Abstract
Clinical expansion of mesenchymal stem cells (MSCs) is hampered by the lack of knowledge regarding how to prevent MSC apoptosis and promote their proliferation in serum-free medium. Our in vitro studies demonstrated that human umbilical cord MSCs (HUCMSCs) underwent apoptosis in the serum-free medium. When HUCMSCs were co-cultured with retinal pigment epithelial cells (ARPE19), however, HUCMSCs exhibited normal growth and morphology in serum-free medium. Their colony formation was promoted by the conditioned medium (CM) of ARPE19 cells on Matrigel. Proteomics analysis showed that pigment epithelium-derived factor (PEDF) was one of the most abundant extracellular proteins in the ARPE19 CM, whereas enzyme-linked immunosorbent assay confirmed that large amounts of PEDF was secreted from ARPE19 cells. Adding anti-PEDF-blocking antibodies to the co-culture of HUCMSCs with ARPE19 cells increased apoptosis of HUCMSCs. Conversely, treatment with PEDF significantly reduced apoptosis and increased proliferation of HUCMSCs in serum-free medium. PEDF was further demonstrated to exert this anti-apoptotic effect by inhibiting P53 expression to suppress caspase activation. In vivo studies demonstrated that co-injection of HUCMSCs with ARPE19 cells in immunocompromised NOD-SCID mice also increased survival and decreased apoptosis of HUCMSCs. PEDF also showed no negative effect on the mesoderm differentiation capability of HUCMSCs. In conclusion, this study is the first to demonstrate that PEDF promotes HUCMSC proliferation and protects them from apoptosis by reducing p53 expression in the serum-free medium. This study provides crucial information for clinical-scale expansion of HUCMSCs.
Introduction
Human umbilical cord mesenchymal stem cells (HUCMSCs) are already applied clinically in stem cell therapy.1, 2, 3 Preclinical experiments on HUCMSCs or their derived tissues in disease models have been reported.3 Differentiated and engrafted HUCMSCs appear to have a successful functional outcome in in vivo rat models for cerebral ischemia,4 Parkinson’s disease, Alzheimer’s disease, multiple sclerosis, retinal disease,5 type 1 and type 2 diabetes, and myogenic disease.2 Furthermore, HUCMSCs exhibit low immunity and immunomodulatory effects, which increases the survival of transplanted cells and decreases the risk of graft-versus-host disease.6, 7 Therefore, they are the ideal stem cells for expansion in clinical cell therapy.
Clinical application of mesenchymal stem cells (MSCs), requires ex vivo MSC expansion to acquire sufficient cell numbers and optimal culture conditions. Expansion using animal-derived growth supplements, such as fetal bovine serum (FBS), involves critical limitations and safety concerns.8 For example, animal-derived (xeno) antigens and infectious agents present in FBS might be transmitted to the recipient of MSC therapy,9, 10, 11, 12, 13, 14, 15 and the composition of FBS is unclear and often inconsistent from lot to lot.16 The first commercially available xeno-free culture medium (Life Technologies stem cell growth medium) formulated for the expansion of human MSC has been approved by the Federal Drug Administration (FDA). However, it is too expensive for large-scale expansion of MSCs for clinical use. Alternative animal product-free media formulations, therefore, must be developed for clinical applications.
Retinal pigment epithelium (RPE) is a monolayer of pigmented, cuboidal epithelial cells that are closely associated with photoreceptor outer segments. The most important functions of the RPE are retinoid metabolism, photoreceptor membrane turnover, and inter-photoreceptor matrix synthesis and maintenance.17 Simply by Transwell-based co-culture with RPE cells, MSCs can be proliferated and differentiated toward an RPE phenotype.17, 18, 19, 20 The RPE cells secrete a variety of cytokines, connective tissue proteins, extracellular matrix proteins, complement factors, proteases and protease inhibitors. These proteins may promote MSC proliferation and differentiation into RPE-like cells. Therefore, the RPE-secreted factors could be used for development of a xeno-free culture medium for MSC expansion.
Mass spectrometry (MS) and label-free quantitation have provided researchers with the ability to accurately measure expression levels in complex mixtures.21 The increase in instrument-sequencing speed has benefited MS/MS spectral counting approaches by improving MS/MS sampling of peptide mixtures. The introduction of high-resolution analyzers (such as FT-Orbitrap) has encouraged the use of methods based on peptide-intensity measurements by greatly facilitating the matching of peptide peaks in different complex maps acquired independently. Nevertheless, the most obvious advantage of the above methods over isotopic labeling techniques is their ease of use at the sample preparation step, as they do not require any preliminary treatment to introduce a label into peptides. Because they are more straightforward, they do not have the disadvantages of labeling methods.22 In this investigation, therefore, we adopted MS/MS coupled with label-free quantitative proteomic analysis that facilitated an effective approach to investigate the key factor for MSC survival in the RPE-secreted proteins.
For clinical expansion of MSCs, it remains elusive how to prevent MSC apoptosis and promote their proliferation in serum-free medium. In this study, we demonstrated that co-culture of HUCMSCs with RPE supported HUCMSC growth and prevented apoptosis under serum-deprived conditions. Using label-free proteomic analysis, we identified PEDF as one of the key secreted proteins from ARPE-19 cell line (ARPE)-conditioned medium that is responsible for the effects mentioned above. We also determined that PEDF-reduced p53 expression to attenuate apoptosis in the HUCMSCs under serum-deprived conditions. These novel findings may provide crucial information for the development of xeno-free culture medium for the clinical expansion of HUCMSCs.
Materials and methods
All experiments involving human samples were approved by the Research Ethics Committee of Buddhist Tzu Chi General Hospital. The methods were carried out according to the approved guidelines. Informed consent was obtained from all subjects.
Cell culture
The human RPE cell line APRE19 was maintained in DMEM/F12 (1:1, Invitrogen, Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS, Biological Industry, Kibbutz, Israel), and penicillin and streptomycin (Thermo Fisher Scientific, Carlsbad, CA, USA). HUCMSCs were maintained in Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen) containing 10% FBS and antibiotics at 37 °C in a 95% air/5% CO2 humidified atmosphere.
Proliferation of HUCMSCs co-cultured with or without ARPE19
HUCMSCs were seeded in a 12-well Transwell bottom well and co-cultured with or without ARPE19 in cell-culture inserts with an 8-μm pore size (BD Bioscience, San Jose, CA, USA) in DMEM/F12 serum-free medium. Proliferation of HUCMSCs co-cultured with or without ARPE19 cell was evaluated after 4–6 days of coculture by using the 2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide (XTT) assay kit (Biological Industries, Beth Haemek, Israel).
Stacking gel-aided purification method
Protein purification was performed using gel-aided sample preparation as previously described.22 The secretomes of ARPE19 cells from concentrated-conditioned medium (CM) samples were run on a self-poured stacking SDS-PAGE gel. The resolving gel portion (0.6 ml H2O, 2.22 ml 1.5 M Tris-HCl (pH 8.8), 90 μl 10% SDS, 6 ml Bis/acrylamide, 90 μl 10% ammonium persulfate and 5 μl TEMED) was poured and set to polymerize for 1 h. The stacking gel portion (2.9 ml H2O, 0.5 ml 1 M Tris-HCl (pH 6.8), 40 μl 10% SDS, 520 μl Bis/acrylamide, 40 μl 10% ammonium persulfate and 4 μl TEMED) was poured next. A custom-made comb (1 mm thickness) was inserted, and the gel was allowed to polymerize for 30 min. A total of 10 μg CM was mixed with 13 μl H2O, 5 μl 4 × SDS sample buffer, and 2 μl 0.5 M DTT and then boiled for 10 min. The gel was run at 55 V. Electrophoresis was stopped after the sample had barely passed into the resolving gel, and the gels were then stained with Coomassie brilliant blue (CBB) R-250.
In-gel digestion
The gel pieces corresponding to the secretomes from ARPE19 were diced into 1 mm3 pieces. The gel slices were washed and dehydrated three times in 25 mM ammonium bicarbonate (ABC) (pH 7.9) and 50 mM ABC/50% acetonitrile. A protein reduction was subsequently performed by incubating 10 mM DTT for 1 h at 56 °C and then alkylating with 50 μl saturated iodoacetic acid (IAA) for 45 min at room temperature in the dark. After two subsequent wash/dehydration cycles, each gel sample was digested with 1 μg sequencing-grade-modified trypsin (Promega), and enough 25 mM ammonium bicarbonate was added to completely saturate the gel. The sample was incubated at 37 °C for an overnight digestion. Following the digestion, peptides were extracted twice in 100 μl 50% ACN in 5% formic acid. The extracted peptides were enriched by OMIX C18 pipet tips (Varian, Lake Forest, CA, USA), to remove the effects of any remaining reagents.
