Abstract
Sarcopenia, the age-related loss of skeletal muscle mass and strength, is a significant cause of morbidity in the elderly and is a major burden on health care systems. Unfortunately, the underlying molecular mechanisms in sarcopenia remain poorly understood. Herein, we utilized top-down proteomics to elucidate sarcopenia-related changes in the fast- and slow-twitch skeletal muscles of aging rats with a focus on the sarcomeric proteome, which includes both myofilament and Z-disc proteins—the proteins that constitute the contractile apparatuses. Top-down quantitative proteomics identified significant changes in the post-translational modifications (PTMs) of critical myofilament proteins in the fast-twitch skeletal muscles of aging rats, in accordance with the vulnerability of fast-twitch muscles to sarcopenia. Surprisingly, age-related alterations in the phosphorylation of Cypher isoforms, proteins that localize to the Z-discs in striated muscles, were also noted in the fast-twitch skeletal muscle of aging rats. This represents the first report of changes in the phosphorylation of Z-disc proteins in skeletal muscle during aging. In addition, increased glutathionylation of slow skeletal troponin I, a novel modification that may help protect against oxidative damage, was observed in slow-twitch skeletal muscles. Furthermore, we have identified and characterized novel muscle type-specific proteoforms of myofilament proteins and Z-disc proteins, including a novel isoform of the Z-disc protein Enigma. The finding that the phosphorylation of Z-disc proteins is altered in response to aging in the fast-twitch skeletal muscles of aging rats opens new avenues for the investigation of the role of Z-discs in age-related muscle dysfunction.
Sarcopenia, the loss of skeletal muscle mass and function (i.e. strength) with aging, is a significant cause of morbidity and disability in the elderly population (1–3). Greater than 35% of persons over 65 years of age and 50% or more of individuals 80 years or older are estimated to be afflicted with sarcopenia (4). Moreover, the sarcopenia-associated burden on healthcare is substantial with an estimated cost of $18.5 billion in the United States in the year 2000 alone (5); and costs are expected to rise because of aging of the population (6). Nevertheless, despite the high prevalence and devastating economic impact, among types of systemic muscle loss and weakness, sarcopenia represents one of the least well understood (1).
Although muscle mass declines with age, largely because of the atrophy and loss of type II (fast-twitch) skeletal muscle fibers whereas type I (slow-twitch) fibers have traditionally been thought to remain relatively unaffected (7, 8), the loss of muscle strength in elderly individuals cannot be entirely explained by the age-associated loss of muscle mass (9). Moreover, even though metabolic perturbations and cell damage resulting from age-related mitochondrial dysfunction undoubtedly contribute to muscle dysfunction with increasing age (10), it is now well-recognized that sarcopenia is associated with a loss of intrinsic contractile function that is due, in part, to alterations in myofilaments (11). Myofilaments are components of sarcomeres, the basic contractile units of muscle; and are composed of the thin filament proteins actin, tropomyosin (Tpm)1, and the troponin complex, which includes troponin I (TnI), troponin T (TnT), and troponin C (TnC), as well as the thick filament proteins myosin heavy chain (MHC), regulatory light chain, essential light chain, and myosin binding protein C. The myofilaments are flanked on either side by protein dense structures known as Z-discs that, together with the myofilaments constitute sarcomeres, which are responsible for mediating muscle contraction at high levels of intracellular Ca2+ (12, 13).
In addition to regulation by Ca2+, which serves as the ultimate trigger for muscle contraction, myofilament protein-protein interactions (and consequently contractile function) are also regulated via the post-translational modification (PTM) of myofilaments, as well as changes in the expression of contractile protein isoforms (12, 13). Evidence in the literature suggests that the oxidative modification of myofilaments may underlie altered contractile function in the skeletal muscles of aging individuals (14). Specifically, the PTM of reactive cysteines in MHC has been shown to increase with age and correlates with decreased actin-activated myosin ATPase activity in aged rat skeletal muscles (15). In agreement with findings in human skeletal muscle (16), we recently identified an age-related decrease in the phosphorylation of RLC in fast-twitch skeletal muscles of aging rats that can account for sarcopenic muscle functional impairments (17). However, a more comprehensive assessment of changes in other contractile proteins in the fast-twitch skeletal muscles of aging individuals remains lacking. Moreover, emerging evidence indicates that slow skeletal muscles may also be vulnerable to sarcopenia at advanced age (18) and, thus, the examination of changes in contractile protein PTMs during the aging process in these muscles, which may contribute to age-related alterations in contractile function, will be imperative for understanding how sarcopenia impacts muscle function in slow-twitch skeletal muscles.
Top-down mass spectrometry (MS)-based proteomics, in which intact proteins are analyzed rather than peptides as in bottom-up proteomics, has supplanted traditional methods of protein analysis as the most powerful method for the comprehensive characterization of proteoforms (a term encompassing the myriad protein forms arising from a single gene, including post-translationally modified forms and those with sequence alterations because of mutations/polymorphisms and/or alternative splicing) (19). The analysis of intact proteoforms in top-down proteomics provides a “bird's eye” view of all protein sequence variations and PTMs, which can subsequently be localized using a variety of tandem MS (MS/MS) techniques, such as collision induced dissociation and electron capture dissociation (ECD) (20–22). Top-down MS in combination with ECD, in particular, is useful for the localization of labile PTMs (e.g. glycosylation and phosphorylation) (23–31), which are preferentially lost when proteins are dissociated with slow heating methods such as collision induced dissociation (24). Moreover, given that PTMs such as phosphorylation and minor sequence variations do not significantly impact the ionization efficiency of intact proteins as they do with peptides (32), top-down proteomics is inherently semiquantitative with respect to the relative abundances of protein proteoforms observed within the same spectrum (20–22). Indeed, our group and others have employed top-down MS to identify proteoform alterations toward a better understanding of the molecular mechanisms of disease (25–27, 33).
