Abstract
Group A Streptococcus (GAS) is among the top 10 causes of infection-related mortality in humans. M protein is the most abundant GAS surface protein, and M1 serotype GAS strains are associated with invasive infections including necrotizing fasciitis and toxic shock syndrome. Here we report that released, soluble M1 protein triggers programmed cell death in macrophages (Mϕ). M1 served as a second signal for caspase-1-dependent NLRP3 inflammasome activation, inducing maturation and release of proinflammatory cytokine IL-1β and macrophage pyroptosis. The structurally dynamic B-repeat domain of M1 was critical for inflammasome activation, which involved K+ efflux and M1 protein internalization by clathrin-mediated endocytosis. Mouse intraperitoneal challenge showed that soluble M1 was sufficient and specific for IL-1β activation, which may represent an early warning to activate host immunity against the pathogen. Conversely, in systemic infection, hyperinflammation associated with M1-mediated pyroptosis and IL-1β release could aggravate tissue injury.
Group A Streptococcus (GAS) causes diseases ranging from common pharyngitis to life-threatening necrotizing fasciitis (NF) and streptococcal toxic shock syndrome (STSS), and is the immunologic trigger for rheumatic heart disease1. With an estimated 500,000 deaths annually, GAS ranks among the ten deadliest human pathogens2. The pathogen expresses numerous surface-bound and secreted virulence factors that subvert innate immune defences1.
M protein, the most abundant protein on the GAS surface3, extends as hair-like fimbriae4, with a structure, function, and immunochemistry unique among known virulence molecules5. Sequence analysis of encoding emm genes suggests >220 variants6; however, only a few M protein/emm types are widespread and associated with systemic infections, with serotype M1/emm1 the most prevalent worldwide7. A globally disseminated serotype M1T1 subclone has been the leading cause of severe invasive GAS infections in recent decades8.
M proteins mediate host epithelial cell adhesion9, and resistance to opsonophagocytosis by binding host components including fibrinogen, C4b-binding protein, and immunoglobulin Fc10. M proteins also block membrane-lytic activities of host antimicrobial peptides and histones by sequestering them away from the bacterial membrane11,12. Soluble M proteins released during infection by neutrophil-derived granule proteases13 can trigger inflammation by forming a supramolecular assembly with fibrinogen that activates neutrophils13,14. Indeed, soluble M1 protein is sufficient in animal models to trigger vascular leakage and tissue injury similar to severe GAS diseases such as NF and STSS13,15. Under physiological conditions, M1 protein is also naturally shed from the GAS surface and is detected at micromolar concentrations in the extracellular medium16, provided it is not degraded by the broad-spectrum GAS cysteine protease SpeB17. In M1T1 GAS and certain other invasive serotypes, mutations in the covR/S (aka csrR/S) two-component regulatory can upregulate several virulence factors while suppressing expression of SpeB18,19. Loss of SpeB expression spares M protein from proteolytic degradation and increases levels of its soluble form. Effects of soluble M1 on macrophages have not previously been investigated.
Macrophages play a central role in pathogen recognition and activation of innate immune and inflammatory responses. Inflammasomes are multimeric cytosolic protein complexes that integrate pathogen-triggered signaling cascades in macrophages, activating caspase-1 and proteolytic processing of pro-IL-1β and pro-IL-18 to their mature cytokine forms, IL-1β and IL-1820. Inflammasomes can activate a rapid pro-inflammatory form of macrophage cell death termed “pyroptosis”, characterized by plasma membrane rupture, release of cytosolic contents, and DNA fragmentation21. Pyroptosis restricts replication of intracellular bacterial pathogens22, and IL-1β signaling promotes immune resistance to several microbes, including GAS23.
Among various inflammasomes, those containing intracellular pattern recognition molecules NACHT, LRR, and pyrin domains-containing protein 3 (NLRP3) are best studied, as their deregulation contributes to inherited autoinflammatory disorders24 and chronic metabolic diseases, including atherosclerosis, type 2 diabetes, gout, and obesity25. To date, two GAS secreted toxins are known to activate macrophage NLRP3 inflammasome-mediated IL-1β signaling and pyroptosis, pore-forming cytolysin streptolysin O (SLO)26 and C3 ADP-ribosyltransferase SpyA27. Consequences of GAS toxin-mediated NLRP3 inflammasome activation range from a protective immune response to exaggerated inflammatory host cell damage.
Here we report that the classical GAS virulence factor M1 protein triggers macrophage cell death in a manner dependent on its B-repeat region. M1 is a second signal for caspase-1-dependent NLRP3 inflammasome activation, leading to IL-1β processing and pyroptotic macrophage cell death. M1 uptake occurs through clathrin-mediated endocytosis accompanied by K+ efflux, both essential events in NLRP3 activation. M1-induced pyroptosis and IL-1β signaling may exert pro-immune and pro-inflammatory effects of potential benefit or harm to the human host depending on the site, stage, and magnitude of infection.
Results
GAS M1 protein triggers macrophage cell death
While probing effects of soluble recombinant M1 protein on host innate immune and inflammatory responses, we surprisingly observed that M1 induced rapid (within 2 h) and dose-dependent loss of plasma membrane integrity in cultured human THP-1 macrophage-like cells (THP-1 Mϕ, as monitored intracellular lactate dehydrogenase (LDH) release (Fig. 1a). This cytolytic phenotype was independent of the previously described13,14 M1-fibrinogen interaction (Supplementary Fig. 1a). Compared to M1, additional recombinant M protein types (M2, M4, M5, M6) induced similar levels of THP-1 Mϕ cytolysis, whereas other M types elicited significantly lower (M22) or markedly lower (M28) cytotoxicity (Fig. 1b). Propidium iodide (PI) uptake, assessed by fluorescence microscopy and quantified by flow cytometry, confirmed significant membrane damage in M1-treated THP-1 Mϕ vs. untreated cells (Fig. 1c-e). Additionally, terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) showed dose-dependent M1-induced DNA fragmentation in THP-1 Mϕ (Fig. 1f, g). To ascertain cell-type specificity of M1-mediated cytotoxicity, we performed LDH assays on THP-1 monocytes (THP-1 Mo), A549 human lung epithelial cells, primary human neutrophils, and primary human peripheral blood mononuclear cells (PBMCs) (Fig. 1h). None of these cells were lysed by M1, nor were freshly isolated human erythrocytes (Supplementary Fig. 1b). Thus, M1 does not cause nonspecific cell membrane permeabilization, but somehow provokes macrophages to undergo a form of programmed cell death.
Figure 1.
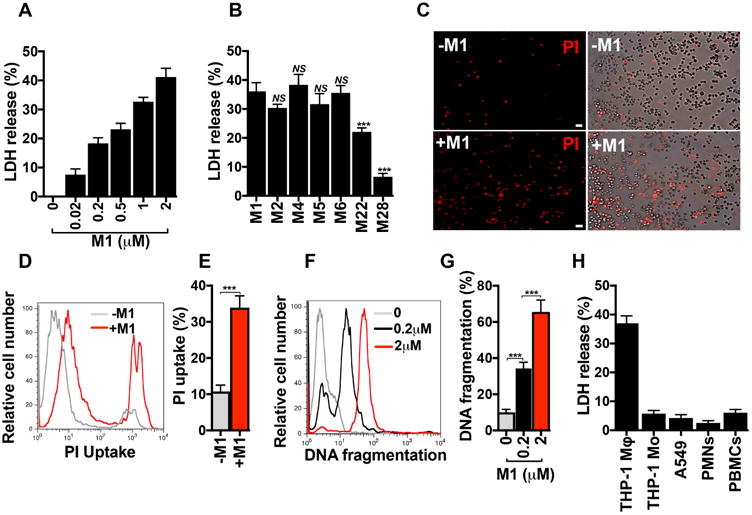
GAS M1 protein promotes cell death in macrophages. a, Percentage of LDH released from THP-1 human macrophages (THP-1 Mϕ after 2 h treatment with increasing concentrations of recombinant purified M1 protein. b, Percentage of LDH released from THP-1 Mϕ incubated for 2 hours with M1 protein (2 μM) or other M proteins (2 μM). c, Fluorescence microscopy of propidium iodide (PI, red) uptake by THP-1 Mϕ that were untreated (-M1, top panels) or treated for 2 h with 2 μM M1 protein (+M1, bottom panels). Fluorescence images are shown on the left and merge of fluorescence and phase contrast images are shown on the right; scale bar = 30 μm. d, e, Flow cytometry analysis of PI uptake by THP-1 Mϕ that were untreated (-M1) or treated for 2 h with 2 μM M1 protein (+M1). A representative histogram of PI fluorescence is shown in panel d, and quantification of PI uptake as measured by percentage of cells with PI uptake (%) is shown in panel e. f, g, FACS analysis of TUNEL assay of THP-1 Mϕ treated with M1 (0.2 or 2 μM) or untreated (0) for 4h. Panel f shows a representative flow cytometry histogram of DNA fragmentation and panel g shows the percentage of cells with DNA fragmentation (%) compared with untreated cells (0). h, LDH release from THP-1 macrophages (THP-1 Mϕ), THP-1 monocytes (THP-1 Mo), A549 human alveolar basal epithelial cells (A549), primary human neutrophils (PMNs), and primary human peripheral blood mononuclear cells (PBMCs) treated for 2 h with M1 (2 μM). Data in panels a, b, e, g and h are plotted as the mean ± SEM and represent three independent experiments performed in triplicate and analyzed by Student's t-test. NS = not significant (P>0.05) and ***P<0.001. Panels c, d, and f, show representative images and flow cytometry histograms of three independent experiments.