LC–MS/MS analysis
Resulting peptides were analyzed by RP-nano-HPLC-ESI-MS/MS using a nanoACQUITY HPLC system (Waters, Milford, MA, USA) coupled to an LTQ-Orbitrap Discovery Hybrid FTMS (Thermo Fisher Scientific, Bremen, Germany). For RP-nano-HPLC-ESI-MS/MS, a sample (2 μl) of the desired peptide digest was loaded into the reverse phase column (Symmetry C18, Waters Corporation, Milford, MA, USA, 5 μm, 180 μm × 20 mm) by an autosampler. After 3 min desalting, the precolumn was switched online with the analytical C18 column (BEH C18, 1.7 μm, 75 μm × 100 mm) equilibrated in 99% solvent A (100% D.I. water, 0.1% formic acid) and 1% solvent B (100% acetonitrile, 0.1% formic acid). Peptides were eluted using a linear acetonitrile gradient from 99% solvent A to 85% solvent B during 45 min at 400 nl min−1 flow rate. The LTQ-Orbitrap (Thermo Finnigan, San Jose, CA, USA) was operated in the data-dependent acquisition mode with the XCalibur software. Survey scan MS was acquired in the Orbitrap in the 400–2000 m/z range with the resolution set to a value of 30 000 and collision energy set at 35%. The four most intense ions per survey scan were selected for CID fragmentation, and the resulting fragments were analyzed in the linear trap mass analyzer (Thermo Finnigan). Dynamic exclusion was employed within 60 s to prevent repetitive selection of the same peptide.
Database search and data quantification
The MS/MS data were processed using MaxQuant v 1.3.0.5 (available via http://www.maxquant.org/). The MaxQuant package integrates a suite of software packages for peak list generation, labeled peptide pairs, SILAC- and XIC-based quantitation, false discovery rates (FDRs) and peptide identification based on a Mascot search. The step-by-step details of this procedure can be found in the reference section.22 Briefly, MaxQuant executed data analysis in four sequential steps. The first step entailed ‘pre-processing and quantitation’ to identify all the peptide pairs without knowing their identity, followed by a Mascot search in the second step. The Mascot search was performed using MaxQuant v 1.3.0.5 against the concatenated forward and reverse versions of UniProt human protein database. Mascot search parameters included the following as variable modifications: enzyme trypsin, one missed cleavage and oxidation of methionine. The maximum allowed mass error tolerances for MS and MS/MS scans were 5 p.p.m. and 0.5 Da, respectively. Only peptides with six amino acid residues were allowed, and proteins that had at least two peptides with ion scores >20 were considered for identification and quantitation. A target-decoy database approach using forward-reverse sequences and contaminants was used to determine the FDR, and the FDR threshold was set to the default values of 1% for both the peptides and the proteins, and posterior error probability of 0.5%.23 Label-free quantification was carried out in MaxQuant software as previously described.24 After MaxQuant analysis, the protein identification and quantification were reported in the table of proteinGroups. The identified proteins in the table of proteinGroups were listed on the column of majority protein IDs. Protein abundance was calculated from the normalized spectral protein intensity (LFQ intensity). The label-free quantification (LFQ) intensity was used to calculate the protein percentage of total protein intensity in each run (protein percentage=LFQ intensity of identified protein/sum of LFQ intensity of total identified proteins). Subsequently, the protein abundance of each protein in CM was calculated by the protein percentage of each protein plus total protein amount measured by protein assay in CM.
In silico analysis of protein sequences
The presence of secretory signal peptide cleavage sites was predicted using SignalP software version 4.1 (http://www.cbs.dtu.dk/services/SignalP/). All identified proteins were queried against this prediction software to look for the signal peptide sequences to verify that these identified proteins had been secreted via classical pathways. A sequence prediction based on SecretomeP version 2.0 (http://www.cbs.dtu.dk/services/SecretomeP) was performed for the prediction of secreted mammalian proteins targeted to non-classical secretion pathways. The transmembrane-domain prediction was performed with TMHMM Server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM/). The identified proteins were annotated to note the presence of a transmembrane helix and classified as either soluble or membrane proteins.
Proliferation of HUCMSCs in the absence or presence of recombinant PEDF or anti-PEDF antibody
A total of 2 × 105 HUCMSCs were seeded on a culture dish with DMEM/F12 serum-free medium in the presence or absence of 100 μg ml−1 rPEDF (Millipore, Billerica, MA, USA) or 5 μg ml−1 polyclonal anti-PEDF-blocking antibody (BioProducts, West Palm Beach, FL, USA) or 5 μg ml−1 non-functional control rabbit IgG (Santa Cruz Biotechnology, Dallas, TX, USA). Cell growth was measured after 7 days of culture.
Enzyme-linked immunosorbent assay
Levels of PEDF in a fresh-culture medium, in conditioned medium of HUCMSCs, in APRE19-conditioned medium and in HUCMSCs co-cultured with ARPE19 collected after 24 h of culture were determined by human enzyme-linked immunosorbent assay (ELISA) kits (PEDF, GeneAsia Biotech, Shanghai, China) according to the manufacturer’s instructions.
Terminal deoxynucleotidyl transferase dUTP nick-end labeling staining
Apoptotic cells were detected with the Click-iT Plus TUNEL Assay Kit (Life Technologies, Waltham, MA, USA) according to the manufacturer’s instructions. The percentage of TUNEL-positive cells was calculated as the number of TUNEL-positive cells divided by the total number of DAPI-positive cells in three non-overlapping areas (2 mm2 per well).
Western blotting
Cell lysates were obtained from HUCMSCs that were serum-starved for 48 h with or without ARPE19 co-culturing. The cell lysates were then loaded onto a 5–20% gradient SDS-polyacrylamide gel, subjected to electrophoresis under reducing conditions, and blotted onto a PVDF membrane (Bio-Rad, Hercules, CA, USA). The blots were blocked with a solution of 3% non-fat dry milk/2PBS/0.1% Tween-20 at room temperature; rinsed twice with PBS/0.1% Tween-20 and incubated with 1:200 diluted polyclonal anti-caspase-3 or anti-cleaved caspase-3, caspase-8 antibody and P53 (St John’s Lab, London, UK), followed by 1:5000 diluted anti-rabbit IgG-HRP (Amersham, GE, Taipei, Taiwan). Detection of actin by an anti-actin antibody (Santa Cruz Biotechnology) was used as a loading control. Membranes were rinsed three times in PBS/0.1% Tween-20. Signals were detected with horseradish peroxidase using an ECL kit (Promega, Fitchburg, WI, USA).
Ex vivo culture of HUCMSCs in the absence or presence of ARPE19-conditioned medium
Lentiviral rRFP-tagged HUCMSCs (2 × 105 cells) were seeded in a Matrigel (BD Bioscience)-coated dish, and cultured with or without conditioned medium from ARPE19 cells (1 × 105 cells) for 7 days. The colony numbers were calculated on the seventh day after the co-culture.
In vivo culture of HUCMSCs in the absence or presence of ARPE19
All animal work was approved by the Institutional Animal Care and Use Committee at Buddhist Tzu Chi General Hospital. Lentiviral rRFP-tagged HUCMSCs (2 × 105 cells) with or without APRE19 cells (1 × 105 cells) were seeded in 0.5 × 0.5 × 0.5 cm3 gelform (Ethicon, Somerville, NJ, USA) soaking in serum-free culture medium for one day, and then the gelform was transplanted subcutaneously into NOD-SCID mice. The gelform was made from porcine gelatin and had the consistency of a sponge-like material. The gelform sponge contained many porous structures, allowing the cells to grow in a 3D manner. After 5 days, the transplanted gelform was removed, and photographs were taken of cells under a fluorescence microscope to count the cells.
To evaluate TUNEL assays under in vivo conditions, DiI (Invitrogen)-stained HUCMSCs with or without ARPE19 cells were embedded at high molecular weight (3000 kD, Hyruan Plus, LG Life Sciences, Seoul, Korea) in serum-free hyaluronan (HA) and injected subcutaneously into NOD-SCID immune-deficient mice. (The mice are non-obese diabetic-severe mice with immune deficiency; the strain name is NOD.CB17-Prkdcscid/JTcu.) Five days after the injection, the transplanted HA-cell mixture was removed and analyzed by the TUNEL assay. Next, apoptotic cells were observed, and cell numbers were counted under a fluorescence microscope.
