Abstract
Mammalian genomes are well established, and highly conserved regions within odorant receptors that are unique from other G-protein coupled receptors have been identified. Numerous functional studies have focused on specific conserved amino acids motifs; however, not all conserved motifs have been sufficiently characterized. Here, we identified a highly conserved 18 amino acid sequence motif within transmembrane domain seven (CAS-TM7) which was identified by aligning odorant receptor sequences. Next, we investigated the expression pattern and distribution of this conserved amino acid motif among a broad range of odorant receptors. To examine the localization of odorant receptor proteins, we used a sequence-specific peptide antibody against CAS-TM7 which is specific to odorant receptors across species. The specificity of this peptide antibody in recognizing odorant receptors has been confirmed in a heterologous in vitro system and a rat-based in vivo system. The CAS-TM7 odorant receptors localized with distinct patterns at each region of the olfactory epithelium; septum, endoturbinate and ectoturbinate. To our great interests, we found that the CAS-TM7 odorant receptors are primarily localized to the dorsal region of the olfactory bulb, coinciding with olfactory epithelium-based patterns. Also, these odorant receptors were ectopically expressed in the various non-olfactory tissues in an evolutionary constrained manner between human and rats. This study has characterized the expression patterns of odorant receptors containing particular amino acid motif in transmembrane domain 7, and which led to an intriguing possibility that the conserved motif of odorant receptors can play critical roles in other physiological functions as well as olfaction.
Keywords: conserved, GPCR, odorant receptor, olfaction, transmembrane
INTRODUCTION
The odorant receptor (OR) is a G-protein coupled receptor (GPCR), a family of receptors with important roles in olfaction. Various ORs are activated by different odor molecules and their structural features are not completely understood.
Since the identification of olfactory-specific seven transmembrane domain receptors by Buck and Axel (1991), and the elucidation of mammalian GPCRs structures by (Palczewski et al., 2000), numerous studies refer to the structural features of GPCRs in characterizing the structure of ORs.
Despite the diversity of ORs, each region has a conserved motif within the OR sequences (Buck and Axel, 1991; Pilpel and Lancet, 1999; Probst et al., 1992). For instance: the LHTPMY in intracellular loop 1 (IC1), MAYDRYVAIC at the end of transmembrane domain 3 (TM3) to beginning of IC2, SY at the end of TM5, KAFSTCASH at the beginning of TM6, and PMLNPFIY at TM7. A number of studies have shown that certain conserved motifs or single amino acid domains within the motif play critical roles in olfactory signaling. A DRY motif included in TM3—highly conserved among rhodopsin-like GPCRs—have been demonstrated to play important roles in G-protein activation and axonal targeting to specific glomerulus (de March et al., 2015; Gaillard et al., 2004; Imai et al., 2006; Nguyen et al., 2007). Furthermore, Kato et al (2008) demonstrated that the serine within the KAFSTC motif is critical to receptor conformational dynamics (Kato et al., 2008). To date, limited studies have focused on the PMLNPFIY motif in TM7 despite its high conservation. These studies imply that a certain conserved motif may have a crucial effect on olfactory signaling and lead to specific features.
Recent studies have shown that ectopically expressed ORs are broadly distributed throughout the human body (Feldmesser et al., 2006; Flegel et al., 2013; Zhang et al., 2007), and several ORs have been demonstrated to play important roles in various non-olfactory peripheral tissues [eg, testis (Flegel et al., 2015; Spehr et al., 2003), prostate (Neuhaus et al., 2009; Spehr et al., 2011), gut (Braun et al., 2007; Kidd et al., 2008), colon (Kaji et al., 2011), blood (Zhao et al., 2013), lung (Kalbe et al., 2016), kidney (Gu et al., 2014), liver (Massberg et al., 2015; Wu et al., 2015), and skin (Busse et al., 2014)]. Fukuda et al. (2006) have screened testicular mRNA expression patterns of ORs in mice using primers designed from the conserved amino acids motifs of ORs (MAYDRYVAIC, KAFSTCSSH, PMLNPFIY), but few studies have been performed at the protein level. Moreover, limited studies have been done to examine the overall OR expression patterns in both olfactory and non-olfactory tissues at the protein level.
A large number of ORs share some conserved amino acid motifs, and each similar region is a part of various groups of ORs. Despite their widespread distribution, none of the ORs that classified according to the similarity of amino acid sequence has been investigated in their expression patterns. When considering an analysis of OR characterization using a motif-based approach, we sorted several ORs out according to common residues they share, and hypothesized that they may have common characteristics as well as specific functions. Here, we showed that characteristics of various ORs could be classified based on its certain conserved sequences, and we also have characterized the expression patterns of ORs containing particular amino acid motif in TM7 in rats. In addition, we identified that these features are not restricted to the olfactory system but are also observed within non-olfactory tissues using a conserved sequence-specific OR antibody. Our study suggests that a conserved amino acid motif in the OR may exert influences on the expression patterns of ORs, implicating some critical roles in olfaction and its mediated behaviors.
MATERIALS AND METHODS
Animals
Male adult Sprague-Dawley rats (5 to 7 week old) were obtained from KOATECH (Korea). All experimental protocols were approved by Institutional Animal Care and Use Committees of DGIST.
Antibodies and Reagents
Antibodies used were: anti-adenylyl cyclase 3 (AC3) (Santa Cruz Biotechnology, USA, #SC588), anti-olfactory marker protein (OMP) (Wako Chemicals, USA, #019-22291), anti-GAPDH (Millipore, USA, #MAB374), anti-FLAG (Sigma, USA, #F1804), peroxidase conjugated anti-mouse secondary antibody (Thermo Fisher Scientific, USA, #31430), peroxidase conjugated anti-rabbit antibody (Thermo Fisher Scientific, #31460), and biotinylated secondary antibody (anti-mouse, rabbit and goat) (Jackson Immuno Research Labs, USA, #715-065-151, #711-065-152, and #705-065-003).
Reagents used were: Poly-D-lysine (Sigma, #P7886), Lipofectamine 2000 (Invitrogen, USA, #11668), puromycin (Sigma, #P3388), Normal Donkey Serum (Jackson Immuno Research Labs, #017-000-121), Protein A/G agarose beads (Calbiochem, USA, #IP05), FLAG magnetic M2 beads (Sigma, #M8823), Vectastain Elite ABC kit (Vector Laboratories, USA, #PK-6100), DAB peroxidase substrate kit (Vector Laboratories, #SK-4100), digitonin (Sigma, #D141), and CellLight ER-RFP, BacMam2.0 (Thermo Fisher Scientific, #C10591).
