Abstract
T helper 17 (Th17) and T regulatory (Treg) cytokines appear to be contributing greatly to colorectal cancer (CRC) development and progression. The aim of the current study was to investigate the expression of Foxp3; IL10; TGFB1; IL17A; IL6 and NOS2 genes in tumor tissue, regional positive lymph nodes and distant metastasis obtained from 26 patients with advanced CRC. Quantitative real-time polymerase chain reaction (qPCR) was performed for mRNA detection by TaqMan gene expression assay. In distant metastasis, IL6 was strongly expressed, over 7.5 fold, followed by Treg-related genes Foxp3; IL10 and TGFB1 in contrast to IL17A and NOS2. The similar pattern of expression was observed in positive regional lymph node in addition to significant down-regulation of NOS2 (RQ = 0.287; p = 0.011) and a trend for the elevation of IL17A. In tumor tissue, Fopx3 was significantly upregulated and Foxp3 mRNA positively correlated with TGFB1 in all investigated tissue types. In tumor tissue, expression of IL17A was correlated with NOS2 (r = 0.68; p = 0.005), while in distant metastasis IL10 was in strong relation with TGFB1 and IL6. In addition, a reverse correlation between IL6 and NOS2 (r = −0.66; p = 0.009), was observed in distant metastasis. The simultaneous expression of given Treg and Th17-related genes found both in the primary tumor and in the regional lymph nodes appears to provide suitable microenvironment sufficient for promoting metastatic growth. The upregulation of Foxp3; IL10, TGFB1 and IL6 might be a transcriptional profile hallmark for colorectal metastases.
Keywords: mRNA, qPCR, Colorectal cancer, Th17, Treg
Introduction
Colorectal cancer (CRC) is one of the most frequent and severe solid tumors in human. Since cancer related inflammation was pointed as the seventh hallmark of cancer [1, 2] and Th1/Th2 paradigm was broken, many studies investigated the significance of CD4+ T cells subset for tumor promotion, progression, or metastasis [3–5]. T helper 17 (Th17) and T regulatory (Treg) cells are abundant in gut associated lymphoid tissue important for the protection of the mucosal surfaces against pathogens and for tolerance to the gut microbiome [6]. Important polarizing cytokines for differentiation into the Th17 subset include TGF-β, IL-6, IL-1β, and IL-23. Human Th17 cells express the transcription factors retinoic acid orphan receptor (ROR)γt that is a critical regulator of IL-17A, IL-17F, and CCR6 expression [7]. Differential cytokines for inducible Treg cells include TGF-β, IL-2 and have been defined by expression of the Forkhead Box P3 (Foxp3) transcription factor. Treg cells are deemed as immunosuppressive CD4+ subset explores different mechanisms including secretion of suppressive cytokines TGF-β and IL-10.
Many studies have reported significant elevation of Treg in cancer patients, although the role of Treg for CRC development and progression is controversial [8–10]. deLeeuw et al. have suggested that Foxp3+ T cells may inhibit tumor-promoting inflammatory responses to gut microbes, which could explain their association with favorable outcomes in CRC [8]. In addition high densities of Foxp3+ Tregs were associated with early T stage and independently predicted improved disease-specific survival in MMR-proficient CRC patients [9]. These studies have suggested that through the suppression of the inflammation driven by bacteria, Foxp3+ Treg prevented carcinogenesis. In case of other cancers, including gastric cancer and hepatocellular carcinoma, high density of Foxp3+ Tregs was associated with poor survival and high recurrences of the gastric cancer [10]. The role of Th17 cells in colorectal cancer was less studied as compared with Treg cells, although their dual role in cancer genesis has been also presented [11]. Previous study showed that in advanced CRCs, the Th17 cells become reduced in the circulation but increased in tumor tissues [12]. Also, the high expression of the Th17 cluster genes (IL17A, RORC) predicts a poor prognosis in patients with colorectal cancer [13].
Obviously Th17-Treg balance is important for CRC development and progression. In addition, recent evidence showed the considerable plasticity between Th17 and Treg cell population according to the local milieu [14]. Respectively, cytokine expression profile of the tumor microenvironment probably is more relevant than the count and type of immune cells. Therefore, the objective of our study was quantification of Treg and Th17-related genes in tumor tissue, regional lymph nodes and distant metastasis in advanced cases of CRC.
Materials and Methods
Subjects
A group 26 advanced CRC patients were included in the present study. All patients were diagnosed in Trakia Hospital, Stara Zagora, Bulgaria and had no previous diagnosis of inflammatory bowel disease or any of the known hereditary cancer syndromes. The histopathology reports for all cancer specimens confirmed adenocarcinomas from poor to well differentiated types. Patients did not receive chemotherapy or radiation therapy before surgery. All included CRC cases were operated on with curative intent.
The patients group was composed by 7 (27%) female and 19 (73%) male. The mean age at diagnosis was 69.89 ± 9.11 yrs. with range 62-85 yrs. No significant differences in age was observed between female and male patients (66.9 ± 6.4 vs. 71.2 ± 10 yrs.; p = 0.3 t-test). Tumor grading and staging was performed according to the tumor–node–metastasis (TNM) classification. In the investigated cohort of advanced CRC, 12 (46%) patients were with 3rd stage and 14 (54%) with 4th stage of CRC. Patients’ demographic and clinicopathologic data are presented in Table 1.
Table 1.