In this study, we used quantitative top-down proteomics to gain insights into age-related molecular changes in fast- and slow-twitch skeletal muscles, with a particular emphasis on changes occurring in the sarcomeric proteome. Interestingly, in addition to the identification of the major muscle type-specific isoforms of myofilament proteins, we detected and characterized several Z-disc proteins, including two isoforms of Cypher, one of which has previously only been detected at the mRNA level, as well as a novel isoform of the protein Enigma. Consistent with the well-established susceptibility of fast-twitch muscle fibers to sarcopenia, top-down quantitative proteomics identified significant changes in the PTMs of several important myofilament proteins in the fast-twitch skeletal muscles of aging rats. Interestingly, although we detected few changes in myofilament protein PTMs in the slow-twitch skeletal muscles of aging rats, a significant increase in the glutathionylation of slow skeletal troponin I (ssTnI), a modification that may help protect against oxidative damage and contribute to differences in the presentation of sarcopenia in fast- and slow-twitch skeletal muscles, was noted. Top-down proteomics further enabled the identification of novel phosphorylated proteoforms of fast- and slow-skeletal troponin I. For the first time, we also provide evidence that the phosphorylation of Z-disc proteins is altered with age in fast-twitch skeletal muscles.
EXPERIMENTAL PROCEDURES
Animals
Male Fisher 344 × Brown Norway F1 hybrid (F344BN) rats aged 6- (n = 12), 24- (n = 12), and 36-months (n = 12) were obtained from the National Institute on Aging colony maintained by Harlan Sprague-Dawley (Indianapolis, IN). Rats were individually housed in clear plastic cages on a 12 h/12 h light/dark cycle with access to food and water ad libitum. Handling and euthanasia were carried out under the guidelines of the University of Wisconsin-Madison Animal Use and Care Committee.
Muscle Measurements
Rats were anesthetized by inhalation of isoflurane, and the gastrocnemius (GAS) and soleus (SOL) muscles from the left hind limb were dissected from origin to insertion and immediately weighed. Muscles were bisected at the mid-belly, embedded in optimal cutting temperature compound (Tissue-Tek; Andwin Scientific, Addison, IL), frozen in liquid N2, and stored at −80 °C for later sectioning. Three consecutive sections (10 μm thick) were cut, starting at the mid-belly, placed on labeled Probe-On Plus microscope slides (Fisher Scientific, Pittsburgh, PA), and stored at −80 °C until use. The first section of each series was stained with hematoxylin and eosin. Sections were photographed using an Olympus BH2 microscope (Olympus, Tokyo, Japan) with an Olympus DP70 digital camera and mid-belly composites of each muscle section were reconstructed by interlacing the images using ImagePro Plus software (Media Cybernetics, Atlanta, GA). For fiber counts, individual muscle fibers were annotated on the composite image of the entire muscle cross-section at the mid-belly, using Adobe Photoshop (Adobe Systems, Inc., San Jose, CA), and total count was tabulated. The whole muscle cross-sectional area (CSA) at the mid-belly was measured by tracing an outline of each muscle using ImagePro Plus. To measure individual muscle fiber CSA, four images (10×) from the H&E sections were captured from GAS and SOL muscles from rats in each age group. A grid with 25 random dots was placed over the images, and the CSA of fibers marked with the dots was measured. Six hundred fibers were measured from each muscle at each age ((4 images per muscle) × (25 fibers per image) × (6 animals per age group) = 600 fibers). GAS and SOL muscles were dissected from the right hind limb as described above, but were immediately flash frozen in liquid N2 and stored at −80 °C for subsequent proteomic analysis.
MHC Isoform and ATPase Staining
For ATPase staining, 10 μm-thick, transverse frozen sections of muscle were cut using a cryostat (−20 °C) and placed on Probe-On Plus slides (Fisher Scientific). Staining for myosin ATPase to determine fiber type distribution was performed according to Hintz et al. (34). Fiber type was determined based on myosin ATPase sensitivity to pH and differences in fiber type staining intensities. Preincubation at pH 10.2 distinguishes between type I (light staining) and type II (dark staining) fibers. At acidic pH (4.5 and 4.25), type I fibers will stain dark. Type II fibers can be further classified into IIa (stain light in both pH 4.5 and 4.25), IIb (stain medium at pH 4.5 but light at pH 4.25), and IIc (stain intermediate at both pH 4.5 and 4.25) fibers. The preincubation solutions were: (1) 20 mm sodium barbital, 18 mm calcium chloride at pH 10.2 for 15 min or (2 and 3) 50 mm sodium acetate, 30 mm sodium barbital at pH 4.5 or 4.25 for 6 min. All sections were then incubated for 30 min at 37 °C in 20 mm sodium barbital, 9 mm calcium chloride, and 2.7 mm ATP at pH 9.4. Sections were subsequently rinsed in 1% calcium chloride (3 × 3 min each), immersed in 2% cobaltum chloride (3 min), rinsed in 10 changes of tap H2O, stained in 1% ammonium sulfide (30 s), washed in tap running H2O (4 min), dehydrated with ethanol, cleared in xylene, and mounted in Permount (Fisher, Fair Lawn, NJ).
Preparation of Sarcomeric Protein-Enriched Extracts
Protein extraction was carried out as previously described (26). Briefly, ∼10 mg of tissue from the GAS (n = 6 for each age group) or SOL (n = 6 for each age group) muscles was homogenized in 100 μl of HEPES extraction buffer containing protease and phosphatase inhibitors (25 mm HEPES pH 7.5, 50 mm NaF, 0.25 mm Na3VO4, 0.25 mm PMSF, 2.5 mm EDTA) using a Teflon pestle (1.5 ml tube rounded tip; Scienceware, Pequannock, NJ). The resulting homogenate was centrifuged at 16,000 × g for 15 min at 4 °C, and the supernatant was discarded. The insoluble pellet was then re-homogenized in 100 μl of TFA extraction buffer (1% trifluoroacetic acid, 1 mm tris(2-carboxyethyl)phosphine) to extract a protein sub-proteome enriched in sarcomeric proteins. After centrifugation (16,000 × g, 4 °C, 25 min), the supernatant was collected and used for on-line top-down proteomic analysis. At least two technical (extraction) replicates were performed per biological replicate.
