Abstract
Nuclear bodies are subnuclear, spheroidal, and membraneless compartments that concentrate specific proteins and/or RNAs. They serve as sites of biogenesis, storage, and sequestration of specific RNAs, proteins, or ribonucleoprotein complexes. Recent studies reveal that a subset of nuclear bodies in various eukaryotic organisms is constructed using architectural long noncoding RNAs (arcRNAs). Here, we describe the unifying mechanistic principles of the construction and function of these bodies, especially focusing on liquid-liquid phase separation induced by architectural molecules that form multiple weakly adhesive interactions. We also discuss three possible advantages of using arcRNAs rather than architectural proteins to build the bodies: position-specificity, rapidity, and economy in sequestering nucleic acid-binding proteins. Moreover, we introduce two recently devised methods to discover novel arcRNA-constructed bodies; one that focuses on the RNase-sensitivity of these bodies, and another that focuses on “semi-extractability” of arcRNAs.
Keywords: architectural RNA, liquid-liquid phase separation, low-complexity domain, multivalency, nuclear body, prion-like domain
NUCLEAR BODIES ARE PHASE-SEPARATED COMPARTMENTS FORMED THROUGH MULTIVALENT INTERMOLECULAR INTERACTIONS
Cellular bodies or condensates are subcellular, membraneless, and spheroidal compartments approximately 0.2 to 2 μm in diameter. Nuclear bodies are cellular bodies within the nucleus, and include the nucleolus, Cajal body, histone locus body, Polycomb body, promyelocytic leukemia body, nuclear speckle, and paraspeckle (Fig. 1). Each nuclear body is defined by enrichment of a specific marker protein and/or RNA, and functions as the site of biogenesis, storage, or sequestration of specific RNAs, proteins, or ribonucleoprotein (RNP) complexes. Most cellular bodies exhibit liquid droplet-like features; the cellular bodies are demixed (phase-separated) from the surrounding nucleoplasm or cytoplasm, they can fuse and become a larger droplet, and shearing forces can deform and break down the droplets to smaller droplets (Banani et al., 2017, 2011; Brangwynne et al., 2011). Such liquid-liquid phase separation (LLPS) is accomplished by molecules that can form multiple intermolecular interactions, a property that is called multivalency, a classical concept in polymer chemistry (Molliex et al., 2015; Nott et al., 2015). Above a threshold concentration, molecules that form multivalent interactions can self-assemble into large oligomers or polymers, often causing LLPS to enable body formation. Indeed, concentrating the key body component at a particular cellular site induces formation of nucleolus, Cajal body, paraspeckle, and histone locus body (Berry et al., 2015; Kaiser et al., 2008; Mao et al., 2011). LLPS is often enabled by proteins that contain a region enriched with a limited number of amino acid types, a domain referred to as low-complexity domain (LCD) (Fig. 2) (Kato et al., 2012; Molliex et al., 2015). LCDs lack a defined three-dimensional structure, which most other classical protein domains possess, and such intrinsically disordered LCDs provide the basis for multivalent weakly adhesive intermolecular interactions, such as electrostatic interactions (e.g., charge, cation-pi, and dipole-dipole), pi stacking interactions, and hydrophobic interactions (Fig. 2) (Brangwynne et al., 2015; Nott et al., 2015; Petri et al., 2012; Reichheld et al., 2017). These weak interactions are short lived and provide little structural order to the peptide chain, consistent with the dynamic nature of phase-separated liquids. Although a cellular body as a whole is often stably maintained over hours or even days, the protein components of the body are usually dynamic, exchanging rapidly with the surrounding nucleoplasm or cytoplasm on timescales of seconds (Fig. 2) (Dundr et al., 2004; Mao et al., 2011).
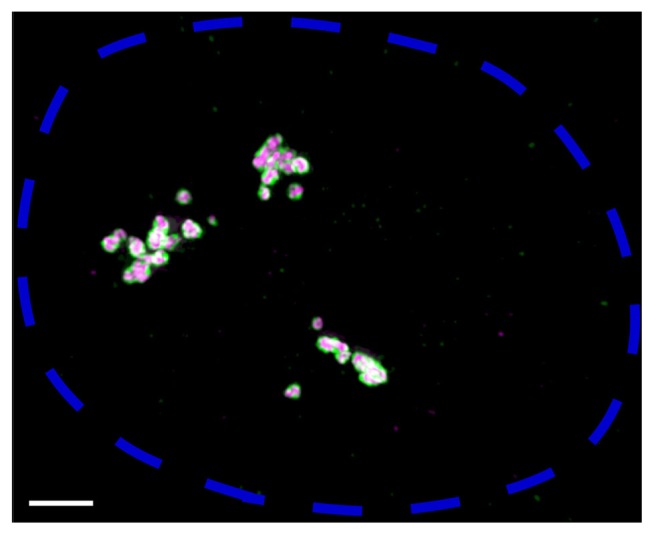
Fig. 1. Super-resolution image of paraspeckle nuclear bodies constructed by NEAT1_2 lncRNA.
Fluorescence in-situ hybridization was performed on HeLa cells to visualize NEAT1_2 5′ and 3′ regions (shown in green) and NEAT1_2 middle region (shown in magenta). A maximum intensity projection image of one nucleus, delineated by a blue dotted line, is shown. Scale bar, 2 μm. Each paraspeckle is estimated to contain about 50 NEAT1_2 molecules in HeLa cells (Chujo et al., 2017). Three clusters of paraspeckles might correspond to the approximate locations of three NEAT1 loci on three copies of chromosome 11 in HeLa cells. 5′ and 3′ regions of NEAT1_2 are located at the “shell” of paraspeckles and the middle region is located at the “core”, as previously reported (Souquere et al., 2010; West et al., 2016).