Demographic and clinicopathologic data of included CRC patients
| Characteristics | Primary tumor tissue samples | Regional lymph node samples | Distant metastasis samples | p-value |
|---|---|---|---|---|
| Sex, n (%) | ||||
| Male | 19 (73) | 13 (68) | 8 (57) | 0.59 |
| Female | 7 (27) | 6 (32) | 6 (43) | |
| Age, years | ||||
| Mean ± s.d. | 69.8 ± 9.2 | 69.6 ± 9.5 | 70.6 ± 7.7 | >0.76 |
| Tumor location, n (%) | ||||
| Colon | 17 (65) | 13 (68) | 10 (71) | 0.92 |
| Rectum | 9 (35) | 6 (32) | 4 (29) | |
| TNM stage, n (%) | ||||
| 3rd stage | 12 (46) | 11 (58) | - | <0.05 |
| 4th stage | 14 (54) | 8 (42) | 14 (100) | |
Informed consent prior to surgical resection was obtained from all individual participants included in the study. The authorization of the studywas given by the Ethics Review Board of the Faculty of Medicine, Trakia University, Stara Zagora, Bulgaria.
Tissue Specimens
Samples from 26 patients with advanced CRC were investigated. These inluded tissue from: 26 primary lessions, 26 non-tumoral mucosa specimens, 19 positive regional lymph nodes and 14 distant metastasis - 10 positive distant lymph node, 2 splenic metastasis and 2 peritoneal metastases. All cases with liver or lung metastasis were excluded. The tissues were harvested immediately after resection and used for RNA isolation. Histopathology reports of the tumor samples showed one (4%) specimen was well-differentiated, 16 (61%) were moderate and 9 (35%) poor-differentiated adenocarcinomas. Primary lesion origin was 17 (65%) colon and 9 (35%) rectal.
RNA Extraction
Total RNA was isolated by a silica-based membrane technology using GeneJet RNA purification kit (Thermo Fisher Scientific Inc.). All samples (30 mg) were homogenized in Lysis Solution supplied with β-mercaptoethanol, according to the manufacturer’s instructions. The total RNA was quantified by spectrophotometrical analysis (GeneQuant 1300 spectrophotometer, GE Healthcare Life Sciences, Switzerland).
Reverse Transcription
Synthesis of cDNA was performed according to manufacturer’s instructions by using RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific Inc.), supplied with random hexamer primers and RevertAid reverse transcriptase enzyme. Incubation conditions for reverse transcription was 5 min at 25°C followed by 1 h at 42°C and termination of the reaction for 5 min at 70°C and was performed on a GeneAmp PCR System 9700 (Applied Biosystems, Foster City, CA, USA).
Quantitative Real-Time Polymerase Chain Reaction
Quantitative real-time polymerase chain reaction (qPCR) was performed on a 7500 Real - Time PCR System (Applied Biosystems, Foster City, CA, USA). The qPCR primers and probes were purchased from ThermoFisher scientific as predesigned inventoried TaqMan gene expression assays for the target genes/genes of interest (GOI) as follows: Foxp3 (Hs01085834_m1); IL10 (Hs00174086_m1); TGFB1 (Hs00998133_m1); IL6 (Hs00985639_m1); NOS2 (Hs01075529_m1); IL17A (Hs00174383_m1); and for the endogenous controls/ reference genes were GAPDH (Hs02758991_g1) and eukaryotic 18S ribosomal RNA (Hs99999901_s1). An aliquot of 5 μl of the RT reaction was amplified in duplicate in final volume of 20 μl using a Maxima Probe qPCR Master Mix with ROX passive reference dye at final concentration 30 nM (Thermo Fisher Scientific Inc) and gene expression assay mix, containing specific forward and reverse primers and labeled probes for target gene and endogenous controls. The thermocycling conditions were: initial 10 min incubation at 95 °C followed by 40 cycles of denaturation for 15 s at 95 °C and annealing/extension for 1 min at 60 °C. PCR data were collected with Sequence Detection System (SDS) software, version 1.3.1.
Statistical Methods and Data Presentation
Statistical analysis was carried out using Statistica software, version 8.0 (StatSoft, Inc. Tulsa, OK, USA). Relative quantitative evaluation of mRNAs was performed by the comparative ddCp method [15] and results are presented as n-fold mean difference (RQ-relative quantity) of target genes relative to calibrator (non-tumoral mucosa) after normalization using the geometric mean of two reference genes.
To compare the differences in gene expression (GE) of each gene among non-tumoral tissue and tumor tissue, regional lymph nodes or distant metastasis, dCp values were used for Student’s t-test. Next, to estimate the degree of the relationship between target genes expression, Pearson correlation analysis was used after log transform the final gene expression results. Also, to determine the percent of samples with alternated gene expression, the RQ-value of particular gene relative to non-tumoral mucosa above 2.0 was accepted as up-regulated gene expression, RQ below 0.5 - as down-regulation and unchanged gene expression was define as 0.5 < RQ < 2.0 The limit of significance for all analyses was defined as a P-value of 0.05.
