Abstract
Ankylosing spondylitis (AS) is an immune-mediated arthritis which primarily affects the spine and sacroiliac joints. Significant progress has been made in discovery of genetic associations with AS by genome-wide association studies (GWAS) over past decade. These findings have uncovered novel pathways involved pathogenesis of the disease and have led to introduction of novel therapeutic treatments for AS. In this Review, we discuss the genetic variations associated with AS identified by GWAS, the major pathways revealed by these AS-associated variations and critical cell types involved in AS development.
Introduction
Ankylosing spondylitis (AS) is a common, chronic, highly heritable, immune-mediated arthritis that affects primarily the spine and sacroiliac joints. It can also affect peripheral joints and extra-articular tissues including the eye, gut and skin. AS is the prototypic form of spondyloarthritis (SpA), a group of rheumatic disorders which share clinical, genetic and radiographic features, and which includes psoriatic arthritis (PsA), reactive arthritis (ReA), arthritis of inflammatory bowel disease (IBD), juvenile-onset spondyloarthritis and undifferentiated Spondyloarthritis (USpA). The diagnosis of AS is determined by clinical and radiographic features as defined by the modified New York (mNYAS) classification criteria.1 This review focuses on mNYAS criteria based studies because multiple lines of evidence demonstrate that classification criteria recently designed to increase sensitivity in early disease2 lack adequate specificity3, 4 and lead to marked increases in genetic heterogeneity.5
The prevalence of AS varies in different countries, being highly correlated with the frequency of the class I major histocompatibility complex (MHC) allele human leucocyte antigen (HLA)-B*27.6 The prevalence of AS in European-descent populations is about 0.55%,7 0.2–0.54% in Han Chinese populations8, 9 and 0.074% in South Africa.10 AS affects men more often than women and the ratio of men with AS to women is about 2–3:1,11, 12, 13 while the ratio in Europe is 3.8:1 and in Asia is 2.3:1.10 In addition, men are generally more likely to have a younger onset age and more often develop extra-articular features such as IBD and psoriasis.
Genetic epidemiology of ankylosing spondylitis
AS has been known to run strongly within families, for a long time. The risk ratio of the sibling or first-degree relative of an AS patient is >52 compared with the general population (non-related individuals) risk.14, 15, 16 It was unclear whether the co-familiality was due to shared environment or genetic factors until the association of HLA-B*27 allele with AS was discovered in the early 1970s.17, 18, 19 The recurrence risk drops rapidly with increasing distance of relationship to the proband (monozygotic (MZ) twins 63%, first-degree relatives 8.2%, second-degree relatives 1.0% and third-degree relatives 0.7% in Europeans;14 first-degree relatives 3.84%, 2nd degree relatives 0.87% and 3rd degree relatives 0.315% in Han Chinese).20 The evidence suggests that AS is a polygenic rather than monogenic disease since the frequency of monogenic disease reduces about half with each increase in distance of relationship to the proband, while frequency in polygenetic disease reduces approximate the square root with each increase in distance of relationship to the proband. Also, the concordance rate in dizygotic twins (DZ, 12.5%), or even in HLA-B*27 positive DZ twins (24–27%), is much lower than in MZ twins (63%), implying the presence of non-HLA*B27 factors, either environmental factors or other non-HLA-B*27 genes influencing disease susceptibility.14, 21
The estimated heritability of AS by twin studies is >90%.21 The variants associated with AS from that study explain 27.82% of AS heritability, with the greatest contribution coming from the MHC (20.44%) and with 7.38% coming from non-MHC loci.22, 23 Disease activity (BASDAI, 51%),24 functional impairment (BASFI, 68%),24 radiographic change (62%),25 and age of symptom onset,26 all additionally show significant heritability in AS.
The co-existence of AS and IBD has been known for a long time.27 Clinically diagnosed IBD presents in 5–10% of the AS patients, and 40–60% of AS patients have developed subclinical inflammation in gut and bowel.28 Moreover, the risk ratios of IBD were 3.0 and 2.1 in first- and second-degree relatives of patients with AS compared with unrelated individuals, respectively.15 These findings suggest that these two diseases may have similar aetiology, and multiple genes shared by these two diseases have been found.22, 29 Studying the heritability captured by the Immunochip SNP microarray, strong co-heritability was observed between AS and Crohn’s disease (40% including and 39% excluding the MHC), ulcerative colitis (33 and 31%) and to a lesser but nonetheless significant extent with psoriasis (27 and 20%) and primary sclerosing cholangitis (23 and 20%).22
HLA-B*27
The association of AS with HLA-B*27 was discovered in the early 1970s17, 18 and it is one of the strongest genetic associations with any common human disease. The prevalence of HLA-B*27 varies in different ethnic groups and populations. The population prevalence of HLA-B*27 is approximately 8% in British,30 4% in black Africans,31 and 3.6–5.7% of Han Chinese.8 In general, the population prevalence of AS parallels the frequency of HLA-B*27 except in West Africans.32 80-95% of AS patients of European ancestry are HLA-B*27 positive.33 Despite the strong association between HLA-B*27 and AS, only 2–5% of HLA-B*27 positive individuals develop AS, suggesting that other factors such as other loci, environmental or stochastic factors also contribute considerably to AS development.6, 34 HLA‐B27 homozygosity moderately increases risk of AS compared with HLA‐B27 heterozygosity.23, 35, 36
The introduction of high-throughput HLA sequencing has revealed that HLA-B*27 is remarkably polymorphic. To date, at least 271 HLA-B*27 subtypes (B*27:01-B27:164) have been reported37 and the number keeps increasing rapidly. Most are too rare to determine whether they are AS-associated or not, but two subtypes have been shown to have neutral associations with AS, HLA-B*27:06 (a common subtype in south-east Asia),38 and HLA-B*27:09 (a rare subtype found primarily on Sardinia).39 HLA-B*27:05 is present in nearly all populations40 and it is suggested to be the possible ancestral allele. HLA-B*27:05 and HLA-B*27:02 are the main subtypes associated with AS in Caucasians, in Asians the main associated subtypes are HLA-B*27:04 and HLA-B*27:07 and in Mediterranean populations is HLA-B*27:02.41 HLA-B*27 positive patients tend to develop AS earlier than HLA-B*27-negative patients.35, 42
Non-HLA-B*27 MHC associations
There are clearly other HLA alleles, and potentially MHC genes, associated with AS. HLA-B*60 was the first non- HLA-B*27 alleles identified to be associated with AS in HLA-B*27 positive patients,43 and was later confirmed in HLA-B*27 positive UK cases.44 In addition to HLA-B*60, HLA-B*61 was identified to be associated with HLA-B*27-negative Taiwan Chinese AS patients.45 Association of HLA-B*39 with HLA-B*27 negative AS patients in Japanese was observed46 in a small cohort, but was not replicated in European-descent cases.44
Though the associations of these HLA-B alleles with AS were identified, the complexity of MHC region makes it is extremely difficult to further study the associations between MHC loci and diseases until new technology and large study cohort are available. Immunochip is an Illumina Infinium SNP microarray with 195 806 SNPs and 718 small insertion–deletions on the array. The chip has dense coverage of the MHC and killer immunoglobulin-like receptor (KIR) loci.47 Dissection of the associations of MHC with diseases has been greatly improved with the help of Immunochip. Other than these HLA-B alleles, HLA-B*51:01, HLA-B*47:01, HLA-B*40:02, HLA-B*13:02 and HLA-B*40:01 were identified to be associated with increasing risk of AS whilst HLA-B*07:02 and HLA-B*57:01 were found to be associated with reduced risk of AS in a study cohort with 9,069 AS cases and 13,578 population controls of European descent using Immunochip.37
Furthermore, non-HLA*B alleles, HLA-A (HLA-A*02:01), HLA-DPB1 and HLA-DRB1 were associated with AS after controlling for the associated HLA-B haplotypes in this study.37 HLA-B51 is also a known risk factor for the spondyloarthritis-related disease, Behcet’s disease, suggesting some shared pathogenic mechanisms with AS. HLA-B*7 has been shown to be highly protective against AS in Chinese (odds ratio after control for HLA-B*27=0.15).48 Another study in the Korean population using Immunochip reported that HLA-C*15:02 allele was associated with AS in addition to HLA-B*27.49
Non-MHC genetic associations
Enabled by the development of high-throughput chip-based genotyping and computational genetics from early 2000s, genetic studies were able 10 years to ago to move to a new era, from family linkage study to genome-wide association studies (GWAS). In this review we consider not only association studies using the GWAS chip but also on customized chips, such as the Illumina Immunochip.47 Table 1 lists the published key AS GWAS/association studies since 2007. The first AS GWAS was conducted by the Wellcome Trust Case Control Consortium (WTCCC) and Australo-Anglo-American Spondylitis Consortium (TASC) in 2007, analysing 14 500 non-synonymous SNPs in 1,000 AS patients and 1500 healthy controls. This study made the first definitive identification of non-MHC susceptibility loci in AS, these being SNPs in endoplasmic reticulum aminopeptidase 1 (ERAP1) and interleukin 23 receptor (IL23R).50
Table 1. Major genome-wide association studies in AS.