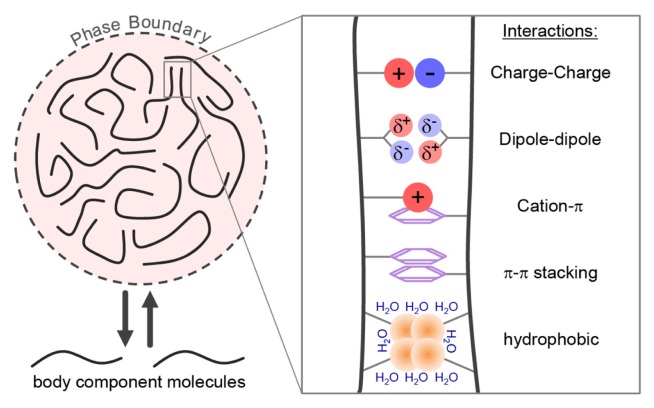
Fig. 2. Multivalency-driven phase separation of cellular bodies.
Most cellular bodies are considered to be phase-separated liquid droplets. Liquid-liquid phase separation is driven by multivalent architectural molecules. Such multivalent interactions can be provided by molecules such as arcRNAs and proteins containing intrinsically disordered, low-complexity domains. Multivalent interactions include charge, dipole-dipole, cation-π, π-π, and hydrophobic interactions. These interactions are very weak and short lived, enabling protein components of most bodies to be highly dynamic, rapidly exchanging with the surrounding nucleoplasm or cytoplasm. By contrast, arcRNA stays inside a nuclear body.
NUCLEAR BODIES BUILT ON ARCHITECTURAL RNAs
The formation of various cellular bodies depends on the specific structural proteins, e.g., Coilin in Cajal bodies or TIA1 in stress granules. A subset of nuclear bodies, however, use specific long noncoding RNA (lncRNA) or pre-mRNA as their scaffolding molecule. These scaffolding lncRNAs and pre-mRNAs are defined as “architectural lncRNAs (arcRNAs)” (Chujo et al., 2016). An RNA qualifies as an arcRNA if: 1) it is localized and enriched in a specific nuclear body, and 2) it constructs and stabilizes the body structure. The latter can be shown by RNA depletion to disrupt the body or artificial RNA tethering to construct the body. Presently, five lncRNAs and one pre-mRNA can firmly be classified as arcRNAs (Table 1). These six established arcRNAs and the nuclear bodies that they build are: 1) mammalian nuclear paraspeckle assembly transcript 1 isoform 2 (NEAT1_2) lncRNA in the paraspeckle (Sasaki et al., 2009; Shevtsov and Dundr, 2011), 2) intergenic spacer (IGS) lncRNA in the amyloid body (Audas et al., 2012), 3) human satellite III (SatIII) lncRNA in the nuclear stress body (Valgardsdottir et al., 2005), 4) histone pre-mRNA in the histone locus body (Shevtsov and Dundr, 2011), 5) Drosophila heat shock RNA (Hsr) omega in the omega speckle (Mallik and Lakhotia, 2009), and 6) fission yeast meiRNA in the Mei2 dot (Yamashita et al., 1998). Non-orthologous arcRNAs are widely used in eukaryotes, from mammals to insects and yeast, suggesting that this class of RNAs was independently selected for massive complex formation during evolution. The molecular, cellular, and physiological functions of these arcRNAs are summarized in Table 1 and detailed in another review (Chujo et al., 2016). In addition to the six established arcRNAs, unidentified arcRNAs are presumed to build several other nuclear bodies including the Sam68 nuclear body (SNB) and DBC1 body, because these bodies are RNase-sensitive, and body construction requires RNA-binding domains of the structural protein of the bodies (Mannen et al., 2016).
Table 1.
The six known nuclear bodies built on arcRNAs
| Nuclear body | arcRNA | Inducing stress or signal | Repetitive sequences in arcRNA | Body proteins | Molecular, cellular, and physiological functions |
|---|---|---|---|---|---|
| Paraspeckle | NEAT1 | Viral infection, hypoxia, MG132, corpus luteum formation, mammary gland development | Fragments of transposable elements | > 60 proteins including PSPC1a, SFPQa, NONO, RBM14a, HNRNPK, FUSa, DAZAP1a, HNRNPH3a, HNRNPA1a, HNRNPR, HNRNPUL1a, TDP-43a, BRG1b, BRMb, BAF155b (Yamazaki and Hirose, 2015) | Sequestration of RNA-binding proteins and transcription factors. Retention of mouse CTN RNA. Suppression of apoptosis under certain stresses, induction of antiviral genes upon viral infection, corpus luteum formation, establishment of pregnancy, mammary gland development, lactation |
| Amyloid body | IGS | Acidosis, heat shock, transcriptional stress | Ribosomal DNA repeats | VHL, DNMT1, POLD1, HSP70, MDM2, RPA40, RPA16, NOL1, NOM1, NOP52, PES1, RRP1B, SENP3 | Immobilization of body proteins, activation of HIF. Arrest of ribosome biogenesis |
| Nuclear stress body | Satellite III | Heat shock, cadmium | Satellite III repeats | SRSF1, SAFB, TDP-43a, HSF1, NFAT5a, BRG1b | Sequestration of RNA-binding proteins and transcription factors |
| Histone locus body | Histone mRNA precursor | Interphase, especially G1 and S phases | Unknown | FLASH, LSM10, LSM11, SLBP, SYMPLEKIN | Processing of histone pre-mRNAs |
| Omega speckle | Hsr omega | Heat shock, amides, ecdysone hormone | Tandem repeats of 280 nt for 10 kb | Nonaa, Sex-lethala, sans fille, PEPa, Hrb87Fa, Hrp40a, Hrb57Aa, ISWIb | Normal development (Nullisomy animals are embryonic lethal or extremely weak) |
| Mei2 dot | meiRNA | Entry into meiosis | U(U/C)AAAC | Mei2, Mmi1 | Sequestration of RNA-binding proteins Progression of meiosis I via derepressing transcription of meiosis-specific genes, and suppressing degradation of meiosis-specific mRNAs |
Proteins containing prion-like domains.
