Calcium-activated chloride channels (CaCCs) encoded by TMEM16A1–3 control neuronal signaling, smooth muscle contraction, airway and exocrine gland secretion, and rhythmic movements of the gastrointestinal system4–7. To understand how CaCCs mediate and control anion permeation to fulfill these physiological functions, knowledge of the mammalian TMEM16A structure and identification of its pore-lining residues is essential. TMEM16A forms a dimer with two pores8,9. Thus far, CaCC structural analyses relied on homology modeling of a fungal homolog nhTMEM16 that functions primarily as a lipid scramblase10–12 and subnanometer-resolution electron cryo-microscopy (cryo-EM)12. Here, we present de novo atomic structures of the transmembrane domains of mouse TMEM16A in nanodiscs and in lauryl maltose neopentyl glycol (LMNG) as determined by single-particle cryo-EM. These structures reveal the ion permeation pore and represent different functional states. The LMNG structure has one Ca2+ ion resolved within each monomer with a constricted pore, likely corresponding to a closed state because CaCC with single Ca2+ occupancy requires membrane depolarization to open13. The nanodisc structure has two Ca2+ ions per monomer and its pore in a closed conformation likely reflects channel rundown, a gradual loss of channel activity following prolonged CaCC activation in 1 mM Ca2+. Structure-based mutagenesis and electrophysiological studies identified ten residues distributed along the pore that interact with permeant anions and affect anion selectivity as well as seven pore-lining residues that cluster near pore constrictions and regulate channel gating. Together, these data clarify the basis of CaCC anion conduction.
Our screen of deletion constructs of the mouse TMEM16A splice variant1 that contains exon ‘a’ but not exons ‘b-d’ identified a C-terminally truncated mutant with high expression and stability (Extended Data Fig. 1a) and channel function (Extended Data Fig. 1b). Its increased Ca2+ sensitivity is reminiscent of gating changes observed in other TMEM16A truncation mutants14,15. Reducing negatively charged lipids via poly-L-lysine (PLL) caused desensitization by reducing Ca2+ sensitivity without altering the plateau current (Extended Data Fig. 1c). This truncated TMEM16A including amino acids 1–903 was purified in detergent followed by reconstitution into lipid nanodiscs (Extended Data Fig. 1d) or kept solubilized in LMNG (Extended Data Fig. 1h) for further analyses. N-[4-(7-diethylamino-4-methyl-3-coumarinyl)phenyl]maleimide (CPM) assay16,17 revealed a similar melting temperature (Tm) for Ca2+-free TMEM16A while stepwise increase of Ca2+ concentration caused Tm to gradually increase for TMEM16A in nanodisc but not for LMNG-solubilized TMEM16A (Extended Data Fig. 1e, i); this stabilizing effect of Ca2+ reached plateau at ~1 μM Ca2+ (Extended Data Fig. 1e), as did channel activation (Extended Data Fig. 1b). The CPM assay thus revealed that TMEM16A in nanodiscs or LMNG differ in their Ca2+ binding characteristics.
Negative stain electron microscopy (EM) of LMNG-solubilized TMEM16A and dodecylmaltoside (DDM):cholesterol hemisuccinate (CHS)-solubilized TMEM16A reconstituted into lipid nanodiscs revealed homogeneous preparations suitable for high resolution structural studies (Extended Data Fig. 1f, g, j, k). Using single-particle cryo-EM we determined the structures of TMEM16A to better than 4 Å nominal resolutions (Extended Data Fig. 2, 3, 4b). Local resolution analysis showed that the core transmembrane domains have the highest (better than 4 Å) resolution and the extracellular and cytoplasmic domains have the poorest (Extended Data Fig. 2f, 3f), suggesting conformational flexibility of these regions. The resolution of the LMNG-solubilized TMEM16A reconstruction is anisotropic because images of particles oriented along certain axes were lacking (Extended Data Fig. 3c, h, i). Because the missing views are within a small angular range, there is no distortion regarding the definition of the transmembrane helices with respect to the membrane. The anisotropy is considerably reduced by adding particles from a dataset in which a monoclonal fragment of antigen binding (Fab) isolated from a human naïve Fab phage library18,19 is bound to TMEM16A (Extended Data Fig. 3b, h, i). The final density maps allowed reliable assignment of most of the side chains in all ten transmembrane segments and enabled building de novo atomic models of the transmembrane segments of dimeric TMEM16A in both nanodisc and LMNG (Fig. 1a, b; Extended Data Fig. 4a, c, d, 5a–f). Cross-validation between the atomic model and the density maps of the transmembrane domains suggested that the average resolutions are around 4 Å (Extended Data Fig. 2g, 3g). The nanodisc-reconstituted TMEM16A is structurally similar to the subnanometer-resolution structure of digitonin-solubilized TMEM16A12 but missing part of the C-terminal cytoplasmic domain owing to the truncation construct used in our study (Extended Data Fig. 5g–i).
Figure 1.

Structure of TMEM16A in nanodiscs. a, EM density map of TMEM16A (sharpened, green and yellow) over a density map with a lower threshold (unsharpened) in light grey for nanodiscs. b, TM1–10 overlayed with EM density map of TMEM16A (sharpened, light grey). c–e, The channel pore (marked in orange) lined with N542 and V595 (in purple) that affect both anion selectivity and channel gating, five other residues that affect channel gating (in magenta) (see Fig. 4) and eight other residues that affect ion selectivity (in green) (see Fig. 3).
To locate the pore, we used the HOLE program20 to delineate potential water-filled profiles in these structures. By testing whether anion selectivity could be altered by mutations of residues lining the potential pore within each monomer or the potential pore at the dimer interface that is associated with lipids, we identified the pore surrounded by transmembrane helices TM3–8 (Fig. 1c–e and Extended Data Fig. 6d). This pore is not exposed to the lipid bilayer as proposed21–24. Similar to a previous study12, our cryo-EM studies revealed a pore too narrow for the passage of permeant anions. It remains possible that some open conformation of the pore is partly lined by lipids.
A comparison of nanodisc-reconstituted and LMNG-solubilized TMEM16A density maps (Fig. 2a, b) revealed differences primarily in the positions and orientations of TM3, TM4, and TM6 (Fig. 2c–f; Extended Data Fig. 7a–f). These pore-lining helices in the TMEM16A channel are also displaced noticeably from the positions of the equivalent helices of the nhTMEM16A lipid scramblase10. We found some helical distortions in the nanodisc-reconstituted TMEM16A starting from G640 and extending towards the lower part of TM6 (Extended Data Fig. 4a, 7a, b). In the LMNG reconstruction, the lower part of the TM6 could not be resolved beyond G640 (Extended Data Fig. 4a, 7d, e). These observations are in agreement with mutagenesis studies implicating G640 as a flexible hinge that affects channel gating13. Additionally, the two structures differ in the orientation of TM3 and in the arrangement of the TM5-TM6 loop that is near the pore entrance and harbors K599 and R617 important for anion selectivity25, and the TM9-TM10 loop that harbors R784 important for anion selectivity25 and packs against the TM5-TM6 loop (Extended Data Fig. 7g–l). We observed densities consistent with the presence of ordered lipids near the dimer interface but not near the pore of both nanodisc-reconstituted and LMNG-solubilized TMEM16A (Fig. 1a; Extended Data Fig. 7b, c, e, f). And the number of ordered lipids observed in the nanodisc structure is greater than that in the LMNG structure. It is possible that lipids may stabilize TMEM16A in the more native environment of nanodiscs.
Figure 2.

Differences in pore-lining helices and Ca2+ ion binding in the two structures. a–b, Density map of nanodisc-reconstituted TMEM16A (unsharpened, green and yellow) overlayed with that of LMNG-solubilized TMEM16A (unsharpened, blue). c, Conformational differences of pore-lining helices. d–f, Comparisons of TM3–8 around the pore (orange asterisk) in two structures. g–i, Two Ca2+ ions (orange spheres) coordinated by five acidic residues and one asparagine (N646) in nanodisc-reconstituted TMEM16A (g, h) versus one Ca2+ ion surrounding by four acidic residues in LMNG-solubilized TMEM16A (i, h) (Methods).
