Abstract
Background and aims
Angiopoietin-like 3 (ANGPTL3) has emerged as a key regulator of lipoprotein metabolism in humans. Homozygous loss of ANGPTL3 function causes familial combined hypolipidemia characterized by low plasma levels of triglycerides (TG), high-density lipoprotein cholesterol (HDL-C), and low-density lipoprotein cholesterol (LDL-C). While known effects of ANGPTL3 in inhibiting lipoprotein lipase and endothelial lipase contribute to the low TG and HDL-C, respectively, the basis of the low LDL-C remains unclear. Our aim was to explore the role of ANGPTL3 in modulating plasma LDL-C.
Methods
We performed RNAi-mediated gene silencing of ANGPTL3 in five mouse models and in human hepatoma cells. We validated results by deleting ANGPTL3 gene using the CRISPR/Cas9 genome editing system.
Results
RNAi-mediated Angptl3 silencing in mouse livers resulted in very low TG, HDL-C and LDL-C, a pattern similar to the human phenotype. The effect was observed in wild-type and obese mice, while in hCETP/apolipoprotein (Apo) B-100 double transgenic mice, the silencing decreased LDL-C and TG, but not HDL-C. In a humanized mouse model (Apobec1−/− carrying human ApoB-100 transgene) deficient in LDL receptor (LDLR), Angptl3 silencing had minimum effect on LDL-C, suggesting the effect being linked to LDLR. This observation is supported by an additive effect on LDL-C between ANGPTL3 and PCSK9 siRNAs. ANGPTL3 gene deletion induced cellular long-chain TG and ApoB-100 accumulation with elevated LDLR and LDLR-related protein (LRP) 1 expression. Consistent with this, ANGPTL3 deficiency by gene deletion or silencing reduced nascent ApoB-100 secretion and increased LDL/VLDL uptake.
Conclusions
Reduced secretion and increased uptake of ApoB-containing lipoproteins may contribute to the low LDL-C observed in mice and humans with genetic ANGPTL3 deficiency.
Keywords: Angiopoietin-like protein 3, ANGPTL3, lipoprotein, LDL receptor, LDLR, low-density lipoprotein, LDL, high-density lipoprotein, HDL, triglyceride, cholesterol
Introduction
Recent genetic studies have highlighted that the angiopoietin-like 3 protein (encoded by the gene ANGPTL3) influences human lipoprotein metabolism [1–3]. Loss-of-function (LoF) mutations in ANGPTL3 are associated with lower plasma TG levels [4], probably due to the role of ANGPTL3 in inhibiting lipoprotein lipase (LPL) [5, 6] and lower HDL-C levels, probably due to the role of ANGPTL3 in inhibiting endothelial lipase (EL) [7]. Using exome sequencing, we identified individuals in whom LoF variants in both copies of ANGPTL3 had extremely low plasma levels of TG, HDL-C, and LDL-C, a phenotype termed familial combined hypolipidema [8]. We and others have also noted that heterozygous individuals with an LoF variant in one copy of ANGPTL3 had intermediate reduced plasma levels of TG, HDL-C and LDL-C and reduced risk for coronary heart disease [9, 10].
ANGPTL3 is a secreted protein that is primarily expressed in liver [11]. It has two domains, the N-terminal coiled-coil region and a C-terminal fibrinogen domain and the intact protein is cleaved at the linker region between the N- and C-terminal domains [5, 12]. It has been shown that the N-terminal domain is a potent inhibitor of LPL activity, and overexpression of the domain is sufficient to increase the TG level in mice [5, 6]. Given the crucial role of LPL-mediated lipolysis in clearance of TG from chylomicrons or very low-density lipoproteins (VLDL) [13], the lack of inhibition of LPL due to the ANGPTL3 LoF mutations largely explains the extremely low TG in both mice and humans with ANGPTL3 LoF variants. ANGPTL3 also inhibits endothelial lipase (EL) [7]. Compared with other members of lipoprotein lipase family, EL has a unique affinity for HDL, as well as a predominant phospholipase activity [14]. As a result, EL emerged as an important regulator for HDL metabolism as confirmed by overexpression or gene deletion analyses in mice [15]. In ANGPTL3 KO mice, EL activity is higher than in WT mice [7]. ANGPTL3 expression significantly correlates with plasma HDL-C level in human subjects [16]. Thus, inhibition of EL by ANGPTL3 is largely consistent with the low HDL-C level observed in individuals carrying the LoF ANGPTL3 mutations [8].
However, it has been unclear how LoF ANGPTL3 mutations cause low plasma LDL-C. Recent studies have shown that immuno-inactivation of ANGPTL3 with monoclonal antibodies reduced plasma cholesterol levels in multiple mouse models and monkeys [17]. Inactivation of ANGPTL3 using an inhibitory antibody was reported to reduce hepatic VLDL-TG (but not VLDL-ApoB) secretion [18], suggesting that this mechanism might contribute to the reduced LDL-C levels. In this study, we used ANGPTL3-specific siRNAs to silence its hepatic expression and confirmed reduced TG, HDL-C, and LDL-C levels in WT, ob/ob, and human cholesteryl ester transfer protein (hCETP)/ApoB-100 transgenic mice. In human hepatoma cells, we demonstrate that ANGPTL3 deficiency both reduced ApoB-100 secretion and enhanced LDL/VLDL uptake. Thus, our results suggest that knockdown of ANGPTL3 expression reduces LDL-C levels through a dual mechanism of reduced ApoB secretion and enhanced ApoB-containing lipoprotein uptake.
Materials and methods
Animals
C57BL/6 (WT), obese (ob/ob) and LDLR+/− mice were obtained from Jackson Labs. The female hCETP/ApoB-100 double transgenic mice (Cat# 3716-F) were obtained from Taconic. LAhB-LDLR_KO (Apobec−/−, hApoB Tg, Ldlr−/−), LAhB-LDLR_H (Apobec−/, hApoB Tg, Ldlr+/−) and LAhB-LDLR_WT (Apobec−/−, hApoB Tg, Ldlr+/+) mice were generated by crossing previously described LA-DKO (Apobec−/−, Ldlr−/−) [19, 20] with human ApoB-100 transgenic mice (obtained from Dr. S. Young). Angptl3 KO mice were obtained from Dr. Kiran Musunuru’s lab [21]. All mice were fed a standard chow diet and maintained on a 12 hour light/12 hour dark cycle. All procedures used in animal studies were approved by the pertinent Institutional Animal Care and Use Committee and were consistent with local, state and federal regulations as applicable.
Liver-specific knockdown studies
Lipidoid formulations of siRNAs were prepared and screened as described previously [22]. Mice received either luciferase or Angptl3 siRNA (Alnylam) via tail vein injection at a dose of 2 mg/kg or otherwise indicated in figures. The time points for injection and blood collection are indicated in figures or supplemental information. At each time point, mice were fasted for 4 h, anaesthetized by isoflurane inhalation, and blood was collected via retro-orbital plexus, followed by centrifugation for plasma separation. Mice were sacrificed at the indicated time points after a 4-hour fast and anaesthesia by isoflurane inhalation. Terminal bleeds were performed and PBS-perfused livers were collected. Livers were flash-frozen in liquid nitrogen.
Western blot and real-time PCR analysis of liver samples
Liver lysates were prepared in PBS with Halt protease inhibitor cocktail (Thermo) by manual dissociation using a Qiagen Tissuelyser II. Protein was quantified by BCA protein assay (Pierce). The liver lysate (50 μg protein) was used for Western analysis. Total RNA was isolated from 50 mg liver using Qiagen Qiasymphony RNA CT 800 protocol. Reverse transcription of total RNA was performed using the first-strand cDNA synthesis kit (GE Healthcare). Real-time quantitative PCR was performed using optimized mouse Angtpl3 primers, or optimized human ANGPTL3 primers, SYBR green master mix (Applied Biosystems) and a 7900 sequence detections system (Applied Biosystems) using default thermal conditions. Relative quantitation was performed using the comparative cycle threshold method, as recommended by the manufacturer. All quantitation was normalized to the endogenous mouse β-actin or GAPDH.
Measurements of mouse plasma lipids, lipoproteins and mouse Angptl3 protein
TG, TC and HDL-C were measured in individual mouse plasma samples enzymatically on a Cobas Mira auto-analyzer (Roche Diagnostic Systems). Non-HDL-C presented in Fig. 1A and Supplementary Fig. 4 was estimated by subtracting the HDL-C from the TC. TG, TC, HDL-C and LDL-C in Fig. 1B, Fig. 2, and Fig. 3C were measured using an Olympus AU400 Chemical Analyzer. Pooled plasma (150 μl) was subjected to fast phase liquid chromatography (FPLC) gel filtration (Pharmacia LKB Biotechnology/GE Healthcare) using two serial superose 6 columns (GE Healthcare). Cholesterol colorimetric plate assays were performed on FPLC fractions using the Cholesterol E reagent (Wako Pure Chemical Industries). Plasma mouse Angptl3 or PCSK9 protein levels were measured using mAngptl3 ELISA kit (R&D Systems) or mPCSK9 ELISA kit (Circulex), respectively, according to the manufacturer’s procedure.
