Abstract
Understanding genetic diversity and population structure is prerequisite to broaden the cultivated base of any crop. In the current investigation, we report discovery of a total of 319 alleles by assaying 81 SSRs on 71 chickpea genotypes. The cluster analysis based on Jaccard coefficient and unweighted neighbor joining algorithm categorized all genotypes into two major clusters. Cultivars grown within the same agro-climatic zones were clustered together, whereas the remaining genotypes particularly advanced breeding lines and accessions assigned to another cluster. Population structure analysis separated the entire collection into two subpopulations (K = 2) and the clustering pattern remained in close agreement with those of distance-based methods. Importantly, we also discovered marker trait association for membrane stability index (MSI) and leaf chlorophyll content measured as SPAD chlorophyll meter reading (SCMR), the two important physiological parameters indicative of heat stress (HS) tolerance in chickpea. Association analysis using both general linear and mixed linear models of the mean phenotypic data of traits recorded in 2016 and 2017 uncovered significant association of NCPGR206 and H2L102 with the MSI trait. Likewise, SSR markers GA9, TR31 and TA113 exhibited significant association with SCMR trait. The genomic regions putatively linked with two traits may be investigated in greater detail to further improve knowledge about the genetic architecture of HS tolerance in chickpea.
Electronic supplementary material
The online version of this article (10.1007/s13205-017-1057-2) contains supplementary material, which is available to authorized users.
Keywords: Chickpea, SSR, Genetic diversity, Population structure, Heat stress, Marker trait-association
Introduction
Globally, chickpea (Cicer arietinum L.) is the third most important grain legume crop after common bean (Phaseolus vulgaris L.) and pea (Pisum sativum L.). A total of 14.2 Mt chickpea is harvested annually from 14.8 m ha area worldwide with an average productivity of 0.96 t ha−1 (FAO 2014). With nearly 80% contribution to global chickpea production, India together with Southern and South-Eastern Asia remains the largest producer (Gaur et al. 2012). Like other grain legumes, chickpea plays a key role in addressing malnutrition related issues by offering plant-based dietary protein coupled with essential micronutrients and vitamins (Graham and Vance 2003). Also, chickpea helps enriching soil fertility through symbiotically fixing the atmospheric nitrogen to soil (Graham and Vance 2003).
Rigorous breeding efforts have led to the development and release of several high-yielding chickpea cultivars adapted to various agro-climatic zones in India (AICRP on Chickpea 2014). Potential yield of chickpea, however, remains unrealized and this may be due to several reasons including the vulnerability of this crop to a range of biotic and abiotic stresses (Jha et al. 2014a). Genetic bottleneck associated with the domestication process followed by human-led selection has caused significant loss in genetic diversity of chickpea (Abbo et al. 2003). At the same time, intensive breeding efforts focusing mainly on the development of high-yielding genotypes have resulted in narrow genetic base of cultivated gene pool, thus rendering these cultivars prone to a variety of biotic and abiotic stresses (Upadhyaya et al. 2008). In this respect, information about the genetic diversity available in crop’s gene pools facilitates broadening the genetic base of chickpea and assists accelerating genetic gains of the crop. To analyze genetic diversity for guiding selection of diverse parental combinations for hybridization, the role of molecular marker has been evident across different crops.
Several attempts have been made to investigate the genetic diversity in chickpea using diverse DNA marker systems such as random amplified polymorphic DNA (RAPD) (Ahmad 1999; Iruela et al. 2002), amplified fragment length polymorphism (AFLP) (Nguyen et al. 2004; Shan et al. 2005), inter-simple sequence repeat (ISSR) (Choudhary et al. 2013a; Aggarwal et al. 2015), simple sequence repeat (SSR) (Upadhyaya et al. 2008; Bharadwaj et al. 2010; Sefera et al. 2011; Keneni et al. 2012; Choudhary et al. 2012a; Ghaffari et al. 2014; Saxena et al. 2014; Hajibarat et al. 2014, 2015; De Giovanni et al. 2017) and single nucleotide polymorphism (SNP) (Bajaj et al. 2015a). Among the various DNA marker systems, SSRs are still preferred by the research community due to their co-dominant inheritance, high reproducibility and greater abundance across the genome (Gupta and Varshney 2000). Assessment of genetic diversity in chickpea so far has primarily focused on accessions of primary gene pool including Cicer reticulatum and C. echinospermum (Upadhyaya et al. 2008; Choudhary et al. 2012a, 2013a). However, studies on population structure and genetic diversity of high-yielding chickpea varieties adapted to different agro-climatic zone of India, along with advanced breeding lines remain limited. In the current study, we here conducted an SSR-based diversity analysis of 44 chickpea varieties cultivated across diverse agro-climatic zones in India along with 27 genotypes that cover germplasm accessions, advanced breeding lines and landraces. Given the fact that up to 53 kg/ha of chickpea yield is lost due to heat stress (HS) (Kalra et al. 2008; Jha et al. 2014a, b), we recorded data on membrane stability index (MSI) and SPAD chlorophyll meter reading (SCMR) in the panel. The MSI and SCMR represent the two key physiological traits related to HS tolerance in chickpea. Finally, we performed association mapping to discover putative MTAs explaining variation for the studied traits.
Materials and methods
A set of 71 desi chickpea genotypes comprising 44 varieties and 27 genotypes including accessions from ICRISAT Hyderabad, advanced breeding lines (from IIPR, Kanpur, and JNKVV, Jabalpur, India) and landraces was used in the present study (see Supplementary File Table 1). Importantly, the panel also involved heat tolerant and sensitive genotypes as reported by previous researchers (Krishnamurthy et al. 2011; Jha and Shil 2015; Gaur et al. 2016).
DNA extraction and SSR analysis
Genomic DNA was extracted from plant leaves following the CTAB method. A total of 120 SSR markers were screened in the collection, of which 81 SSRs yielded polymorphic fragments. The SSRs used here are reported previously by different research groups (Winter et al. 1999, 2000; Sethy et al. 2003, 2006; Gaur et al. 2011; Choudhary et al. 2012b) and correspond to all eight linkage groups (LGs) of chickpea (Table 1).
Table 1.
Allele number (Na), gene diversity (He) and polymorphism information content recorded in 71 genotypes with 81 SSR markers
| No. | Marker | PIC | No. of alleles (Na) | Gene diversity (He) | LG group | References |
|---|---|---|---|---|---|---|
| 1 | CakTpSSR03637 | 0.526 | 3 | 0.59 | – | – |
| 2 | Cak TpSSR2543 | 0.508 | 3 | 0.57 | – | – |
| 3 | NCPGR149 | 0.73 | 6 | 0.77 | – | Gaur et al. (2011) |