Chromatin remodeling complex protein.
COMMONALITIES OF ArcRNAs
ArcRNAs share four common characteristics that are closely connected with the benefits of using RNA, but not protein, as the architectural molecules of nuclear bodies. These benefits will be discussed below.
First, the expression of arcRNAs is temporarily upregulated by specific stimuli (Table 1). For example, NEAT1_2 expression is enhanced upon cellular stresses such as proteasome inhibition, viral infection, or hypoxia, and NEAT1_2 promotes cell survival under such adverse conditions (Choudhry et al., 2014; Hirose et al., 2014; Imamura et al., 2014). NEAT1_2 is also transcriptionally upregulated during and required for mammalian corpus luteum formation and mammary gland development (Nakagawa et al., 2014; Standaert et al., 2014). Moreover, NEAT1_2 is upregulated in many cancer tissues and variously regulates cancer progression (Adriaens et al., 2016; Chakravarty et al., 2014; Choudhry et al., 2014; Mello et al., 2017).
Second, arcRNAs sequestrate various regulatory proteins, such as RNA-binding proteins, DNA-binding proteins, and E3 ubiquitin ligases, thereby affecting gene expression patterns. For example, upon NEAT1_2 upregulation and paraspeckle number increase, nucleoplasmic SFPQ protein is sequestrated into paraspeckles, reducing SFPQ-mediated transcription of target genes such as ADARB2 (Hirose et al., 2014). Hypoxia and acidosis upregulate IGS28 lncRNA, and IGS28 sequestrates Von-Hippel-Lindau (VHL), an E3 ligase that targets hypoxia-inducible factor (HIF), allowing HIF to evade degradation and activate its target genes (Audas et al., 2012; Mekhail et al., 2004).
Third, all six arcRNA-constructed nuclear bodies are assembled at the site of arcRNA transcription (Fig. 3) (Audas et al., 2012; Clemson et al., 2009; Dangli et al., 1983; Liu et al., 2006; Rizzi et al., 2004; Shimada et al., 2003).
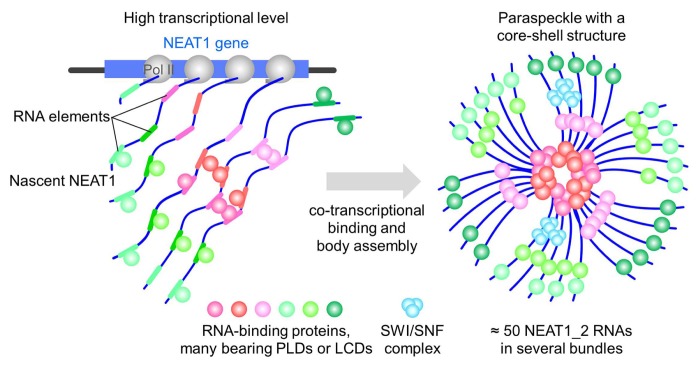
Fig. 3. Molecular mechanism of paraspeckle formation.
NEAT1_2 transcription is coupled with the binding of specific proteins to putative RNA elements embedded in NEAT1_2 sequences. Paraspeckle assembly initiates at NEAT1_2 transcription sites, via PSPs that stabilize NEAT1_2 and/or promote interactions with other PSPs, such as several PLD-containing PSPs and SWI/SNF complexes. A high transcriptional level of NEAT1_2 promotes assembly of paraspeckle by concentrating NEAT1_2 at its transcription site. Each paraspeckle contains approximately 50 NEAT1_2 molecules that are organized in several bundles, forming a core-shell spheroidal structure.
Fourth, most arcRNAs are transcribed from or include repetitive sequences. IGS and SatIII lncRNAs are transcribed from the ribosomal DNA repeat intergenic regions and peri-centromeric SatIII repeats, respectively (Audas et al., 2012; Rizzi et al., 2004). Drosophila Hsr omega contains tandem repeats of 280 nt for a stretch of approximately 10 kb (Garbe et al., 1986). The abnormal expansion of repetitive sequence in some RNA species can give an architectural role to the RNA, which often results in neurodegenerative diseases such as myotonic dystrophy type 1 (DM1) (Wojciechowska and Krzyzosiak, 2011). In DM1 patients with abnormally long CTG repeats in the DMPK gene, CUG RNA foci are aberrantly formed and sequestrate muscle-blind family splicing regulatory proteins, thereby affecting splicing patterns of hundreds of mRNAs (Charizanis et al., 2012; Miller et al., 2000). Interestingly, repeat expansion of CTG or CAG DNA creates repetitive RNAs that form multivalent base-pairing, causing purified RNA to undergo a sol-gel transition in vitro (Jain and Vale, 2017).