These two structures showed a surprising difference near the putative Ca2+ binding sites even though both structures were determined in the presence of 1 mM calcium. Consistent with previous studies10,26,27, the acidic residues E650, E698, E701, E730 and D734 group together to form two Ca2+ binding sites in nanodisc-reconstructed TMEM16A (Fig. 2g, h). Comparisons of the maps and models suggest that these residues are well organized in nanodisc-reconstructed TMEM16A, but less so in LMNG-solubilized TMEM16A (Fig. 2h, i). The difference maps between the experimental map and the map from the refined structure that do not contain Ca2+ ions showed a clear omit density that we assigned as a bound Ca2+ ion, for LMNG-solubilized TMEM16 at σ = 13 and for nanodisc-reconstituted TMEM16A at σ = 8. The location of this bound Ca2+ ion matches well with one of the two bound Ca2+ ions in the crystal structure of nhTMEM16A10. In the nanodisc structure, there is another omit density (σ = 10) nearby that may correspond to the second bound Ca2+ ion (Fig. 2h). In contrast, in the LMNG structure, no other significant omit density was found nearby, suggesting there is only one Ca2+ ion bound (Fig. 2h). Notably, E650 on the lower half of TM6 is not resolved in LMNG-solubilized TMEM16A (Fig. 2h; Extended Data Fig. 4a, 7d). Our mutagenesis studies showing that alanine substitution of E650 raises the Ca2+ concentration for half maximal activation (EC50) and reduces the Hill coefficient from two to one13,26 indicate that this residue in TM6 is critical for Ca2+ binding. It is therefore understandable that the more flexible TM6 harboring E650 results in partial Ca2+ occupancy of LMNG-solubilized TMEM16A (Fig. 2h, i), raising the possibility that some of the conformational differences between the two structures could reflect different channel conformations. It appears that the native-like lipid environment of nanodiscs allowed stabilization of TM6 so that two Ca2+ ions can be fully coordinated by their binding sites (Fig. 2g, h).
Based on our structures (see Extended Data Fig. 8 for structural determination details), mutagenesis of residues lining the pore revealed that a subset of these residues affected anion selectivity (Fig. 3a–c). Besides R511 on TM3 and K599 on the TM5-TM6 loop important for anion selectivity25, and K584 on TM5 that partially accounts for the selectivity for anions over cations28 (Fig. 3b), we tested alanine substitutions of 24 other pore-lining residues including N542 and D550 on TM4, N587 and V595 on TM5, Q705 and F712 on TM7, and S635 on TM6 (Fig. 3a, b and Extended Data Fig. 6d). The permeability ratios determined by replacing external Cl− with I− or SCN− were reduced by 40% by S635A but increased by 40–130% by the other six mutations (Fig. 3d–f). Whereas the side chains of Q705 and F712 do not appear to be pointing into the pore, it remains an open question whether they may reorient and face the pore as the channel opens. These ten pore-lining residues identified thus far for their roles in anion selectivity (Fig. 3c) may reflect an extended selectivity filter, or multiple pore regions for permeant ion interactions.
Figure 3.
The pore of TMEM16A in nanodiscs. a, b, The solvent-accessible (blue mesh) pore lined with residues important for anion selectivity (salmon sticks, labeled in b) and those without detected effects (blue backbone) (N and P values are given in Methods and Extended Data Fig. 6d). c, Pore radius along the Z axis. For those residues not completely resolved, a solid line and a dotted line based on positioning two rotamers bracket the pore radius estimate. d, Representative recordings under bi-ionic conditions (repeated 7 times with similar results). e–f, Permeability ratios for iodide/chloride (e), or thiocyanate/chloride (f) (Methods) assessed via one-way ANOVA followed by Bonferroni post-hoc test; **** designates p < 0.0001; mean ± SEM.
We noticed narrow constrictions of the pore of LMNG-solubilized TMEM16A (Fig. 4a–c), likely representing a stable closed conformation. To test whether residues facing the pore affect channel gating possibly by altering the relative stability of the open versus closed states, we examined alanine substitutions of 21 residues including N542 and I546 on TM4, Y589 and I592 on TM5, and F708 on TM7. These five alanine mutations increased the apparent Ca2+ sensitivity whereas alanine substitution of V595 on TM5 and L639 on TM6 decreased the apparent Ca2+ sensitivity (Fig. 4d–f and Extended Data Fig. 9c). In contrast to the pore-lining residues important for anion selectivity that are spread over more than 25 Å along the length of the pore (Fig. 3c), these seven residues affecting the Ca2+ dependence of channel activation are all located within ~10 Å from the pore constrictions (Fig. 4c), indicating that these residues at the constricted pore influence the stability of the channel conformation important for gating.
Figure 4.
The pore of LMNG-solubilized TMEM16A. a, b, The solvent-accessible (red mesh) pore lined with residues affecting channel gating (red sticks, labeled in b) and those without effects (dark blue backbone). c, Pore radius (salmon; pore radius of nanodisc-reconstituted TMEM16A in green) along the Z axis. For those residues not completely resolved, a solid line and a dotted line bracket the pore radius estimate (Methods). d, Representative recordings of inside-out patch exposed to increasing Ca2+ levels (repeated 8 times with similar results). e, Normalized currents fit to the Hill equation. The Hill coefficient for L639A is 2.34 ± 0.11. f, EC50 values (N and P values are given in Methods and Extended Data Fig. 9c) compared via one-way ANOVA followed by Bonferroni post-hoc test; **** designates p < 0.0001; mean ± SEM.
TMEM16A-CaCC likely have multiple open states2,13 and closed states13 (Fig. 5a). With the same anion concentration on both sides of the membrane, occupancy of the first Ca2+ binding site allows the channel to open when the membrane potential is depolarized to positive values13, whereas CaCC with double Ca2+ occupancy activates in a voltage independent manner13. The currents conducted by the C-terminally truncated TMEM16A also displayed voltage dependence at 30 nM Ca2+ but not 1 mM Ca2+ (Fig. 5b). Given the single Ca2+ occupancy of TMEM16A in LMNG, this structure is in a closed conformation probably because of the lack of depolarization. Moreover, we found that the K584Q mutation altered the ion selectivity in low but not high Ca2+ (Extended Data Fig. 9a, b), suggestive of multiple open states with differences in the permeation pathway2 – a finding of potential relevance to the different mutagenesis results8,9,28 regarding this pore-lining residue (Fig. 3a–c). The nanodisc-reconstituted TMEM16A structure likely corresponds to a closed conformation due to channel rundown following prolonged activation in 1 mM Ca2+ (Fig. 5c–e)12. Intrigued by the involvement of the hydrophobic V595 in anion selectivity, we tested multiple substitutions and found bulky basic residues to be less effective than alanine in enhancing the relative permeability of large anions (Extended Data Fig. 6a–d). These studies underscore the importance of structure-based functional analyses.
Figure 5.
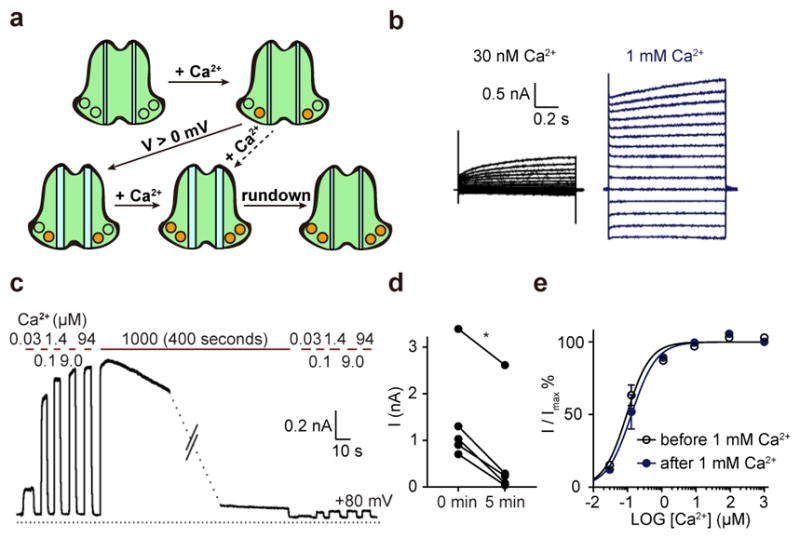
Different conformations of TMEM16A. a, Schematic showing the dimeric CaCC in a calcium-free closed conformation (top left panel), CaCC with single calcium occupancy that remains closed (top right panel) until depolarization causes it to open (bottom left panel), CaCC with double calcium occupancy that opens in a voltage-independent manner (bottom middle panel), and channel rundown following prolonged activation (bottom right panel). b–e, TMEM16A with C-terminal truncation yielded voltage-dependent current in 30 nM Ca2+ but voltage-independent current in 1 mM Ca2+ (b), while prolonged activation causes channel rundown (c) (N = 5; p = 0.03) (d) without significant change in Ca2+ sensitivity (EC50 = 0.11 ± 0.02 μM before exposure to 1 mM Ca2+, EC50 = 0.15 ± 0.04 μM after exposure to EC50 = 0.15 ± 0.04 μM, p = 0.13, N = 4, two-tailed Wilcoxon test) (e); mean ± SEM.