| Study | Year | Discovery cohort (case/control) | Coverage/variant types | Ethnicity | No. of non-MHC variants reaching genome-wide significance |
|---|---|---|---|---|---|
| Burton et al. (WTCCC & TASC)46 | 2007 | 1000/1500 | Non-synonymous variants only | European | 2 |
| Reveille et al. (TASC)30 | 2010 | 2053/5140 | GWAS | European | 4 |
| Evans et al. (TASC)47 | 2011 | 3023/8779 | GWAS | European | 8 |
| Lin et al.49 | 2011 | 1965/4301 | GWAS | Han Chinese | 3 |
| Cortes et al. (IGAS)20 | 2013 | 10,619/15,145 | Illumina Immunochip | European, East Asian and Latin American | 24 |
| Robinson et al.48 | 2016 | 5040/21,133 | Illumina Exomechip | European | 2 |
| Ellinghaus et al.19 | 2016 | 8726/34,213 | Illumina Immunochip | European and East Asian | 113 |
Abbreviation: MHC, major histocompatibility complex.
A subsequent and comprehensive GWAS on a larger cohort of 2,053 AS patients and 5,140 controls of European ancestry was performed by TASC in 2010.33 This study replicated the previous findings of ERAP1 and IL23R. Moreover, additional loci, including Anthrax Toxin Receptor 2 (ANTXR2), Interleukin 1 Receptor Type 2 (IL1R2) and two intergenic regions on 2p15 and 21q22 were also found to be associated with AS at genome-wide significance. The association with intergenic regions suggests that they may harbour non-coding functional elements or be involved in regulating the expression of other genes. Further studies are needed to investigate their roles in AS pathogenesis.
In 2011, TASC and WCCC2 published the largest AS GWAS to date, involving a cohort of 3,023 AS patients and 9,141 health controls of European descent.51 This study identified several new associations, with its larger cohort. For the first time, RUNX3, LTBR-TNFRSF1A and IL12B were reported to be associated with AS at genome-wide significance level. It also replicated the HLA-B*27, ERAP1, IL23R, 2p15, 21q22 and KIF21B findings.
In 2013, International Genetics of Ankylosing Spondylitis Consortium (IGAS) conducted a case–control AS association study on Immunochip which has dense coverage in immune loci and MHC region. This cohort contains 10 619 AS cases and 15 145 controls in populations of European, East Asian and Latin American ancestry.23 IGAS identified 13 new associations of AS with replication of 11 previous findings other than HLA-B*27. These new associations are with SNPs nearby or within IL6R, FCGR2A, UBE2E3, GPR35, BACH2, ZMIZ1, NKX2-3, SH2B3, GPR65, SULT1A1, NOS2, TYK2 and ICOSLG.
In 2016 an AS association study using the Illumina Exomechip was conducted in a cohort of 5,040 AS patients and 21 133 healthy controls of European descent.52 Technically, this is not a genome-wide study but an exome-wide study. Exomechip has dense coverage of low-frequency and rare variants in addition to common coding variants. Due to the limited power of detecting rare variants and incomplete coverage of rare variants, this study only identified two novel loci (USP8 and CDKAL1) associated with AS at genome-wide significance and confirmed 11 known AS associations.52
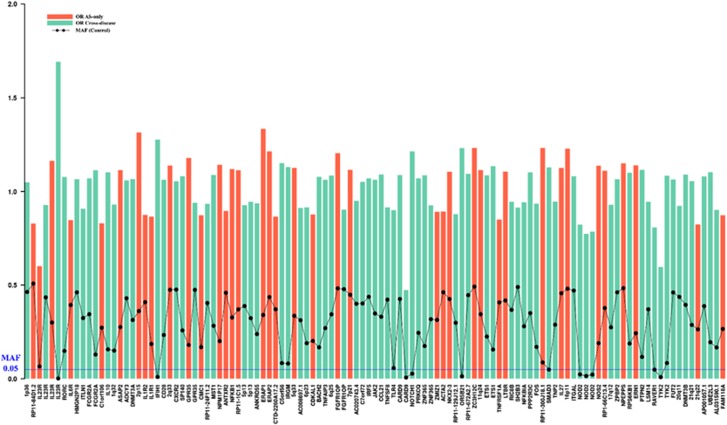
More recently, the largest AS case–control association study on Immunochip identified 113 AS-associated genome-wide significant variants by combining case cohorts from five related diseases (Crohn’s disease, psoriasis, primary sclerosing cholangitis and ulcerative colitis).22 This study has 8,726 AS cases and 34 213 health controls, and another 43 536 cases in other four diseases on Immunochip. 39 out of these variants are GWS significant in AS-only cohort, while additional 74 non-MHC GWS variants were identified in pleiotropy analysis with other four diseases then confirmed as being independently AS-associated. The larger sample size of this study increased its power to detect loci in disease-specific analyse, and to an even greater extent in analyses of pleiotropic genetic effects. It identified 17 new loci in AS missed in the original Immunochip study simply through the use of a much larger number of controls. The new AS-associated loci are ITLN1, CTLA4, CMC1, NPM1P17, NFKB1, CDKAL1, FGFR10P, 6p22, 7p21, ACTA2, 11q24, PPP2R3C, CORO1A, 16p11, ERN1, PTPN2 and FAM118A (for which suggestive association had already been identified in the AS Exomechip study). Odds ratios and minor allele frequencies in controls of 113 non-MHC variants identified in this study are illustrated in Figure 1. Given its large size, this study was sufficiently powered to identify several rare variants, including four variants with MAF < 1% and seven with MAF 1–5%. Notably, all the rare variants were identified by pleiotropic analysis with multiple diseases.
