Abstract
We report the biological functions of a zebrafish homologue of RING-finger protein 152 (rnf152) during embryogenesis. rnf152 was initially identified as a brain-enriched E3 ligase involved in early embryogenesis of zebrafish. Expression of rnf152 was ubiquitous in the brain at 24 hpf but restricted to the eyes, midbrain-hindbrain boundary (MHB), and rhombomeres at 48 hpf. Knockdown of rnf152 in zebrafish embryos caused defects in the eyes, MHB, and rhombomeres (r1–7) at 24 hpf. These defects in rnf152-deficient embryos were analyzed by whole-mount in situ hybridization (WISH) using neuroD, deltaD, notch1a, and notch3 probes. NeuroD expression was abolished in the marginal zone, outer nuclear layer (ONL), inner nuclear layer (INL), and ganglion cell layer (GCL) of the eyes at 27 hpf. Furthermore, deltaD and notch1a expression was remarkably reduced in the ONL, INL, subpallium, tectum, cerebellum, and rhombomeres (r1–7) at 24 hpf, whereas notch3 expression was reduced in the tectum, cerebellum, and rhombomeres at 24 hpf. Finally, we confirmed that expression of Notch target genes, her4 and ascl1a, also decreased significantly in these areas at 24 hpf. Thus, we propose that Rnf152 is essential for development of the eyes, midbrain and hindbrain, and that Delta-Notch signaling is involved.
Keywords: delta-notch signaling, neurogenesis, neuroD, rnf152, zebrafish embryo
INTRODUCTION
Ubiquitination is an important cellular process that affects protein homeostasis; it also controls complex processes during embryogenesis (Ro et al., 2015). Abnormal ubiquitination can result in the development of major human neurodegenerative diseases such as Parkinson’s and Alzheimer’s (Anuppalle et al., 2013). Ubiquitination involves three enzymes, ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), and ubiquitin ligase (E3), which modify proteins and affect their localization and stability (Morreale and Walden, 2016). Among the three enzymes, E3 ligases are critical for determining substrate specificity. E3 ligases are classified into two major subtypes based on their structural characteristics: RING domain-containing and HECT domain-containing E3 ligases (Metzger et al., 2014). Furthermore, discrete ubiquitin chains on a substrate dictate whether the protein will alter its cellular localization or undergo proteasomal degradation (Swatek and Komander, 2016).
As a member of the RING domain-containing E3 ligase family, RNF152 is an important regulator of proteins functioning downstream of mechanistic target of rapamycin complex 1(mTORC1) signaling (Deng et al., 2015). In particular, mechanistic target of rapamycin (mTOR) is a serine/threonine kinase that regulates cellular growth and proliferation during the early stages of neurogenesis (Garza-Lombó et al., 2016). RNF152 targets RagA GTPase for K-63-mediated ubiquitination, which activates its inhibitor GATOR1, a GAP complex for Rag GTPase, thereby inactivating mTORC1 signaling (Deng et al., 2015). Although mTORC1 activity is critical in neonatal neuronal stem cells in the subventricular zone during neurogenesis (Hartman et al., 2013), the biological functions of RNF152 in embryogenesis have not yet been addressed.
Thus, we conducted molecular genetic studies on Rnf152 using zebrafish embryos to define the expression patterns, biological functions, and transcriptional regulation of rnf152. We discovered that rnf152 was maternally and zygotically expressed at early and late embryonic stages. Expression of rnf152 was found in the midbrain and hindbrain boundary (MHB), rhombomeres (r1–7), and eyes at 48 hpf (hours post fertilization). In addition, overexpression and knockdown studies demonstrated that Rnf152 was involved in development of the eyes, MHB, and hindbrain. Furthermore, molecular marker studies using neuroD, deltaD, notch1a, and notch3 showed that Rnf152 might be an upstream regulator of Delta-Notch signaling during embryogenesis in zebrafish. Based on these observations, we describe the functions of rnf152 in embryogenesis.
MATERIALS AND METHODS
Rnf152 sequence analysis
Phylogenetic analysis was performed to identify evolutionary relationship between zebrafish Rnf152 (NP_001014380) with human RNF152 (NP_775828), chimpanzee RNF152 (XP_001143948), mouse RNF152 (NP_848894), chicken RNF152 (NP_001291963), and Xenopus Rnf152 (XP_004915369). Neighbor-joining phylogenetic tree was generated with the online available software, MEGA7.0 (https://www.megasoftware.net). Information about amino acids sequence for human, chimpanzee, mouse, chicken, Xenopus, and zebrafish was collected from available proteins in NCBI, https://www.ncbi.nlm.nih.gov/protein/.
Zebrafish maintenance and embryo generation
Wild type synchronized embryos were produced and maintained at 28.8°C (Westerfield, 2000). Embryos were collected with natural breeding in the cycle of 10-hour dark/14-hour light and morphological confirmation was done as described in Kimmel et al. (1995). Embryos were treated with 0.2 mM Phenylthiourea (PTU) after 9 hpf to stop melanogenesis.
RNA preparation, cDNA synthesis, and RT-PCR
Total RNA was isolated from various stages of zebrafish embryos using R&A-BLUE® Total RNA Extraction kit (iNtRON Biotechnology) and cDNA was synthesized with M-MLV Reverse Transcriptase (Enzynomics) with Oligo (dT)20 primer. The primers were used for PCR to analyze the zebrafish rnf152 specific template (235 bp) in different stages of embryos, forward primer; TCTCCCATCTCCCAGATG and reverse primer; AGACCGTCATGTCCTAGA, for β-actin (500 bp), as an internal control for this experiment, Forward Primer; GAGGAGCACCCCGTCCTGC and Reverse Primer; GATGGCT GGAACAGGGCC. After the completion of PCR reaction, every set of reaction was confirmed with running 1% agarose gel in TE buffer using gel electrophoresis and sent for sequencing to SolGent Co. Ltd after cloning in pGEMT-easy vector. The protocols were established and applied as previously in our laboratory (Anuppalle et al., 2017).
