Abstract
The fidelity of DNA replication relies on three error avoidance mechanisms acting in series: nucleotide selectivity of replicative DNA polymerases, exonucleolytic proofreading, and post-replicative DNA mismatch repair (MMR). MMR defects are well known to be associated with increased cancer incidence. Due to advances in DNA sequencing technologies, the past several years have witnessed a long-predicted discovery of replicative DNA polymerase defects in sporadic and hereditary human cancers. The polymerase mutations preferentially affect conserved amino acid residues in the exonuclease domain and occur in tumors with an extremely high mutation load. Thus, a concept has formed that defective proofreading of replication errors triggers the development of these tumors. Recent studies of the most common DNA polymerase variants, however, suggested that their pathogenicity may be determined by functional alterations other than loss of proofreading. In this review, we summarize our current understanding of the consequences of DNA polymerase mutations in cancers and the mechanisms of their mutator effects. We also discuss likely explanations for a high recurrence of some but not other polymerase variants and new ideas for therapeutic interventions emerging from the mechanistic studies.
Keywords: DNA polymerase ε, DNA polymerase δ, mutator, cancer, proofreading
1. Prehistory
The idea that cancer may be caused by error-prone variants of replicative DNA polymerases dates back to the early 1970s. A hypothesis proposed by Larry Loeb and colleagues posited that the infidelity of DNA replication could be responsible for the multiple cellular changes associated with tumor initiation and progression [1]. Alterations in replicative DNA polymerases that increase the rate of base pairing errors were regarded as the most obvious source of such infidelity. In the 40+ years that followed, it has been established that DNA replication in eukaryotic cells is accomplished by a concerted action of three DNA polymerases, Polα, Polδ and Polε [2,3], and the high fidelity of synthesis relies on accurate nucleotide selection by these enzymes, exonucleolytic proofreading by Polδ and Polε, and post-replicative DNA mismatch repair (MMR) [4–6]. MMR defects had been recognized as the cause of hereditary colorectal cancer (CRC) predisposition in Lynch syndrome almost 25 years ago [7] and were soon shown to be widespread in sporadic cancers. In contrast, although defects in DNA polymerase selectivity or proofreading produce a mutator phenotype in model eukaryotic organisms [8–12] and accelerate tumorigenesis in mice [13–16], the association of replicative DNA polymerase mutations with cancer in humans has escaped the spotlight until very recently.
Prior to the release of The Cancer Genome Atlas (TCGA) sequencing data on CRC in 2012, reports of Polδ or Polε mutations in cancer cells had appeared in three publications. In the 1990s, two groups addressed the prevalence of mutations in the POLD1 gene encoding the catalytic subunit of Polδ in human CRC cell lines and sporadic colon tumors [17,18], with one study focusing only on the exonuclease domain area [17] and the other analyzing the entire coding region [18]. Among 12 cell lines and seven tumor samples analyzed, 10 changes were found in the amino acid sequence of Polδ. With the exception of one, these changes did not represent polymorphisms commonly observed in healthy people. However, the cells lines with POLD1 mutations were also defective in MMR, leaving uncertainty as to whether the polymerase mutations played a role in the tumor formation and a general consensus that the MMR defect was the likely culprit. This view was not challenged until 2010, when functional studies in yeast of a Polδ variant (Polδ-R689W) found in the MSH6-defective CRC cell line DLD-1 revealed its exceptionally strong mutator properties, and biochemical analysis showed that Polδ-R689W is a highly error-prone DNA polymerase [19]. This study provided the first indication of the functional importance of a replicative DNA polymerase mutation present in human cancer cells. In 2011, analysis of selected exons of POLD1 and POLE encoding the catalytic subunit of Polε in a larger collection of tumor samples identified a Polε exonuclease domain variant, F367S, in a rectal tumor [20]. It was the first Polε mutation to be reported in human disease. The revelation was soon to come that replicative DNA polymerase mutations are common in certain tumor types and are often responsible for the genomic instability that leads to the development of these tumors.
2. The era of genome sequencing: discovery of Poε and Polδ mutations in hypermutated cancers
With advancing DNA sequencing technologies has come the ability to perform large-scale studies of human tumor DNA in order to better understand cancers at the genomic level. In 2012, TCGA published the results of a comprehensive genomic study of colorectal carcinoma, including exome sequencing of 224 tumor samples [21]. This analysis revealed a distinct subset of so-called hypermutated tumors (>10 mutations per 106 bases) comprising ~16% of all sporadic cases. A majority of these showed microsatellite instability (MSI) indicative of MMR deficiency, but the most hypermutated tumors (>100 mutations per 106 bases) were, strikingly, all microsatellite stable (MSS) and contained mutations in POLE. Mutations in POLD1 were also observed. However, in contrast to the POLE-mutant tumors, all tumors with POLD1 mutations were MSI, in line with the view that the POLD1 variants could be neutral passenger changes resulting from the high mutation rate in MMR-deficient cells. The following year, TCGA reported the results of analysis of over 370 endometrial cancers (EC), which similarly showed that a fraction of tumors was hypermutated, and tumors with the highest mutation frequency were MSS and contained mutations in POLE [22]. A separate study specifically addressing the prevalence of POLD1 and POLE exonuclease domain mutations in sporadic EC also reported a high frequency of POLE changes in hypermutated MMR-proficient tumors [23].
Shortly after the discovery of POLE mutations in sporadic hypermutated CRC, germline mutations in POLE and POLD1 were found to be responsible for a high-penetrance colorectal cancer predisposition syndrome [24]. The POLD1 mutation carriers were also predisposed to EC and, likely, brain tumors. The causative role of two germline variants, POLE-L424V and POLD1-S478N, has been convincingly demonstrated by co-segregation of the alleles with the cancer phenotype, and additional POLD1 variants potentially altering the polymerase properties were found in patients whose clinical characteristics suggested genetic predisposition [24]. Similar to the sporadic POLE-mutant CRC and EC, tumors from carriers of germline POLE and POLD1 mutations were MSS and showed a high number of base substitution mutations.
Following these breakthroughs, multiple studies utilizing either whole-exome analysis or targeted sequencing of the DNA polymerase genes reported somatic POLE and, less frequently, POLD1 mutations in sporadic CRC and EC [25–53]. Several thousands of colorectal and endometrial tumor samples have been analyzed to date, producing an impressive list of more than 200 distinct POLE mutations and more than 80 POLD1 mutations. The POLE mutations are observed at a highly variable frequency, with some constituting frequently recurring hotspots. Several POLD1 mutations were also observed more than once. The available data suggests that at least 6% of colorectal tumors and 7% of endometrial tumors carry POLE mutations, and at least 4% of both colorectal and endometrial tumors carry POLD1 mutations. The exact frequency of these mutations in cancers is uncertain, because many studies limited the search for mutations to the exonuclease domains or even selected exons, and studies employing whole-exome approaches can potentially underestimate the actual number of mutations. Somatic POLE and POLD1 mutations have also been reported, albeit less frequently, in other tumor types, including breast, ovarian, brain, pancreas, lung, and prostate [54,55]. Notably, somatic mutations in POLE have been found to occur as early events in the development of brain tumors in children with constitutional mismatch repair deficiency [56,57]. The list of germline replicative DNA polymerase mutations detected in families with hereditary cancer predisposition has also grown and now comprises at least eight distinct POLE variants and at least seven POLD1 variants [24,58–66], although good evidence for co-segregation with the disease only exists for POLE-L424V [24], POLE-N363K [63], POLE-Y458F [64], POLD1-S478N [24] and POLD1-L474P [59]. The originally discovered POLE-L424V mutation appeared to be highly recurrent, with incidence reported in over 20 families with hereditary cancers [24,58–62], and POLD1-S478N and POLD1-L474P have also been seen repeatedly [24,59–61].
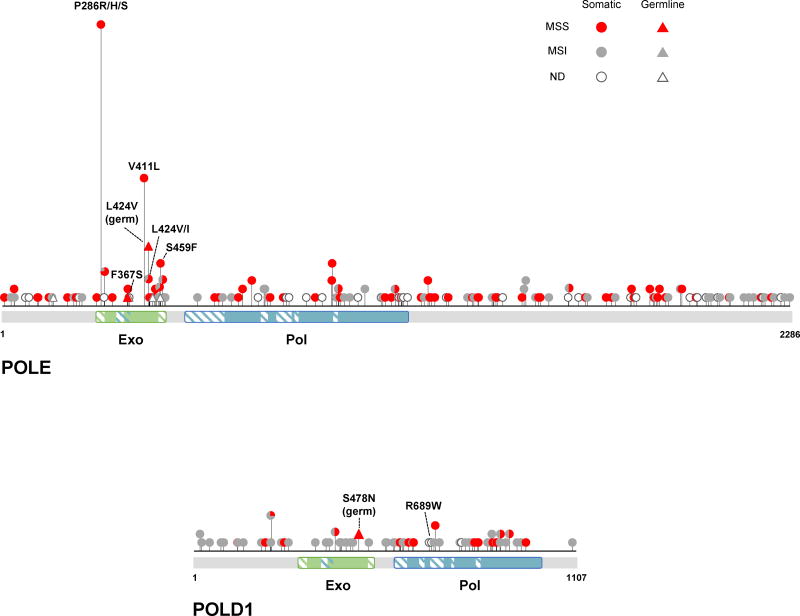
The location of CRC- and EC-associated variants in the POLE and POLD1 proteins is shown schematically in Figure 1. There are several notable characteristics of these variants. First, the vast majority occur in tumors in a heterozygous state in which both the mutant and wild-type alleles are present. Second, POLE is altered much more frequently than POLD1 in hypermutated MSS tumors, where the polymerase variants are strongly suspected to play a causative role. Most somatic POLD1 mutations are found in MSI tumors. Third, POLE but not POLD1 mutations tend to preferentially affect the exonuclease domain of the polymerase. Fourth, in both sporadic and hereditary cancers, some mutations are observed at a vastly greater frequency than others. It is likely that many of these observations are related to the effects imposed by the mutations on Polε and Polδ, and, subsequently, on the various cellular transactions involving these enzymes. With the exception of the germline mutations, for which the causative role in cancer could be unequivocally established by genetic analysis of large multigenerational families [24], the identification of functionally significant somatic variants is not straightforward and has been a subject of much speculation. At the frequency of mutation observed in hypermutated tumors, almost every gene is expected to be impacted, and some tumors have been reported to contain up to 10 non-silent replicative DNA polymerase mutations. In the following sections, we discuss currently available data on the functional consequences of cancer-associated Polε and Polδ variants, the mechanisms underlying their mutator effects, and the likely reasons for the preferential occurrence of some but not other polymerase variants in sporadic and hereditary cancers.