In this study, we identify for the first time pore-lining residues of CaCC based on structural analyses of TMEM16A as well as mutagenesis of these highly conserved pore-lining residues (Extended Data Fig. 10). Unlike previous efforts searching among positively charged residues for those involved in anion permeation12,25,28, we tested residues facing water-filled potential pore structures to delineate the permeation pathway based on the involvement of pore-lining residues in anion selectivity. For ion permeation and channel gating, we classify pore-lining residues into two groups: one group is distributed along the entire pore and is important for anion selectivity, and the other group is clustered together and is important for gating. For the first group, we validated the ability of ten pore-lining residues to interact with permeant ions by showing that mutations of these residues alter anion selectivity in this study and in two previous studies25,28. Visualization of the pore-lining residues and functional assessments of their role in anion selectivity thus firmly establishes their pore assignment. For the second group, we demonstrate a role for seven residues facing the constricted pore in channel gating, indicating that channel opening likely involves displacement of these residues that affect the relative stability of the open and closed states. Our study provides an essential framework to guide further studies of gating and anion permeation of calcium-activated chloride channels.
METHODS
Protein expression and purification
Truncated mouse TMEM16A was C-terminally fused to a 3C consensus sequence, a Strep-tag II peptide, and a GFP moiety and expressed in HEK293 GnTi− cells using the BacMam system as described previously29,30.
Protein purification and sample processing were carried out at 4°C. For nanodisc-reconstituted preparations, approximately 10 g of cell pellet (from about 1 L culture) was lysed by stirring for 40 min in 200 ml hypotonic buffer containing 50 mM TrisNO3 (pH 9.0) supplemented with 0.1 mg/ml DNase, 1× complete protease inhibitor cocktail (Roche) and 1 mM phenylmethylsulfonyl (PMSF). The membrane fraction was collected by centrifugation at 30,000g for 30 min, and then homogenized with a Dounce homogenizer in extraction buffer containing 50 mM TrisNO3 (pH 9.0), 150 mM KNO3, and 10 mM CaCl2 supplemented with 0.1 mg/ml DNase, 1× complete protease inhibitor cocktail, and 1 mM PMSF. Protein was extracted in 200 ml extraction buffer plus 0.5% n-dodecyl-b-d-maltopyranoside (DDM) and 0.1% cholesteryl hemisuccinate (CHS) with gentle stirring for 2 h. The insoluble fraction was removed by centrifugation at 30,000g for 30 min. The recombinant protein was affinity purified with an anti-GFP nanobody immobilized on CNBr-activated sepharose resin (GE Healthcare) in wash buffer containing 10 mM TrisNO3 (pH 9.0), 150 mM KNO3, 1 mM CaCl2, 0.05% DDM, and 0.01% CHS supplemented with a 0.1 mg/ml lipid mixture containing 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoylsn-glycero-3-phosphoethanolamine (POPE) and 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (POPS) at a ratio of 3:1:1. The purified protein was recovered by incubation with 1.5 CV wash buffer containing 0.5 mM dithiothreitol (DTT) and 50 mg 3C protease overnight. To reconstitute the protein in nanodiscs, the lipid was prepared as described previously in buffer containing 10 mM TrisNO3 and 150 mM KNO3. After purification, the protein sample was mixed with MSP2N2 and soy PC (Avanti) at a molar ratio of TMEM16A monomer: MSP2N2: soy PC = 1:4:100. The mixture was allowed to equilibrate for 1 h and Bio-beads SM2 (Bio-Rad) were added to the mixture three times within 24 h to gradually remove detergents from the system. Afterwards, the sample was filtered through a 0.45 μm filter, and reconstituted protein was separated on a Superdex-200 column in column buffer containing 10 mM TrisNO3 (pH 9.0), 150 mM KNO3, and 1 mM CaCl2. The peak fraction was collected and concentrated to 0.6–0.7 mg/ml using a 100 kDa MWCO Amicon Ultra filter device (Millipore). The lipid compositions used for protein purification and nanodisc reconstitutions are chosen based on previous studies30,31.
The same purification procedures were used to prepare LMNG solubilized preparations except 5 mM LMNG was in the extraction buffer and 0.02 mM LMNG was used in the wash buffer instead of DDM and CHS. After digestion by 3C, the protein was collected and immediately separated on a Superdex-200 column in column buffer containing 10 mM TrisNO3 (pH 9.0), 150 mM KNO3 and 1 mM CaCl2. The peak fractions were collected and concentrated to 1 mg/ml using 100-kDa Amicon Ultra filter device.
N-[4-(7-diethylamino-4-methyl-3-coumarinyl)phenyl] maleimide (CPM) assay of TMEM16A stability as function of calcium concentration
To assess the stability of TMEM16A in the presence of a range of Ca2+ concentrations via CPM assay16, we dialyzed TMEM16A in nanodiscs or LMNG in solutions containing buffered calcium made (at pH7.5 buffered with HEPES) by combining Ca(OH)2 and EGTA to a ratio predicted by CaBuf softward (KU Leuven). Calcium concentrations of the solutions containing dialyzed TMEM16A protein preparations were subsequently measured directly against calcium standards, using a Fluo-8 fluorescent assay in a BioTek Synergy H4 plate reader. MSP proteins have no cysteines and empty nanodiscs generate no CPM fluorescence signals17, so the CPM signal measured via a fluoromax fluorometer (excitation at 387 nm, emission at 463 nm) provides a readout of cysteine exposure upon unfolding of TMEM16A at temperature ranging from 25°C to 75°C.
Isolation of monoclonal Fab (fragment antigen binding) for TMEM16A
To obtain Fabs to be used as fiducial markers for cryo-EM studies, TMEM16A was reconstituted into nanodiscs to provide a native-like lipid bilayer environment for presentation of the calcium-activated chloride channel to the recombinant Fab library. Once reconstituted, TMEM16A in nanodiscs was used for in-solution selections. A subtractive (nanodisc alone) and competitive panning strategy was used to find selective binders for TMEM16A, from our fully human naïve Fab phage library18,19. The selections yielded 23 clones with a signal 4 times higher than the background in an ELISA assay. Out of these 23 clones, 3 clones showed a very specific signal for TMEM16A and almost no signal for empty nanodiscs. Clone 2F11 was then expressed and purified, and complex formation with TMEM16A was tested with size exclusion chromatography. Fab(2F11) and TMEM16A indeed formed a complex which was also demonstrated by two-dimensional class averages of electron microscopy negative stain images (Extended Data Fig. 3b).
Electrophysiology
HEK293 cells were maintained at 37°C and 5% CO2 in Dulbecco’s Modified Eagle’s Medium supplemented with 10% Fetal Bovine Serum and 1% penicillin/streptomycin, and were passaged upon reaching confluency (every 2–4 days) by digestion with 0.05% Trypsin-EDTA. 24 hours before recording, cells were transiently transfected with TMEM16A constructs using Lipofectamine 2000 (Invitrogen). On the morning before recording, cells were re-plated onto poly-L-lysine coated coverslips using trypsin and allowed to settle for at least 1 hour.