Figure 1.
Odds ratios and minor allele frequencies in controls of 113 non-MHC AS-associated variants identified in Ellinghaus et al.22 Each bar and dot represents a GWS variant. The variants are labelled by nearby genes. The red and green bars respectively represent odds ratio of AS-associated variants from AS-only case–control analyses, and analyses leveraging pleiotropy. Black dot represents minor allele frequency in healthy controls. The blue line indicates 5% minor allele frequency.
Thus far, most AS GWAS or case–control association studies have been performed in white European cohorts. The only GWAS yet carried out in the Han Chinese population, reported in 2011, identified two novel genome-wide significant non-MHC loci (2p15, 5q14.3 and 12q12), and replicated the known HLA-B*27 and 2p15 loci.53 The two novel non-MHC loci were not replicated in the IGAS Immunochip study (either in European descent or a large Chinese case–control analysis), nor the cross-disease study mentioned above, and are thus likely to be false positives.
There is clearly great need for a large GWAS to be performed in Han Chinese population to leverage the difference in genetic makeup of east-Asians to identify further genes and pinpoint AS-genetic associations. For example, it has been shown that the IL23R SNPs associated with AS in European descent populations are not associated with AS in Han Chinese.54 Subsequent studies have shown that this is likely because the key European-associated variant, rs11209026, is not polymorphic in east Asians. Rather, a different variant (rs76418789, G149R) is AS-associated in east Asians.55 This variant shows weak association in European descent populations too but that effect is difficult to see because of the strength of association of other variants at this locus, notably rs11209026 (R381Q).
There is significant gender bias in AS and women are generally more resistant to getting AS than men. In addition, differences in recurrent risk between the offspring of affected mothers and fathers have been observed. These findings imply loci of X-chromosome may contribute to disease susceptibility. Despite multiple AS GWAS having been performed on autosomes, there are only two AS association studies on X chromosome, each in small cohorts. Thus far, no association or linkage of the X-chromosome with AS have been found.56, 57 Therefore, the sex bias in AS hasn’t been explained by X-chromosome–encoded genetic effects, although no formal high-density SNP analysis of this chromosome has yet been reported.
Identified AS susceptibility loci are involved in numerous immunomodulatory pathways affecting innate and adaptive immunity, such as antigen presentation and binding, TNF-α/NF-κB activation and signalling, IL-23R signalling, lymphocyte development and activation, IL-1 cluster genes, G-protein coupled receptor and IL-17/IL-22 mediated immunity. Though there are over 100 AS-associated loci that have been identified, the underlying mechanism by which they are involved remain unclear for most of these loci. There are extensive ongoing studies try to pinpoint the causative variants of AS, understand their roles in AS, and develop new therapeutic treatment for AS. Among these pathways, MHC class 1 presentation and IL-23 pathway are major pathways involved in AS development and also the best studied.
Aminopeptidase in AS
ERAP1 is one of the three aminopeptidases (other two are ERAP2 and NPEPPS) have been shown to be associated with AS. Association of ERAP1 with AS was firstly identified by WTCCC in European Caucasian population in 2007.50 It was a big leap in AS research since it is one of the two first AS-associated non-MHC genes. This association has been widely replicated different ethnics groups, including (not exclusively) Han Chinese,53, 54 Korean,58 white European ancestry59, 60 and Iranian populations,61 suggesting common ERAP1 variants are involved in AS rather than diverse rare variants. A recent study investigated haplotypes generated from five common ERAP1 SNPs—rs2287987 (M349V), rs30187 (K528R), rs10050860 (D575N), rs17482078 (R725Q) and rs27044 (Q730E), and further confirmed that the association of ERAP1 with AS is primarily contributed by individual common ERAP1 genotypes, rather than by haplotypes.62
ERAP1 operates in AS by acting as a molecular ruler, trimming peptides to optimal length (10-16 residues) for presentation by HLA class 1 molecules.63 The main ERAP1 variant rs30187 only shows association with AS when HLA-B*27 or HLA-B*40 is present.37 Interaction between ERAP1 and HLA-B*27 in AS indicates that peptide trimming and presentation contribute to disease susceptibility. Multiple variants in ERAP1 have been found to influence the risk of AS. The non-synonymous SNP rs30187 (K528R) is directly associated with AS.51 An independent association is observed with rs10050860 (D575N), but this is in a very strong LD with a range of other SNPs and it is unclear if itself is directly disease associated.51 According to ERAP1 enzyme structure studies, ERAP1 variants that are associated with AS are commonly located in the active site of enzyme, on the inner surface of the C-terminal cavity and junctions.64 The SNP rs30187 is located at the junction domain and it could indirectly affect specificity or enzymatic activity by altering the conformational change between open and closed forms.65 Indeed, the protective alleles of rs30187 and rs17482078 (R725Q) are reported to be associated with 40% reduction in enzyme activities of peptide trimming in vitro using recombinant ERAP1 protein, whereas rs10050860 showed no effect.51
These studies show that disease-associated coding variants in ERAP1 are gain-of-function. The further molecular and immunological mechanisms by which these variants influence AS risk are the subject of active research and have been reviewed elsewhere. It is also clear that ERAP1 expression is strongly affected by genetic variation at the locus, something first reported by Myriad Genetics66 and subsequently confirmed by others.67, 68 Thus it is likely that the genetic variants influencing AS risk at this locus do so by a combination of effects on ERAP1 expression and function.
Apart from ERAP1, polymorphisms of ERAP2 are also found to be associated with AS in multiple studies.50, 69 Particularly, the protective allele (G) of rs2248374 affects the exon 10 splicing site and produces a truncated mRNA. This truncated mRNA is degraded leading to a complete absence of ERAP2. Unlike ERAP1, ERAP2 plays a role in both HLA-B27 positive and negative AS.70 Though further studies are needed to understand precise mechanism of how ERAP1 and ERAP2 affect AS status, this finding together with the protective effects of rs30187 implies that inhibiting these proteins may be potential treatment for AS.
NPEPPS encodes puromycin-sensitive aminopeptidase or alanine aminopeptidase, which is expressed in the cytosol. NPEPPS knockdown has been shown to influence cell surface HLA Class I expression, either increasing or decreasing cell surface expression depending on the HLA Class I antigen involved, and indirectly, through studies of HLA Class I complex stability, to influence the peptide pool available for presentation.71
IL-23 pathway
IL23R was first reported to have association with AS in 2007 and this finding revealed the involvement of IL-23 pathway in AS pathogenesis.50 It encodes the receptor for a proinflammatory cytokine IL-23 which plays important roles in activation of various proinflammatory cells, including Th17 T cells, gamma-delta T cells, NK cells, mast cells, Paneth cells and others.
