encoded antigen detected by antibodies from AIDS patients. Science. 1985;230:810–13. doi: 10.1126/science.2997921. [DOI] [PubMed] [Google Scholar]
- 130.Kirchhoff F, Greenough TC, Brettler DB, Sullivan JL, Desrosiers RC. Absence of intact Nef sequences in a long-term survivor with nonprogressive HIV-1 infection. N. Engl. J. Med. 1995;332:228–32. doi: 10.1056/NEJM199501263320405. [DOI] [PubMed] [Google Scholar]
- 131.Guy B, Kieny MP, Riviere Y, Le Peuch C, Dott K, et al. HIV F/3′ ORF encodes a phosphorylated GTP-binding protein resembling an oncogene product. Nature. 1987;330:266–69. doi: 10.1038/330266a0. [DOI] [PubMed] [Google Scholar]
- 132.Garcia JV, Miller AD. Serine phosphorylation-independent downregulation of cell-surface CD4 by Nef. Nature. 1991;350:508–11. doi: 10.1038/350508a0. [DOI] [PubMed] [Google Scholar]
- 133.Schaefer MR, Wonderlich ER, Roeth JF, Leonard JA, Collins KL. HIV-1 Nef targets MHC-I and CD4 for degradation via a final common β-COP-dependent pathway in T cells. PLOS Pathog. 2008;4:e1000131. doi: 10.1371/journal.ppat.1000131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Schwartz O, Maréchal V, Le Gall S, Lemonnier F, Heard J-M. Endocytosis of major histocompatibility complex class I molecules is induced by the HIV-1 Nef protein. Nat. Med. 1996;2:338–42. doi: 10.1038/nm0396-338. [DOI] [PubMed] [Google Scholar]
- 135.Stumptner-Cuvelette P, Morchoisne S, Dugast M, Le Gall S, Raposo G, et al. HIV-1 Nef impairs MHC class II antigen presentation and surface expression. PNAS. 2001;98:12144–49. doi: 10.1073/pnas.221256498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Geyer M, Fackler OT, Peterlin BM. Structure–function relationships in HIV-1 Nef. EMBO Rep. 2001;2:580–85. doi: 10.1093/embo-reports/kve141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Roeth JF, Williams M, Kasper MR, Filzen TM, Collins KL. HIV-1Nef disrupts MHC-I trafficking by recruiting AP-1 to the MHC-I cytoplasmic tail. J. Cell Biol. 2004;167:903–13. doi: 10.1083/jcb.200407031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chaudhuri R, Lindwasser OW, Smith WJ, Hurley JH, Bonifacino JS. Downregulation of CD4 by human immunodeficiency virus type 1 Nef is dependent on clathrin and involves direct interaction of Nef with the ap2 clathrin adaptor. J. Virol. 2007;81:3877–90. doi: 10.1128/JVI.02725-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wildum S, Schindler M, Münch J, Kirchhoff F. Contribution of Vpu, Env, and Nef to CD4 down-modulation and resistance of human immunodeficiency virus type 1-infected T cells to superinfection. J. Virol. 2006;80:8047–59. doi: 10.1128/JVI.00252-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Usami Y, Wu Y, Göttlinger HG. SERINC3 and SERINC5 restrict HIV-1 infectivity and are counteracted by Nef. Nature. 2015;526:218–23. doi: 10.1038/nature15400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Rosa A, Chande A, Ziglio S, De Sanctis V, Bertorelli R, et al. HIV-1 Nef promotes infection by excluding SERINC5 from virion incorporation. Nature. 2015;526:212–17. doi: 10.1038/nature15399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wang J-K, Kiyokawa E, Verdin ETD. The Nef protein of HIV-1 associates with rafts and primes T cells for activation. PNAS. 2000;72:1337–40. [PubMed] [Google Scholar]
- 143.Mangino G, Percario ZA, Fiorucci G, Vaccari G, Acconcia F, et al. HIV-1 Nef induces proinflammatory state in macrophages through its acidic cluster domain: involvement of TNF alpha receptor associated factor 2. PLOS ONE. 2011;6:e22982. doi: 10.1371/journal.pone.0022982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sauter D, Hotter D, Van Driessche B, Stürzel CM, Kluge SF, et al. Differential regulation of NF-κB-mediated proviral and antiviral host gene expression by primate lentiviral Nef and Vpu proteins. Cell Rep. 2015;10:586–99. doi: 10.1016/j.celrep.2014.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Williams SA, Chen L-F, Kwon H, Ruiz-Jarabo CM, Verdin E, Greene WC. NF-κB p50 promotes HIV latency through HDAC recruitment and repression of transcriptional initiation. EMBO J. 2006;25:139–49. doi: 10.1038/sj.emboj.7600900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Simmons A, Aluvihare V, McMichael A. Nef triggers a transcriptional program in T cells imitating single-signal T cell activation and inducing HIV virulence mediators. Immunity. 2001;14:763–77. doi: 10.1016/s1074-7613(01)00158-3. [DOI] [PubMed] [Google Scholar]
- 147.Querido E, Morrison MR, Chu-Pham-Dang H, Thirlwell SW, Boivin D, et al. Identification of three functions of the adenovirus E4orf6 protein that mediate p53 degradation by the E4orf6-E1B55K complex. J. Virol. 2001;75:699–709. doi: 10.1128/JVI.75.2.699-709.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–36. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 149.Edwards MR, Johnson B, Mire CE, Xu W, Shabman RS, et al. The Marburg virus VP24 protein interacts with Keap1 to activate the cytoprotective antioxidant response pathway. Cell Rep. 2014;6:1017–25. doi: 10.1016/j.celrep.2014.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Felsani A, Mileo AM, Paggi MG. Retinoblastoma family proteins as key targets of the small DNA virus oncoproteins. Oncogene. 2006;25:5277–85. doi: 10.1038/sj.onc.1209621. [DOI] [PubMed] [Google Scholar]
- 151.Pavletich NP, Lee J-O, Russo AA. Structure of the retinoblastoma tumour-suppressor pocket domain bound to a peptide from HPV E7. Nature. 1998;391:859–65. doi: 10.1038/36038. [DOI] [PubMed] [Google Scholar]
- 152.Iaquinta PJ, Lees JA. Life and death decisions by the E2F transcription factors. Curr. Opin. Cell Biol. 2007;19:649–57. doi: 10.1016/j.ceb.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]