For patch clamp electrophysiology, coverslips were transferred into a recording bath solution containing 140 mM NaCl, 10 mM HEPES, and 5 mM EGTA, with pH adjusted to 7.2 with NaOH and osmolarity adjusted to 305–315 mOsm with mannitol. All recordings were made at room temperature. For whole cell recordings, intracellular solutions contained 140 mM NaCl, 10 mM HEPES, 1 mM CaCl2 and 2 mM MgCl2 with pH adjusted to 7.2 with NaOH and osmolarity adjusted to 310 mOsm with mannitol. For anion selectivity measurements, recordings from HEK293 cells transiently expressing wild-type or mutant TMEM16A under bi-ionic conditions using whole-cell patch-clamp and subjected to one-second ramp protocols between −80 and +80 mV. Extracellular solutions are exchanged between voltage ramp commands and contain 140 mM NaCl, 140 mM NaI, or 140 mM NaSCN. Permeability ratios for iodide/chloride ions (2.70 ± 0.09, N = 28 for WT; 3.73 ± 0.12, N = 11 for N542A; 5.32 ± 0.25, N = 9 for D550A; 4.85 ± 0.16, N = 7 for N587A; 4.00 ± 0.16, N = 9 for V595A; 1.82 ± 0.08, N = 7 for S635A; 4.65 ± 0.06, N = 5 for Q705A; 4.45 ± 0.10, N = 6 for F712A), or thiocyanate/chloride ions (5.47 ± 0.21, N = 28 for WT; 8.99 ± 0.24, N = 8 for N542A; 13.33 ± 0.66, N = 9 for D550A; 9.10 ± 0.29, N = 7 for N587A; 9.96 ± 0.30, N = 9 for V595A; 3.24 ± 0.26, N = 7 for S635A; 13.36 ± 0.32, N = 5 for Q705A;10.78 ± 0.34, N = 6 for F712A), are calculated by fitting the GHK equation to currents recorded using voltage ramps (Extended Data Fig. 6d). For Fig. 3d, the reversal potential (at 0 nA) is −46 mV for WT and −57 mV for N587A in external SCN−, −27 mV for WT and −40 mV for N587A in external I−, −2 mV for WT and 0 mV for N587A in external Cl−.
For inside out patch recording, intracellular solutions consisted of normal bath solution supplemented with 2 mM MgCl2. Intracellular solutions with varying calcium concentrations were generated by buffering calcium ions with EGTA at pH 7.2 and were made by combining Ca(OH)2 and EGTA to a ratio predicted by CaBuf software (KU Leuven) and then adding 140 mM NaCl or NMDG-Cl and 10 mM HEPES to the desired volume. Calcium concentrations were subsequently measured directly against calcium standards using a Fluo-8 or Fluo-8FF fluorescent assay in a BioTek Synergy H4 plate reader. To assess calcium sensitivity of channel activation via recordings from inside-out patches excised from HEK293 cells transiently expressing wild-type or mutant TMEM16A, the membrane potential was held at +60 mV, and patches were exposed to intracellular solutions containing 140 mM NaCl and increasing concentrations of free Ca2+. Solutions (a)–(f) in Fig. 4d contained 150 nM, 300 nM, 400 nM, 600 nM, 5.5 μM, and 1 mM free Ca2+, respectively. Ca2+-dependent currents from wild-type (WT) and mutant TMEM16A channels were normalized to their maximum values and fit to the Hill equation. The EC50 values in nM (796 ± 66, N = 10 for WT; 295 ± 24, N = 10 for N542A; 148 ± 31, N = 6 for I546A; 192 ± 13, N = 6 for Y589A; 252 ± 17, N = 6 for I592A; 2036 ± 52, N = 7 for V595A; 2008 ± 63, N = 7 for L639A; 205 ± 30, N = 8 for F708A) were compared using one-way ANOVA followed by the Bonferroni post-hoc test for significance (Extended Data Fig. 9c).
External or internal solutions using alternate anions were made by replacing NaCl with the indicated sodium salt at equimolar concentrations. Patch pipettes were pulled from 1.5/0.86 (OD/ID) glass and polished to 2–2.5 MΩ resistance (inside-out patch) or 3–5 MΩ (whole cell patch). Perfusion exchange was performed using a VM-8 perfusion apparatus with Octaflow software (ALA Scientific). Data were collected at 10 kHz sampling rate and low pass filtered online at 1 kHz. Recordings were made using an Axon Instruments Multiclamp 700 with Digidata 1440 and were collected into pClamp10 software. All patch clamp seals were allowed to reach at least 3 GΩ resistance before patch rupture, but typical seal resistance usually exceeded 10 GΩ. All recordings were made using a 1 M KCl agar bridge to prevent baseline fluctuation at the reference electrode.
For Data analysis, all offline data analysis for patch clamp recording was performed using Graphpad Prism 6, Clampfit 10, and Microsoft Excel. Permeability ratios were determined from bi-ionic conditions using a reduced form of the Goldman-Hodgkin-Katz voltage equation:
where PX represents relative permeability of ion species “X” and F, R and T have their usual thermodynamic meanings. Concentration-dependence curves for Ca2+ were generated by fitting data to an equation of the form:
Where I/IMAX denotes current normalized to the maximum amplitude in the highest [Ca2+] tested, KD denotes the dissociation constant for that ion and nH denotes the Hill coefficient. Statistical analysis for data acquired using patch clamp employed one-way ANOVA followed by Bonferroni post-hoc tests for statistical significance. p < 0.01 was used as a threshold for significance.
Electron microscopy
For negative staining, 2.5 μl of purified TMEM16A at a concentration of 0.02 mg/ml were placed on home-made continuous carbon grids glow discharged for 30 sec and coated with 0.75% (w/v) uranyl formate for 30 sec. Grids were imaged on a Tecnai T12 microscope (FEI Company) operated at 120 kV with a 4k×4k scintillator-based charge-coupled device camera UltraScan 4000, (Gatan). A nominal magnification of × 52,000, corresponding to a pixel size of 2.21 Å on the specimen, and a defocus around −1.5 μm were used for image recording.
For cryo-EM, 2.5 μl of purified TMEM16A at a concentration of approximately 0.5 mg/ml were applied to holey carbon grids (Quantifoil 400 mesh Cu R1.2/1.3) glow-discharged for 30 sec. After 30 sec incubation on the grids at 4°C under 100% humidity, grids were blotted with Whatman #4 filter paper for 6–8 sec and plunge-frozen in liquid ethane cooled by liquid nitrogen using a FEI Mark III Vitrobot. Grids were transferred to an FEI Titan Krios electron microscope equipped with a field emission electron source and operated at 300 kV at the Howard Hughes Medical Institute Cryo-EM facility at Janelia Research Campus.
Images were recorded using SerialEM32 in super-resolution mode using a Gatan K2 Summit direct electron detector (Gatan) at a calibrated magnification of 29,000X, yielding a physical pixel size of 1.02 Å (0.51 Å super resolution pixel size). A dose rate of 10 electrons per physical pixel per second and an exposure of 8 sec were used for data collection with 0.2 sec subframes to give a total dose of 80 electrons per Å2.
Data processing
For negative-stain data, Simplified Application Managing Utilities for EM Labs (SAMUEL) scripts33 were used for particle picking and 2D classification. In this package, 2D classification was carried out by ten cycles of correspondence analysis, k-means classification, and multi-reference alignment using SPIDER operations ‘CA S’, ‘CL KM,’ and ‘AP SH’34.
For Cryo-EM data, drift correction was performed using MotionCor235 and images were binned 2×2 by Fourier cropping to a pixel size of 1.02 Å. The contrast transfer function (CTF) was estimated using CTFFIND436 (LMNG) or GCTF37 (Nanodisc) using motion-corrected sums without dose-weighting. Motion-corrected sums with dose-weighting were used for all other image processing. RELION (1.4 and 2.1beta1, in which the bug on gold standard refinement procedure was fixed)38 was used for 2D classification, 3D classification, and refinement procedures.