M1 protein promotes IL-1β signalling in macrophages
GAS can trigger cell death in macrophages by pyroptosis27, but no role for M1 protein in this process has been reported. Pyroptosis is accompanied by release of characteristic inflammatory cytokines, most prominently IL-1β. Soluble M1 was sufficient to induce dose-dependent release of IL-1β from THP-1 Mϕ, as detected by ELISA (Fig. 2a). Since ELISA could not discriminate between pro-IL-1β and mature IL-1β, we assessed signaling activity using HEK-Blue™ IL-1β reporter cells (Fig. 2b), confirming dose-dependent release of mature IL-1β upon M1 treatment. M2, M4, M5, and M6 proteins triggered similar release of functional IL-1β from THP-1 Mϕ, while decreased IL-1β activity was seen with M22 or M28 exposure (Fig. 2c), paralleling cytolytic effects.
Figure 2. M1 protein provides a second signal that specifically triggers IL-1β signalling.
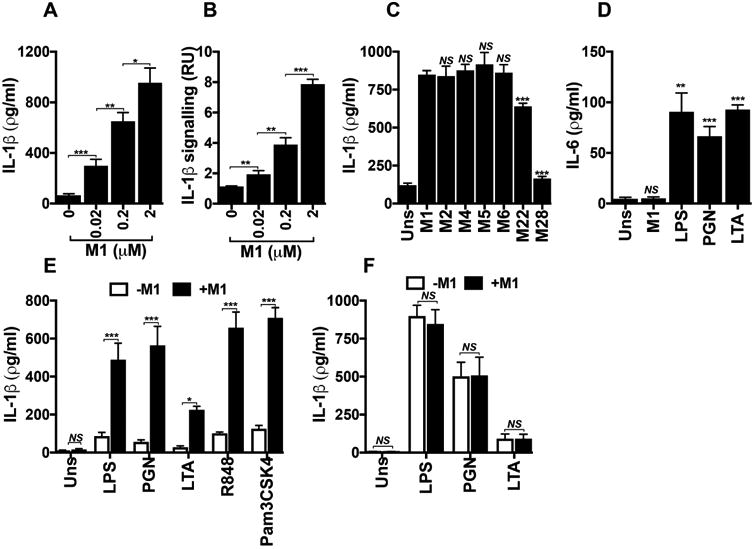
Total IL-1β (a) and the processed form of IL-1β (b) produced by THP-1 Mϕ incubated for 2 h with increasing concentrations of M1 protein, as measured by ELISA or by HEK-Blue™ IL-1R reporter cells, respectively. c, Comparison of total IL-1β measured by ELISA from THP-1 Mϕ incubated for 2 h with M1 (2 μM) or other M proteins (2 μM). d, IL-6 production assessed by ELISA from THP-1 Mϕ that were unstimulated (Uns) or stimulated for 15 h with M1 (0.02 μM), lipopolysaccharide (LPS, 100 ng ml-1), peptidoglycan (PGN, 1 μg ml-1), or lipoteichoic acid (LTA, 1 μg ml-1). IL-1β release measured by ELISA from mouse BMDMs (e) or human PBMCs (f) that were unprimed (Uns) or primed for 16 h with different TLR agonists: LPS (10 ng ml-1), PGN (1 μg ml-1), LTA (1 μg ml-1), R848 (1 μg ml-1), or Pam3CSK4 (10 ng ml-1). BMDMs and PBMCs were then untreated (-M1) or treated with 2 μM of M1 protein (+M1) for 2 h. Data in panels a-f are plotted as the mean ± SEM and represent three independent experiments performed in triplicate and analyzed by Student's t-test. NS = not significant (P>0.05), * P<0.05, ** P<0.01 and *** P<0.001),
M1 protein did not stimulate THP-1 Mϕ to release IL-6, a cytokine activated in response to toll-like receptor (TLR) signaling but unrelated to the pyroptosis machinery (Fig. 2d). In contrast, classical activators of TLR signaling (e.g., lipopolysaccharide, peptidoglycan, lipoteichoic acid) triggered IL-6 release (Fig. 2d) under assay conditions yielding similar macrophage viabilities (Supplementary Fig. 2). Thus, soluble M1 protein was sufficient to specifically induce IL-1β signaling, with pyroptosis the likely cell death pathway concurrently activated in macrophages.
M1 activates the NLRP3 inflammasome and pyroptosis in a caspase-1-dependent manner
Synthesis, processing, and release of mature IL-1β by macrophages depend upon two independent signals28. An inflammatory signal is provided by pathogen-associated molecular patterns (PAMPs) or danger-associated molecule patterns (DAMPs) recognized by specific receptors, including TLRs. These first signals, known as priming signals, induce transcription and synthesis of pro-IL-1β and initiate expression of inflammasome components29. A second signal is subsequently required for inflammasome assembly and activation, resulting in maximal generation of mature IL-1β. In our experiments with cultured THP-1 Mϕ, phorbol 12-myristate 13-acetate (PMA) was used to differentiate cells into the macrophage phenotype. PMA provided signal 1, as IL-1β transcript increased over 1000-fold compared to undifferentiated THP-1 Mo (Supplementary Fig. 3), and we hypothesized that M1 was a second signal required for efficient inflammasome activation. Murine bone marrow-derived macrophages (BMDMs) primed with a variety of TLR stimuli, but not unprimed BMDMs, responded to M1 protein by releasing IL-1β (Fig. 2e). In contrast, signal 1 alone was sufficient to trigger IL-1β in human primary PBMCs, and addition of M1 did not further boost release of the cytokine (Fig. 2f). This observation correlates with recent studies that demonstrate human monocytes engage an alternative inflammasome pathway, wherein TLR agonists by themselves trigger IL-1β in a process independent of K+ efflux and pyroptosis30.
M1 protein strongly induced IL-1β release from primed WT BMDMs but not primed casp1/11-/- BMDMs, which cannot form functional inflammasomes (Fig. 3a). Nigericin, an ionophore and established canonical NLRP3 inflammasome activator31, served as a positive control in this experiment. Pretreatment of THP-1 Mϕ with Ac-YVAD-cmk, a specific and irreversible pharmacological inhibitor of caspase-1, nearly abolished M1-mediated release of IL-1β (Fig. 3b) and macrophage cytolysis (Fig. 3c). Thus, M1 protein-triggered IL-1β release and pyroptosis are caspase-1-dependent.
Figure 3. M1 B-repeat region is essential for activation of the NLRP3 inflammasome and pyroptosis in a caspase-1-dependent manner requiring potassium efflux.