DNA constructs
Full length genomic DNA sequences of ORs were obtained from rat (Olr1493) mouse (Olfr394) tissue and HEK293 cells (human ORs) using specific primers (Supplementary Table S1). The primers included restriction enzyme sequences of Mlu1 and Not1 to facilitate subcloning into a pCI mammalian expression vector. pCI vector containing N-terminal Rho tags and RTP1S-pCI constructs were a kind gift from Dr. Hiroaki Matsunami (Univ. of Duke, USA). The FLAG sequence was attached in front of a Rho sequence at the N-terminus of ORs to allow for detection of surface expression. DNA construct of FLAG-β2AR was a kind gift from Robert Lefkowitz (Univ. of Duke, USA). All constructs were performed DNA sequencing to confirm its identity.
Human samples
Human turbinate tissues were obtained during turbinate surgery from patients. The turbinate tissues were obtained from a patient who underwent partial turbinectomy with an informed consent before surgery. The procedures were approved by the Institutional Review Board of Seoul St. Mary’s Hospital, the Catholic University of Korea (KC08TISS0341).
Cell culture
The Hana3A cell stock was a kind gift from Dr. Hiroaki Matsunami, and maintained in DMEM (Corning, USA) containing 10% fetal bovine serum (Tissue culture biologicals, USA) and penicillin streptomycin (Gibco, USA). Puromycin (Sigma) was added to the feeding medium at a concentration of 1 μg/ml when culturing.
Olfactory sensory neurons (OSNs) were obtained from Sprague-Dawley rats as previously described with some modifications (Ronnett et al., 1991). Cells were plated on tissue culture dishes coated with 25 μg/ml of laminin (BD Bioscience, USA) in modified Eagle’s medium containing D-valine (MDV, Gibco). Cultures were placed in a humidified 37°C incubator receiving 5% CO2. On 2 days in vitro every day thereafter, cells are fed with MDV containing 15% dialyzed fetal bovine serum (Gibco), gentamicin, kanamycin and 2.5 ng/ml nerve growth factor. Two days prior to use, the culture medium was changed to medium without nerve growth factor.
DNA transfection
Hana3A cells were grown from a density of 3.5 × 105/ml to the poly-D-lysine (Sigma) coated plate 24 h prior to the transfection. Every OR was transfected onto Hana3A cells with pCI-RTP1s to promote surface expression at the ratio of 5:1 with Lipofectamine 2000 reagent (Invitrogen, USA). 24 h of post-transfection, cells were prepared for the experiments.
Generation of polyclonal antibodies to an OR peptide
A synthetic peptide (NH2-VTPMLNPFIYSLRNRDC-OH) was generated to the predicted protein sequence of the OR (amino acid 278–296 of olfactory receptor Olr1493, protein_id “NP001006610.1”) based on its antigenicity and other considerations. A terminal cysteine residue was added to the peptide sequence which is necessary to conjugate with the carrier proteins to induce immune response in generating antigen. We used two common carrier protein, keyhole limpet hemocyanin (KLH) and bovine serum albumin (BSA).
Rabbit polyclonal antiserum to the peptide was generated by Zymed (USA). The resulting antisera were antigen affinity purified using affinity columns by AbClon (Korea).
Histology
Tissue preparation
The Sprague-Dawley rats were anesthetized using a mixture of Zoletil and Rompun solution (9:1 ratio, 1 μl/g, i.m.) and then transcardially perfused with cold saline (0.9% NaCl) and fixed with prechilled 4% paraformaldehyde in phosphate-buffered saline (PBS, pH7.4). The OE and OB were post-fixed overnight at 4°C.
Post-fixed OE tissues were treated with a decalcification solution (Calci-Clear Rapid, National diagnostics, GA, USA) for 4 h. After D.W. washing, OE tissues underwent dehydration with ethanol solutions for 2 h (70%, 85%, 95% and 100%), and then soaked in xylene for 4 h, followed by embedding in paraffin over 4 h.
The OB tissues of rat were sunken in 30% (wt/vol) sucrose overnight for cryo-embedding. Tissues were embedded in optimal cutting temperature (OCT) compound (Leica, Germany), and stored at −80°C until use.
The paraffin-molded samples were sectioned (5 μm) with a rotary microtome (Leica, Germany, RM2235), and cryo-embedded OB tissues were serial sectioned using cryostat at 12 μm (Thermo Scientific, Microm HM550). All sliced tissues were attached to the micro slide glasses (Muto Pure Chemicals, Japan).
Immunohistochemistry
OE, OB and testis tissue sections were boiled with citrate buffer (0.01 M citric acid monohydrate, pH 6.0) for 5 min to retrieved antigen and permeated using 3% hydrogen peroxide (H2O2) for 30 min. The section slides were incubated with 5% normal donkey serum (NDS) blocking solution and labeled with primary antibodies (OR; 1:500, AC3; 1:500 and OMP; 1:5000 dilution in 5% NDS solution). Secondary antibody incubation was 1 h at room temperature with biotin conjugated anti goat, rabbit, mouse (1:500 dilution in 5% NDS solution) and then detected using an avidin-biotin-peroxidase complex method.
Imaging
Cell and tissue samples were visualized with a microscope (Nikon-Eclipse-90i; Nikon Corp., Japan) and captured with a Nikon digital camera (DS-Ri1). All images were analyzed and reorganized with Image J software (NIH, USA).
Immunoblot analysis
Cell lysates underwent BCA assay for protein-concentration estimation, and divided into 500 μg for each purpose. For OR detection, lysates were incubated with 5x SDS sample buffer (250 mM Tris-HCl pH6.8, 10% SDS, 30% glycerol, 5% β-mercaptoethanol, 0.02% bromophenol blue) at room temperature for 30 min without boiling. For FLAG-sequence detection, lysates were gently agitated with anti-FLAG® M2 magnetic beads (Sigma) for 2 h at room temperature. Immunoprecipitated Flag-tagged proteins were detected using an anti-FLAG mouse monoclonal antibody (1:1000 dilution in 3% BSA/TBS). 100 μg of tissue lysates (rat and human) were separated in a 10% SDS-PAGE gel and transferred to a PVDF (polyvinylidene difluoride) membrane (Milipore).
All PVDF membranes of cells and tissues samples were blocked with 5% skim milk in TBS for 1 h at room temperature, and incubated with primary antibodies (OR, 1:500 dilution). Secondary antibody incubation was 1 h at room temperature with HRP-conjugated anti-mouse (1:5000 dilution in blocking solution for FLAG and GAPDH) and anti-rabbit (1:5000 dilution for OR).
Immunocytochemistry