On-Line Liquid Chromatography (LC)-MS Profiling
Protein extracts prepared from aging rat GAS and SOL muscles were separated using a nanoACQUITY LC system (Waters, Milford, MA) equipped with a home-packed PLRP column (PLRP-S, 200 mm × 500 μm, 10 μm, 1000 Å; Varian, Lake Forest, CA) and a gradient going from 20% B to 90% B (solvent A: 0.10% formic acid in water; solvent B: 0.10% formic acid in a 50:50 mixture of acetonitrile and ethanol) over 40 min at a flow rate of 8 μl/min. The nanoACQUITY LC system was coupled on-line with an impact II quadrupole-time-of-flight (q-TOF) mass spectrometer (Bruker Daltonics, Billerica, MA). Spectra were collected using a scan rate of 1 Hz over a 500–3000 m/z range. Sarcomeric protein separation and the relative abundances of modified and un-modified protein proteoforms were highly reproducible across replicates (supplemental Figs. S1–S2).
Fraction Collection
LC separation of GAS and SOL extracts, and fraction collection of both known and unknown proteins, was carried out as previously described (26), with minor modifications. Briefly, GAS and SOL extracts containing the sarcomeric sub-proteomes were separated using a 2D-nano-LC system (Eksigent, Redwood City, CA) equipped with a home-packed PLRP column and a gradient going from 20% B to 90% B over 40 min (solvent A: 0.10% formic acid in water; solvent B: 0.10% formic acid in a 50:50 mixture of acetonitrile and ethanol) at a flow rate of 12.5 μl/min. The 2D-nano-LC system was coupled on-line with a linear ion trap (LTQ) mass spectrometer (Thermo Scientific). After LC separation, a small portion of the sample (∼5% of the total amount) was ionized by electrospray ionization through a 25–30 μm i.d. tip and analyzed by LTQ/MS to track protein elution from the column. The remaining ∼95% of the sample was collected on ice for comprehensive protein analysis by off-line MS/MS.
High-Resolution MS/MS Protein Characterization
The collected fractions were analyzed using a 7T LTQ/Fourier transform ion cyclotron resonance (FT-ICR) mass spectrometer (LTQ/FT Ultra, Thermo Scientific, Bremen, Germany) equipped with an automated chip-based nano-electrospray ionization source (Triversa NanoMate; Advion Bioscience, Ithaca, NY, USA) as described previously (24–27). The sample was introduced into the mass spectrometer using a spray voltage of 1.3 kV. Ion transmission into the LTQ and, subsequently, the FT-ICR cell, was optimized to achieve maximum ion signal. The resolving power of the FT-ICR was set at 200,000 (at 400 m/z). The number of accumulated charges for a full scan in the LTQ, FT-ICR cell, MSn FT-ICR cell, and ECD were 3 × 104, 8 × 106, 8 × 106 and 8 × 106, respectively. For MS/MS experiments, the protein molecular ions of the individual charge states were first isolated and subsequently fragmented using ECD. The energy, delay, and duration parameters for ECD, were determined on a case-by-case basis to achieve optimal fragmentation of precursor ions. Typically between 1000 and 5000 scans were averaged for MS/MS experiments to ensure the collection of high quality tandem mass spectra for data analysis.
Experimental Design and Statistical Rationale
Muscle and body weight measurements in rats aged 6-, 24-, and 36-months were performed with 12 animals (biological replicates) per group. Quantitative top-down proteomics analysis was carried out with 6 animals (biological replicates) per age group with two technical replicates per sample to ensure accurate measurement of proteoform relative abundances in the fast- and slow-twitch skeletal muscles of individual animals.
R was used for the statistical analysis. For each protein, significant differences among the 6-, 24-, and 36-month age groups were calculated using the Kruskal-Wallis rank sum test and the resulting p values were adjusted using the Benjamini-Hochberg algorithm. For groups with significant differences, pairwise differences between the 6-, 24-, and 36-month-old age groups were determined using a post-hoc two-sample Wilcoxon (Mann-Whitney) test. p values less than 0.05 were considered statistically significant.
All mass and tandem mass spectra were analyzed with in-house developed MASH Suite Pro software (35), which is an integrated software with MS-Align+ (36), using a signal-to-noise ratio threshold of 2.5 and a minimum fit of 60%. A 15 ppm cut-off was used for fragment ion assignments and all program-processed data was manually validated. For protein identification, MASH Suite Pro was used to create a mass list from the raw MS/MS data, and the list was subsequently searched against a rat database generated from the UniProtKB/Swiss-Prot knowledgebase (Rattus norvegicus, proteome ID UP000002494, 29,979 protein sequences) using the MS-Align+ search engine incorporated into MASH Suite Pro (35). The maximum number of unexpected changes was set to 2 and the acceptance criteria was an E-value <0.01. The use of both collisionally activated dissociation and ECD tandem mass spectra for various charge states of the same protein resulted in unique protein identification with significantly lower statistical p values and a higher number of assigned fragments correlating directly to a higher confidence level in protein identification. All protein identifications were validated manually. Monoisotopic masses are reported for both intact proteins and fragment ions.
For relative proteoform quantification, the intensities of individual proteoforms were obtained from deconvoluted mass spectra using the MassList function in DataAnalysis software (Bruker Daltonics), and the relative percentages of each species were calculated as described previously (25–27). Based on these percentages, the total protein phosphorylation (Ptotal) of each phosphorylated protein was calculated using the following equation as previously published (25–27):
| (Eq. 1) |
Minor oxidation products and non-covalent adducts were taken into consideration in the quantification.
Characterization of Novel Enigma Isoform
The exon structure of the human PDLIM7 gene, which encodes the Enigma protein, was obtained from Ensembl (www.ensembl.org). The amino acid sequences for Enigma coding exons were generated using the Translate Tool available at ExPASy (web.expasy.org/translate/).
RESULTS
Both Fast- and Slow-twitch Skeletal Muscles Display Signs of Sarcopenia
F344BN rats were chosen as this strain is recommended by the National Institute on Aging as a model of healthy aging for age-related research (37). The primary advantage of this model system is the very low incidence of age-related pathologies in this rat strain (38), which aids the identification of true molecular signatures of sarcopenia without the confounding influence of the other comorbidities.