Multiple variants involved with IL-23 pathway genes, such as TYK2, JAK2, IL12B, IL6R, IL27, PTGER4 and CARD9 have also been found to be associated with AS.23, 72, 73 TYK2 is a member of Janus kinases (JAKs) protein families and it plays important roles in transducing a range of signals, including IL-23, IL-10, IL-6, IFN-α and -β and IL-12 signals.74 JAK2 encodes the component of JAK-STAT signalling pathway, which is the downstream pathway of IL-23R. IL12B encodes IL-12p40, one of the subunits of IL-23 heterodimer. CARD9 mediates the downstream signalling of Dectin-1 which occurs in response particularly to fungal stimulation, leading to expression of pro-inflammatory cytokines.75, 76 As the receptor for IL-6, IL-6R is involved in the IL-6 signalling pathway which affects T helper (Th)17 cell differentiation amongst other things. Activation of Prostaglandin E2 receptor 4 (EP4), the protein product of PTGER4 promotes development of Th17 cells by decreasing IL-12p70 and increases IL-23 expression level.77 IL-27 is in the same IL-12 family with IL-23. It has widespread immunological effects, including promotion of Th1 differentiation, and inhibition of Th2 cell and Th17 differentiation.78 Its expression is increased in response to LPS interacting with TLR4, which is itself AS-associated, and by prostaglandin E2 (reviewed in Wang et al.).79
Multiple genes in this pathway show pleiotropic effect in different diseases. For example, IL23R is known to be associated with AS-related diseases including IBD80 and psoriasis.81 Variants in TYK2 are associated with AS, IBD, psoriasis, type 1 diabetes, multiple sclerosis and rheumatoid arthritis.82 Multiple genes related with IL23R pathway are shared between IBD and AS, including IL23R, IL12B, TYK2, JAK2, IL27 and CARD9, highlighting that the two diseases probably share similar aetiology through IL-23R pathway.29
IL-23 was reported to have higher expression level in the terminal ileum of AS patients and CD patients83 and mouse model studies have revealed that overexpression of IL-23 alone can cause spondyloarthritis.53, 84 Sherlock et al identified a population of IL-23-responsive cells from entheseal expressed IL-17 and IL-22 that developed in response to the IL-23 treatment.84 Clinical paw swelling scores were reduced by inhibiting IL-17 or IL-22, especially by inhibiting both. Over-expression of IL-22 caused disease but overexpression of IL-17 did not. Other animal models of AS, both rat and mouse, have also been shown to be significantly affected by IL-23 pathway targeted interventions.85 Clinical trials of therapeutic treatments targeting IL-23 pathway shows promising results, with secukinumab being widely used now in treatment of AS, and multiple other agents in development and trial. It will be very interesting to see in humans the differential effect of targeting IL-23, IL-22 and IL-17, which is hard to predict from genetic studies alone.
Critical cell types involved in AS
Similar to other complex trait associated polymorphisms, the vast majority of AS GWS variants lie within intergenic regions and therefore raise a great challenge to unravel the mechanisms through which they operate. For instance, AS-associated loci 2p15 and 21q22 are located in intergenic regions which makes it is difficult to pinpoint the associated genes, let alone to elucidate the underlying biological function and involved pathways. Nevertheless, recent studies have indicated that disease-associated variants often contribute through effects on gene regulatory regions in specific cell types or tissues rather than directly modifying the protein function, such as regulating gene expression level by expression quantitative trait loci (eQTL) effect or by altering the accessibility of chromatin.72, 86 Though the DNA sequence is the same among all the cells and tissues, the regulatory effect of a variant could be different between cell types and tissues. In addition, with the rapidly increasing availability of epigenetic and gene expression data in hundreds of cell types or tissue types, disease-associated variants can be further investigated in the cell types which they primarily operate.
Various cell types have been suggested to be involved in AS, including CD4+, CD8+ and regulatory T cells,87 Th17 cell,88 natural killer (NK) cells,89 gamma-delta cells,90 innate lymphoid cells (particularly ILC3 subset),91 B cells,92 mast cells93 and intestinal Paneth cells.83 These studies indirectly reveal cell types which AS-associated variants predominantly function. However, understandably they have mainly focused on circulating immune cells and tissues that are easily accessed.
A role for NK and CD8 T-cells has been suggested both by evidence that these KIR3DL2 bearing cells of these types interact with HLA-B27 dimers leading to IL-17 production.89 Further, AS is associated with TBX21 which encodes T-bet, a transcription factor with effects on differentiation and function of multiple immunological cell types. NK and CD8 T-cells expressing T-bet are increased in AS cases, risk variants of TBX21 lead to increased expression in AS cases, and TBX21 knockout mouse are resistant to the development of spondyloarthritis and IBD in the Skg mouse model.94
To identify the key cell types involved in the mechanism by which AS-associated genes influence its aetiopathogenesis, we recently integrated AS-genetic and cell-type specific data of epigenomes, transcriptomes and proteomes in a wide range of cell types.95 This analysis suggested that AS-associated variants primarily operate through effects in CD8+ T cells, CD4+ T cells, NK cells, regulatory T cells, monocytes, as well as gastrointestinal cells. These findings are consistent with a previous study which performed a similar epigenetic analysis with predicted causal AS SNPs.96, 97 In addition, AS-associated variants were shown to be enriched in regulatory elements, especially in enhancer regions, in related immune cells. Moreover, the AS upregulated genes show different enrichment bias in both gene and protein expression compared with AS downregulated genes. The AS upregulated genes are significantly enriched in monocytes, whereas AS downregulated genes showed enrichment in CD8+ T cell and NK cells in mRNA and protein expression.
The strong co-familiality between AS and IBD, the shared susceptibility loci, especially IL-23 pathway genes, suggest that gut microbial is involved in AS development. Differences in microbial signature in the terminal ileum of AS patients compared with healthy individuals have been demonstrated.98 Our finding that the genes specifically enriched in gut cells are also enriched in ‘response to bacterium’ and other similar Gene Ontologies further supports that AS status is influenced by the interaction among the host genetics, the intestinal microbiome and the immune response. Over-expression of the anti-microbial peptides alpha-defensin, phospholipase A2 and lysozyme by Paneth cells in human AS cases provides further support for the involvement of these cells and the gut mucosal immune system in AS pathogenesis.99 Bone cell types showed relative weak enrichment signals, consistent with the hypothesis that bone changes in AS are secondary to inflammatory processes rather than driven by bone-specific genetic effects. However, data on more AS-relevant tissues and cell types, such as enthesial and synovial cell populations, innate lymphoid cells and Paneth cells, is needed to identify more AS-relevant cell types and to further investigate the specific role of AS-associated variants in these cell types.
Challenges
The two main remaining challenges in immunogenetic research in AS are to identify the variants responsible for the large proportion of heritability of the disease that remains unexplained, and determining the functional mechanisms underpinning those genetic associations. The sample size of GWAS that have been performed to date are relatively modest in comparison with the far larger studies that have been funded in other major immune-mediated diseases that have similar or lower disease prevalence, such as type 1 diabetes, IBD, and multiple sclerosis.100 Further large scale GWAS are clearly indicated.
In addition to discovering more genetic variants associations with AS, another challenge is fine mapping the casual variants through the associated signals. About 70% of the AS-associated SNPs are located in non-coding regions, making them difficult to be interpreted via traditional protein coding theory. Furthermore, these variants may function in specific cell types or tissues. Therefore, integrating cell-type specific multi-omics data, such as transcriptomic, epigenetic and proteomics, is necessary.
Besides genetic factors, only limited studies of epigenetic factors have been reported in AS.95, 101 Larger more systematic studies controlling for the profound effects of age, cigarette smoking and tissue type are required to properly investigate the role of epigenetic factors and AS, and also how genetic factors operate through epigenetic mechanisms to influence disease.