Figure 1. POLE and POLD1 mutations reported in CRC and EC.
A schematic of the POLE and POLD1 proteins is shown with the location of cancer-associated variants indicated by lollipops. Only variants identified in studies where the entire coding sequence of POLE or POLD1 was analyzed [21,22,24,25,27,29,35,41–43,45–51,61–64] are included to show an unbiased distribution. The height of each lollipop corresponds to the number of times the mutation has been reported. A description of individual mutations is provided in Supplemental Table 1. Note a concentration of POLE variants in the exonuclease domain and a more even distribution of POLD1 variants throughout the protein. The MMR status of the tumor in which the polymerase mutation was found is indicated by color. MSS, microsatellite stable; MSI, microsatellite instable; ND, not determined. Exo, exonuclease domain; Pol, polymerase domain. Hatched boxes indicate conserved motifs.
3. The proofreading deficiency paradigm
The genome of hypermutated tumors is flooded by mutations, most of which probably play no role in the tumor development. However, analysis of the CRC exome sequencing data published by TCGA [21] immediately revealed that the POLE mutations in MSS hypermutated tumors non-randomly hit highly conserved amino acid residues in the exonuclease domain. Along with the discovery of POLE and POLD1 exonuclease domain mutations in hereditary CRC [24], this finding strongly suggested that loss of proofreading activity of replicative DNA polymerases is responsible for the high level of genome instability in these cancers. This concept was met with substantial excitement and spread quickly among basic and clinical scientists [67–69] despite the paucity of data demonstrating that a proofreading defect is the main consequence of the mutations. The newly characterized hereditary CRC predisposition syndrome was termed Polymerase Proofreading-Associated Polyposis (PPAP) [67]. A variety of theoretical approaches have been used to support the idea of defective proofreading in the variant polymerases [23,24,62–64, 67,70,71]. These included analysis of amino acid residue conservation, location within or close to conserved exonuclease motifs and in respect to available crystal structures of orthologous enzymes, and in silico prediction tools. Published data on mutator phenotypes or biochemical defects resulting from similar mutations in model organisms were also considered. It is of note that in the majority of cases, the experiments in model organisms cited as evidence of functional significance used a different DNA polymerase (e.g., Polδ rather than Polε), often a different amino acid substitution, and sometimes not the same amino acid residue [23,24,63,64,67,70–72]. In most cases, the results of these analyses led the authors to conclude that the mutations were likely to affect proofreading. However, two observations were difficult to reconcile with the view that the pathogenicity of the POLE and POLD1 mutations results from their adverse effects on proofreading. First, it remained puzzling and unexplained by the in silico analysis why some mutations are seen more frequently than others. Second, alterations of catalytic residues known to inactivate proofreading in model organisms are rarely, if ever, seen in human cancers. Clues to these puzzles, along with the need to revisit the proofreading deficiency paradigm, were suggested by recent functional studies that we review below.
4. Lessons from functional analysis of cancer-associated Poε and Polδ variants
4.1. Biochemical studies
Reduction in exonuclease activity has been demonstrated for seven Polε variants mapping to the exonuclease domain [29]. These include the P286R and V411L variants most frequently observed in sporadic cancers, the recurrent germline variant L424V, a less frequent somatic variant S459F, as well as P286H, F367S and L424I that have so far been observed in only one or two tumors each. These experiments were performed with a purified fragment of the catalytic subunit of Polε containing both DNA polymerase and exonuclease active sites. The exonuclease activity was impaired to varying degrees by the mutations and ranged from 5% to 42% of the corresponding wild-type protein activity. For five of these variants, a reduction in the fidelity of in vitro DNA synthesis was also demonstrated and was proportional to the extent of exonuclease deficiency [29]. Remarkably, however, no correlation was observed between the severity of the proofreading defect and the frequency at which the Polε variants are seen in cancers. This observation raises a possibility, which is discussed further below, that the exonuclease domain variants increase cancer risk via mechanisms more complicated than loss of proofreading.
Although Polε exonuclease domain variants attracted much attention, at present, the most comprehensively characterized cancer-associated variant is Polδ-R689W, which maps to the DNA polymerase domain and was one of the first polymerase mutations discovered in cancer cells [18]. In addition to being present in the hypermutated MSH6-deficient CRC cell lines DLD-1 [18] and HCT15 [26] derived from the same tumor, it was reported in two other sporadic tumors [54] that are not hypermutated. All this together would place Polδ-R689W in a category of variants that are considered insignificant by much of the current literature. However, biochemical studies performed initially with the yeast analog of Polδ-R689W [19,73], and, most recently, with the four-subunit human Polδ-R689W [74] showed a profound defect in nucleotide selectivity. DNA synthesis catalyzed by both human Polδ-R689W and its yeast mimic was extremely error-prone despite wild-type levels of exonuclease activity. These results indicated that POLD1 mutations seen in sporadic tumors can be highly significant. They also showed that DNA polymerase mutations occurring in MMR-deficient tumors can be significant and act synergistically with the MMR defects to promote hypermutation. It is important to note, however, that the CRC cell lines carrying Polδ-R689W are deficient only in MSH6-dependent MMR. Severe DNA polymerase fidelity defects may be incompatible with full inactivation of MMR resulting from a loss of MLH1 or MSH2, as discussed elsewhere [71]. Finally, the studies of Polδ-R689W demonstrated that functionally important mutations can occur outside the exonuclease domain and affect nucleotide selectivity rather than proofreading.
4.2. In vivo effects in model systems
Yeast Saccharomyces cerevisiae or Schizosaccharomyces pombe has been commonly used to assess the effects of cancer-associated DNA polymerase mutations in vivo. Yeast genes encoding the catalytic subunits of Polε and Polδ show a high degree of similarity to the human POLE and POLD1 genes, respectively. Therefore, a common approach involves creating a mimic of a tumor-associated mutation in the chromosomal DNA polymerase gene and determining the effect on replication fidelity inside the cell by measuring the spontaneous mutation rate. Although the significance of many human mutations was claimed to be confirmed by mutagenesis assays in yeast [23,24,59,60,67,71,72], to our knowledge, in only ten cases listed in Table 1 were the variants actually modeled by creating an analogous amino acid substitution in the corresponding polymerase. Most of these are POLD1 mutations. Out of several hundred somatic POLE mutations found in tumors, evidence for functional significance in vivo has been reported only for the most common variant, P286R [33]. Despite this limited analysis, the experiments in yeast provided several important insights.
Table 1.
Cancer-associated Pol ε and Pol δ variants, for which mutator effects have been assessed in cell-based assays.
| Human mutation | Domain | Cancer type | Mutation origin |
Mutation in model organism |
Mutator effect |
Reference |
|---|---|---|---|---|---|---|
| Modeled in S. cerevisiae | ||||||
| POLE-P286R | Exo | CRC, EC, pancreas, ovary, brain | Somatic | pol2-P301R | Yes | [33] |
| POLD1-D316N | Exo | gastric | Somatic1 | pol3-D321N | Yes | [80] |
| POLD1-C319Y | Exo | multiple myeloma, brain | Somatic | pol3-C324Y | Yes | [80] |
| POLD1-D402N | Exo | prostate | Somatic | pol3-D407N | Yes | [80] |
| POLD1-R506H | Exo | CRC | Unknown | pol3-R511H | No2 | [19] |
| POLD1-L606M | Pol | brain | Somatic | pol3-L612M | Yes | [12] |
| POLD1-R689W | Pol | CRC, liver | Somatic | pol3-R696W | Yes | [19] |
| POLD1-D316G | Exo | CRC, EC | Germline3 | pol3-D321G | Yes | [80] |
| Modeled in S. pombe | ||||||
| POLE-W347C | Exo | Melanoma | Germline4 | pol2-F348C | Yes | [65] |
| POLD1-S478N | Exo | CRC, EC | Germline | pol3-C462N | Yes | [24] |
| Modeled in human cells | ||||||
| POLD1-R689W | Pol | CRC | Sporadic | POLD1-R689W | Yes | [74] |
POLD1-D316N has been reported in a single tumor as a subclonal mutation (allele frequency 0.13) [101]. Its contribution to the tumor development in uncertain.
The yeast mimic of POLD1-R506H conferred a very weak mutator phenotype detectable only in the presence of a MMR defect [19].
POLD1-D316G was identified in a family with apparent predisposition to multiple cancers [60]. Co-segregation of the mutation with the disease has not been comprehensively studied.
POLE-W347C was identified in a family with strong predisposition to cutaneous melanoma [65]. Co-segregation of the mutation with the disease has not been unequivocally established.