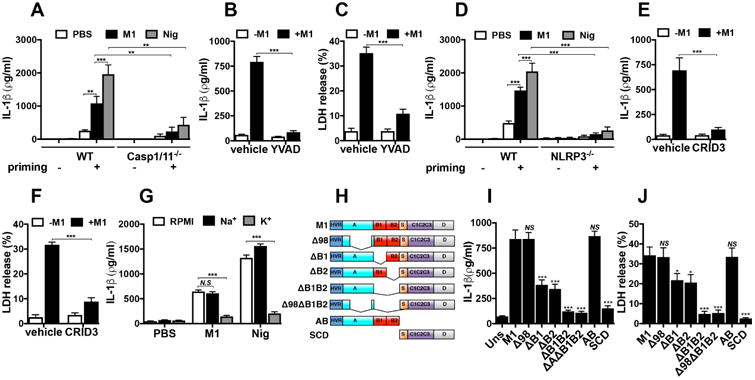
Comparison of IL-1β production measured by ELISA from WT and casp1/11-/- (a) or from WT and nlrp3-/- (d) BMDMs incubated 16 h with (priming +) or without (priming -) LPS (10 ng ml-1), and then treated for 2 h with M1 protein (2 μM) or nigericin (20 μM). Production of IL-1β and LDH release from THP-1 Mϕ pretreated for 1 h with Ac-YVAD-cmk (b, c) or CRID3 (e, f), which are caspase-1 and NLRP-3 inhibitors, respectively, before stimulation with M1 (2 μM) for 2 h. DMSO and PBS were used as vehicle for Ac-YVAD-cmk and CRID3, respectively. g, Release of IL-1β as measured by ELISA from THP-1 Mϕ incubated with M1 (2 μM) or nigericin (20 μM) for 2 h in RPMI with potassium- (K+ media) or sodium- (Na+ media) rich media. h, Schematic representation of M1 constructs, with domains denoted: M1 (mature M1 protein, residues 42-453), Δ98 (Δ98-125), ΔB1 (Δ133-161), ΔB2 (Δ162-189), ΔB1ΔB2 (Δ133-189), Δ98ΔB1ΔB2 (Δ98-125, Δ133-189), AB (residues 4-194), SCD (residues 195-453). Comparison of THP1 Mϕ treated with M1 (2 μM) and M1 truncated proteins (2 μM) for 2 h, after which time supernatants were collected and analyzed for IL-1β by ELISA (i) and LDH release (j). Data in panels a-g and i-j are plotted as the mean ± SEM and represent three independent experiments performed in triplicate and analyzed by Student's t-test. NS = not significant (P>0.05), * P<0.05, ** P<0.01 and ***P<0.001).
We found that BMDMs from NLRP3-deficient mice failed to secrete significant IL-1β after M1 stimulation (Fig. 3d). As expected, NLRP3-/- cells stimulated with nigericin also produced minimal IL-1β (Fig. 3d). CRID3, a powerful and specific NLRP3 inhibitor32, also blocked macrophage release of IL-1β (Fig. 3e) and pyroptosis (Fig. 3f) in response to M1 protein. A drop in cytosolic K+ serves as a common step required to activate the NLRP3 inflammasome33. M1-mediated macrophage IL-1β production was markedly reduced when the gradient for K+ efflux was eliminated using K+-rich media (Fig. 3g). This reduction resembled that observed with nigericin (Fig. 3g), whose NLRP3 activation is also mechanistically dependent on K+ efflux. M1 thus activates the NLRP3 inflammasome in a scenario involving K+ efflux, activation of caspase-1, processing of pro-IL-1β, and secretion of mature IL-1β, all accompanied by macrophage pyroptosis.
The B-repeat domain of M1 is critical for NLRP3 inflammasome activation
To map the domain(s) of M1 protein required for NLRP3 inflammasome activation, we expressed and purified a set of previously described M1 truncations14,34, depicted schematically in Fig. 3h. Quantifying both IL-1β production and cytotoxicity (LDH release) from THP-1 Mϕ, we found the N-terminal AB fragment of M1 protein induced similar levels of IL-1β (Fig. 3i) and LDH release as intact M1 protein (Fig. 3j). In contrast, the C-terminal SCD fragment did not induce inflammasome activation and pyroptosis. Deleting the majority of the A region (Δ98) failed to block M1-induced macrophage IL-1β and LDH release, but eliminating either the B1 or B2 repeat region led to significantly less inflammasome activation, with a deletion of both B regions (ΔB1ΔB2) yielding an even greater reduction (Fig. 3i, j). Thus, the B-repeat region of M1 protein plays a key functional role in NLRP3 inflammasome activation, IL-1β release, and macrophage pyroptosis.
M1 uptake by macrophages is required for NLRP3 inflammasome activation
To determine if macrophage uptake of M1 was required to stimulate IL-1β, THP-1 Mϕ were treated with well-characterized inhibitors of endocytic pathways. Neither wortmannin, a covalent inhibitor of phosphoinositide 3-kinases used to block macrophage macropinocytosis, nor methyl-β-cyclodextrin, a pharmacological inhibitor of caveolae/lipid-raft mediated endocytosis, altered M1-induced IL-1β production (Fig. 4a). In contrast, treatment with Pitstop-2, a small molecule that selectively binds the C-terminal domain of clathrin to inhibit clathrin pit-mediated dynamics and endocytosis (CME), clearly reduced macrophage IL-1β release and processing upon M1 exposure, as measured by ELISA (Fig. 4a) and HEK-Blue™ IL-1β reporter cells (Fig. 4b).
Figure 4. M1 uptake is required for inflammasome activation.
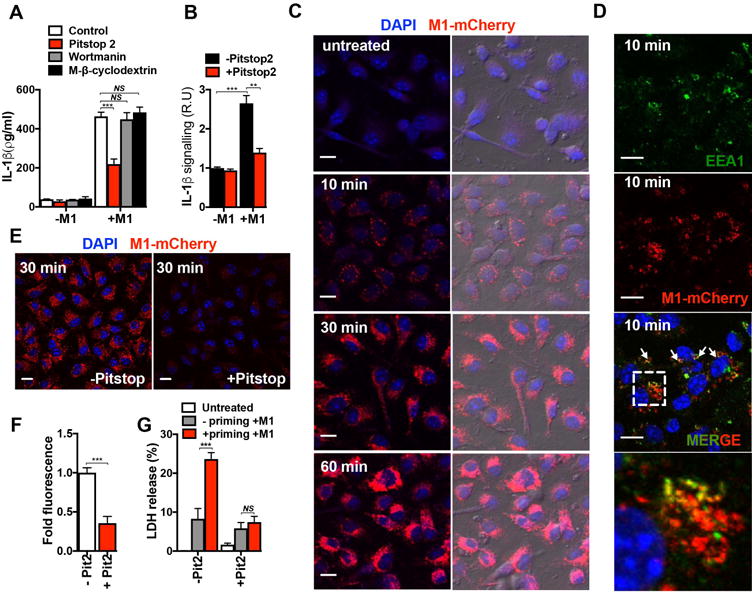
a, Production of IL-1β measured by ELISA from THP1-Mϕ that were untreated (Control) or pretreated with wortmannin or methyl-β-cyclodextrin for 30 min or with Pitstop-2 for 10 min prior to stimulation with M1 protein (2 μM) for 1 h. b, Production of processed IL-1β in THP1-Mϕ that were either untreated or pretreated with Pitstop-2 for 10 min and then treated with M1 (2 μM) for 1 h. c, Confocal microscopy of BMDMs untreated (-M1) or incubated with M1-mCherry protein (2 μM) for the indicated times. DAPI (blue) and M1-mCherry (red) merged images are shown in the left panels, and a merge of fluorescence and phase contrast images are shown in the right panels; scale bar = 10 μm. d, Confocal microscopy images of BMDMs incubated for 10 min with M1-mCherry protein (2 μM) and then fixed and stained with anti-EEA1 antibody. The colocalization of M1-mCherry (red) and EEA1 (green) is indicated by white arrows in the merged image. At bottom is a magnification of the dashed boxed region in the merged image; scale bar = 10μm. Panel e, shows confocal microscopy images of BMDMs treated with M1-mCherry for 30 min in the presence (+Pitstop) or in the absence (-Pitstop) of Pitstop-2. Panel f, shows quantification of M1-mCherry fluorescence (intensity of M1-mCherry fluorescence of each treatment group and shown as relative fluorescence in arbitrary units). DAPI (blue) and M1-mCherry (red) merged images are shown; scale bar = 10μm. g, LDH release from BMDMs pretreated (+ priming) or untreated (- priming) with LPS for 4 h and incubated without (-M1) or with (+M1) M1-mCherry (2 μM) in the presence (+ Pit 2) or absence (- Pit 2) of Pitstop-2 (100 % represents total cytolysis). Data in panels a, b, and g are plotted as the mean ± SEM from three independent experiments performed in triplicate. Panels c, e, and d show representative images of three and two independent experiments, respectively. Data in panel f is plotted as the mean ± SEM, representing the fluorescence of at least 100 cells from two independent experiments. Data in panels a, b, f and g were analyzed by Student's t-test. NS = not significant (P>0.05), **P<0.01 and ***P<0.001).