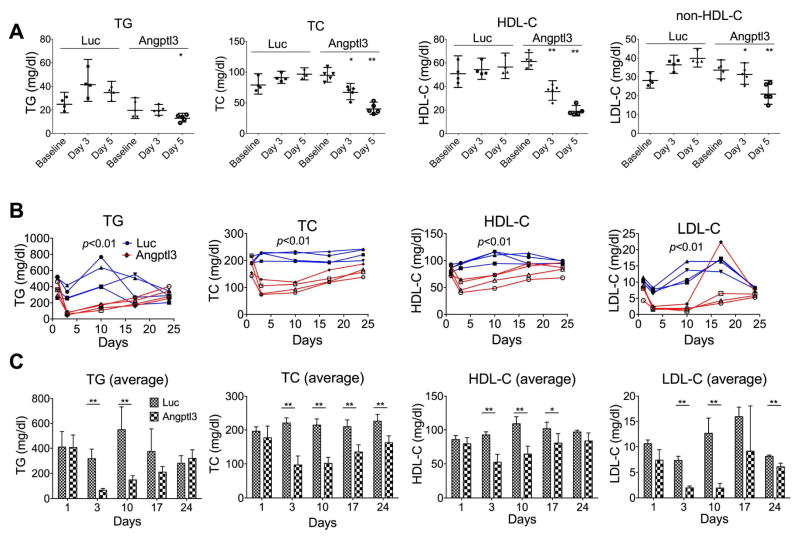
Fig. 1.
Silencing of hepatic Angptl3 expression induced combined hypolipidemia phenotype in WT and obese mice.
(A) WT mice were injected with luciferase (Luc) or Angptl3 siRNAs (2 mg/kg). Blood samples were collected before injection (baseline), and on day 3 and 5 after injection. TG, TC, and HDL-C levels were measured. Average and 95% CI were calculated from 4 or 5 mice of each group. p values (t-test) were calculated by comparing the changes (%) (day 3/5 vs. baseline) of Angptl3 siRNA with those of Luc control. (B and C) ob/ob mice were injected with Luc or Angptl3 siRNAs (1 mg/kg). Blood samples were collected one day before injection (baseline), and on day 3, 10, 14 and 24 after injection. TG, TC, HDL-C and LDL-C were measured. TG, TC, HDL-C and LDL-C levels from individual mouse (B) and averages of each group (C) are shown. Standard deviations were calculated from 4 mice of each group. p values (ANOVA) in (B) were calculated by comparing Angptl3 siRNA with Luc siRNA groups. p values (t-test) in (C) were calculated by comparing Angptl3 siRNA with Luc siRNA groups at the days indicated.
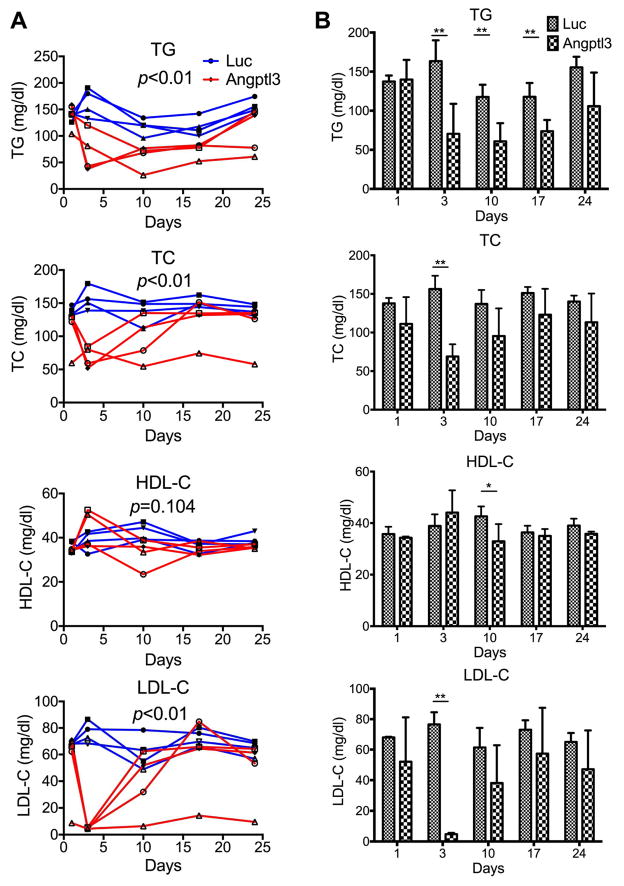
Fig. 2.
Silencing of hepatic Angptl3 lowered LDL-C, but not HDL-C, level in hCETP/ApoB-100 mice.
hCETP/ApoB-100 mice were injected with Luc or Angptl3 siRNAs. Blood samples were collected one day before injection (baseline), and on day 3, 10, 14 and 24 after injection. TG, TC, HDL-C and LDL-C were measured. TG, TC, HDL-C and LDL-C levels from individual mouse (A) and averages of each group (B) are shown. Standard deviations were calculated from 4 mice of each group. p values (ANOVA) in (A) were calculated by comparing the Angptl3 siRNA with Luc siRNA groups. p values (t-test) in (B) were calculated by comparing Angptl3 siRNA with Luc siRNA groups at the days indicated.
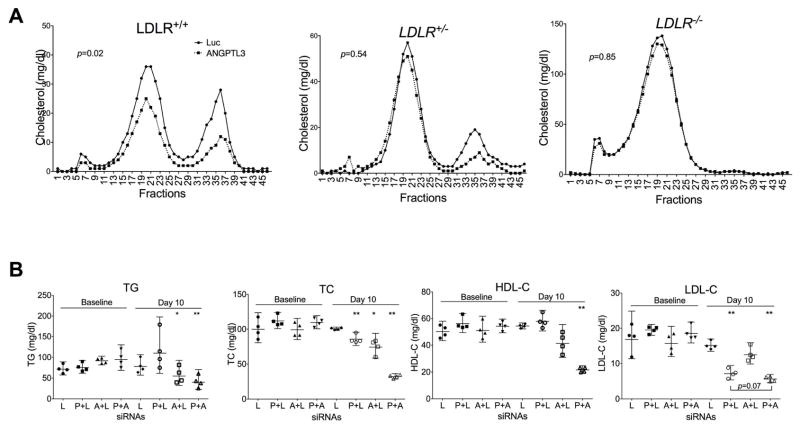
Fig. 3.
The effect of Angptl3 silencing on LDL-C is linked to LDLR.
(A) Angptl3 silencing had no significant effect on LDL-C in mice deficient in LDLR. LAhB-LDLR_WT (LDLR+/+), LAhB-LDLR_H (LDLR+/−) and LAhB-LDLR_KO (LDLR−/−) mice were injected with luciferase (Luc) or Angptl3 siRNAs (as in Fig. 1A). Pooled plasma was analyzed with FPLC. The cholesterol levels were measured in each fraction. p values (t-test) were calculated by comparing Angptl3 siRNA with Luc control groups. (B) An additive effect of Angptl3 and PCSK9 siRNAs on LDL-C. LDLR+/− mice were injected with Luc, PCSK9, Angptl3 or PCSK9 plus Angptl3 siRNAs twice with 7 days interval at the total dose of 1 mg/kg. Blood samples were collected on day 10 following the first injection. Plasma samples were used to measure TG, TC, HDL-C and LDL-C. The average and 95% CI were calculated from 4 mice of each group. Equal amount of total and PCSK9/Angptl3 siRNAs (0.5 mg/kg) were used. Luc control siRNA was added to make equal total amount siRNAs when PCSK9 or Angptl3 siRNAs was used individually. p values (t-test) were calculated by comparing the groups at day 10 with their corresponding baselines. L, Luciferase siRNA; P, PCSK9 siRNA; A, Angptl3 siRNA.
Cell culture, siRNA transfection, total RNA extraction from cells, and RT-PCR/real-time RT-PCR
Huh7 and HepG2 cells were cultured in DMEM medium (Invitrogen) supplemented with 10% fetal bovine serum (FBS). Two siRNAs (siRNA1 and siRNA2) targeting human ANGPTL3 were used for transfection. siRNA1 was from Alnylam and siRNA2 was purchased from Qiagen. Transfection of the siRNAs to cells was performed as previously described [23] but using RNAiMAX (Invitrogen). The transfected cells were used for different assays as described below. Cell duplicates treated under the same condition were used for cell counting. Culture media of the transfected cells with lipoprotein-deficient FBS for ~12 h was used for Western analysis with ApoB monoclonal antibody (Santa Cruz) and albumin antibody (Bethyl). Total RNA extraction was achieved with TRIzol (Invitrogen) followed by DNase I treatment according to the manufacturer’s instructions (Promega). cDNA was obtained using the SuperScript first-strand synthesis system (Invitrogen). RT-PCR/real-time RT-PCRs were carried out with the primers specific for human ANGPTL3 and β-actin.