Whole-mount in situ hybridization (WISH)
After sequence confirmation, cloned rnf152 construct was linearized with restriction enzyme and anti-sense DIG-labeled probe was synthesized with T7 RNA polymerase and sense probe was also synthesized as well to provide WISH in a negative manner, protocol was adapted from the Roche instructions. Embryos were fixed in 4% paraformaldehyde (PFA) overnight. Embryos over 24 hpf were treated with proteinase K. WISH was performed as described in (Thisse and Thisse, 2004) with minor laboratory modification. Two color WISH was performed as previously done in the laboratory (Anuppalle et al., 2017). Images were captured when embryos were in 90% glycerol in PBST solution with Leica MZ16.
Overexpression of rnf152 mRNA
We selected a positive strand of rnf152 ORF (675bp) and primers were designed as; Forward Primer; CGGAATTCATGT GCAACAGCCACGATTT, Reverse Primer; GCCTCGAGTCCAC AGGAAATAATAGTGA. After amplification, cloning was performed in pcGlobin2 vector (Ro et al., 2004). After confirming the sequence, the construct was linearized and applied for the synthesis of rnf152 capped mRNA with mMessage mMachine® High Yield Capped RNA Transcription Kit (Ambion® Applied Biosystems) and purified capped mRNA was injected (200 pg) in 1- or 2- cell stage of zebrafish embryos, phenol red dye with distilled water was injected as vehicle control in similar volume. Phenotypes were observed in every 6 hours and images were captured at 24 hpf.
Knock-down analysis
As we identified that zebrafish pre-mRNA has only a single exon, so a splice modifying Morpholino was not a possibility and we designed a translational blocking Morpholino; GCTC TGGGACAAGCTATCCATCGTC. For a specificity control, we designed 5′ mismatch oligo; GCTgTcGGAgAAcCTATCCATCcTC. We collected rnf152 Morpholino and its 5′ mismatch control from www.gene-tool.com. commercially available oligoes. Injected in synchronized 1- or 2-cell zebrafish embryos and incubated in E3 medium at 28.8°C incubator, we investigated phenotype after every 3 h. Methods for morpholino injection were adopted from previously done in laboratory (Yoo et al., 2017).
RESULTS
Zebrafish Rnf152 is an orthologue of human RNF152
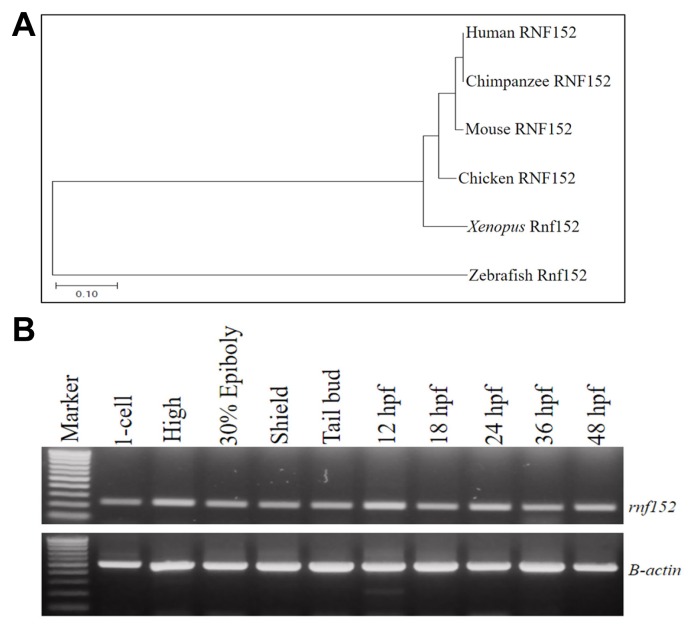
The neighbor-joining phylogenetic relationships between the amino acid sequence of zebrafish Rnf152 and those of homologues from other species were analyzed using Phylogenetic and Molecular Evolutionary Analysis MEGA software package (version 7.0) (Kumar et al., 2016). Rnf152 (NP_001014380) comprised 198 amino acid residues. The chimpanzee, mouse, chicken, and Xenopus RNF152 clade shared a common ancestor with zebrafish Rnf152 (Fig. 1A). Zebrafish Rnf152 was 57.6% homologous with human RNF152, and 55% homologous with chimpanzee, mouse, chicken, and Xenopus RNF152 (Supplementary Fig. S1). Zebrafish rnf152 was located on linkage group (LG) 2 within NC_007113.7 (www.ncbi.nlm.nih.gov). Based on the phylogenic relationship between zebrafish Rnf152 and human RNF152, molecular and functional analysis of zebrafish Rnf152 during early embryogenesis might provide important insights on the physiological function of human RNF152.
Fig. 1. (A) Phylogenetic analysis for Rnf152 in higher vertebrates.
Neighbor-joining method of MEGA7 was applied to derive the phylogenetic tree upon the amino acid sequences of human (NP_775828), chimpanzee (XP_001143948), mouse (NP_848894), chicken (NP_001291963), Xenopus (XP_004915369) and zebrafish (NP_001014380) which were collected from NCBI database. The scale bar denotes the branch distance in the figure. (B) RT-PCR analysis for temporal expression pattern of zebrafish rnf152. rnf152 was expressed both maternally and zygotically in zebrafish embryos. β-actin (500bp) was served as internal control (lower panel). Early and late stages of the zebrafish embryos abundantly expressed rnf152. Amplicon size of rnf152 was 235bp and equal amount of cDNA was in each reaction of all the tested stages with the endogenous control [No. of experiments (n) =3].