Transiently expressed ORs in Hana3A cells were seeded at a density of 1.5 × 105/ml onto poly-D-lysine-coated glass slides 24 h prior to the experiment. Cells were rinsed with PBS twice and fixed directly with 4% PFA for 20 min at room temperature without permeabilization to examine surface expression of receptor. After three times washing with PBS, cells were incubated with 0.01% of H2O2 solution that dissolved in methanol for 30 min to reduce non-specific reaction of DAB. Cells were washed with PBS-T (0.01% Triton X-100) thrice and blocked with 4% normal donkey serum in PBS for 1 h at room temperature. OR antibody-incubation was performed overnight at 4°C at a dilution of 1:200 in blocking solution, and 1:100 dilution for FLAG antibody (Sigma). The next day, cells were incubated for 15 min at room temperature before PBS-T washing. Biotinylated secondary antibody was added to the cells at a dilution of 1:500 in blocking solution, followed by cell staining with avidin-biotin-peroxidase complexes using Vectastain Elite ABC kit (Vector Laboratories). After PBS washing, peroxidase activity was examined in 0.05% DAB solution (Vector Laboratories) for several minutes. Cells were mounted with aqua mount solution (Polysciences, USA), and visualized under the microscope (Nikon/eclipse 90i).
Membrane protein isolation from the cells and tissue
Cells
ORs expressed in the plasma membrane of Hana3A cells were isolated according to the previous study with some modifications (Pal et al., 2015). Cells were washed with 1 x TBS and harvested by scraping with TBS (150 mM NaCl, 100 mM Tris, pH 7.5) containing 2 mM EDTA and 1 mM EGTA. Cells were centrifuged directly at 500 x g for 5 min at 4°C, and washed with 1 x TBS twice. Cells were frozen in liquid nitrogen and lysed with lysis buffer (1% digitonin in TBS with protease inhibitors) for 2 h with gentle agitation at 4°C. Membrane protein lysates were obtained from centrifugation for 10 min at 16,000 x g at 4°C.
Tissues
Rat and human tissues were stored at −80°C. We used protocols as previously described to fractionate intracellular and membrane proteins following serial extraction steps (Sherman and Lesne, 2011). Specifically, intracellular proteins were obtained from lysates which were generated with TNT lysis buffer composed of 50 mM Tris-Cl, pH 7.6, 0.01% NP-40, 150 mM NaCl, 2 mM EDTA, 0.1% SDS and added protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific). Membrane proteins were gathered after intracellular proteins were obtained by lysis with RIPA buffer composed of 50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.5% Triton X-100, 1 mM EDTA, 3% SDS, and protease and phosphatase inhibitor cocktail. All lysates were centrifuged at 16,100 × g for 90 min to clarify.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 7 (GraphPad Software, Inc., USA).
RESULTS
Conserved amino acid motif of odorant receptors (ORs)
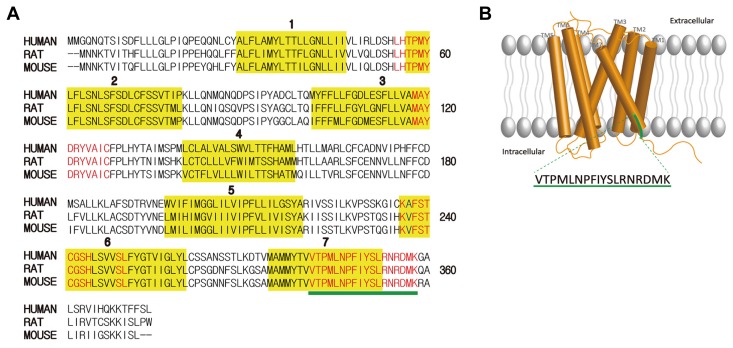
To identify common conserved motifs of ORs, three conserved motifs: i) MAYDRYVAIC, ii) KAFSTCASH, and iii) PMLNPFIY were used to search for ORs that possesses highly conserved region with a BLASTP analysis (Altschul et al., 1997). The amino acid sequences of ORs from BLASTP searches were aligned using a Constraint-based Multiple Alignment Tool (COBALT). The result represented conserved area within OR amino acid sequences, and the 18 amino acids at the TM7 containing PMLNPFIY were highly conserved (data not shown). 18 amino acids of ORs in three species including human were analyzed to find the most highly conserved sequence. The illustration of the most highly conserved motif with ORs of rats, mice and humans as identified using CLUSTAL O (1.2.4) multiple sequence alignment tool are presented in Fig. 1A. Conserved motifs were determined by thorough analysis of highly similar 18 amino acids stretches in TM7 among human, mouse and rat. Through the analysis, we have identified that these ORs mostly contain VTPMLNPFIYSLRNKDMK and VTPMLNPFIYSLRNRDMK (Supplementary Fig. S1).
Fig. 1. Conserved motifs of odorant receptors (ORs).
(A) Sequence alignment of three representative ORs from Human, rat, and mouse [NP_003544.2, NP_667218.1, NP_001006610.1] that have most identical sequences with conserved motif. Conserved motifs were indicated as red color, and the transmembrane domains were boxed in yellow color and numbered. 18 amino acid conserved residues were underlined in green color. (B) Cartoon of an OR sitting in the cellular plasma membrane. Seven transmembrane domains are numbered. The location of the conserved motif at the TM7-C terminal was represented in green colored line and its amino acid sequences are indicated.
OR antibody targeting the peptide sequence PMLNPFIY specifically detects ORs of human and rat
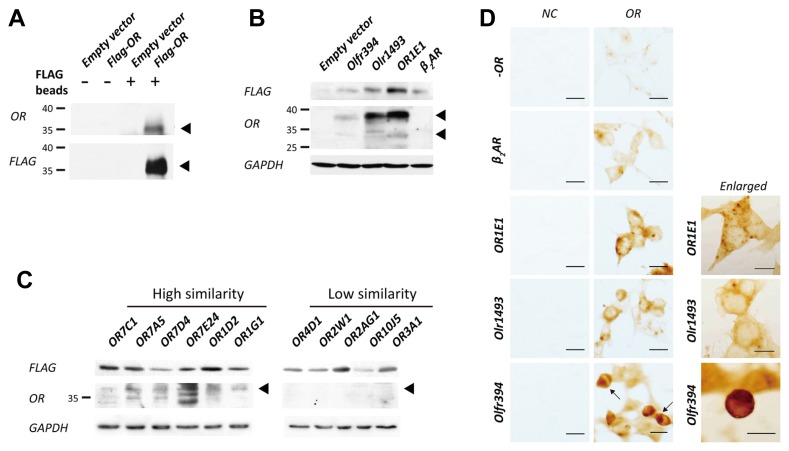
To examine how the conserved motif exerts influence on the functional characteristics of ORs, we generated a TM7-specific peptide antibody targeting the conserved 18 amino acid sequence; we call this region CAS-TM7 (Conserved Amino acid Sequences in TM7). The terminal amino acids M (methionine) and K (lysine) of the motif were replaced with C (cysteine) to allow peptide conjugation to carrier proteins, keyhole limpet hemocyanin (KLH), when generating suitable peptide antigen. Next, we verified if the CAS-TM7 OR antibody recognizes ORs sharing the similar consensus within TM7. We selected and organized ORs according to similarity of the reference peptide sequence, VTPMLNPFIYSLRNRDC (Table 1), and confirmed using the heterologous in vitro system. We first examined if the CAS-TM7 OR antibody is capable of detecting OR1E1 which contains a fully identical CAS-TM7 motif. OR1E1 was transiently expressed in heterologous HEK293 cells and tagged with the N-terminus FLAG sequence (DYKDDDDK) (Fig. 2A). We isolated FLAG-tagged ORs within the cell lysate using M2 FLAG beads and examined its expression using a FLAG-specific antibody versus CAS-TM7-specific OR antibody. The size of the OR band was roughly 35 kDa, similar to the size of the band detected with the FLAG antibody. Since ORs found within the plasma membrane are difficult to detect, we used a lysis buffer with digitonin, a mild non-ionic detergent (Figs. 2B and 2C). The ORs (OR1E1, Olfr394, and Olr1493) that share an identical peptide sequence within the target region resulted in roughly 35 kDa signals, which were similar to the size seen Fig. 2A, however, there are some slight variations among species (Fig. 2B). As seen in Fig. 2B, the antibody seemed capable of detecting human OR relatively well compared to other species, especially mouse OR, which was only weakly detected. To determine the antibodies OR-detection range, we checked its immunoreactivity with human ORs containing similar TM7-motifs with between one (OR7C1) and seven (OR3A1) different amino acids within the targeted peptide sequence (Fig. 2C), and also checked a non-olfactory GPCR, β2-Adrenergic receptor (Fig. 2B). ORs that have highly similar amino acid sequences with the conserved motif showed specific bands near 35 kDa, but as a relative smear for all ORs other than OR1E1. However, the sequence specific antibody hardly detected ORs with low similarity to the conserved motif.
Table 1.
Result of BLASTP with selected conserved sequence of ORs
| Conserved motif in TM7 to C-terminal | |
| VTPMLNPFIYSLRNRDMK | |
|
| |
| Same identity | |
| Olr1493 | VTPMLNPFIYSLRNRDMK |
| Olfr394 | VTPMLNPFIYSLRNRDMK |
| OR1E1 | VTPMLNPFIYSLRNRDMK |
| OR1A1 | VTPMLNPFIYSLRNRDMK |
|
| |
| High similarity | |
| OR7C1 |