Pulse-chase metabolic labeling with [35S] methionine/cysteine and [125I]-labeled human LDL uptake experiments
For the pulse-chase experiments, Huh7 and HepG2 cells were transfected and split as above. Before labeling, the cells were washed with methionine- and cysteine-free DMEM three times and then pulse-labeled for 30 min with 20 μCi of [35S] methionine/cysteine labeling mix (PerkinElmer Life Sciences) in the same medium plus 10% dialyzed FBS. The cells were washed with normal DMEM four times and chased with the medium plus 10% lipoprotein-deficient FBS for 20 and 40 min. The media collected at 20 and 40 min were used for immunoprecipitation with ApoB monoclonal antibody and precipitates were analyzed with Western blotting and liquid scintillation counting.
For the [125I]-labeled LDL [24] (bti/Alfa Aesar) uptake experiments, the siRNA-transfected Huh7 and HepG2 cells were split as described above. The cells were washed with serum-free media, and then incubated in the media with lipoprotein-deficient FBS for ~12 h. After wash, the cells were incubated in the same media but with [125I]-labeled LDL for 20 min. After three times washes, the cells were harvested and total cell lysates were prepared for liquid scintillation counting. Cell duplicates treated at the same condition but without [125I]-labeled LDL were used for cell counting.
ANGPTL3 gene deletion using CRISPR/Cas9 genome editing, ApoB-100 measurement and LDL uptake assays
Three single guide RNA (sgRNA) oligoes that target exon 1 of human ANGPTL3 gene were subcloned into lentiGuide-Puro vector. The vector together with accessory plasmids for lentivirus assembly was transfected into 293T cells for lentivirus production [25, 26]. Packaged viruses were used to transduce the Cas9-expressing Huh7 cells for ~16 h. Subsequently, the cells were cultured in presence of puromycin (5 μg/ml) for five days before splitting for assays. To confirm the ANGPTL3 gene deletion, two PCR reactions with different backward primers were performed using the genomic DNA purified from the stable Huh7 cells as above. The PCR products with backward primer 1 were used for T7 endonuclease I (New England Biolabs) assay as described before [27] and CRISPR deep DNA sequencing.
Cells for ApoB-100 measurement assay were cultured for ~16 h in serum-free medium, then the amount of ApoB-100 in medium was measured using ELISA kit (MABTECH) according to the manufacturer’s instructions. To measure LDL-uptake, the Huh7 cells were incubated in serum-free medium for 4 h followed by incubation in serum-free medium containing Dil-LDL [28] (5 μg/ml, Invitrogen) for 1 hr. The cells were then fixed with 4% paraformaldehyde and co-stained with DAPI for imaging. Using automated high-content microscopy Cellomics Array Scan VTI, images including ~20,000 cells were taken and cellular intensity of Dil-LDL was quantified.
Individual ANGPTL3 KO and scramble control cell clones were obtained by growing single cells in 96-well plates. The media ApoB-100 measurement in the time course experiments with individual clones were performed as above. The Dil-LDL and Dil-VLDL uptake experiments with individual clones were carried out as above but visualized under confocal microscope.
Total cell lipid extract preparation and metabolite mass spectrometry analysis
ANGPTL3 KO and scramble control Huh7 cells were grown in DMEM media with 10% FBS. The cells were washed with cold PBS (no Mg2+/no Ca2+) and immediately 800μL of pre-cooled isopropanol (HPLC grade) was added. The cells were then scraped and transferred to a 1.5mL tube and kept at 4°C for 1hr. The extract was vortexed and centrifuged (9,000 × g, 4°C, 10 min). Supernatant was used for the metabolite mass spectrometry analysis as previously described [29].
Statistic analysis
Data of each study group were represented as average ± standard deviation or average ± 95% confidence interval (CI). Statistic comparisons of groups were performed using Prism 7 (GraphPad) with the student’s t-test or analysis of variance (ANOVA). Detailed group comparisons were described in individual figure legends. p values of ≤ 0.05 were considered to be statistically significant (*, p<0.05; **, p <0.01).
Results
RNAi silencing of hepatic Angptl3 expression reduced LDL-C in WT and ob/ob mice
To investigate the role of ANGPTL3 in regulating LDL-C, we first used siRNAs to silence hepatic Angptl3 expression in WT mice. We assumed that silencing might induce a phenotype that mimics human familial combined hypolipidemia as previously described [8]. WT mice were injected with Angptl3 or luciferase control siRNAs. Angptl3-specific siRNA effectively reduced hepatic Angptl3 expression (~95% reduction) (Supplementary Table 1). Angptl3 silencing (Fig. 1A) markedly reduced plasma TG (~72%), TC (~80%), HDL-C (~81%), and non-HDL-C (~78%) on day 5 as compared with the control siRNA. As a validation to the Angptl3 silencing, we performed FPLC gel filtration analysis of the pooled plasma from WT or Angptl3 KO mice [21]. The results show that HDL-C and LDL-C decreased by ~50% and ~40%, respectively, in the KO mice as compared with the WT (Supplementary Fig. 1A). The data indicate that inhibition of hepatic Angptl3 expression is sufficient to induce the combined hypolipidemia in mice.
Angptl3 was shown to be intimately associated with hyperlipidemia of obese mice [30]. We tested whether Angptl3 silencing with siRNA could reduce the hyperlipidemia, especially the LDL-C level, in ob/ob mice. Towards this end, we injected siRNAs targeting Angptl3 or luciferase into obese mice. Plasma Angptl3 levels substantially decreased by day 3 (by ~97%) and were still reduced on day 24 (~56%) (Supplementary Fig. 2). Plasma lipid results (Fig. 1B) show that Angptl3 siRNA not only reduced TG and HDL-C levels, but also strongly reduced LDL-C levels (on day 3 and 10, LDL-C levels were decreased by ~73% and ~84%, respectively). Striking LDL-C reduction was also observed in the FPLC analysis (combined of day 3 and 10) (Supplementary Fig. 1B). These data indicate that Angptl3 silencing reduces all plasma lipids, especially the LDL-C, in ob/ob mice, a pattern similar to what is seen in humans and mice with LoF mutations in ANGPTL3 [8, 30].
Angptl3 silencing reduced LDL-C in ‘humanized’ hCETP/ApoB-100 transgenic mice
To better model the potential human effects of Angptl3 silencing on LDL-C, we made use of the hCETP/ApoB-100 double transgenic mouse model, which has been shown to possess a human-like lipoprotein distribution and a higher level of LDL-C [31]. Consistent with previous findings, hCETP/ApoB-100 mice showed much higher LDL-C (~5.7 times more than that of the WT) and lower HDL-C (~46% of the WT) (compare Supplementary Fig. 1A with C). siRNAs targeting Angptl3 or luciferase control were injected into these mice. Analysis of plasma Angptl3 protein concentration again showed a drastic reduction (by ~97%) by day 3 (Supplementary Fig. 3). Angptl3 silencing reduced TG and TC levels but had relatively little effect on HDL-C levels (Fig. 2). Angptl3 silencing markedly reduced LDL-C (by ~93%) on day 3, but the LDL-C level rebounded by 10 days.
Reduction in LDL-C by hepatic Angptl3 silencing is associated with an intact LDL receptor
To investigate whether LDLR might contribute to the low level of LDL-C, we turned to the Apobec1−/− mice carrying human ApoB-100 transgene (LahB-LDLR_WT, ldlr+/+) and the same mice that are haploinsufficient (LahB-LDLR_H, ldlr+/−) or completely insufficient (LahB-LDLR_KO, ldlr−/−) for LDLR [19, 20]. These mouse models are also more humanized and have considerably higher levels of LDL-C. For all these mouse strains, Angptl3-specific siRNA effectively reduced hepatic Angptl3 expression (>95% reduction) (see Supplementary Table 1).
In the LahB-LDLR_WT mice, Angptl3 siRNA reduced levels of TG (~39%), TC (~29%), HDL-C (~40%), and non-HDL-C (~21%). However, in the LahB-LDLR_H and LahB-LDLR_KO mice, the effect of Angptl3 silencing on non-HDL-C was greatly diminished (Supplementary Fig. 4). The most notable changes in the cholesterol levels were found in FPLC gel filtration analysis of the pooled plasma from the mice injected with Angptl3 or luciferase siRNAs. Cholesterol profiles (Fig. 3A) show that Angptl3 siRNA markedly reduced the levels of LDL-C (fractions 15–25) and HDL-C (fractions 31–39) in the LAhB-LDLR_WT mice. The peak fractions for LDL-C and HDL-C from the Angptl3 siRNA-treated mice were reduced by ~40% and ~60%, respectively. In LahB-LDLR_H mice, Angptl3 silencing still reduced HDL-C, but its effect on LDL-C was greatly diminished (~64% and ~18% reduction in peak fractions, respectively) (Fig. 3A). However, in LahB-LDLR_KO mice, the effect of Angptl3 silencing on LDL-C was barely detectable (only ~8% reduction in the peak fractions) (Fig. 3A). In both LahB-LDLR_H and LAhB-LDLR_KO mice, Angptl3 siRNA was highly effective in decreasing TG levels (~48% and ~52% reductions, respectively, as compared with the LahB-LDLR_WT, ~39%) (Supplementary Fig. 4). These data suggest that an intact LDLR is partially responsible for Angptl3 knockdown to reduce LDL-C, but not TG, and implicate LDLR in the mechanism of the LDL-C reduction.