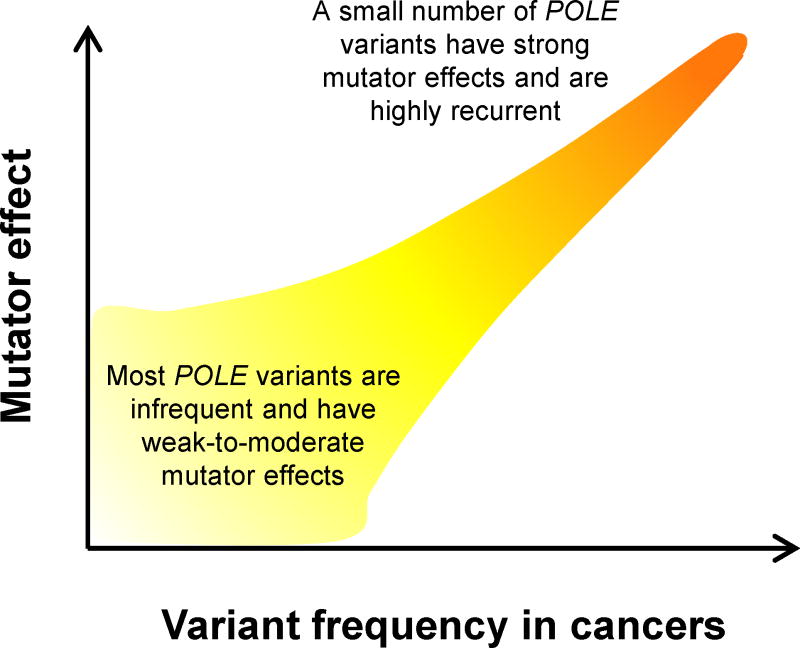
First, they revealed that exceptionally powerful mutators are seen recurrently in tumors. For example, the mutator effect of the yeast POLE-P286R analog exceeds that of any previously studied Polε mutation by an order of magnitude [33]. Likewise, the mutator effect of the POLD1-R689W mimic greatly exceeds that of any known eukaryotic mutator allele [19]. We have proposed previously that the frequent occurrence of POLE-P286R in tumors (Figure 1) is due to its unusually strong mutator effect, which leads to a greater cancer risk [33]. In support of this hypothesis, our recent studies of several other cancer-associated Polε variants showed that their mutator effects are highly variable, and a strong correlation exists between the mutator effect in yeast and the variant frequency in tumors (S. R. Barbari, D. P. Kane, E. A. Moore and P. V. Shcherbakova, manuscript in preparation). A model emerging from these studies suggests that there is a large number of relatively infrequent polymerase variants with weak-to-moderate mutator effects that are collectively responsible for the majority of hypermutated tumors (~70% in the case of CRC and EC). The remaining 30% are driven by a small number of strong mutators that are highly recurrent (Figure 2).
Figure 2. The frequency at which a Polε mutation is seen in tumors correlates with its mutator effect.
The figure illustrates the relationship between the incidence of individual POLE variants in sporadic tumors and their mutator effects deduced from in vivo functional assays.
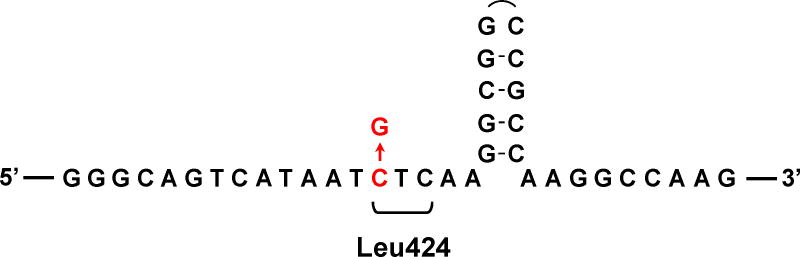
Interestingly, we found that the striking predominance of the POLE-L424V variant in the spectrum of germline cancer-causing mutations (Figure 1) is related not to its mutator effect, which is modest (S. R. Barbari, D. P. Kane, E. A. Moore and P. V. Shcherbakova, manuscript in preparation), but apparently to the genomic DNA sequence context that makes this site a mutational hotspot. The base substitution (C→G) occurs in close proximity of a GC-rich palindromic sequence with a strong potential for hairpin structure formation (Figure 3). We have shown previously that such DNA sequences present an obstacle for Polδ and Polε ([75]; X. Xing and P. V, Shcherbakova, unpublished) and promote mutations in the nearby region, particularly C→G transversions dependent on translesion synthesis DNA polymerase ζ [75]. We hypothesize that the location of the codon for Leu424 at this at-risk sequence explains the fact that not only it is the most frequently seen germline DNA polymerase mutation, but it has also been repeatedly reported as a de novo germline variant [58,59] and several times as a somatic mutation in sporadic tumors [22,29,30,43].
Figure 3. A possible hairpin DNA structure adjacent to the site of POLE-L424V mutation.
The genomic DNA sequence context is shown for the recurrent C→G mutation in the POLE gene that leads to an L424V amino acid substitution. The sequence presented is for the non-transcribed DNA strand. The codon for Leu424 is indicated, with the mutation highlighted in red.
The second revelation from the in vivo functional studies is that mutator effects of cancer-associated Polε variants greatly exceed the effects expected from loss of proofreading, which in the case of the P286R variant is by two orders of magnitude [33]. Thus, the mutations must impact the polymerase in some additional ways, which at present remain uncharacterized. It is likely that these additional defects, and not the loss of proofreading per se, determine the pathogenicity of POLE mutations. Indeed, the variant frequency in tumors correlates with the severity of the mutator effect in vivo (Figure 2) and not with the degree to which proofreading is impaired [29]. Therefore, the mutagenic potential is separable from the effects on proofreading, and the magnitude of the mutator effect in cell-based assays seems to be a better predictor of cancer risk.
Third, the in vivo assays demonstrated the functional significance of many POLD1 mutations (Table 1), including the ones found in MMR-deficient tumors. Mutations affecting both the exonuclease and the polymerase domain were found to be significant. Perhaps an interesting clue to the differential tissue-specific roles of Polε and Polδ in tumorigenesis is provided by the following observation. Over 20 different mutator versions of Polδ have been artificially created in S. cerevisiae by either site-directed or random mutagenesis, including a dozen with amino acid substitutions in the exonuclease domain and some with documented exonuclease defects [9,12,76–79]. None of these mutations have been seen among thousands of sporadic and hereditary cancer cases analyzed. However, an experiment where strong mutator variants of Polδ were selected for by the ability to mutate a single given chromosomal site within 12–13 cell generations [80] produced a collection of eight variants in the proofreading domain, four of which have now been seen in human cancers (Table 1). Interestingly, sporadic cancers with these mutations included gastric, brain and prostate tumors, as well as multiple myeloma, but not CRC or EC. Thus, Polδ exonuclease domain mutations may preferentially contribute to pathogenesis of a different subset of cancer types, similar to earlier findings in mice [14,15].
Although the yeast-based assays help pinpoint potentially significant DNA polymerase variants, ultimately, establishing the pathogenic nature of a mutant allele requires the demonstration of the mutator effect in human cells. To date, this has only been done for the very first cancer-associated mutation discovered, POLD1-R689W [74]. In this assay, the mutant allele was stably overexpressed in a human cell line carrying wild-type endogenous DNA polymerase genes, and the mutation rate was measured at a chromosomal reporter gene. Both the mutator effect and the specificity of nucleotide misincorporation previously observed with the yeast POLD1-R689W analog have been recapitulated in the human cell system. These experiments validated the use of the yeast model and also established a precedent and a simple strategy for functional analysis of cancer-associated DNA polymerase mutations in human cells. In addition to confirming the pathogenicity of variants identified as mutators in yeast, the use of human cell-based assays may be necessary for assessing the impact of mutations that affect poorly conserved amino acid residues.
4.3. Expression of a mutator phenotype does not require loss of heterozygosity
Replicative DNA polymerase variants are typically present in tumors in the heterozygous state. In patients with germline POLD1 or POLE mutations, loss of heterozygosity is not required for the tumor development [24]. DNA sequence analysis of sporadic tumors with POLD1 or POLE mutations almost always shows the presence of both wild-type and mutant alleles. While the subclonal nature of the mutation could be responsible for the wild-type signal in some cases, all cell lines established from hypermutated tumors are heterozygous for the DNA polymerase mutations [18,20,26,32]. The heterozygous state was mimicked in the yeast system for several Polε and Polδ variants ([19,33,73], S. R. Barbari, D. P. Kane, E. A. Moore and P. V. Shcherbakova, manuscript in preparation). All of them caused a significant mutator effect in the presence of the wild-type allele, although reduced compared to that seen in homozygous diploids, consistent with participation of both the mutant and wild-type polymerases in DNA synthesis. This is in contrast to most DNA repair genes implicated in cancer, e.g. MMR genes, where loss of both alleles is required to produce a mutator phenotype. Thus, functional analysis of DNA polymerase variants should perhaps primarily address their ability to increase the mutation rate in the heterozygous state. While this is easily achieved in yeast, human cell-based assays where the wild-type and mutant alleles are expressed at comparable levels have yet to be developed.
Curiously, loss or inactivation of the second allele has been reported in a few tumors with functionally significant POLE mutations, and at least one example illustrates that this could have consequences for the manifestation of the disease. Two tumors in the TCGA CRC study [21] carried a recurrent S459F variant, for which exonuclease deficiency has been demonstrated in vitro [29]. One of these tumors also contained a nonsense mutation at codon 150 of the POLE gene, which presumably inactivated the second allele. Although both tumors were hypermutated, the heterozygous tumor developed in a 57-year-old patient and showed a total of ~1,800 genomic mutations, while the patient with the additional nonsense mutation was diagnosed at 35, and the tumor had almost 10,000 mutations. Studies of additional similar cases are required to determine whether loss of heterozygosity or the presence of second hits in POLE could be an important prognostic marker.
The predominantly heterozygous state of Polε and Polδ mutations has implications for the regulation of mutator activity in tumor cells. A constantly high mutation rate might be disadvantageous to the tumor cells because of the accumulation of deleterious mutations. While many tumors carry exceptionally strong mutators (exemplified by Polε-P286R and Polδ-R689W), their effects are buffered by the presence of wild-type enzymes in the heterozygous cells. At the same time, the mutator effects depend greatly on the ratio of the wild-type and mutator enzymes [19]. We hypothesized previously that variations in expression level of the wild-type and mutant alleles may allow for both transient spikes of hypermutation that promote tumor growth and subsequent suppression of the mutator phenotype that helps maintain fitness [19].