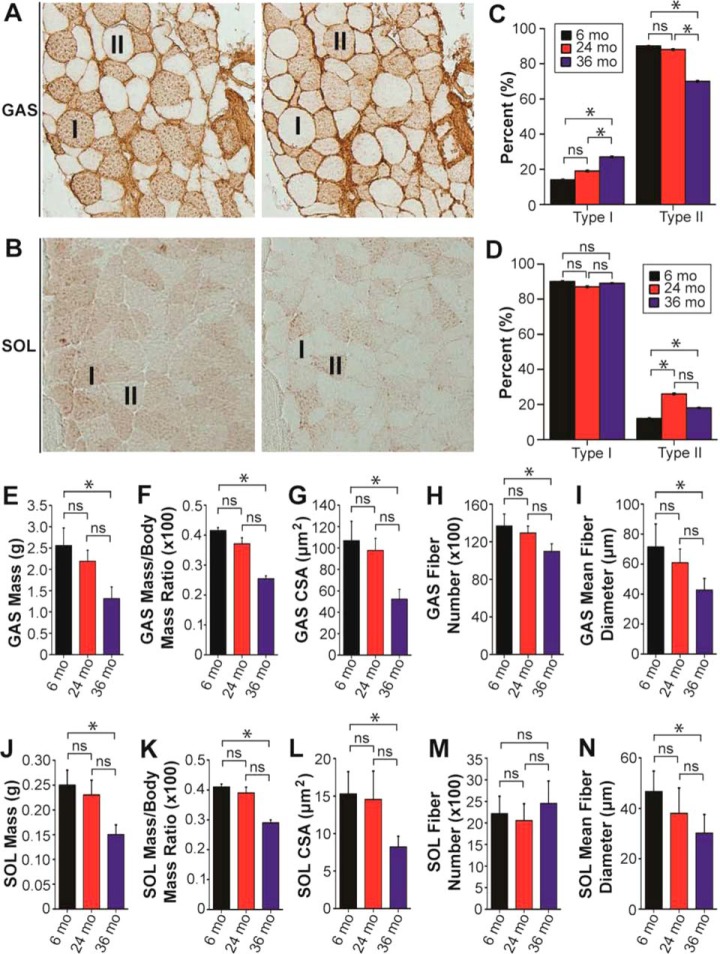
To assess potential sarcopenia-related changes in fast and slow skeletal muscles, the GAS and SOL muscles of aging F344BN rats were selected for analysis because of the reportedly high proportion of fast- and slow-twitch muscle fibers in these muscles, respectively (39). The high proportion of fast- and slow-twitch muscle fibers in the GAS and SOL muscles, respectively, was confirmed using two independent methods, immunohistochemical staining for type I and type II MHC (Fig. 1A–1D), as well as staining for ATPase activity (supplemental Fig. S3, supplemental Results).
Fig. 1.
Sarcopenia affects both fast- and slow-twitch skeletal muscles at advanced age. A, Representative serial sections from the GAS muscle of a 6-month-old F344BN rat stained with antibodies against type I MHC (left) and type II MHC (right). “I” and “II” denote fibers expressing type I and type II MHC, respectively. B, Representative serial sections from the SOL muscle of a 6-month-old F344BN rat stained with antibodies against type I MHC (left) and type II MHC (right). “I” and “II” denote fibers expressing type I and type II MHC, respectively. C, Graph showing quantification of age-dependent changes in the percentage of type I and type II MHC-expressing fibers in the GAS muscles of aging rats. D, Graph showing quantification of age-dependent changes in the percentage of type I and type II MHC-expressing fibers in the SOL muscles of rats in different age groups. Graphs showing the muscle mass (E), muscle mass-to-body mass ratio (F), and cross-sectional area (CSA) at the muscle mid-belly (G) for GAS muscles isolated from rats in different age groups. Graphs showing the quantification of fiber number (H) and mean fiber diameter (I) for GAS muscles from rats in different age groups. Graphs showing the muscle mass (J), muscle mass-to-body mass ratio (K), and CSA at the muscle mid-belly (L) for SOL muscles isolated from rats in different age groups. Graphs showing quantification of the fiber (M) and mean fiber diameter (N) for SOL muscles isolated from rats in different age groups. n = 12 for all panels. Each data point is the mean and error bar the S.E. ns, not significant. *p < 0.05.
Measurement of the mass of GAS muscles from rats in different age groups revealed a marked decrease in muscle mass in 36-month-old rats in comparison to those from 6-month-old animals (Fig. 1E). Similarly, normalization to the total body mass showed a significant decrease in the GAS mass-to-body mass ratio in the 36-month-old rats (Fig. 1F). In addition to decreased GAS muscle mass, whole muscle cross sections from the GAS muscles of 36-month-old rats displayed a marked decrease in CSA in comparison to those from 6-month-old rats (Fig. 1G). Further examination of the isolated GAS muscles revealed a significant decrease in both the number of muscle fibers and the mean fiber diameter in muscles from 36-month-old rats (Fig. 1H, 1I), indicating that the observed decreases in GAS mass and CSA are the combined result of muscle fiber loss and a reduction in the size of the remaining fibers.
Like sarcopenia-associated changes in GAS muscles, the masses of SOL muscles, as well as the SOL mass-to-body mass ratios, were also significantly decreased in 36-month-old rats in comparison to 6-month-old rats (Fig. 1J, 1K). SOL muscles from 36-month-old rats also had significantly reduced CSAs in comparison to muscles from 6-month-old animals (Fig. 1L). However, unlike in GAS muscles, which displayed a marked decrease in fiber number at advanced age (Fig. 1H), fiber number in SOL muscles was not significantly different between age groups (Fig. 1M). The mean fiber diameter was significantly decreased in SOL muscles from 36-month-old animals in comparison to those from 6-month-old rats (Fig. 1N), signifying that the noted decreases in the masses and CSAs of SOL muscles are almost exclusively the result of muscle fiber atrophy with little or no contributions from fiber loss.
Identification of Proteins in Sarcomeric Protein-Enriched Extracts
To characterize age-related changes in the fast- and slow-twitch skeletal muscles of aging F344BN rats, we employed a top-down proteomics strategy consisting of several steps (Fig. 2). These steps include isolation of a sarcomeric-protein enriched fraction from both fast- and slow-twitch skeletal muscles; sarcomeric protein separation by 1D reverse phase LC; on-line sarcomeric proteoform profiling on a high-resolution q-TOF mass spectrometer; fraction collection of sarcomeric proteins concomitant with on-line proteoform profiling, followed by in-depth proteoform characterization off-line on a high-resolution FT-ICR mass spectrometer; and integrated analysis of age-dependent changes in proteoform relative abundances and modifications.