For the LMNG dataset, around 4000 particles were manually picked and classified by 2D classification in SAMUEL to generate the templates for automatic particle picking with samautopick.py. 533,545 particles were identified after manually inspection of auto-picked particles. These particles were subjected to 2D classification, using RELION 1.4. A total of 342,875 particles from 2D classes that show clear structural features were combined and binned 4 to a pixel size of 4.08 Å for 3D classification. The crystal structure of nhTMEM16A (PDB:4WIS) was converted to a density map with e2pdb2mrc.py39 and low pass filtered to 60 Å as the initial model for 3D classification and refinement using RELION 1.4 without applied symmetry. After 3D classification, images without binning with a pixel size of 1.02 Å were used for further auto-refinement with a C2 symmetry. Two of five classes, which gave maps with resolution better than 5 Å (4.6 Å and 4.9 Å), were combined together for another round of 3D refinement to generate a 4.0 Å map. The resolution was further improved to 3.8 Å by adding particles from other three classes.
The TMEM16A with Fab bound dataset was processed with the same strategy as for TMEM16A in LMNG. Starting with 338,705 particles from automatic particle picking and 2D classification by RELION, 132,444 particles from 4 classes that present clear side views were combined together for 3D auto-refinement. Then the Fab density for each particle was subtracted with script projection_subtraction.py (written by Danniel Arsanow in Cheng lab at UCSF) based on final data.star file containing Eular angle information from 3D refinement. The 132,444 subtracted particles without Fab density were combined with the 342,875 particles from all five 3D classes of TMEM16A in LMNG dataset and processed for 3D auto-refinement. After refinement have converged, the final data set was subjected to ‘post-processing’ in RELION, in which a soft mask was calculated and applied to the two half-maps before calculating the corrected FSC and the resolution was estimated using FSC=0.143 criterion40 on the corrected FSC curves in Fourier space in which the influences of the mask were removed. During this step, temperature-factor estimation and map sharpening were also performed to get final map at resolution of 3.4 Å.
For the Nanodisc dataset, particle picking performed with the Gautomatch program (developed by Dr. Kai Zhang, MRC Laboratory of Molecular Biology, Cambridge, UK, http://www.mrc-lmb.cam.ac.uk/kzhang/Gautomatch/) with templates from 2D classes of the LMNG dataset generated 927,414 particles in total. All particles were extracted and binned 4 (pixel size 4.08 Å) and 2D classified using RELION. 341,875 particles selected from good 2D classes were un-binned with a pixel size of 1.02 Å, and were used for 3D refinement using an initial model low pass filtered from the LMNG structure that had been low pass filtered to a resolution of 60 Å, giving rise to a 5.5 Å map. This 5.5 Å map was low pass filtered to 10 Å as the initial model to apply for 3D classification without applied symmetry for all particles with pixel size of 4.08 Å. Two of seven classes gave a map with improved resolution (4.7 Å and 5.1 Å respectively) after 3D auto-refinement with C2 symmetry. These two classes were combined and another 3D auto-refinement was run to generate the map at a resolution of 4.6 Å. Similar to LMNG dataset, the ‘post-processing’ in RELION was applied to the final data set and the final map was generated at resolution of 3.8 Å and used for model building.
Directional FSC is calculated using home-made script (see below). Local resolution estimates were calculated with unsharpened raw density maps using ResMap41.
Model building
Ab initio model building was carried out in COOT42 for all transmembrane helices and small parts of soluble domains. Initial placement of TM helices was made based on sequence alignment with the nhTMEM16A (PDB accession 4WIS)10. For major part of soluble domain, the resolutions are insufficient for assignments. For these parts, we used homology model that was generated with swiss-model based on a sequence alignment with the crystal structure of nhTMEM16A10, and refined against the EM map on MDFF43. For the transmembrane region, the model was refined both in real space with Phenix.real_space_refine44 and also in Fourier space with REFMAC45, followed by further manual adjustment in COOT. This process was repeated until Ramachandran validation was satisfied. Side chain assignments were further validated by using difference densities calculated by subtracting a simulated map based on the Ca only model from the experimental map. The model building following similar procedure was carried out independently by four individuals and cross-validated to ensure the accuracy of assignments. Additional validation of the final model was carried out using MolProbity46 and EMRinger47.
For cross-validation, the final model was refined against one of the half maps generated in RELION. FSC curves were then calculated between the refined model and half map 1 (‘work,’ used in test refinement), the refined model and half map 2 (‘free,’ not used in test refinement), and the refined model and the summed map. There is no notable gap between ‘work’ and ‘free’ FSC curves, indicating little effect of over-fitting. In this process, the final model was displaced and then refined against the half-map (work). Then this model was used for calculation of cross-validation. Using FSC = 0.5 criterion, the resolution estimated from cross-validation is about 4.5 Å for nanodisc-reconstituted TMEM16A and 3.8 Å for LMNG-solubilized TMEM16A.
HOLE20 was used to calculate the pore profile shown in Fig. 3 and Fig. 4. The narrowest point is defined as the location of the pore with the smallest radius. For those pore-lining residues that are not completely resolved, a solid line and a dotted line are drawn based on positioning two rotamers that will provide the largest and smallest estimate for the pore radius at that location. The program PyMol48 was used to calculate surface representation of electrostatic potentials.
Difference map calculation
We used “omit density” to assign the position of one or two Ca2+ ions. It was calculated by subtracting a map calculated based on the refined atomic model without any ligand from the experimental density map, i.e. density – model. In this difference map, any experimental map features not included in the atomic model, such as densities contributed by bound ions, appear as positive densities. The map was calculated using PHENIX44 with the command line option “phenix.real_space_diff_map” available in the 2017-07-07 nightly build: first a theoretical map was calculated from LMNG and nanodisc structures at a resolution of 4 Å and 3.8 Å resolution, respectively, corresponding to the resolution at which the model refinement was performed. The electron scattering factors were used. Then the calculated map was subtracted from the respective experimental electron density maps. The difference maps clearly show the presence of one Ca2+ ion in the LMNG structure and two in the nanodisc one. Ca2+ ion positions were assigned based on the residual density between the actual experimental data and the density calculated from the model and, with a threshold of σ = 7 and σ = 10 for densities corresponding to nanodisc and LMNG respectively (Fig. 2h).
Calculation of directional Fourier shell correlation (dFSC)
The Fourier shell correlation (FSC) between two half-maps was calculated as described previously49,50, except conical shells were used instead of spherical shells to allow calculation of the FSC in different directions. Cones with an apex angle of 20° were sampled on a 500-point Fibonacci spherical grid where the apex of a particular cone is at the center of the sphere and the central axis of that cone lay on the vector joining the center of the sphere with a grid point on the surface of the sphere. This analysis resulted in 500 1D directional FSC (dFSC) curves, which were averaged to generate a global average dFSC curve. A 3D representation of the dFSC analysis was generated by constructing a sphere from the cones oriented in their respective directions and setting each conical shell of each cone to its respective dFSC value. dFSC values were averaged in regions of overlap between cones and a 3 pixel × 3 pixel × 3 pixel median filter was applied three times to the 3D dFSC map in UCSF Chimera51 to reduce noise. The resolution in different directions was determined as the spatial frequency with a dFSC value of 0.143, the same criterion as generally used for the FSC52. The trend in anisotropy of resolution that is observed at FSC = 0.143 is also observed at other FSC thresholds irrespective of the shape of the cones used in dFSC calculations.