| 4 | NCPGR231 | 0.693 | 5 | 0.74 | LG4 | Gaur et al. (2011) |
| 5 | NCPCR234 | 0.709 | 5 | 0.75 | – | Gaur et al. (2011) |
| 6 | NCPGR136 | 0.663 | 5 | 0.71 | LG1 | Gaur et al. (2011) |
| 7 | CESSR172 | 0.421 | 3 | 0.53 | LG2 | Choudhary et al. (2012b) |
| 8 | STMS10 | 0.581 | 3 | 0.66 | LG3 | Winter et al. (1999) |
| 9 | GA105 | 0.307 | 2 | 0.38 | LG3 | Winter et al. (2000) |
| 10 | ICCeM018 | 0.415 | 3 | 0.53 | LG3 | Gujaria et al. (2011) |
| 11 | TR19 | 0.711 | 7 | 0.75 | LG2 | Winter et al. (2000) |
| 12 | TA2 | 0.696 | 5 | 0.74 | LG4 | Winter et al. (1999) |
| 13 | TS54 | 0.812 | 8 | 0.83 | LG4 | Winter et al. (2000) |
| 14 | NCPGR76 | 0.371 | 2 | 0.49 | LG6 | Gaur et al. (2011) |
| 15 | CESSR164 | 0.479 | 4 | 0.57 | LG4 | Choudhary et al. (2012b) |
| 16 | CESSR114 | 0.374 | 2 | 0.50 | LG4 | Choudhary et al. (2012b) |
| 17 | TS53 | 0.762 | 7 | 0.79 | LG5 | Winter et al. (2000) |
| 18 | NCPGR139 | 0.633 | 6 | 0.68 | LG6 | Gaur et al. (2011) |
| 19 | TA176 | 0.685 | 7 | 0.72 | LG6 | Winter et al. (1999) |
| 20 | H2L102 | 0.436 | 3 | 0.51 | LG5 | Choudhary et al. (2012b) |
| 21 | TAAS | 0.7 | 5 | 0.74 | LG5 | Winter et al. (2000) |
| 22 | NCPGR199 | 0.528 | 4 | 0.60 | LG4 | Gaur et al. (2011) |
| 23 | NCPGR200 | 0.802 | 7 | 0.83 | LG6 | Gaur et al. (2011) |
| 24 | TA80 | 0.749 | 6 | 0.78 | LG 6 | Winter et al. (2000) |
| 25 | GA102 | 0.528 | 3 | 0.60 | LG7 | – |
| 26 | H4F07 | 0.493 | 5 | 0.53 | LG6 | Gujaria et al. (2011) |
| 27 | CESSR432 | 0.37 | 3 | 0.47 | LG5 | Choudhary et al. (2012b) |
| 28 | NCPGR56 | 0.411 | 3 | 0.46 | LG5 | Sethy et al. (2006) |
| 29 | NCPGR 238 | 0.352 | 2 | 0.46 | LG6 | Gaur et al. (2011) |
| 30 | GA9 | 0.682 | 5 | 0.73 | LG6 | Winter et al. (2000) |
| 31 | H5A04 | 0.515 | 4 | 0.57 | LG6 | Choudhary et al. (2012b) |
| 32 | NCPGR202 | 0.632 | 5 | 0.69 | LG6 | Gaur et al. (2011) |
| 33 | CESSR45 | 0.413 | 3 | 0.49 | LG1 | Choudhary et al. (2012b) |
| 34 | CESSR159 | 0.352 | 3 | 0.42 | LG1 | Choudhary et al. (2012b) |
| 35 | TR31 | 0.506 | 3 | 0.59 | LG3 | Winter et al. (1999) |
| 36 | CakTpSSR02719 | 0.545 | 4 | 0.61 | – | – |
| 37 | CakTpSSR04076 | 0.395 | 3 | 0.51 | – | – |
| 38 | CakTpSSR03090 | 0.609 | 4 | 0.67 | – | – |
| 39 | NCPGR165 | 0.649 | 4 | 0.70 | LG1 | Gaur et al. (2011) |
| 40 | ICCM0297 | 0.55 | 3 | 0.63 | LG1 | Nayak et al. (2010) |
| 41 | TA113 | 0.59 | 6 | 0.64 | LG1 | Winter et al. (1999) |
| 42 | TAA60 | 0.54 | 3 | 0.61 | LG2 | Winter et al. (2000) |
| 43 | GA6 | 0.785 | 7 | 0.81 | LG2 | – |
| 44 | NCPGR40 | 0.656 | 5 | 0.71 | LG2 | Sethy et al. (2006) |
| 45 | TA64 | 0.801 | 6 | 0.83 | LG3 | Winter et al. (1999) |
| 46 | NCPGR12 | 0.431 | 3 | 0.54 | LG3 | Sethy et al. (2003) |
| 47 | NCPGR274 | 0.534 | 4 | 0.61 | LG3 | Gaur et al. (2011) |
| 48 | NCPGR220 | 0.533 | 4 | 0.60 | LG3 | Gaur et al. (2011) |
| 49 | NCPGR232 | 0.664 | 5 | 0.71 | LG5 | Gaur et al. (2011) |
| 50 | NCPGR46 | 0.58 | 3 | 0.65 | LG6 | Sethy et al. (2006) |
| 51 | NCPGR156 | 0.381 | 3 | 0.50 | LG6 | Gaur et al. (2011) |
| 52 | NCPGR155 | 0.375 | 2 | 0.50 | LG6 | Gaur et al. (2011) |
| 53 | STMS25 | 0.391 | 3 | 0.51 | LG7 | Winter et al. (1999) |
| 54 | H5G12 | 0.375 | 2 | 0.50 | LG7 | Choudhary et al. (2012b) |
| 55 | TA140 | 0.454 | 3 | 0.54 | LG7 | Winter et al. (2000) |
| 56 | CESSR43 | 0.375 | 2 | 0.50 | LG1 | Choudhary et al. (2009) |
| 57 | NCPGR33 | 0.596 | 4 | 0.67 | LG1 | Sethy et al. (2006) |
| 58 | NCPGR225 | 0.362 | 2 | 0.47 | LG3 | Gaur et al. (2011) |
| 59 | H1B04 | 0.547 | 5 | 0.61 | LG3 | Choudhary et al. (2012b) |
| 60 | TA106 | 0.797 | 8 | 0.82 | LG6 | Winter et al. (2000) |
| 61 | NCPGR93 | 0.699 | 5 | 0.74 | LG6 | Sethy et al. (2006) |
| 62 | GA26 | 0.614 | 4 | 0.68 | LG6 | Winter et al. (2000) |
| 63 | STMS7 | 0.385 | 3 | 0.49 | LG5 | Winter et al. (1999) |
| 64 | NCPGR6 | 0.541 | 4 | 0.60 | LG1 | Sethy et al. (2003) |
| 65 | TA8 | 0.636 | 5 | 0.69 | LG1 | Winter et al. (1999) |
| 66 | CESSR433 | 0.369 | 2 | 0.49 | LG1 | Choudhary et al. (2012b) |
| 67 | CESSR139 | 0.364 | 3 | 0.46 | LG1 | Choudhary et al. (2012b) |
| 68 | H2B061 | 0.371 | 2 | 0.49 | LG2 | Gujaria et al. (2011) |
| 69 | NCPGR193 | 0.433 | 3 | 0.54 | LG2 | Gaur et al. (2011) |
| 70 | NCPGR13 | 0.298 | 2 | 0.36 | LG2 | Sethy et al. (2003) |
| 71 | NCPGR110 | 0.491 | 3 | 0.56 | LG2 | Gaur et al. (2011) |
| 72 | TR7 | 0.447 | 3 | 0.55 | LG6 | Winter et al. (2000) |
| 73 | CESSR105 | 0.39 | 3 | 0.50 | LG3 | Choudhary et al. (2012b) |
| 74 | NCPGR267 | 0.347 | 2 | 0.45 | LG6 | Gaur et al. (2011) |
| 75 | NCPGR206 | 0.352 | 3 | 0.44 | LG6 | Gaur et al. (2011) |
| 76 | NCPGR255 | 0.415 | 3 | 0.53 | LG7 | Gaur et al. (2011) |
| 77 | NCPGR52 | 0.701 | 4 | 0.75 | LG7 | Sethy et al. (2006) |
| 78 | TA110 | 0.74 | 6 | 0.78 | LG2 | Winter et al. (2000) |
| 79 | NCPGR41 | 0.556 | 3 | 0.63 | LG7 | Sethy et al. (2006) |
| 80 | TA18 | 0.367 | 3 | 0.44 | LG7 | Winter et al. (2000) |
| 81 | TA180 | 0.69 | 5 | 0.74 | LG7 | Winter et al. (2000) |
PCR analysis
PCR was carried out in a 10 µl reaction mixture that contained 5.9 µl of sterilized distilled water, 1.00 µl template DNA (25 ng), 0.5 µl of forward and 0.5 µl of reverse primer (5 µM), 1.00 µl 10 × PCR buffer (10 mM Tris–Hcl, 50 mM KCl, pH 8.3), 1.00 µl dNTP mix (0.2 mM each of dATP, dGTP, dCTP and dTTP) and 0.1 µl Taq polymerase (5 U/µl) (Thermo Fisher Scientific Mumbai, India, Pvt. Ltd.) using G-40402 thermo cycler (G-STORM, Somerset, UK). A touchdown PCR profile was used for amplification with initial denaturation at 94 °C for 5 min followed by 10 cycles of touch down 61–51 °C, 30 s at 94 °C, annealing for 30 s at 61 °C (the annealing temperature for each cycle being reduced by 1 °C per cycle) and extension for 30 s at 72 °C. This was accompanied by 40 cycle of denaturation at 94 °C for 30 s, annealing at 51 °C for 30 s, elongation at 72 °C for 45 s, and 10 min of final extension at 72 °C. Amplified fragments were resolved in 3% agarose gel using 0.5 × TBE running buffer and images were analyzed with Quantity one software (Bio-Rad, CA 94547, USA).
Genetic diversity and population structure analysis
The genetic diversity parameters viz., number of alleles per locus (Na), gene diversity (He) and polymorphism information content (PIC) were analyzed by Power Marker v. 3.25 (Liu and Muse 2005). With 1000 bootstrap value, neighborhood joining tree analysis was performed with DARwin v. 6.0.13 (Perrier and Jacquemoud-Collet 2006). DARwin v. 6.0.13 was also used for factorial analysis, while principal coordinate analysis (PCoA) was performed using GenAlEx v. 6.502 (Peakall and Smouse 2012). To determine population structure (Q) and the subpopulation (K) in the given set, model-based analysis was conducted with STRUCTURE v 2.3.4 (Pritchard et al. 2000). By applying admixture model, five independent runs were conducted with 200000 Markov Chain Monte Carlo (MCMC) iterations for each K value ranging from 1 to 10 with a burn-in length of 200. In parallel, the best K value was obtained according to the ΔK method of Evanno et al. (2005) by processing the STRUCTURE results by STRUCTURE HARVESTER (Earl and von Holdt 2012) (http://taylor0.biology.ucla.edu).