Fig. 2.
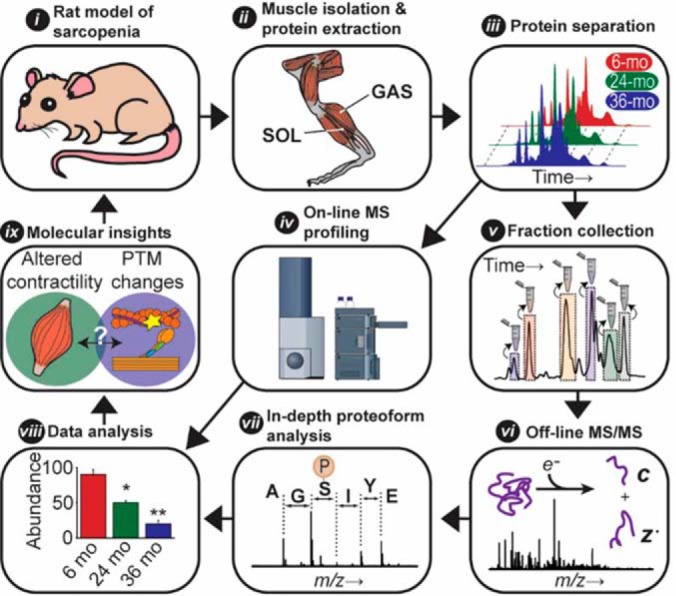
Overview of top-down proteomics strategy employed for the quantification and in-depth characterization of sarcomeric proteins extracted from fast- (GAS) and slow-twitch (SOL) muscles isolated from 6-, 24-, and 36-month-old F344BN rats. This strategy includes: i) an NIA approved rat model of healthy aging; ii) isolation of a sarcomeric-protein enriched fraction from both fast- and slow-twitch skeletal muscles; iii) protein separation by 1D liquid chromatography (LC); iv) rapid on-line proteoform profiling on a high-resolution q-TOF mass spectrometer; v) fraction collection of sarcomeric proteins concomitant with on-line proteoform profiling; vi) off-line MS/MS on an ultra-high-resolution FT-ICR mass spectrometer; vii) in-depth proteoform characterization; viii) integrated analysis of changes in proteoform relative abundances and modifications; and ix) correlation of age-dependent changes in fast- and slow-twitch muscles with proteomics findings to gain insights into the molecular mechanisms underlying sarcopenic phenotype.
First, we sought to identify proteins present in sarcomeric protein-enriched extracts prepared from the fast- and slow-twitch skeletal muscles of aging F344BN rats. Following MS/MS analysis of fraction collected proteins and protein identification using MASH Suite Pro (35), we were able to identify several sarcomeric and non-sarcomeric proteins in extracts prepared from the GAS muscles of aging rats (supplemental Table S1, supplemental Fig. S4). Identified fast-twitch skeletal muscle-specific sarcomeric proteins included an isoform of fast skeletal troponin T (fsTnT), fast skeletal troponin I (fsTnI), α-tropomyosin (αTpm), fast skeletal myosin essential light chain 1 (MLC-1F), fast skeletal myosin essential light chain 3 (MLC-3F), and fast skeletal troponin C (fsTnC) (supplemental Table S1, supplemental Figs. S4–S11). The fast skeletal isoform of the myosin regulatory light chain (MLC-2F) in rat, which has previously been characterized by our group (17), was identified in sarcomeric protein-enriched extracts prepared from rat GAS muscles (supplemental Table S1, supplemental Fig. S4). We also detected a 21,153.90 Da protein (supplemental Fig. S12), which was identified as an isoform of the PDZ/LIM domain-containing protein Enigma (supplemental Fig. S13). Comprehensive sequencing of this protein revealed that it is a novel isoform of the Enigma protein (nEnigma) produced via alternative splicing of exons 1 (non-coding), 2–4, and 6–9 (Fig. 3, supplemental Table S2, supplemental Figs. S12–S17, supplemental Results).
Fig. 3.
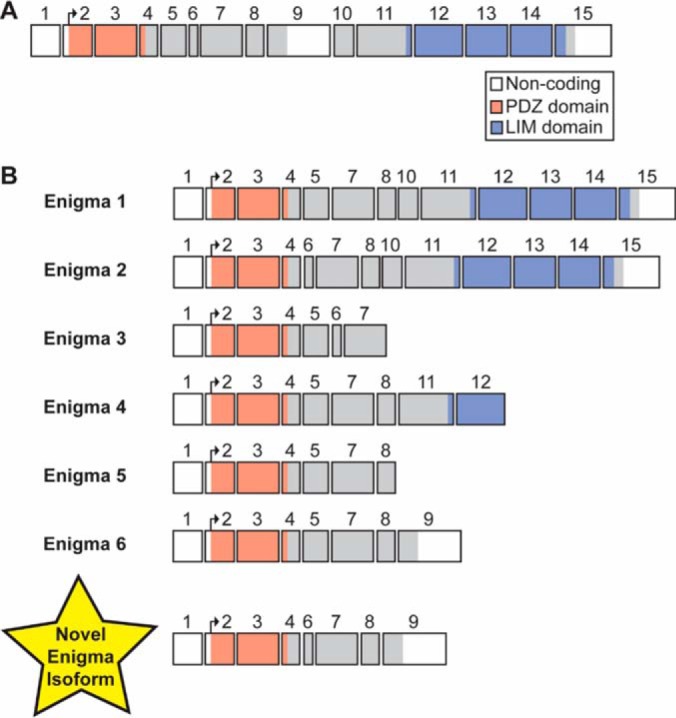
Exon structure of PDLIM7 gene and Enigma protein isoforms. A, The exon structure of the Enigma gene (PDLIM7), which is comprised of 15 exons (14 of which are coding exons). Regions of each exon corresponding to protein coding sequence are colored and the portions encoding the PDZ and LIM domains are indicated. B, The exon structure of human Enigma splice isoforms (Enigma 1-Enigma 6) from the UniProtKB/Swiss-Prot database are shown. The exon structure of the novel Engima (nEnigma) isoform identified in the GAS muscles of F344BN rats herein is shown at the bottom of the panel. This isoform is comprised of exons 1 (non-coding), 2–4, and 6–9.