Extended Data
Extended Data Figure 1. TMEM16A protein purification and negative staining.
a, Western blot (bottom) of nine TMEM16A constructs with different N-terminal and/or C-terminal truncations (diagramed, top). Construct 5 corresponding to mouse TMEM16A residues 1–903 was selected for this study for the absence of the smaller fragment of ~30 kD on the western blot as well as its high expression. b, Top, representative trace of inside-out patch from HEK293 cells transiently transfected with wild-type TMEM16A (WT) or Construct 5. The membrane potential was held at +60 mV, and patches were exposed to intracellular solutions containing 140 mM NaCl and 150 nM, 300 nM, 400 nM, 600 nM, 1.8 μM, or 1 mM free Ca2+. Repeated independently four times with similar results. Bottom, normalized chloride currents were fit to the Hill equation. EC50 for Ca2+ sensitivity is 178 ± 14 nM for construct 5 (four independent experiments) and 796 ± 66 nM for WT (ten independent experiments; p < 0.0001, see Extended Data Fig. 9c). c, Top, poly-L-lysine (PLL, 30 μg/mL) treatment for 30 sec to reduce PIP2 and other lipids with negatively charged head groups caused desensitization of TMEM16A with C-terminal truncation (a.a. 1–903) in excised inside-out patch exposed to 150 mM NaCl on both sides of the membrane, as evident from the reduction of Ca2+ sensitivity. Repeated independently six times with similar results. Bottom, following the PLL treatment the current amplitudes were reduced at 30 nM Ca2+ and 100 nM Ca2+ (“Inst” for the “instantaneous” current amplitude at the start of depolarization from a holding potential of 0 mV to +100 mV, p = 0.02 from two-way ANOVA between “Pre” and “post” PLL; “Overall” for the current amplitude at the end of depolarization, p = 0.004 from two-way ANOVA between “Pre” and “Post”; 6 independent experiments) but not at 1 μM Ca2+ (Sidak’s multiple comparisons, p > 0.99 and p = 0.73 for “Inst” and “Overall”, respectively). Mean ± SEM are shown in b and c. d, Size-exclusion chromatography of TMEM16A reconstituted into lipid nanodiscs with MSP2N2. The peak fractions corresponding to nanodisc-reconstituted TMEM16A (16A) and free MSP2N2 are indicated. The 16A peak fraction was examined by SDS-polyacrylamide gel electrophoresis (SDS-PAGE). TMEM16A and MSP2N2 (MSP) monomers are approximately 105 kDa and 46 kDa, respectively. The faint band at 210 kDa may correspond to incompletely disassociated TMEM16A dimers. e, CPM analysis16,17 of nanodisc-reconstituted TMEM16A in 0, 71 nM, 293 nM, 782 nM, 4120 nM or 1 mM Ca2+. f, Raw micrographs of nanodisc-reconstituted TMEM16A examined by negative-stain EM. g, 2D class averages of particles from negative-stain EM of TMEM16A reconstituted into nanodiscs h, Size-exclusion chromatography of TMEM16A solubilized in LMNG. The peak fraction was examined by SDS-PAGE. i, CPM analysis of LMNG-solubilized TMEM16A in 0, 83 nM, 333 nM, 1122 nM, 5290 nM or 1 mM Ca2+. j, Raw micrographs of LMNG-solubilized TMEM16A examined by negative-stain EM. Both micrographs, (f) and (j), showed mono-dispersed and homogeneous particles. k, 2D class averages of particles from negative-stain EM of TMEM16A solubilized in LMNG.
Extended Data Figure 2. Cryo-EM analysis of TMEM16A reconstituted in nanodiscs.
a, A representative cryo-EM micrograph of nanodisc-reconstituted TMEM16A. Green circles indicate individual particles. b, Representative 2D class averages from boxed particles with 256-pixel box size (261.12 Å). c, Eular angle distribution of all particles included in the final 3D reconstruction. The size of the spheres is proportional to the number of particles seen from that specific orientation. d, FSC curves of two independently refined maps before (blue) and after (red) post-processing in RELION. Curves with resolution corresponding to FSC = 0.143 are shown. e, Planar slices through the unsharpened EM density map at different levels along the channel symmetry axis. f, Local resolution of TMEM16A as estimated by RELION and shown with pseudo-color representation of resolution. g, Cross-validation using FSC curves of the density map calculated from the refined model versus half map 1 (work), versus half map 2 (free), and versus summed map. h, Directional FSC from different Fourier cones. Each curve indicates a different direction. i, Calculated resolution from different views. The directions are indicated as x, y, and z in the 3D resolution map. The highest and lowest resolutions are labeled with red and blue circles respectively. The green circle shows global average resolution.
Extended Data Figure 3. Cryo-EM analysis of TMEM16A solubilized in LMNG.
a, A representative cryo-EM micrograph of LMNG-solubilized TMEM16A. Green circles indicate individual particles. b, Representative 2D class averages from boxed particles with a 256 pixels box size (261.12 Å) and TMEM16A in complex with Fabs (bottom row, with the two right panels showing particles after subtraction of densities for Fabs). c, Eular angle distribution of all particles included in the final 3D reconstruction. The size of the spheres is proportional to the number of particles visualized from that specific orientation. d, FSC curves of two independently refined maps before (blue) and after (red) post-processing in RELION. Curves with resolution corresponding to FSC = 0.143 are shown. e, Planar slices through the unsharpened EM density map at different levels along the channel symmetry axis. f, Local resolution of TMEM16A as estimated by RELION and shown with pseudo-color representation of resolution. g, Cross-validation using FSC curves of the density map calculated from the refined model versus half map 1 (work), versus half map 2 (free), and versus summed map. h, Directional FSC (dFSC) from different Fourier cones. Each curve indicates a different direction. dFSC for TMEM16A alone in LMNG in grey (average in yellow); dFSC for combination of TMEM16A alone and with Fabs bound in LMNG in purple (average in red). i, Calculated resolution from different views (grey for combination of TMEM16A alone and with Fabs bound, yellow for TMEM16A alone). The directions are indicated as x, y, and z in the 3D resolution map. The highest and lowest resolutions are labeled with red and blue circles respectively. The green circle shows global average resolution.
Extended Data Figure 4. Cryo-EM densities of the ten transmembrane helices of TMEM16A, summary of cryo-EM data collection and processing, and summaries of sidechain assignments.
a, Representative cryo-EM densities of the ten transmembrane helices (TM1-TM10) of nanodisc-reconstituted TMEM16A (right) or LMNG-solubilized TMEM16A (left) are superimposed on the corresponding atomic model. The EM densities are shown in blue meshes for nanodisc-reconstituted TMEM16A, or green meshes for LMNG-solubilized TMEM16A, and the model is show as sticks and colored according to atom type (C: light grey; N: blue; O: red; S: yellow). b, Summary of cryo-EM data collection and model refinement. c, Summary of sidechain assignment of TMEM16A in nanodiscs. d, Summary of sidechain assignment of TMEM16A in LMNG.
Extended Data Figure 5. Atomic models of TMEM16A in two conformations.
a–c, Ribbon diagrams of TMEM16A reconstituted in nanodiscs (in green and yellow) with lipids (in red), overlayed with EM density map (sharpened, in light grey). Two Ca2+ ions (orange spheres) are present in each monomer. d–f, Ribbon diagrams of TMEM16A solubilized in LMNG (in blue) with lipids (in red), overlayed with EM density map (sharpened, in light grey). One Ca2+ ion (orange sphere) is present in each monomer g–i, EM densities of nanodisc-reconstituted TMEM16A (unsharpened, in green and yellow) overlayed with digitonin-solubilized TMEM16A12 (in grey).
Extended Data Figure 6. Anion selectivity depends on residues lining the pore surrounded by TM3–8 but not TM10 residues at the dimer interface.
a, Bi-ionic conditions for assessing the effect of the V595L mutation on permeability ratios. b, Effects of different substitutions of V595 on the permeability ratio PI-/PCl- (2.70 ± 0.09, N = 28 for WT; 4.00 ± 0.16, N = 9 for V595A; 3.49 ± 0.21, N = 6 for V595K; 3.81 ± 0.07, N = 7 for V595L; 3.48 ± 0.14, N = 7 for V595R). c, Effects of different substitutions of V595 on the permeability ratio PSCN-/PCl- (5.47 ± 0.21, N = 28 for WT; 9.96 ± 0.30, N = 9 for V595A; 6.64 ± 0.58, N = 6 for V595K; 7.86 ± 0.37, N = 7 for V595L; 6.30 ± 0.37, N = 7 for V595R). d, Permeability ratios determined in bi-ionic conditions for TMEM16A mutants. The exact n values (independent experimental samples from individually recorded HEK293 cells) are given for every experiment. The P-values are generated after a Dunnett’s posthoc test following one-way ANOVA. For these multiplicity adjusted P-values, values smaller than 0.0001 cannot be estimated precisely; Prism’s documentation suggests this approach is the most rigorous and conservative way to generate a P-value from a multiple comparison test53.