Results
Gene Expression in Tumor Tissue
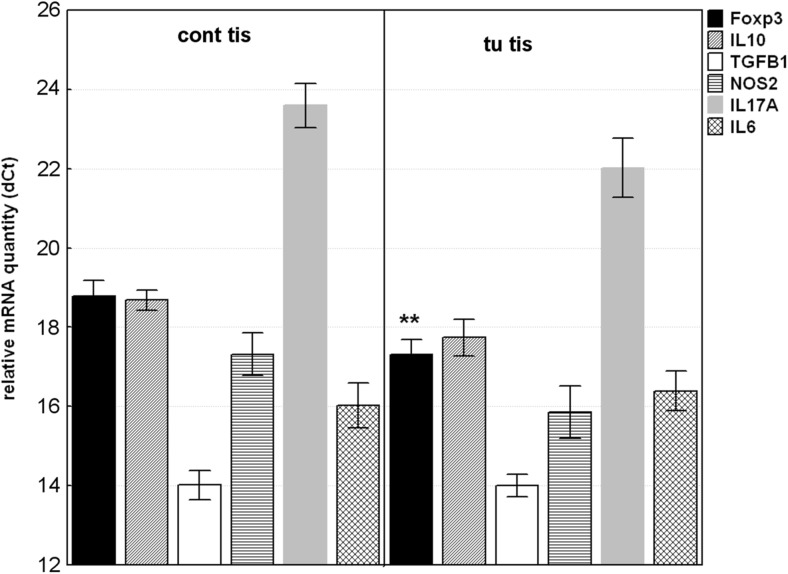
Foxp3 mRNA level in tumor tissues was significantly upregulated compared to adjacent non-tumoral tissues with mean RQ = 2.783 (p = 0.0096). IL10, IL17A and NOS2 mRNA were also upregulated in contrast to IL6 and TGFB1, without reaching the significance (Fig. 1). It should be noted that IL17A mRNA was undetectable in 2 (8%) of tumor tissues and in 10 (38%) of non-tumoral mucosa (χ2 = 6.933; pc = 0.021).
Fig. 1.
Relative mRNA quantity of Foxp3, IL10, TGFB1; IL6; NOS2 and IL17A genes in tumor tissue (tu tis) and non-tumoral mucosa (cont tis). The data are expressed as delta CT values normalized to geometric mean of two reference genes (GAPDH and 18srRNA); therefore a higher value represents a lower expression level. Statistical significance: ** p < 0.01
Furthermore, when RQ value above 2.0 in tumor samples calibrated to non-tumoral samples was accepted for upregulated gene expression, the high percentage of tumor specimens showed upregulation of Foxp3 (58%), IL17A (53%) and NOS2 (42%).
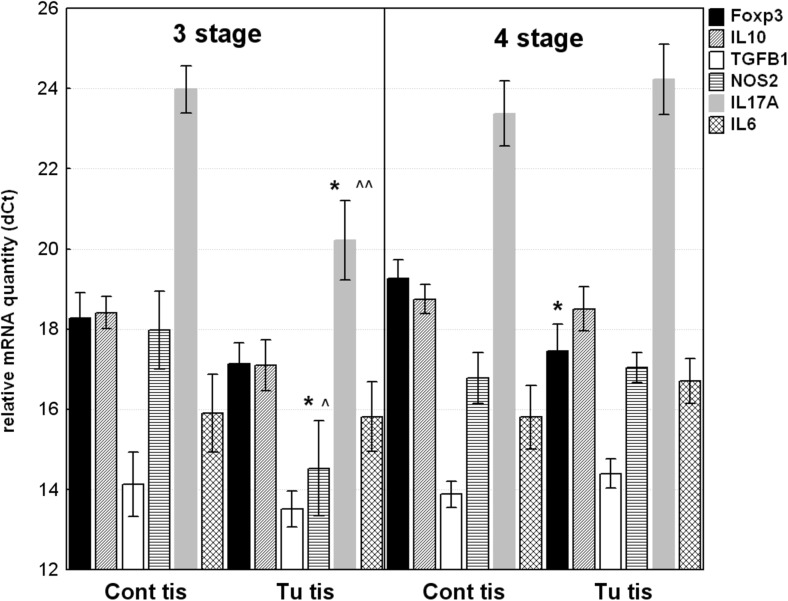
In comparison between gene expression in tumoral tissue from advanced CRC patients with distant metastasis (4th stage) and without distant metastasis (3rd stage), a significant difference in Th17-raleted genes was observed (Fig. 2). IL17A and NOS2 mRNAs were over 10-fold higher in tumoral tissue samples from patients with 3rd stage CRC (RQ = 13.555; p = 0.0241 and RQ = 10.9; p = 0.0397, respectively), while in 4th stage IL17A and NOS2 mRNAs were in lower quantity.
Fig. 2.
Relative mRNA quantity of Foxp3, IL10, TGFB1; IL6; NOS2 and IL17A genes in tumor tissue samples (tu tis) and non-tumoral mucosa (cont tis) from 3rd and 4th stages of CRC. The data are expressed as delta CT values normalized to geometric mean of two reference genes (GAPDH and 18srRNA); therefore a higher value represents a lower expression level. Statistical significance: * p < 0.05 – compared to non-tumoral mucosal counterparts; ^^ p < 0.01; ^p < 0.05 - compared to tumoral tissue from 4th stage
IL17A and NOS2 were expressed at very distinct levels within tumor tissues from patients with 3rd and 4th stages of CRC. IL17A was overexpressed in 3rd stage tumors compared to 4th stage with mean RQ = 16.15; p = 0.007. The expression of NOS2 was also higher in 3rd stage than in 4th stage with RQ = 5.7; p = 0.076. Contrary, gene expression of all investigated genes was in nearly equal level in the cancer free mucosa adjacent to tumor tissue.