Silencing of Angptl3 and PCSK9 are additive in reducing LDL-C
To gain more evidence for the LDLR involvement, we made use of PCSK9-specific siRNAs. We hypothesized that PCSK9 silencing, by increasing LDLR activity, would enhance the Angptl3 siRNA effect on LDL-C levels. To this end, LDLR+/− mice were injected with PCSK9 and Angptl3 siRNAs individually or together along with luciferase siRNA. Both PCSK9 and Angptl3 siRNAs significantly decreased their corresponding mRNA levels without affecting their counterparts (Supplementary Fig. 5). Similar to the results presented in Fig. 1, Angptl3 siRNA reduced the levels of TG (by ~38%), HDL-C (by ~18%), LDL-C (by ~20%), and TC (by ~25%) (Fig. 3B). In contrast, the PCSK9 siRNA specifically decreased LDL-C (by ~63%) and TC (by ~24%) levels without affecting TG and HDL-C. The combined injection of both Angptl3 and PCSK9 siRNAs significantly reduced TC (by ~71%) and LDL-C (by ~70%) levels and the reductions are greater than those from Angptl3 siRNA or PCSK9 siRNA alone.
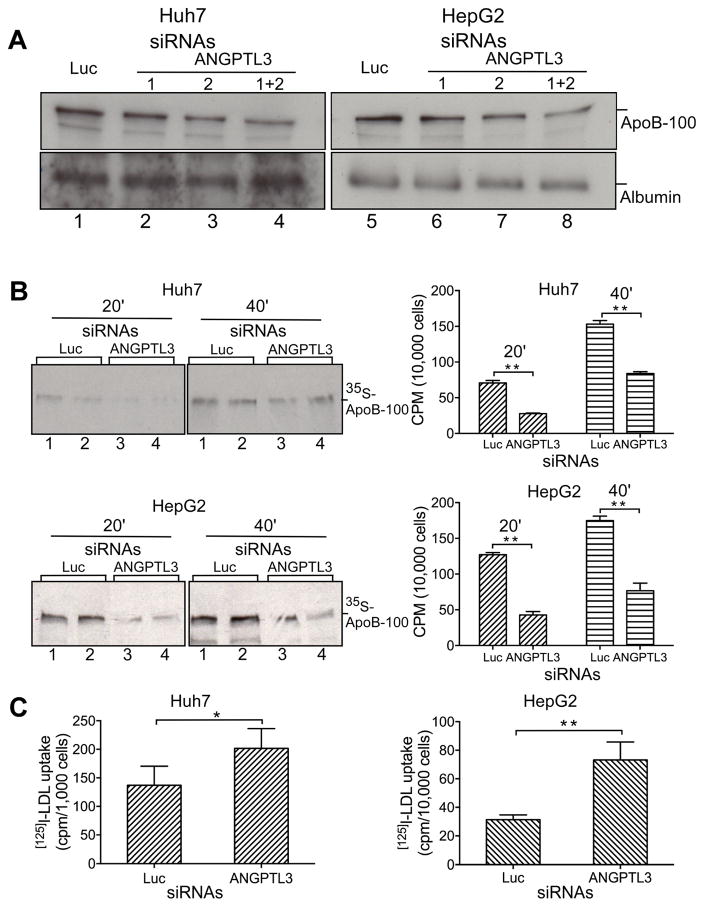
ANGPTL3 silencing in Huh7 and HepG2 cells reduced ApoB-100 secretion and enhanced LDL uptake
To obtain more insights into the LDL-C reduction induced by ANGPTL3 silencing, we used Huh7 and HepG2 cells for in vitro analysis. We transfected two siRNAs specific for human ANGPTL3, either individually or together, along with luciferase siRNA into Huh7 and HepG2 cells. RT-PCR analyses indicate that both siRNAs efficiently decreased ANGPTL3 mRNA levels, especially when both siRNAs were used together (Supplementary Table 2). To examine the effect of ANGPTL3 silencing on the ApoB-100 dynamics, we measured ApoB-100 level in culture media. The data (Fig. 4A) show that ANGPTL3 siRNAs reduce ApoB-100 accumulation in media in both Huh7 and HepG2 cells. A marked decrease was observed when the two ANGPTL3 siRNAs were used simultaneously (lanes 4 and 8).
Fig. 4.
ANGPTL3 silencing in Huh7 and HepG2 cells reduced ApoB-100 secretion and enhanced LDL uptake.
(A) ANGPTL3 silencing reduced the accumulation of ApoB-100 in culture media. Huh7 and HepG2 cells were transfected with Luc or ANGPTL3 siRNAs as indicated. Equivalent amount of culture media (based on cell numbers) from the transfected cells were analyzed by Western blotting with ApoB (top) or albumin (bottom) antibodies. (B) ANGPTL3 silencing reduced nascent ApoB-100 secretion. Huh7 and HepG2 cells were transfected with Luc or ANGPTL3 siRNAs (1+2) as in (A). The transfected cells were pulse-labeled with [35S] labeling mix, and chased for 20 and 40 min. Equivalent amount of culture media were used for immunoprecipitation with ApoB antibodies and precipitates were separated with SDS-PAGE and directly exposed with X-ray film (left panel), or analyzed by scintillation counting for quantification (right panel). (C) ANGPTL3 silencing increased LDL uptake. Huh7 and HepG2 cells were transfected with Luc or ANGPTL3 siRNAs (1+2) as in (A). The transfected cells were incubated with fresh media containing [125I]-labeled LDL. Total cell lysates were prepared and measured by scintillation counting. Averages and standard deviations were calculated from three independent experiments. p values (t-test) were determined by comparing ANGPTL3 siRNA with Luc control groups.
The decreased accumulation of ApoB-100 in culture media could result from reduced secretion or/and increased uptake of ApoB-containing lipoproteins. To confirm the impact of ANGPTL3 silencing on the ApoB-100 secretion, the above siRNA-transfected cells (with two ANGPTL3 siRNAs) were pulse-labeled with [35S]-methionine/cysteine to monitor the newly synthesized protein. ApoB-100 in the media was purified by immunoprecipitation with ApoB-specific antibody and the precipitates were analyzed with Western blotting and scintillation counting. The results (Fig. 4B) show that ANGPTL3 silencing greatly reduced the secretion of newly synthesized ApoB-100 into the media both from Huh7 (by ~61% and ~45%) and HepG2 (by ~66% and ~56% at 20 and 40 min, respectively) cells.
To examine whether ANGPTL3 silencing might also affect LDL uptake, the siRNA-transfected cells were incubated in the media containing [125I]-labeled LDL. Whole cell lysates were prepared for scintillation counting. The results (Fig. 4C) indicate that ANGPTL3 silencing increased LDL uptake both in Huh7 and HepG2 cells (by ~47% and ~133%, respectively). Thus, our data indicate that ANGPTL3 silencing in Huh7 and HepG2 cells reduces secretion of ApoB-100 and increases LDL uptake.
ANGPTL3 gene knockout in Huh7 cells reduced ApoB-100 secretion and enhanced LDL uptake
To further validate the above results, we deleted ANGPTL3 in Huh7 cells using the CRISPR/Cas9 genome editing [25] and investigated the effect on ApoB secretion and LDL uptake. T7 endonuclease I assay [27] (Supplementary Fig. 6A) and CRISPR deep DNA sequencing (Supplementary Fig. 7) confirmed the gene deletion. ANGPTL3 gene deletion significantly reduced ApoB-100 level in culture media by ~13% as compared with the scramble control cells. As controls, LDLR deletion increased the media ApoB-100 by ~100% and ApoB-100 deletion reduced it by ~92% (Supplementary Fig. 6C, left panel). Furthermore, ANGPTL3 deletion markedly increased LDL uptake (by ~62%). As controls, the LDLR deletion dramatically reduced LDL uptake, while ApoB-100 deletion did not affect the uptake (Supplementary Fig. 6C, right panel) (see Supplementary Fig. 6B for LDLR deletion validation).