COMMONALITIES OF PROTEIN COMPONENTS OF ArcRNA-CONSTRUCTED NUCLEAR BODIES
ArcRNA-constructed nuclear bodies contain various protein components, and there are several commonalities between the body proteins (Fig. 3). First, many are RNA-binding proteins, as expected given the requirement of RNA to build the bodies. Second, arcRNA-constructed nuclear bodies are often enriched with RNA-binding proteins that also contain a form of LCD called a prion-like domain (PLD) (Table 1) (Chujo et al., 2016; King et al., 2012; Naganuma et al., 2012). The PLD is rich in amino acids such as tyrosine, asparagine, glutamine, and glycine, and can form protein–protein interactions. Some of the PLD-containing RNA-binding proteins may bridge RNPs to form large nuclear bodies. Indeed, each paraspeckle contains about 50 NEAT1_2 molecules, and the PLDs of paraspeckle proteins such as FUS and RBM14 are required to construct paraspeckles (Chujo et al., 2017; Hennig et al., 2015). Moreover, recombinant RBM14 or FUS can form hydrogel in a PLD-dependent manner (Hennig et al., 2015). These results suggest that PLDs of RBM14 and FUS may induce phase separation to form paraspeckles in the nucleoplasm. Also, the PLD of HNRNPD is required to construct the SNB, and PLD-containing proteins (e.g., Nona and Hrb87F) are enriched in the Drosophila omega speckle (Chujo et al., 2016; Mannen et al., 2016). The third group of proteins that characterizes many arcRNA-constructed nuclear bodies is chromatin remodeling complex proteins (Table 1). For instance, mammalian paraspeckle and human nuclear stress body contain and require SWI/SNF chromatin remodeling complex for their construction (Kawaguchi et al., 2015). Moreover, the Drosophila ISWI chromatin remodeling complex is required for the formation of the omega speckle (Onorati et al., 2011). The SWI/SNF complex facilitates protein–protein interactions of paraspeckle proteins. Surprisingly, paraspeckle formation does not require ATPase activity of the SWI/SNF catalytic protein, demonstrating that the ability of the SWI/SNF complex to promote paraspeckle formation is independent of its well-known chromatin remodeling function (Kawaguchi et al., 2015).
WHY RNA IS USED AS THE ARCHITECTURAL BIOMOLECULE IN VARIOUS NUCLEAR BODIES
ArcRNAs are widely used as the architectural molecule of nuclear bodies in a variety of eukaryotes, suggesting that RNA was evolutionarily adopted as the scaffold molecule in some cases. In this section, we discuss three possible reasons why RNA is used as the architectural molecule of nuclear bodies: position-specificity, rapidity, and economy in sequestering nucleic acid-binding proteins.
First, the use of RNA enables position-specific nuclear body formation, which enables gene regulation at a specific nuclear region. To induce LLPS to form membraneless subcellular bodies, it is necessary to raise the concentration of the key scaffold molecules above a threshold. In the nucleus, certain homogeneous RNAs can be highly concentrated at the site of transcription, if the transcription level is high enough (Fig. 1 and 3). In fact, all six known nuclear bodies with arcRNA scaffolds are formed at the site of arcRNA transcription (Audas et al., 2012; Clemson et al., 2009; Dangli et al., 1983; Liu et al., 2006; Rizzi et al., 2004; Shimada et al., 2003). Moreover, by simply repeating a specific scaffold sequence in one RNA molecule, the scaffold sequence can easily be enriched in one place, as in the case of SatIII (containing repeats of 158 nt), Hsr omega (containing repeats of 280 nt), and DM1-related RNA with triplet repeat expansion (Garbe et al., 1986; Moyzis et al., 1987; Taneja et al., 1995). In addition, RNA can increase the local concentration of specific RNA-binding proteins that the RNA interacts with. As many RNA-binding proteins bear PLDs (Table 1), arcRNA can increase the local concentration of PLD-containing RNA-binding proteins to promote PLD-mediated LLPS to construct a massive complex. For example, PLD-containing RNA-binding protein FUS forms high-order assemblies at a low concentration in the presence but not in the absence of RNAs (Schwartz et al., 2013). Through nuclear body formation at the transcription sites of arcRNAs, arcRNAs can regulate the local nucleoplasmic concentration of freely available regulatory proteins by sequestrating them into the nuclear body, which may eventually regulate local gene expression.
Second, RNA enables rapid and reversible sequestration of nucleic acid-binding proteins. Interestingly, all the known nuclear bodies with arcRNA scaffolds form or increase in size or number upon specific stimuli (Table 1). When such stimuli are no longer present, arcRNAs can be quickly degraded and proteins trapped in the bodies disperse into the nucleoplasm, rapidly allowing gene expression to return to normal. For example, Drosophila Hsr omega level increases several fold after 1 hour of heat shock, inducing various RNA-binding proteins to be sequestrated into omega speckles. When the heat shock is no longer present, the level of Hsr omega returns to normal within about 2 hours (Hogan et al., 1994; Prasanth et al., 2000). Sequestrating bodies can rapidly appear because this only requires the transcription of arcRNA, and they can rapidly disappear because most RNAs are unstable with a relatively high turnover. Stress-induced temporary sequestration of nucleic acid-binding proteins, rather than the destruction and regeneration of these proteins, is beneficial for cells in terms of energy and the availability of proteasomes and ribosomes, as well as enabling a rapid response.