Extended Data Figure 7.
Comparisons of extracellular loops and lipids in nanodisc-resonstituted and LMNG-solubilized TMEM16A. a–c, Lipids (in red) in the nanodisc-reconstituted TMEM16A (in green and yellow, overlayed with EM density map in light grey) (b, c), with helical distortions of TM6 near G640 (a). d–f, Lipids (in red) in LMNG-solubilized TMEM16A (in blue, overlayed with EM density map in light grey) (e, f), with the lower half of TM6 beyond G640 disordered and hence absent from the reconstruction (d). g–i, Extracellular domains of nanodisc-reconstituted TMEM16A (unsharpened, in green and yellow) overlayed with those of LMNG-solubilized TMEM16A (unsharpened, in blue). j–l, Extracellular TM5-TM6 and TM9-TM10 loops in ribbon diagrams for nanodisc-reconstituted TMEM16A (in green and yellow) overlayed with those of LMNG-solubilized TMEM16A (in blue).
Extended Data Figure 8.
Data processing of TMEM16A in nanodisc (a) or LMNG (b). a, Data processing of nanodisc-reconstituted TMEM16A. Particle picking was performed with Gautomatch with templates from 2D classes from the LMNG dataset and generated 927,414 particles in total. All particles were extracted and binned 4 (pixel size is 4.08 Å) and then 2D classified. 341,875 particles from good 2D classes were used in 3D refinement with an initial model from the LMNG structure low pass filtered to 60 Å, giving rise to a 5.5 Å map. The 5.5 Å map was then low pass filtered to 10 Å as the initial model for 3D classification without applied symmetry for all particles with a 1.02 Å pixel size. Of the seven classes, two classes (15.92% and 11.15% of the 927,414 particles) gave maps with improved resolution (4.7 Å and 5.1 Å respectively) after 3D auto-refinement with C2 symmetry. These two classes were combined together yielding a total of 251,851 particles, and another 3D auto-refinement was run to generate the unmasked map at a resolution of 4.6 Å. The map was then masked to get the final map at resolution of 3.8 Å. b, Data processing of LMNG-solubilized TMEM16A. Approximately 4000 particles were manually picked and classified by 2D classification in SAMUEL to generate the templates for automatic particle picking with samautopick.py. 533,545 particles were identified after manually inspection. The crystal structure of nhTMEM16A10 (PDB: 4WIS) was converted to mrc with e2pdb2mrc.py and low pass filtered to 60 Å as the initial model. 44 of 200 2D classes were used for 3D auto-refinement with C2 symmetry. Since 3D classification failed for further separation, the reported resolution of the final map was 3.8 Å. To reduce anisotropy due to underrepresentation of side views, this dataset was merged with another dataset for Fabs bound to TMEM16A in LMNG. Starting with 338,705 particles from automatic particle picking, 4 of 40 2D classes (132,444 particles) were used for 3D auto-refinement. Then the Fab density for each particle was subtracted. The 132,444 subtracted particles without Fab density were combined with the 342,875 particles from all 5 classes of TMEM16A in LMNG dataset with resolution of 3.8 Å and processed for 3D auto-refinement to generate the unmasked map with resolution of 3.9 Å. This map was then masked to get the final map at resolution of 3.4 Å. Pixel sizes are shown in parenthesis for each class.
Extended Data Figure 9.
Multiple open and closed states of TMEM16A calcium-activated chloride channel (CaCC) and involvement of pore-lining residues in channel gating. a, Reduction from 150 mM NaCl to 15 mM NaCl in the intracellular solution containing 1 μM or 1 mM Ca2+ caused identical shift of reversal potential of wildtype TMEM16A but not K584Q mutant channels in excised inside-out patch held at +80 mV and subjected to a ramp to −80 mV. Repeated independently 8 times for WT with similar results, and 5 times for K584Q with similar results. b, The K584Q mutation altered the permeability ratio PNa+/PCl- at 1 μM but not 1 mM Ca2+. N = 8 for WT, N = 5 for K584Q. c, Calcium sensitivity of channel activation of wildtype and mutant TMEM16A channels (number of independent experiments and P values are given in this table). Permeability ratios for mutants were compared to those of wild-type (WT) using one-way ANOVA followed by the Bonferroni post-hoc test for significance; **** designates p < 0.0001; *** designates p < 0.001; ** designates p < 0.005; data are presented as mean ± SEM.
Extended Data Figure 10.
Sequence alignment of TMEM16A homologues. a, Sequences of TMEM16A homologues were analyzed by Clustal omega. Conserved residues are highlighted. Transmembrane helices are indicated above the sequences. The residues that were shown in this study (D550, N587, S635, Q705 and F712) and previous studies (R511, K584 and K599) to be crucial for selectivity are marked in orange and blue respectively. The residues (I546, Y589, I592, L639 and F708) that were shown in this study to be critical for gating are marked in green. The residues (N542 and V595) that contributed to both selectivity and gating property is marked in purple. The residues (E650, E698, E701, E730 and D734) important for Ca2+ binding are marked in red. b, Sequence alignment of mouse TMEM16A and nhTMEM16. Conserved residues are highlighted. Transmembrane helices of TMEM16A are indicated above the sequence.
Acknowledgments
We thank R. MacKinnon, T. Xiao and W. Wang for advice on protein purification, B. Kobilka and A. Koehl for help on CPM assay, Z. Yu and his colleagues at the HHMI Janelia Cryo-EM Facility for help with data acquisition, and J. Wang at Tsinghua University for discussions on model building. We also thank Peng Jin for help with data presentation and colleagues in our laboratories for discussion. This work was supported by grants from the NIH (R01GM098672 and S100D0020054 to Y.C., R01NS069229 to L.Y.J., R35NS097227 to Y.N.J., R01HL080050 and R01DC007664 to D.L.M., P41CA196276 and P50GM111126 to C.S.C., K99DA041500 to C.J.P.) and from the American Heart Association (M.L.), and by a UCSF Breakthrough Biomedical Research (Y.C.). S. Dang is supported by a Human Frontier Science Program (HFSP) Postdoctoral Fellowship. J.Z. is supported by Banting Postdoctoral Fellowship from the Canadian Institute of Health Research. L.Y.J., Y.N.J. and Y.C. are Investigators with the Howard Hughes Medical Institute.
Footnotes
Data Availability
Cryo-EM density maps of TMEM16A have been deposited to the Electron Microscopy Data Bank (EMDB) under the accession number EMD-7095 and EMD-7096. Particle image stack after motion correction related to TMEM16A has been deposited in the Electron Microscopy Public Image Archive (http://www.ebi.ac.uk/pdbe/emdb/empiar/) under accession number EMPIAR-10123 for nanodisc-reconstituted TMEM16A and EMPIAR-10124 for LMNG-solubilized TMEM16A. Atomic coordinates for TMEM16A have been deposited in the Protein Data Bank (PDB) under the accession number 6BGI and 6BGJ. All electrophysiological data generated or analyzed during this study are included in this published article. All other data are available from the corresponding authors upon reasonable request.
Author Contributions
S.D., S.F. and J.T. designed and performed most of the biochemical and cryo-EM experiments. C.J.P., J.T., W.Y., L.Q. and T.X. performed mutagenesis and electrophysiology experiments. K.Z. and J.T. isolated the Fab that binds TMEM16A, M.L. and J.T. performed systematic screens of deletion constructs and purification schemes. S.D., D.B., M.L. and S.F. built de novo models. J.Z. developed methods to calculate directional resolution. L.Y.J., Y.N.J., D.L.M., C.S.C. and Y.C. supervised experiments and data analysis. All authors contributed to manuscript preparations.
The authors declare no competing financial interest.