Differentiation of HUCMSCs after various treatments
After various treatments (co-cultured with ARPE19, cultured with APRE19-conditioned medium, adding rPEDF) for 7 days, HUCMSCs were sent for a differentiation assay to measure adipogenesis, osteogenesis and chondrogenesis.
Induction of adipogenesis
The resulting HUCMSCs were seeded in a 12-well plate at a density of 5 × 104 cells per well with adipogenic medium (DMEM supplemented with 10% FBS, 1 μmol l−1 dexamethasone (Sigma-Aldrich), 5 μg ml−1 insulin (Sigma-Aldrich), 0.5 mmol l−1 isobutylmethylxanthine (Sigma-Aldrich) and 60 μmol l−1 indomethacin (Sigma-Aldrich)). They were then allowed to grow for 14 days, the medium was changed every 3 days, and then cells were stained with Oil Red O (Sigma-Aldrich). After washing twice with PBS, lipids in the sample were extracted with 1 ml 100% isopropanol (Sigma-Aldrich) and shaken gently for 5 min. The concentration of the lipids was measured based on absorbance at 510 nm. The quantity of lipids in each sample was measured in triplicate.
Induction of osteogenesis
HUCMSCs were seeded in a 12-well plate at a density of 1 × 104 cells per well and grown with osteogenic medium (DMEM supplemented with 10% FBS, 0.1 μmol l−1 dexamethasone, 10 mmol l−1 β-glycerol phosphate (Sigma-Aldrich) and 50 μmol l−1 ascorbate (Sigma-Aldrich)), which was changed every 3 days. The cells were allowed to grow for 14 days and were then stained with Alizarin red (Sigma-Aldrich). For quantification of the staining, aliquots (150 μl) of the supernatant after multiple treatments as previously reported25 were read in triplicate at 405 nm in a 96-well (Costar; Corning, Lowell, MA, USA) opaque-walled, transparent-bottomed plates (Costar; Corning).
Induction of chondrogenesis
A total of 1.5 × 104 HUCMSCs were seeded in 6-well-plate for the above treatments. After 7 days, the HUCMSCs were seeded in a 15 ml conical tube at a density of 1 × 106 cells cm−2 and grown in chondrogenic media consisting DMEM, 10% FBS, 10 ng ml−1 transforming growth factor-β1 (Pepro Tech, Rocky Hill, NJ, USA), 50 μg ml−1 ascorbic acid-2-phospate (Sigma-Aldrich) and 6.25 μg ml−1 of insulin (Sigma-Aldrich). The media was changed every 3 days. The cells were incubated with the chondrogenic media at 37 °C with 5% CO2 for 3 weeks. After fixing in paraformaldehyde (Bionovas, Toronto, ON, Canada), the pellets were sliced into 5-μm sections, mounted on slides and stained by immunohistochemistry using type 2 collagen (Sigma-Aldrich).
Statistical analysis
All statistical analyses were conducted with SPSS version 20 software (IBM, Armonk, NY, USA). Statistical significance was calculated using t-tests, and significance was set at P<0.05. All values are expressed as the mean± s.d. ANOVA with the post hoc Bonferroni test was used for multiple group comparisons.
Results
Proliferation of HUCMSCs co-cultured with ARPE19 cells in a serum-free medium was not inhibited
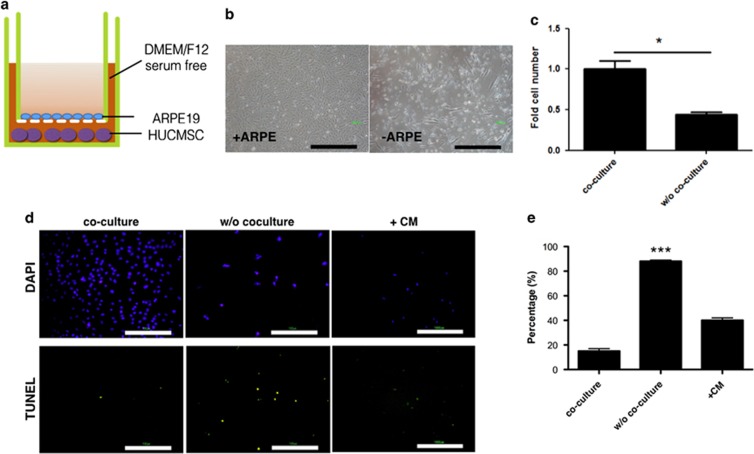
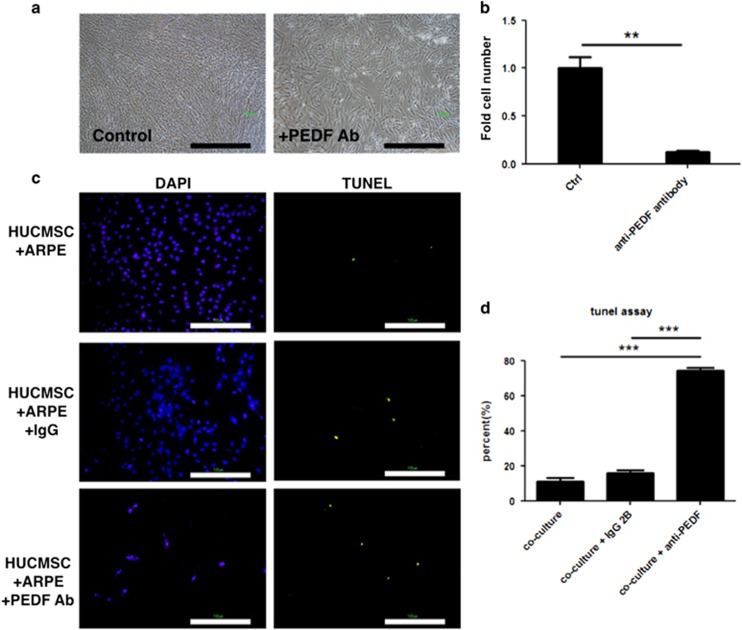
A co-culture experiment was designed to explore the effect of factors secreted by RPE cells on HUCMSCs in vitro (Figure 1). ARPE19 cells were seeded on Matrigel-coated Transwell inserts, and HUCMSCs were seeded on bottom dishes in a DMEM/F12 serum-free medium (Figure 1a). After 7 days, the co-cultured HUCMSCs were collected, and the number of cells was recorded. Co-culturing with RPE allowed HUCMSCs to maintain proliferation (Figure 1b and c). Without co-culturing with RPE, however, there was reduced proliferation of HUCMSCs and more apoptotic cell death, as indicated by the increase of TUNEL-positive cells compared with co-culture conditions or without co-culture but with addition of ARPE19-conditioned medium (Figure 1d and e).
Figure 1.
Proliferation of human umbilical cord mesenchymal stem cells (HUCMSCs) co-cultured with ARPE19 retina pigment epithelium cells under serum-starved conditions was unaffected. (a) Illustration of co-culturing HUCMSCs with ARPE19 cells. HUCMSCs were seeded on the bottom of each dish and co-cultured with ARPE19 cells seeded on culture inserts with Matrigel in a serum-free medium. (b) Phase-contrast photos of HUCMSCs in 12-well Transwells either cultured alone or co-cultured with ARPE19 cells under serum-starved conditions. Scale bar=1000 μm. (c) Quantification of HUCMSC cell growth alone or co-cultured with ARPE19 cells. *P<0.05. (d) Apoptotic cell death was examined by TUNEL assays and visualized as green spots on day 6 of culturing HUCMSC with or without co-culture or with addition of CM. Scale bar=100 μm. (e) The ratio (percentage) of TUNEL-positive to DAPI-positive HUCMSCs either cultured alone, co-cultured with ARPE19 cells, or without co-culture but with addition of CM. The results were from three independent experiments and are expressed as the mean±s.d. CM, conditioned medium. ***P<0.001, compared to co-culture and CM group.
Factors secreted from ARPE19 cells enhanced HUCMSC colony formation
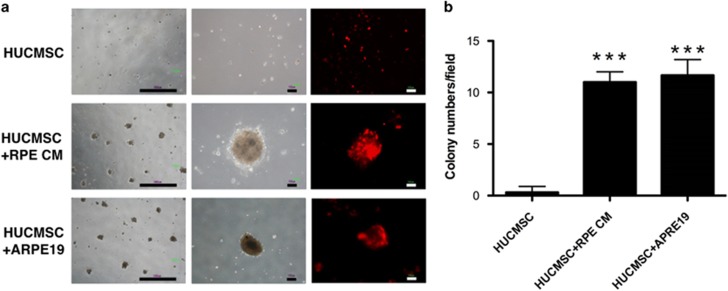
An ex vivo culture system with Matrigel was used to evaluate whether the factors secreted from ARPE19 might promote the growth of HUCMSCs in serum-free medium. When co-cultured with ARPE19 cells in the Transwell system or without co-culturing but with addition of ARPE19-conditioned medium, HUCMSCs formed colonies on the Matrigel-coated dish. Without co-culturing with ARPE19 cells or addition of conditioned medium, HUCMSCs could not form colonies (Figure 2a and b).