Third, it is more economical to use RNAs than proteins to sequestrate a massive number of various nucleic acid-binding proteins. The formation of a nuclear body requires the accumulation of specific proteins at specific sites in the nucleus. Therefore, it is beneficial for the architectural molecules to be able to trap a massive number of proteins. One protein can usually be captured by one or several protein domains containing tens to hundreds of amino acids, whereas one protein can be captured by only 4–17 nt of RNA (Lunde et al., 2007; Prikryl et al., 2011). Thus, one protein comprising 100 amino acids can capture only one or two proteins, whereas one RNA molecule comprising 100 nt may capture 5–20 proteins. One arcRNA molecule, for instance a 23-kb NEAT1_2 RNA molecule, may capture thousands of proteins. To date, more than 60 protein species have been identified as components of paraspeckles (Yamazaki and Hirose, 2015). The majority of these are RNA-binding proteins and transcription factors. Such collective binding and sequestration of various nucleic acid-binding proteins helps to regulate the overall gene expression. Also, whereas longer polypeptides of random sequences can easily aggregate and become toxic to cells, longer RNAs of random sequences remain soluble due to the charged phosphate backbone. This may be one reason most proteins are shorter than 2,000 amino acids, whereas many lncRNAs and mRNA precursors comprise 10,000 nt. A longer architectural molecule enables multivalent interaction with a massive number of nucleic acid-binding proteins, which facilitates the regulation of gene expression. Whereas body proteins rapidly exchange in and out of the body, arcRNA can stay inside the body (Mao et al., 2011), which is presumably due to the high molecular weight of arcRNA and the great number of interactions it makes. Such soluble, highly multivalent, and static features may make arcRNAs competent architectural molecules. RNA hardly forms insoluble aggregates and the reading frames of lncRNAs do not need to remain translatable; therefore, the sequence of an architectural lncRNA is likely to be only constrained by the requirement to form the correct RNA–protein interaction, and an arcRNA has flexibility to add and change sequences. Such advantages would allow lncRNAs to increase the diversity of their binding partners and the combinations of proteins that are incorporated into nuclear bodies, enabling the formation of a wide variety of nuclear bodies for faster adaptation to environmental changes.
METHODS TO DISCOVER NOVEL ArcRNA-BUILT NUCLEAR BODIES
Formation of known arcRNA-dependent nuclear bodies is initiated by expression of arcRNAs induced during specific stresses and developmental stages. So, considering that there are over 20,000 lncRNAs with unknown functions (Hon et al., 2017), it is likely that arcRNA-dependent nuclear bodies remain to be identified in various cellular contexts. Here, we introduce two recently reported methods to discover novel nuclear bodies with arcRNA scaffolds: 1) RNase-sensitivity screening of nuclear bodies (Mannen et al., 2016), and 2) transcriptome-wide screening of semi-extractable RNA (Chujo et al., 2017).
The structures of known arcRNA-constructed nuclear bodies can be disrupted by RNase treatment of the permeabilized cells. Thus, in combination with a venus-tagged cDNA expression library, one can seek proteins that form foci that are disrupted upon RNase treatment. In the reported RNase-sensitivity screen, first, a venus-tagged cDNA expression library of 10,432 proteins was used to select 463 proteins that formed foci within the nucleus (Hirose and Goshima, 2015). Then, the cells were permeabilized with weak detergent, followed by treatment with RNases, identifying 32 proteins in HeLa cells that form RNase-sensitive foci in the nucleus. Many of the identified proteins were known components of various nuclear bodies. However, six proteins were new components of the SNB, one of which, HNRNPD, was required for SNB formation. This study also showed that both the RNA-binding domain and the PLD of HNRNPD are required for the SNB formation; the PLD of HNRNPD enabled HNRNPD–HNRNPD and HNRNPD–Sam68 interactions. These results collectively demonstrate that SNB formation requires (a) putative arcRNA(s), RNA–protein interactions, and protein–protein interactions. The molecular entity of SNB arcRNA remains to be identified.
Transcriptome-wide screening of semi-extractable RNAs looks for RNAs that are unusually difficult to extract from cells. This method originated from a serendipitous finding that cellular NEAT1_2 is extremely difficult to extract using conventional RNA extraction methods. A complete extraction of cellular NEAT1_2 requires either needle shearing or heating of the cell lysates in an RNA extraction reagent such as TRIzol, a property that was named semi-extractability. One feature of phase-separated liquid droplets is that such droplets can be broken down by shearing forces. So, requirement of needle shearing to extract NEAT1_2 suggests that after lysing cells with TRIzol, paraspeckles might still somehow remain as phase-separated droplets, unless sheared with a needle or heated. Semi-extractability of NEAT1_2 was largely dependent on the FUS protein, especially its PLD, which also indicates that LLPS of paraspeckles is the cause of semi-extractability. As LLPS and PLD are often linked to nuclear body formation, it was hypothesized that unidentified semi-extractable RNAs might also form nuclear bodies. Using transcriptome-wide identification of semi-extractable RNAs in HeLa cells, 45 semi-extractable RNAs were identified, of which the top 10 abundant RNAs all showed predominant localization in nuclear body-like foci. Of these, three RNAs (CCAT1 lncRNA, LINC00473 lncRNA, and ADARB2 pre-mRNA) were confirmed to form foci not only at the transcription site, but also outside of transcription sites, strongly suggesting formation of novel nuclear bodies. To firmly establish these RNAs as arcRNAs, body marker proteins need to be identified, and disintegration of body marker foci upon RNA depletion needs to be confirmed. As this semi-extractable RNA screening can be performed relatively easily, this method is useful to find novel arcRNAs in various cellular contexts. Taken together, the two approaches above suggest the presence of unidentified arcRNAs in the human transcriptome. Identification and characterization of the additional arcRNAs will unveil the commonality in this class of RNA in terms of the sequence elements and modes of action.