Gene Expression in Regional Lymph Nodes
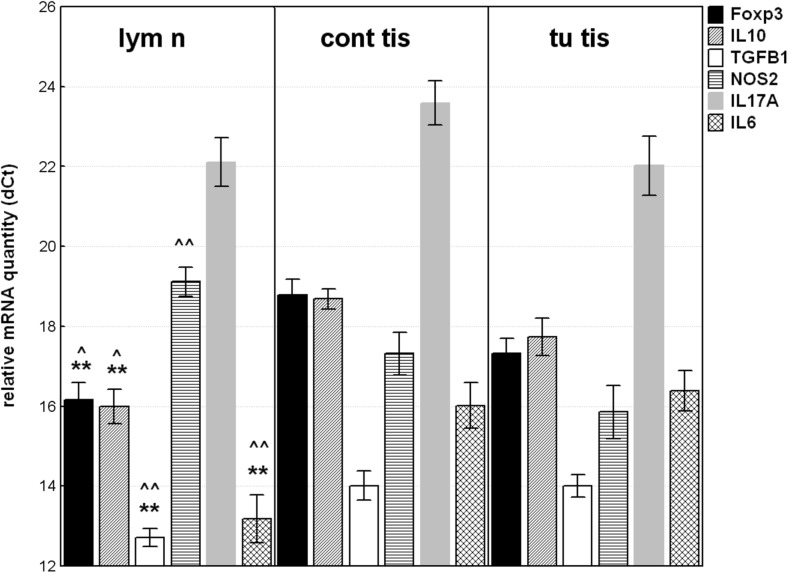
The highest gene expression in regional lymph node was observed for IL6 followed by IL10; Foxp3 and TGFB1 compared to normal mucosa. IL17A was undetectable in 2 (10.5%) of samples and was up-regulated without reaching the statistical significance. Opposite, NOS2 mRNA level was significantly lower in regional lymph node than in non-tumoral mucosa with RQ = 0.29; p = 0.011 (Fig. 3a).
Fig. 3.
Relative mRNA quantity of Foxp3, IL10, TGFB1; IL6; NOS2 and IL17A genes in regional lymph node (lym n) compared to their non-tumoral mucosal counterparts (cont tis) or to tumoral tissue (tu tis). The data are expressed as delta CT values normalized to geometric mean of two reference genes (GAPDH and 18srRNA); therefore a higher value represents a lower expression level. Statistical significance: * p < 0.05; ** p < 0.01-compared to compared to non-tumoral mucosal counterparts; ^^ p < 0.01; ^p < 0.05 - compared to tumoral tissue
Respectively, Foxp3 was upregulated in 16 (84%) of lymph node samples (individual RQ above 2.0, calibrated to paired normal mucosa), followed by IL10 and IL6 (79% and 74%, respectively). In contrast, NOS2 was down-regulated in 16 (84%) of lymph node samples (individual RQ below 0.5).
In addition, when gene expression in regional lymph node was compared to gene expression in tumor tissue (Fig. 3b), the mRNA level of IL6 was significantly higher by 9.2-fold (p = 0.00017), while NOS2 was downregulated in the equal ratio (RQ = 0.104; p = 0.0002). The level of IL10; TGFB1 and Foxp3 mRNAs were also significantly elevated in lymph node than in tumor tissue.
Gene Expression in Distant Metastasis
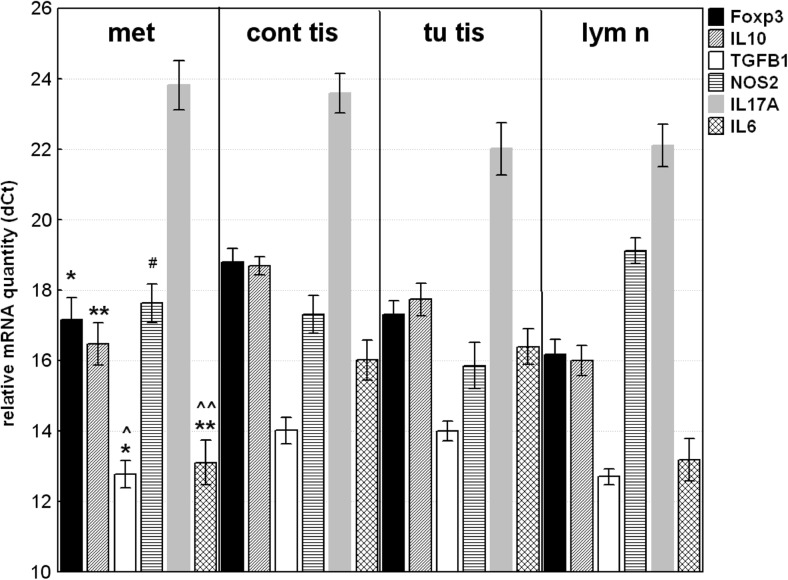
The relative gene expression of all investigated Treg-related genes (Foxp3; IL10; TGFB) and IL6 was significantly higher in distant metastasis compared to the non-tumoral mucosa in contrast to IL17A and NOS2 (Fig. 4a). RQ data above 2.0 (upregulation) was observed in 12 (86%) of distant metastasis for TGFB1 and IL10 genes, and in 10 (71%) specimens for Foxp3 and IL6, calibrated to non-tumoral mucosa. Contrary, NOS2 was downregulated in 8 (57%) or unchanged in 6 (43%) of samples, and none of the investigated distant metastasis shows gene expression above 2.0.
Fig. 4.
Relative mRNA quantity of Foxp3, IL10, TGFB1; IL6; NOS2 and IL17A genes in distant metastasis (met) compared to their non-tumoral mucosal counterparts (cont tis); to tumor tissue (tu tis) or to lymph node (lym n). The data are expressed as delta CT values normalized to geometric mean of two reference genes (GAPDH and 18srRNA); therefore a higher value represents a lower expression level. Statistical significance: * p < 0.05; ** p < 0.01 compared to compared to non-tumoral mucosal counterparts; ^^ p < 0.01; ^p < 0.05 - compared to tumoral tissue; # p < 0.05 - compared to regional lymph node
Comparing gene expression in distant metastasis to that in tumor tissue (Fig. 4b), the significantly higher the mRNA levels of IL6 (RQ = 9.77; p = 0.0003) and TGFB1 (RQ = 2.34; p = 0.014) were detected. The similar trend was observed for IL10 mRNA (RQ = 2.41; p = 0.12). While, NOS2 and IL17A mRNA were in lower quantity (RQ = 0.293 and RQ = 0.287) in distant metastasis, without reaching statistical significance (p = 0.073 and p = 0.11).