Figure 2.
Ex vivo growth of human umbilical cord mesenchymal stem cells (HUCMSCs) alone, co-cultured with ARPE19 cells, or with condition ARPE19 medium. (a) Phase-contrast and fluorescence microscopic images of HUCMSCs grown on Matrigel after 7 days with or without co-culture with ARPE19 or with conditioned ARPE19 medium. Scale bar=1000 μm (left panel) and 100 μm (middle and right panels). (b) HUCMSC colony numbers in microscopic field. ***P<0.001 compared with the group without co-culture.
Identification and quantification of the ARPE19 secretome
To investigate which secreted protein is responsible for the anti-apoptotic effect, we used label-free LC–MS/MS analysis to identify and quantify the ARPE19 secretome. Quantitative data were analyzed by the non-parametric MaxQuant to determine relative protein abundance. A total of 158 non-redundant proteins were identified and quantified. Among these proteins, 77 were predicted to be extracellular proteins (classical secreted protein, non-classical secreted protein and transmembrane protein) by SignalP 4.1, SecretomeP 2.0 and TMHMM 2.0 (Table 1). Pigment epithelium-derived factor (PEDF) was the fourth most relatively abundant protein of the 77 predicted secreted proteins. Because PEDF can activate the ERK and AKT signaling pathways in MSCs,26 it was selected to be the candidate for investigation of the anti-apoptosis mechanism in HUCMSCs.
Table 1. Extracellular proteins of ARPE19 identified by secretomic analysis.
| Uniprot# | Protein name | SP | TM | Unique peptide | Score |
|---|---|---|---|---|---|
| P08195 | 4F2 cell-surface antigen heavy chain | — | Y | 2 | 12.4 |
| Q13443 | ADAM 9 | Y | Y | 2 | 21.6 |
| O00468 | Agrin | Y | — | 26 | 162.6 |
| P05067 | Amyloid beta A4 protein | Y | Y | 3 | 30.0 |
| Q06481 | Amyloid-like protein 2 | Y | Y | 2 | 11.5 |
| P07355 | Annexin A2 | Y | — | 13 | 163.7 |
| O75882 | Attractin | — | Y | 6 | 98.9 |
| P61769 | Beta-2-microglobulin | Y | — | 4 | 67.7 |
| P43251 | Biotinidase | Y | — | 2 | 13.6 |
| P80723 | Brain acid soluble protein 1 | — | Y | 3 | 18.1 |
| P55287 | Cadherin-11 | Y | Y | 2 | 16.9 |
| P19022 | Cadherin-2 | Y | Y | 19 | 123.9 |
| P27797 | Calreticulin | Y | — | 3 | 25.7 |
| O94985 | Calsyntenin-1 | Y | Y | 8 | 67.7 |
| Q9UI42 | Carboxypeptidase A4 | Y | — | 7 | 53.7 |
| P07339 | Cathepsin D | Y | — | 2 | 18.6 |
| Q13740 | CD166 antigen | Y | Y | 5 | 67.2 |
| P20908 | Collagen alpha-1(V) chain | Y | Y | 2 | 20.8 |
| P05997 | Collagen alpha-2(V) chain | Y | Y | 3 | 20.6 |
| P01024 | Complement C3 | Y | Y | 6 | 79.5 |
| Q9ULV4 | Coronin-1C | — | Y | 2 | 11.6 |
| P01034 | Cystatin-C precursor | Y | — | 2 | 12.6 |
| P81605 | Dermcidin | Y | — | 3 | 23.4 |
| Q14126 | Desmoglein-2 | Y | Y | 6 | 73.9 |
| B2RKV3 | Dipeptidyl-peptidase 7 | Y | — | 2 | 20.3 |
| P04843 | Dolichyl-diphosphooligosaccharide—protein glycosyltransferase subunit 1 | Y | Y | 3 | 32.6 |
| Q14118 | Dystroglycan | Y | Y | 2 | 25.6 |
| Q14118 | Extracellular matrix protein 1 | Y | Y | 3 | 43.4 |
| P35555 | Fibrillin-1 | Y | Y | 5 | 42.2 |
| P35556 | Fibrillin-2 | Y | Y | 3 | 29.7 |
| P02751 | Fibronectin | Y | — | 11 | 182.7 |
| O95633 | Follistatin-related protein 1 | Y | — | 5 | 79.5 |
| P09382 | Galectin-1 | Y | — | 5 | 50.5 |
| Q08380 | Galectin-3-binding protein | Y | — | 3 | 38.7 |
| Q92820 | Gamma-glutamyl hydrolase | Y | — | 2 | 15.2 |
| P35052 | Glypican-1 | Y | — | 4 | 41.7 |
| P08069 | Insulin-like growth factor-binding protein 7 | Y | Y | 2 | 23.4 |
| P05556 | Integrin beta-1 | Y | Y | 3 | 19.1 |
| Q16363 | Laminin subunit alpha-4 | Y | — | 4 | 38.4 |
| P07942 | Laminin subunit beta-1 | Y | — | 8 | 71.4 |
| P61626 | Lysozyme C | Y | — | 2 | 14.3 |
| Q9Y4K0 | Lysyl oxidase homolog 2 | Y | — | 4 | 56.2 |
| P01033 | Metalloproteinase inhibitor 1 | Y | — | 2 | 19.2 |
| P60174 | Triosephosphate isomerase | Y | — | 4 | 29.5 |
| Q969H8 | Myeloid-derived growth factor | Y | — | 3 | 46.4 |
| P26022 | Pentraxin-related protein PTX3 | Y | — | 5 | 63.7 |
| P15692 | Vascular endothelial growth factor A | Y | — | 3 | 42.2 |
| Q96FE7 | Phosphoinositide-3-kinase-interacting protein 1 | Y | Y | 2 | 16.5 |
| P36955 | Pigment epithelium-derived factor | Y | — | 12 | 162.6 |
| P05155 | Plasma protease C1 inhibitor | Y | — | 8 | 81.6 |
| P05121 | Plasminogen activator inhibitor 1 | Y | — | 17 | 177.3 |
| Q6UX71 | Plexin domain-containing protein 2 | Y | Y | 3 | 34.0 |
| P23560 | Brain-derived neurotrophic factor | Y | — | 6 | 69.8 |
| Q07954 | Prolow-density lipoprotein receptor-related protein 1 | Y | Y | 5 | 66.1 |
| P41222 | Prostaglandin-H2 D-isomerase precursor | Y | — | 2 | 13.1 |
| P07237 | Protein disulfide-isomerase | Y | — | 3 | 19.4 |
| Q15084 | Protein disulfide-isomerase A6 | Y | — | 2 | 20.6 |
| P06703 | Protein S100-A6 | Y | — | 3 | 25.4 |
| O14522 | Receptor-type tyrosine-protein phosphatase kappa | Y | — | 5 | 67.7 |
| Q99969 | Retinoic acid receptor responder protein 2 | Y | — | 4 | 34.0 |
| O75326 | Semaphorin-7A | Y | — | 3 | 20.2 |
| P02768 | Serum albumin | Y | Y | 15 | 145.1 |
| P11166 | Solute carrier family 2, facilitated glucose transporter member 1 | Y | Y | 3 | 19.8 |
| P09486 | SPARC | Y | — | 6 | 119.3 |
| O00391 | Sulfhydryl oxidase 1 | Y | Y | 11 | 97.0 |
| P00441 | Superoxide dismutase | Y | — | 3 | 23.4 |
| P31431 | Syndecan-4 | Y | Y | 10 | 182.7 |
| Q9UI42 | Carboxypeptidase A4 | Y | — | 6 | 119.3 |
| P07585 | Decorin | Y | — | 2 | 32.8 |
| P07996 | Thrombospondin-1 | Y | — | 9 | 100.1 |
| O43897 | Tolloid-like protein 2 | Y | — | 2 | 12.4 |
| Q15582 | Transforming growth factor-beta-induced protein ig-h3 | Y | Y | 7 | 71.4 |
| P07093 | Glia-derived nexin | Y | — | 2 | 12.4 |
| Q6EMK4 | Vasorin | Y | Y | 5 | 67.7 |
| Q12907 | Vesicular integral-membrane protein VIP36 | Y | Y | 2 | 21.1 |
ARPE19 cells secreted large quantities of PEDF
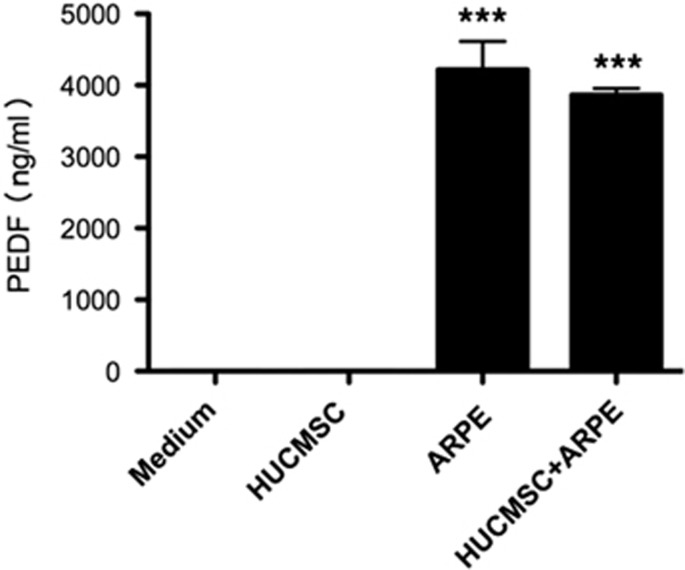
The secreted proteins from ARPE19 cells that affect HUCMSC proliferation were investigated by label-free LC–MS/MS analysis. PEDF was selected to be the target protein in this study because of its anti-apoptosis effect.27 The abundance of PEDF protein in the CM of APRE19 cells was determined and confirmed by ELISA. Up to 4000 ng ml−1 of PEDF was secreted from 2 × 105 ARPE19 cells (Figure 3). Thus, the effect of PEDF on HUCMSCs was examined first.