To visualize M1 protein uptake into macrophages, we fused M1 protein and fluorescent mCherry (M1-mCherry). Confocal microscopy revealed progressive uptake of M1-mCherry into macrophages over 1 h (Fig. 4c). No significant difference in M1 uptake was observed between primed cells and unprimed BMDMs, indicating that M1 endocytosis proceeded independently of inflammasome signal one (Supplementary Fig. 4). In most treated macrophages, M1-mCherry assembled into intracellular speckles as early as 10 min, after which the number and size of the speckles increased (30 min), eventually reaching a diffuse distribution throughout the cytosolic compartment (1 h), suggesting a rapid and dynamic cellular uptake process (Fig. 4c). Neither macrophage uptake nor intracellular speckle formation was observed with recombinant mCherry alone (Supplementary Fig. 5). Intracellular speckles were also observed with a fusion construct between M1ΔB1ΔB2 protein and mCherry (Supplementary Fig. 6), suggesting that macrophage internalization of M1 occurs independently of its B-repeat region.
M1-mCherry co-localized with early endosome marker EEA1 in as early as 10 min (Fig. 4d). Mander's coefficients analysis indicated a reciprocal degree of overlapping signal between the M1-mCherry and EEA1 signals, and vice versa (Supplementary Fig. 7), suggesting that M1 is internalized through a specific endocytic pathway. Incubation of macrophages with CME inhibitor Pitstop-2 prevented formation of M1-mCherry intracellular speckles (Fig. 4e). Single cell analysis confirmed a significant (∼65%) decrease in M1-mCherry signal in Pitstop-2 treated cells (Fig. 4f), indicating that macrophage M1 protein uptake is strongly dependent on CME. Pitstop-2 also markedly decreased LDH release and cytotoxicity in primed BMDMs exposed to recombinant M1 (Fig. 4g), confirming that CME helps to activate the NLRP3 inflammasome and pyroptosis.
Contribution of M1 expressed by GAS to IL-1β production and pyroptosis
As soluble M1 was sufficient to induce IL-1β release, we examined whether M1 protein expressed by live GAS contributes to inflammasome activation and pyroptosis. When incubated with THP-1 Mϕ, a WT M1T1 GAS bacterial strain induced significantly more cytolysis (Fig. 5a) and IL-1β release (Fig. 5b), at multiplicities of infection ranging from 1 to 20, than an isogenic M1-protein deficient (ΔM1) mutant strain. Correspondingly, supernatants from WT M1T1 GAS-infected macrophages contained higher levels of processed IL-1β, as detected by IL-1β reporter cells, compared to supernatants from ΔM1 GAS-infected macrophages (Fig. 5c). Restoring M1 expression by plasmid complementation of the ΔM1 mutant (GASΔM1comp) yielded a very similar profile of cytolysis (Fig. 5a), IL-1β production (Fig. 5b) and IL-1β processing (Fig. 5c) to the WT parent strain across the different MOIs tested. Flow cytometry experiments of macrophages infected with FITC-labeled M1T1 GAS WT or isogenic mutant ΔM1 strains showed a very minor reduction in bacterial invasion of macrophages infected with the latter (Supplementary Fig. 8).
Figure 5. M1 action as natively expressed on GAS in vitro and in vivo and as soluble protein in vivo.
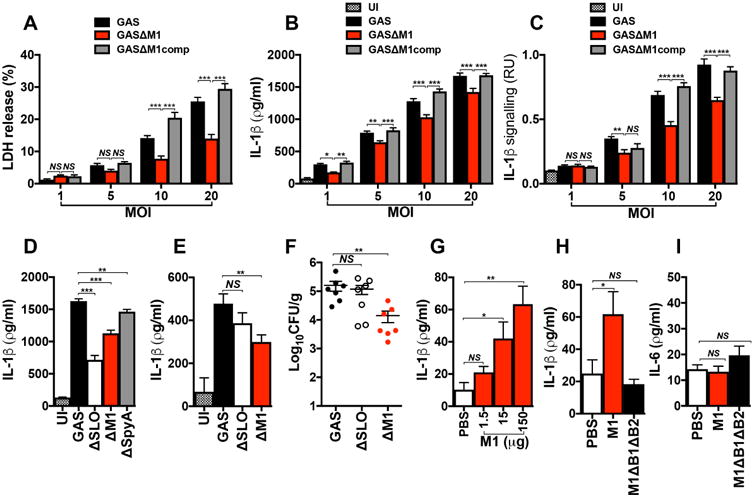
THP1-Mϕ were coincubated with WT (GAS), isogenic Δemm1 mutant (GASΔM1) or M1-complemented (GASΔM1comp) strains at different multiplicities of infection (MOI). After 2 h of infection, macrophage supernatants were collected and analyzed for detection of LDH (a), total IL-1β (b) and mature IL-1β (c). d, Comparison of IL-1β secretion from THP1-Mϕ coincubated for 2 h with WT GAS or isogenic Δemm1 (GASΔM1), Δslo (GASΔSLO) and ΔspyA (GASΔSpyA) mutant strains. Uninfected macrophages were used as negative control (UI). IL-1β production was analyzed by ELISA and the presence of mature IL-1β by HEK-Blue™ IL-1R reporter cells. IL-1β detection (e) and bacteria recovered (CFU) (f) from peritoneal lavage fluid of wild type C57BL/6 mice 6 h after intraperinoneal infection with 1 × 108 CFU of WT or isogenic ΔM1 or ΔSLO mutant strains. Control group mice were injected with PBS and used as negative control (UI). Detection of IL-1β (g and h) and IL-6 (i) from peritoneal lavage fluid of WT C57BL/6 mice, 4 h after injections with different concentrations of M1 protein (g), and with 150 μg of purified M1 or ΔB1ΔB2 proteins (h and i). Control group animals were injected with PBS. Cytokine quantification was performed by ELISA. Data in panels a-i are plotted as the mean ± SEM. Panels a, b, c, and d represent three independent experiments performed in triplicate. In panels e and f N=7, in panel g N=6 and in panels h and i N=10 and represent the combination of 2 independent experiments. Results in panels a-c were analyzed by Two-way ANOVA multiple comparisons and panels d-i were analyzed by Student's t-test. NS = not significant (P>0.05), * P<0.05, ** P<0.01 and ***P<0.001).
GAS expresses at least two other protein virulence factors that activate the NLRP3 inflammasome: SLO and SpyA. We compared release of IL-1β from macrophages infected with GAS M1T1 WT or isogenic ΔM1, ΔSLO, or ΔSpyA mutants (Fig. 5d). Macrophages infected with the three isogenic mutant strains had significantly lower IL-1β production than those infected with the WT strain, however IL-1β release was most attenuated in macrophages infected with ΔSLO or ΔM1 strains. Natively expressed M1 protein thus plays an important role in stimulating IL-1β processing and pyroptosis in macrophages.
M1 stimulates release of IL-1β in vivo
To corroborate our in vitro findings, we compared induction of IL-1β by GAS WT and isogenic ΔM1 and ΔSLO mutant strains during intraperitoneal infection of mice. While production of IL-1β from ΔSLO-infected mice did not differ from WT-infected animals, the ΔM1 strain induced significantly less IL-1β (Fig. 5e). Of note, there was a significant decrease in bacterial load recovered from the peritoneal fluid of mice infected with GAS ΔM1 mutant strain compared to GAS WT- or ΔSLO-infected mice (Fig. 5f). These results support a crucial role of M1 protein for full GAS virulence and its contribution to stimulation of IL-1β signaling in vivo.
We next injected increasing concentrations of M1 into the peritoneum of WT C57BL/6 mice, and quantified IL-1β levels in the peritoneal fluid 4 h later. M1 indeed triggered IL-1β production in vivo in a dose-dependent manner (Fig. 5g). Consistent with our findings using cultured macrophages, removal of the B1 and B2 repeat domains (ΔB1ΔB2) from M1 protein eliminated its ability to stimulate IL-1β in vivo following peritoneal injection (Fig. 5h). Moreover, M28 protein, which did not induce pyroptosis in vitro (Figs. 1b, 1c), did not stimulate production of IL-1β in mice when injected as a recombinant protein (Supplementary Fig. 9). Injection of intact M1 or ΔB1ΔB2 mutant proteins did not stimulate release of inflammasome-independent cytokine IL-6 (Fig. 5i), in contrast to the high levels of IL-6 produced upon injection of TLR agonist LPS (Supplementary Fig. 10). Together these results indicate a specific role for M1 protein in vivo as a second signal for inflammasome activation.
Discussion
IL-1β signaling and inflammasome activation are important in the complex suite of innate immune responses elicited during GAS infection, wherein robust tissue inflammation is a hallmark of tissue invasion 23. Both canonical NLRP3 inflammasome and non-canonical (bacterial protease-induced) IL-1β signaling occur in macrophages responding to a GAS encounter35. Our studies indicate that the classical GAS virulence factor M1 protein makes an independent and important contribution to GAS NLRP3 inflammasome activation, IL-1β processing, and macrophage pyroptosis, thereby joining pore-forming toxin SLO26 and ADP-ribosylating toxin SpyA27 as known inflammasome activators produced by the pathogen.