IMPORTANT QUESTIONS TO BE ADDRESSED
Recent studies have greatly advanced our mechanistic understanding of the formation and function of arcRNA-constructed nuclear bodies. However, many important questions remain to be answered, including the following:
What are the fundamental rules of RNA sequences and/or RNA structures that enable LLPS of the nuclear bodies? Are they clusters of binding sites for PLD-containing RNA-binding proteins?
There are more than 20,000 human lncRNAs with unknown functions. Are there novel, uncharacterized arcRNAs that function during specific cellular stresses and developmental stages?
There is a large gap between the molecular functions of arcRNA-constructed nuclear bodies (e.g., protein sequestration) and the physiological functions of the bodies (e.g., organ development). How does the molecular role of the nuclear bodies link to the physiological functions?
REFERENCES
- Adriaens C., Standaert L., Barra J., Latil M., Verfaillie A., Kalev P., Boeckx B., Wijnhoven P.W., Radaelli E., Vermi W., et al. p53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat Med. 2016;22:861–868. doi: 10.1038/nm.4135. [DOI] [PubMed] [Google Scholar]
- Audas T.E., Jacob M.D., Lee S. Immobilization of proteins in the nucleolus by ribosomal intergenic spacer noncoding RNA. Mol Cell. 2012;45:147–157. doi: 10.1016/j.molcel.2011.12.012. [DOI] [PubMed] [Google Scholar]
- Banani S.F., Lee H.O., Hyman A.A., Rosen M.K. Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol. 2017;18:285–298. doi: 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry J., Weber S.C., Vaidya N., Haataja M., Brangwynne C.P. RNA transcription modulates phase transition-driven nuclear body assembly. Proc Natl Acad Sci USA. 2015;112:E5237–5245. doi: 10.1073/pnas.1509317112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne C.P., Eckmann C.R., Courson D.S., Rybarska A., Hoege C., Gharakhani J., Julicher F., Hyman A.A. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324:1729–1732. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- Brangwynne C.P., Mitchison T.J., Hyman A.A. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc Natl Acad Sci USA. 2011;108:4334–4339. doi: 10.1073/pnas.1017150108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne C.P., Tompa P., Pappu R.V. Polymer physics of intracellular phase transitions. Nat Phys. 2015;11:899–904. [Google Scholar]
- Chakravarty D., Sboner A., Nair S.S., Giannopoulou E., Li R., Hennig S., Mosquera J.M., Pauwels J., Park K., Kossai M., et al. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat Commun. 2014;5:5383. doi: 10.1038/ncomms6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charizanis K., Lee K.Y., Batra R., Goodwin M., Zhang C., Yuan Y., Shiue L., Cline M., Scotti M.M., Xia G., et al. Muscleblind-like 2-mediated alternative splicing in the developing brain and dysregulation in myotonic dystrophy. Neuron. 2012;75:437–450. doi: 10.1016/j.neuron.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhry H., Albukhari A., Morotti M., Hider S., Moralli D., Smythies J., Schodel J., Green C.M., Camps C., Buffa F., et al. Tumor hypoxia induces nuclear paraspeckle formation through HIF-2alpha dependent transcriptional activation of NEAT1 leading to cancer cell survival. Oncogene. 2014;34:4546. doi: 10.1038/onc.2014.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chujo T., Yamazaki T., Hirose T. Architectural RNAs (arcRNAs): A class of long noncoding RNAs that function as the scaffold of nuclear bodies. Biochim et Biophys Acta. 2016;1859:139–146. doi: 10.1016/j.bbagrm.2015.05.007. [DOI] [PubMed] [Google Scholar]
- Chujo T., Yamazaki T., Kawaguchi T., Kurosaka S., Takumi T., Nakagawa S., Hirose T. Unusual semi-extractability as a hallmark of nuclear body-associated architectural noncoding RNAs. EMBO J. 2017;36:1447–1462. doi: 10.15252/embj.201695848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson C.M., Hutchinson J.N., Sara S.A., Ensminger A.W., Fox A.H., Chess A., Lawrence J.B. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangli A., Grond C., Kloetzel P., Bautz E.K. Heatshock puff 93 D from Drosophila melanogaster: accumulation of a RNP-specific antigen associated with giant particles of possible storage function. EMBO J. 1983;2:1747–1751. doi: 10.1002/j.1460-2075.1983.tb01652.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M., Hebert M.D., Karpova T.S., Stanek D., Xu H., Shpargel K.B., Meier U.T., Neugebauer K.M., Matera A.G., Misteli T. In vivo kinetics of Cajal body components. J Cell Biol. 2004;164:831–842. doi: 10.1083/jcb.200311121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbe J.C., Bendena W.G., Alfano M., Pardue M.L. A Drosophila heat shock locus with a rapidly diverging sequence but a conserved structure. J Biol Chem. 1986;261:16889–16894. [PubMed] [Google Scholar]
- Hennig S., Kong G., Mannen T., Sadowska A., Kobelke S., Blythe A., Knott G.J., Iyer K.S., Ho D., Newcombe E.A., et al. Prion-like domains in RNA binding proteins are essential for building subnuclear paraspeckles. J Cell Biol. 2015;210:529–539. doi: 10.1083/jcb.201504117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose T., Goshima N. Genome-wide co-localization screening of nuclear body components using a fluorescently tagged FLJ cDNA clone library. Methods Mol Biol. 2015;1262:155–163. doi: 10.1007/978-1-4939-2253-6_9. [DOI] [PubMed] [Google Scholar]