Figure 3.
Quantification of pigment epithelium-derived factor (PEDF) by enzyme-linked immunosorbent assay (ELISA) in conditioned medium of human umbilical cord mesenchymal stem cells (HUCMSCs), HUCMSCs co-cultured with ARPE19, and ARPE19 cells. A cultured medium was used as a negative control. The results were from three independent experiments and are expressed as the mean±s.d.
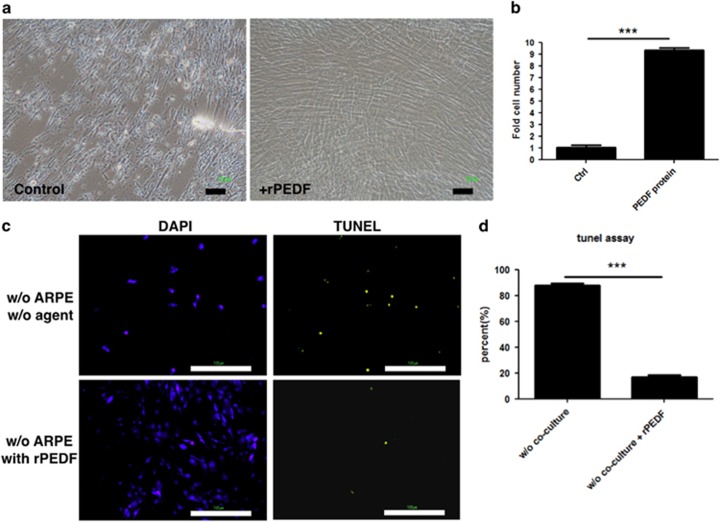
PEDF exerted an anti-apoptotic effect on HUCMSCs in serum-free medium
Recombinant PEDF protein (rPEDF) and anti-PEDF antibody were used to evaluate the effects of PEDF on the proliferation and apoptosis of HUCMSCs co-cultured with ARPE19 cells in serum-free medium. The rPDEF (50 μg ml−1) increased HUCMSC proliferation (Figure 4a and b), and simultaneously decreased HUCMSC apoptosis, as demonstrated by the TUNEL assay (Figure 4c and d). Conversely, the anti-PEDF antibody blocked HUCMSC proliferation and increased HUCMSC apoptosis (Figure 5a and b). Ratios (percentage) of TUNEL-positive to DAPI-positive HUCMSCs with or without the addition of IgG1 or anti-PEDF antibody are shown in Figure 5c and d.
Figure 4.
Pigment epithelium-derived factor (PEDF) rescued serum-starved human umbilical cord mesenchymal stem cell (HUCMSC) from death. (a) Phase-contrast photos of HUCMSCs not co-cultured with ARPE19 cells. Adding recombinant PEDF (rPEDF) resulted in more cells (right panel) than not adding rPEDF (left panel). Scale bar=100 μm. (b) Quantification of HUCMSC cell growth with or without rPEDF. ***P<0.001. (c) TUNEL assays examined apoptotic cell death and were visualized as green spots on day 6 of culture. Scale bar=100 μm. (d) The ratio of TUNEL-positive to DAPI-positive HUCMSCs (percentage) with or without rPEDF. The results were from three independent experiments and are expressed as the mean±s.d. ***P<0.001.
Figure 5.
Pigment epithelium-derived factor (PEDF)-blocking antibody inhibited proliferation of human umbilical cord mesenchymal stem cells (HUCMSCs) co-cultured with ARPE19 cells in serum-starved medium. (a) Phase-contrast photos of HUCMSCs co-cultured with ARPE19 cells, with (right panel) and without (left panel) adding PEDF blocking antibody. Scale bar=1000 μm. (b) Quantification of HUCMSC cell numbers in a. ***P<0.001. (c) Apoptotic cell death was examined by TUNEL assays and visualized as green spots on day 6 of culture. Scale bar=100 μm. (d) Ratio (percentage) of TUNEL-positive to DAPI-positive HUCMSCs co-cultured with ARPE and with the addition of IgG1 or anti-PEDF antibody. The results were from three independent experiments and are expressed as the mean±s.d. ***P<0.001.
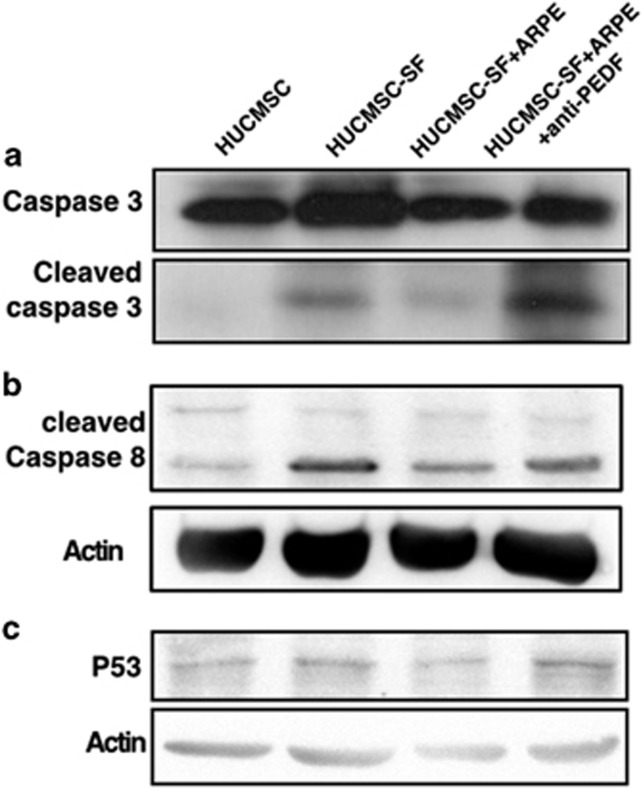
The PEDF-mediated signaling pathway causing these anti-apoptotic effects in HUCMSCs was then studied. Cleaved caspase-3 was detected by western blotting after anti-PEDF antibody stimulation of HUCMSCs (Figure 6a). Caspase-9, caspase-8 and cytochrome c (the proteins upstream of caspase-3) were investigated, and caspase-8 was found to be the regulatory protein (Figure 6b). In addition, p53 was markedly increased after anti-PEDF antibody stimulation of HUCMSCs under serum-starved conditions (Figure 6c).
Figure 6.
P53-activated caspase-8 and caspase-3. Western blot detected cleaved caspase-3 (a), cleaved caspase-8 (b) and P53 (c) in human umbilical cord mesenchymal stem cells (HUCMSCs) with different culture conditions (normal medium, serum-free (SF) medium, co-culturing with ARPE with or without adding anti-PEDF antibody) with a specific antibody (display of cropped blots).