Since IL-1R deficiency in mice36 or treatment with IL-1R antagonist Anakinra in mice or humans35 increase susceptibility to severe GAS infection, NLRP3 detection of M1 protein and other GAS factors appears on balance to serve a critical role in host defense against the organism. However, during advanced stages of systemic infection with invasive M1 GAS, IL-1β release and pyroptosis elicited by M1 may contribute to hyperinflammation and associated pathologies10,13,15. During invasive GAS M1T1 disease, pathoadaptive covR/S mutations that shut off SpeB expression may be selected for and are associated with hypervirulence18,19. Clinical epidemiological studies found that SpeB expression and activity were significantly higher in GAS serotype M1T1 isolates from non-severe invasive infections than isolates from severe cases17. Silencing of SpeB activity may allow for increased presence of soluble, released M1 protein during severe GAS infections. Indeed, the presence of soluble, released M1 has been documented in biopsies of patients with NF and SSTS13.
Our in vitro experiments examining macrophages infected with live WT or M1-deficient GAS bacterial strains show a significant contribution of natively expressed M1 protein in the induction of IL-1β and pyroptosis. In our studies, the presence of M1 was not a strong factor in establishing initial interaction with macrophages compared to its known role in adhesion or invasion of epithelial cell types, emphasizing the dominance of M1 action on the inflammasome as compared to other known M1 properties in our assays. Production of IL-1β during GAS infection in vivo was greater when M1 was present, although the bacterial load was significantly lower in the absence of the protein. However, purified M1 protein alone injected in vivo was sufficient to induce the production of IL-1β, suggesting that soluble M1 induces IL-1β signaling independently of its other well-known virulence properties.
In contrast to the two signals required in vitro for M1-induction of IL-1β, M1 alone was sufficient to stimulate IL-1β response in vivo. We hypothesize that in the latter case, macrophages are intrinsically stimulated by cellular processes such as metabolic activation elicited by the thioglycollate pre-treatment in our in vivo experiments, or alternatively that M1 may activate host-derived pro-inflammatory factors facilitating signal 1 priming. The later mechanism is consistent with recent studies, which have shown that keratinocytes recognize soluble M1 as a PAMP to release interleukin-8, growth-related oncogene-alpha, migration inhibitory factor, and other inflammatory response alarms37. Another study reported that M1 protein can synergize with heparin-binding protein to interact with TLR2 on monocytes and promote release of IL-1β and IL-638, which differs somewhat from our findings showing no effect of M1 protein on IL-6 production in vitro or in vivo. This difference likely reflects the source of the protein in each study: purified recombinant M1 protein in our present work vs. M1 protein purified from GAS supernatants in the aforementioned study. Our studies suggest that M1 perse is not a direct TLR agonist as proposed earlier; however, during GAS infection, release of M1 protein complexed with PGN or LTA to serve as PAMPs may contribute to the activation of TLRs and consequent accelerated cytokine production.
CME, which constitutes the major and best-characterized endocytic pathway, carries out the continuous uptake of essential nutrients, antigens, growth factors, and pathogens. Our study demonstrates that CME contributes to M1 uptake, and that a specific clathrin inhibitor blocks M1 uptake to reduce, NLRP3 activation, IL-1β production and pyroptosis. Human immunodeficiency virus and hepatitis C virus were recently shown to activate the inflammasome by CME-dependent mechanisms39. Furthermore, muramyl dipeptide, a bacterially derived agonist of the NOD2 receptor that induces caspase-1 activation through the NLRP3 inflammasome, is internalized by macrophages through CME40. The mechanism by which M1 is targeted into clathrin-coated pits is at present unclear, but warrants further study as it may reveal mechanistic commonalities for inflammasome activation. Because the first step of internalization through CME usually involves binding of a ligand to a specific membrane receptor, our studies suggest that M1 is recognized by a host factor expressed by macrophages.
The M1 B-repeat region serves an essential function in GAS pathogenesis, as it is responsible for M1 binding to host fibrinogen (Fg) and formation of supramolecular M1-Fg complexes that induce neutrophil activation and transition to a proinflammatory state14,41. Based on these previous studies and taking into account our new findings, a potential scenario during GAS invasive infection is that released, soluble M1 protein first activates infiltrating neutrophils through the M1-Fg supramolecular complex, resulting in the recruitment and influx of macrophages to the site. Released, soluble M1 would then act in a Fg-independent manner on these macrophages, causing pyroptotic cell death. Our studies provide evidence that the M1 B-repeat region is a multifunctional domain that plays an essential role in M1-induced inflammation.
Comparison of the inflammasome-activating effect of M1 to other M proteins suggests that the activation of NLRP3 and the consequent cell death is not unique to the M1 serotype, but is not a general property of all M protein serotypes. Due to the pronounced hypervariability of the N-terminal domain (B-repeats included) across M proteins, a single common pattern for the M proteins that activates IL-1β is unlikely. However, it has recently been recognized by x-ray crystallography that the three-dimensional structure of different M proteins in complex with host factors may reveal conserved sequence patterns hidden within hypervariability42.
K+ efflux is a common mechanism required for NLRP3 activation33, and we confirmed that M1-induced inflammasome activation also requires K+ efflux. Further studies are required to understand the exact molecular mechanism that triggers K+ efflux and downstream mechanisms of NLRP3 activation upon M1 uptake. Components or byproducts from the depolymerization of clathrin-coated pits, the subsequent transition from early to late endosomes, or the final fusion with recycling lysosomes are potentially important for the molecular mechanism of M1-induced NLRP3 activation. Indeed, the NLRP3 inflammasome may detect internal membrane perturbations and thereby respond to many different stimuli, which all have in common the ability to induce lysosomal destabilization43.
In summary, we propose a model for GAS M1-dependent inflammasome activation and pyroptosis in macrophages (Fig. 6). Upon GAS tissue invasion, bacteria-derived PAMPs (e.g. PGN or LTA) or host-derived DAMPs generated upon tissue injury are recognized by the macrophage through different mechanisms, including TLRs; these first signals activate the transcription of genes encoding NLRP3 inflammasome components and pro-IL-1β. M1 may be released from the GAS surface by cell wall turnover, host neutrophil proteases, or both13,16,17, and is taken up by macrophages via clathrin-mediated endocytosis (CME) to serves as a second signal required for NLRP3 activation. Internalization of M1 activates NLRP3 through an unknown subcellular molecular intermediate or event, e.g. lysosomal destabilization, triggering K+ efflux and promoting inflammasome assembly by recruitment of NLRP3, its well-characterized apoptosis-associated speck-like accessory protein (ASC), and caspase-1, resulting in the maturation and secretion of the pro-inflammatory cytokine IL-1β, DNA damage, membrane disruption, and pyroptosis in macrophages. In sum, this work reveals an unexpected function in GAS molecular pathogenesis for the M protein, the most abundant protein on the GAS surface with multifaceted roles in virulence and as a target of host immunity.
Figure 6. Model of M1 protein-mediated NLRP3 inflammasome activation and pyroptosis.