4.4. Mutational signature of cancer-associated polymerase variants
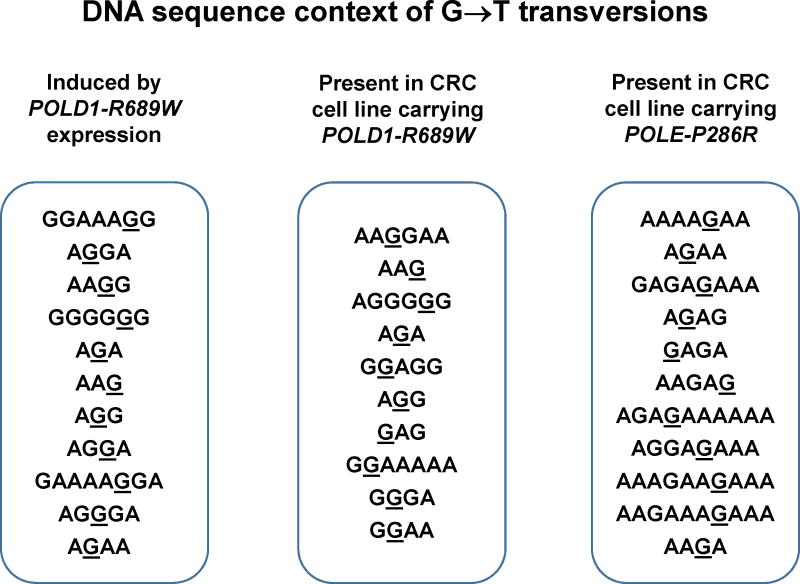
While the frequency of all types of base substitutions is elevated in tumors with Polε and Polδ exonuclease domain variants, a disproportionally large increase in GC→TA transversions with a particular preference for AGA/TCT sequence context has been noted [23,24,29,81]. The high fraction of GC→TA transversions has even been proposed as a criterion for the identification of functionally significant DNA polymerase mutations [29]. However, the mechanism through which the various Polε and Polδ variants would uniformly produce the same mutational signature, as well as the reasons for the preferential mutability of AGA/TCT sequences, remain unclear. Importantly, the mutational spectra of tumor genomes represent the outcome of multiple DNA maintenance processes and may not necessarily reflect the specificity of the polymerase variants. An alternative approach is to analyze individual signatures of the mutator polymerases by expressing them in cultured human cells and determining the spectrum of mutation they induce, which was recently done for Polδ-R689W [74]. Despite the location of Arg689 in the DNA polymerase rather than exonuclease domain, synthesis by Polδ-R689W showed the notorious high frequency of GC→TA transversions with a striking sequence context specificity. All GC→TA transversions occurred in polypurine/polypurimidine tracts (up to eight consecutive purines in one strand). Remarkably, the same context specificity of GC→TA transversions was observed for genomic mutations present in the CRC cell line carrying this Polδ polymerase domain variant and in another hypermutated CRC cell line carrying the Polε exonuclease domain variant P286R ([74]; Figure 4). Thus, the previously described AGA/TCT motif in fact represents a variation of this more general sequence context of DNA replication errors, which is not specific for exonuclease domain variants. The information obtained from such experimental assessment of DNA polymerase signatures in the cellular environment will be useful for tracking the activity of cancer-associated polymerase variants in human tumors.
Figure 4. Mutator DNA polymerases present in cancer cells induce GC→TA transversions in polypurine/polypyrimidine tracts.
Left, DNA sequence context of G→T transversions induced by introduction of the POLD1-R689W allele into HCT116 cells lacking DNA polymerase mutations. Middle and right, DNA sequence context of G→T transversions present in the genomes of CRC cell lines HCT15 (POLD1-R689W) and HCC2998 (POLE-P286R). The mutated base is underlined. Randomly picked transversions are shown to demonstrate that all of them occur in polypurine/polypyrimidine sequences. Data are from [74].
5. Mechanisms of the ultramutator phenotype
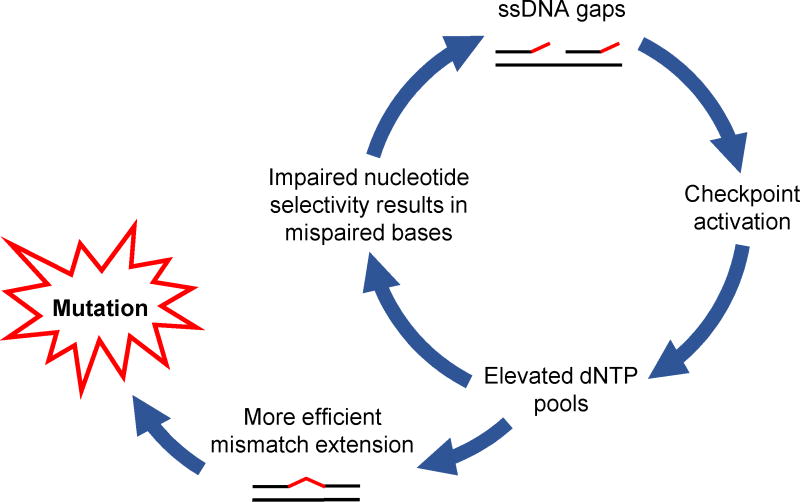
As discussed in the previous sections, many cancer-associated Polε and Polδ mutations modeled in yeast confer very strong mutator phenotypes much exceeding those of previously characterized DNA polymerase mutants. The mechanism of this unusual mutator effect is best understood for the yeast Polδ-R696W, which mimics the human polymerase domain variant R689W. The yeast Polδ-R696W has dramatically reduced nucleotide selectivity but poor mismatch extension capacity [19]. This results in frequent misincorporations that impede DNA synthesis and result in checkpoint activation, which, in turn, leads to expansion of dNTP pools [73]. The increase in intracellular dNTP levels promotes extension of the mismatched primer termini and also further increases the likelihood of incorrect base insertion by an already error-prone polymerase, ultimately resulting in a catastrophic accumulation of mutations ([73]; Figure 5). Studies in yeast suggested that Polε polymerase domain variants could act through the same mechanism [82], although this has not been demonstrated for any cancer-associated Polε mutations. The human Polδ-R689W, however, has impaired nucleotide selectivity and poor mismatch extension ability, being nearly identical to its yeast mimic in this respect [74]. Whether its infidelity is similarly augmented by upregulation of dNTP synthesis is yet to be determined.
Figure 5. Vicious circle model for mutagenesis caused by the yeast analog of Polδ-R689W.
(modified from [73]). A mutation in the DNA polymerase domain that impairs nucleotide selectivity results in mismatched primer termini that are not efficiently extended, leading to the accumulation of single-stranded DNA gaps. These gaps trigger a checkpoint response that results in the upregulation of ribonucleotide reductase and, consequently, an expansion of intracellular dNTP pools. Elevated dNTP pools allow for more efficient mismatch extension, leaving a mispaired base in the newly synthesized DNA, and also promote further misinsertions that continue to fuel this mutagenic pathway.
An apparently different case is presented by the Polε exonuclease domain variants. Although they show a various degree of exonuclease deficiency [29], the magnitude of their mutator effect in yeast suggests a mechanism distinct from the loss of proofreading [33]. Our recent studies suggested that the hypermutability is not caused by the expansion of dNTP pools either (S. Sharma, A. Chabes and P. V. Shcherbakova, unpublished). Unraveling the mystery of this ultramutator effect, which drives the genomic instability in many human cancers, is a high priority for the nearest future. Possible clues are provided by the following observations. Polε exonuclease deficiency results in a very small increase in the mutation rate in both yeast and human cells [8,83,84], even though the fidelity of purified Polε in vitro is strongly affected by the inactivation of proofreading [85,86]. It has been suggested that the majority of Polε errors are corrected in cells by extrinsic mechanisms, for example, by the exonuclease activity of Polδ [2,87]. On the other hand, many Polε exonuclease domain mutations found in cancers, and particularly P286R, were predicted to affect DNA binding [23,24,63]. The altered interaction of Polε variants with DNA could potentially reduce the efficiency of extrinsic proofreading, in addition to the intrinsic exonuclease defect, which would provide one possible explanation of the ultramutator phenotype.
6. Therapeutic implications of DNA polymerase deficiency
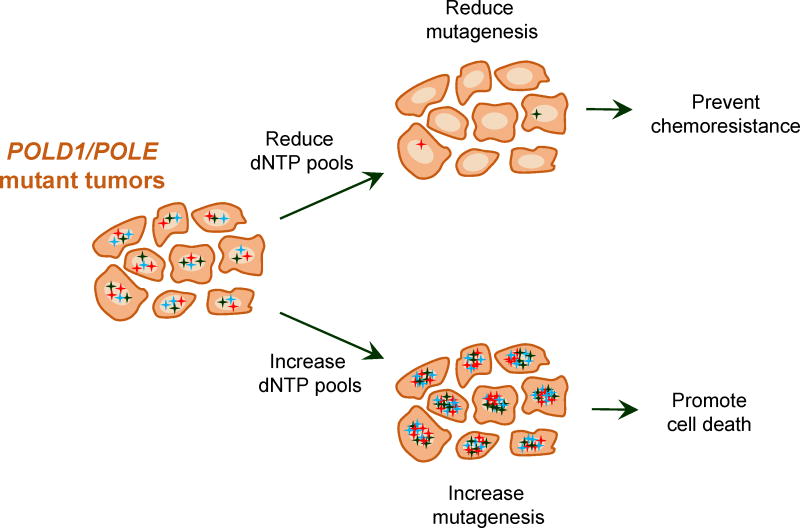
Patients with hypermutated POLE-mutant endometrial cancers have an excellent prognosis with nearly 100% progression-free survival after surgery [22,28,31,34,38,41,88–91]. A significantly better survival has also been noted for POLE-mutant CRC [44]. Recent studies suggest this could be due to the high immunogenicity of the tumors [31,34,38,41,89,90], which likely results from the hypermutation increasing the number of neoepitopes that can be recognized by the immune system [37,40–42,44,92,93]. The improved survival suggests that, while the hypermutated POLE tumors are often of higher grade, they should be classified separately and could be treated less aggressively [36,94]. Other hypermutated tumors such as melanomas and lung cancers are also highly immunogenic [95,96]. Consequently, hypermutated tumors, including rare relapses of POLE-mutant EC, have responded well to immunotherapy [42,93,95– 97]. While further studies are needed, this may indicate that immunotherapy alone, if necessary, could replace radiation and chemotherapy after surgery in these cases. We refer the reader to other, more comprehensive, reviews of this topic [91,98–100] and would like to finish by discussing additional possible therapeutic approaches suggested by mechanistic studies in model systems. In yeast, the mutator effects of both exonuclease and polymerase domain variants of Polε and Polδ are highly sensitive to even small fluctuations of dNTP levels ([73,82]; section 5). Mutagenesis can be reduced to wild-type levels when dNTP pools are low and increases catastrophically when dNTP pools are high. At the same time, the mutation rate in the wild-type strains is barely affected by the size of dNTP pools. While the details of dNTP metabolism may differ in yeast and human cells, the sensitivity of mutator polymerases to dNTP levels is likely to be conserved. Figure 6 illustrates how this property can be exploited for cancer therapy. Because the number of mutations in hypermutated tumors is likely just below the fitness threshold [56], therapies which increase dNTP pools could push the tumors past this threshold. Normal cells would not be affected because of high nucleotide selectivity of the wild-type polymerases. Conversely, inhibition of dNTP synthesis would reduce mutagenesis and, subsequently, the ability of the tumor to adapt and develop resistance to therapy. Such approaches could be particularly valuable for tumors that carry mild mutator alleles and might not be hypermutated enough for immunotherapy to be efficient. These insights underscore the importance of mechanistic studies in locating the Achilles’ heel of the DNA polymerase-mutant tumors, especially given the fact that the mechanisms through which the exonuclease domain variants cause hypermutability are not yet fully understood.