Uncategorized References
- 1.Caputo A, et al. TMEM16A, a membrane protein associated with calcium-dependent chloride channel activity. Science (New York, N Y) 2008;322:590–594. doi: 10.1126/science.1163518. [DOI] [PubMed] [Google Scholar]
- 2.Schroeder BC, Cheng T, Jan YN, Jan LY. Expression cloning of TMEM16A as a calcium-activated chloride channel subunit. Cell. 2008;134:1019–1029. doi: 10.1016/j.cell.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang YD, et al. TMEM16A confers receptor-activated calcium-dependent chloride conductance. Nature. 2008;455:1210–1215. doi: 10.1038/nature07313. [DOI] [PubMed] [Google Scholar]
- 4.Hartzell C, Putzier I, Arreola J. Calcium-activated chloride channels. Annual review of physiology. 2005;67:719–758. doi: 10.1146/annurev.physiol.67.032003.154341. [DOI] [PubMed] [Google Scholar]
- 5.Huang F, Wong X, Jan LY. International Union of Basic and Clinical Pharmacology. LXXXV: Calcium-Activated Chloride Channels. Pharmacol Rev. 2012;64:1–15. doi: 10.1124/pr.111.005009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pedemonte N, Galietta LJ. Structure and function of TMEM16 proteins (anoctamins) Physiological reviews. 2014;94:419–459. doi: 10.1152/physrev.00039.2011. [DOI] [PubMed] [Google Scholar]
- 7.Oh U, Jung J. Cellular functions of TMEM16/anoctamin. Pflugers Arch. 2016;468:443–453. doi: 10.1007/s00424-016-1790-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lim NK, Lam AK, Dutzler R. Independent activation of ion conduction pores in the double-barreled calcium-activated chloride channel TMEM16A. The Journal of general physiology. 2016;148:375–392. doi: 10.1085/jgp.201611650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeng G, Aggarwal M, Yu WP, Chen TY. Independent activation of distinct pores in dimeric TMEM16A channels. The Journal of general physiology. 2016;148:393–404. doi: 10.1085/jgp.201611651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brunner JD, Lim NK, Schenck S, Duerst A, Dutzler R. X-ray structure of a calcium-activated TMEM16 lipid scramblase. Nature. 2014;516:207–212. doi: 10.1038/nature13984. [DOI] [PubMed] [Google Scholar]
- 11.Brunner JD, Schenck S, Dutzler R. Structural basis for phospholipid scrambling in the TMEM16 family. Curr Opin Struct Biol. 2016;39:61–70. doi: 10.1016/j.sbi.2016.05.020. [DOI] [PubMed] [Google Scholar]
- 12.Paulino C, et al. Structural basis for anion conduction in the calcium-activated chloride channel TMEM16A. Elife. 2017;6 doi: 10.7554/eLife.26232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peters CJ, Tien J, Bethel NP, Qi L, Chen T, Wang L, Jan YN, Grabe M, Jan LY. The sixth transmembrane segment as the primary gating machineray of the TMEM16A calcium-activated chloride channel. 2017 doi: 10.1016/j.neuron.2018.01.048. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scudieri P, Sondo E, Caci E, Ravazzolo R, Galietta LJ. TMEM16A-TMEM16B chimaeras to investigate the structure-function relationship of calcium-activated chloride channels. Biochem J. 2013;452:443–455. doi: 10.1042/BJ20130348. [DOI] [PubMed] [Google Scholar]
- 15.Scudieri P, Musante I, Gianotti A, Moran O, Galietta LJ. Intermolecular Interactions in the TMEM16A Dimer Controlling Channel Activity. Sci Rep. 2016;6:38788. doi: 10.1038/srep38788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alexandrov AI, Mileni M, Chien EY, Hanson MA, Stevens RC. Microscale fluorescent thermal stability assay for membrane proteins. Structure. 2008;16:351–359. doi: 10.1016/j.str.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 17.Ashok Y, Jaakola VP. Nanodisc-Tm: Rapid functional assessment of nanodisc reconstituted membrane proteins by CPM assay. MethodsX. 2016;3:212–218. doi: 10.1016/j.mex.2016.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duriseti S, et al. Antagonistic anti-urokinase plasminogen activator receptor (uPAR) antibodies significantly inhibit uPAR-mediated cellular signaling and migration. The Journal of biological chemistry. 2010;285:26878–26888. doi: 10.1074/jbc.M109.077677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu S, et al. Fabs enable single particle cryoEM studies of small proteins. Structure. 2012;20:582–592. doi: 10.1016/j.str.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smart OS, Neduvelil JG, Wang X, Wallace BA, Sansom MS. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J Mol Graph. 1996;14:354–360. 376. doi: 10.1016/s0263-7855(97)00009-x. [DOI] [PubMed] [Google Scholar]
- 21.Whitlock JM, Hartzell HC. A Pore Idea: the ion conduction pathway of TMEM16/ANO proteins is composed partly of lipid. Pflugers Arch. 2016;468:455–473. doi: 10.1007/s00424-015-1777-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whitlock JM, Hartzell HC. Anoctamins/TMEM16 Proteins: Chloride Channels Flirting with Lipids and Extracellular Vesicles. Annual review of physiology. 2017;79:119–143. doi: 10.1146/annurev-physiol-022516-034031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu K, et al. Identification of a lipid scrambling domain in ANO6/TMEM16F. Elife. 2015;4:e06901. doi: 10.7554/eLife.06901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang TYK, Hartzell HC, Tajkhorshid E. Lipids and ions traverse the membrane by the same physical pathway in the nhTMEM16 scramblase. Elife. 2017 doi: 10.7554/eLife.28671. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peters CJ, et al. Four basic residues critical for the ion selectivity and pore blocker sensitivity of TMEM16A calcium-activated chloride channels. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:3547–3552. doi: 10.1073/pnas.1502291112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tien J, et al. A comprehensive search for calcium binding sites critical for TMEM16A calcium-activated chloride channel activity. Elife. 2014;3 doi: 10.7554/eLife.02772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu K, Duran C, Qu Z, Cui YY, Hartzell HC. Explaining calcium-dependent gating of anoctamin-1 chloride channels requires a revised topology. Circ Res. 2012;110:990–999. doi: 10.1161/CIRCRESAHA.112.264440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang H, et al. TMEM16F forms a Ca2+-activated cation channel required for lipid scrambling in platelets during blood coagulation. Cell. 2012;151:111–122. doi: 10.1016/j.cell.2012.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goehring A, et al. Screening and large-scale expression of membrane proteins in mammalian cells for structural studies. Nature protocols. 2014;9:2574–2585. doi: 10.1038/nprot.2014.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whicher JR, MacKinnon R. Structure of the voltage-gated K(+) channel Eag1 reveals an alternative voltage sensing mechanism. Science (New York, N Y. 2016;353:664–669. doi: 10.1126/science.aaf8070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao Y, Cao E, Julius D, Cheng Y. TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Nature. 2016;534:347–351. doi: 10.1038/nature17964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 33.Liao M, Cao E, Julius D, Cheng Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature. 2013;504:107–112. doi: 10.1038/nature12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frank J, et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 35.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nature methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang K. Gctf: Real-time CTF determination and correction. J Struct Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang G, et al. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 40.Scheres SH, Chen S. Prevention of overfitting in cryo-EM structure determination. Nature methods. 2012;9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nature methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trabuco LG, Villa E, Mitra K, Frank J, Schulten K. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure. 2008;16:673–683. doi: 10.1016/j.str.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brown A, et al. Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr D Biol Crystallogr. 2015;71:136–153. doi: 10.1107/S1399004714021683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barad BA, et al. EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy. Nature methods. 2015;12:943–946. doi: 10.1038/nmeth.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.DeLano WL. The PyMOL molecular graphics system on world wide web. 2002 http://www.pymol.org.
- 49.Saxton WO, Baumeister W. The correlation averaging of a regularly arranged bacterial cell envelope protein. J Microsc. 1982;127:127–138. doi: 10.1111/j.1365-2818.1982.tb00405.x. [DOI] [PubMed] [Google Scholar]
- 50.van Heel M, Stoffler-Meilicke M. Characteristic views of E. coli and B. stearothermophilus 30S ribosomal subunits in the electron microscope. EMBO J. 1985;4:2389–2395. doi: 10.1002/j.1460-2075.1985.tb03944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 52.Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 53.Wright SP. Adjusted P-values for simultaneous inference. Biometrics. 1992;48:1005–1013. [Google Scholar]