Whilst the main goal of genetic studies in AS is clearly to inform research ultimately leading to therapeutic or preventative approaches for the disease, given the high heritability of AS and its clinical manifestations, it is possible that genetic risk prediction may prove valuable in the disease. Genetic risk scores have been shown to have high discriminatory capacity between AS and healthy controls.5 Further development of these approaches using whole of GWAS data may prove even more informative given their ability to capture a higher proportion of disease heritability than analyses restricted to genome-wide significant loci.102 This may prove useful in identifying people either at high risk of disease to enable preventative approaches, or to distinguish that set of patients with suggestive early disease who truly have axial spondyloarthritis rather than non-inflammatory causes of back pain.103 Given the evidence that delay in implementing effective therapy is linked with worse outcome in AS,104 reducing the current substantial diagnostic delay in the disease would be a valuable contribution.
Conclusions
GWAS is a hypothesis-free approach for testing the associations between hundreds of thousands of variants and phenotype. To date, at least 113 non-MHC variants have been identified as well as HLA-B*27 and these findings provide a wealth of new genetic information for AS, critical clues on biological pathways of development of AS and contributed to the development of new treatments. As mentioned above, at least several treatments targeting genes in IL-23R pathway, which was first uncovered in WTCCC GWAS, have been approved in AS or under clinical trials. In addition to genetic studies, functional studies of genetic variants in AS-relevant cell types will be necessary for understanding AS pathogenesis and developing novel treatments.
Acknowledgments
MAB is funded by a National Health and Medical Research Council Senior Principal Research Fellowship.
Footnotes
The authors declare no conflict of interest.
References
- van der Linden S, Valkenburg HA, Cats A. Evaluation of diagnostic criteria for ankylosing spondylitis. A proposal for modification of the New York criteria. Arthritis Rheum 1984; 27: 361–368. [DOI] [PubMed] [Google Scholar]
- Rudwaleit M, Landewe R, van der Heijde D, Listing J, Brandt J, Braun J et al. The development of Assessment of SpondyloArthritis international Society classification criteria for axial spondyloarthritis (part I): classification of paper patients by expert opinion including uncertainty appraisal. Ann Rheum Dis 2009; 68: 770–776. [DOI] [PubMed] [Google Scholar]
- Akkoc N, Khan MA. Looking into the new ASAS classification criteria for axial spondyloarthritis through the other side of the glass. Curr Rheumatol Rep 2015; 17: 515. [DOI] [PubMed] [Google Scholar]
- Robinson PC, Wordsworth BP, Reveille JD, Brown MA. Axial spondyloarthritis: a new disease entity, not necessarily early ankylosing spondylitis. Ann Rheum Dis 2013; 72: 162–164. [DOI] [PubMed] [Google Scholar]
- Thomas GP, Willner D, Robinson PC, Cortes A, Duan R, Rudwaleit M et al. Genetic diagnostic profiling in axial spondyloarthritis: a real world study. Clin Exp Rheumatol 2017; 35: 229–233. [PubMed] [Google Scholar]
- Braun J, Bollow M, Remlinger G, Eggens U, Rudwaleit M, Distler A et al. Prevalence of spondylarthropathies in HLA-B27 positive and negative blood donors. Arthritis Rheum 1998; 41: 58–67. [DOI] [PubMed] [Google Scholar]
- Akkoc N, Khan MA. Overestimation of the prevalence of ankylosing spondylitis in the Berlin study: comment on the article by Braun et al. Arthritis Rheum 2005; 52: 4048–4049 author reply 4049–4050. [DOI] [PubMed] [Google Scholar]
- Zeng QY, Chen R, Darmawan J, Xiao ZY, Chen SB, Wigley R et al. Rheumatic diseases in China. Arthritis Res Ther 2008; 10: R17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SC, Liao Z, Yu DT, Chan ES, Zhao L, Gu J. Epidemiology of spondyloarthritis in the People's Republic of China: review of the literature and commentary. Semin Arthritis Rheum 2007; 37: 39–47. [DOI] [PubMed] [Google Scholar]
- Dean LE, Jones GT, MacDonald AG, Downham C, Sturrock RD, Macfarlane GJ. Global prevalence of ankylosing spondylitis. Rheumatology (Oxford) 2014; 53: 650–657. [DOI] [PubMed] [Google Scholar]
- Lee W, Reveille JD, Davis JC Jr., Learch TJ, Ward MM, Weisman MH. Are there gender differences in severity of ankylosing spondylitis? Results from the PSOAS cohort. Ann Rheum Dis 2007; 66: 633–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonen A, van der Heijde D, Landewe R, Guillemin F, Spoorenberg A, Schouten H et al. Costs of ankylosing spondylitis in three European countries: the patient's perspective. Ann Rheum Dis 2003; 62: 741–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masi AT, Wilkins WR. Does male:female sex ratio in ankylosing spondylitis change with age? J Rheumatol 1996; 23: 947–948. [PubMed] [Google Scholar]
- Brown MA, Laval SH, Brophy S, Calin A. Recurrence risk modelling of the genetic susceptibility to ankylosing spondylitis. Ann Rheum Dis 2000; 59: 883–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thjodleifsson B, Geirsson AJ, Bjornsson S, Bjarnason I. A common genetic background for inflammatory bowel disease and ankylosing spondylitis: a genealogic study in Iceland. Arthritis Rheum 2007; 56: 2633–2639. [DOI] [PubMed] [Google Scholar]
- Geirsson AJ, Kristjansson K, Gudbjornsson B. A strong familiality of ankylosing spondylitis through several generations. Ann Rheum Dis 2010; 69: 1346–1348. [DOI] [PubMed] [Google Scholar]
- Caffrey MF, James DC. Human lymphocyte antigen association in ankylosing spondylitis. Nature 1973; 242: 121. [DOI] [PubMed] [Google Scholar]
- Brewerton DA, Hart FD, Nicholls A, Caffrey M, James DC, Sturrock RD. Ankylosing spondylitis and HL-A 27. Lancet 1973; 1: 904–907. [DOI] [PubMed] [Google Scholar]
- Schlosstein L, Terasaki PI, Bluestone R, Pearson CM. High association of an HL-A antigen, W27, with ankylosing spondylitis. N Engl J Med 1973; 288: 704–706. [DOI] [PubMed] [Google Scholar]
- ZM L The Genetic Risk and Genome-wide Linkage and Association Study. 2010.