Figure 6. Modulation of dNTP pools in hypermutated tumor cells as a potential therapeutic avenue.
Tumor cells with replicative DNA polymerase defects have a high rate of mutation (designated by multicolor stars). Reducing intracellular dNTP pools would improve the polymerase fidelity, thereby reducing mutagenesis and decreasing the possibility that the tumor cells will produce drug-resistant clones. Increasing dNTP pools would further increase the already high mutation rate, bringing it to a level incompatible with cell viability.
Supplementary Material
Acknowledgments
We thank Garrett Barbari for creative help in the preparation of Figure 1 and Youri Pavlov for critically reading the manuscript. Research in the laboratory of P.V.S. was supported by the National Institutes of Health grant ES015869 and Nebraska Department of Health and Human Services LB506 grants. S.R.B. is supported by the Cancer Biology Training Grant T32CA009476 from the National Cancer Institute.
Abbreviations
- Pol
DNA polymerase
- MMR
DNA mismatch repair
- TCGA
The Cancer Genome Atlas
- CRC
colorectal cancer
- EC
endometrial cancer
- MSI
microsatellite instability
- MSS
microsatellite stable
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
The authors declare no conflict of interest.
References
- 1.Loeb LA, Springgate CF, Battula N. Errors in DNA replication as a basis of malignant changes. Cancer Res. 1974;34:2311–2321. [PubMed] [Google Scholar]
- 2.Pavlov YI, Shcherbakova PV. DNA polymerases at the eukaryotic fork-20 years later. Mutat. Res. - Fundam. Mol. Mech. Mutagen. 2010;685:45–53. doi: 10.1016/j.mrfmmm.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lujan SA, Williams JS, Kunkel TA. DNA polymerases divide the labor of genome replication. Trends Cell Biol. 2016;26:640–654. doi: 10.1016/j.tcb.2016.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morrison A, Johnson AL, Johnston LH, Sugino A. Pathway correcting DNA replication errors in Saccharomyces cerevisiae. EMBO J. 1993;12:1467–1473. doi: 10.1002/j.1460-2075.1993.tb05790.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kunkel TA. DNA replication fidelity. J. Biol. Chem. 2004;279:16895–16898. doi: 10.1074/jbc.R400006200. [DOI] [PubMed] [Google Scholar]
- 6.Ganai RA, Johansson E. DNA replication—a matter of fidelity. Mol. Cell. 2016;62:745–755. doi: 10.1016/j.molcel.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 7.Lynch HT, Snyder CL, Shaw TG, Heinen CD, Hitchins MP. Milestones of Lynch syndrome: 1895–2015. Nat. Rev. Cancer. 2015;15:181–194. doi: 10.1038/nrc3878. [DOI] [PubMed] [Google Scholar]
- 8.Morrison A, Bell JB, Kunkel TA, Sugino A. Eukaryotic DNA polymerase amino acid sequence required for 3’ to 5’ exonuclease activity. Proc. Natl. Acad. Sci. U. S. A. 1991;88:9473–9477. doi: 10.1073/pnas.88.21.9473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simon M, Giot L, Faye G. The 3’ to 5’ exonuclease activity located in the DNA polymerase δ subunit of Saccharomyces cerevisiae is required for accurate replication. EMBO J. 1991;10:2165–70. doi: 10.1002/j.1460-2075.1991.tb07751.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Venkatesan RN, Hsu JJ, Lawrence NA, Preston BD, Loeb LA. Mutator phenotypes caused by substitution at a conserved motif A residue in eukaryotic DNA polymerase. J. Biol. Chem. 2006;281:4486–4494. doi: 10.1074/jbc.M510245200. [DOI] [PubMed] [Google Scholar]
- 11.Pursell ZF, Isoz I, Lundström E-B, Johansson E, Kunkel TA. Yeast DNA polymerase ε participates in leading-strand DNA replication. Science. 2007;317:127–30. doi: 10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li L, Murphy KM, Kanevets U, Reha-Krantz LJ. Sensitivity to phosphonoacetic acid: A new phenotype to probe DNA polymerase δ in Saccharomyces cerevisiae. Genetics. 2005;170:569–580. doi: 10.1534/genetics.104.040295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldsby RE, Lawrence NA, Hays LE, Olmsted EA, Chen X, Singh M, Preston BD. Defective DNA polymerase-δ proofreading causes cancer susceptibility in mice. Nat. Med. 2001;7:638–639. doi: 10.1038/88963. [DOI] [PubMed] [Google Scholar]
- 14.Goldsby RE, Hays LE, Chen X, Olmsted EA, Slayton WB, Spangrude GJ, Preston BD. High incidence of epithelial cancers in mice deficient for DNA polymerase δ proofreading. Proc. Natl. Acad. Sci. U. S. A. 2002;99:15560–5. doi: 10.1073/pnas.232340999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Albertson TM, Ogawa M, Bugni JM, Hays LE, Chen Y, Wang Y, Treuting PM, Heddle JA, Goldsby RE, Preston BD. DNA polymerase ε and δ proofreading suppress discrete mutator and cancer phenotypes in mice. Proc. Natl. Acad. Sci. U. S. A. 2009;106:17101–4. doi: 10.1073/pnas.0907147106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Venkatesan RN, Treuting PM, Fuller ED, Goldsby RE, Norwood TH, Gooley TA, Ladiges WC, Preston BD, Loeb LA. Mutation at the polymerase active site of mouse DNA polymerase δ increases genomic instability and accelerates tumorigenesis. Mol. Cell. Biol. 2007;27:7669–82. doi: 10.1128/MCB.00002-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.da Costa LT, Liu B, El-Deiry W, Hamilton SR, Kinzler KW, Vogelstein B, Markowitz S, Willson JK, de la Chapelle A, Downey KM, et al. Polymerase δ variants in RER colorectal tumours. Nat. Genet. 1995;9:10–11. doi: 10.1038/ng0195-10. [DOI] [PubMed] [Google Scholar]
- 18.Flohr T, Dai JC, Büttner J, Popanda O, Hagmüller E, Thielmann HW. Detection of mutations in the DNA polymerase δ gene of human sporadic colorectal cancers and colon cancer cell lines. Int. J. Cancer. 1999;80:919–29. doi: 10.1002/(sici)1097-0215(19990315)80:6<919::aid-ijc19>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 19.Daee DL, Mertz TM, Shcherbakova PV. A cancer-associated DNA polymerase δ variant modeled in yeast causes a catastrophic increase in genomic instability. Proc. Natl. Acad. Sci. U. S. A. 2010;107:157–162. doi: 10.1073/pnas.0907526106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoshida R, Miyashita K, Inoue M, Shimamoto A, Yan Z, Egashira A, Oki E, Kakeji Y, Oda S, Maehara Y. Concurrent genetic alterations in DNA polymerase proofreading and mismatch repair in human colorectal cancer. Eur. J. Hum. Genet. 2011;19:320–325. doi: 10.1038/ejhg.2010.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cancer Genome Atlas Research Network. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Church DN, Briggs SEW, Palles C, Domingo E, Kearsey SJ, Grimes JM, Gorman M, Martin L, Howarth KM, Hodgson SV, Kaur K, Taylor J, Tomlinson IPM. DNA polymerase ε and δ exonuclease domain mutations in endometrial cancer. Hum. Mol. Genet. 2013;22:2820–8. doi: 10.1093/hmg/ddt131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palles C, Cazier J-B, Howarth KM, Domingo E, Jones AM, Broderick P, Kemp Z, Spain SL, Guarino E, Guarino Almeida E, Salguero I, Sherborne A, Chubb D, Carvajal-Carmona LG, Ma Y, Kaur K, Dobbins S, Barclay E, Gorman M, Martin L, Kovac MB, Humphray S, Lucassen A, Holmes CC, Bentley D, Donnelly P, Taylor J, Petridis C, Roylance R, Sawyer EJ, Kerr DJ, Clark S, Grimes J, Kearsey SE, Thomas HJW, McVean G, Houlston RS, Tomlinson I. Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat. Genet. 2013;45:136–44. doi: 10.1038/ng.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seshagiri S, Stawiski EW, Durinck S, Modrusan Z, Storm EE, Conboy CB, Chaudhuri S, Guan Y, Janakiraman V, Jaiswal BS, Guillory J, Ha C, Dijkgraaf GJP, Stinson J, Gnad F, Huntley MA, Degenhardt JD, Haverty PM, Bourgon R, Wang W, Koeppen H, Gentleman R, Starr TK, Zhang Z, Largaespada DA, Wu TD, de Sauvage FJ. Recurrent R-spondin fusions in colon cancer. Nature. 