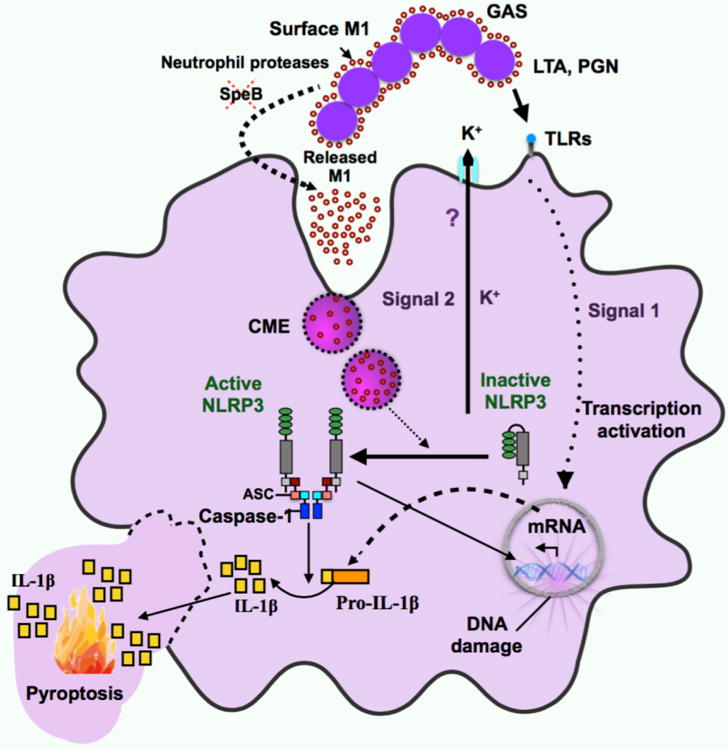
M1-mediated activation of the NLRP3 inflammasome requires two signals. The first signal is essential for stimulating increased transcriptional levels of inflammasome components, and arises from the binding of pathogen-associated molecular patterns (PAMP) or damage-associated molecular patterns (DAMP) molecules, such as peptidoglycan (PGN) or lipoteichoic acid (LTA) on the GAS cell wall, to specific receptors, such as toll-like receptors (TLRs). Increased availability of released M1 (second signal) is likely to occur after down-regulation of the bacterial protease SpeB, by the activity of neutrophil proteases, or both. M1 is taken up by macrophages through clathrin-mediated endocytosis (CME), after interaction with a potential surface receptor. Once internalized, M1 activates the assembly and activation of the NLRP3 inflammasome and triggers potassium efflux (K+ efflux) through an unknown mechanism (?). The assembly of the inflammasome leads to the processing of pro-IL-1β to mature IL-1β, and results in DNA damage and pyroptotic cell death in macrophages.
Methods
Reagents and inhibitors
Lipopolysaccharide from E. coli Serotype EH100(Ra) was purchased from Enzo. Lipoteichoic acid from Bacillus subtilis, peptidoglycan from Staphylococcus aureus, R848, and Pam3CSK4 were purchased from Invivogen. Ac-YVAD-CMK (inhibitor of caspase-1) and CRID 3 (inhibitor of NLRP3) were purchased from Enzo and Tocris, respectively. Recombinant mCherry was purchased from BioVision. Pitstop-2 was purchased from Abcam. Wortmannin, methyl-β-cyclodextrin, ATP, nigericin and human fibrinogen were purchased from Sigma.
Cell isolation and culture
Bone marrow-derived macrophages were generated as previously described44 from femurs and tibias of 8 to 12-week-old male or female WT C57BL/6 (Jackson Laboratory), casp-1/11-/- (provided by R. Flavell), or nlrp3-/-(provided by J. Bertin) mice. Human THP-1 monocytes cell line was provided and authenticated by the ATCC and stored at the UCSD cell culture facility, which routinely does surveillance mycoplasma testing of its facility. THP-1 cells were cultured in RPMI medium supplemented with 10% fetal bovine serum (FBS), 0.05 mM 2-mercaptoethanol, 0.2% D-glucose, 10 mM HEPES, and 1 mM sodium pyruvate. Human THP-1 Mϕ were differentiated for 24 h with 25 nM phorbol myristate acetate (PMA) (Fisher). Primary human neutrophils, peripheral blood mononuclear cells, and red blood cells were isolated from small volumes of whole blood from healthy volunteers after informed consent following a protocol for simple phlebotomy approved by the UCSD IRB/Human Research Protection Program; cell isolation was achieved using 1-step® Polymorph according to manufacturer's instructions (Accurate Chemical and Scientific Corporation). A549 human epithelial cells were cultured in 10% FBS-RPMI medium. All experiments with THP-1 Mϕ and BMDMs were performed in 2% FBS-RPMI medium, except for the experiment in which potassium efflux was inhibited. In this experiment, cells were stimulated in K+-rich medium (serum free buffer with 10 mM HEPES, 150 mM KCl, 5 mM NaH2PO4, 150 mM NaCl, 1 mM MgCl2, 1 mM CaCl2, 1% BSA, pH 7.4) or in Na+-medium (serum free buffer with 10 mM HEPES, 150 mM NaCl, 5 mM KH2PO4, 1 mM MgCl2, 1 mM CaCl2, 1% BSA, pH 7.4).
Animal experiments
The UCSD Institutional Animal Care and Use Committee approved all animal use and procedures. In compliance with ethical guidelines to minimize the number of animals used, we used a minimum of five mice for each data point (except where indicated in the figure legends) to ensure statistical power. All mice were randomly distributed into the different groups as indicated in the corresponding figure legend. No blinding was performed in our animal work. For GAS infection, 8 to 10-week-old C57Bl/6 female mice were injected intraperitoneally (i.p.) with 100 μL PBS as control group or with 100 μL PBS containing 1 × 108 CFU of various GAS strains. For soluble M protein injection, 8 to 10-week-old C57Bl/6 male or female mice were i.p. injected with 2 mL of 3% thioglycolate (Difco) to elicit peritoneal macrophages. At 72 h post-treatment, mice were injected with different amounts of M protein in 100 μL PBS. At 4h (M protein treatment) or 6 h (bacterial infection), mice were euthanized with isoflurane followed by cervical dislocation. Post-mortem, the peritoneal fluid was collected by lavage with 3 mL sterile PBS, followed by massage. Cytokines and bacterial counts in the peritoneal fluid collected were quantified by ELISA and CFU, respectively.
Bacterial strains and culture conditions
GAS M1T1 5448 was originally isolated from a patient with necrotizing fasciitis and toxic shock44. The isogenic in-frame allelic exchange knock-out mutant 5448Δemm1 (ΔM1) and its complemented strain have been described previously45 as well as the knock-out mutants 5448Δslo (ΔSLO)46 and 5448 ΔspyA (ΔSpyA)27. All GAS strains were routinely propagated at 37°C on Todd-Hewitt agar (Difco) or in static liquid cultures of Todd-Hewitt broth. Where appropriate, strains were grown in medium supplemented with 5 μg/mL erythromycin or 2 μg/mL chloramphenicol.
Infection of macrophages with GAS
THP-1 Mϕ were seeded at 5 × 105 cells in 600 μl of RPMI-2% FBS in a 12-well plate. Macrophages were infected with overnight bacterial cultures diluted in 100 μl of RPMI-2% FBS at an MOI of 1, 5, 10 or 20. Plates were centrifuged at 600 × g for 5 min to facilitate bacterial contact with macrophages. Cells were incubated at 37°C for 2 h. Supernatants were collected by centrifugation (600 × g for 5 min at room temperature) and analyzed for LDH release, total IL-1β production, and IL-1β signaling. For GAS invasion experiments, stationary phase bacteria were labeled with fluorescein isothiocyanate (FITC) through 30 min incubation with 0.2 mg/ml FITC, covered on ice, and washed with PBS. PMA-differentiated THP-1 Mϕ were coincubated with FITC-labeled bacteria at MOI 10 in RPMI containing 2% FBS for 2 h. Macrophages were dissociated with trypsin-EDTA, washed in PBS supplemented with 0.5% FBS, and immediately run on FACSCalibur (BD Biosciences). Ten thousand events were collected, and samples were gated for live macrophages based on unlabeled bacteria-infected controls. Flow cytometry data were analyzed using FlowJo v. 9.4.10 (Tree Star, Inc.)
LDH assay and ELISA
Cell culture supernatants were harvested by centrifugation at 500 × g for 5 min at 20°C and analyzed with a Cytotox 96® Non-Radioactive Cytotoxicity Assay (Promega). Procedures and calculations of LDH release percentage (% LDH) were according to manufacturer's directions. Cell culture supernatants were assayed for human or murine IL-β or IL-6 with ELISA kits following the manufacturer's instructions (R&D Systems).
IL-1p signalling assay
Stably transfected HEK-Blue IL-1p reporter cells (Invivogen) (50,000 cells per well in 96-well plates), were stimulated for 16 h at 37°C in 5% CO2 with 50 μL of supernatants from macrophages previously treated or infected under the conditions that each experiment indicates. After 18 h, supernatants from the HEK-Blue cells were analyzed for secreted alkaline phosphatase activity by the addition of 50 μL of HEK-Blue IL-1p reporter cell supernatants onto 150 μL of Quanti-Blue™ reagent (Invivogen) and monitoring the optical density at 620 nm via EnSpire plate reader (PerkinElmer).