Identified slow-twitch skeletal muscle-specific sarcomeric proteins in extracts prepared from aging rat SOL muscles included slow skeletal troponin T isoform 1 (ssTnT1), slow skeletal troponin I (ssTnI), slow skeletal myosin essential light chain (MLC-1S), and troponin C (ssTnC), as well as the slow skeletal/ventricular isoforms of the myosin essential (MLC-1V) and regulatory (MLC-2S) light chains (supplemental Table S3, supplemental Figs. S4, S18–S24).
In addition to skeletal muscle-type-specific sarcomeric protein isoforms, several sarcomeric proteins were also commonly identified in extracts prepared from both GAS and SOL muscles (supplemental Tables S1, S3; supplemental Fig. S4). These included two isoforms of the PDZ/LIM domain protein Cypher and β-Tpm (βTpm) (supplemental Tables S1, S2; supplemental Figs. S4, S25–S28). Not surprisingly, the α isoform of skeletal actin was also among the identified proteins (supplemental Tables S1, S3). Interestingly, although one of the identified Cypher isoforms corresponded to the previously observed Cypher2s isoform identified by Huang et al. (UniProtKB/Swiss-Prot accession number A0A0G2JXR0; supplemental Tables S1, S3; supplemental Fig. S27) (40), the second isoform corresponded to an undesignated isoform of Cypher that contains exons 1–3, 5–6, and 8–10 and has, to the best of our knowledge, not previously been detected at the protein level (termed Cypher4s) here (supplemental Table S4, supplemental Fig. S26) (41).
Sarcopenic Fast-Twitch Skeletal Muscles are Characterized by Age-Related Changes in Sarcomeric Protein Proteoforms
Quantitative top-down proteomics analysis revealed age-associated changes in the relative abundances of sarcomeric protein proteoforms in the fast-twitch skeletal muscles of aging rats (Fig. 4, supplemental Results). Moreover, changes in the phosphorylation of both myofilament and Z-disc proteins were noted (Fig. 6A, supplemental Results).
Fig. 4.
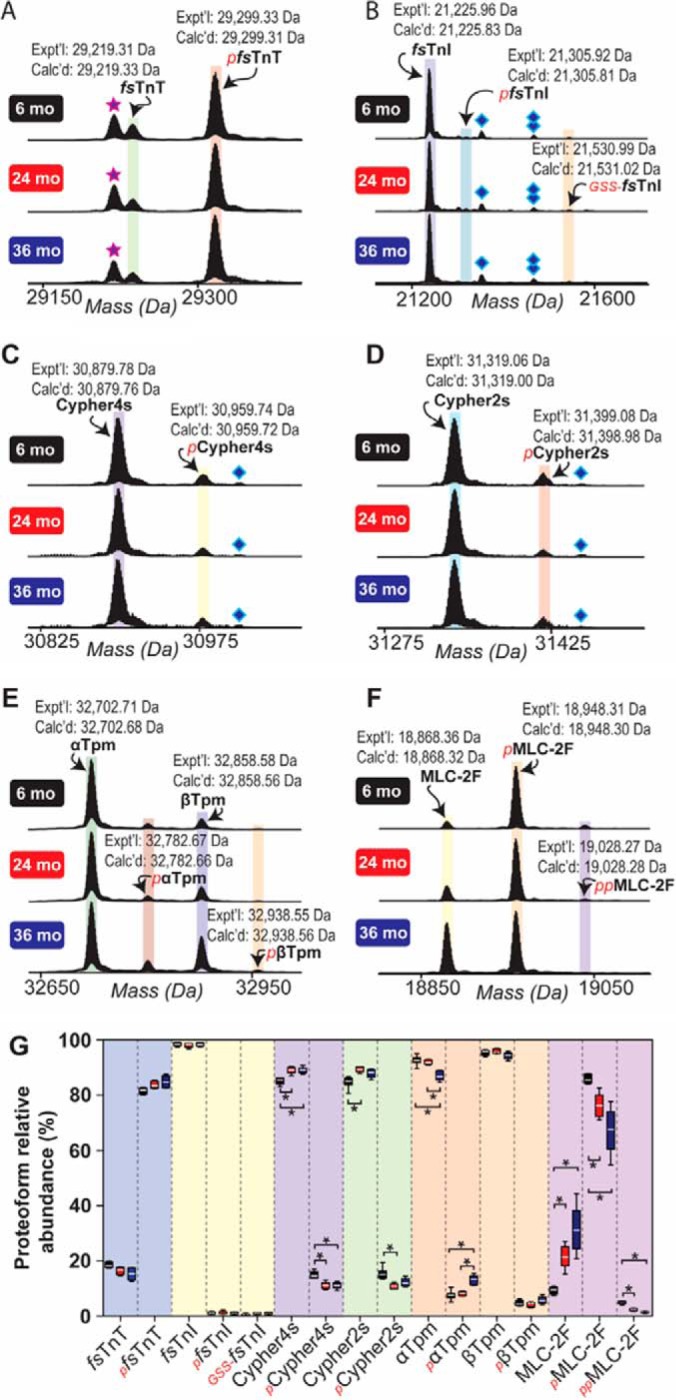
Top-down high-resolution MS-based quantification of sarcomeric protein proteoform relative abundances in fast-twitch GAS muscles isolated from 6-, 24-, and 36-month-old rats. Representative mass spectra showing fsTnT (A), fsTnI (B), Cypher4s (C), Cypher2s (D), Tpm (E), and MLC-2F (F). Star represents H3PO4 loss from pfsTnT. Diamond and double diamond represent protein associated non-covalently with one and two TFA molecules, respectively. G, Graph showing the relative quantification of proteoform relative abundances in GAS muscles isolated from rats in different age groups. Quantification is based on n = 6 animals for all age groups. Expt'l, experimentally determined relative molecular mass. Calc'd, calculated relative molecular mass based on sequence in the UniProtKB/Swiss-Prot database. *p < 0.05.
Fig. 6.