- Brown MA, Kennedy LG, MacGregor AJ, Darke C, Duncan E, Shatford JL et al. Susceptibility to ankylosing spondylitis in twins: the role of genes, HLA, and the environment. Arthritis Rheum 1997; 40: 1823–1828. [DOI] [PubMed] [Google Scholar]
- Ellinghaus D, Jostins L, Spain SL, Cortes A, Bethune J, Han B et al. Analysis of five chronic inflammatory diseases identifies 27 new associations and highlights disease-specific patterns at shared loci. Nat Genet 2016; 48: 510–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Genetics of Ankylosing Spondylitis CInternational Genetics of Ankylosing Spondylitis CCortes A International Genetics of Ankylosing Spondylitis CHadler J International Genetics of Ankylosing Spondylitis CPointon JP International Genetics of Ankylosing Spondylitis CRobinson PC International Genetics of Ankylosing Spondylitis CKaraderi T et al. Identification of multiple risk variants for ankylosing spondylitis through high-density genotyping of immune-related loci. Nat Genet 2013; 45: 730–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamersma J, Cardon LR, Bradbury L, Brophy S, van der Horst-Bruinsma I, Calin A et al. Is disease severity in ankylosing spondylitis genetically determined? Arthritis Rheum 2001; 44: 1396–1400. [DOI] [PubMed] [Google Scholar]
- Brophy S, Hickey S, Menon A, Taylor G, Bradbury L, Hamersma J et al. Concordance of disease severity among family members with ankylosing spondylitis? J Rheumatol 2004; 31: 1775–1778. [PubMed] [Google Scholar]
- Brown MA, Brophy S, Bradbury L, Hamersma J, Timms A, Laval S et al. Identification of major loci controlling clinical manifestations of ankylosing spondylitis. Arthritis Rheum 2003; 48: 2234–2239. [DOI] [PubMed] [Google Scholar]
- Mielants H, Veys EM, Goemaere S, Goethals K, Cuvelier C, De Vos M. Gut inflammation in the spondyloarthropathies: clinical, radiologic, biologic and genetic features in relation to the type of histology. A prospective study. J Rheumatol 1991; 18: 1542–1551. [PubMed] [Google Scholar]
- Crane AM, Bradbury L, van Heel DA, McGovern DP, Brophy S, Rubin L et al. Role of NOD2 variants in spondylarthritis. Arthritis Rheum 2002; 46: 1629–1633. [DOI] [PubMed] [Google Scholar]
- Parkes M, Cortes A, van Heel DA, Brown MA. Genetic insights into common pathways and complex relationships among immune-mediated diseases. Nat Rev Genet 2013; 14: 661–673. [DOI] [PubMed] [Google Scholar]
- Brown MA, Pile KD, Kennedy LG, Calin A, Darke C, Bell J et al. HLA class I associations of ankylosing spondylitis in the white population in the United Kingdom. Ann Rheum Dis 1996; 55: 268–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum J, Ziff M. The rarity of ankylosing spondylitis in the black race. Arthritis Rheum 1971; 14: 12–18. [DOI] [PubMed] [Google Scholar]
- Brown MA, Jepson A, Young A, Whittle HC, Greenwood BM, Wordsworth BP. Ankylosing spondylitis in West Africans—evidence for a non-HLA-B27 protective effect. Ann Rheum Dis 1997; 56: 68–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Australo-Anglo-American Spondyloarthritis C, Reveille JD, Sims AM, Danoy P, Evans DM, Leo P et al. Genome-wide association study of ankylosing spondylitis identifies non-MHC susceptibility loci. Nat Genet 2010; 42: 123–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reveille JD, Arnett FC. Spondyloarthritis: update on pathogenesis and management. Am J Med 2005; 118: 592–603. [DOI] [PubMed] [Google Scholar]
- Jaakkola E, Herzberg I, Laiho K, Barnardo MC, Pointon JJ, Kauppi M et al. Finnish HLA studies confirm the increased risk conferred by HLA-B27 homozygosity in ankylosing spondylitis. Ann Rheum Dis 2006; 65: 775–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MA, Kushner I, Braun WE, Zachary AA, Steinberg AG. HLA—B27 homozygosity in ankylosing spondylitis: relationship to risk and severity. Tissue Antigens 1978; 11: 434–438. [DOI] [PubMed] [Google Scholar]
- Cortes A, Pulit SL, Leo PJ, Pointon JJ, Robinson PC, Weisman MH et al. Major histocompatibility complex associations of ankylosing spondylitis are complex and involve further epistasis with ERAP1. Nat Commun 2015; 6: 7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasution AR, Mardjuadi A, Kunmartini S, Suryadhana NG, Setyohadi B, Sudarsono D et al. HLA-B27 subtypes positively and negatively associated with spondyloarthropathy. J Rheumatol 1997; 24: 1111–1114. [PubMed] [Google Scholar]
- D'Amato M, Fiorillo MT, Carcassi C, Mathieu A, Zuccarelli A, Bitti PP et al. Relevance of residue 116 of HLA-B27 in determining susceptibility to ankylosing spondylitis. Eur J Immunol 1995; 25: 3199–3201. [DOI] [PubMed] [Google Scholar]
- Sheehan NJ. HLA-B27: what's new? Rheumatology (Oxford) 2010; 49: 621–631. [DOI] [PubMed] [Google Scholar]
- Taurog JD. The mystery of HLA-B27: if it isn't one thing, it's another. Arthritis Rheum 2007; 56: 2478–2481. [DOI] [PubMed] [Google Scholar]
- Feldtkeller E, Khan MA, van der Heijde D, van der Linden S, Braun J. Age at disease onset and diagnosis delay in HLA-B27 negative vs. positive patients with ankylosing spondylitis. Rheumatol Int 2003; 23: 61–66. [DOI] [PubMed] [Google Scholar]
- Robinson WP, van der Linden SM, Khan MA, Rentsch HU, Cats A, Russell A et al. HLA-Bw60 increases susceptibility to ankylosing spondylitis in HLA-B27+ patients. Arthritis Rheum 1989; 32: 1135–1141. [DOI] [PubMed] [Google Scholar]
- Brown M, Bunce M, Calin A, Darke C, Wordsworth P. HLA-B associations of HLA-B27 negative ankylosing spondylitis: comment on the article by Yamaguchi et al. Arthritis Rheum 1996; 39: 1768–1769. [DOI] [PubMed] [Google Scholar]
- Wei JC, Tsai WC, Lin HS, Tsai CY, Chou CT. HLA-B60 and B61 are strongly associated with ankylosing spondylitis in HLA-B27-negative Taiwan Chinese patients. Rheumatology (Oxford) 2004; 43: 839–842. [DOI] [PubMed] [Google Scholar]
- Yamaguchi A, Tsuchiya N, Mitsui H, Shiota M, Ogawa A, Tokunaga K et al. Association of HLA-B39 with HLA-B27-negative ankylosing spondylitis and pauciarticular juvenile rheumatoid arthritis in Japanese patients. Evidence for a role of the peptide-anchoring B pocket. Arthritis Rheum 1995; 38: 1672–1677. [DOI] [PubMed] [Google Scholar]