Analysis of MSI and SCMR in the panel
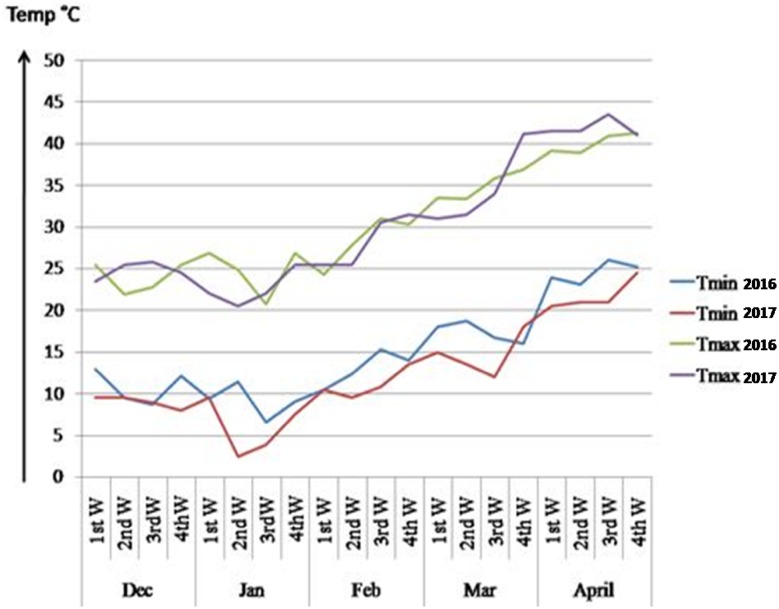
The data on these two traits were recorded at reproductive stage from 71 genotypes for consecutive 2 years (2016 and 2017) under late sown condition. The mean weekly temperature during crop growth is shown in Fig. 1. The genotypes were grown in augmented design along with three checks in five blocks. The third leaf from the top of each genotype was selected for recording observations on MSI and SCMR. The MSI was estimated based on electrolyte leakage under stress (Sullivan 1972) and was calculated using the following formula given by Blum and Ebercon (1979).MSI = 100 − membrane injury index (MII),where MII is calculated as a ratio of C1 and C2, with C1 and C2 representing the electrolytes measured at 40 and 80 °C, respectively. Similarly, SCMR is used as an indirect measure of leaf chlorophyll content. Three SCMR measurements were recorded in third fully expanded young leaf from the top at reproductive stage using a non-destructive, portable SPAD-502 chlorophyll, meter and averaged subsequently.
Fig. 1.
Mean weekly minimum and maximum day temperature recorded during crop growing period under late sown condition
Statistical analysis
Analysis of variance (ANOVA) was performed for year-wise data for the two traits using SAS v.9.2. A combined analysis of variation over years was carried out for partitioning the phenotypic variance into year, genotype, and error variances. To study the genotypic variation in the given panel of genotypes, various descriptive statistics including mean, median, standard deviation (SD), standard error, coefficient of variation and broad sense heritability were computed based on the phenotypic data recorded for the two traits in the genotypes.
Association mapping to detect significant MTAs
The phenotypic data on MSI and SCMR traits and the genotypic data were analyzed to discover significant MTAs. Both models viz., general linear (GLM) and mixed linear (MLM) based on Q and Q + K matrix, respectively, were employed. TASSEL v. 3.0 (Bradbury et al. 2007; Zhang et al. 2010) was used to detect MTAs, and P = 0.05 was considered as a significance threshold.
Putative candidate gene analysis
To find the possible candidate genes that correspond to the reported MTAs and the putative proteins encoded by these, we performed BLASTn search for the associated SSRs against the reference genome sequence of chickpea (CDC frontier) (Varshney et al. 2013). In parallel, the proteins were predicted for the corresponding sequences using InterPro (IPR).
Results
SSR analysis
In the current study, genotyping of 71 chickpea genotypes with 81 SSRs provided a total of 319 alleles with an average of 3.9 alleles per marker (Table 1). The number of alleles ranged from 1 to 8, and the maximum number of alleles was obtained for the markers TS53, TA106 and TS54 (Table 1). The PIC values ranged from 0.29 to 0.81 with an average of 0.53. Similarly, gene diversity varied between 0.3 and 0.8 with a mean value of 0.6. Interestingly, a comparative (decade wise) analysis of different cultivars released over the last 50 years suggested an overall improvement of genetic diversity based on estimated gene diversity with the values ranging between 0.36 and 0.83 (Table 2).
Table 2.
Comparison of undertaken released chickpea cultivars as per the period of release
| Period of release | No. of genotypes/varieties | Total no. of alleles | Avg. no. of alleles/marker | PIC |
|---|---|---|---|---|
| All genotypes | 71 | 319 | 3.9 | 0.53 |
| All cultivars | 44 | 299 | 3.7 | 0.51 |
| 2001–2012 | 9 | 242 | 2.9 | 0.47 |
| 1990–2000 | 15 | 267 | 3.3 | 0.48 |
| 1980–1990 | 15 | 261 | 3.2 | 0.47 |
| Before 1970 | 5 | 213 | 2.6 | 0.43 |
Genetic diversity analysis and relationships among the chickpea genotypes
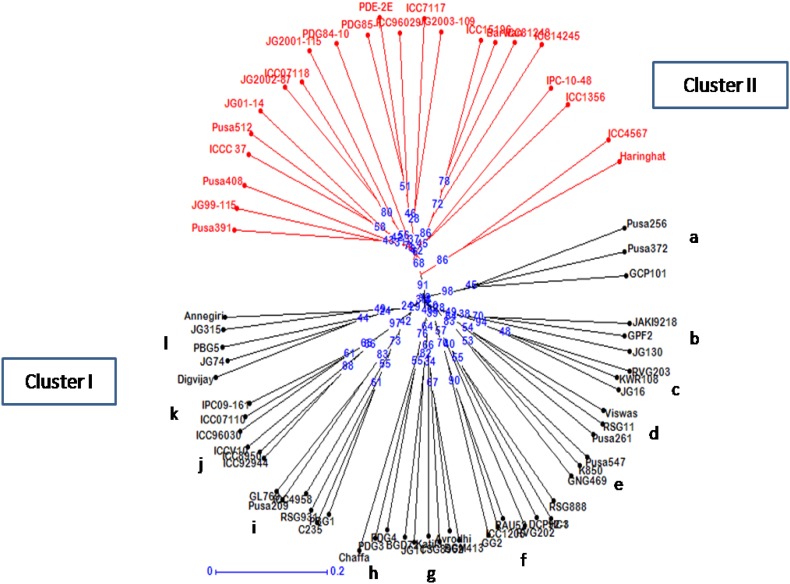
The cluster analysis based on unweighted neighbor joining method clearly separated 71 genotypes into two major clusters (Fig. 2). Cluster I contained 41 released cultivars that are widely adapted to various agro-climatic zones in India, along with six improved lines and one landrace (Katila). The second cluster harbored 19 advanced breeding lines, one landrace Barwan and three released cultivars viz., Pusa 512, Pusa 391 and Pusa 408. Further, examination at sub-cluster level suggested that most of the genotypes harbored within the same sub-cluster are recommended for cultivation in the same agro-climatic zone of India. For instance, cultivars Pusa 256 and Pusa 372 belonging to sub-cluster (a) were released for both North West Plain Zone (NWPZ) and Central Zone (CZ). Likewise, RSG 11 and Pusa 261 existing in sub-cluster (d) were released for NWPZ. The majority of varieties belonging to sub-clusters (e)–(i) were released for NWPZ (see Fig. 2). Importantly, KWR 108 and JG 16 from sub-cluster (c) were released for North East Plain Zone (NEPZ) of India. While sub-cluster (b) containing cultivars JAKI 9218 and JG130 and sub-cluster (j) containing ICCV 10 and ICCV 92944 were released for CZ and South Zone (SZ). Similarly, most of the cultivars existing in sub-cluster (l) were released for CZ.
Fig. 2.