2012;488:660–664. doi: 10.1038/nature11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abaan OD, Polley EC, Davis SR, Zhu YJ, Bilke S, Walker RL, Pineda M, Gindin Y, Jiang Y, Reinhold WC, Holbeck SL, Simon RM, Doroshow JH, Pommier Y, Meltzer PS. The exomes of the NCI-60 panel: A genomic resource for cancer biology and systems pharmacology. Cancer Res. 2013;73:4372–4382. doi: 10.1158/0008-5472.CAN-12-3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao S, Choi M, Overton JD, Bellone S, Roque DM, Cocco E, Guzzo F, English DP, Varughese J, Gasparrini S, Bortolomai I, Buza N, Hui P, Abu-Khalaf M, Ravaggi A, Bignotti E, Bandiera E, Romani C, Todeschini P, Tassi R, Zanotti L, Carrara L, Pecorelli S, Silasi D-A, Ratner E, Azodi M, Schwartz PE, Rutherford TJ, Stiegler AL, Mane S, Boggon TJ, Schlessinger J, Lifton RP, Santin AD. Landscape of somatic single-nucleotide and copy-number mutations in uterine serous carcinoma. Proc. Natl. Acad. Sci. U. S. A. 2013;110:2916–21. doi: 10.1073/pnas.1222577110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Church DN, Stelloo E, Nout Ra, Valtcheva N, Depreeuw J, ter Haar N, Noske A, Amant F, Tomlinson IPM, Wild PJ, Lambrechts D, Jurgenliemk-Schulz IM, Jobsen JJ, Smit VTHBM, Creutzberg CL, Bosse T. Prognostic Significance of POLE Proofreading Mutations in Endometrial Cancer. JNCI J. Natl. Cancer Inst. 2014;107 doi: 10.1093/jnci/dju402. dju402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shinbrot E, Henninger EE, Weinhold N, Covington KR, Schultz N, Chao H, Doddapaneni H, Muzny DM, Gibbs RA, Sander C, Pursell ZF, Wheeler DA, Read C. Exonuclease mutations in DNA polymerase ε reveal replication strand specific mutation patterns and human origins of replication. Genome Res. 2014;24:1740–1750. doi: 10.1101/gr.174789.114.1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stenzinger A, Pfarr N, Endris V, Penzel R, Jansen L, Wolf T, Herpel E, Warth A, Klauschen F, Kloor M, Roth W, Bläker H, Chang-Claude J, Brenner H, Hoffmeister M, Weichert W. Mutations in POLE and survival of colorectal cancer patients - link to disease stage and treatment. Cancer Med. 2014;3:1527–1538. doi: 10.1002/cam4.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meng B, Hoang LN, McIntyre JB, Duggan MA, Nelson GS, Lee CH, Köbel M. POLE exonuclease domain mutation predicts long progression-free survival in grade 3 endometrioid carcinoma of the endometrium. Gynecol. Oncol. 2014;134:15–19. doi: 10.1016/j.ygyno.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 32.Mouradov D, Sloggett C, Jorissen RN, Love CG, Li S, Burgess AW, Arango D, Strausberg RL, Buchanan D, Wormald S, O’Connor L, Wilding JL, Bicknell D, Tomlinson IPM, Bodmer WF, Mariadason JM, Sieber OM. Colorectal cancer cell lines are representative models of the main molecular subtypes of primary cancer. Cancer Res. 2014;74:3238–3247. doi: 10.1158/0008-5472.CAN-14-0013. [DOI] [PubMed] [Google Scholar]
- 33.Kane DP, Shcherbakova PV. A common cancer-associated DNA polymerase ε mutation causes an exceptionally strong mutator phenotype, indicating fidelity defects distinct from loss of proofreading. Cancer Res. 2014;74:1895–1901. doi: 10.1158/0008-5472.CAN-13-2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Billingsley CC, Cohn DE, Mutch DG, Stephens JA, Suarez AA, Goodfellow PJ. Polymerase ε (POLE) mutations in endometrial cancer: Clinical outcomes and implications for Lynch syndrome testing. Cancer. 2015;121:386–394. doi: 10.1002/cncr.29046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van De Wetering M, Francies HE, Francis JM, Bounova G, Iorio F, Pronk A, Van Houdt W, Van Gorp J, Taylor-Weiner A, Kester L, McLaren-Douglas A, Blokker J, Jaksani S, Bartfeld S, Volckman R, Van Sluis P, Li VSW, Seepo S, Sekhar Pedamallu C, Cibulskis K, Carter SL, McKenna A, Lawrence MS, Lichtenstein L, Stewart C, Koster J, Versteeg R, Van Oudenaarden A, Saez-Rodriguez J, Vries RGJ, Getz G, Wessels L, Stratton MR, McDermott U, Meyerson M, Garnett MJ, Clevers H. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 2015;161:933–945. doi: 10.1016/j.cell.2015.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Talhouk A, McConechy MK, Leung S, Li-Chang HH, Kwon JS, Melnyk N, Yang W, Senz J, Boyd N, Karnezis AN, Huntsman DG, Gilks CB, McAlpine JN. A clinically applicable molecular-based classification for endometrial cancers. Br. J. Cancer. 2015;113:299–310. doi: 10.1038/bjc.2015.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Howitt BE, Shukla SA, Sholl LM, Ritterhouse LL, Watkins JC, Rodig S, Stover E, Strickland KC, D’Andrea AD, Wu CJ, Matulonis UA, Konstantinopoulos PA. Association of polymerase e-mutated and microsatellite-instable endometrial cancers with neoantigen load, number of tumor-infiltrating lymphocytes, and expression of PD-1 and PD-L1. JAMA Oncol. 2015;1:1–5. doi: 10.1001/jamaoncol.2015.2151. [DOI] [PubMed] [Google Scholar]
- 38.Stelloo E, Bosse T, Nout RA, MacKay HJ, Church DN, Nijman HW, Leary A, Edmondson RJ, Powell ME, Crosbie EJ, Kitchener HC, Mileshkin L, Pollock PM, Smit VT, Creutzberg CL. Refining prognosis and identifying targetable pathways for high-risk endometrial cancer; a TransPORTEC initiative. Mod. Pathol. 2015;28:836–44. doi: 10.1038/modpathol.2015.43. [DOI] [PubMed] [Google Scholar]
- 39.Kang SY, Park CK, Chang DK, Kim JW, Son HJ, Cho YB, Yun SH, Kim HC, Kwon M, Kim KM. Lynch-like syndrome: Characterization and comparison with EPCAM deletion carriers. Int. J. Cancer. 2015;136:1568–1578. doi: 10.1002/ijc.29133. [DOI] [PubMed] [Google Scholar]
- 40.Bellone S, Centritto F, Black J, Schwab C, English D, Cocco E, Lopez S, Bonazzoli E, Predolini F, Ferrari F, Silasi D-A, Ratner E, Azodi M, Schwartz PE, Santin AD. Polymerase ε (POLE) ultra-mutated tumors induce robust tumor-specific CD4+ T cell responses in endometrial cancer patients. Gynecol. Oncol. 2015;138:1–7. doi: 10.1016/j.ygyno.2015.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wong A, Kuick CH, Wong WL, Tham JM, Mansor S, Loh E, Jain S, Vikas NN, Tan SH, Chan SH, Li ST, Chew SH, Hong W, Ngeow J. Mutation spectrum of POLE and POLD1 mutations in South East Asian women presenting with grade 3 endometrioid endometrial carcinomas. Gynecol. Oncol. 2016;141:113–120. doi: 10.1016/j.ygyno.2015.12.031. [DOI] [PubMed] [Google Scholar]
- 42.Mehnert JM, Panda A, Zhong H, Hirshfield K, Damare S, Lane K, Sokol L, Stein MN, Rodriguez-rodriquez L, Kaufman HL, Ali S, Ross JS, Pavlick DC, Bhanot G, White EP, Dipaola RS, Lovell A, Cheng J, Ganesan S. Immune activation and response to pembrolizumab in POLE-mutant endometrial cancer. J. Clin. Invest. 2016;126:1–7. doi: 10.1172/JCI84940.expression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Köbel M, Meng B, Hoang LN, Almadani N, Li X, Soslow RA, Gilks CB, Lee C-H. Molecular analysis of mixed endometrial carcinomas shows clonality in most cases. Am. J. Surg. Pathol. 2016;40:166–80. doi: 10.1097/PAS.0000000000000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Domingo E, Freeman-Mills L, Rayner E, Glaire M, Briggs S, Vermeulen L, Fessler E, Medema JP, Boot A, Morreau H, van Wezel T, Liefers G-J, Lothe RA, Danielsen SA, Sveen A, Nesbakken A, Zlobec I, Lugli A, Koelzer VH, Berger MD, Castellví-Bel S, Muñoz J, The Epicolon consortium. de Bruyn M, Nijman HW, Novelli M, Lawson K, Oukrif D, Frangou E, Dutton P, Tejpar S, Delorenzi M, Kerr R, Kerr D, Tomlinson I, Church DN. Somatic POLE proofreading domain mutation, immune response, and prognosis in colorectal cancer: a retrospective, pooled biomarker study. Lancet Gastroenterol. Hepatol. 2016;1:207–216. doi: 10.1016/S2468-1253(16)30014-0. [DOI] [PubMed] [Google Scholar]
- 45.Ahn S, Ahmad AA, Kim J, Kim D, Kim J, Kim TW, Park I, Yu C, Jang SJ. The somatic POLE P286R mutation defines a unique subclass of colorectal cancer featuring hypermutation, representing a potential genomic biomarker for immunotherapy. Oncotarget. 2016;7:68638–68649. doi: 10.18632/oncotarget.11862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Giannakis M, Mu XJ, Shukla SA, Qian ZR, Cohen O, Nishihara R, Bahl S, Cao Y, Amin-Mansour A, Yamauchi M, Sukawa Y, Stewart C, Rosenberg M, Mima K, Inamura K, Nosho K, Nowak JA, Lawrence MS, Giovannucci EL, Chan AT, Ng K, Meyerhardt JA, Van Allen EM, Getz G, Gabriel SB, Lander ES, Wu CJ, Fuchs CS, Ogino S, Garraway LA. Genomic correlates of immune-cell infiltrates in colorectal carcinoma. Cell Rep. 2016;15:857–865. doi: 10.1016/j.celrep.2016.03.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lim B, Mun J, Kim J-H, Kim CW, Roh SA, Cho D-H, Kim YS, Kim S-Y, Kim JC, Lim B, Mun J, Kim J-H, Wook Kim C, Roh SA, Cho D-H, Kim YS, Kim S-Y, Kim JC. Genome-wide mutation profiles of colorectal tumors and associated liver metastases at the exome and transcriptome levels. Oncotarget. 2015;6:22179–22190. doi: 10.18632/oncotarget.4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Giannakis M, Hodis E, Jasmine Mu X, Yamauchi M, Rosenbluh J, Cibulskis K, Saksena G, Lawrence MS, Qian ZR, Nishihara R, Van Allen EM, Hahn WC, Gabriel SB, Lander ES, Getz G, Ogino S, Fuchs CS, Garraway LA. RNF43 is frequently mutated in colorectal and endometrial cancers. Nat. Genet. 2014;46:1264–6. doi: 10.1038/ng.3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kothari N, Teer JK, Abbott AM, Srikumar T, Zhang Y, Yoder SJ, Brohl AS, Kim RD, Reed DR, Shibata D. Increased incidence of FBXW7 and POLE proofreading domain mutations in young adult colorectal cancers. Cancer. 