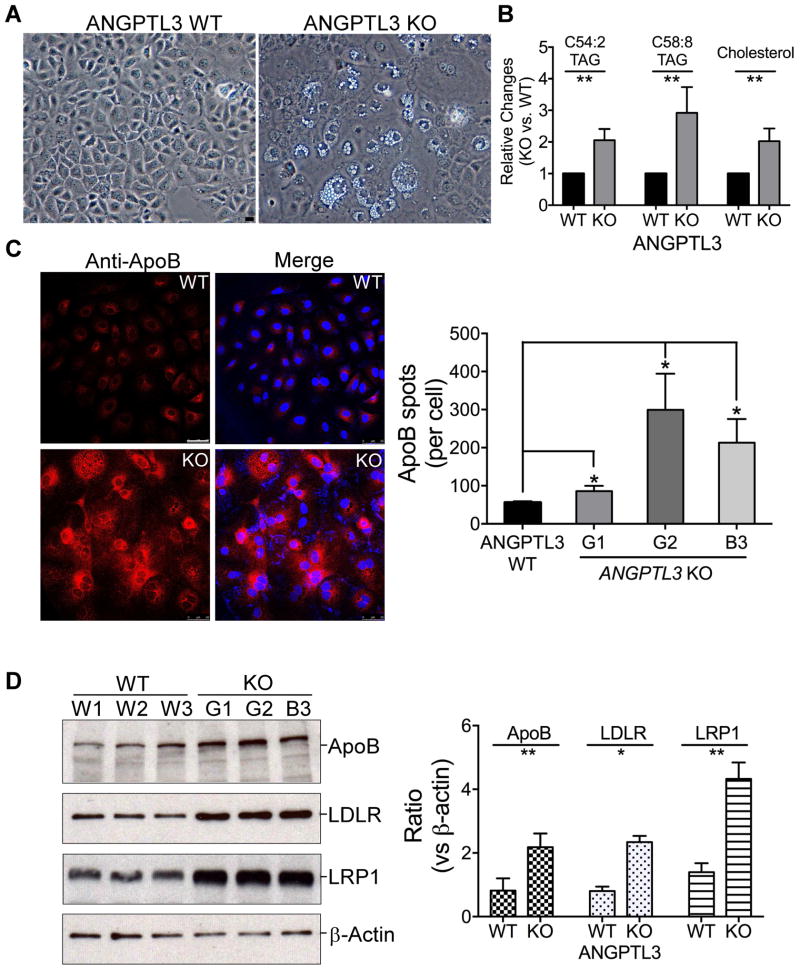
To further analyze the clonal effect on ApoB secretion and LDL uptake, we isolated multiple individual clones from the ANGPTL3 KO and control cells as above. The deletion of ANGPTL3 gene induced dramatic lipid accumulation (Fig. 5A). We then used metabolite mass spectrometry to systematically examine the lipid content in these cells [29]. Consistent with the above observation, the cellular level of TG (or TAG as labeled in Fig. 5B and Supplementary Fig. 8) and cholesterol dramatically increased in ANGPTL3 KO cells. The top abundant long-chain C54:2, C58:8 TGs, and cholesterol increased by ~100%, ~192% and ~100%, respectively, (Fig. 5B). But for the short-chain TGs and diglycerides (or DAG as labeled in Supplementary Fig. 8) decreased in the KO cells (Supplementary Fig. 8), indicating that the ANGPTL3 deletion interrupted cellular TG metabolism. Consistently, confocal analysis shows that the deletion of ANGPTL3 gene also induced cellular ApoB accumulation (increased by ~121% in the KO cells) (Fig. 5C). Western analysis further confirmed the ApoB accumulation (increased by 1.6 times) (Fig. 5D, top).
Fig. 5.
Deletion of ANGPTL3 gene in Huh7 cells induced cellular long-chain TG and ApoB accumulation, and increased LDLR/LRP1 expression.
(A) ANGPTL3 gene deletion induced lipid droplet formation. Representative phase images of individual ANGPTL3 scramble control (WT) and KO Huh7 clones. (B) ANGPTL3 gene deletion induced long-chain TG accumulation. Total cell lipids from the cells as in (A) were analyzed by metabolite mass spectrometry. Two top abundant long-chain TG, C54:2 and C58:8 TAG as well as cholesterol are presented. See Supplementary Fig. 8 for more data. (C) ANGPTL3 gene deletion induced ApoB accumulation. The cells as in (A) were stained with anti-ApoB antibody, and analyzed with confocal microscope (Left). Images were quantified with ImageJ from 100–200 cells (Right). (D) ANGPTL3 gene deletion increased ApoB-100, LDLR and LRP1 expression. Extracts of the cells as in (C) were analyzed with Western blotting and detected with the antibodies as indicated (Left). The blots were quantified with ImageJ and the averages and standard deviations were calculated from three clones (Right). p values (t-test) were calculated by comparing the results from the KO cells with those from the control cells. Bars, 50 μm.
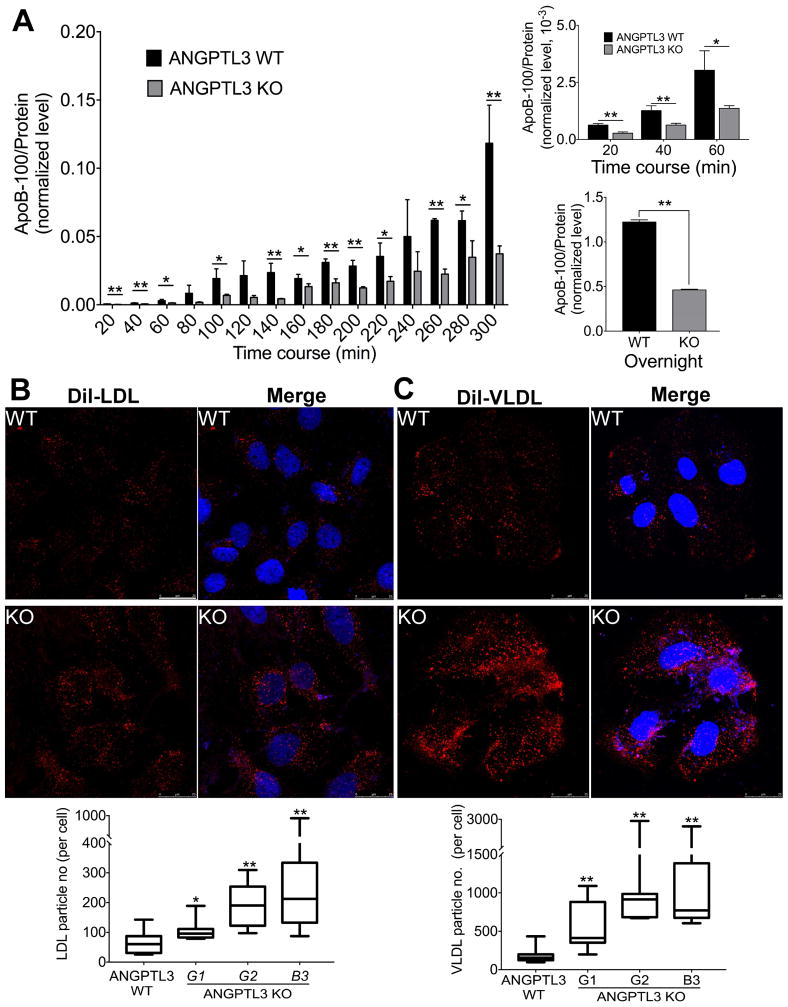
We monitored the presence of ApoB in culture media in a time course experiment with three ANGPTL3 KO and control clones. Consistent with the above results, the deletion of ANGPTL3 gene greatly reduced the ApoB accumulation at all time points (Fig. 6A, left panel). Dramatic decrease of ApoB accumulation was found with overnight incubation (~62% decrease) (right panel, bottom). Importantly, the low ApoB levels at the early stage of the time course may suggest a reduced ApoB secretion rate in the ANGPTL3 KO clones as compared with the controls (right panel, top).
Fig. 6.
Deletion of ANGPTL3 gene in Huh7 cells reduced ApoB-100 accumulation in culture media and enhanced LDL/VLDL uptake.
(A) Three individual ANGPTL3 scramble control (WT) and KO Huh7 clones were used for time course experiments to monitor the ApoB presence in culture media using ELISA kit (left panel). Right panel, top, highlights of the media ApoB-100 levels between 20 and 60 mins from the time course experiments on the left panel. Right panel, Bottom, the media ApoB levels after overnight (~12 h) incubation. (B) and (C) Individual clones from the cells as in A were used for Dil-LDL (B) and Dil-VLDL (C) uptake. Top, Representative images of the uptake assays from ANGPTL3 KO and control cells. Bottom, Quantification of the uptake assays. The averages and deviations were calculated from three clones of the control cells (~200 cells) and KO cells (100–200 cells). Cell Dil-LDL/Dil-VLDL particles were quantified using ImageJ. p values (t-test) were calculated by comparing the KO cells with the control cells. Bar, 25 μm.
To evaluate LDL uptake, the above clones were incubated with Dil-LDL, the cells were then fixed and examined under confocal microscope. The results show that the deletion of ANGPTL3 gene indeed increased LDL uptake in three individual clones (by ~61%, ~1.8 and ~2.5 times) (Fig. 6B). Similarly, the clones were also tested for VLDL uptake. Surprisingly, the ANGPTL3 deletion drastically increased the Dil-VLDL uptake (by ~2, 4 and 5 times), which is much stronger than the LDL uptake (Fig. 6C). Consistent with these data, Western analysis shows that the deletion of ANGPTL3 gene strongly induced the expression of LDLR and LRP1 (Fig. 5D). The elevated LDLR and LRP1 explain the increased uptake of LDL and VLDL. Taken together, the data from the ANGPTL3 gene deletion are fully consistent with those from Angptl3 silencing, indicating that the ablation of ANGPTL3 expression reduces the ApoB secretion and increases LDL/VLDL uptake.