rnf152 transcripts are expressed in the eyes, MHB, and rhombomeres (r1–7) at 48 hpf
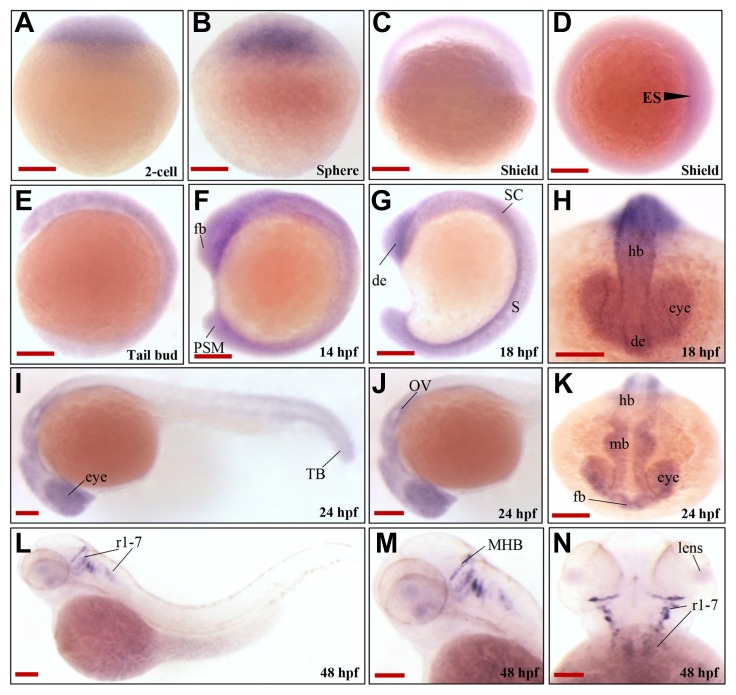
The temporal expression pattern of rnf152 during embryonic development was examined using RT-PCR. The rnf152 transcript was detected at the 1-cell stage, and its expression persisted until 48 hpf (Fig. 1B), indicating that it was expressed both maternally and zygotically. To further define the spatiotemporal expression pattern of rnf152, whole-mount in situ hybridization (WISH), which employed an rnf152-specific antisense RNA probe, was performed using zebrafish embryos at various developmental stages. The maternal transcript was ubiquitously distributed from the 1-cell stage to the gastrula stage (Figs. 2A and 2D), where it uniformly localized along the anteroposterior axis until mid-somitogenesis (Figs. 2E and 2F). Although rnf152 expression was restricted to the diencephalon and spinal cord at 18 hpf (Figs. 2G and 2H), it localized to the developing eyes, forebrain, midbrain, hindbrain, and presomitic mesoderm of posterior somites at 24 hpf (Figs. 2I and 2K). At 48 hpf, rnf152 expression was exclusively detected in the retina, midbrain-hindbrain boundary (MHB), and rhombomeres (r1–7) (Figs. 2L and 2N), indicating that rnf152 might be required for proper development of specific regions in the brain during embryogenesis.
Fig. 2. Spatiotemporal expression patterns of zebrafish rnf152.
(A–C) Lateral view; rnf152 was abundantly expressed in the zebrafish embryos at early stages. (D) Image from the animal pole of shield stage demonstrated thin germ ring with embryonic shield (arrowhead). (E–G) Lateral view of the embryos at tail bud, 14 hpf and 18 hpf. (H) Dorsal view of the embryo at 18 hpf found rnf152 transcripts in the eyes, CNS and spinal cord. (I&J) Lateral and (K) anterior view of the embryos at 24 hpf showed rnf152 transcripts were present in the eyes and CNS and some parts of posterior region. (L&M) Lateral view, (N) dorsal view of the embryos at 48 hpf demonstrated that rnf152 transcripts were in the eyes, midbrain and hindbrain boundary (MHB), and rhombomeres. All embryos were collected synchronously from WT zebrafish for WISH analysis at the corresponding stages. Abbreviations: de- Diencephalon, ES- Embryonic shield, Fb-Forebrain, hb-hind brain, PSM- presomitic mesoderm, ret-eye, SC- spinal cord (n=3). Scale bars A-N: 50 μm.
rnf152 is required for proper development of the eyes, MHB, and rhombomeres (r1–7) during embryogenesis
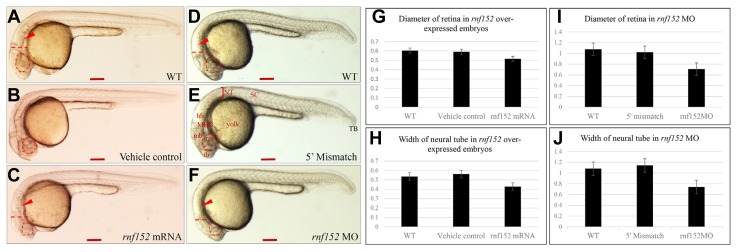
To examine the biological functions of Rnf152, its expression was forced by microinjecting the rnf152 transcript into zebrafish embryos at the 1- to 2-cell stage. Although there were no significant morphological defects until 24 hpf, we found three major changes in Rnf152-overexpressing embryos (Fig. 3C): an indistinctive midbrain-hindbrain boundary (MHB), a diminished hindbrain (r1–7), and a smaller eye compared with vehicle control and WT embryos (Figs. 3A and 3B). The graph depicts the diameter of the eye (Fig. 3G) and the width of the neural tube in rnf152 overexpressing, vehicle control, and WT embryos (Fig. 3H). Furthermore, we knocked down rnf152 with gene-specific morpholino antisense oligonucleotides and observed that the diameter of the eye was significantly smaller, whereas the MHB was undistinguishable and rhombomeres (r1–7) were smaller (Fig. 3F), than comparable structures in 5′ mismatch control (Fig. 3E) and WT (Fig. 3D) embryos at 24 hpf. We also measured the diameter of the eye (I) and the width of the neural tube (J) in knocked down embryos and compared these parameters with those in 5′ mismatch control and WT embryos, confirming that both the overexpression and knockdown of rnf152 significantly reduced the size of the eyes and the width of the neural tube (Figs. 3C and 3F). These results indicate that Rnf152 is required for the proper development of the eyes and neural tube.
Fig. 3. Overexpression and knockdown of rnf152 in zebrafish embryos.