- Hirose T., Virnicchi G., Tanigawa A., Naganuma T., Li R., Kimura H., Yokoi T., Nakagawa S., Benard M., Fox A.H., et al. NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol Biol Cell. 2014;25:169–183. doi: 10.1091/mbc.E13-09-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan N.C., Traverse K.L., Sullivan D.E., Pardue M.L. The nucleus-limited Hsr-omega-n transcript is a polyadenylated RNA with a regulated intranuclear turnover. J Cell Biol. 1994;125:21–30. doi: 10.1083/jcb.125.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon C.C., Ramilowski J.A., Harshbarger J., Bertin N., Rackham O.J., Gough J., Denisenko E., Schmeier S., Poulsen T.M., Severin J., et al. An atlas of human long non-coding RNAs with accurate 5′ ends. Nature. 2017;543:199–204. doi: 10.1038/nature21374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura K., Imamachi N., Akizuki G., Kumakura M., Kawaguchi A., Nagata K., Kato A., Kawaguchi Y., Sato H., Yoneda M., et al. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol Cell. 2014;53:393–406. doi: 10.1016/j.molcel.2014.01.009. [DOI] [PubMed] [Google Scholar]
- Jain A., Vale R.D. RNA phase transitions in repeat expansion disorders. Nature. 2017;546:243–247. doi: 10.1038/nature22386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser T.E., Intine R.V., Dundr M. De novo formation of a subnuclear body. Science. 2008;322:1713–1717. doi: 10.1126/science.1165216. [DOI] [PubMed] [Google Scholar]
- Kato M., Han T.W., Xie S., Shi K., Du X., Wu L.C., Mirzaei H., Goldsmith E.J., Longgood J., Pei J., et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi T., Tanigawa A., Naganuma T., Ohkawa Y., Souquere S., Pierron G., Hirose T. SWI/SNF chromatin-remodeling complexes function in noncoding RNA-dependent assembly of nuclear bodies. Proc Natl Acad Sci USA. 2015;112:4304–4309. doi: 10.1073/pnas.1423819112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King O.D., Gitler A.D., Shorter J. The tip of the iceberg: RNA-binding proteins with prion-like domains in neurodegenerative disease. Brain Res. 2012;1462:61–80. doi: 10.1016/j.brainres.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.L., Murphy C., Buszczak M., Clatterbuck S., Goodman R., Gall J.G. The Drosophila melanogaster Cajal body. J Cell Biol. 2006;172:875–884. doi: 10.1083/jcb.200511038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunde B.M., Moore C., Varani G. RNA-binding proteins: modular design for efficient function. Nat Rev Mol Cell Biol. 2007;8:479–490. doi: 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallik M., Lakhotia S.C. RNAi for the large non-coding hsromega transcripts suppresses polyglutamine pathogenesis in Drosophila models. RNA Biol. 2009;6:464–478. doi: 10.4161/rna.6.4.9268. [DOI] [PubMed] [Google Scholar]
- Mannen T., Yamashita S., Tomita K., Goshima N., Hirose T. The Sam68 nuclear body is composed of two RNase-sensitive substructures joined by the adaptor HNRNPL. J Cell Biol. 2016;214:45–59. doi: 10.1083/jcb.201601024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y.S., Sunwoo H., Zhang B., Spector D.L. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat Cell Biol. 2011;13:95–101. doi: 10.1038/ncb2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekhail K., Gunaratnam L., Bonicalzi M.E., Lee S. HIF activation by pH-dependent nucleolar sequestration of VHL. Nat Cell Biol. 2004;6:642–647. doi: 10.1038/ncb1144. [DOI] [PubMed] [Google Scholar]
- Mello S.S., Sinow C., Raj N., Mazur P.K., Bieging-Rolett K., Broz D.K., Imam J.F.C., Vogel H., Wood L.D., Sage J., et al. Neat1 is a p53-inducible lincRNA essential for transformation suppression. Genes Dev. 2017;31:1095–1108. doi: 10.1101/gad.284661.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.W., Urbinati C.R., Teng-Umnuay P., Stenberg M.G., Byrne B.J., Thornton C.A., Swanson M.S. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000;19:4439–4448. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molliex A., Temirov J., Lee J., Coughlin M., Kanagaraj A.P., Kim H.J., Mittag T., Taylor J.P. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015;163:123–133. doi: 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyzis R.K., Albright K.L., Bartholdi M.F., Cram L.S., Deaven L.L., Hildebrand C.E., Joste N.E., Longmire J.L., Meyne J., Schwarzacher-Robinson T. Human chromosome-specific repetitive DNA sequences, novel markers for genetic analysis. Chromosoma. 1987;95:375–386. doi: 10.1007/BF00333988. [DOI] [PubMed] [Google Scholar]