Discussion
In this study, we focused on determining the mechanism by which LoF ANGPTL3 mutations cause low LDL-C. We used RNAi-mediated silencing approach to target hepatic Angptl3 expression in multiple mouse models. We found that hepatic Angptl3 silencing is sufficient to reduce LDL-C levels. Our analysis using human hepatoma cells showed that ANGPTL3 silencing and deletion reduced ApoB-100 secretion and increased LDL/VLDL uptake. Thus, we suggest that the reduction in LDL-C as a result of reduced ANGPTL3 is explained by both reduced hepatic ApoB secretion and increased hepatic LDL uptake.
The low levels of TG and LDL-C seen in the individuals carrying the ANGPTL3 LoF mutations provide important human genetic validation of ANGPTL3 as a potential therapeutic target [4, 8]. Thus, inhibition of ANGPTL3 expression could provide an attractive approach for treatment of hyperlipidemia. Indeed, administration of ANGPTL3 siRNAs (this study) as well as monoclonal antibody and antisense oligonucleotides targeting ANGPTL3 dramatically decreased the plasma TG, LDL-C and HDL-C levels in mouse models, monkeys, and humans [9, 10, 17, 18]. As described earlier, the obese mouse could be an ideal model to test this. In this study we observed even stronger effect in those mice. ANGPTL3 is highly associated with hyperlipidemia in the obese mice and mutation of Angptl3 is sufficient to convert the hyperlipidemia to hypolipidemia in the mutant mouse strain [30].
However, exploitation of ANGPTL3 as a therapeutic target is complicated by the finding that it also reduces the HDL-C level [8], which was shown to be inversely associated with coronary heart disease [32]. The relationship between HDL-C and ANGPTL3 has been validated by various studies in both humans and mice [7]. Mice have a different lipoprotein metabolism as evidenced by the high level of HDL-C, which constitutes ~80% of TC, whereas in humans HDL-C only takes about 20–30% [31]. It is well-known that mice lack CETP activity [33], and double transgenic expression of human CETP and ApoB-100 in hCETP/ApoB-100 mice profoundly shifts the lipoprotein cholesterol distribution that is similar to human normolipidemia [31]. Surprisingly, using the hCETP/ApoB-100 mice we found that down-regulation of Angptl3 had no obvious effect on the HDL-C level, while still maintains a strong activity in reducing the LDL-C level. CETP plays a central role in the HDL catabolism by mediating the exchange of neutral lipids between HDL and VLDL/LDL [34]. The CETP protein level is inversely correlated with HDL-C level and transgenic expression of hCETP markedly reduced the HDL-C in mice [31, 35]. As described earlier, ANGPTL3 modulates HDL-C presumably through the inhibition of EL activity [7]. The resistance of hCETP/ApoB-100 mice to Angptl3 silencing perhaps largely comes from the much stronger activity of hCETP on the HDL-C level.
A previous study reported that immuno-inactivation of extracellular Angptl3 reduced hepatic VLDL-TG, but not VLDL-apoB-100, secretion [17, 18]. In this study, we confirmed in two different human hepatoma cell lines using two different methods of ANGPTL3 silencing that reduced ApoB-100 secretion. Importantly, the Angptl3 antibody was used to target the circulating Angptl3 and it did not affect liver Angptl3 expression [17, 18], whereas our approach silenced Angptl3 expression intracellularly. This difference could explain the different results. Intracellular ANGPTL3 may be involved in the VLDL assembly in the endoplasmic reticulum, which is supported by our recent findings that ANGPTL3 interacts with a large number of cytoplasmic and endoplasmic reticulum proteins. Thus, ANGPTL3 silencing or null expression may interfere with VLDL assembly and decrease its secretion, whereas antibody inhibition of extracellular ANGPTL3 may not. Furthermore, we used two different human hepatoma cell lines to show reduction in ApoB-100 secretion, whereas the previous work was done in mice.
We also found that ANGPTL3 silencing enhanced LDL/VLDL uptake. A previous report suggested, but did not demonstrate experimentally, that antibody inhibition of ANGPTL3 may alter the content of VLDL particles secreted from the livers, which are then more rapidly cleared via a non-canonical pathway [18]. Our experimental results are consistent with the concept that reduced ANGPTL3 leads to accelerated uptake of ApoB-containing lipoproteins by liver cells. It should be mentioned that, regardless of intracellular activity, both immuno-inactivation of Angptl3 and hepatic Angptl3 silencing would increase the LPL activity. Higher lipolytic activity of LPL would change the content of TG in the LDL/VLDL particles, which may also be cleared more rapidly. The mechanism of these effects remains uncertain and will require additional studies.
Our finding of a dual role for ANGPTL3 in altering VLDL secretion and LDL uptake is consistent with lipoprotein kinetics tracer studies in humans deficient for ANGPTL3 [8]. In a study comparing humans carrying two ANGPTL3 LoF alleles, heterozygous null, and wild-type participants in a single family, VLDL ApoB production rate was significantly lower and LDL ApoB fractional catabolic rate higher among those carrying two LoF alleles.
In summary, we investigated the effect of Angptl3 silencing on LDL-C levels in five different mouse models and on mechanisms of LDL regulation in two different human hepatoma cell lines. Angptl3 silencing in mouse livers induced the combined hypolipidemia phenotype, including reduction in LDL-C levels. ANGPTL3 silencing in human hepatoma cell lines reduced ApoB-100 secretion and enhanced LDL/VLDL uptake. Both mechanisms may help to explain the reduction of LDL-C levels in mice silenced for Angptl3 and the low plasma LDL-C levels observed in individuals carrying LoF ANGPTL3 mutations. Further studies are needed to deepen our understanding about the molecular mechanisms of both of these effects.
Supplementary Material
Supplementary Fig. 1. FPLC analysis of plasma cholesterol profiles. (A). The plasma cholesterol profiles of Angptl3 WT and KO mice. Pooled plasma samples were from 12 WT and 12 KO mice. (B) and (C). The plasma cholesterol profiles of ob/ob mice (as in Fig. 1B) and hCETP/ApoB-100 transgenic mice (as in Fig. 2), respectively. Pooled plasma samples were from the mice injected with luciferase (Luc) or Angptl3 siRNAs at day 3 and 10.
Supplementary Fig. 2. Expression analysis of Angptl3 from obese mice injected with Luc or Angptl3 siRNAs. Blood samples from the mice as in Fig. 1B were used for mouse Angptl3 ELISA analysis. Protein levels of individual mouse (A) and group average (B) are the ratios of Angptl3 levels at the indicated days after injection over those at the pre-dose.
Supplementary Fig. 3. Expression analysis of Angptl3 in hCETP/ApoB-100 transgenic mice injected with Luc or Angptl3 siRNAs. Blood samples from the mice as in Fig. 2 were used for mouse Angptl3 ELISA analysis. Protein levels of individual mouse (A) and group average (B) are the ratios of Angptl3 levels at the indicated days after injection over those at the pre-dose.
Supplementary Fig. 4. The effect of Angptl3 silencing on LDL-C level is linked to LDLR. LAhB- LDLR_WT (LDLR+/+) (A), LAhB- LDLR_H (LDLR+/−) (B) and LAhB- LDLR_KO (LDLR−/−) (C) mice were injected with Luc or Angptl3 siRNAs. Blood samples were collected before the injection (baseline) and on Day 5 after the injection, and analyzed as in Fig. 1A. The Y axis represents the changes (%) (day 5 vs. baseline) of Angptl3 siRNA and Luc siRNA. p values (t-test) were calculated by comparing the changes of Angptl3 siRNA with those of Luc siRNA.
Supplementary Fig. 5. Angptl3 or PCSK9 levels from the mice injected with Luc, Angptl3 or/and PCSK9 siRNAs. LDLR+/− mice were injected with Luc, PCSK9, Angptl3 or PCSK9 plus Angptl3 siRNAs twice with 7 days interval at the total dose of 1 mg/kg. Blood samples were collected on day 10 following the first injection (as in Fig. 3B). The mouse livers (day 10) and blood samples (day 7 and 10) from the mice were used for real-time RT-PCR and ELISA analyses for Angptl3 and PCSK9 mRNA and protein expression, respectively. The mRNA levels (A) are the ratios of Angptl3 (right) or PCSK9 (left) over GAPDH. The protein levels (B) are the ratios of plasma Angptl3 (right) and PCSK9 (left) levels from the injection combinations over those in PBS control. N=4 for each group, n=2 for PBS. L=Luciferase siRNA. P=PCSK9 siRNA. A= Angptl3 siRNA.