(A) WT zebrafish embryo, (B) vehicle control injected with the same volume of phenol red dye in distilled water as the volume of the mRNAs injected, and (C) rnf152 mRNA injected embryo. Microinjection of rnf152 mRNA (200 pg) into embryos at 1- or 2 cell stages for overexpression of rnf152. All images are in lateral view (n = 3). (D) WT, (E) 5′ mismatch MO, and (F) rnf152 MO that 5 ng of rnf152 morpholino was injected into embryos at 1-cell stage for knockdown of rnf152. All images (D–F) are in lateral view of embryos at 24 hpf. (G–J) Graphs showing diameter of the eyes and width of the neural tubes. (G) Diameter of the eyes, (H) width of the neural tube of WT, vehicle control, and rnf152 overexpressed embryos. (I) Diameter of the eyes, (J) width of the neural tube of WT, 5′ mismatch control, and rnf152 MO. Red arrowheads indicate the rhombomeres (r1–7) while dotted red lines indicate the width of the neural tube. Area of the eyes was marked with red dotted lines. The graphs were obtained using ImageJ, error bars depicting standard errors. Unpaired t-test was executed to obtained p-values, which is p<0.0001 (n=3). Abbreviations: fb- forebrain, hb- hindbrain, mb- midbrain, NT- neural tube, ret- eye, SC- spinal cord, and TB- tail bud. Scale bars A–F: 50 μm.
neuroD expression pattern was significantly altered in the eyes and brain of rnf152 morphants
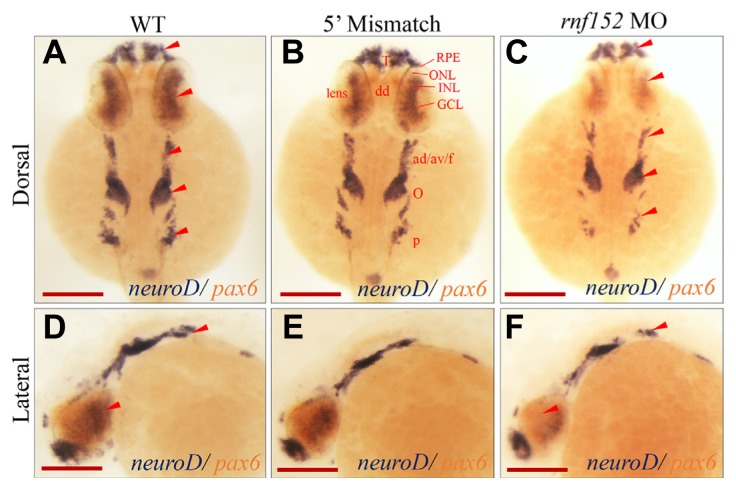
To identify the molecular elements in developing tissues affected by knockdown of rnf152, we selected molecular markers for the eyes, MHB, and rhombomeres. NeuroD is a marker for the inner and outer layers of the eyes at 48 hpf, although its expression is also observed in the central nervous system and lateral line in zebrafish embryos (Thomas et al., 2012). NeuroD is crucial in mitotic cells, where it serves as an important connector of cell cycle exit and cell fate determination in zebrafish (Ochocinska et al., 2017). To examine whether the knockdown of rnf152 affected any cellular events, including neuroD expression during development of the eyes and brain, two color WISH analysis using neuroD-and pax6-probes was performed in rnf152-deficient embryos at 27 hpf. Surprisingly, neuroD expression was significantly reduced in the telencephalon, dorsal diencephalon, eyes, ganglion, octaval/statoacustic (III) ganglion, posterior lateral line ganglion, and pancreatic bud of rnf152-deficient embryos at 27 hpf (Figs. 4C and 4F), but was unaffected in the indicated areas of WT (Figs. 4A and 4D) and 5′ mismatch MO control (Figs. 4B and 4E) embryos. In particular, neuroD expression was completely abolished from the ONL, INL, and GCL of the eyes in rnf152-deficient embryos at 27 hpf. On the other hand, knockdown of rnf152 did not affect pax6 expression in rnf152-deficient embryos at 27 hpf (Figs. 4C and 4F). Furthermore, WISH analysis using a rx2-specific probe to detect modifications in the optic vesicles of rnf152 deficient embryos at 12, 15, and 18 hpf found no significant changes in the optic vesicles and eyes after knockdown (Supplementary Fig. S4). These results strongly support the role of Rnf152 in tissue-specific development of the eyes, midbrain, and hindbrain at developmental stages later than 18 hpf during zebrafish embryogenesis.
Fig. 4. Knock-down of rnf152 decreased the level of neuroD in the telencephalon, dorsal diencephalon, eyes, octaval/statoacustic ganglia, and posterior lateral line ganglia.
Two color WISH analysis with neuroD- and pax6–specific probes detected changes in their transcripts in the retinal layers of embryos at 27 hpf. (A, D) WT, (B, E) 5′ mismatch MO control, and (C, F) rnf152 MO. neuroD transcripts (dark blue) were abundant in the INL and ONL of eyes, telencephalon, dorsal diencephalon, octaval/statoacustic ganglia and posterior lateral line ganglia of WT as well as the 5′ mismatch control, but remarkably reduced in rnf152 MO. Levels of pax6 transcripts (tomato color) in the telencephalon, dorsal diencephalon, hindbrain, anterior spinal cord, and eyes of the WT were similar those of rnf152 MO at 27 hpf. Red arrowheads indicates the areas of significant differences in the level of neuroD transcripts between WT and rnf152 MO. Abbreviations: ad/av/f- Anterodorsal/anteroventral lateral line/facial placodes/ganglia, dd- Dorsal diencephalon, GCL- Ganglion cell layer, INL- Inner nuclear layer, o- Octaval/statoacustic ganglia. ONL- outer nuclear layer, p- Posterior lateral line ganglia, RPE- retinal pigment epithelium, T- Telencephalon (n = 3). Scale bars (A–F): 50 μm.