2016 doi: 10.1002/cncr.30082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Le Gallo M, O’Hara AJ, Rudd ML, Urick ME, Hansen NF, O’Neil NJ, Price JC, Zhang S, England BM, Godwin AK, Sgroi DC, Hieter P, Mullikin JC, Merino MJ, Bell DW. Exome sequencing of serous endometrial tumors identifies recurrent somatic mutations in chromatin-remodeling and ubiquitin ligase complex genes. Nat. Genet. 2012;44:1310–1315. doi: 10.1038/ng.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stadler ZK, Battaglin F, Middha S, Hechtman JF, Tran C, Cercek A, Yaeger R, Segal NH, Varghese AM, Reidy-Lagunes DL, Kemeny NE, Salo-Mullen EE, Ashraf A, Weiser MR, Garcia-Aguilar J, Robson ME, Offit K, Arcila ME, Berger MF, Shia J, Solit DB, Saltz LB. Reliable detection of mismatch repair deficiency in colorectal cancers using mutational load in next-generation sequencing panels. J. Clin. Oncol. 2016;34:2141–2147. doi: 10.1200/JCO.2015.65.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nowak JA, Yurgelun MB, Bruce JL, Rojas-Rudilla V, Hall DL, Shivdasani P, Garcia EP, Agoston AT, Srivastava A, Ogino S, Kuo FC, Lindeman NI, Dong F. Detection of mismatch repair deficiency and microsatellite instability in colorectal adenocarcinoma by targeted next generation sequencing. J. Mol. Diagnostics. 2016;19:1–8. doi: 10.1016/j.jmoldx.2016.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jesinghaus M, Pfarr N, Endris V, Kloor M, Volckmar A-L, Brandt R, Herpel E, Muckenhuber A, Lasitschka F, Schirmacher P, Penzel R, Weichert W, Stenzinger A. Genotyping of colorectal cancer for cancer precision medicine: Results from the IPH Center for Molecular Pathology. Genes. Chromosomes Cancer. 2016;55:505–521. doi: 10.1002/gcc. [DOI] [PubMed] [Google Scholar]
- 54.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio Cancer Genomics Portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Forbes SA, Beare D, Gunasekaran P, Leung K, Bindal N, Boutselakis H, Ding M, Bamford S, Cole C, Ward S, Kok CY, Jia M, De T, Teague JW, Stratton MR, McDermott U, Campbell PJ. COSMIC: Exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43:D805–D811. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shlien A, Campbell BB, de Borja R, Alexandrov LB, Merico D, Wedge D, Van Loo P, Tarpey PS, Coupland P, Behjati S, Pollett A, Lipman T, Heidari A, Deshmukh S, Avitzur N, Meier B, Gerstung M, Hong Y, Merino DM, Ramakrishna M, Remke M, Arnold R, Panigrahi GB, Thakkar NP, Hodel KP, Henninger EE, Göksenin aY, Bakry D, Charames GS, Druker H, Lerner-Ellis J, Mistry M, Dvir R, Grant R, Elhasid R, Farah R, Taylor GP, Nathan PC, Alexander S, Ben-Shachar S, Ling SC, Gallinger S, Constantini S, Dirks P, Huang A, Scherer SW, Grundy RG, Durno C, Aronson M, Gartner A, Meyn MS, Taylor MD, Pursell ZF, Pearson CE, Malkin D, Futreal PA, Stratton MR, Bouffet E, Hawkins C, Campbell PJ, Tabori U. Combined hereditary and somatic mutations of replication error repair genes result in rapid onset of ultra-hypermutated cancers. Nat. Genet. 2015;47:257–262. doi: 10.1038/ng.3202. [DOI] [PubMed] [Google Scholar]
- 57.Erson-Omay EZ, Cağlayan AO, Schultz N, Weinhold N, Omay SB, Özduman K, Köksal Y, Li J, Serin Harmanc A, Clark V, Carrión-Grant G, Baranoski J, Cağlar C, Barak T, Coskun S, Baran B, Köse D, Sun J, Bakırcıoğlu M, Moliterno Gunel J, Pamir MN, Mishra-Gorur K, Bilguvar K, Yasuno K, Vortmeyer A, Huttner AJ, Sander C, Günel M. Somatic POLE mutations cause an ultramutated giant cell high-grade glioma subtype with better prognosis. Neuro. Oncol. 2015;0:1–9. doi: 10.1093/neuonc/nov027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Elsayed FA, Kets CM, Ruano D, van den Akker B, Mensenkamp AR, Schrumpf M, Nielsen M, Wijnen JT, Tops CM, Ligtenberg MJ, Vasen HF, Hes FJ, Morreau H, van Wezel T. Germline variants in POLE are associated with early onset mismatch repair deficient colorectal cancer. Eur. J. Hum. Genet. 2015;23:1080–1084. doi: 10.1038/ejhg.2014.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Valle L, Hernández-Illán E, Bellido F, Aiza G, Castillejo A, Castillejo MI, Navarro M, Seguí N, Vargas G, Guarinos C, Juarez M, Sanjuán X, Iglesias S, Alenda C, Egoavil C, Segura Á, Juan MJ, Rodriguez-Soler M, Brunet J, González S, Jover R, Lázaro C, Capellá G, Pineda M, Soto JL, Blanco I. New insights into POLE and POLD1 germline mutations in familial colorectal cancer and polyposis. Hum. Mol. Genet. 2014;23:3506–3512. doi: 10.1093/hmg/ddu058. [DOI] [PubMed] [Google Scholar]
- 60.Bellido F, Pineda M, Aiza G, Valdés-Mas R, Navarro M, Puente DA, Pons T, González S, Iglesias S, Darder E, Piñol V, Soto JL, Valencia A, Blanco I, Urioste M, Brunet J, Lázaro C, Capellá G, Puente XS, Valle L. POLE and POLD1 mutations in 529 kindred with familial colorectal cancer and/or polyposis: review of reported cases and recommendations for genetic testing and surveillance. Genet. Med. 2015;18:325–332. doi: 10.1038/gim.2015.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chubb D, Broderick P, Frampton M, Kinnersley B, Sherborne A, Penegar S, Lloyd A, Ma YP, Dobbins SE, Houlston RS. Genetic diagnosis of high-penetrance susceptibility for colorectal cancer (CRC) is achievable for a high proportion of familial CRC by exome sequencing. J. Clin. Oncol. 2015;33:426–432. doi: 10.1200/JCO.2014.56.5689. [DOI] [PubMed] [Google Scholar]
- 62.Spier I, Holzapfel S, Altmüller J, Zhao B, Horpaopan S, Vogt S, Chen S, Morak M, Raeder S, Kayser K, Stienen D, Adam R, Nürnberg P, Plotz G, Holinski-Feder E, Lifton RP, Thiele H, Hoffmann P, Steinke V, Aretz S. Frequency and phenotypic spectrum of germline mutations in POLE and seven other polymerase genes in 266 patients with colorectal adenomas and carcinomas. Int. J. Cancer. 2015;137:320–331. doi: 10.1002/ijc.29396. [DOI] [PubMed] [Google Scholar]
- 63.Rohlin A, Zagoras T, Nilsson S, Lundstam U, Wahlström J, Hultén L, Martinsson T, Karlsson GB, Nordling M. A mutation in POLE predisposing to a multi-tumour phenotype. Int. J. Oncol. 2014;45:77–81. doi: 10.3892/ijo.2014.2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hansen MF, Johansen J, Bjørnevoll I, Sylvander AE, Steinsbekk KS, Sætrom P, Sandvik AK, Drabløs F, Sjursen W. A novel POLE mutation associated with cancers of colon, pancreas, ovaries and small intestine. Fam. Cancer. 2015;14:437–448. doi: 10.1007/s10689-015-9803-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aoude LG, Heitzer E, Johansson P, Gartside M, Wadt K, Pritchard AL, Palmer JM, Symmons J, Gerdes AM, Montgomery GW, Martin NG, Tomlinson I, Kearsey S, Hayward NK. POLE mutations in families predisposed to cutaneous melanoma. Fam. Cancer. 2015;14:621–628. doi: 10.1007/s10689-015-9826-8. [DOI] [PubMed] [Google Scholar]
- 66.Wimmer K, Beilken A, Nustede R, Ripperger T, Lamottke B, Ure B, Steinmann D, Reineke-Plaass T, Lehmann U, Zschocke J, Valle L, Fauth C, Kratz CP. A novel germline POLE mutation causes an early onset cancer prone syndrome mimicking constitutional mismatch repair deficiency. Fam. Cancer. 2016 doi: 10.1007/s10689-016-9925-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Briggs S, Tomlinson I. Germline and somatic polymerase ε and δ mutations define a new class of hypermutated colorectal and endometrial cancers. J. Pathol. 2013;230:148–153. doi: 10.1002/path.4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Seshagiri S. The burden of faulty proofreading in colon cancer. Nat. Genet. 2013;45:121–2. doi: 10.1038/ng.2540. [DOI] [PubMed] [Google Scholar]
- 69.Church JM. Polymerase proofreading-associated polyposis: A new, dominantly inherited syndrome of hereditary colorectal cancer predisposition. Dis. Colon Rectum. 2014;57:396–397. doi: 10.1097/DCR.0000000000000084. [DOI] [PubMed] [Google Scholar]
- 70.Heitzer E, Tomlinson I. Replicative DNA polymerase mutations in cancer. Curr. Opin. Genet. Dev. 2014;24:107–113. doi: 10.1016/j.gde.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rayner E, van Gool IC, Palles C, Kearsey SE, Bosse T, Tomlinson I, Church DN. A panoply of errors: polymerase proofreading domain mutations in cancer. Nat. Rev. Cancer. 2016;16:71–81. doi: 10.1038/nrc.2015.12. [DOI] [PubMed] [Google Scholar]