- Cortes A, Brown MA. Promise and pitfalls of the Immunochip. Arthritis Res Ther 2011; 13: 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi L, Wang J, Guo X, Espitia MG, Chen E, Assassi S et al. Profiling of HLA-B alleles for association studies with ankylosing spondylitis in the Chinese population. Open Rheumatol J 2013; 7: 51–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Bang SY, Lee S, Lee HS, Shim SC, Kang YM et al. An HLA-C amino-acid variant in addition to HLA-B*27 confers risk for ankylosing spondylitis in the Korean population. Arthritis Res Ther 2015; 17: 342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellcome Trust Case Control CAustralo-Anglo-American Spondylitis CWellcome Trust Case Control CBurton PR Wellcome Trust Case Control CClayton DG Wellcome Trust Case Control CCardon LR Wellcome Trust Case Control CCraddock N et al. Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat Genet 2007; 39: 1329–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans DM, Spencer CC, Pointon JJ, Su Z, Harvey D, Kochan G et al. Interaction between ERAP1 and HLA-B27 in ankylosing spondylitis implicates peptide handling in the mechanism for HLA-B27 in disease susceptibility. Nat Genet 2011; 43: 761–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson PC, Leo PJ, Pointon JJ, Harris J, Cremin K, Bradbury LA et al. Exome-wide study of ankylosing spondylitis demonstrates additional shared genetic background with inflammatory bowel disease. npj Genomic Med 2016; 1: 16008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamopoulos IE, Tessmer M, Chao CC, Adda S, Gorman D, Petro M et al. IL-23 is critical for induction of arthritis, osteoclast formation, and maintenance of bone mass. J Immunol 2011; 187: 951–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson SI, Wu X, Liu Y, Wei M, Danoy PA, Thomas G et al. Association of ERAP1, but not IL23R, with ankylosing spondylitis in a Han Chinese population. Arthritis Rheum 2009; 60: 3263–3268. [DOI] [PubMed] [Google Scholar]
- Davidson SI, Jiang L, Cortes A, Wu X, Glazov EA, Donskoi M et al. Brief report: high-throughput sequencing of IL23R reveals a low-frequency, nonsynonymous single-nucleotide polymorphism that is associated with ankylosing spondylitis in a Han Chinese population. Arthritis Rheum 2013; 65: 1747–1752. [DOI] [PubMed] [Google Scholar]
- Hoyle E, Laval SH, Calin A, Wordsworth BP, Brown MA. The X-chromosome and susceptibility to ankylosing spondylitis. Arthritis Rheum 2000; 43: 1353–1355. [DOI] [PubMed] [Google Scholar]
- Zhang G, Luo J, Bruckel J, Weisman MA, Schumacher HR, Khan MA et al. Genetic studies in familial ankylosing spondylitis susceptibility. Arthritis Rheum 2004; 50: 2246–2254. [DOI] [PubMed] [Google Scholar]
- Bang SY, Kim TH, Lee B, Kwon E, Choi SH, Lee KS et al. Genetic studies of ankylosing spondylitis in Koreans confirm associations with ERAP1 and 2p15 reported in white patients. J Rheumatol 2011; 38: 322–324. [DOI] [PubMed] [Google Scholar]
- Maksymowych WP, Inman RD, Gladman DD, Reeve JP, Pope A, Rahman P. Association of a specific ERAP1/ARTS1 haplotype with disease susceptibility in ankylosing spondylitis. Arthritis Rheum 2009; 60: 1317–1323. [DOI] [PubMed] [Google Scholar]
- Pimentel-Santos FM, Ligeiro D, Matos M, Mourao AF, Sousa E, Pinto P et al. Association of IL23R and ERAP1 genes with ankylosing spondylitis in a Portuguese population. Clin Exp Rheumatol 2009; 27: 800–806. [PubMed] [Google Scholar]
- Mahmoudi M, Jamshidi AR, Amirzargar AA, Farhadi E, Nourijelyani K, Fallahi S et al. Association between endoplasmic reticulum aminopeptidase-1 (ERAP-1) and susceptibility to ankylosing spondylitis in Iran. Iran J Allergy Asthma Immunol 2012; 11: 294–300. [PubMed] [Google Scholar]
- Roberts AR, Appleton LH, Cortes A, Vecellio M, Lau J, Watts L et al. ERAP1 association with ankylosing spondylitis is attributable to common genotypes rather than rare haplotype combinations. Proc Natl Acad Sci USA 2017; 114: 558–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang SC, Momburg F, Bhutani N, Goldberg AL. The ER aminopeptidase, ERAP1, trims precursors to lengths of MHC class I peptides by a ‘molecular ruler’ mechanism. Proc Natl Acad Sci USA 2005; 102: 17107–17112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TT, Chang SC, Evnouchidou I, York IA, Zikos C, Rock KL et al. Structural basis for antigenic peptide precursor processing by the endoplasmic reticulum aminopeptidase ERAP1. Nat Struct Mol Biol 2011; 18: 604–U118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochan G, Krojer T, Harvey D, Fischer R, Chen L, Vollmar M et al. Crystal structures of the endoplasmic reticulum aminopeptidase-1 (ERAP1) reveal the molecular basis for N-terminal peptide trimming. Proc Natl Acad Sci USA 2011; 108: 7745–7750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inc MG. Gene Variants and Use Thereof, In: USPTO (ed.) 2007.
- Harvey D, Pointon JJ, Evans DM, Karaderi T, Farrar C, Appleton LH et al. Investigating the genetic association between ERAP1 and ankylosing spondylitis. Hum Mol Genet 2009; 18: 4204–4212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantino F, Talpin A, Evnouchidou I, Kadi A, Leboime A, Said-Nahal R et al. ERAP1 gene expression is influenced by nonsynonymous polymorphisms associated with predisposition to spondyloarthritis. Arthritis Rheumatol 2015; 67: 1525–1534. [DOI] [PubMed] [Google Scholar]
- Tsui FW, Haroon N, Reveille JD, Rahman P, Chiu B, Tsui HW et al. Association of an ERAP1 ERAP2 haplotype with familial ankylosing spondylitis. Ann Rheum Dis 2010; 69: 733–736. [DOI] [PubMed] [Google Scholar]
- Robinson PC, Costello ME, Leo P, Bradbury LA, Hollis K, Cortes A et al. ERAP2 is associated with ankylosing spondylitis in HLA-B27-positive and HLA-B27-negative patients. Ann Rheum Dis 2015; 74: 1627–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Kwak H, Ahn K. Cytosolic aminopeptidases influence MHC class I-mediated antigen presentation in an allele-dependent manner. J Immunol 2009; 183: 7379–7387. [DOI] [PubMed] [Google Scholar]
- Edwards SL, Beesley J, French JD, Dunning AM. Beyond GWASs: illuminating the dark road from association to function. Am J Hum Genet 2013; 93: 779–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson SI, Liu Y, Danoy PA, Wu X, Thomas GP, Jiang L et al. Association of STAT3 and TNFRSF1A with ankylosing spondylitis in Han Chinese. Ann Rheum Dis 2011; 70: 289–292. [DOI] [PubMed] [Google Scholar]
- Dendrou CA, Cortes A, Shipman L, Evans HG, Attfield KE, Jostins L et al. Resolving TYK2 locus genotype-to-phenotype differences in autoimmunity. Sci Transl Med 2016; 8: 363ra149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeibundGut-Landmann S, Gross O, Robinson MJ, Osorio F, Slack EC, Tsoni SV et al. Syk- and CARD9-dependent coupling of innate immunity to the induction of T helper cells that produce interleukin 17. Nat Immunol 2007; 8: 630–638. [DOI] [PubMed] [Google Scholar]
- Gross O, Gewies A, Finger K, Schafer M, Sparwasser T, Peschel C et al. Card9 controls a non-TLR signalling pathway for innate anti-fungal immunity. Nature 2006; 442: 651–656. [DOI] [PubMed] [Google Scholar]