The main difference in gene expression pattern between distant metastasis and regional lymph node (Fig. 4c) concerns NOS2 gene. A significantly higher expression of NOS2 in metastasis specimens (RQ = 2.802; p = 0.025) was detected. IL17A and Foxp3 mRNAs were in lower level in distant metastasis compared to lymph node (RQ = 0.305; p = 0.073 and RQ = 0.499; p = 0.176, respectively).
The Gene Expression Pattern in Tumor Tissue, Regional Lymph Node and Distant Metastasis Compared to Non-tumoral Mucosa
The overall differences in gene expression pattern in tumor tissue, regional lymph node and distant metastasis compared to non-tumoral mucosa are shown in Table 2. Foxp3 and IL10 genes were up-regulated in all investigated tissue types (tumor tissue, regional lymph nodes and distant metastasis). IL6 and TGFB1 were significantly upregulated in regional lymph nodes and distant metastasis, in contrast to tumor tissue. Gene expression of NOS2 and IL17A was down-regulated in distant metastasis, opposite to primary tumor tissue where a trend for elevation of IL17A mRNA was observed. The reduced NOS2 mRNA and elevated IL17A mRNA quantity in regional lymph nodes were detected.
Table 2.
Relative gene expression in tumor tissue, regional lymph node and distant metastasis compared to non-tumoral mucosa
| Mean ∆Cp ± s.d. | RQ (95%CI) vs. non-tumoral tissue | ||||||
|---|---|---|---|---|---|---|---|
| Genes | Non-tumoral mucosa | Tumor tissue | Regional lymph node | Distant metastasis | Tumor | Regional lymph node | Distant metastasis |
| Foxp3 | 18.79 ± 1.9 | 17.32 ± 1.8 | 16.17 ± 1.9 | 17.17 ± 2.3 | 2.78 (1.9–6.1) p = 0.0096 | 6.15 (2.8–10.3) p = 0.000056 | 3.07 (1.8–10.2) p = 0.026 |
| IL10 | 18.69 ± 0.9 | 17.74 ± 1.5 | 15.99 ± 1.4 | 16.47 ± 2.2 | 1.93 (0.9–2.7) p = 0.08 | 6.47 (2.6–14.0) p = 0.000019 | 4.65 (1.6–11.4) p = 0.004 |
| TGFB1 | 14.02 ± 1.8 | 14.00 ± 1.4 | 12.71 ± 1.0 | 12.78 ± 1.5 | 1.01 (0.6–2.2) p = 0.98 | 2.47 (1.5–4.7) p = 0.007 | 2.36 (1.8–5.5) p = 0.036 |
| IL6 | 16.02 ± 2.7 | 16.39 ± 2.5 | 13.18 ± 2.6 | 13.10 ± 2.4 | 0.77 (0.2–1.1) p = 0.63 | 7.14 (2.1–14.4) p = 0.001 | 7.55 (2.8–41.9) p = 0.002 |
| NOS2 | 17.32 ± 2.6 | 15.86 ± 3.2 | 19.12 ± 1.6 | 17.63 ± 2.0 | 2.75 (0.6–3.2) p = 0.09 | 0.29 (0.1–0.3) p = 0.011 | 0.81 (0.2–1.0) p = 0.7 |
| IL17A | 23.59 ± 2.1 | 22.02 ± 3.5 | 22.11 ± 2.5 | 23.82 ± 2.6 | 2.97 (0.5–4.7) p = 0.14 | 2.8 (0.7–3.7) p = 0.087 | 0.85 (0.1–1.5) p = 0.801 |
s.d. standard deviation, RQ relative quantity; 95% CI- confidential interval
Correlation Analysis
Correlation analysis (Table 3) reveals a significant moderate positive relationship between gene expression of Foxp3 and TGFB1 in all investigated tissue samples, in tumor tissue (r = 0.57; p = 0.006), in lymph node (r = 0.52; p = 0.04) and in distant metastasis (r = 0.74; p = 0.002). In respect to other Treg-related gene IL10, a significant positive relationship with Foxp3 and with TGFB1 was detected just in distant metastasis, in contrast to primary tumor and regional lymph node samples. In tumor tissue, TGFB1 was also in relation to IL6 (r = 0.43; p = 0.048). The relation in gene expression between Th17-related genes IL17A and NOS2 was observed just in tumor tissue (r = 0.68; p = 0.005). In regional lymph node, the expression of NOS2 mRNA was in the moderate reverse correlation with Foxp3 mRNA. According to IL6 gene expression, moderate to strong positive correlation was detected between IL6 and IL10 in regional lymph node (r = 0.75; p = 0.019) and in distant metastasis (r = 0.83; p < 0.001), in contrast to tumor tissue. Additionally, a negative correlation (r = −0.665; p = 0.009) between IL6 and NOS2 gene expression in distant metastasis was observed.
Table 3.