- 72.Mertz TM, Harcy V, Roberts SA. Risks at the DNA replication fork: Effects upon carcinogenesis and tumor heterogeneity. Genes (Basel) 2017;8:46. doi: 10.3390/genes8010046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mertz TM, Sharma S, Chabes A, Shcherbakova PV. Colon cancer-associated mutator DNA polymerase δ variant causes expansion of dNTP pools increasing its own infidelity. Proc. Natl. Acad. Sci. U. S. A. 2015;112:E2467–76. doi: 10.1073/pnas.1422934112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mertz TM, Baranovskiy AG, Wang J, Tahirov TH, Shcherbakova PV. Nucleotide selectivity defect and mutator phenotype conferred by a colon cancer-associated DNA polymerase δ mutation in human cells. Oncogene. 2017 Apr 3; doi: 10.1038/onc.2017.22. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Northam MR, Moore EA, Mertz TM, Binz SK, Stith CM, Stepchenkova EI, Wendt KL, Burgers PMJ, Shcherbakova PV. DNA polymerases ζ and Rev1 mediate error-prone bypass of non-B DNA structures. Nucleic Acids Res. 2014;42:290–306. doi: 10.1093/nar/gkt830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jin YH, Obert R, Burgers PM, Kunkel TA, Resnick MA, Gordenin DA. The 3’-->5’ exonuclease of DNA polymerase δ can substitute for the 5’ flap endonuclease Rad27/Fen1 in processing Okazaki fragments and preventing genome instability. Proc. Natl. Acad. Sci. U. S. A. 2001;98:5122–5127. doi: 10.1073/pnas.091095198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Herr AJ, Ogawa M, Lawrence NA, Williams LN, Eggington JM, Singh M, Smith RA, Preston BD. Mutator suppression and escape from replication error-induced extinction in yeast. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1002282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tran HT, Degtyareva NP, Gordenin DA, Resnick MA. Genetic factors affecting the impact of DNA polymerase δ proofreading activity on mutation avoidance in yeast. Genetics. 1999;152:47–59. doi: 10.1093/genetics/152.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pavlov YI, Shcherbakova PV, Kunkel TA. In vivo consequences of putative active site mutations in yeast DNA polymerases α, ε, δ, and ζ. Genetics. 2001;159:47–64. doi: 10.1093/genetics/159.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Murphy K, Darmawan H, Schultz A, da Silva EF, Reha-Krantz LJ. A method to select for mutator DNA polymerase δs in Saccharomyces cerevisiae. Genome. 2006;49:403–410. doi: 10.1139/G05-106. [DOI] [PubMed] [Google Scholar]
- 81.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SAJR, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Børresen-Dale A-L, Boyault S, Burkhardt B, Butler AP, Caldas C, Davies HR, Desmedt C, Eils R, Eyfjörd JE, Foekens JA, Greaves M, Hosoda F, Hutter B, Ilicic T, Imbeaud S, Imielinski M, Imielinsk M, Jäger N, Jones DTW, Jones D, Knappskog S, Kool M, Lakhani SR, López-Otín C, Martin S, Munshi NC, Nakamura H, Northcott PA, Pajic M, Papaemmanuil E, Paradiso A, Pearson JV, Puente XS, Raine K, Ramakrishna M, Richardson AL, Richter J, Rosenstiel P, Schlesner M, Schumacher TN, Span PN, Teague JW, Totoki Y, Tutt ANJ, Valdés-Mas R, van Buuren MM, van ‘t Veer L, Vincent-Salomon A, Waddell N, Yates LR, Zucman-Rossi J, Futreal PA, McDermott U, Lichter P, Meyerson M, Grimmond SM, Siebert R, Campo E, Shibata T, Pfister SM, Campbell PJ, Stratton MR. Signatures of mutational processes in human cancer. Nature. 2013;500:415–21. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Williams LN, Marjavaara L, Knowels GM, Schultz EM, Fox EJ, Chabes A, Herr AJ. dNTP pool levels modulate mutator phenotypes of error-prone DNA polymerase ε variants. Proc. Natl. Acad. Sci. U. S. A. 2015;112:E2457–66. doi: 10.1073/pnas.1422948112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tran HT, Gordenin DA, Resnick MA. The 3’-->5’ exonucleases of DNA polymerases δ and and ε the 5’-->3’ exonuclease Exo1 have major roles in postreplication mutation avoidance in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:2000–7. doi: 10.1128/MCB.19.3.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Agbor AA, Göksenin AY, LeCompte KG, Hans SH, Pursell ZF. Human Pol ε-dependent replication errors and the influence of mismatch repair on their correction. DNA Repair (Amst) 2013;12:954–963. doi: 10.1016/j.dnarep.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shcherbakova PV, Pavlov YI, Chilkova O, Rogozin IB, Johansson E, Kunkel TA. Unique error signature of the four-subunit yeast DNA polymerase ε. J. Biol. Chem. 2003;278:43770–43780. doi: 10.1074/jbc.M306893200. [DOI] [PubMed] [Google Scholar]
- 86.Korona DA, Lecompte KG, Pursell ZF. The high fidelity and unique error signature of human DNA polymerase ε. Nucleic Acids Res. 2011;39:1763–1773. doi: 10.1093/nar/gkq1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Flood CL, Rodriguez GP, Bao G, Shockley AH, Kow YW, Crouse GF. Replicative DNA polymerase δ but not ε proofreads errors in cis and in trans. PLOS Genet. 2015;11:e1005049. doi: 10.1371/journal.pgen.1005049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Billingsley CC, Cohn DE, Mutch DG, Hade EM, Goodfellow PJ. Prognostic significance of POLE exonuclease domain mutations in high-grade endometrioid endometrial cancer on survival and recurrence. Int. J. Gynecol. Cancer. 2016;26:933–938. doi: 10.1097/IGC.0000000000000681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hussein YR, Weigelt B, Levine DA, Schoolmeester JK, Dao LN, Balzer BL, Liles G, Karlan B, Köbel M, Lee C-H, Soslow RA. Clinicopathological analysis of endometrial carcinomas harboring somatic POLE exonuclease domain mutations. Mod. Pathol. 2015;28:505–514. doi: 10.1038/modpathol.2014.143. [DOI] [PubMed] [Google Scholar]
- 90.Santin AD, Bellone S, Centritto F, Schlessinger J, Lifton R. Improved survival of patients with hypermutation in uterine serous carcinoma. Gynecol. Oncol. Reports. 2015;12:3–4. doi: 10.1016/j.gore.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.van Gool IC, Bosse T, Church DN. POLE proofreading mutation, immune response and prognosis in endometrial cancer. Oncoimmunology. 2016;5:e1072675. doi: 10.1080/2162402X.2015.1072675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.van Gool IC, Eggink FA, Freeman-Mills L, Stelloo E, Marchi E, de Bruyn M, Palles C, Nout RA, de Kroon CD, Osse EM, Klenerman P, Creutzberg CL, Tomlinson IPM, Smit VTHBM, Nijman HW, Bosse T, Church DN. POLE proofreading mutations elicit an anti-tumor immune response in endometrial cancer. Clin. Cancer Res. 2015;21:3347–3356. doi: 10.1158/1078-0432.CCR-15-0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Santin AD, Bellone S, Buza N, Choi J, Schwartz PE, Schlessinger J, Lifton RP. Regression of chemotherapy-resistant Polymerase epsilon (POLE) ultra-mutated and MSH6 hyper-mutated endometrial tumors with nivolumab. Clin. Cancer Res. 2016 doi: 10.1158/1078-0432.CCR-16-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Uppendahl L, Mullany SA, Winterhoff B. Molecular characterization of endometrial cancer and therapeutic implications. Curr. Opin. Obstet. Gynecol. 2017;29:35–39. doi: 10.1097/GCO.0000000000000342. [DOI] [PubMed] [Google Scholar]
- 95.Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, Walsh La, Postow MA, Wong P, Ho TS, Hollmann TJ, Bruggeman C, Kannan K, Li Y, Elipenahli C, Liu C, Harbison CT, Wang L, Ribas A, Wolchok JD, Chan TA. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N. Engl. J. Med. 2014:2189–2199. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, Lee W, Yuan J, Wong P, Ho TS, Miller ML, Rekhtman N, Moreira AL, Ibrahim F, Bruggeman C, Gasmi B, Zappasodi R, Maeda Y, Sander C, Garon EB, Merghoub T, Wolchok JD, Schumacher TN, Chan TA. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Johanns TM, Miller CA, Dorward IG, Tsien C, Chang E, Perry A, Uppaluri R, Ferguson C, Schmidt RE, Dahiya S, Ansstas G, Mardis ER, Dunn GP. Immunogenomics of hypermutated glioblastoma: A patient with germline POLE deficiency treated with checkpoint blockade immunotherapy. Cancer Discov. 2016;6:1230–1236. doi: 10.1158/2159-8290.CD-16-0575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gargiulo P, Della Pepa C, Berardi S, Califano D, Scala S, Buonaguro L, Ciliberto G, Brauchli P, Pignata S. Tumor genotype and immune microenvironment in POLE-ultramutated and MSI-hypermutated Endometrial Cancers: New candidates for checkpoint blockade immunotherapy? Cancer Treat. Rev. 2016;48:61–68. doi: 10.1016/j.ctrv.2016.06.008. [DOI] [PubMed] [Google Scholar]
- 99.Nelson BH, McAlpine JN. The more tumors change, the more they stay tame: Do T cells keep POLE ultramutated endometrial carcinomas in check? Gynecol. Oncol. 2015;138:1–2. doi: 10.1016/j.ygyno.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 100.Snyder A, Wolchok JD. Successful treatment of a patient with glioblastoma and a germline POLE mutation: Where next? Cancer Discov. 2016;6:1210–1211. doi: 10.1158/2159-8290.CD-16-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cancer Genome Atlas Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–9. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.