|
| OR1C1 |

|
| OR7D4 |

|
| OR1D2 |

|
| OR1E3 |

|
| OR7A5 |

|
| OR1A2 |
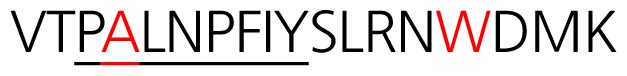
|
| OR1G1 |

|
| OR7E24 |

|
|
| |
| Low similarity | |
| OR4D1 |

|
| OR5V1 |

|
| OR4Q3 |

|
| OR6Q1 |

|
| OR2W1 |

|
| OR10J5 |

|
| OR2AG1 |

|
| OR3A1 |

|
| β2AR |

|
Peptide Similarity Analysis with 18 letters of peptide sequence VTPMLNPFIYSLRNRDMK by using BLASP program (BLASTP 2.6.1+(protein-protein BLAST), and the database is Reference protein (refseq_protein)-http://www.ncbi.nlm.nih.gov/BLAST/). The table showed ORs that contain peptide identity with conserved motif, and different sequences were indicated with red letter.
Fig. 2. Transient heterologous expression of N-terminally FLAG tagged ORs in Hana3A cells.
(A) N-terminally FLAG tagged ORs were captured by M2-magnetic FLAG beads and confirmed through immunoblotting with sequence specific FLAG and OR antibody. Both results showed OR expression at the same size (indicated with black arrows), and compared with empty vector which does not contatin FLAG-OR sequence to support its specificity. (B) The ORs that have peptide sequence VTPMLNPFIYSLRNRDMK in three species were specifically detected with CAS-TM7 OR antibody, but not beta 2 adrenergic receptor (β2AR). (C) As shown in Table 1, the ORs that have high similarity with peptide sequence VTPMLNPFIYSLRNRDMK were specifically detected by CAS-TM7 OR antibody, but ORs that have low similarity were not detected. (D) Immunocytochemistry of surface-expressed ORs with CAS-TM7 OR antibody. Every result was compared with empty vector (−OR) and β2AR to support specificity of OR antibody (Microscope objectives; 20x, scale bars represent 50 μm). The enlarged version of figures of each OR were shown directly next to each OR. Unlike OR1E1 and Olr1493, most cells that express Olfr394 was retained at the endoplasmic reticulum (ER), and hardly surface-expressed (indicated with black arrows). (Microscope objectives 40x-Olfr394; scale bar represents 50 μm, Microscope objectives; 60x – OR1E1 and Olr1493, scale bars represent 50 μm).
To determine if the CAS-TM7 antibody detects surface-expressed ORs, we performed immunocytochemical analysis using Hana3A cells heterologous expressing ORs (Fig. 2D). The OR1E1 and Olr1493 expressed on the plasma membrane of the Hana3A cells were well detected (Fig. 2D), but Olfr394—shown to be expressed within the endoplasmic reticulum (ER) (indicated with arrows)—is rarely found at the plasma membrane (Fig. 2D). To explain our observation of OR retention in the ER, we examined localization of FLAG signal from FLAG tagged ORs in Hana3A cells. FLAG-OR and ER-RFP constructs were co-transfected and visualized 24 h of post-transfection (Supplementary Fig. S3A). Interestingly, each OR showed different localization in the Hana3A cells, and we analyzed the portion of surface-expressed and ER-retained ORs (Supplementary Fig. S3B). The rate of surface expressed OR1E1 and Olr1493 came around 50%, on the other hands, only about 17% of Olfr394 were surfaced expressed showing most of them retained in the ER (Supplementary Fig. S3A-Olfr394). Based on these results, it is able to infer the reason why Olfr394 expression showed a relatively weak band in Fig. 2B.
CAS-TM7 ORs are detected in an in vivo system
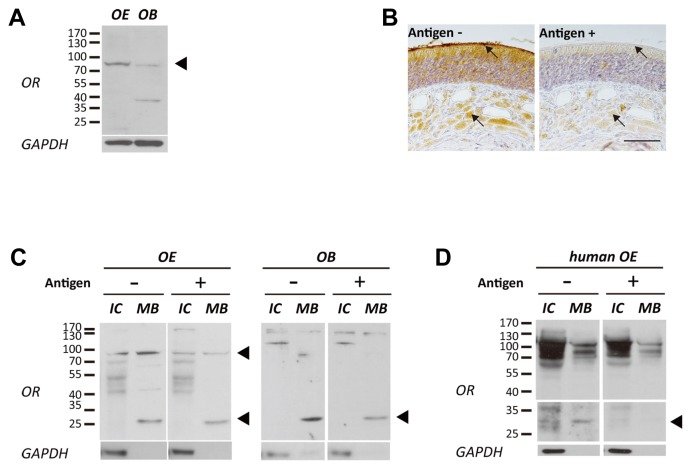
Since the OR antibody presumably detected ORs containing CAS-TM7 in heterologous in vitro system, we also checked whether it is capable of recognizing ORs endogenously expressed in an in vivo system as well. Immunoblot analysis data indicated that ORs containing CAS-TM7 were well-detected in the OE of rats and humans (Figs. 3A and 3D) but not in the OE of mouse (data not shown), corresponding to the in vitro results.
Fig. 3. Expression pattern of PMLNPFIY motif conserved OR in vivo tissues.
(A) The ORs were specifically detected at the size of over 70 kDa in whole lysate of rat OE and OB (Black arrow head indicated). (B) Histological expression pattern of ORs that having target sequences. ORs were observed at the olfactory cilia and nerve bundles of OE tissue (Black arrow points cilia and nerve bundle, Left panel). The specific signals were disappeared after peptide blocking (Antigen conc.;100 ng/ml; Right panel; Scale bar represents 50 μm). (C) Membrane bound (MB) proteins of rat OE and OB were fractionated from intracellular (IC) proteins. Expression of ORs at each sample was confirmed with CAS-TM7 OR antibody which is pre-blocked with or without antigen (Antigen conc.; 1 ng/ml). The intensity of OR specific bands were decreased after pre-incubated with antigen peptide (VTPMLNPFIYSLRNRDC) (Black arrow heads indicate decreased OR bands at OE and OB). (D) Fractionated membrane bound (MB) proteins of human OE also showed expression of ORs at 25~35 kDa and over 70 kDa, and these bands became smear after incubation with peptide pre-blocked CAS-TM7 OR antibody (Antigen conc.; 1 ng/ml).
To characterize the tissue-specific localization of OR expression, we conducted an immunohistochemical analysis in rat adult OE tissue using CAS-TM7 OR specific antibody. ORs were abundantly detected at the olfactory cilia layer and nerve bundles (Fig. 3B). Also, the immunoreactivity (IR) at the olfactory cilia layer and within nerve bundles completely disappeared when the antibody was pre-incubated with an antigen peptide (VTPMLNPFIYSLRNRDC), supporting its specificity (Fig. 3B, right panel). Our results demonstrate that ORs containing CAS-TM7 were expressed in mammalian olfactory tissues and it is feasible to use this CAS-TM7 specific peptide antibody to successfully detect ORs sharing similar conserved amino acid motifs.
To better verify the specific cellular localization of OR proteins, membrane fractionated rat OE and OB lysate were reacted with OR antibody (Fig. 3C). Immunoblot results showed two specific bands (over 70kDa and 25~35kDa) at membrane bound (MB) proteins of OE which has a different pattern with intracellular (IC) proteins, and the intensity of both bands decreased when pre-reacted with antigen antibody (Supplementary Figs. S2A and S2B, and Fig. 3C). However, ORs were only detected at 25~35kDa in membrane bound proteins from the OB. The specific bands for ORs in OE, OB and primary cultured OSNs at same size (ie, 35 kDa), were confirmed through immunoprecipitation and coincided with in vitro results (Supplementary Fig. S2C).