Correlation analysis between gene expression of target genes in tumor tissues; regional lymph nodes and distant metastasis
| Gene pair | Tumor tissue | Regional lymph node | Distant metastasis | |||
|---|---|---|---|---|---|---|
| Correlation coefficient | p-value | Correlation coefficient | p-value | Correlation coefficient | p-value | |
| Foxp3- IL10 | 0.3538 | n.s. | −0.5483 | n.s. | 0.6315 | 0.015 |
| Foxp3- TGFB1 | 0.5704 | 0.006 | 0.5174 | 0.04 | 0.7417 | 0.002 |
| Foxp3-IL6 | 0.2484 | n.s. | −0.1698 | n.s. | 0.2431 | n.s. |
| Foxp3-NOS2 | −0.1209 | n.s. | −0.5525 | 0.033 | 0.2659 | n.s. |
| Foxp3-IL17A | 0.0233 | n.s. | 0.0158 | n.s. | −0.0581 | n.s. |
| IL10-TGFB1 | 0.3377 | n.s. | −0.556 | n.s. | 0.8266 | <0.001 |
| IL10-IL6 | −0.2105 | n.s. | 0.7530 | 0.019 | 0.8314 | <0.001 |
| IL10-NOS2 | 0.3799 | n.s. | 0.6372 | n.s. | −0.3799 | n.s. |
| IL10-IL17A | 0.3721 | n.s. | −0.2461 | n.s. | 0.3208 | n.s. |
| TGFB1-IL6 | 0.4255 | 0.048 | −0.0497 | n.s. | 0.4827 | n.s. |
| TGFB1-NOS2 | −0.1561 | n.s. | −0.4016 | n.s. | 0.0008 | n.s. |
| TGFB1-IL17A | 0.2980 | n.s. | 0.5085 | n.s. | 0.4209 | n.s. |
| IL6-NOS2 | −0.1751 | n.s. | 0.1214 | n.s. | −0.6648 | 0.009 |
| IL6-IL17A | −0.1850 | n.s. | 0.1552 | n.s. | −0.0581 | n.s. |
| NOS2-IL17A | 0.6809 | 0.005 | 0.0231 | n.s. | −0.2316 | n.s. |
n.s. non-significant
Discussion
The aim of the current study was to investigate the Treg and Th17-related gene expression in three different types of tissues tightly involved in colorectal cancerogenesis among advanced CRC cases. We found that Treg-related genes Foxp3; IL10 and TGFB1 as well as IL6 were strongly expressed in distant metastasis in contrast to IL17A and NOS2. The similar pattern of expression of the investigated genes was observed in regional lymph node in addition to significant down-regulation of NOS2 and a trend for the elevation of IL17A. In contrast, in primary tumor tissue we observed significant elevation for Fopx3 mRNA simultaneously with a trend for elevation of Th17-related genes (IL17A and NOS2) and IL10.
These results confirm that Treg phenotype contribute significantly to CRC progression. Previous studies demonstrated increased number of Treg cell in peripheral blood, tumors and regional lymph node of colorectal cancer patients [16–18]. However, conflicting data exist about the prognostic value of intratumoral Foxp3+ cells in human cancer. Some studies associated high density of regulatory T-cells in tumor tissue with improved survival in CRC patients [19]. However, the Foxp3 expression levels were not necessarily associated to Treg alone and FoxP3+ cells might be a heterogeneous cell population, also could be produced from tumor cells. Kim M et al. had demonstrated that Foxp3 expression mediated by cancer cells but not by Treg cells, contribute to CRC progression and prognosis [20]. Our data show that high mRNA Foxp3 expression is a common (could be accepted as a hallmark for) not only for tumor tissue but also for lymph node and metastases, which suppose a pivotal role of this transcription factor for tumor grow and invasion/spread, in addition to maintain tumor escape from immune surveillance. For example, Foxp3 expression can convert CD4+ T helper cells to TGF-β producing regulatory T cells which inhibit cell-mediated immunity via the release of suppressive factors such as IL-10 [21]. Collectively we can suggest that high Foxp3 mRNA expression in tumor tissue, lymph node and metastasis is essential for the immunosuppressive phenotype in advanced CRC.
However, probably our more pronouncing findings concern the relation between high Foxp3 and Th17 related genes (IL17A and NOS2). In a view of the recent studies consider the plasticity between different T cell lineages; including Th17-Treg we could suggest that the observed elevation of IL17A mRNA simultaneously with TGFB1, IL10 and IL6 genes in tumor tissue and regional lymph node could be associated with Foxp3+ Treg transdifferentiating into Th17-like Treg cells which can synthesize IL-17 cytokine and retained immunosuppressive phenotype. Some studies demonstrated accumulation of high levels of IL-17 + Foxp3 + CD4+ T cells in the colitic microenvironment and associated colon carcinoma [22]. The main cytokines discussed as regulators of Treg/Th17 plasticity includes IL-1b; IL-6, IL-23 and TGF-β. Generally it is accepted that the high concentrations of Th17-generating cytokines are able to convert Foxp3+ Treg cells into IL-17 secreting Th cells in vitro. Our previous studies have shown a significant upregulation of Foxp3 and IL23A in tumor CRC tissue [23] and elevated IL10 mRNA on systemic and local level in CRC patients [24]. In present study, focused on advanced CRC cases, we were not able to present a significant correlation between IL6 and IL17A expression in any of investigated tissue samples. IL-6 can also act in concert with TGF-β which favors Th17 phenotype [25], but the presence of high Foxp3 and IL-10 signaling can affect the differentiation process. We could assume that overexpression IL6 in metastatic tissue is most likely related to its growth-promoting, antiapoptotic and proangiogenic properties than to promoting Th17 phenotype, in contrast to regional lymph node. Also, we must note that the observed gene expression profile could be changed during or within the time/ duration and the growth of metastasis. Probably the predominant Treg phenotype may convert to Th17 in distant metastasis after stimulation with favorable microenvironmental factors. Obviously in tumor tissue it is possible to differentiate subpopulations which cannot be found in peripheral blood. Duhen et al., demonstrated for the first time that Treg cells can differentiate into specialized subsets that mirror different Th cell subsets phenotypically with unique specificities and immunomodulatory functions. During inflammatory response, this process depended on co-localization of Th and Treg subset in concrete environment [26].