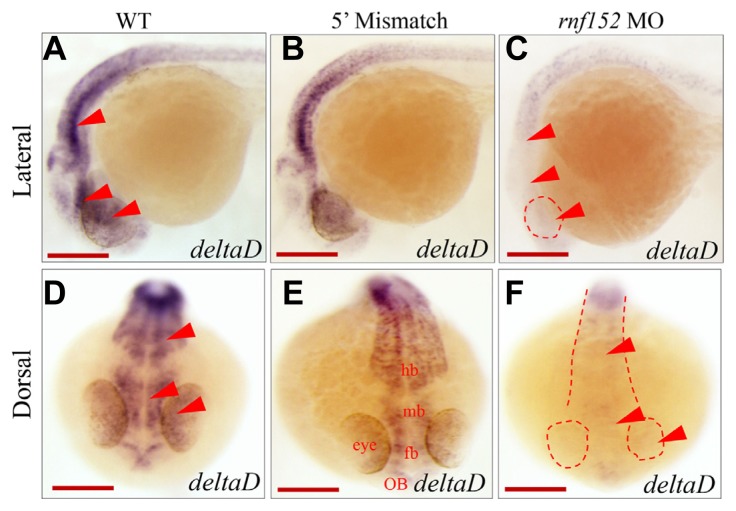
deltaD expression significantly decreased in rnf152 morphants
Notch-Delta signaling determines cell fate in the eyes, particularly the decision of neurons or Müller glia to differentiate or not differentiate (Taylor et al., 2015). In addition, deltaD and deltaC, as well as notch1a and notch 3, were expressed in the ciliary marginal zone in larval and juvenile zebrafish (Raymond et al., 2006). Thus, we investigated whether the expression patterns of the delta and notch genes matched that of neuroD found in rnf152-deficient embryos (Fig. 4).
We examined deltaD expression in rnf152-deficient embryos at 24 hpf. DeltaD expression was reduced in the eyes, forebrain, MHB, and hindbrain in rnf152-deficient embryos (Figs. 5C and 5F). Although deltaD expression was also significantly reduced in the midbrain of rnf152-deficient embryos (Figs. 5C and 5F), it was unaffected in WT (Figs. 5A and 5D) and 5′ mismatch control (Figs. 5B and 5E) embryos. WISH analysis further confirmed that deltaC expression in rnf152 deficient embryos was elevated in the outer plexiform layer, ONL, INL, and inner plexiform layer of the eye but significantly reduced in the brain at 24 hpf (Supplementary Figs. S2C and S2F) due to the fact that deltaC and deltaD are inversely regulated in the eyes and brain of zebrafish embryos (Wright et al., 2011). Thus, deltaD expression might be positively regulated by Rnf152 in the eyes and brain of the developing zebrafish embryos at 24 hpf.
Fig. 5. Level of deltaD transcripts was reduced in rnf152 MO.
WISH analysis of rnf152 MO using deltaD as a probe. (A, D) WT embryos, (B, E) embryos injected with 5′ mismatch as control, and (C, F) rnf152 MO. deltaD transcripts were abundant in the ventral midbrain and rhombomeres of the brain but low in the eyes of WT (A, D) and 5′ mismatch (B, D). However, level of deltaD transcripts was significantly reduced in the respected tissues (C, F). Red arrowheads indicate the eye, midbrain, and hindbrain (n = 3). Scale bars (A–F): 50 μm.
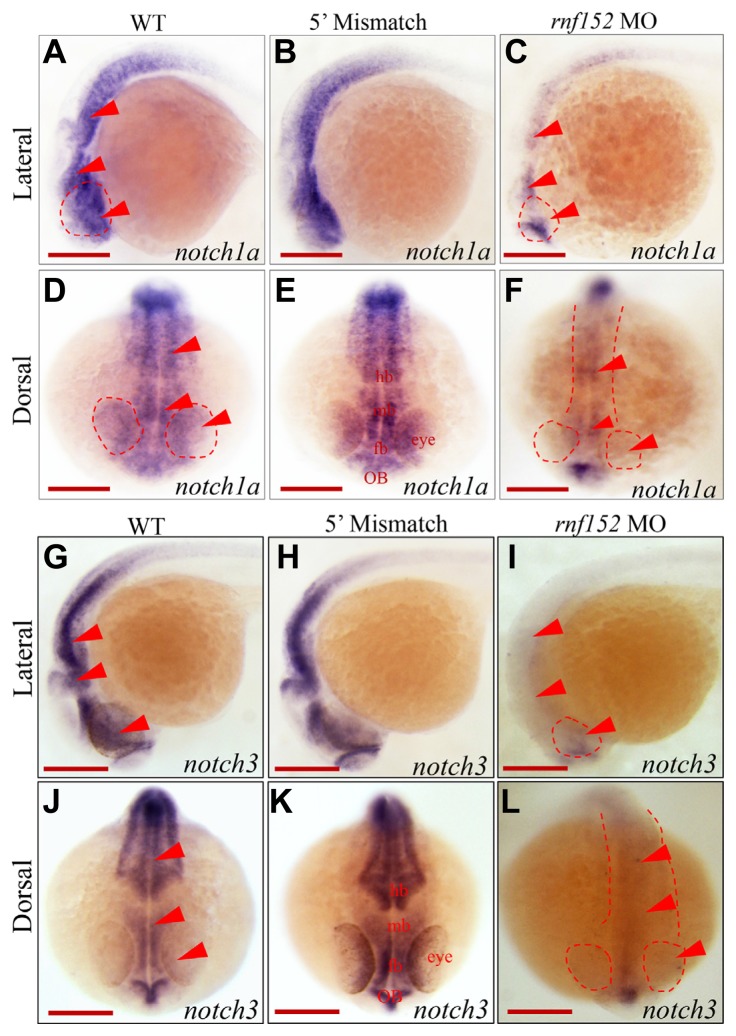
Knockdown of rnf152 decreased expression of Notch downstream genes
We showed that neuroD expression decreased in the telencephalon, octaval/statoacustic (III) ganglion, posterior lateral line ganglion, and dorsal diencephalon, but was abolished in the ONL, INL, and GCL of the eyes in rnf152-deficient embryos at 27 hpf (Figs. 4C and 4F). DeltaD expression was also abolished in the eyes and brain at 24 hpf (Figs. 5C and 5F). Various members of the Notch receptor family are expressed in the zebrafish brain (Banote et al., 2016). Notch receptors also serve as molecular switches between neurogenesis and the self-renewal of neural stem cells (Zhou et al., 2010). We initially selected three members of the Notch receptor family, notch1a, notch1b, and notch3, and examined their spatiotemporal expression patterns in rnf152 deficient embryos at 24 hpf by WISH analysis.