Neighbour joining phylogenetic tree using SSR marker data in 71 chickpea genotypes. Bootstrap values are indicated at the node of each cluster
Structure analysis
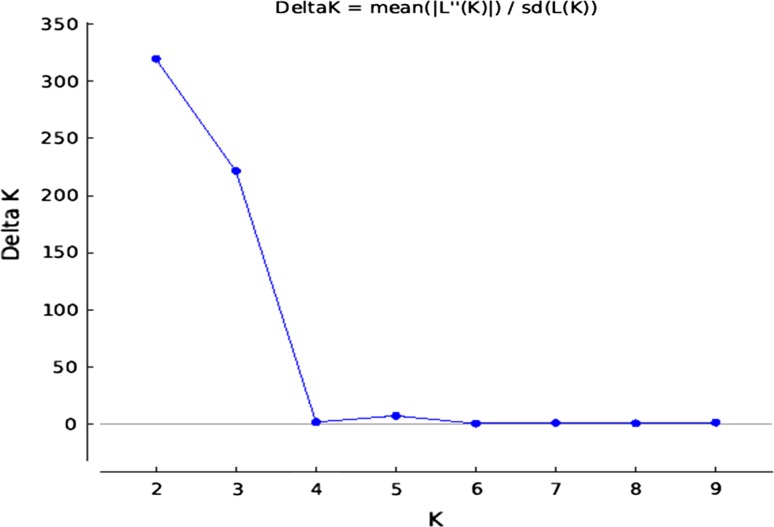
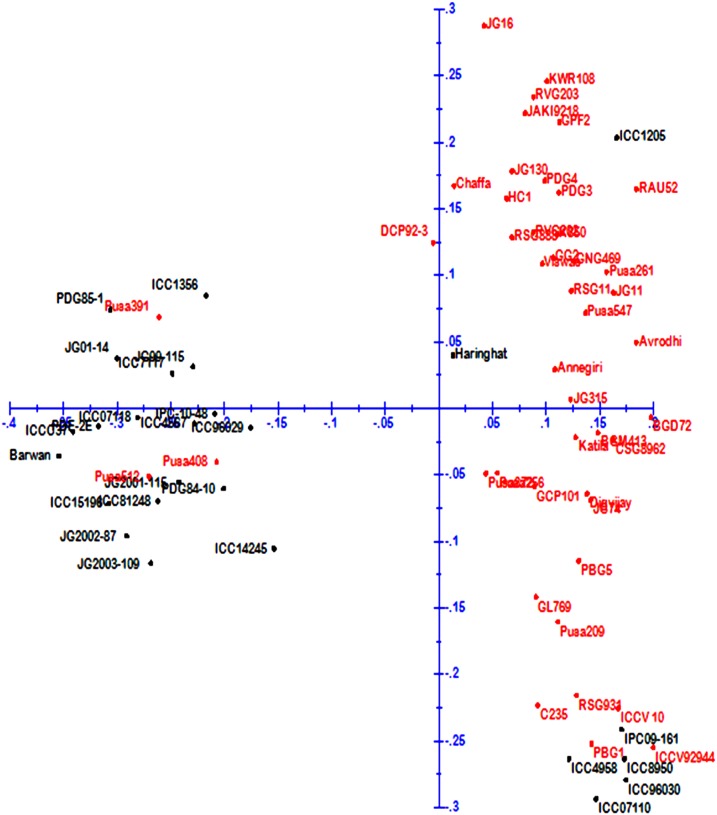
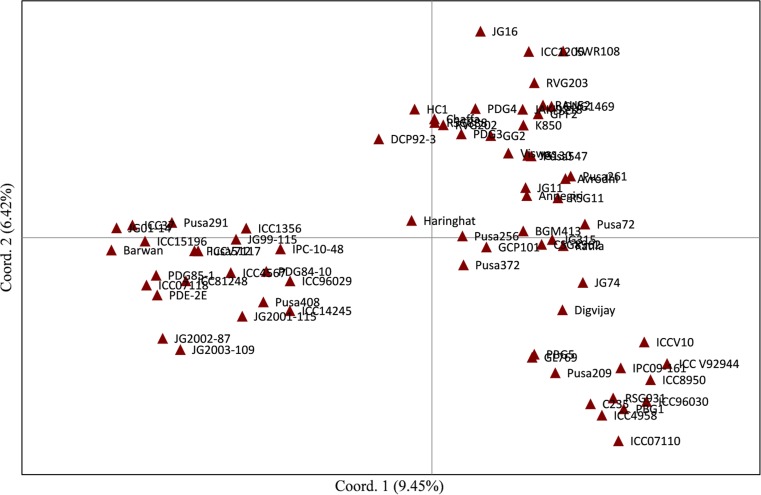
We dissected the population structure of the germplasm panel using Bayesian-based clustering approach with K values ranging from 1 to 10, which led the genotypes clustering into two distinct subpopulations (see Fig. 1 in Supplementary File). This pattern corroborated with the highest peak found at K = 2 (Fig. 3). The first subpopulation comprised the released cultivars, whereas the rest of the genotypes remained in the second subpopulation representing predominantly the advanced breeding lines and germplasm accessions. The clustering pattern inferred from factorial analysis agreed with the results obtained from STRUCTURE analysis. The genotypes existing in subpopulation of STRUCTURE are contained in quadrants I and IV of factorial analysis (Fig. 4), whereas the genotypes in subpopulation 2 of STRUCTURE were represented by quadrants II and III. Similarly, as shown in Fig. 5, two clusters were also recorded in PCoA with the PC1 and PC2 explaining 9.45 and 6.42%, respectively of the total variance.
Fig. 3.
Relationship between K and ΔΚ based on STRUCTURE analysis of chickpea genotypes based on SSR marker data
Fig. 4.
Factorial analysis of 71 genotypes based on 81 SSR markers
Fig. 5.
PCoA analysis of 71 genotypes, PC1 explaining 9.45% and PC2 explaining 15.87% of variance, respectively
Analysis of MSI and SCMR
Combined ANOVA of MSI and SCMR recorded over 2 years suggested significant genetic difference among the genotypes (Table 3). For the two traits, values for the range, median, standard error, and coefficient of variation are given in Table 4. The frequency distributions for the two traits are shown in (Figs. 2 and 3 see in Supplementary File).
Table 3.
Combined ANOVA for MSI and SCMR traits under late sown conditions (2016 and 2017)
| Source of variation | df | MSI 2016–2017 | SCMR 2016–2017 |
|---|---|---|---|
| Mean of squares | Mean of squares | ||
| MSI | SCMR | ||
| Year | 1 | 59.67 | 47.082935 |
| Genotype | 70 | 155.8** | 158.325869** |
| Error | 70 | 53.26 | 80.862299 |
| h2 (Broad sense) | 0.7417 | 0.4893 |
**P < 0.01
Table 4.
Descriptive statistics for two traits recorded in 71 genotypes
| Trait | Minimum | Maximum | Mean | Median | Standard deviation | Mean standard error | CV% |
|---|---|---|---|---|---|---|---|
| MSI (%) | 19.56 | 75.3 | 41.4 | 37.4 | 15.1 | 1.8 | 35.09 |
| SCMR | 11.1 | 57.1 | 27.3 | 24.9 | 9 | 1.07 | 22.67 |
Association analysis
We attempted to discover MTAs through analyzing the genotyping data of 81 SSRs and two traits with GLM and MLM approaches. Concerning analysis of MSI (year 2016) with GLM, the markers NCPGR267, NCPGR206, H2L102, TA64, NCPGR156, and NCPGR238 showed significant associations, with phenotypic variation (PV) ranging from 6.1 to 20.2% (Table 5). On the other hand, analysis with MLM approach for MSI (year 2016) uncovered significant association of five SSR markers (NCPGR267, NCPGR206, H2L102, TS53 and NCPGR156) with the trait accounting for PVs in the range of 8.5 and 22.2% (Table 6). Importantly, association of three SSR markers (NCPGR267, H2L102 and NCPGR206) with MSI trait was evident by both GLM and MLM in 2 years (2016 and 2017).
Table 5.
Significant MTA and their effects on MSI and SCMR traits by Q GLM approach of association mapping
| Trait | Year | Marker | LG group | P value | PV% |
|---|---|---|---|---|---|
| MSI | 2016 | NCPGR267 | LG5 | 0.0004 | 17 |
| MSI | NCPGR206 | LG2 | 0.00252 | 12.6 | |
| MSI | H2L102 | LG6 | 0.04448 | 11.2 | |
| MSI | TA64 | LG6 | 0.02062 | 20.2 | |
| MSI | NCPGR156 | LG6 | 0.01094 | 15.2 | |
| MSI | NCPGR238 | LG6 | 0.03756 | 6.1 | |
| MSI | 2017 | NCPGR 267 | LG5 | 0.00139 | 13.8 |
| MSI | H2L102 | LG6 | 0.01282 | 14.7 | |
| MSI | NCPGR206 | LG2 | 0.0423 | 5.9 | |
| MSI | Mean phenotypic | NCPGR206 | LG2 | 0.00866 | 9.7 |
| MSI | Data of 2016–2017 | H2L102 | LG6 | 0.02105 | 13.4 |
| SCMR | 2016 | GA9 | LG5 | 0.01191 | 19.7 |
| SCMR | CESSR172 | LG1 | 0.00422 | 17.7 | |
| SCMR | TA 113 | LG1 | 0.03007 | 17.7 | |
| SCMR | NCPGR199 | LG1 | 0.03453 | 15.6 | |
| SCMR | CESSR114 | LG3 | 0.03455 | 9.4 | |
| SCMR | ICCM0297 | LG5 | 0.0205 | 10.8 | |
| SCMR | NCPGR267 | LG5 | 0.03812 | 6 | |
| SCMR | 2017 | TR31 | LG2 | 0.04308 | 6.5 |
| SCMR | CESSR433 | LG4 | 0.04755 | 5.5 | |
| SCMR | Mean phenotypic | TA113 | LG1 | 0.00697 | 22.2 |
| SCMR | Data of 2016–2017 | GA9 | LG5 | 0.00856 | 20.7 |
P = 0.05
Table 6.