- Boniface K, Bak-Jensen KS, Li Y, Blumenschein WM, McGeachy MJ, McClanahan TK et al. Prostaglandin E2 regulates Th17 cell differentiation and function through cyclic AMP and EP2/EP4 receptor signaling. J Exp Med 2009; 206: 535–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura T, Takeda A, Hamano S, Miyazaki Y, Kinjyo I, Ishibashi T et al. Two-sided roles of IL-27: induction of Th1 differentiation on naive CD4+ T cells versus suppression of proinflammatory cytokine production including IL-23-induced IL-17 on activated CD4+ T cells partially through STAT3-dependent mechanism. J Immunol 2006; 177: 5377–5385. [DOI] [PubMed] [Google Scholar]
- Wang Q, Liu J. Regulation and Immune Function of IL-27. Adv Exp Med Biol 2016; 941: 191–211. [DOI] [PubMed] [Google Scholar]
- Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science 2006; 314: 1461–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cargill M, Schrodi SJ, Chang M, Garcia VE, Brandon R, Callis KP et al. A large-scale genetic association study confirms IL12B and leads to the identification of IL23R as psoriasis-risk genes. Am J Hum Genet 2007; 80: 273–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diogo D, Bastarache L, Liao KP, Graham RR, Fulton RS, Greenberg JD et al. TYK2 protein-coding variants protect against rheumatoid arthritis and autoimmunity, with no evidence of major pleiotropic effects on non-autoimmune complex traits. PLoS ONE 2015; 10: e0122271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia F, Bombardieri M, Principato A, Giardina A, Tripodo C, Porcasi R et al. Overexpression of interleukin-23, but not interleukin-17, as an immunologic signature of subclinical intestinal inflammation in ankylosing spondylitis. Arthritis Rheum 2009; 60: 955–965. [DOI] [PubMed] [Google Scholar]
- Sherlock JP, Joyce-Shaikh B, Turner SP, Chao CC, Sathe M, Grein J et al. IL-23 induces spondyloarthropathy by acting on ROR-gammat+ CD3+CD4-CD8- entheseal resident T cells. Nat Med 2012; 18: 1069–1076. [DOI] [PubMed] [Google Scholar]
- Benham H, Rehaume LM, Hasnain SZ, Velasco J, Baillet AC, Ruutu M et al. Interleukin-23 mediates the intestinal response to microbial beta-1,3-glucan and the development of spondyloarthritis pathology in SKG mice. Arthritis Rheumatol 2014; 66: 1755–1767. [DOI] [PubMed] [Google Scholar]
- Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat Genet 2010; 42: 1118–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szalay B, Meszaros G, Cseh A, Acs L, Deak M, Kovacs L et al. Adaptive immunity in ankylosing spondylitis: phenotype and functional alterations of T-cells before and during infliximab therapy. Clin Dev Immunol 2012; 2012: 808724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Goodall JC, Hill Gaston JS. Frequency and phenotype of peripheral blood Th17 cells in ankylosing spondylitis and rheumatoid arthritis. Arthritis Rheum 2009; 60: 1647–1656. [DOI] [PubMed] [Google Scholar]
- Chan AT, Kollnberger SD, Wedderburn LR, Bowness P. Expansion and enhanced survival of natural killer cells expressing the killer immunoglobulin-like receptor KIR3DL2 in spondylarthritis. Arthritis Rheum 2005; 52: 3586–3595. [DOI] [PubMed] [Google Scholar]
- Kenna TJ, Davidson SI, Duan R, Bradbury LA, McFarlane J, Smith M et al. Enrichment of circulating interleukin-17-secreting interleukin-23 receptor-positive gamma/delta T cells in patients with active ankylosing spondylitis. Arthritis Rheum 2012; 64: 1420–1429. [DOI] [PubMed] [Google Scholar]
- Ciccia F, Guggino G, Rizzo A, Saieva L, Peralta S, Giardina A et al. Type 3 innate lymphoid cells producing IL-17 and IL-22 are expanded in the gut, in the peripheral blood, synovial fluid and bone marrow of patients with ankylosing spondylitis. Ann Rheum Dis 2015; 74: 1739–1747. [DOI] [PubMed] [Google Scholar]
- Niu XY, Zhang HY, Liu YJ, Zhao D, Shan YX, Jiang YF. Peripheral B-cell activation and exhaustion markers in patients with ankylosing spondylitis. Life Sci 2013; 93: 687–692. [DOI] [PubMed] [Google Scholar]
- Noordenbos T, Yeremenko N, Gofita I, van de Sande M, Tak PP, Canete JD et al. Interleukin-17-positive mast cells contribute to synovial inflammation in spondylarthritis. Arthritis Rheum 2012; 64: 99–109. [DOI] [PubMed] [Google Scholar]
- Lau MC, Keith P, Costello ME, Bradbury LA, Hollis KA, Thomas R et al. Genetic association of ankylosing spondylitis with TBX21 influences T-bet and pro-inflammatory cytokine expression in humans and SKG mice as a model of spondyloarthritis. Ann Rheum Dis 2017; 76: 261–269. [DOI] [PubMed] [Google Scholar]
- Hao J, Liu Y, Xu J, Wang W, Wen Y, He A et al. Genome-wide DNA methylation profile analysis identifies differentially methylated loci associated with ankylosis spondylitis. Arthritis Res Ther 2017; 19: 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farh KK, Marson A, Zhu J, Kleinewietfeld M, Housley WJ, Beik S et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 2015; 518: 337–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin M, Codner D, Hasan SM, Scherer SW, O'Rielly DD, Rahman P. Integrated genomics identifies convergence of ankylosing spondylitis with global immune mediated disease pathways. Sci Rep 2015; 5: 10314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello ME, Ciccia F, Willner D, Warrington N, Robinson PC, Gardiner B et al. Brief Report: Intestinal Dysbiosis in Ankylosing Spondylitis. Arthritis Rheumatol 2015; 67: 686–691. [DOI] [PubMed] [Google Scholar]
- Ciccia F, Bombardieri M, Rizzo A, Principato A, Giardina AR, Raiata F et al. Over-expression of paneth cell-derived anti-microbial peptides in the gut of patients with ankylosing spondylitis and subclinical intestinal inflammation. Rheumatology (Oxford) 2010; 49: 2076–2083. [DOI] [PubMed] [Google Scholar]
- Visscher PM, Wray NR, Zhang Q, Sklar P, McCarthy MI, Brown MA et al. 10 Years of GWAS Discovery: Biology, Function, and Translation. Am J Hum Genet 2017; 101: 5–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai NS, Chou JL, Chen GCW, Liu SQ, Lu MC, Chan MWY. Association between cytokines and methylation of SOCS-1 in serum of patients with ankylosing spondylitis. Mol Biol Rep 2014; 41: 3773–3780. [DOI] [PubMed] [Google Scholar]
- Speed D, Balding DJ. MultiBLUP: improved SNP-based prediction for complex traits. Genome Res 2014; 24: 1550–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Linden S, Akkoc N, Brown MA, Robinson PC, Khan MA. The ASAS criteria for axial spondyloarthritis: strengths, weaknesses, and proposals for a way forward. Curr Rheumatol Rep 2015; 17: 62. [DOI] [PubMed] [Google Scholar]
- Haroon N, Inman RD, Learch TJ, Weisman MH, Lee M, Rahbar MH et al. The impact of tumor necrosis factor alpha inhibitors on radiographic progression in ankylosing spondylitis. Arthritis Rheum 2013; 65: 2645–2654. [DOI] [PMC free article] [PubMed] [Google Scholar]