ARPE19 cells protected HUCMSCs from apoptosis when co-transplanted in vivo
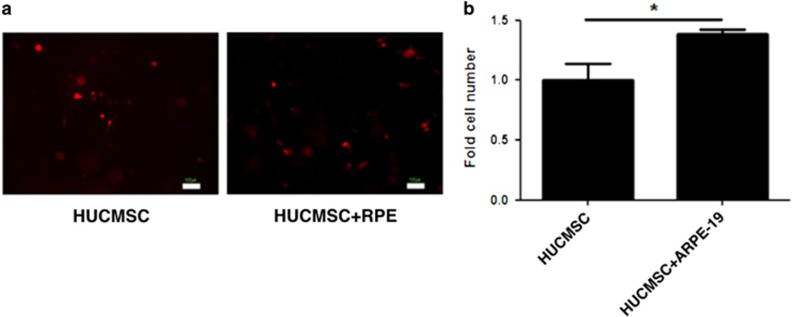
To determine whether APRE19 cells could inhibit the apoptosis of HUCMSCs in vivo, 105 HUCMSCs were co-transplanted without (0) or with (2 × 105) ARPE19 cells in gelform subcutaneously into NOD-SCID immune-deficient mice. Five days after transplantation, the number of HUCMSCs co-transplanted with ARPE19 cells was significantly greater than that of HUCMSCs without co-transplantation with ARPE19 cells (Figure 7) (P<0.05).
Figure 7.
In vivo growth of human umbilical cord mesenchymal stem cells (HUCMSCs) alone or co-cultured with ARPE19 cells. HUCMSCs were co-transplanted with or without retinal pigment epithelium (RPE) on gelform into NOD-SCID mice. (a) Fluorescence microscopy images were taken 5 days after the transplantation. Scale bar=100 μm. (b) Quantification of HUCMSCs on the transplanted gelform. *P<0.05.
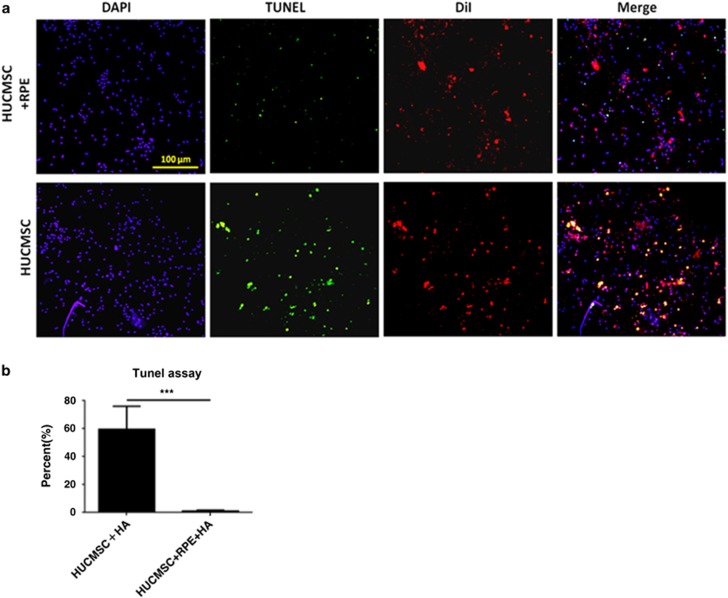
In an alternate method, HUCMSCs (DiI-labeled) and ARPE19 cells were embedded in high molecular weight HA (3000 kD) under serum-free conditions and then transplanted into the back of NOD-SCID mice. Five days after transplantation, the HA was taken out from the mice. TUNEL assays showed more apoptotic cells in the HUCMSCs-only group than in those co-injected with ARPE19 cells in HA (P<0.001, Figure 8).
Figure 8.
In vivo rescue of apoptotic human umbilical cord mesenchymal stem cells (HUCMSCs) co-cultured with ARPE19 cells in serum-free hyaluronan (HA). (a) HUCMSCs co-cultured with (upper panel) or without (lower panel) ARPE19 cells were embedded in serum-free 3000 kD HA and then transplanted into the subcutaneous region of NOD-SCID mice. The TUNEL assay was evaluated 5 days after the transplantation. (b) Ratio (percentage) of TUNEL-positive to DAPI-positive HUCMSCs (stained with DiI) with or without APRE19 cells in 3000 kD HA was shown. The results were from three independent experiments and are expressed as the mean±s.d. ***P<0.001. Scale bar=100 μm.
Co-culture with ARPE19, adding ARPE19-conditioned medium or rPEDF affected the differentiation ability of HUCMSCs in serum-free medium
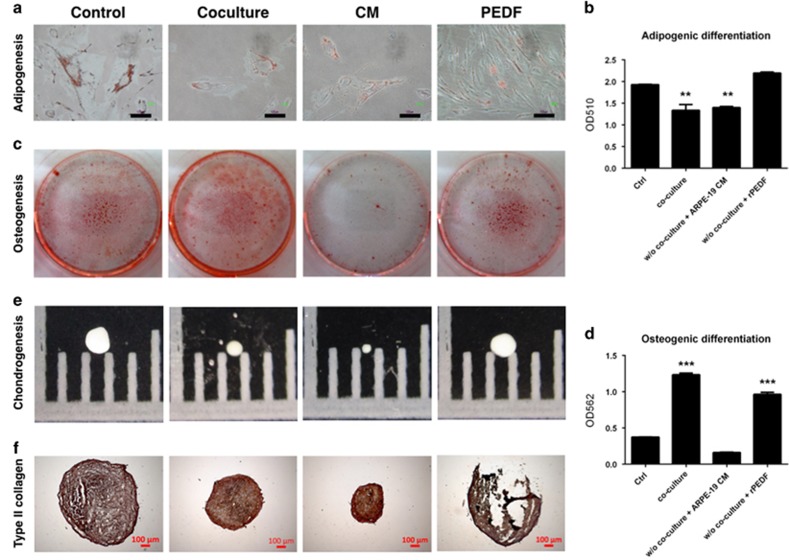
Decreased adipogenesis of HUCMSCs was noted in the co-cultured group and CM group, but there was no effect in the PEDF group (Figure 9a and b). Increased osteogenesis of HUCMSCs was noted in the co-culture group and PEDF group compared with the control and CM groups (Figure 9c and d). When measuring chondrogenesis, the PEDF group had the same pellet size as the control. The co-culture and CM groups formed smaller pellets than the control (Figure 9e). They all stained positive for type 2 collagen, a chondrocyte marker (Figure 9f).
Figure 9.
Effect of ARPE19 co-culture, conditioned medium (CM), and pigment epithelium-derived factor (PEDF) on the differentiation of human umbilical cord mesenchymal stem cells (HUCMSCs). Adipogenesis potential of HUCMSCs under various conditions was shown by (a) Oil Red O stain, and the amount of staining at day 14 was measured by (b) spectrophotometry at OD510. Osteogenesis of HUCMSCs under various conditions was shown by (c) Alizarin Red stain, and the amount of staining at day 14 was measured by (d) spectrophotometry at OD562. Chondrogenesis of HUCMSCs under various conditions was shown by (e) pellet formation at day 21 and (f) immunohistochemical staining of type 2 collagen. Scale bar=100 μm. **P<0.01,***P<0.001.
Co-culture with ARPE19, conditioned medium or rPEDF increased proliferation and reduced apoptosis of HUCMSCs in serum-free medium
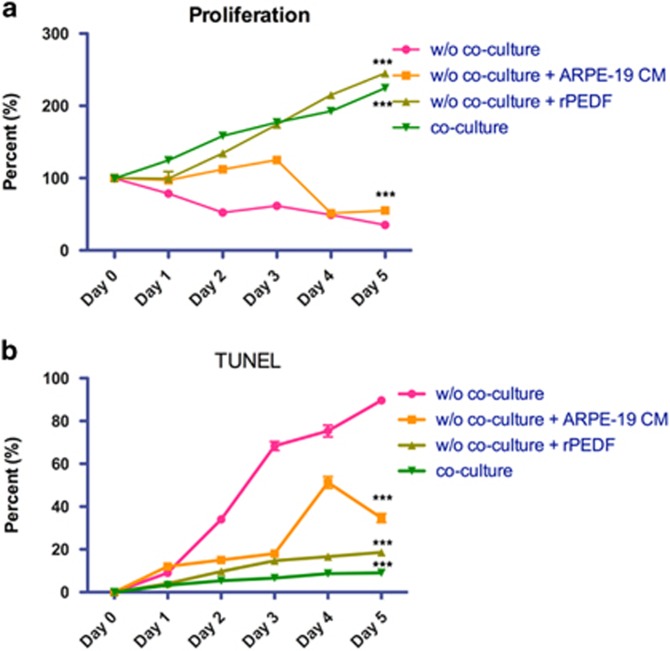
To determine the effects of co-culture, APRE19 CM, and rPEDF on proliferation and apoptosis of HUCMSCs with serum-free medium, proliferation (XTT assay) and apoptosis (TUNEL assay) were measured in a 5-day time course. During the 5-day time course, co-culture, CM and rPEDF were found to significantly increase proliferation (Figure 10a) and decrease apoptosis (Figure 10b) of HUCMSCs.