Significant MTA and their effects on MSI and SCMR traits by Q + K MLM approach of association mapping
| Trait | Year | Marker | LG group | P value | PV% |
|---|---|---|---|---|---|
| MSI | 2016 | NCPGR267 | LG5 | 0.00112 | 16.5 |
| MSI | NCPGR206 | LG2 | 0.01758 | 8.5 | |
| MSI | H2L102 | LG6 | 0.02399 | 14.3 | |
| MSI | TS 53 | LG7 | 0.03182 | 22.2 | |
| MSI | NCPGR 156 | LG6 | 0.0365 | 12.8 | |
| MSI | 2017 | NCPGR267 | LG5 | 0.00268 | 13.8 |
| MSI | H2L 102 | LG6 | 0.02129 | 14.7 | |
| MSI | NCPGR206 | LG2 | 0.04754 | 5.9 | |
| MSI | Mean phenotypic | NCPGR206 | LG2 | 0.02589 | 7.5 |
| MSI | Data of 2016–2017 | NCPGR267 | LG5 | 0.00132 | 16 |
| MSI | H2L102 | LG6 | 0.01711 | 15.5 | |
| SCMR | 2016 | ICCM 0297 | LG5 | 0.04255 | 9.5 |
| SCMR | CESSR 172 | LG1 | 0.04478 | 12.2 | |
| SCMR | 2017 | TR31 | LG2 | 0.02917 | 7.6 |
| SCMR | Mean phenotypic | TR31 | LG2 | 0.03894 | 6.8 |
| SCMR | Data of 2016–2017 | GA9 | LG5 | 0.04221 | 17.5 |
P = 0.05
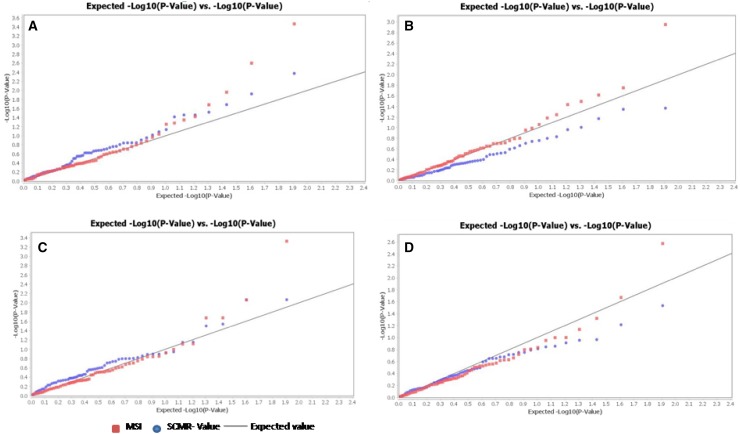
Likewise for SCMR data (2016), GLM revealed that the SSRs GA9, CESSR172, TA113, NCPGR199, CESSR114, ICCM0297 and NCPGR267 had significant association with the trait. In case of MLM, association of two SSR markers ICCM0297 and CESSR172 with the SCMR could be established. In 2017, GLM analysis suggested that TR31 and CESSR433 markers had significant association with SCMR (P values 0.04308 and 0.04755). The significant association of the SSR marker TR31 with SCMR trait was obtained in both MLM and GLM in the year 2017. Two important markers NCPGR206 and H2L102 showed consistently significant MTA for MSI trait recorded exclusively in the year 2016 and 2017 and mean data of 2016 and 2017 given the both GLM and MLM analysis. The quantile–quantile (Q–Q) plot of these calculated P values is depicted in Fig. 6 considering both GLM and MLM and the markers showed association with the studied traits by deviating from the null expectation (Fig. 6). The significant MTA between the marker GA9 and SCMR trait was evident by both methods across 2 years. The SSR markers viz., NCPGR206, H2L102 and GA9 showing stable MTAs with MSI and SCMR traits could be potentially harnessed in future chickpea breeding for improving HS tolerance.
Fig. 6.
a, b Quantile–Quantile (Q–Q) plots for MSI and SCMR (tested by GLM). c, d Quantile–Quantile (Q–Q) plots for MSI and SCMR (tested by MLM) witnessing significant association of markers with the given traits by deviating from the expected null distribution of P values, assuming no associations, represented as solid line; distribution of P values observed
Candidate gene identification
The SSR markers showing significant MTAs were BLASTed against the Kabuli genome (CDC frontier) sequence of chickpea (Varshney et al. 2013) for gene prediction. For MSI, two proteins including ATPase/AAA-type/ATP-binding protein, and zinc finger/C2H2-type were predicted from the sequence showing homology with the SSR markers showing association with the trait. Similarly for SCMR, five proteins viz., Domain of unknown function DUF1084, DNA glycosylase/AP lyase, H2TH DNA-binding, zinc finger, Nitrilase/cyanide hydratase and apolipoprotein N-acyltransferase and Aquaporin NIP were predicted (Table 7).
Table 7.
Putative candidate gene underlying the significant MTAs and their putative functions
| Trait | Marker | Candidate gene | LG group | P value | PV% | Marker effect | Putative proteins (based on InterPro) |
|---|---|---|---|---|---|---|---|
| MSI | NCPGR267 | Ca_05237 | CaLG6 | 0.00112 | 16.5 | 13.54 | ATPase, AAA-type, ATP binding |
| MSI | H2L102 | Scaffold | – | 0.02399 | 14.3 | – | – |
| MSI | TS53 | Scaffold | – | 0.03182 | 22.2 | – | – |
| MSI | NCPGR156 | Ca_18128 | CaLG2 | 0.0365 | 12.8 | – | – |
| MSI | NCPGR238 | Ca_14617 | CaLG6 | 0.03756 | 6.1 | 6.97364 | Zinc finger, RING-type, zinc ion binding |
| SCMR | GA9 | Ca_05159 | CaLG5 | 0.01191 | 19.7 | 3.83725 | Domain of unknown function DUF 1084 |
| SCMR | CESSR172 | Ca_02896 | CaLG1 | 0.00422 | 17.7 | 9.12119 | DNA glycosylase/AP lyase, H2TH DNA-binding |
| SCMR | TA113 | Ca_02459 | CaLG1 | 0.03007 | 17.7 | 7.19934 | Zinc finger, C2H2-type, Zinc ion binding |
| SCMR | NCPGR199 | Scaffold | – | 0.03453 | 15.6 | – | – |
| SCMR | CESSR 114 | Ca_07065 | CaLG1 | 0.03455 | 9.4 | – | Nitrilase/cyanide hydratase and apolipoprotein N-acyltransferase |
| SCMR | ICCM 0297 | – | – | 0.0205 | 10.8 | – | – |
| SCMR | NCPGR267 | Ca_05237 | CaLG6 | 0.03812 | 6 | – | ATPase, AAA-type, ATP binding |
| SCMR | TR31 | Ca_21333 | CaLG3 | 0.04308 | 6.5 | 4.9576 | Aquaporin NIP, Major intrinsic protein |
| SCMR | CESSR433 | Ca_14730 | CaLG1 | 0.04755 | 5.5 | – | Putative uncharacterized protein |
Discussion
Knowledge about the genetic diversity within crop’s gene pool is a prerequisite for improving gains in breeding programs of crops (Upadhyaya et al. 2008; Ghaffari et al. 2014). Introduction of novel alleles to the cultivated pool holds the potential to break the yield ceiling (Choudhary et al. 2012a). In this respect, molecular markers such as SSR are promising in discerning genetic diversity at DNA level. Also, the marker systems have been efficiently recruited for identifying important MTAs to accelerate trait improvement in chickpea (Hüttel et al. 1999; Udupa et al. 1999; Upadhyaya et al. 2008; Bharadwaj et al. 2011; Sefera et al. 2011; Choudhary et al. 2012a). In the present study, a total of 319 alleles with an average of 3.9 alleles per locus were recovered from 71 chickpea genotypes assayed with 81 SSRs. The results suggested the presence of moderate level of genetic diversity in the studied panel. This finding showed agreement with the previous reports (Ghaffari et al. 2014; Hajibarat et al. 2015), which also suggested lesser genetic diversity among cultivated chickpea genotypes. A moderate value of average PIC and lower count for average alleles per locus noted here might be due to inclusion of cultivated genotypes. Inclusion of various chickpea wild species and landraces could produce higher PIC value and higher number of allele count per locus as suggested by Upadhyaya et al. (2008) and Ghaffari et al. (2014).
The level of heterozygosity ranged from 0.3 (GA105) to 0.83 (TS54) with a mean of 0.6, which was in accordance with the values obtained by Upadhyaya et al. (2008) and Hajibarat et al. (2015). A positive association between number of alleles and PIC value observed in this study was congruent with the results reported previously by other researchers in chickpea (Upadhyaya et al. (2008) and Sefera et al. (2011). Increasing trends of genetic diversity based on PIC values calculated in the cultivars released in the early decades of seventies and post decades of eighties reflected in the our study were similar with the results obtained by Sefera et al. (2011). Additionally, Thudi et al. (2016) have also evidenced increase in genetic diversity based on the presence of higher copy number variations (CNVs) in chickpea varieties released after the year 2002. A decade wise analysis of 59 Indian pigeonpea varieties with 60 SSR markers showed a near constant genetic diversity over the last 50 years (Bohra et al. 2017). A trend of depicting progressive increase in genetic diversity was reported in rice using SSR analysis of varieties released from 1970 to 2000 in India (Choudhary et al. 2013b). Moreover, results of UPGMA and sub-cluster analysis indicated limited genetic diversity among the cultivars released for the same agro-climatic zone.