Supplementary Fig. 6. Confirmation of ANGPTL3 and LDLR gene deletions. (A) PCR reactions were performed with the primers for ANGPTL3 gene deletion in Huh7 cells for ANGPTL3 gene deletion using the genomic DNA from the ANGPTL3 KO (lane 2) and scramble control (lane 1) Huh7 cells. PCR products were purified and used for T7 endonuclease I assay (lanes 3 and 4). (B) Whole cell extracts from scramble control and LDLR KO, as well as PCSK9 KO Huh7 cells (see below), were used for Western analysis with the antibodies against LDLR and β-actin. (C) The Huh7 cells with ANGPTL3 gene deletion or scramble control were analyzed for ApoB-100 accumulation in culture media (left) and Dil-LDL uptake (right). The Huh7 cells with LDLR and APOB gene deletions were used as controls. Averages and standard deviations were calculated from 6 samples.
Supplementary Fig. 7. Confirmation of ANGPTL3 gene deletion in Huh7 cells using CRISPR deep sequencing. Purified PCR products as in Supplementary Fig. 6A were used for CRISPR deep DNA sequencing. (A) Sequence alignments of PCR amplicons (Supplementary Fig. 6A) from ANGPTL3 scramble control Huh7 cells with ANGPTL3 reference sequence. (B) Sequence alignments of PCR amplicons from ANGPTL3 KO Huh7 cells.
Supplementary Fig. 8. Deletion of ANGPTL3 gene in Huh7 cells induced cellular long-chain TG and cholesterol accumulation. Total lipid extracts of ANGPTL3 scramble control (WT) and KO Huh7 clones were used for metabolite mass spectrometry analysis. Averages and standard deviations were measured from three WT clones and three KO clones (each with three duplicates). p values (t-test) were calculated by comparing the results from ANGPTL3 KO cells with those from ANGPTL3 WT cells. TAG, triacylglycerol. DAG, diacylglycerol. MAG, monoacylglycerol.
Highlights.
Angptl3 silencing induced combined hypolipidemia in WT and obese mice.
Angptl3 silencing reduced plasma TG and LDL-C, but not HDL-C, in hCETP/ApoB-100 transgenic mice.
The reduction of LDL-C from Angptl3 silencing is linked to LDLR in Apobec1−/−/ApoB-100 transgenic mice deficient in LDLR.
ANGPTL3 silencing and KO in human hepatoma cells reduced nascent ApoB-100 secretion and increased LDL/VLDL uptake.
A dual mechanism of reduced secretion and increased uptake of ApoB-containing lipoproteins may contribute to the low LDL-C caused by genetic ANGPTL3 deficiency.
Acknowledgments
Financial support
This work was supported in part by the National Heart, Lung, and Blood Institute (R37-HL055323 and RC2-HL101864 to DJR; R33-HL120781 to SK and CC). SK is supported by the Ofer and Shelly Nemirovsky Research Scholar award from the Massachusetts General Hospital (MGH), the Howard Goodman Fellowship from MGH, the Donovan Family Foundation, and a grant from Foundation Leducq. YXX was in part supported by American Heart Association SDG grant (11SDG7670007) (2011–2015).
We thank our animal care staff (Edwige Edouard, Aisha Faruqi, and Mao-Sen Sun), Debra Cromley, and Antonino Picataggi for excellent technical support. We also thank the Penn Vector Core for the generation of the AAV constructs and vector particles. We also thank Nicolas Kuperwasser for the contribution at the early stage of this project.
Footnotes
Conflict of interest
Dr. Kathiresan has served on Scientific Advisory Boards for Alnylam, Regeneron, and Ionis Pharmaceuticals. The other authors have nothing to disclose.
Author contributions
X.Y.X., R.V., Q.W., P.J., F.K., R.D., and K.S. designed research; X.Y.X., R.V., Y.H., Q.W., P.J., A.L. A.D., K.T., and X.W. performed research; X.Y.X., R.V., Y.H., Q.W., P.J., A.L., A.D., K.T., X.W., K.M., C.B.C., C.C., F.K., R.D., and K.S. analyzed data; X.Y.X., R.V., Y.H., Q.W., R.D., and K.S. wrote the paper.
The statement of conflict of interest is included in the main text of the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Musunuru K, Kathiresan S. Genetics of coronary artery disease. Annual review of genomics and human genetics. 2010;11:91–108. doi: 10.1146/annurev-genom-082509-141637. [DOI] [PubMed] [Google Scholar]
- 2.Bauer RC, Stylianou IM, Rader DJ. Functional validation of new pathways in lipoprotein metabolism identified by human genetics. Curr Opin Lipidol. 2011;22(2):123–128. doi: 10.1097/MOL.0b013e32834469b3. [DOI] [PubMed] [Google Scholar]
- 3.Mattijssen F, Kersten S. Regulation of triglyceride metabolism by Angiopoietin-like proteins. Biochim Biophys Acta. 2012;1821(5):782–789. doi: 10.1016/j.bbalip.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 4.Romeo S, Yin W, Kozlitina J, Pennacchio LA, Boerwinkle E, Hobbs HH, Cohen JC. Rare loss-of-function mutations in ANGPTL family members contribute to plasma triglyceride levels in humans. The Journal of clinical investigation. 2009;119(1):70–79. doi: 10.1172/JCI37118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ono M, Shimizugawa T, Shimamura M, Yoshida K, Noji-Sakikawa C, Ando Y, Koishi R, Furukawa H. Protein region important for regulation of lipid metabolism in angiopoietin-like 3 (ANGPTL3): ANGPTL3 is cleaved and activated in vivo. The Journal of biological chemistry. 2003;278(43):41804–41809. doi: 10.1074/jbc.M302861200. [DOI] [PubMed] [Google Scholar]
- 6.Lee EC, Desai U, Gololobov G, et al. Identification of a new functional domain in angiopoietin-like 3 (ANGPTL3) and angiopoietin-like 4 (ANGPTL4) involved in binding and inhibition of lipoprotein lipase (LPL) The Journal of biological chemistry. 2009;284(20):13735–13745. doi: 10.1074/jbc.M807899200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shimamura M, Matsuda M, Yasumo H, et al. Angiopoietin-like protein3 regulates plasma HDL cholesterol through suppression of endothelial lipase. Arterioscler Thromb Vasc Biol. 2007;27(2):366–372. doi: 10.1161/01.ATV.0000252827.51626.89. [DOI] [PubMed] [Google Scholar]
- 8.Musunuru K, Pirruccello JP, Do R, et al. Exome sequencing, ANGPTL3 mutations, and familial combined hypolipidemia. The New England journal of medicine. 2010;363(23):2220–2227. doi: 10.1056/NEJMoa1002926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dewey FE, Gusarova V, Dunbar RL, et al. Genetic and Pharmacologic Inactivation of ANGPTL3 and Cardiovascular Disease. N Engl J Med. 2017;377(3):211–221. doi: 10.1056/NEJMoa1612790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graham MJ, Lee RG, Brandt TA, et al. Cardiovascular and Metabolic Effects of ANGPTL3 Antisense Oligonucleotides. N Engl J Med. 2017;377(3):222–232. doi: 10.1056/NEJMoa1701329. [DOI] [PubMed] [Google Scholar]
- 11.Miida T, Hirayama S. Impacts of angiopoietin-like proteins on lipoprotein metabolism and cardiovascular events. Current opinion in lipidology. 2010;21(1):70–75. doi: 10.1097/MOL.0b013e328333269e. [DOI] [PubMed] [Google Scholar]
- 12.Liu J, Afroza H, Rader DJ, Jin W. Angiopoietin-like protein 3 inhibits lipoprotein lipase activity through enhancing its cleavage by proprotein convertases. The Journal of biological chemistry. 2010;285(36):27561–27570. doi: 10.1074/jbc.M110.144279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Young SG, Zechner R. Biochemistry and pathophysiology of intravascular and intracellular lipolysis. Genes Dev. 2013;27(5):459–484. doi: 10.1101/gad.209296.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi SY, Hirata K, Ishida T, Quertermous T, Cooper AD. Endothelial lipase: a new lipase on the block. J Lipid Res. 2002;43(11):1763–1769. doi: 10.1194/jlr.r200011-jlr200. [DOI] [PubMed] [Google Scholar]
- 15.Ishida T, Choi S, Kundu RK, Hirata K, Rubin EM, Cooper AD, Quertermous T. Endothelial lipase is a major determinant of HDL level. The Journal of clinical investigation. 2003;111(3):347–355. doi: 10.1172/JCI16306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robciuc MR, Tahvanainen E, Jauhiainen M, Ehnholm C. Quantitation of serum angiopoietin-like proteins 3 and 4 in a Finnish population sample. J Lipid Res. 2010;51(4):824–831. doi: 10.1194/jlr.M002618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gusarova V, Alexa CA, Wang Y, et al. ANGPTL3 blockade with a human monoclonal antibody reduces plasma lipids in dyslipidemic mice and monkeys. J Lipid Res. 2015 doi: 10.1194/jlr.M054890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, Gusarova V, Banfi S, Gromada J, Cohen JC, Hobbs HH. Inactivation of ANGPTL3 reduces hepatic VLDL-triglyceride secretion. J Lipid Res. 2015;56(7):1296–1307. doi: 10.1194/jlr.M054882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Powell-Braxton L, Veniant M, Latvala RD, Hirano KI, Won WB, Ross J, Dybdal N, Zlot CH, Young SG, Davidson NO. A mouse model of human familial hypercholesterolemia: markedly elevated low density lipoprotein cholesterol levels and severe atherosclerosis on a low-fat chow diet. Nature medicine. 1998;4(8):934–938. doi: 10.1038/nm0898-934. [DOI] [PubMed] [Google Scholar]