Figure 10.
Effect of ARPE19 co-culture, conditioned medium (CM), and pigment epithelium-derived factor (PEDF) on the proliferation and apoptosis of human umbilical cord mesenchymal stem cells (HUCMSCs). (a) The proliferation of HUCMSCs under various conditions from day 0 to day 5. (b) The percentage of apoptosis of HUCMSCs under various conditions from day 0 to day 5. *P<0.05, ***P<0.001 compared with non-co-cultured group.
Discussion
We demonstrated that ARPE19-secreted factors could support HUCMSC proliferation and differentiation in serum-free medium. Label-free LC–MS/MS analysis of ARPE19 secretomes showed that PEDF was one of the most abundant secreted proteins in the CM. We find that PEDF may be the key factor that increases HUCMSC proliferation in serum-free medium. Using functional assays, we demonstrated that PEDF protected HUCMSCs from apoptosis and enhanced their proliferation in serum-free culture medium in vitro and in vivo, and anti-PEDF antibody exerted opposite effects. The anti-apoptotic mechanism was mediated by inhibition of p53-dependent apoptosis.
Human bone marrow stromal cells (hBMSCs) can differentiate into an RPE phenotype when they are co-cultured with pig RPE in a Transwell system, suggesting that the soluble factors in the co-culture system can facilitate lineage differentiation of hBMSCs into RPE-like cells.19 These soluble factors not only promote lineage differentiation of hBMSCs into RPE-like cells but also support hBMSC growth in the serum-free culture medium. Like hBMSCs, HUCMSCs have multipotent differentiation potential and can differentiate into RPE-like cells. Our data also demonstrated that the soluble factors from ARPE19 improved HUCMSC colony formation, and supported HUCMSC growth and morphology in serum-free medium. Thus, the soluble factors from RPE cells may contain some important factors that protect HUCMSCs from apoptotic cell death and enhance their proliferation in serum-free medium.
Using label-free secretome analysis, we demonstrated that PEDF was one of the most abundant secreted proteins in the ARPE19 CM. PEDF is a 50 kDa molecule known as serpin F128, 29 and is mainly secreted from ARPE19. PEDF has various biological functions, including anti-inflammatory and anti-apoptotic effects in neuroretina.30, 31 It sustains the retinal microenvironment and structure, and promotes differentiation of primitive retinal cells32 and retinoblastoma cells.33 It stimulates several signaling pathways, including Ras, NF-kB,34 FAS/FASL,35 PPAR-gamma and p53.36 Recently, PEDF was reported to activate ERK1/2 and maintain the growth of serum-starved hESCs.37 When 100 ng ml−1 rPEDF was added to serum-starved hESCs, ERK1/2 was activated; the ERK1/2 inhibitor PD98059 inhibited the growth of hESCs. In addition, PEDF can activate ERK and AKT signaling pathways in hMSCs.26 The proliferative effects of PEDF on hESCs are like those on hMSCs. These data support our findings that PEDF has a significant role in protecting HUCMSCs from apoptosis and in enhancing their proliferation.
Under serum-starved conditions, cells undergo apoptotic changes.38 The receptor and mitochondrial pathways are involved in this process.39 Fas or TNF-α activation trigger caspase-8 to initiate the subsequent caspase-3 apoptotic reaction. In our present investigation, cleavage of caspase-8 and caspase-3 increased after PEDF-blocking antibody was used to stimulate HUCMSCs co-cultured with ARPE19 cells. In addition, p53 protein levels were highly increased in the HUCMSCs after PEDF neutralization under serum-starved conditions. Conversely, adding rPEDF could ‘rescue’ apoptosis of HUCMSCs under serum-starved conditions. Lastly, an animal model also showed decreased apoptosis of HUCMSCs when they were co-injected with ARPE19 cells. Taken together, our data demonstrate that PEDF can inhibit serum-deprivation-induced apoptosis of HUCMSCs via inhibition of p53-dependent apoptosis.
Recently, PEDF was reported to facilitate apoptotic cell death of induced pluripotent stem cells (iPSCs).40 The study found that PEDF-induced p38 MAPK-dependent cleavage of multiple caspases could lead to apoptosis of iPSCs after PEDF stimulation. Although their results were contrary to our observations, this phenomenon allowed for iPSC-induced teratoma elimination. The differences between iPSCs and HUCMSCs may be due to the nature of different cell lines and culture conditions.
A primary limitation of the current study is the possibility that the secreted factors of ARPE19 may contain lipids and carbohydrates, which may also affect apoptosis and proliferation of HUCMSCs.
In conclusion, our novel findings indicate that PEDF can support proliferation and differentiation, and reduce apoptosis of serum-starved HUCMSCs. These results also suggest that PEDF may be a critical supplement to xeno-free culture medium in the quest for clinical expansion of human MSCs.
Acknowledgments
This study was supported by a Buddhist Tzu Chi General Hospital research grant ‘TCRDI 104-01-03, TCRD 104-08’ (Hualien, Taiwan). We thank Dr Kuo CS for proofreading the manuscript.
Author contributions
D-CD performed all biological assays and prepared the manuscript; Y-TW helped plan all experiments and prepared the manuscript; and R-KT analyzed all experimental results and edited the manuscript.
Footnotes
Publisher’s note:
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The authors declare no conflict of interest.