Population structure analysis unravels the existing genetic diversity across the collection. In recent years, previous studies have shed light into the genetic structure of chickpea using SSR and SNP markers (Kujur et al. 2013; Diapari et al. 2014; Thudi et al. 2014; Bajaj et al. 2015a; De Giovanni et al. 2017). In our analysis, the patterns arising from STRUCTURE were consistent with those obtained from distance-based clustering methods (NJ and PCoA analyses). Of the two major clusters, released varieties grouped within one cluster, while the other group retained advanced breeding lines and accessions. A previous report in chickpea divided the entire genotypes into two distinct groups: desi and kabuli types (Upadhyaya et al. 2008). More recently, population structure analysis of 103 chickpea accessions yielded three distinct subpopulations (De Giovanni et al. 2017).
Elucidating the genetic architecture of complex traits in crops is crucial to accelerated trait improvement and association mapping has emerged as a robust technique in this respect (Huang and Han 2014). In chickpea, genome-wide genetic variants showing association with a range of agriculturally important traits including drought, HS, zinc and iron content, 100 seed weight, seed coat color, seed protein content, flowering time were discovered by employing association genetics (Thudi et al. 2014; Diapari et al. 2014; Bajaj et al. 2015b; Upadhyaya et al. 2015, 2016a, b). Stress conditioned by heat is reported to cause severe deterioration in chickpea yield (see Jha et al. 2017). In the context, identification of DNA markers that could enable rapid selection of the traits underlying HS tolerance is of great significance. The MSI and SCMR traits have been extensively studied as indicators to improve HS tolerance in several crops such as sorghum (Sullivan 1972), wheat (Talukder et al. 2014), etc. Our observation of significant genetic variation for these two traits is in line with the earlier findings documented in chickpea with respect to MSI and SCMR under HS (Jha et al. 2015; Thudi et al. 2014). High heritability was recorded for MSI (74.1%) in comparison to SCMR (48.9%).
To the best of our knowledge, this study is the first to report MTAs for MSI and SCMR in chickpea, two important physiological parameters associated with HS tolerance. Importantly, we found three SSR markers NCPGR206, NCPGR267, and H2L102 that consistently showed significant association with MSI. Similarly, the SSRs TR31 and GA9 on LG2 and LG5, respectively, had significant MTA. Considering MSI and chlorophyll content as important physiological parameters for drought tolerance in wheat, Elshafei et al. (2013) identified five SRAP markers linked to chlorophyll content QTL with PV up to 53%. The authors also reported another five SRAP markers linked with the QTL underlying MSI trait, which explained up to 44% PV. Recently, five QTLs associated with plasma membrane damage trait were mapped in wheat on chromosomes 7A, 7B and 7D based on the analysis of a bi-parental mapping population under HS (Talukder et al. 2014). Also, seven QTLs explaining PV up to 30.8% were identified on chromosomes 6A, 7A, 1B and 1D in wheat for SCMR chlorophyll content under HS. An earlier study aimed to dissect traits relevant to drought and HS in chickpea revealed more than 300 MTAs, the majority of which were related to 100-seed weight trait. Importantly, the MTAs for two important traits viz., 100-seed weight and δ13C coincided with the “QTL-hotspot” of Varshney et al. (2014) on LG4.
Limited studies have been performed that describe the candidate genes and their corresponding function for membrane stability and chlorophyll-related traits in relation to HS tolerance in plants (Jespersen et al. 2017). In our current study, the candidate gene Ca_05237 underlying MSI and SCMR-associated NCPGR267 marker region was predicted to encode ATPase/AAA protein. Overexpression of ZmSKD1 gene encoding putative ATPase/AAA protein exhibiting drought tolerance has been recorded in tobacco (Xia et al. 2013). Given the reported involvement of zinc finger proteins (ZNP) in regulation of abiotic stress tolerance in plants, the markers NCPGR238 and TA113 associated with MSI and SCMR were found to reside within the genomic region that encodes zinc finger or RING-type protein. Role of zinc finger or RING-type protein in conferring HS tolerance has also been reported (Liu et al. 2015; Zhang et al. 2012). Contribution ofaquaporins towards plant’s tolerance to abiotic stresses including HS has been documented in various crops (Afzal et al. 2016; Obaid et al. 2016), and we observed SCMR-associated marker TA113 coincided with genomic region encoding aquaporin NIP/major intrinsic protein. Likewise, SCMR-associated CESSR172 marker region was predicted to encode DNA glycosylase/AP lyase protein, which is involved in imparting tolerance to abiotic stresses as elucidated from its higher expression in Arabidopsis (Chen et al. 2012). However, we advocate further confirmation for the MTAs established in the present study in other genetic backgrounds prior to deploying these DNA markers in breeding chickpea for HS tolerance.
Conclusion
We conclude that the genetic diversity among cultivars and advanced breeding lines along with accessions could be exploited for broadening the genetic base of chickpea and breeding superior chickpea cultivars. After validation of the preliminary results of association mapping for MSI and SCMR might be helpful for screening of HS tolerance genotype. The putatively linked genomic regions may be investigated in greater detail for better understanding of HS tolerance in chickpea.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors acknowledge support from Indian Council of Agricultural Research (ICAR), India. The authors also acknowledge support from Dr. P. S. Basu for providing instruments for taking physiological data.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Electronic supplementary material
The online version of this article (10.1007/s13205-017-1057-2) contains supplementary material, which is available to authorized users.
References
- Abbo S, Berger J, Turner NC. Evolution of cultivated chickpea: four bottlenecks limit diversity and constrain adaptation. Funct Plant Biol. 2003;30:1081–1087. doi: 10.1071/FP03084. [DOI] [PubMed] [Google Scholar]
- Afzal Z, Howton TC, Sun Y, Mukhtar MS. The roles of aquaporins in plant stress responses. J Dev Biol. 2016;4:1–22. doi: 10.3390/jdb4010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggarwal H, Rao A, Kumar A, Rana JS, Naik PK, Chhokar V. Assessment of genetic diversity among 125 cultivars of chickpea (Cicer arietinum L.) of Indian origin using ISSR markers. Turk J Bot. 2015;39:218–226. doi: 10.3906/bot-1401-80. [DOI] [Google Scholar]
- Ahmad F. Random amplified polymorphic DNA (RAPD) analysis reveals genetic relationships among the annual Cicer species. Theor Appl Genet. 1999;98:657–663. doi: 10.1007/s001220051117. [DOI] [Google Scholar]
- AICRP on Chickpea (2014): Project Coordinator’s Report, 2014–2015. Indian Institute of Pulses Research, Kanpur, India
- Bajaj D, Das S, Badoni S, Kumar V, Singh M, Bansal KC, Tyagi AK, Parida S. Genome-wide high-throughput SNP discovery and genotyping for understanding natural (functional) allelic diversity and domestication patterns in wild chickpea. Sci Rep. 2015;5:12468. doi: 10.1038/srep12468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajaj D, Das S, Upadhyaya HD, Ranjan R, Badoni S, Kumar V, Tripathi S, Gowda CL, Sharma S, Singh S, Tyagi AK, Parida SK. A genome wide combinatorial strategy dissects complex genetic structure of seed coat color in chickpea. Front Plant Sci. 2015;6:979. doi: 10.3389/fpls.2015.00979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharadwaj C, Chauhan SK, Rajguru G, Srivastava R, Satyavathi TC, Yadav S, Rizvi AH, Solanki RK. Diversity analysis of chickpea (Cicer arietinum L.) using STMS markers. Indian J Agric Sci. 2010;80:947–951. [Google Scholar]
- Bharadwaj C, Srivastava R, Chauhan SK, Satyavathi CT, Kumar J, Faruqui A, Yadav S, Rizvi AH, Kumar T. Molecular diversity and phylogeny in geographical collection of chickpea (Cicer sp.) accessions. J Genet. 2011;90:e94–e100. [PubMed] [Google Scholar]
- Blum A, Ebercon A. Cell membrane stability as a measure of drought and heat tolerance in wheat. Crop Sci. 1979;21:43–47. doi: 10.2135/cropsci1981.0011183X002100010013x. [DOI] [Google Scholar]