Except for the telencephalon, notch1a expression was markedly lower in the eyes and brain of rnf152-deficient embryos at 24 hpf (Figs. 6C and 6F) than in WT (Figs. 6A and 6D) and 5′ mismatch control (Figs. 6B and 6E) embryos. However, there was no change in notch1b expression in rnf152-deficient embryos compared with WT and 5′ mismatch control embryos (Supplementary Fig. S3). In addition, except for the olfactory bulbs (Figs. 6I and 6L), notch3 expression was significantly lower in the midbrain and rhombomeres (r1–7) than in the WT (Figs. 6G and 6J) and 5′ mismatch MO control (Figs. 6H and 6K) embryos. These results clearly indicate that delta and notch family genes might be downstream targets of Rnf152 in specific regions of the brain in zebrafish embryos during development.
Fig. 6. Knock-down of rnf152 down-regulated transcription of notch1a and notch3.
WISH analysis of rnf152 MO using notch1a and notch3 as probes. (A, D) notch1a transcripts in the eye, forebrain, midbrain, and hindbrain of WT embryos at 24 hpf. (B, E) Embryos injected with 5′ mismatch at 24 hpf showed similar patterns to those of WT embryos. (C, F) rnf152 MO shows remarkable reduction of notch1a transcripts in the eyes, forebrain, midbrain, and hindbrain at 24 hpf. Eyes, mid-hindbrain were shown in red arrowheads. Transcripts of notch3 in (I, L) rnf152 MO, (G, J) WT, and (H, K) 5′ mismatch control at 24 hpf. notch3 transcripts were present in the olfactory bulbs, midbrain, and hindbrain in WT and 5′ mismatch control whereas they were significantly diminished in the corresponding areas of rnf152 MO. Red arrowheads indicate eyes, mid-hindbrain tissues (n = 3). Scale bars (A–L): 50 μm.
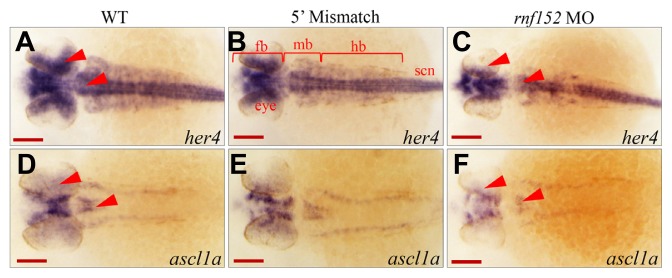
Among the her genes in zebrafish (Weizsäcker, 1994), her4 mediates Notch signaling during neurogenesis (Takke et al., 1999). Transcription of her4 is activated by a constitutively active notch1a variant encoded by the Notch intracellular domain, and abnormal expression of her4 decreases the number of islet-1-positive cells (Takke et al., 1999; Wettstein et al., 1997). On the other hand, a recent study reported that neuroD represses Notch signaling, which positively regulates transcription of ascl1a in photoreceptor progenitors (Tayler et al., 2015). Thus, we used WISH analysis to investigate her4 as a downstream gene of Notch signaling to determine whether its expression pattern was affected in rnf152-deficient embryos. As shown in Fig. 7C, her4 expression was hardly visible in the eyes and was significantly decreased in the MHB; however, it was abundant at these sites in control embryos (Figs. 7A and 7B). However, knockdown of rnf152 did not affect her4 expression in the spinal cord (Fig. 7C). On the other hand, ascl1a expression in the eyes and midbrain of rnf152-deficient embryos (Fig. 7F) was lower than that in WT and 5′ mismatch control (Figs. 7D and 7E) embryos. Thus, rnf152 might be required for maintenance of notch1a expression, as well as for the proper expression of her4 in the eyes and MHB, but dispensable in the spinal cord, where her4 expression might be dependent upon other Notch receptors or Notch-related genes besides Notch1a and Notch3 (Chen and Corliss, 2004; Hegde et al., 2008; Hwang et al., 2009; Mumm and Kopan, 2000). Taken together, these results indicate that Rnf152 regulates transcription of ascl1a, which is independent of neuroD expression.
Fig. 7. rnf152 knock-down reduced expression of ascl1a and her4 in zebrafish embryos.
(A–F) Dorsal view of embryos at 29 hpf. WISH analysis with her4 detected the transcripts in the eyes, forebrain, mid-hindbrain, and spinal cord of the WT (A) and 5′ mismatch control (B) while they disappeared in the eye and rhombomeres of rnf152 MO (C). Red arrowheads indicate her4 transcripts in the eyes and anterior rhombomeres (n = 3). WISH analysis with ascl1a found that ascl1a transcripts in the eyes, forebrain and mid-hindbrain of the two control groups but were significantly reduced in those of rnf152 MO. Red arrowheads indicate the eyes and rhombomeres (n = 3). Abbreviations: fb- forebrain, mb- midbrain, hb- hindbrain, ret- retina. Scale bars (A–F): 50 μm.
DISCUSSION
We report here that neuroD expression was hardly visible in the ONL, INL, and GCL of the eyes, and was reduced in the telencephalon, octaval/statoacustic (III) ganglion, posterior lateral line ganglion, and dorsal diencephalon, in rnf152 morphants at 27 hpf (Fig. 4). A previous study demonstrated neuroD expression in the cranial ganglia, diencephalon, epiphysis, retina, rhombomeres, mid-hindbrain, telencephalon, and spinal cord of zebrafish embryos at 48 hpf (Rauch et al., 2003). NeuroD is a pro-neural bHLH transcription factor that is responsible for regeneration and genesis of photoreceptor cells via Delta-Notch signaling in zebrafish embryos at 72 hpf (Taylor et al., 2015). NeuroD negatively regulates expression of molecules downstream of Notch signaling, which affects expression of Notch targets at 72 hpf (Taylor et al., 2015). Unexpectedly, we found that Notch signaling was abolished in the absence of Rnf152, even when neuroD expression was severely impaired in the rnf152 morphants. Thus, the results indicate that Rnf152 is required for expression of neuroD and for maintenance of Delta-Notch signaling, which might fine-tune differentiation of the anterior neural tissue, including the eyes. In addition, it is possible that knockdown of neuroD alone in the absence of functional Rnf152 might not be enough to de-repress Notch signaling. Identification of the molecular relationship between NeuroD and Rnf152 will unveil the role of Rnf152 in Delta-Notch signaling.