Also, there are several evidences for the importance of additional mechanisms for Treg/Th17 plasticity including epigenetic changes and metabolism control mediated by HIF-1 [27] or iNOS [28]. Obermajer et al. have been clearly demonstrated that stability of effector Th17 cell function as well as the development of human Th17 cells from naive, effector, and memory CD4+ T cell precursors requires NOS2 expression [28]. They reported that the local expression of NOS2 positively correlates with Th17 responses in patients with ovarian cancer. Here, we present an elevation of IL17A and NOS2 mRNA levels and a significant positive correlation between gene expression of IL17A and NOS2 in tumor tissue of advanced CRC simultaneously with a significant downregulation of NOS2 and up-regulation of IL17A in regional lymph nodes. An explanation of this result could be the differentiation of Th17-like Treg subpopulation and cytokine production in these Th17-like Treg cells is qualitatively and quantitatively different from that observed in effector Th17 cells.
We must mention some limitations in our present study. First, we investigated the gene expression on mRNA level and the observed results does not necessary be confirm on protein level and does not necessary correlate with enzyme activity in case of iNOS. Secondary, we did not phenotype immune cells in tumor tissue. In attempt to detect gene expression to the conditions closer to the physiological state in patients, we used three different types of specimens surgically removed by advanced cases of CRC. Finally, we also must note the relatively small number of available specimens.
Despite the above limitation, we may conclude: i) the upregulation of Foxp3; IL10, TGFB1 and IL6 might be a transcriptional profile hallmark for colorectal metastases; ii) in addition, simultaneously Treg and Th17-related gene expression in primary tumor and in regional lymph node might provide suitable microenvironment sufficient for promoting metastatic growth.
Acknowledgements
This work was supported by Grants: №1/2016 and №2/2017 from the Fund for Scientific and Mobile project from Faculty of Medicine at the Trakia University, Stara Zagora, Bulgaria.
Funding
This work was supported by Grants: №1/2016 and №2/2017 from the Fund for Scientific and Mobile project from Faculty of Medicine at the Trakia University, Stara Zagora, Bulgaria.
Compliance with Ethical Standards
Conflict of Interest
The authors declare they have no conflict of interest.
Ethical Approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
References
- 1.Colotta F, Allavena P, Sica A, Garlanda C, Mantovani A. Cancer-related inflammation, the seventh hallmark of cancer: links to genetic instability. Carcinogenesis. 2009;30:1073–1081. doi: 10.1093/carcin/bgp127. [DOI] [PubMed] [Google Scholar]
- 2.Mantovani A. Cancer: inflaming metastasis. Nature. 2009;457:36–37. doi: 10.1038/457036b. [DOI] [PubMed] [Google Scholar]
- 3.Dobrzanski MJ. Expanding roles for CD4 T cells and their subpopulations in tumor immunity and therapy. Front Oncol. 2013;3:63. doi: 10.3389/fonc.2013.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Betts G, Jones E, Junaid S, El-Shanawany T, Scurr M, Mizen P, Kumar M, Jones S, Rees B, Williams G, Gallimore A, Godkin A. Suppression of tumour-specific CD4+ T cells by regulatory T cells is associated with progression of human colorectal cancer. Gut. 2012;61:1163–1171. doi: 10.1136/gutjnl-2011-300970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Omenetti S, Pizarro TT. The Treg/Th17 Axis: a dynamic balance regulated by the gut microbiome. Front Immunol. 2015;6:639. doi: 10.3389/fimmu.2015.00639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manel N, Unutmaz D, Littmanet DR. The differentiation of human T(H)-17 cells requires transforming growth factor-beta and induction of the nuclear receptor RORγt. Nat Immunol. 2008;9:641–649. doi: 10.1038/ni.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Leeuw RJ, Kost SE, Kakal JA, Nelson BH. The prognostic value of FoxP3+ tumor-infiltrating lymphocytes in cancer: a critical review of the literature. Clin Cancer Res. 2012;18:3022–3029. doi: 10.1158/1078-0432.CCR-11-3216. [DOI] [PubMed] [Google Scholar]
- 9.Frey DM, Droeser RA, Viehl CT, Zlobec I, Lugli A, Zingg U, Oertli D, Kettelhack C, Terracciano L, Tornillo L. High frequency of tumor-infiltrating FOXP3 (+) regulatory T cells predicts improved survival in mismatch repair-proficient colorectal cancer patients. Int J Cancer. 2010;126:2635–2643. doi: 10.1002/ijc.24989. [DOI] [PubMed] [Google Scholar]