- Naganuma T., Nakagawa S., Tanigawa A., Sasaki Y.F., Goshima N., Hirose T. Alternative 3′-end processing of long noncoding RNA initiates construction of nuclear paraspeckles. EMBO J. 2012;31:4020–4034. doi: 10.1038/emboj.2012.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S., Shimada M., Yanaka K., Mito M., Arai T., Takahashi E., Fujita Y., Fujimori T., Standaert L., Marine J.C., et al. The lncRNA Neat1 is required for corpus luteum formation and the establishment of pregnancy in a subpopulation of mice. Development. 2014;141:4618–4627. doi: 10.1242/dev.110544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott T.J., Petsalaki E., Farber P., Jervis D., Fussner E., Plochowietz A., Craggs T.D., Bazett-Jones D.P., Pawson T., Forman-Kay J.D., et al. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol Cell. 2015;57:936–947. doi: 10.1016/j.molcel.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onorati M.C., Lazzaro S., Mallik M., Ingrassia A.M., Carreca A.P., Singh A.K., Chaturvedi D.P., Lakhotia S.C., Corona D.F. The ISWI chromatin remodeler organizes the hsromega ncRNA-containing omega speckle nuclear compartments. PLoS Genet. 2011;7:e1002096. doi: 10.1371/journal.pgen.1002096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petri M., Frey S., Menzel A., Gorlich D., Techert S. Structural characterization of nanoscale meshworks within a nucleoporin FG hydrogel. Biomacromolecules. 2012;13:1882–1889. doi: 10.1021/bm300412q. [DOI] [PubMed] [Google Scholar]
- Prasanth K.V., Rajendra T.K., Lal A.K., Lakhotia S.C. Omega speckles - a novel class of nuclear speckles containing hnRNPs associated with noncoding hsr-omega RNA in Drosophila. J Cell Sci. 2000;113(Pt 19):3485–3497. doi: 10.1242/jcs.113.19.3485. [DOI] [PubMed] [Google Scholar]
- Prikryl J., Rojas M., Schuster G., Barkan A. Mechanism of RNA stabilization and translational activation by a pentatricopeptide repeat protein. Proc Natl Acad Sci USA. 2011;108:415–420. doi: 10.1073/pnas.1012076108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichheld S.E., Muiznieks L.D., Keeley F.W., Sharpe S. Direct observation of structure and dynamics during phase separation of an elastomeric protein. Proc Natl Acad Sci USA. 2017;14:E4408–E4415. doi: 10.1073/pnas.1701877114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzi N., Denegri M., Chiodi I., Corioni M., Valgardsdottir R., Cobianchi F., Riva S., Biamonti G. Transcriptional activation of a constitutive heterochromatic domain of the human genome in response to heat shock. Mol Biol Cell. 2004;15:543–551. doi: 10.1091/mbc.E03-07-0487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y.T., Ideue T., Sano M., Mituyama T., Hirose T. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci USA. 2009;106:2525–2530. doi: 10.1073/pnas.0807899106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz J.C., Wang X., Podell E.R., Cech T.R. RNA seeds higher-order assembly of FUS protein. Cell Rep. 2013;5:918–925. doi: 10.1016/j.celrep.2013.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevtsov S.P., Dundr M. Nucleation of nuclear bodies by RNA. Nat Cell Biol. 2011;13:167–173. doi: 10.1038/ncb2157. [DOI] [PubMed] [Google Scholar]
- Shimada T., Yamashita A., Yamamoto M. The fission yeast meiotic regulator Mei2p forms a dot structure in the horse-tail nucleus in association with the sme2 locus on chromosome II. Mol Biol Cell. 2003;14:2461–2469. doi: 10.1091/mbc.E02-11-0738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souquere S., Beauclair G., Harper F., Fox A., Pierron G. Highly ordered spatial organization of the structural long noncoding NEAT1 RNAs within paraspeckle nuclear bodies. Mol Biol Cell. 2010;21:4020–4027. doi: 10.1091/mbc.E10-08-0690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standaert L., Adriaens C., Radaelli E., Van Keymeulen A., Blanpain C., Hirose T., Nakagawa S., Marine J.C. The long noncoding RNA Neat1 is required for mammary gland development and lactation. RNA. 2014;20:1844–1849. doi: 10.1261/rna.047332.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneja K.L., McCurrach M., Schalling M., Housman D., Singer R.H. Foci of trinucleotide repeat transcripts in nuclei of myotonic dystrophy cells and tissues. J Cell Biol. 1995;128:995–1002. doi: 10.1083/jcb.128.6.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valgardsdottir R., Chiodi I., Giordano M., Cobianchi F., Riva S., Biamonti G. Structural and functional characterization of noncoding repetitive RNAs transcribed in stressed human cells. Mol Biol Cell. 2005;16:2597–2604. doi: 10.1091/mbc.E04-12-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West J.A., Mito M., Kurosaka S., Takumi T., Tanegashima C., Chujo T., Yanaka K., Kingston R.E., Hirose T., Bond C., et al. Structural, super-resolution microscopy analysis of paraspeckle nuclear body organization.J. Cell Biol. 2016;214:817–830. doi: 10.1083/jcb.201601071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojciechowska M., Krzyzosiak W.J. Cellular toxicity of expanded RNA repeats, focus on RNA foci. Hum Mol Genet. 2011;20:3811–3821. doi: 10.1093/hmg/ddr299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A., Watanabe Y., Nukina N., Yamamoto M. RNA-assisted nuclear transport of the meiotic regulator Mei2p in fission yeast. Cell. 1998;95:115–123. doi: 10.1016/s0092-8674(00)81787-0. [DOI] [PubMed] [Google Scholar]
- Yamazaki T., Hirose T. The building process of the functional paraspeckle with long non-coding RNAs. Front Biosci. 2015;7:1–41. doi: 10.2741/715. [DOI] [PubMed] [Google Scholar]