Confocal and Fluorescence Microscopy
Bone marrow-derived macrophages (BMDMs) were seeded on coverslips in 24-well plates. Cells were primed with LPS (10 ng ml-1) for 4 h. As appropriate, cells were pretreated with Pitstop-2 (25 μM) or with the vehicle control (DMSO) for 10 min and then stimulated with M1-mCherry protein (2 μM) for 10, 30, or 60 min. After these treatments, cells were washed × 2 with PBS and fixed with 4% paraformaldehyde for 20 min. After fixation, cells were washed × 3 with PBS, and cover slips were mounted with ProLong® Gold antifade reagent containing DAPI (Life Technologies). To quantify the M1-mCherry signal, in-focus planes were analyzed with ImageJ (v2.0.0., NIH) as previously described47. An outline of the cell was drawn and measurements of cell area, mean fluorescence, and integrated density were recorded. For local background subtraction, the mean fluorescence of four regions surrounding each cell was also recorded. The total corrected cellular fluorescence was calculated as TCCF = integrated density – (area of selected cell × mean fluorescence of background readings) and reported as relative fluorescence. For colocalization analysis, BMDMs were incubated with 2 μM of purified M1 protein resuspended in RPMI media (no phenol red, Gibco) for 10 min. After treatment, cells were washed × 2 with PBS to remove unbound protein, and then fixed with 4% paraformaldehyde for 20 min. After fixation, cells were washed × 3 with PBS and then permeabilized with PBS+0.2% Triton X-100 for 30 min. Coverslips were then blocked for 1 hr in 3% BSA/1X PBS+0.02% sodium azide. Staining was performed by incubation with anti-EEA1 antibody (BD biosciences), resuspended in 3% BSA/1X PBS+0.02% sodium azide for 1 hr. Coverslips were then washed × 3 with PBS and incubated for 1 h with 0.4 μg/ml AlexaFluor-488®-conjugated anti-mouse secondary antibody (Life Technologies). Coverslips were again washed × 3 with PBS and stained with 1 μg/ml of Hoechst dye (Life Technologies). After 3 additional PBS washes, coverslips were mounted onto slides using ProLong® Gold antifade reagent (Life Technologies). Colocalization analysis was performed with ImageJ (v2.0.0., NIH) using the Coloc2 plugin. Cells were visualized using an Olympus FV1000 confocal microscope or a Zeiss Axio Observer.D1 fluorescence microscope.
Propidium iodide uptake assay
THP-1 Mϕ were incubated in the presence or absence of purified M1 protein (2 μM) for 2 h in RPMI containing 2% FBS. Cells were washed × 3 with PBS, incubated in 200 μL propidium iodide (PI) staining solution (1% BSA and 3 UM PI, Invitrogen), and were visualized using a Zeiss Axio Observer.D1 microscope with an AxioCamMR3 camera. To quantify the number of cells that had taken up PI, THP-1 Mϕ were collected by incubating the cells for 5 min at 37 °C with cold PBS containing 5 mM EDTA, were washed × 3 with PBS, incubated in 500 μL PI staining solution, and analyzed by flow cytometry. Mean fluorescence intensity was calculated from a total of 10,000 cells; macrophages treated with PBS alone were used as a negative control. Flow cytometry data were analyzed with FlowJo v. 9.4.10 (Tree Star, Inc.).
TUNEL assay
THP-1 Mϕ were incubated for 4 h in the presence or absence of purified M1 protein (0.2 μM or 2 μM). Cells were detached from the well by incubation for 5 min at 37 °C with cold PBS containing 5 mM EDTA, washed once with PBS, and assayed for the percentage of cells with DNA fragmented by the TUNEL technique (APO-BrdU TUNEL assay kit; Invitrogen) according to the manufacturer's instructions. DNA fragmentation-fluorescence intensity was measured by flow cytometry for a total of 10,000 cells. Macrophages treated with PBS were used as a negative control. Flow cytometry data was analyzed with FlowJo v. 9.4.10 (Tree Star, Inc.).
Protein expression and purification
Procedures for the construction and expression of M1 protein, truncated variants of M1 protein, and other M protein types have been previously described14,34, except for M1-mCherry and M1ΔB1ΔB2-mCherry proteins. For M1-mCherry, the coding sequence of intact mCherry protein (residues 1-236) was cloned into the pET28-M1 vector25. The PCR product was amplified with primers 5′XhoIm-Cherry (5′-ccgctcgagggaggaagcatggtgagcatggtgagcaagggcgagaggg-3′) + 3′XhoIm-Cherry (5′-ccgctcgagcttgtacagctcgtccatgccgcc-3′), with mCherry fused to the C-terminus of M1. For M1AB1AB2-mCherry, PCR product was amplified with primers 5′M1dB1B2-cherry(EcoRI) (5′-ccgccgaattcttagaaacgattactagagaacaagagattaatcg-3′) and 3′ M1dB1B2-cherry(EcoRI) (5′-ccgccgaattctctttgtctatcccaacttgttgattcctttgcta-3′) from pET28-M1mCherry vector. Protein constructs were expressed in E. coli BL21 (DE3) cells. Bacteria were grown in LB containing 34mgml1 kanamycin at 37 °C until mid-logarithmic phase and then induced at room temperature with 1 mM isopropyl P-D-1-thiogalactopyranoside, and grown for a further 18 h. Bacteria were harvested by centrifugation and re-suspended in lysis buffer (300 mM NaCl, 100 mM Tris-HCl, pH 8, 10 mM imidazole) with protease inhibitors (Complete Tablet, Roche). Bacteria were lysed using an EmulsiFlex-C5 (Avestin; 20,000 psi with three passes). His-tagged M1 constructs were then purified as previously reported34 by Ni2+-NTA affinity and Q-Sepharose anion exchange chromatography. Purified proteins were dialyzed against 2 l of PBS buffer at 4 °C overnight (3500 MWCO tubing, Spectrum Laboratories). Proteins were aliquoted and flash frozen in liquid N2 for storage at -80 °C.
Statistical analysis
Data shown were collected from three independent experiments in triplicates, unless otherwise indicated. Data were combined and represented as mean ± SEM. Results were either analyzed by umpaired Student's t-test or by Two-way ANOVA using GraphPad Prism version 7. P values < 0.05 were considered statistically significant. Flow cytometry histograms and microscopy images are representative of at least 2 independent experiments.
Data availability
The data that support the findings of this study are available from the corresponding author upon request.
Supplementary Material
Acknowledgments
We would like to thank the members of the P.G. and V.N. labs for their valuable input. This work was supported by NIH grants AI096837 (P.G. and V.N), AI077780 (V.N.), and AI52430 (H.M.H.) A.R. is a San Diego IRACDA Program fellow supported by NIGMS/NIH award K12GM068524, N.J.G. is supported by the UCSD Genetics Training Program (T32 GM008666) and the UCSD Global Health Institute, and C.N.L. is a recipient of an A.P. Giannini Foundation Postdoctoral Research Fellowship.
Footnotes
Author contributions: J.A.V., P.G., and V.N. formulated the original hypothesis, designed the study, and analyzed results. J.A.V., A.R., N.J.G., C.N.L., N.G., and S.R.A. performed and optimized experiments. H.M.H. provided novel reagents. J.A.V., P.G., and V.N. wrote the manuscript, and all authors reviewed the manuscript, data, and conclusions before submission.
Competing interests: The authors declare no competing financial interests.