- Bohra A, Jha R, Pandey G, Patil PG, Saxena RK, Singh IP, Singh D, Mishra RK, Mishra A, Singh F, Varshney RK, Singh NP. New hypervariable SSR markers for diversity analysis, hybrid purity testing and trait mapping in Pigeonpea [Cajanus cajan (L.) Millspaugh] Front Plant Sci. 2017;8:1–15. doi: 10.3389/fpls.2017.00377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23:2633–2635. doi: 10.1093/bioinformatics/btm308. [DOI] [PubMed] [Google Scholar]
- Chen H, Chu P, Zhou Y, Li Y, Liu J, Ding Y, Tsang EWT, Jiang L, Wu K, Huang S. Overexpression of AtOGG1, a DNA glycosylase/AP lyase, enhances seed longevity and abiotic stress tolerance in Arabidopsis. J Expt Bot. 2012;63:4107–4121. doi: 10.1093/jxb/ers093. [DOI] [PubMed] [Google Scholar]
- Choudhary S, Sethy NK, Shokeen B, Bhatia S. Development of chickpea EST-SSR markers and analysis of allelic variation across related species. Theor Appl Genet. 2009;118:591–608. doi: 10.1007/s00122-008-0923-z. [DOI] [PubMed] [Google Scholar]
- Choudhary P, Khanna SM, Jain PK, Bharadwaj C, Kumar J, Lakhera PC, Srinivasan R. Genetic structure and diversity analysis of the primary gene pool of chickpea using SSR markers. Genet Mol Res. 2012;11:891–905. doi: 10.4238/2012.April.10.5. [DOI] [PubMed] [Google Scholar]
- Choudhary S, Gaur R, Gupta S, Bhatia S. EST-derived genic molecular markers: development and utilization for generating an advanced transcript map of chickpea. Theor Appl Genet. 2012;124:1449–1462. doi: 10.1007/s00122-012-1800-3. [DOI] [PubMed] [Google Scholar]
- Choudhary P, Khanna SM, Jain PK, Bharadwaj C, Kumar J, Lakhera PC, Srinivasan R. Molecular characterization of primary gene pool of chickpea based on ISSR markers. Biochem Genet. 2013;51:306–322. doi: 10.1007/s10528-012-9564-7. [DOI] [PubMed] [Google Scholar]
- Choudhary G, Ranjitkumar N, Surapaneni M, Deborah DA, Vipparla A, Anuradha G, Siddiq EA, Vemireddy LR. Molecular genetic diversity of major Indian cultivars over decadal periods. PLoS One. 2013;8:e66197. doi: 10.1371/journal.pone.0066197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Giovanni C, Pavan S, Taranto F, De Rienzo V, Miazzi MM, Marcotrigiano AR, Mangini G, Montemurro C, Ricciardi L, Lotti C. Genetic variation of a global germplasm collection of chickpea (Cicer arietinum L.) including Italian accessions at risk of genetic erosion. Physiol Mol Biol Plants. 2017;23:197. doi: 10.1007/s12298-016-0397-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diapari M, Sindhu A, Bett K, Deokar A, Wartkentin TD, Taran B. Genetic diversity and association mapping of iron and zinc concentrations in chickpea (Cicer arietinum L.) Genome. 2014;57:459–468. doi: 10.1139/gen-2014-0108. [DOI] [PubMed] [Google Scholar]
- Earl DA, von Holdt BM. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour. 2012;4:359–361. doi: 10.1007/s12686-011-9548-7. [DOI] [Google Scholar]
- Elshafei AA, Saleh M, Al-Doss AA, Moustafa KA, Al-Qurainy FH, Barakat MN. Identification of new SRAP markers linked to leaf chlorophyll content, flag leaf senescence and cell membrane stability traits in wheat under water-stressed condition. AJCS. 2013;7:887–893. doi: 10.1556/ABiol.66.2015.1.8. [DOI] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- FAOSTAT (2014) FAO statistics division. http://faostat3.fao.org/compare/E. Accessed 27 2016
- Gaur R, Sethy NK, Choudhary S, Shokeen B, Gupta V, Bhatia S. Advancing the STMS genomic resources for defining new locations on the intraspecific genetic linkage map of chickpea (Cicer arietinum L.) BMC Genom. 2011;12:117. doi: 10.1186/1471-2164-12-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur PM, Jukanti AK, Varshney RK. Impact of genomic technologies on chickpea breeding strategies. Agronomy. 2012;2:199–221. doi: 10.3390/agronomy2030199. [DOI] [Google Scholar]
- Gaur PM, Samineni S, Sajja SB, Varshney RK, Chaturvedi SK, Jayalakshmi V, Babbar A, Mannur DM, Kumar AGV (2016) Enhancing resilience of chickpea to climate change. In: Symposium on Physiological approaches to enhance productivity in pulses under changing climate, April 2016, JNKVV, Jabalpur India, pp 18–23
- Ghaffari P, Talebi R, Keshavarz F. Genetic diversity and geographical differentiation of Iranian landrace, cultivars and exotic chickpea lines as revealed by morphological and microsatellite markers. Physiol Mol Biol Plant. 2014;20:225–233. doi: 10.1007/s12298-014-0223-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham PH, Vance CP. Legumes: importance and constraints to greater use. Plant Physiol. 2003;131:872–877. doi: 10.1104/pp.017004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gujaria N, Kumar A, Dauthal P, Dubey A, Hiremath P, BhanuPrakash A, Farmer A, Bhide M, Shah T, Gaur PM, Upadhyaya HD, Bhatia S, Cook DR, May GD, Varshney RK. Development and use of genic molecular markers (GMMs) for construction of a transcript map of chickpea (Cicer arietinum L.) Theor Appl Genet. 2011;122:1577–1589. doi: 10.1007/s00122-011-1556-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta PK, Varshney RK. The development and use of microsatellite markers for genetics and plant breeding with emphasis on bread wheat. Euphytica. 2000;1(13):163–185. doi: 10.1023/A:1003910819967. [DOI] [Google Scholar]
- Hajibarat Z, Saidi A, Hajibarat Z, Talebi R. Genetic diversity and population structure analysis of landrace and improved chickpea (Cicer arietinum) genotypes using morphological and microsatellite markers. Environ Expt Biol. 2014;12:161–166. [Google Scholar]
- Hajibarat Z, Saidi A, Hajibarat Z, Talebi R. Characterization of genetic diversity in chickpea using SSR markers, start codon targeted polymorphism (SCoT) and conserved DNA-derived polymorphism (CDDP) Physiol Mol Biol Plants. 2015;21:365–373. doi: 10.1007/s12298-015-0306-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Han B. Natural variations and genome-wide association studies in crop plants. Annu Rev Plant Biol. 2014;65:531–551. doi: 10.1146/annurev-arplant-050213-035715. [DOI] [PubMed] [Google Scholar]
- Hüttel B, Winter P, Weising K. Sequence tagged microsatellite site markers for chickpea (Cicer arietinum L.) Genome. 1999;42:210–217. doi: 10.1139/g98-122. [DOI] [PubMed] [Google Scholar]
- Iruela M, Rubio J, Cubero JI, Gil J, Milan T. Phylogenetic analysis in the genus Cicer and cultivated chickpea using RAPD and ISSR markers. Theor Appl Genet. 2002;104:643–651. doi: 10.1007/s001220100751. [DOI] [PubMed] [Google Scholar]
- Jespersen D, Belanger FC, Huang B. Candidate genes and molecular markers associated with heat tolerance in colonial Bentgrass. PLoS One. 2017;12(2):e0171183. doi: 10.1371/journal.pone.0171183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha UC, Shil S. Association analysis of yield contributing traits of chickpea genotypes under high temperature condition. Trends Biosci. 2015;8:2335–2341. [Google Scholar]
- Jha UC, Chaturvedi SK, Bohra A, Basu PS, Khan MS, Debmalya B. Abiotic stresses, constraints and improvement strategies in chickpea. Plant Breed. 2014;133:163–178. doi: 10.1111/pbr.12150. [DOI] [Google Scholar]
- Jha UC, Bohra A, Singh NP. Heat stress in crop plants: its nature, impacts and integrated breeding strategies to improve heat tolerance. Plant Breed. 2014;133:679–701. doi: 10.1111/pbr.12217. [DOI] [Google Scholar]
- Jha UC, Basu PS, Singh DK. Genetic variation and diversity analysis of chickpea genotypes based on quantitative traits under high temperature stress. Intl J Bioresour Stress Manag. 2015;6:700–706. [Google Scholar]
- Jha UC, Bohra A, Jha R, Parida S. Integrated ‘omics’ approaches to sustain major global grain legume productivity under heat stress. Plant Breed. 2017 [Google Scholar]
- Kalra N, Chakraborty D, Sharma A, Rai HK, Jolly M, Chander S, Kumar PR, Bhadraray S, Barman D, Mittal RB, Lal M, Sehgal M. Effect of temperature on yield on some winter crops in northwest India. Curr Sci. 2008;94:82–88. [Google Scholar]
- Keneni G, Bekele E, Imtiaz M, Dagne K, Getu E, Assefa F. Genetic diversity and population structure of Ethiopian chickpea (Cicer arietinum L.) germplasm accessions from different geographical origins as revealed by microsatellite markers. Plant Mol Biol Rep. 2012;30:654–665. doi: 10.1007/s11105-011-0374-6. [DOI] [Google Scholar]
- Krishnamurthy L, Gaur PM, Basu PS, Chaturvedi SK, Tripathi S, Vadez V, Varshney RK, Gowda CLL. Large genetic variation for heat tolerance in the reference collection of chickpea (Cicer arietinum L.) germplasm. Plant Genet Res. 2011;9:59–69. doi: 10.1017/S1479262110000407. [DOI] [Google Scholar]