Furthermore, we also examined the expression of ORs in the human OE tissue to confirm the results showing relatively strong IR against human ORs as described in the Fig. 2. The number of specific bands detected in human OE was different than those detected in rats. Noticeably, the size of the most abundant OR band was similar to that of rat ORs (Fig. 3D). In addition, the intensity of these specific bands decreased after peptide blocking (Fig. 3D, right panel).
Histological localization of ORs with a conserved PMLNPFIY sequence
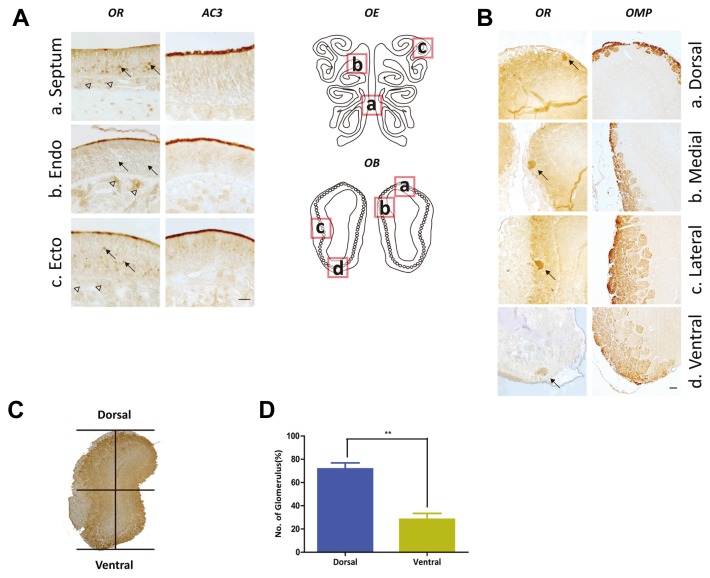
Since the 1990s, there have been conflicting reports about the regional expression of ORs within the olfactory tissue. Vassar R et al. reported a topological map with their expression pattern of OR within the OE could be segregated into 4 zones (Vassar et al., 1993). This corresponds to the glomerulus map in the OB. Since the projection of olfactory sensory neurons (OSNs) expressing a certain OR gene converge into the same glomeruli in the OB, the OB map formation appears to be dependent on the OR (Feinstein and Mombaerts, 2004; Serizawa et al., 2006). Thus, we assumed that the ORs containing conserved amino acid sequences may display similar expression patterns in the olfactory tissue. To examine the histological expression patterns of ORs containing the PMLNPFIY motif, we performed immunohistochemical analysis using rat OE and OB tissues (Fig. 4). The CAS-TM7 ORs were expressed in OSNs at septum, endoturbinate and ectoturbinate regions of the OE. The ORs appeared to be expressed most abundantly at the olfactory cilia layer where ORs used to be located, and the olfactory cilia layer was verified using AC3 antibody (Fig. 4A).
Fig. 4. Expression pattern of sequence specific ORs at OE and OB of rat.
Local information of sequence specific OR expression at rat OE and OB. (A) (a–c) CAS-TM7 ORs were expressed at cilia layer of rat OE septum(a), endoturbinate(b) and ectoturbinate(c) like AC3 (cilia marker protein). Black arrows indicated that CAS-TM7 ORs expressed at cell body of OSNs in the ventral part of septum(a) and ectoturbinate( c). Also, arrow heads pointed at axon bundles of endoturbinate(b) where expressed CAS-TM7 ORs. (B) (a–d) CAS-TM7 ORs were detected at rat glomerulus layer of OB (confirmed with OMP, right panels). The CAS-TM7 ORs that localized at the axon terminus of OSNs were detected at the glomerulus of OB with CAS-TM7 antibody (indicated with black arrows) (Scale bar represents 50 μm). (C–D) Localization of OR expressed glomerulus was analyzed by dividing OB area horizontally. The ORs mostly expressed at the dorsal part of OB (two-tailed t-test, P < 0.05).
Besides the major role of ORs (ie, odorant detection), they have played various roles within the OSNs. Previous studies have indicated that ORs at the axonal part of OSNs are responsible for innervation into each glomeruli at OB (Barnea et al., 2004; Strotmann et al., 2004). We identified the expression pattern of ORs both at the dendritic and axonal part of OSNs (Figs. 4A and 4B). Thereafter, we found that CAS-TM7 OR antibody detected only a limited number of glomeruli among large number of glomerulus which were labeled by the antibody against olfactory marker protein (OMP), a protein found in mature olfactory sensory neurons in various vertebrate species (Margolis et al., 1980). It may reflect that OSNs express limited number of PMLNPFIY conserved ORs (Fig. 4B).
The CAS-TM7 ORs project to the dorsal part of rat OB
Interestingly, we observed differential expression of ORs at each region of OE; those of septum and ectoturbinate were similar but that within the endoturbinate was different. We examined expression of ORs not only in the cilia layer but also in the cell bodies of OSNs at the ectoturbinate and ventral region of septum, which have been classified as zone 3 or 4 in the OE (Miyamichi et al., 2005). In contrast to this pattern, ORs expressed at endoturbinate area (zone 1 or 2 of OE) were predominantly located at the cilia layer and axon bundles, but not within cell bodies (Fig. 4A). These distinct expression patterns may be connected to a topographical map in the OB (Miyamichi et al., 2005). Although numerous studies have demonstrated the important role of ORs in local projection of OSNs to the OB, few studies have been done about the effect of specifically conserved amino acid sequences of ORs in OSN targeting for topographical formation of OB map.
To characterize the effect of CAS-TM7 in OSN projection to specific glomeruli of the OB, we examined local expression of OSN axon terminals in the glomeruli using the CAS-TM7 antibody and analyzing regions of the OR positive glomerulus. We evenly divided the coronal-sectioned OB horizontally into two regions referred to as dorsal and ventral (Fig. 4C). We found that a large number of OR-positive glomerulus were located at the dorsal area. (Fig. 4D). These results demonstrate that OSNs expressing CAS-TM7 ORs may project their axons to distinct areas in the OB, especially the dorsal region.
Broad distribution of ORs in non-olfactory tissue
We next investigated localization of CAS-TM7 ORs in non-olfactory tissues. De la Cruz et al. (2009) showed significant correlation of ectopic expressions in human and chimpanzee suggesting that evolutionary constrained ectopic expression of ORs (De la Cruz et al., 2009). We assumed that ectopic expression of rat ORs would display similar patterns to human ORs. Then, we examined expression of ORs in non-olfactory tissues of rats; testis, liver, kidney, and muscle, according to the our survey which shows CAS-TM7 ORs are most abundantly expressed ectopically in the testis, liver and muscle (Table 2).
Table 2.
Ectopically expressed human ORs that have conserved PMLNPFIY motif.
| ORs | Ectopic expression | Motif at TM7-C-terminal | Motif at TM7-C-terminal (Ortholog gene in rat) | ||
|---|---|---|---|---|---|
|
| |||||
| Tissue/Cell type | Putative function/Signaling | References | |||
| OR1E1 | Cortex/Tongue | Dysregulation in Parkinson’s Disease/Not determined | Garcia-Esparcia et al. (2013)/Gaudin et al. (2001) | VTPMLNPFIYSLRNRDMK | VTPMLNPFIYSLRNRDMK |
| OR1A1 | Enterochromaffin cells/Liver | Serotonin secretion not determined | Braun et al. (2007)/Ichimura et al. (2008) | VTPMLNPFIYSLRNRDMK | VTPMLNPFIYSLRNRDMK |
| OR1A2 | Liver | Not determined | Ichimura et al. (2008) |