References
- 1.Walker MJ, et al. Disease manifestations and pathogenic mechanisms of group A Streptococcus. Clin Microbiol Rev. 2014;27:264–301. doi: 10.1128/CMR.00101-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carapetis JR, Steer AC, Mulholland EK, Weber M. The global burden of group A streptococcal diseases. Lancet Infect Dis. 2005;5:685–694. doi: 10.1016/S1473-3099(05)70267-X. [DOI] [PubMed] [Google Scholar]
- 3.Severin A, et al. Proteomic analysis and identification of Streptococcus pyogenes surface-associated proteins. J Bacteriol. 2007;189:1514–1522. doi: 10.1128/JB.01132-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Phillips GN, Jr, Flicker PF, Cohen C, Manjula BN, Fischetti VA. Streptococcal M protein: alpha-helical coiled-coil structure and arrangement on the cell surface. Proc Natl Acad Sci U S A. 1981;78:4689–4693. doi: 10.1073/pnas.78.8.4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ghosh P. The nonideal coiled coil of M protein and its multifarious functions in pathogenesis. Adv Exp Med Biol. 2011;715:197–211. doi: 10.1007/978-94-007-0940-9_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanderson-Smith M, et al. A systematic and functional classification of Streptococcus pyogenes that serves as a new tool for molecular typing and vaccine development. J Infect Dis. 2014;210:1325–1338. doi: 10.1093/infdis/jiu260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steer AC, Law I, Matatolu L, Beall BW, Carapetis JR. Global emm type distribution of group A streptococci: systematic review and implications for vaccine development. Lancet Infect Dis. 2009;9:611–616. doi: 10.1016/S1473-3099(09)70178-1. [DOI] [PubMed] [Google Scholar]
- 8.Zhu L, et al. A molecular trigger for intercontinental epidemics of group A Streptococcus. J Clin Invest. 2015;125:3545–3559. doi: 10.1172/JCI82478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okada N, Liszewski MK, Atkinson JP, Caparon M. Membrane cofactor protein (CD46) is a keratinocyte receptor for the M protein of the group A streptococcus. Proc Natl Acad Sci U S A. 1995;92:2489–2493. doi: 10.1073/pnas.92.7.2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oehmcke S, Shannon O, Morgelin M, Herwald H. Streptococcal M proteins and their role as virulence determinants. Clin Chim Acta. 2010;411:1172–1180. doi: 10.1016/j.cca.2010.04.032. [DOI] [PubMed] [Google Scholar]
- 11.LaRock CN, et al. Group A streptococcal M1 protein sequesters cathelicidin to evade innate immune killing. Cell Host Microbe. 2015;18:471–477. doi: 10.1016/j.chom.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dohrmann S, et al. Group A streptococcal M1 protein provides resistance against the antimicrobial activity of histones. Sci Rep. 2017;7:43039. doi: 10.1038/srep43039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herwald H, et al. M protein, a classical bacterial virulence determinant, forms complexes with fibrinogen that induce vascular leakage. Cell. 2004;116:367–379. doi: 10.1016/s0092-8674(04)00057-1. [DOI] [PubMed] [Google Scholar]
- 14.Macheboeuf P, et al. Streptococcal M1 protein constructs a pathological host fibrinogen network. Nature. 2011;472:64–68. doi: 10.1038/nature09967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soehnlein O, et al. Neutrophil primary granule proteins HBP and HNP1-3 boost bacterial phagocytosis by human and murine macrophages. J Clin Invest. 2008;118:3491–3502. doi: 10.1172/JCI35740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akesson P, Schmidt KH, Cooney J, Bjorck L. M1 protein and protein H: IgGFc- and albumin-binding streptococcal surface proteins encoded by adjacent genes. Biochem J. 1994;300(Pt 3):877–886. doi: 10.1042/bj3000877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kansal RG, McGeer A, Low DE, Norrby-Teglund A, Kotb M. Inverse relation between disease severity and expression of the streptococcal cysteine protease, SpeB, among clonal M1T1 isolates recovered from invasive group A streptococcal infection cases. Infect Immun. 2000;68:6362–6369. doi: 10.1128/iai.68.11.6362-6369.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sumby P, Whitney AR, Graviss EA, DeLeo FR, Musser JM. Genome-wide analysis of group A streptococci reveals a mutation that modulates global phenotype and disease specificity. PLoS Pathog. 2006;2:e5. doi: 10.1371/journal.ppat.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cole JN, Barnett TC, Nizet V, Walker MJ. Molecular insight into invasive group A streptococcal disease. Nat Rev Microbiol. 2011;9:724–736. doi: 10.1038/nrmicro2648. [DOI] [PubMed] [Google Scholar]
- 20.Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell. 2002;10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 21.Miao EA, Rajan JV, Aderem A. Caspase-1-induced pyroptotic cell death. Immunol Rev. 2011;243:206–214. doi: 10.1111/j.1600-065X.2011.01044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Franchi L, Munoz-Planillo R, Nunez G. Sensing and reacting to microbes through the inflammasomes. Nat Immunol. 2012;13:325–332. doi: 10.1038/ni.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.LaRock CN, Nizet V. Inflammasome/IL-1β responses to streptococcal pathogens. Front Immunol. 2015;6:518. doi: 10.3389/fimmu.2015.00518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Broderick L, De Nardo D, Franklin BS, Hoffman HM, Latz E. The inflammasomes and autoinflammatory syndromes. Annu Rev Pathol. 2015;10:395–424. doi: 10.1146/annurev-pathol-012414-040431. [DOI] [PubMed] [Google Scholar]
- 25.Wen H, Ting JP, O'Neill LA. A role for the NLRP3 inflammasome in metabolic diseases--did Warburg miss inflammation? Nat Immunol. 2012;13:352–357. doi: 10.1038/ni.2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harder J, et al. Activation of the Nlrp3 inflammasome by Streptococcus pyogenes requires streptolysin O and NF-κB activation but proceeds independently of TLR signaling and P2X7 receptor. J Immunol. 2009;183:5823–5829. doi: 10.4049/jimmunol.0900444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin AE, et al. A group A Streptococcus ADP-ribosyltransferase toxin stimulates a protective interleukin 1β-dependent macrophage immune response. MBio. 2015;6:e00133. doi: 10.1128/mBio.00133-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bauernfeind FG, et al. Cutting edge: NF-κB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. J Immunol. 2009;183:787–791. doi: 10.4049/jimmunol.0901363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo H, Callaway JB, Ting JP. Inflammasomes: mechanism of action, role in disease, and therapeutics. Nat Med. 2015;21:677–687. doi: 10.1038/nm.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaidt MM, et al. Human monocytes engage an alternative inflammasome pathway. Immunity. 2016;44:833–846. doi: 10.1016/j.immuni.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 31.Mariathasan S, et al. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature. 2006;440:228–232. doi: 10.1038/nature04515. [DOI] [PubMed] [Google Scholar]
- 32.Coll RC, et al. A small-molecule inhibitor of the NLRP3 inflammasome for the treatment of inflammatory diseases. Nat Med. 2015;21:248–255. doi: 10.1038/nm.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Munoz-Planillo R, et al. K(+) efflux is the common trigger of NLRP3 inflammasome activation by bacterial toxins and particulate matter. Immunity. 2013;38:1142–1153. doi: 10.1016/j.immuni.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McNamara C, et al. Coiled-coil irregularities and instabilities in group A Streptococcus M1 are required for virulence. Science. 2008;319:1405–1408. doi: 10.1126/science.1154470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.LaRock CN, et al. IL-1β is an innate immune sensor of microbial proteolysis. Science Immunol. 2016;1:eaah3539. doi: 10.1126/sciimmunol.aah3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hsu LC, et al. IL-1β-driven neutrophilia preserves antibacterial defense in the absence of the kinase IKKβ. Nat Immunol. 2011;12:144–150. doi: 10.1038/ni.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Persson ST, Wilk L, Morgelin M, Herwald H. Vigilant keratinocytes trigger pathogen-associated molecular pattern signaling in response to streptococcal M1 protein. Infect Immun. 2015;83:4673–4681. doi: 10.1128/IAI.00887-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pahlman LI, et al. Streptococcal M protein: a multipotent and powerful inducer of inflammation. J Immunol. 2006;177:1221–1228. doi: 10.4049/jimmunol.177.2.1221. [DOI] [PubMed] [Google Scholar]
- 39.Chattergoon MA, et al. HIV and HCV activate the inflammasome in monocytes and macrophages via endosomal Toll-like receptors without induction of type 1 interferon. PLoS Pathog. 2014;10:e1004082. doi: 10.1371/journal.ppat.1004082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marina-Garcia N, et al. Clathrin- and dynamin-dependent endocytic pathway regulates muramyl dipeptide internalization and NOD2 activation. J Immunol. 2009;182:4321–4327. doi: 10.4049/jimmunol.0802197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stewart C, et al. Coiled-coil destabilizing residues in the group A Streptococcus M1 protein are required for functional interaction. Proc Natl Acad Sci U S A. 2016;113:9515–9520. doi: 10.1073/pnas.1606160113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buffalo CZ, et al. Conserved patterns hidden within group A Streptococcus M protein hypervariability recognize human C4b-binding protein. Nat Microbiol. 2016;1:16155. doi: 10.1038/nmicrobiol.2016.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hornung V, et al. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat Immunol. 2008;9:847–856. doi: 10.1038/ni.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chatellier S, et al. Genetic relatedness and superantigen expression in group A streptococcus serotype M1 isolates from patients with severe and nonsevere invasive diseases. Infect Immun. 2000;68:3523–3534. doi: 10.1128/iai.68.6.3523-3534.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lauth X, et al. M1 protein allows Group A streptococcal survival in phagocyte extracellular traps through cathelicidin inhibition. J Innate Immun. 2009;1:202–214. doi: 10.1159/000203645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Timmer AM, et al. Streptolysin O promotes group A Streptococcus immune evasion by accelerated macrophage apoptosis. J Biol Chem. 2009;284:862–871. doi: 10.1074/jbc.M804632200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McCloy RA, et al. Partial inhibition of Cdk1 in G 2 phase overrides the SAC and decouples mitotic events. Cell Cycle. 2014;13:1400–1412. doi: 10.4161/cc.28401. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon request.