DeltaD is a Notch ligand expressed in differentiating neurons of the CNS and in the eyes of zebrafish embryos at 24 hpf (Banote et al., 2016). In agreement with these observations, we found that deltaD expression was abundant in the eyes and CNS of WT, but markedly reduced in these tissues in rnf152-deficient, embryos at 24 hpf (Figs. 5C and 5F). On the other hand, deltaC expression was elevated in rnf152-deficient embryos at 24 hpf (Supplementary Fig. S2). Considering that DeltaC and DeltaD are Notch ligands in the eyes and brain, and that DeltaC counteracts DeltaD in zebrafish embryos at 16 hpf (Wright et al., 2011), we propose that Rnf152 might be an upstream regulator of deltaD, and that elevated deltaC expression in the rnf152 morphants might be caused by reduction of deltaD expression.
NeuroD2 is a transcription factor harboring a Cdc20 recognition motif and destruction box (D-box) that regulates presynaptic development in the brain (Yang et al., 2009). The major mitotic E3 ubiquitin ligase, Cdc20-anaphase-promoting complex, activates presynaptic differentiation by promoting protein degradation, which negatively regulates presynaptic development (Yang et al., 2010). Given that NeuroD2 expression is inversely correlated with that of Cdc20, the cellular level of NeuroD2 is regulated by the ubiquitin-proteasome system (Yang et al., 2009).
The E3 ligase gene, Mib1, regulates neurogenesis in the developing spinal cord via Delta-Notch signaling (Kang et al., 2013). Furthermore, expression of Notch receptor family members such as notch1a and notch3 was also reduced in rnf152-deficient embryos at 24 hpf. Transcription of Notch downstream targets, her4 and ascl1a, was also compromised by depletion of Rnf152 at 24 hpf. In this context, it is plausible that Rnf152 functions in these tissues by targeting molecules such as NeuroD, Delta, and Notch. Considering that both autosomal dominant and autosomal recessive mutations in proteins of the Notch signaling pathway cause several human disorders, including Alagille syndrome and spondylocostal dysostosis (Lasky and Wu, 2005), the discovery of novel Notch regulators increases our understanding of Notch signaling. We propose that Rnf152 is a novel Notch regulator, and that it might have therapeutic potential with respect to diseases caused by aberrant Notch activation.
Supplementary data
ACKNOWLEDGEMENTS
We thank Professor Hyunju Ro at Chungnam National University for his critical reading of this manuscript, and the Zebrafish Organogenesis Mutant Bank (ZOMB), Korea for providing the zebrafish lines. This work was funded by the National Research Foundation of Korea Government Grants (NRF-2014R1A1A2058288) and Brain Korea 21 plus.
Footnotes
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Anuppalle M., Maddirevula S., Huh T.R., Rhee M. Ubiquitin proteasome system networks in the neurological disorder. Anim Cells Syst. 2013;17:383–387. [Google Scholar]
- Anuppalle M., Maddirevula S., Kumar A., Huh T.L., Choe J., Rhee M. Notch and retinoic acid signaling are required for the spatiotemporal distribution of prune2 mRNA in the zebrafish embryonic development. Gene Exp Patterns. 2017;11:23–24. 45–51. doi: 10.1016/j.gep.2017.03.002. [DOI] [PubMed] [Google Scholar]
- Banote R.K., Edling M., Eliassen F., Kettunen P., Zetterberg H., Abramsson A. β-Amyloid precursor protein-b is essential for Mauthner cell development in the zebrafish in a Notch-dependent manner. Dev Biol. 2016;413:26–38. doi: 10.1016/j.ydbio.2016.03.012. [DOI] [PubMed] [Google Scholar]
- Chen W.B., Corliss D.C. Three modules of zebrafish Mind bomb work cooperatively to promote Delta ubiquitination and endocytosis. Dev Biol. 2004;267:361–373. doi: 10.1016/j.ydbio.2003.11.010. [DOI] [PubMed] [Google Scholar]
- Deng L., Jiang C., Chen L., Jin J., Wei J., Zhao L., Chen M., Pan W., Xu Y., Chu H., et al. The ubiquitination of RagA GTPase by RNF152 negatively regulates mTORC1 activation. Mol Cell. 2015;58:804–818. doi: 10.1016/j.molcel.2015.03.033. [DOI] [PubMed] [Google Scholar]
- Garza-Lombó C., Gonsebat M.E. Mammalian target of rapamycin: its role in early neural development and in adult and aged brain function. Front Cell Neurosci. 2016;10:157. doi: 10.3389/fncel.2016.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman N.W., Lin T.V., Zhang L., Paquelet G.E., Feliciano D.M., Bordey A. mTORC1 targets the translational repressor 4E-BP2, but not S6 kinase 1/2, to regulate neural stem cell self-renewal in vivo. Cell Rep. 2013;5:433–444. doi: 10.1016/j.celrep.2013.09.017. [DOI] [PubMed] [Google Scholar]
- Hegde A., Qiu N.C., Qiu X.H., Ho S.H., Tay K.Q., George J., Ng F.S., Govindarajan K.R., Gong Z.Y., Mathavan S., et al. Genomewide expression analysis in zebrafish mind bomb alleles with pancreas defects of different severity identifies putative notch responsive genes. PLos One. 2008;3:e1479. doi: 10.1371/journal.pone.0001479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J., Kim H.S., Seok J.W., Kim J.D., Koun S., Park S.Y., Lee J., Kim K.S., Chang K.T., Ryoo Z.Y., et al. Transcriptome analysis of the zebrafish mind bomb mutant. Mol Genet Genomics. 2009;281:77–85. doi: 10.1007/s00438-008-0395-5. [DOI] [PubMed] [Google Scholar]