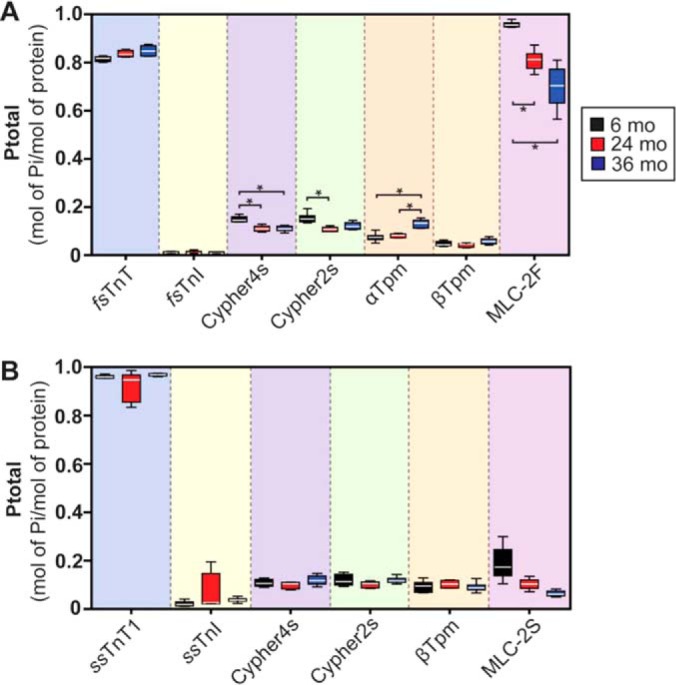
MS-based quantification of sarcomeric protein phosphorylation in fast- and slow-twitch skeletal muscles isolated from 6-, 24-, and 36-month-old rats. A, Quantification of sarcomeric protein phosphorylation in GAS muscles from rats in different age groups. B, Quantification of sarcomeric protein phosphorylation in SOL muscles from aging F344BN rats. Quantification is based on n = 6 individuals for all age groups. *p < 0.05.
Specifically, we detected minor novel proteoforms presumably corresponding to phosphorylated fsTnI (pfsTnI) and glutathionylated fsTnI (GSS-fsTnI) based on accurate mass measurement (ΔM = +79.96 Da and ΔM = +305.03 Da versus un-phosphorylated fsTnI for pfsTnI and GSS-fsTnI, respectively) (Fig. 4B). However, neither the relative abundances of these proteoforms, nor total fsTnI phosphorylation, changed with age in fast-twitch GAS muscles of aging rats (Figs. 4, 6A, supplemental Results). Phosphorylated proteoforms of Cypher4s and Cypher2s were also detected and quantified (Fig. 4C–4D, 4G, supplemental Results). Quantitative analysis revealed that, whereas the phosphorylation of Cypher2s was decreased in the GAS muscles of rats at 24-months of age (compared with that in muscles from 6-month-old animals), the phosphorylation of Cypher4s was decreased at both 24- and 36-months of age in comparison to that in muscles from 6-month-old animals (Figs. 4C–4D, 4G, 6A, supplemental Results).
A peak presumably corresponding to a phosphorylated proteoform of nEnigma was detected in sarcomeric protein-enriched extracts prepared from F344BN rat GAS muscles, although quantification of the relative abundance of this proteoform was not possible because of significant overlap with peaks corresponding to fsTnI (supplemental Fig. S12). In addition to age-related changes in the phosphorylation of Cypher isoforms, the phosphorylation of αTpm was also significantly increased in GAS muscles at 36-months of age compared with that in muscles from 6- and 24-month-old animals (Figs. 4E, 6A, supplemental Results). In agreement with our results and those of others (16, 17), a significant age-dependent decrease in the phosphorylation of MLC-2F was observed in the GAS muscles of aging rats (Figs. 4F–4G, 6A; supplemental Results).
Few Sarcomeric Protein Proteoform Alterations Occur in the Slow-twitch Muscles of Rats with Advanced Sarcopenia
Given that age-related changes in the morphology of slow-twitch SOL muscles were observed in 36-month-old F344BN rats (Fig. 1), next, we sought to determine whether sarcomeric protein PTMs are also altered with age in this muscle group. Quantitative top-down proteomics analysis uncovered relatively few age-related changes in the relative abundances of sarcomeric protein proteoforms in the SOL muscles of aging rats (Fig. 5; supplemental Results). Notably, in contrast to age-related changes in sarcomeric protein phosphorylation detected in GAS muscles, the phosphorylation of detected sarcomeric proteins in the SOL muscles of aging rats did not change with age (Fig. 6B, supplemental Results). Unlike in the GAS muscles of aging F344BN rats, we detected a significant increase in the relative abundance of the glutathionylated proteoform of ssTnI (GSS-ssTnI) in slow-twitch SOL muscles of 36-month-old rats (relative to that in muscles from 6- and 24-month-old animals) (Fig. 5B, 5G; supplemental Results). Although there was a trend toward decreased phosphorylation of MLC-2S with age, this trend did not reach statistical significance (Figs. 5F–5G, 6B; supplemental Results).
Fig. 5.