- 10.Shang B, Liu Y, Jiang SJ, Liu Y. Prognostic value of tumor-infiltrating FoxP3+ regulatory T cells in cancers: a systematic review and meta-analysis. Sci Rep. 2015;5:15179. doi: 10.1038/srep15179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amicarella F, Muraro MG, Hirt C, Cremonesi E, Padovan E, Mele V, Governa V, Han J, Huber X, Droeser RA, Zuber M, Adamina M, Bolli M, Rosso R, Lugli A, Zlobec I, Terracciano L, Tornillo L, Zajac P, Eppenberger-Castori S, Trapani F, Oertli D, Iezzi G. Dual role of tumour-infiltrating T helper 17 cells in human colorectal cancer. Gut. 2017;66:692–704. doi: 10.1136/gutjnl-2015-310016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang J, Xu K, Wu J, Luo C, Li Y, Wu X, Gao H, Feng G, Yuan BZ. The changes of Th17 cells and the related cytokines in the progression of human colorectal cancer. BMC Cancer. 2012;12:418. doi: 10.1186/1471-2407-12-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tosolini M, Kirilovsky A, Mlecnik B, Fredriksen T, Mauger S, Bindea G, Berger A, Bruneval P, Fridman WH, Pagès F, Galon J. Clinical impact of different classes of infiltrating T cytotoxic and helper cells (Th1, Th2, Treg, Th17) in patients with colorectal cancer. Cancer Res. 2011;71:1263–1271. doi: 10.1158/0008-5472.CAN-10-2907. [DOI] [PubMed] [Google Scholar]
- 14.Brucklacher-Waldert V, Carr EJ, Linterman MA, Veldhoen M. Cellular plasticity of CD4+ T cells in the intestine. Front Immunol. 2014;5:488. doi: 10.3389/fimmu.2014.00488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-(Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Ling KL, Pratap SE, Bates GJ, Singh B, Mortensen NJ, George BD, Warren BF, Piris J, Roncador G, Fox SB, Banham AH, Cerundolo V. Increased frequency of regulatory T cells in peripheral blood and tumour infiltrating lymphocytes in colorectal cancer patients. Cancer Immun. 2007;7:7. [PMC free article] [PubMed] [Google Scholar]
- 17.Colombo MP, Piconese S. Regulatory-T-cell inhibition versus depletion: the right choice in cancer immunotherapy. Nat Rev Cancer. 2007;7:880–887. doi: 10.1038/nrc2250. [DOI] [PubMed] [Google Scholar]
- 18.Deng L, Zhang H, Luan Y, Zhang J, Xing Q, Dong S, Wu X, Liu M, Wang S. Accumulation of Foxp3+ T regulatory cells in draining lymph nodes correlates with disease progression and immune suppression in colorectal cancer patients. Clin Cancer Res. 2010;16:4105–4112. doi: 10.1158/1078-0432.CCR-10-1073. [DOI] [PubMed] [Google Scholar]
- 19.Salama P, Phillips M, Grieu F, Morris M, Zeps N, Joseph D, Platell C, Iacopetta B. Tumor-infiltrating FOXP3+ T regulatory cells show strong prognostic significance in colorectal cancer. J Clin Oncol. 2009;27:186–192. doi: 10.1200/JCO.2008.18.7229. [DOI] [PubMed] [Google Scholar]
- 20.Kim M, Grimmig T, Grimm M, Lazariotou M, Meier E, Rosenwald A, Tsaur I, Blaheta R, Heemann U, Germer CT, Waaga-Gasser AM, Gasser M. Expression of Foxp3 in colorectal cancer but not in Treg cells correlates with disease progression in patients with colorectal cancer. PLoS One. 2013;8:e53630. doi: 10.1371/journal.pone.0053630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miossec P, Kolls JK. Targeting IL-17 and TH17 cells in chronic inflammation. Nat Rev Drug Discov. 2012;11:763–776. doi: 10.1038/nrd3794. [DOI] [PubMed] [Google Scholar]
- 22.Kryczek I, Wu K, Zhao E, Wei S, Vatan L, Szeliga W, Huang E, Greenson J, Chang A, Roliński J, Radwan P, Fang J, Wang G, Zou W. IL-17+ regulatory T cells in the microenvironments of chronic inflammation and cancer. J Immunol. 2011;186:4388–4395. doi: 10.4049/jimmunol.1003251. [DOI] [PubMed] [Google Scholar]
- 23.Stanilov N, Miteva L, Mintchev N, Stanilova S. High expression of Foxp3, IL-23p19 and survivin mRNA in colorectal carcinoma. Int J Color Dis. 2009;24:151–157. doi: 10.1007/s00384-008-0588-8. [DOI] [PubMed] [Google Scholar]
- 24.Miteva LD, Stanilov NS, Deliysky TS, Stanilova SA. Significance of -1082A/G polymorphism of IL10 gene for progression of colorectal cancer and IL-10 expression. Tumour Biol. 2014;35:12655–12664. doi: 10.1007/s13277-014-2589-2. [DOI] [PubMed] [Google Scholar]
- 25.Kimura A, Kishimoto T. IL-6: regulator of Treg/Th17 balance. Eur J Immunol. 2010;40:1830–1835. doi: 10.1002/eji.201040391. [DOI] [PubMed] [Google Scholar]
- 26.Duhen T, Duhen R, Lanzavecchia A, Sallusto F, Campbell DJ. Functionally distinct subsets of human FOXP3+ Treg cells that phenotypically mirror effector Th cells. Blood. 2012;119:4430–4440. doi: 10.1182/blood-2011-11-392324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barbi J, Pardoll D, Pan F. Metabolic control of the Treg/Th17 axis. Immunol Rev. 2013;252:52–77. doi: 10.1111/imr.12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Obermajer N, Wong JL, Edwards RP, Chen K, Scott M, Khader S, Kolls JK, Odunsi K, Billiar TR, Kalinski P. Induction and stability of human Th17 cells require endogenous NOS2 and cGMP-dependent NO signaling. J Exp Med. 2013;210:1433–1445. doi: 10.1084/jem.20121277. [DOI] [PMC free article] [PubMed] [Google Scholar]