|
|
| OR8B3 | Liver | Not determined | Ichimura et al. (2008) |

|

|
| OR7A5 | Testis (spermatozoa)/Tongue | Not determined | Veitinger et al. (2011)/Durzyński et al. (2005) |

|

|
| OR7C1 | Prostate, testis/Brain | Not determined | Flegel et al. (2013) |

|
|
| OR1C1 | Testis | Not determined | Flegel et al. (2013) |

|
|
| OR1D2 | Testis (spermatozoa)/Smooth muscle cells | Chemotaxis/modulate HASMC contractility and the secretion of cytokines | Spehr et al. (2003)/Kalbe et al. (2016) |

|
|
| OR7E24 | Primordial germ cells | Not determined | Goto et al. (2001) |

|
|
| OR4D1 | Testis (spermatozoa) | Not determined | Veitinger et al. (2011) |

|
|
Several known ectopic expressed ORs have conserved PMLNPFIY motif in their TM7, and related references were presented in the table.
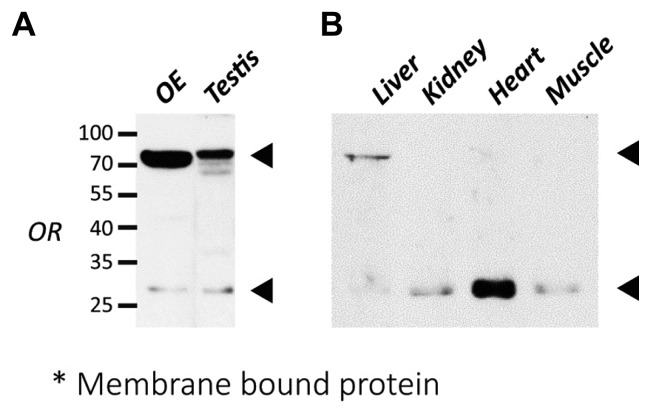
To detect functional ORs expressed in the plasma membrane, we obtained membrane protein lysates from each rat tissue by membrane fractionation, and later confirmed using immunoblot analysis. There were two specific bands observed in the testis and liver (roughly 25–35 kDa and >70 kDa), similar to what was observed in the OE. On the other hand, only one band around 25–35 kDa was observed in the kidney, heart, and muscle, which is comparable to the predicted molecular weight of OR (Figs. 5A and 5B).
Fig. 5. Broad distribution of ORs expression in non-olfactory tissues.
(A) Western blot analysis of OR expression in the testis of rat. CAS-TM7 ORs expression patterns were similar to the OE which observed two specific bands at the size of 25~35 kDa and over 70 kDa. (B) Examination of expression of ORs in the non-olfactory tissues of rat. The expression pattern of ORs in the liver was similar to the OE and testis which showed two specific bands. However, ORs in other tissues such as kidney, heart, and liver showed only one specific band at the size of 25~35 kDa in the membrane fractionated lysates. OR expressions were indicated with arrows.
From these results, we observed strong correlation in the ectopic expression patterns of human and rat ORs because of the evolutionary maintained amino acid motif.
DISCUSSION
The fact that amino acid sequences in a specific system have been evolutionarily conserved across species implies that a certain conserved motif may have critical roles in such system. ORs have several conserved motifs, but their specific roles within the olfactory system have been incompletely identified. In this study, we characterized a highly conserved motif containing PMLNPFIY around TM7 to the C-terminus of ORs of mouse, rat and even human. This conservation may provide insight into its significant functional roles within the olfactory system as previously suggested (Gaillard et al., 2004; Imai et al., 2006; Kato et al., 2008; de March et al., 2015; Nguyen et al., 2007).
Since the OE and OB are the primary places for detecting and discriminating the variety of odorants information, analyzing the topographical location of each OR has been considered as fundamental to understanding olfactory sensory signaling. Thus, our finding that CAS-TM7 ORs within the endoturbinate are predominantly expressed at the cilia layer and axon bundles suggest that these ORs can be regarded as functional ORs (Serizawa et al., 2004). Moreover, the axon terminals of the CAS-TM7 ORs projected to the glomerulus at the dorsal region of OB as previously defined (Miyamichi et al., 2005), suggesting the CAS-TM7 ORs may play a critical role in the processing of odorant information. Intriguingly, our immunohistochemistry results showed that CAS-TM7 ORs within the ectoturbinate and septum were mostly found in the cell body of the OSNs, indicating that the CAS-TM7 ORs may be found within immature neurons because CAS-TM7 ORs were not observed at the ventral region of OB. Hanchate and colleagues demonstrated that immature OSNs express multiple OR genes compared to mature OSNs that express one gene among about 1000 OR genes of mice. We assumed that CAS-TM7 ORs may not be selected to express mainly in the mature neurons in the septum and ectoturbinate of OE (Hanchate et al., 2015), suggesting the involvement of the CAS-TM7 ORs in sorting of OSNs and glomerulus mapping.
It has been known that OR-derived cAMP signals regulate targeting of OSNs along the anterior-posterior (A-P) axis of OB, however, the dorsal-ventral (D-V) projection is correlated with the anatomic location of OSNs within the OE (Miyamich et al., 2005; Ressler et al., 1994; Takeuchi et al., 2010; Vassar et al., 1993). Therefore, we can conclude that topographic region of CAS-TM7 ORs within OE and also their projection to dorsal OB are fated. Interestingly, the dorsal region of OB is known to be related to the innate response of rodents which is critical for survival (Kobayakawa et al., 2007). Considering the significant function of dorsal OB, we additionally assumed that some ORs of this region should be evolutionarily maintained to effectively perform their role. Our finding suggests the topographical fate of CAS-TM7 OR expression, which corresponds well to our further hypothesis.
From our investigation of the ectopic expression patterns of CAS-TM7 ORs in the non-olfactory system, we confirmed the expression of CAS-TM7 in germ cells where the first ectopically expressed ORs were discovered. Numerous studies have already shown that OR proteins are substantially expressed in mature spermatozoa of testis in various species (Asai et al., 1996; Flegel et al., 2015; Neuhaus et al., 2006; Vanderhaeghen et al., 1993; Walensky et al., 1995). Messenger RNA localization of testicular ORs in early spematozoa was identified (Fukuda and Touhara, 2006), but little is known at the protein level in the developing spermatocytes. We first demonstrated the presence of CAS-TM7 OR proteins in developing spermatocytes (data not shown). Since previous studies elucidated that sertoli cells interact with germ cells to support development of spermatocytes and movement of spermatozoa by releasing cytokines and neurotropins (Park et al., 2001; Persson et al., 1990), we assumed that ORs expressed in germ cells modulate signals from sertoli cells and such functions may be utterly different from that of the OSNs. Thus, the CAS-TM7 ORs may play additional roles in spermatogenesis.
Taken together, we found that specific conserved amino acid sequences in ORs, i.e, CAS-TM7 ORs, seemed evolutionarily constrained. The expression pattern in the OE and OB were also distinct, implying a critical role in odorant-information processing. Although we could not cover all ORs containing conserved amino acid sequences at TM7, we can still suggest a critical effect of the PMLNPFIY conserved motif in the formation of certain ORs features.
Supplementary data
ACKNOWLEDGMENTS
This work was supported by DGIST Convergence Science Center (16-BD-0402).
Footnotes