- Kujur A, Bajaj D, Saxena MS, Tripathi S, Upadhyaya HD, Gowda CLL, Singh S, Jain M, Tyagi AK, Parida SK. Functionally relevant microsatellite markers from chickpea transcription factor genes for efficient genotyping applications and trait association mapping. DNA Res. 2013;20:355–374. doi: 10.1093/dnares/dst015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Muse SV. Power marker: an integrated analysis environment for genetic marker analysis. Bioinformatics. 2005;21:2128–2129. doi: 10.1093/bioinformatics/bti282. [DOI] [PubMed] [Google Scholar]
- Liu Q, Wang Z, Xu X, Zhang H, Li C. Genome-wide analysis of C2H2 zinc-finger family transcription factors and their responses to abiotic stresses in poplar (Populus trichocarpa) PLoS One. 2015;10:e0134753. doi: 10.1371/journal.pone.0134753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak SN, Zhu H, Varghese N, Datta S, Choi HK, Horres R, Jüngling R, Singh J, Kishor PB, Sivaramakrishnan S, Hoisington DA, Kahl G, Winter P, Cook DR, Varshney RK. Integration of novel SSR and gene-based SNP marker loci in the chickpea genetic map and establishment of new anchor points with Medicago truncatula genome. Theor Appl Genet. 2010;120:1415–1441. doi: 10.1007/s00122-010-1265-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TT, Taylor PW, Redden RJ, Ford R. Genetic diversity estimates in Cicer using AFLP analysis. Plant Breed. 2004;123:173–179. doi: 10.1046/j.1439-0523.2003.00942.x. [DOI] [Google Scholar]
- Obaid AY, Sabir JS, Atef A, Liu X, Edris S, El-Domyati FM, Mutwakil MZ, Gadalla NO, Hajrah NH, Al-Kordy MA, Hall N, Bahieldin A, Jansen RK. Analysis of transcriptional response to heat stress in Rhazyastricta. BMC Plant Biol. 2016;16:252. doi: 10.1186/s12870-016-0938-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peakall R, Smouse PE. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics. 2012;28:2537–2539. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrier X, Jacquemoud-Collet JP (2006) DARwin Software. Paris: Centre de Cooperation Internationale en Recherche Agronomique Pour le De’veloppement (CIRAD)
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena MS, Bajaj D, Kujur A, Das S, Badoni S, Kumar V, Singh M, Bansal KC, Tyagi AK, Parida SK. Natural allelic diversity, genetic structure and linkage disequilibrium pattern in wild chickpea. PLoS One. 2014;9:e107484. doi: 10.1371/journal.pone.0107484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sefera T, Abebie B, Gaur PM, Assefa K, Varshney RK. Characterization and genetic diversity analysis of selected chickpea cultivars of nine countries using simple sequence repeat (SSR) markers. Crop Pasture Sci. 2011;62:177–187. doi: 10.1071/CP10165. [DOI] [Google Scholar]
- Sethy NK, Shokeen B, Bhatia S. Isolation and characterization of sequence-tagged microsatellite sites markers in chickpea (Cicer arietinum L.) Mol Ecol Notes. 2003;3:428–430. doi: 10.1046/j.1471-8286.2003.00472.x. [DOI] [Google Scholar]
- Sethy NK, Shokeen B, Edwards KJ, Bhatia S. Development of microsatellite markers and analysis of intra specific genetic variability in chickpea (Cicer arietinum L.) Theor Appl Genet. 2006;1(12):1416–1428. doi: 10.1007/s00122-006-0243-0. [DOI] [PubMed] [Google Scholar]
- Shan F, Clarke HC, Plummer JA, Yan G, Siddique KHM. Geographical patterns of genetic variation in the world collection of wild annual Cicer characterized by amplified fragment length polymorphisms. Theor Appl Genet. 2005;110:381–391. doi: 10.1007/s00122-004-1849-8. [DOI] [PubMed] [Google Scholar]
- Sullivan CY. Mechanisms of heat and drought resistance in grain sorghum and methods of measurement. In: Rao NGP, House LR, editors. Sorghum in the seventies. New Delhi: Oxford and IBH publishing Co.; 1972. [Google Scholar]
- Talukder SK, Babar MA, Vijayalakshmi K, Poland J, Prasad PV, Bowden R, Fritz A. Mapping QTL for the traits associated with heat tolerance in wheat (Triticum aestivum L.) BMC Genet. 2014;15:97. doi: 10.1186/s12863-014-0097-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thudi M, Upadhyaya HD, Rathore A, Gaur PM, Krishnamurthy L, Roorkiwal M, Nayak SN, Chaturvedi SK, Basu PS, Gangarao NV, Fikre A, Kimurto P, Sharma PC, Sheshashayee MS, Tobita S, Kashiwagi J, Ito O, Killian A, Varshney RK. Genetic dissection of drought and heat tolerance in chickpea through genome-wide and candidate gene-based association mapping approaches. PLoS One. 2014;9:e96758. doi: 10.1371/journal.pone.0096758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thudi M, Chitikineni A, Liu X, He W, Roorkiwal M, Yang W, Jian J, Doddamani D, Gaur PM, Rathore A, Samineni S, Saxena RK, Xu D, Singh NP, Chaturvedi SK, Zhang G, Wang J, Datta SK, Xu X, Varshney RK. Recent breeding programs enhanced genetic diversity in both desi and kabuli varieties of chickpea (Cicer arietinum L.) Sci Rep. 2016;6:38636. doi: 10.1038/srep38636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udupa SM, Robertson LD, Weigand F, Baum M, Kahl G. Allelic variation at (TAA)n microsatellite loci in a world collection of chickpea (Cicer arietinum L.) germplasm. Mol Genet Genomics. 1999;261:354–363. doi: 10.1007/s004380050976. [DOI] [PubMed] [Google Scholar]
- Upadhyaya HD, Dwivedi SL, Baum M, Varshney RK, Udupa SM, Gowda CLL, Hoisington D, Singh S. Genetic structure, diversity, and allelic richness in composite collection and reference set in chickpea (Cicer arietinum L.) BMC Plant Biol. 2008;8:106. doi: 10.1186/1471-2229-8-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyaya HD, Bajaj D, Das S, Saxena MS, Badoni S, Kumar V, Tripathi S, Gowda CL, Sharma S, Tyagi AK, Parida SK. A genome-scale integrated approach aids in genetic dissection of complex flowering time trait in chickpea. Plant Mol Biol. 2015;89:403–420. doi: 10.1007/s11103-015-0377-z. [DOI] [PubMed] [Google Scholar]
- Upadhyaya HD, Bajaj D, Narnoliya L, Das S, Kumar V, Gowda CL, Sharma S, Tyagi AK, Parida SK. Genome-wide scans for delineation of candidate genes regulating seed-protein content in chickpea. Front Plant Sci. 2016;7:302. doi: 10.3389/fpls.2016.00302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyaya HD, Bajaj D, Das S, Kumar V, Gowda CL, Sharma S, Tyagi AK, Parida SK. Genetic dissection of seed-iron and zinc concentrations in chickpea. Sci Rep. 2016;6:24050. doi: 10.1038/srep24050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney RK, Song C, Saxena RK, Azam S, Yu S, Sharpe AG, Cannon S, et al. Draft genome sequence of chickpea (Cicer arietinum) provides a resource for trait improvement. Nat Biotechnol. 2013;31:240–246. doi: 10.1038/nbt.2491. [DOI] [PubMed] [Google Scholar]
- Varshney RK, Thudi M, Nayak SN, Gaur PM, Kashiwagi J, Krishnamurthy L, Jaganathan D, Koppolu J, Bohra A, Tripathi S, Rathore A, Jukanti AK, Jayalakshmi V, Vemula A, Singh SJ, Yasin M, Sheshshayee MS, Viswanatha KP. Genetic dissection of drought tolerance in chickpea (Cicer arietinum L.) Theor Appl Genet. 2014;127:445–462. doi: 10.1007/s00122-013-2230-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter P, Pfaff T, Udupa SM, Huttel B, Sharma PC, Sahi S, Arreguin-Espinoza R, Weigand F, Muehlbauer FJ, Kahl G. Characterization and mapping of sequence-tagged microsatellite sites in the chickpea (Cicer arietinum L.) genome. Mol Gen Genet. 1999;262:90–101. doi: 10.1007/s004380051063. [DOI] [PubMed] [Google Scholar]
- Winter P, Benko-Iseppon AM, Hüttel B, et al. A linkage map of chickpea (Cicer arietinum L.) genome based on recombinant inbred lines from a C. arietinum × C. reticulatum cross: localization of resistance genes for fusarium wilt races 4 and 5. Theor Appl Genet. 2000;101:1155–1163. doi: 10.1007/s001220051592. [DOI] [Google Scholar]
- Xia Z, Wei Y, Sun K, Wu J, Wang Y, et al. The maize AAA-type protein SKD1 confers enhanced salt and drought stress tolerance in transgenic tobacco by interacting with lyst-interacting protein 5. PLoS One. 2013;8:e69787. doi: 10.1371/journal.pone.0069787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Ersoz E, Lai CQ, Todhunter RJ, Tiwari HK, Gore MA, Bradbury PJ, Yu J, Arnett DK, Ordovas JM. Mixed linear model approach adapted for genome-wide association studies. Nat Genet. 2010;42:355–360. doi: 10.1038/ng.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Li J, Liu A, Zou J, Zhou X, et al. Expression profile in rice panicle: insights into heat response mechanism at reproductive stage. PLoS One. 2012;7:e49652. doi: 10.1371/journal.pone.0049652. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.