References
- El Omar R, Beroud J, Stoltz JF, Menu P, Velot E, Decot V. Umbilical cord mesenchymal stem cells: the new gold standard for mesenchymal stem cell-based therapies? Tissue Eng Part B Rev 2014; 20: 523–544. [DOI] [PubMed] [Google Scholar]
- Ding DC, Chang YH, Shyu WC, Lin SZ. Human umbilical cord mesenchymal stem cells: a new era for stem cell therapy. Cell Transplant 2015; 24: 339–347. [DOI] [PubMed] [Google Scholar]
- Fan CG, Zhang QJ, Zhou JR. Therapeutic potentials of mesenchymal stem cells derived from human umbilical cord. Stem Cell Rev 2011; 7: 195–207. [DOI] [PubMed] [Google Scholar]
- Ding DC, Shyu WC, Chiang MF, Lin SZ, Chang YC, Wang HJ et al. Enhancement of neuroplasticity through upregulation of beta1-integrin in human umbilical cord-derived stromal cell implanted stroke model. Neurobiol Dis 2007; 27: 339–353. [DOI] [PubMed] [Google Scholar]
- Ramsden CM, Powner MB, Carr AJ, Smart MJ, da Cruz L, Coffey PJ. Stem cells in retinal regeneration: past, present and future. Development 2013; 140: 2576–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manochantr S, U-pratya Y, Kheolamai P, Rojphisan S, Chayosumrit M, Tantrawatpan C et al. Immunosuppressive properties of mesenchymal stromal cells derived from amnion, placenta, Wharton's jelly and umbilical cord. Intern Med J 2013; 43: 430–439. [DOI] [PubMed] [Google Scholar]
- Ding DC, Chou HL, Chang YH, Hung WT, Liu HW, Chu TY. Characterization of HLA-G and related immunosuppressive effects in human umbilical cord stroma-derived stem cells. Cell Transplant 2016; 25: 217–228. [DOI] [PubMed] [Google Scholar]
- Halme DG, Kessler DA. FDA regulation of stem-cell-based therapies. N Engl J Med 2006; 355: 1730–1735. [DOI] [PubMed] [Google Scholar]
- Heiskanen A, Satomaa T, Tiitinen S, Laitinen A, Mannelin S, Impola U et al. N-glycolylneuraminic acid xenoantigen contamination of human embryonic and mesenchymal stem cells is substantially reversible. Stem Cells 2007; 25: 197–202. [DOI] [PubMed] [Google Scholar]
- Ivan A, Indrei LL. Emergence of transmissible disorders, a continuous process—a new type of viral meningoencephalitis. Rev Med Chir Soc Med Nat Iasi 2000; 104: 51–55. [PubMed] [Google Scholar]
- Mackensen A, Drager R, Schlesier M, Mertelsmann R, Lindemann A. Presence of IgE antibodies to bovine serum albumin in a patient developing anaphylaxis after vaccination with human peptide-pulsed dendritic cells. Cancer Immunol Immunother 2000; 49: 152–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvaggi TA, Walker RE, Fleisher TA. Development of antibodies to fetal calf serum with arthus-like reactions in human immunodeficiency virus-infected patients given syngeneic lymphocyte infusions. Blood 1997; 89: 776–779. [PubMed] [Google Scholar]
- Spees JL, Gregory CA, Singh H, Tucker HA, Peister A, Lynch PJ et al. Internalized antigens must be removed to prepare hypoimmunogenic mesenchymal stem cells for cell and gene therapy. Mol Ther 2004; 9: 747–756. [DOI] [PubMed] [Google Scholar]
- Sundin M, Ringden O, Sundberg B, Nava S, Gotherstrom C, Le Blanc K. No alloantibodies against mesenchymal stromal cells, but presence of anti-fetal calf serum antibodies, after transplantation in allogeneic hematopoietic stem cell recipients. Haematologica 2007; 92: 1208–1215. [DOI] [PubMed] [Google Scholar]
- Tuschong L, Soenen SL, Blaese RM, Candotti F, Muul LM. Immune response to fetal calf serum by two adenosine deaminase-deficient patients after T cell gene therapy. Hum Gene Ther 2002; 13: 1605–1610. [DOI] [PubMed] [Google Scholar]
- Shih DT, Burnouf T. Preparation, quality criteria, and properties of human blood platelet lysate supplements for ex vivo stem cell expansion. N Biotechnol 2015; 32: 199–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Lauro S, Rodriguez-Crespo D, Gayoso MJ, Garcia-Gutierrez MT, Pastor JC, Srivastava GK et al. A novel coculture model of porcine central neuroretina explants and retinal pigment epithelium cells. Mol Vis 2016; 22: 243–253. [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Crespo D, Di Lauro S, Singh AK, Garcia-Gutierrez MT, Garrosa M, Pastor JC et al. Triple-layered mixed co-culture model of RPE cells with neuroretina for evaluating the neuroprotective effects of adipose-MSCs. Cell Tissue Res 2014; 358: 705–716. [DOI] [PubMed] [Google Scholar]
- Duan P, Xu H, Zeng Y, Wang Y, Yin ZQ. Human bone marrow stromal cells can differentiate to a retinal pigment epithelial phenotype when co-cultured with pig retinal pigment epithelium using a transwell system. Cell Physiol Biochem 2013; 31: 601–613. [DOI] [PubMed] [Google Scholar]
- Vossmerbaeumer U, Ohnesorge S, Kuehl S, Haapalahti M, Kluter H, Jonas JB et al. Retinal pigment epithelial phenotype induced in human adipose tissue-derived mesenchymal stromal cells. Cytotherapy 2009; 11: 177–188. [DOI] [PubMed] [Google Scholar]
- Han X, Aslanian A, Yates JR 3rd. Mass spectrometry for proteomics. Curr Opin Chem Biol 2008; 12: 483–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen YT, Wang JS, Tsai SH, Chuan CN, Wu JJ, Liao PC. Label-free proteomic analysis of environmental acidification-influenced Streptococcus pyogenes secretome reveals a novel acid-induced protein histidine triad protein A (HtpA) involved in necrotizing fasciitis. J Proteomics 2014; 109: 90–103. [DOI] [PubMed] [Google Scholar]
- Aryal UK, Stockel J, Krovvidi RK, Gritsenko MA, Monroe ME, Moore RJ et al. Dynamic proteomic profiling of a unicellular cyanobacterium Cyanothece ATCC51142 across light-dark diurnal cycles. BMC Syst Biol 2011; 5: 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luber CA, Cox J, Lauterbach H, Fancke B, Selbach M, Tschopp J et al. Quantitative proteomics reveals subset-specific viral recognition in dendritic cells. Immunity 2010; 32: 279–289. [DOI] [PubMed] [Google Scholar]
- Ding DC, Wu KC, Chou HL, Hung WT, Liu HW, Chu TY. Human infrapatellar fat pad-derived stromal cells have more potent differentiation capacity than other mesenchymal cells and can be enhanced by hyaluronan. Cell Transplant 2015; 24: 1221–1232. [DOI] [PubMed] [Google Scholar]
- Li F, Song N, Tombran-Tink J, Niyibizi C. Pigment epithelium-derived factor enhances differentiation and mineral deposition of human mesenchymal stem cells. Stem Cells 2013; 31: 2714–2723. [DOI] [PubMed] [Google Scholar]
- Kawaguchi T, Yamagishi S, Itou M, Okuda K, Sumie S, Kuromatsu R et al. Pigment epithelium-derived factor inhibits lysosomal degradation of Bcl-xL and apoptosis in HepG2 cells. Am J Pathol 2010; 176: 168–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filleur S, Nelius T, de Riese W, Kennedy RC. Characterization of PEDF: a multi-functional serpin family protein. J Cell Biochem 2009; 106: 769–775. [DOI] [PubMed] [Google Scholar]
- Steele FR, Chader GJ, Johnson LV, Tombran-Tink J. Pigment epithelium-derived factor: neurotrophic activity and identification as a member of the serine protease inhibitor gene family. Proc Natl Acad Sci USA 1993; 90: 1526–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Zhang H, Zhuang W, Yuan G, Sun T, Jiang X et al. PEDF and PEDF-derived peptide 44mer protect cardiomyocytes against hypoxia-induced apoptosis and necroptosis via anti-oxidative effect. Sci Rep 2014; 4: 5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Subramanian P, Shen D, Tuo J, Becerra SP, Chan CC. Pigment epithelium-derived factor reduces apoptosis and pro-inflammatory cytokine gene expression in a murine model of focal retinal degeneration. ASN Neuro 2013; 5: e00126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funatsu H, Yamashita H, Nakamura S, Mimura T, Eguchi S, Noma H et al. Vitreous levels of pigment epithelium-derived factor and vascular endothelial growth factor are related to diabetic macular edema. Ophthalmology 2006; 113: 294–301. [DOI] [PubMed] [Google Scholar]
- Tombran-Tink J, Chader GG, Johnson LV. PEDF: a pigment epithelium-derived factor with potent neuronal differentiative activity. Exp Eye Res 1991; 53: 411–414. [DOI] [PubMed] [Google Scholar]
- Tombran-Tink J, Barnstable CJ. PEDF: a multifaceted neurotrophic factor. Nat Rev Neurosci 2003; 4: 628–636. [DOI] [PubMed] [Google Scholar]
- Volpert OV, Zaichuk T, Zhou W, Reiher F, Ferguson TA, Stuart PM et al. Inducer-stimulated Fas targets activated endothelium for destruction by anti-angiogenic thrombospondin-1 and pigment epithelium-derived factor. Nat Med 2002; 8: 349–357. [DOI] [PubMed] [Google Scholar]
- Ho TC, Chen SL, Yang YC, Liao CL, Cheng HC, Tsao YP. PEDF induces p53-mediated apoptosis through PPAR gamma signaling in human umbilical vein endothelial cells. Cardiovasc Res 2007; 76: 213–223. [DOI] [PubMed] [Google Scholar]
- Gonzalez R, Jennings LL, Knuth M, Orth AP, Klock HE, Ou W et al. Screening the mammalian extracellular proteome for regulators of embryonic human stem cell pluripotency. Proc Natl Acad Sci USA 2010; 107: 3552–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon S, Ki SM, Park SE, Kim MJ, Hyung B, Lee NK et al. Anti-apoptotic effects of human Wharton’s jelly-derived mesenchymal stem cells on skeletal muscle cells mediated via secretion of XCL1. Mol Ther 2016; 24: 1550–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton BR, Reutens DC, Sobey CG. Apoptotic mechanisms after cerebral ischemia. Stroke 2009; 40: e331–e339. [DOI] [PubMed] [Google Scholar]
- Kanemura H, Go MJ, Nishishita N, Sakai N, Kamao H, Sato Y et al. Pigment epithelium-derived factor secreted from retinal pigment epithelium facilitates apoptotic cell death of iPSC. Sci Rep 2013; 3: 2334. [DOI] [PMC free article] [PubMed] [Google Scholar]