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai H., Kasai H., Matsuda Y., Yamazaki N., Nagawa F., Sakano H., Tsuboi A. Genomic structure and transcription of a murine odorant receptor gene: differential initiation of transcription in the olfactory and testicular cells. Biochem Biophys Res Commun. 1996;221:240–247. doi: 10.1006/bbrc.1996.0580. [DOI] [PubMed] [Google Scholar]
- Barnea G., O’Donnell S., Mancia F., Sun X., Nemes A., Mendelsohn M., Axel R. Odorant receptors on axon termini in the brain. Science. 2004;304:1468. doi: 10.1126/science.1096146. [DOI] [PubMed] [Google Scholar]
- Braun T., Voland P., Kunz L., Prinz C., Gratzl M. Enterochromaffin cells of the human gut: sensors for spices and odorants. Gastroenterology. 2007;132:1890–1901. doi: 10.1053/j.gastro.2007.02.036. [DOI] [PubMed] [Google Scholar]
- Buck L., Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- Busse D., Kudella P., Gruning N.M., Gisselmann G., Stander S., Luger T., Jacobsen F., Steinstrasser L., Paus R., Gkogkolou P., et al. A synthetic sandalwood odorant induces wound-healing processes in human keratinocytes via the olfactory receptor OR2AT4. J Invest Dermatol. 2014;134:2823–2832. doi: 10.1038/jid.2014.273. [DOI] [PubMed] [Google Scholar]
- De la Cruz O., Blekhman R., Zhang X., Nicolae D., Firestein S., Gilad Y. A signature of evolutionary constraint on a subset of ectopically expressed olfactory receptor genes. Mol Biol Evol. 2009;26:491–494. doi: 10.1093/molbev/msn294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de March C.A., Kim S.K., Antonczak S., Goddard W.A.3rdGolebiowski J. G protein-coupled odorant receptors: From sequence to structure. Protein Sci. 2015;24:1543–1548. doi: 10.1002/pro.2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durzynski L., Gaudin J.C., Myga M., Szydlowski J., Gozdzicka-Jozefiak A., Haertle T. Olfactory-like receptor cDNAs are present in human lingual cDNA libraries. Biochem Biophys Res Commun. 2005;333:264–272. doi: 10.1016/j.bbrc.2005.05.085. [DOI] [PubMed] [Google Scholar]
- Feinstein P., Mombaerts P. A contextual model for axonal sorting into glomeruli in the mouse olfactory system. Cell. 2004;117:817–831. doi: 10.1016/j.cell.2004.05.011. [DOI] [PubMed] [Google Scholar]
- Feldmesser E., Olender T., Khen M., Yanai I., Ophir R., Lancet D. Widespread ectopic expression of olfactory receptor genes. BMC Genomics. 2006;7:121. doi: 10.1186/1471-2164-7-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flegel C., Manteniotis S., Osthold S., Hatt H., Gisselmann G. Expression profile of ectopic olfactory receptors determined by deep sequencing. PLoS One. 2013;8:e55368. doi: 10.1371/journal.pone.0055368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flegel C., Vogel F., Hofreuter A., Schreiner B.S., Osthold S., Veitinger S., Becker C., Brockmeyer N.H., Muschol M., Wennemuth G., et al. Characterization of the Olfactory Receptors Expressed in Human Spermatozoa. Front Mol Biosci. 2015;2:73. doi: 10.3389/fmolb.2015.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda N., Touhara K. Developmental expression patterns of testicular olfactory receptor genes during mouse spermatogenesis. Genes Cells. 2006;11:71–81. doi: 10.1111/j.1365-2443.2005.00915.x. [DOI] [PubMed] [Google Scholar]
- Gaillard I., Rouquier S., Chavanieu A., Mollard P., Giorgi D. Amino-acid changes acquired during evolution by olfactory receptor 912–93 modify the specificity of odorant recognition. Hum Mol Genet. 2004;13:771–780. doi: 10.1093/hmg/ddh086. [DOI] [PubMed] [Google Scholar]
- Garcia-Esparcia P., Schluter A., Carmona M., Moreno J., Ansoleaga B., Torrejon-Escribano B., Gustincich S., Pujol A., Ferrer I. Functional genomics reveals dysregulation of cortical olfactory receptors in Parkinson disease: novel putative chemoreceptors in the human brain. J Neuropathol Exp Neurol. 2013;72:524–539. doi: 10.1097/NEN.0b013e318294fd76. [DOI] [PubMed] [Google Scholar]
- Gaudin J.C., Breuils L., Haertle T. New GPCRs from a human lingual cDNA library. Chem Senses. 2001;26:1157–1166. doi: 10.1093/chemse/26.9.1157. [DOI] [PubMed] [Google Scholar]
- Goto T., Salpekar A., Monk M. Expression of a testis-specific member of the olfactory receptor gene family in human primordial germ cells. Mol Hum Reprod. 2001;7:553–558. doi: 10.1093/molehr/7.6.553. [DOI] [PubMed] [Google Scholar]
- Gu X., Karp P.H., Brody S.L., Pierce R.A., Welsh M.J., Holtzman M.J., Ben-Shahar Y. Chemosensory functions for pulmonary neuroendocrine cells. Am J Respir Cell Mol Biol. 2014;50:637–646. doi: 10.1165/rcmb.2013-0199OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanchate N.K., Kondoh K., Lu Z., Kuang D., Ye X., Qiu X., Pachter L., Trapnell C., Buck L.B. Single-cell transcriptomics reveals receptor transformations during olfactory neurogenesis. Science. 2015;350:1251–1255. doi: 10.1126/science.aad2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura A., Kadowaki T., Narukawa K., Togiya K., Hirasawa A., Tsujimoto G. In silico approach to identify the expression of the undiscovered molecules from microarray public database: identification of odorant receptors expressed in non-olfactory tissues. Naunyn Schmiedebergs Arch Pharmacol. 2008;377:159–165. doi: 10.1007/s00210-007-0255-6. [DOI] [PubMed] [Google Scholar]
- Imai T., Suzuki M., Sakano H. Odorant receptor-derived cAMP signals direct axonal targeting. Science. 2006;314:657–661. doi: 10.1126/science.1131794. [DOI] [PubMed] [Google Scholar]
- Kaji I., Karaki S., Kuwahara A. Effects of luminal thymol on epithelial transport in human and rat colon. Am J Physiol Gastrointest Liver Physiol. 2011;300:G1132–1143. doi: 10.1152/ajpgi.00503.2010. [DOI] [PubMed] [Google Scholar]
- Kalbe B., Knobloch J., Schulz V.M., Wecker C., Schlimm M., Scholz P., Jansen F., Stoelben E., Philippou S., Hecker E., et al. Olfactory Receptors Modulate Physiological Processes in Human Airway Smooth Muscle Cells. Front Physiol. 2016;7:339. doi: 10.3389/fphys.2016.00339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato A., Katada S., Touhara K. Amino acids involved in conformational dynamics and G protein coupling of an odorant receptor: targeting gain-of-function mutation. J Neurochem. 2008;107:1261–1270. doi: 10.1111/j.1471-4159.2008.05693.x. [DOI] [PubMed] [Google Scholar]
- Kidd M., Modlin I.M., Gustafsson B.I., Drozdov I., Hauso O., Pfragner R. Luminal regulation of normal and neoplastic human EC cell serotonin release is mediated by bile salts, amines, tastants, and olfactants. Am J Physiol Gastrointest Liver Physiol. 2008;295:G260–272. doi: 10.1152/ajpgi.00056.2008. [DOI] [PubMed] [Google Scholar]
- Kobayakawa K., Kobayakawa R., Matsumoto H., Oka Y., Imai T., Ikawa M., Okabe M., Ikeda T., Itohara S., Kikusui T., et al. Innate versus learned odour processing in the mouse olfactory bulb. Nature. 2007;450:503–508. doi: 10.1038/nature06281. [DOI] [PubMed] [Google Scholar]
- Margolis F. A marker protein for the olfactory chemoreceptor neuron. In: Bradshaw R.A., Schneider D., editors. Proteins of the nervous system. Newyork, America: Raven Press; 1980. pp. 59–84. [Google Scholar]