- Kang K., Lee D., Hong S., Park S.G., Song M.R. The E3 Ligase Mind Bomb-1 (Mib1) Modulates Delta-Notch Signaling to Control Neurogenesis and Gliogenesis in the Developing Spinal Cord. J Biol Chem. 2013;288:2580–2592. doi: 10.1074/jbc.M112.398263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel C.B., Ballard W.W., Kimmel S.R., Ullmann B., Schilling T.F. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetic analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasky J.L., Wu H. Notch signaling, brain development, and human disease. Pediatr Res. 2005;57:104R–109R. doi: 10.1203/01.PDR.0000159632.70510.3D. [DOI] [PubMed] [Google Scholar]
- Morreale F.E., Walden H. Snapshot: types of ubiquitin ligase. Cell. 2016;165:248–248.e1. doi: 10.1016/j.cell.2016.03.003. [DOI] [PubMed] [Google Scholar]
- Metzger M.B., Pruneda J.N., Klevit R.E., Weissman A.M. RING-type E3 ligases: Master manipulators of E2 ubiquitinconjugating enzymes and ubiquitination. Biochim Biophys Acta. 2014;1843:47–60. doi: 10.1016/j.bbamcr.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumm J.S., Kopan R. Notch signaling: from the outside in. Dev Biol. 2000;228:151–165. doi: 10.1006/dbio.2000.9960. [DOI] [PubMed] [Google Scholar]
- Ochocinska M.J., Hitchcock P.F. Dynamic expression of the basic helix-loop-helix transcription factor NeuroD in the rod and cone photoreceptor lineage in the retina of the embryonic and larval zebrafish. J Comp Neurol. 2017;501:1–12. doi: 10.1002/cne.21150. [DOI] [PubMed] [Google Scholar]
- Rauch G.J., Lyons D.A., Middendorf I., Friedlander B., Arana N., Reyes T., Talbot W.S. Submission and curation of gene expression data. ZFIN Direct Data Submission. 2003 http://zfin.org.
- Raymond P.A., Barthel L.K., Bernardos R.L., Perkowski J.J. Molecular characterization of retinal stem cells and their niches in adult zebrafish. BMC Dev Biol. 2006;6:36. doi: 10.1186/1471-213X-6-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ro H., Soun K., Kim E.-J., Rhee M. Novel vector systems optimized for injecting in vitro-synthesized mRNA into zebrafish embryos. Mol Cells. 2004;17:373–376. [PubMed] [Google Scholar]
- Ro H., Hui T.L., Rhee M. Ubiquitin conjugation system for the body axes specification in vertebrates. Animal Cells Sys. 2015;19:87–95. [Google Scholar]
- Swatek K.N., Komander D. Ubiquitin modifications. Cell Res. 2016;26:399–422. doi: 10.1038/cr.2016.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takke C., Dornseifer P., Weizsäcker v.E., Campos-Ortega J.A. her4, a zebrafish homologue of the Drosophila neurogenic gene E(spl), is a target of NOTCH signaling. Development. 1999;126:1811–1821. doi: 10.1242/dev.126.9.1811. [DOI] [PubMed] [Google Scholar]
- Taylor S.M., Alvarez-Delfin K., Saade C.J., Thomas J.L., Thummel R., Fadool J.M., Hitchcock P.F. The bHLH transcription factor NeuroD governs photoreceptor genesis and regeneration through Delta-Notch signaling. Invest Ophthalmol Vis Sci. 2015;56:7496–7515. doi: 10.1167/iovs.15-17616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse B., Thisse C. Fast release clones: a high throughput expression analysis. ZFIN Direct Data Submission 2004. 2004 (Available from: http://zfin.org)
- Thomas J.L., Ochocinska M.J., Hitchcock P.F., Thummel R. Using the Tg(nrd:egfp)/albino Zebrafish Line to Characterize In Vivo Expression of neurod. PLoS One. 2012;7:e29128. doi: 10.1371/journal.pone.0029128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wettstein D.A., Turner D.L., Kintner C. The Xenopus homolog of Drosophila Suppressor of Hairless mediates Notch signaling during primary neurogenesis. Development. 1997;124:693–702. doi: 10.1242/dev.124.3.693. [DOI] [PubMed] [Google Scholar]
- Weizsäcker E.V. Inaugural dissertation. Universität zu Koln; 1994. Molekulargenetische Untersuchungen an sechs Zebrafischgenen mit Homologie zur Enhancer of split-Genfamilie von Drosophila. [Google Scholar]
- Westerfield M. The Zebrafish Book. A guide for the laboratory use of Zebrafish (Danio rerio) 4th ed. University of Oregon Press; Eugene: 2000. [Google Scholar]
- Wright J.G., Giudicelli F., Soza-Ried C., Hanisch A., Ariza-McNaughton L., Lewis J. DeltaC and DeltaD interact as notch ligands in the zebrafish segmentation clock. Development. 2011;138:2947–2956. doi: 10.1242/dev.066654. [DOI] [PubMed] [Google Scholar]
- Yang Y., Kim A.H., Yamada T., Wu B., Bilimoria P.M., Ikeuchi Y., Iglesia N.D.L., Shen J., Bonni A. A Cdc20-APC Ubiquitin Signaling Pathway Regulates Presynaptic Differentiation. Science. 2009;326:575–578. doi: 10.1126/science.1177087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Kim A.H., Bonni A. The Dynamic ubiquitin ligase duo: Cdh1-APC and Cdc20-APC regulate neuronal morphogenesis and connectivity. Curr Opin Neurobiol. 2010;20:92–99. doi: 10.1016/j.conb.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo K.W., Maddirevula S., Kumar A., Ro H., Huh T.L., Rhee M. Sinup is essential for the integrity of centrosomes and mitotic spindles in zebrafish embryos. Animal Cells Sys. 2017;21:93–99. doi: 10.1080/19768354.2017.1308438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z.D., Kumari U., Xiao Z.C., Tan E.K. Notch as a molecular switch in neural stem cells. IUBMB Life. 2010;62:618–623. doi: 10.1002/iub.362. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.