- 20.Millar JS, Maugeais C, Fuki IV, Rader DJ. Normal production rate of apolipoprotein B in LDL receptor-deficient mice. Arteriosclerosis, thrombosis, and vascular biology. 2002;22(6):989–994. doi: 10.1161/01.atv.0000018304.30943.06. [DOI] [PubMed] [Google Scholar]
- 21.Stitziel NO, Khera AV, Wang X, et al. ANGPTL3 Deficiency and Protection Against Coronary Artery Disease. J Am Coll Cardiol. 2017;69(16):2054–2063. doi: 10.1016/j.jacc.2017.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akinc A, Zumbuehl A, Goldberg M, et al. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nature biotechnology. 2008;26(5):561–569. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu YX, Liu L, Caffaro CE, Hirschberg CB. Inhibition of Golgi apparatus glycosylation causes endoplasmic reticulum stress and decreased protein synthesis. J Biol Chem. 2010;285(32):24600–24608. doi: 10.1074/jbc.M110.134544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bilheimer DW, Eisenberg S, Levy RI. The metabolism of very low density lipoprotein proteins. I. Preliminary in vitro and in vivo observations. Biochim Biophys Acta. 1972;260(2):212–221. doi: 10.1016/0005-2760(72)90034-3. [DOI] [PubMed] [Google Scholar]
- 25.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11(8):783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343(6166):84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cho SW, Kim S, Kim Y, Kweon J, Kim HS, Bae S, Kim JS. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2014;24(1):132–141. doi: 10.1101/gr.162339.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stephan ZF, Yurachek EC. Rapid fluorometric assay of LDL receptor activity by DiI-labeled LDL. J Lipid Res. 1993;34(2):325–330. [PubMed] [Google Scholar]
- 29.Mascanfroni ID, Takenaka MC, Yeste A, et al. Metabolic control of type 1 regulatory T cell differentiation by AHR and HIF1-alpha. Nat Med. 2015;21(6):638–646. doi: 10.1038/nm.3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koishi R, Ando Y, Ono M, Shimamura M, Yasumo H, Fujiwara T, Horikoshi H, Furukawa H. Angptl3 regulates lipid metabolism in mice. Nature genetics. 2002;30(2):151–157. doi: 10.1038/ng814. [DOI] [PubMed] [Google Scholar]
- 31.Grass DS, Saini U, Felkner RH, Wallace RE, Lago WJ, Young SG, Swanson ME. Transgenic mice expressing both human apolipoprotein B and human CETP have a lipoprotein cholesterol distribution similar to that of normolipidemic humans. Journal of lipid research. 1995;36(5):1082–1091. [PubMed] [Google Scholar]
- 32.Emerging Risk Factors C. Di Angelantonio E, Sarwar N, Perry P, Kaptoge S, Ray KK, Thompson A, Wood AM, Lewington S, Sattar N, Packard CJ, Collins R, Thompson SG, Danesh J. Major lipids, apolipoproteins, and risk of vascular disease. JAMA. 2009;302(18):1993–2000. doi: 10.1001/jama.2009.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schaefer EJ. Effects of cholesteryl ester transfer protein inhibitors on human lipoprotein metabolism: why have they failed in lowering coronary heart disease risk? Current opinion in lipidology. 2013;24(3):259–264. doi: 10.1097/MOL.0b013e3283612454. [DOI] [PubMed] [Google Scholar]
- 34.Tall AR. Plasma cholesteryl ester transfer protein. Journal of lipid research. 1993;34(8):1255–1274. [PubMed] [Google Scholar]
- 35.Agellon LB, Walsh A, Hayek T, Moulin P, Jiang XC, Shelanski SA, Breslow JL, Tall AR. Reduced high density lipoprotein cholesterol in human cholesteryl ester transfer protein transgenic mice. The Journal of biological chemistry. 1991;266(17):10796–10801. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1. FPLC analysis of plasma cholesterol profiles. (A). The plasma cholesterol profiles of Angptl3 WT and KO mice. Pooled plasma samples were from 12 WT and 12 KO mice. (B) and (C). The plasma cholesterol profiles of ob/ob mice (as in Fig. 1B) and hCETP/ApoB-100 transgenic mice (as in Fig. 2), respectively. Pooled plasma samples were from the mice injected with luciferase (Luc) or Angptl3 siRNAs at day 3 and 10.
Supplementary Fig. 2. Expression analysis of Angptl3 from obese mice injected with Luc or Angptl3 siRNAs. Blood samples from the mice as in Fig. 1B were used for mouse Angptl3 ELISA analysis. Protein levels of individual mouse (A) and group average (B) are the ratios of Angptl3 levels at the indicated days after injection over those at the pre-dose.
Supplementary Fig. 3. Expression analysis of Angptl3 in hCETP/ApoB-100 transgenic mice injected with Luc or Angptl3 siRNAs. Blood samples from the mice as in Fig. 2 were used for mouse Angptl3 ELISA analysis. Protein levels of individual mouse (A) and group average (B) are the ratios of Angptl3 levels at the indicated days after injection over those at the pre-dose.
Supplementary Fig. 4. The effect of Angptl3 silencing on LDL-C level is linked to LDLR. LAhB- LDLR_WT (LDLR+/+) (A), LAhB- LDLR_H (LDLR+/−) (B) and LAhB- LDLR_KO (LDLR−/−) (C) mice were injected with Luc or Angptl3 siRNAs. Blood samples were collected before the injection (baseline) and on Day 5 after the injection, and analyzed as in Fig. 1A. The Y axis represents the changes (%) (day 5 vs. baseline) of Angptl3 siRNA and Luc siRNA. p values (t-test) were calculated by comparing the changes of Angptl3 siRNA with those of Luc siRNA.
Supplementary Fig. 5. Angptl3 or PCSK9 levels from the mice injected with Luc, Angptl3 or/and PCSK9 siRNAs. LDLR+/− mice were injected with Luc, PCSK9, Angptl3 or PCSK9 plus Angptl3 siRNAs twice with 7 days interval at the total dose of 1 mg/kg. Blood samples were collected on day 10 following the first injection (as in Fig. 3B). The mouse livers (day 10) and blood samples (day 7 and 10) from the mice were used for real-time RT-PCR and ELISA analyses for Angptl3 and PCSK9 mRNA and protein expression, respectively. The mRNA levels (A) are the ratios of Angptl3 (right) or PCSK9 (left) over GAPDH. The protein levels (B) are the ratios of plasma Angptl3 (right) and PCSK9 (left) levels from the injection combinations over those in PBS control. N=4 for each group, n=2 for PBS. L=Luciferase siRNA. P=PCSK9 siRNA. A= Angptl3 siRNA.
Supplementary Fig. 6. Confirmation of ANGPTL3 and LDLR gene deletions. (A) PCR reactions were performed with the primers for ANGPTL3 gene deletion in Huh7 cells for ANGPTL3 gene deletion using the genomic DNA from the ANGPTL3 KO (lane 2) and scramble control (lane 1) Huh7 cells. PCR products were purified and used for T7 endonuclease I assay (lanes 3 and 4). (B) Whole cell extracts from scramble control and LDLR KO, as well as PCSK9 KO Huh7 cells (see below), were used for Western analysis with the antibodies against LDLR and β-actin. (C) The Huh7 cells with ANGPTL3 gene deletion or scramble control were analyzed for ApoB-100 accumulation in culture media (left) and Dil-LDL uptake (right). The Huh7 cells with LDLR and APOB gene deletions were used as controls. Averages and standard deviations were calculated from 6 samples.
Supplementary Fig. 7. Confirmation of ANGPTL3 gene deletion in Huh7 cells using CRISPR deep sequencing. Purified PCR products as in Supplementary Fig. 6A were used for CRISPR deep DNA sequencing. (A) Sequence alignments of PCR amplicons (Supplementary Fig. 6A) from ANGPTL3 scramble control Huh7 cells with ANGPTL3 reference sequence. (B) Sequence alignments of PCR amplicons from ANGPTL3 KO Huh7 cells.
Supplementary Fig. 8. Deletion of ANGPTL3 gene in Huh7 cells induced cellular long-chain TG and cholesterol accumulation. Total lipid extracts of ANGPTL3 scramble control (WT) and KO Huh7 clones were used for metabolite mass spectrometry analysis. Averages and standard deviations were measured from three WT clones and three KO clones (each with three duplicates). p values (t-test) were calculated by comparing the results from ANGPTL3 KO cells with those from ANGPTL3 WT cells. TAG, triacylglycerol. DAG, diacylglycerol. MAG, monoacylglycerol.