- Massberg D., Simon A., Haussinger D., Keitel V., Gisselmann G., Conrad H., Hatt H. Monoterpene (-)-citronellal affects hepatocarcinoma cell signaling via an olfactory receptor. Arch Biochem Biophys. 2015;566:100–109. doi: 10.1016/j.abb.2014.12.004. [DOI] [PubMed] [Google Scholar]
- Miyamichi K., Serizawa S., Kimura H.M., Sakano H. Continuous and overlapping expression domains of odorant receptor genes in the olfactory epithelium determine the dorsal/ventral positioning of glomeruli in the olfactory bulb. J Neurosci. 2005;25:3586–3592. doi: 10.1523/JNEUROSCI.0324-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhaus E.M., Mashukova A., Barbour J., Wolters D., Hatt H. Novel function of beta-arrestin2 in the nucleus of mature spermatozoa. J Cell Sci. 2006;119:3047–3056. doi: 10.1242/jcs.03046. [DOI] [PubMed] [Google Scholar]
- Neuhaus E.M., Zhang W., Gelis L., Deng Y., Noldus J., Hatt H. Activation of an olfactory receptor inhibits proliferation of prostate cancer cells. J Biol Chem. 2009;284:16218–16225. doi: 10.1074/jbc.M109.012096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen M.Q., Zhou Z., Marks C.A., Ryba N.J., Belluscio L. Prominent roles for odorant receptor coding sequences in allelic exclusion. Cell. 2007;131:1009–1017. doi: 10.1016/j.cell.2007.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal K., Badgandi H., Mukhopadhyay S. Studying G protein-coupled receptors: immunoblotting, immunoprecipitation, phosphorylation, surface labeling, and cross-linking protocols. Methods Cell Biol. 2015;127:303–322. doi: 10.1016/bs.mcb.2014.12.003. [DOI] [PubMed] [Google Scholar]
- Palczewski K., Kumasaka T., Hori T., Behnke C.A., Motoshima H., Fox B.A., Le Trong I., Teller D.C., Okada T., Stenkamp R.E., et al. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- Park C., Choi W.S., Kwon H., Kwon Y.K. Temporal and spatial expression of neurotrophins and their receptors during male germ cell development. Mol Cells. 2001;12:360–367. [PubMed] [Google Scholar]
- Persson H., Ayer-Le Lievre C., Soder O., Villar M.J., Metsis M., Olson L., Ritzen M., Hokfelt T. Expression of beta-nerve growth factor receptor mRNA in Sertoli cells downregulated by testosterone. Science. 1990;247:704–707. doi: 10.1126/science.2154035. [DOI] [PubMed] [Google Scholar]
- Pilpel Y., Lancet D. The variable and conserved interfaces of modeled olfactory receptor proteins. Protein Sci. 1999;8:969–977. doi: 10.1110/ps.8.5.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst W.C., Snyder L.A., Schuster D.I., Brosius J., Sealfon S.C. Sequence alignment of the G-protein coupled receptor superfamily. DNA Cell Biol. 1992;11:1–20. doi: 10.1089/dna.1992.11.1. [DOI] [PubMed] [Google Scholar]
- Ressler K.J., Sullivan S.L., Buck L.B. Information coding in the olfactory system: evidence for a stereotyped and highly organized epitope map in the olfactory bulb. Cell. 1994;79:1245–1255. doi: 10.1016/0092-8674(94)90015-9. [DOI] [PubMed] [Google Scholar]
- Ronnett G.V., Hester L.D., Snyder S.H. Primary culture of neonatal rat olfactory neurons. J Neurosci. 1991;11:1243–1255. doi: 10.1523/JNEUROSCI.11-05-01243.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serizawa S., Miyamichi K., Sakano H. One neuron-one receptor rule in the mouse olfactory system. Trends Genet. 2004;20:648–653. doi: 10.1016/j.tig.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Serizawa S., Miyamichi K., Takeuchi H., Yamagishi Y., Suzuki M., Sakano H. A neuronal identity code for the odorant receptor-specific and activity-dependent axon sorting. Cell. 2006;127:1057–1069. doi: 10.1016/j.cell.2006.10.031. [DOI] [PubMed] [Google Scholar]
- Sherman M.A., Lesne S.E. Detecting abeta*56 oligomers in brain tissues. Methods Mol Biol. 2011;670:45–56. doi: 10.1007/978-1-60761-744-0_4. [DOI] [PubMed] [Google Scholar]
- Spehr J., Gelis L., Osterloh M., Oberland S., Hatt H., Spehr M., Neuhaus E.M. G protein-coupled receptor signaling via Src kinase induces endogenous human transient receptor potential vanilloid type 6 (TRPV6) channel activation. J Biol Chem. 2011;286:13184–13192. doi: 10.1074/jbc.M110.183525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spehr M., Gisselmann G., Poplawski A., Riffell J.A., Wetzel C.H., Zimmer R.K., Hatt H. Identification of a testicular odorant receptor mediating human sperm chemotaxis. Science. 2003;299:2054–2058. doi: 10.1126/science.1080376. [DOI] [PubMed] [Google Scholar]
- Strotmann J., Levai O., Fleischer J., Schwarzenbacher K., Breer H. Olfactory receptor proteins in axonal processes of chemosensory neurons. J Neurosci. 2004;24:7754–7761. doi: 10.1523/JNEUROSCI.2588-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi H., Inokuchi K., Aoki M., Suto F., Tsuboi A., Matsuda I., Suzuki M., Aiba A., Serizawa S., Yoshihara Y., et al. Sequential arrival and graded secretion of Sema3F by olfactory neuron axons specify map topography at the bulb. Cell. 2010;141:1056–1067. doi: 10.1016/j.cell.2010.04.041. [DOI] [PubMed] [Google Scholar]
- Vanderhaeghen P., Schurmans S., Vassart G., Parmentier M. Olfactory receptors are displayed on dog mature sperm cells. J Cell Biol. 1993;123:1441–1452. doi: 10.1083/jcb.123.6.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassar R., Ngai J., Axel R. Spatial segregation of odorant receptor expression in the mammalian olfactory epithelium. Cell. 1993;74:309–318. doi: 10.1016/0092-8674(93)90422-m. [DOI] [PubMed] [Google Scholar]
- Veitinger T., Riffell J.R., Veitinger S., Nascimento J.M., Triller A., Chandsawangbhuwana C., Schwane K., Geerts A., Wunder F., Berns M.W., et al. Chemosensory Ca2+ dynamics correlate with diverse behavioral phenotypes in human sperm. J Biol Chem. 2011;286:17311–17325. doi: 10.1074/jbc.M110.211524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walensky L.D., Roskams A.J., Lefkowitz R.J., Snyder S.H., Ronnett G.V. Odorant receptors and desensitization proteins colocalize in mammalian sperm. Mol Med. 1995;1:130–141. [PMC free article] [PubMed] [Google Scholar]
- Wu C., Jia Y., Lee J.H., Kim Y., Sekharan S., Batista V.S., Lee S.J. Activation of OR1A1 suppresses PPAR-gamma expression by inducing HES-1 in cultured hepatocytes. Int J Biochem Cell Biol. 2015;64:75–80. doi: 10.1016/j.biocel.2015.03.008. [DOI] [PubMed] [Google Scholar]
- Zhang X., De la Cruz O., Pinto J.M., Nicolae D., Firestein S., Gilad Y. Characterizing the expression of the human olfactory receptor gene family using a novel DNA microarray. Genome Biol. 2007;8:R86. doi: 10.1186/gb-2007-8-5-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W., Ho L., Varghese M., Yemul S., Dams-O’Connor K., Gordon W., Knable L., Freire D., Haroutunian V., Pasinetti G.M. Decreased level of olfactory receptors in blood cells following traumatic brain injury and potential association with tauopathy. J Alzheimers Dis. 2013;34:417–429. doi: 10.3233/JAD-121894. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.