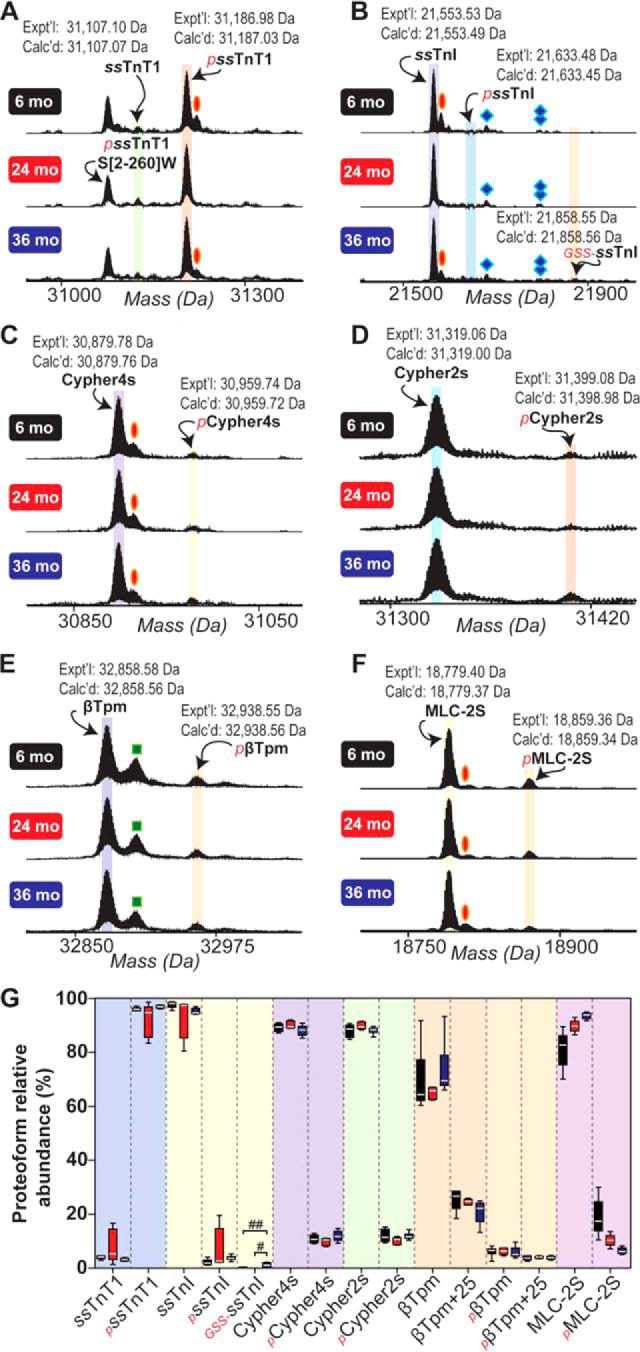
Top-down high-resolution MS-based quantification of sarcomeric protein proteoform relative abundances in slow-twitch SOL muscles isolated from 6-, 24-, and 36-month-old rats. Representative mass spectra showing ssTnT1 (A), ssTnI (B), Cypher4s (C), Cypher2s (D), βTpm (E), and MLC-2S (F). Oval, diamond, double diamond, and square represent peaks corresponding to oxidation, protein associated non-covalently with one TFA molecule, protein associated non-covalently with two TFA molecules, and a proteoform exhibiting a mass difference of +25 Da versus βTpm, respectively. G, Graph showing the relative quantification of proteoform relative abundances in SOL muscles isolated from rats in different age groups. Quantification is based on n = 6 animals for all age groups. Expt'l, experimentally determined relative molecular mass. Calc'd, calculated relative molecular mass based on sequence in the UniProtKB/Swiss-Prot database. #p < 0.001; ##p < 0.0001.
DISCUSSION
Sarcopenia Affects Both Fast- and Slow-twitch Skeletal Muscles in Aging Rats
Consistent with previous reports of age-related fast-skeletal muscle fiber atrophy (7, 8, 37, 42), GAS muscles from 36-month-old rats displayed characteristic signs of sarcopenia, including significant decreases in muscle mass and CSA at the muscle mid-belly (Fig. 1E–1G). These changes were, in part, attributable to a decrease in the mean fiber diameter (Fig. 1I), which is consistent with the well-established sarcopenia-associated atrophy of type II (fast-twitch) muscle fibers (7, 8, 42). Interestingly, although it has traditionally been thought that sarcopenia-associated fiber atrophy affects primarily type II (fast-twitch) fibers with relatively little impact on type I (slow-twitch) fibers (7, 8), recent evidence has suggested that slow-twitch fibers are also vulnerable to age-related atrophy, particularly at advanced age when sarcopenia becomes severe (18).
In agreement with these findings, we identified significant decreases in the masses and CSAs (at the muscle mid-belly) of SOL muscles from 36-month-old rats in comparison to muscles from 6-month-old animals (Fig. 1J–1L). Moreover, as in the GAS muscles (Fig. 1I), the mean fiber diameter was significantly decreased in SOL muscles from 36-month-old rats (Fig. 1N), indicating that the atrophy of slow-twitch fibers likely contributes to sarcopenic changes in slow-twitch skeletal muscles. Although it is possible that these results could be because of atrophy of a small population of fast-twitch fibers in the SOL, based on the relatively small proportion of type II fibers in this muscle (Fig. 1D; supplemental Fig. S3) (39), it is unlikely that this is the reason for age-related decreases in the masses and CSAs of SOL muscles. Instead, it is more likely that slow-twitch muscle fibers, like fast-twitch muscle fibers, are vulnerable to age-related atrophy.
In addition to age-related fiber atrophy, we also observed marked fiber loss in the GAS muscles of 36-month-old rats (Fig. 1H), in accordance with previous studies (42, 43). Notably, even though fibers in the SOL muscles of 36-month-old animals showed signs of atrophy (Fig. 1N), no change in total fiber number was detected as the animals aged (Fig. 1M). These findings suggest that, although slow-twitch fibers might indeed be vulnerable to sarcopenia-associated atrophy, these fibers may also be protected from age-related fiber loss. Whether type I (slow-twitch) fibers are truly protected from age-related cell death will necessitate a better understanding of the molecular mechanisms underlying sarcopenia-associated fiber loss, which, to date, remains incompletely understood (44).
Age-Related Changes in Myofilament Protein Proteoforms in Fast- and Slow-Twitch Skeletal Muscles
Previous studies from our lab, as well as the labs of others, have shown that MLC-2F phosphorylation declines with age in the fast-twitch skeletal muscles (16, 17). In agreement with these findings, we observed a significant decrease in MLC-2F phosphorylation with increasing age in the fast-twitch GAS muscles of aging F344BN rats (Figs. 4, 6A). Interestingly, the phosphorylation of MLC-2S in the slow-twitch skeletal muscles of rats did not change with age (Figs. 5, 6B). This finding is surprising given evidence in the literature suggesting that the phosphorylation of MLC-2S increases in aging muscles (45). The reason for this discrepancy may be related to the method employed for the quantification of protein phosphorylation (top-down proteomics herein versus ProQ Diamond staining in the aforementioned study). We have previously shown that rat MLC-2F is phosphorylated at two N-terminal Ser residues, namely Ser14 and Ser15 (17). At least one of these sites is conserved across all striated muscle-specific isoforms of MLC-2 (note that Ser14/15 in MLC-2F are analogous to Ser22/23 in MLC-2S in rat), even in different species (46). Functionally, phosphorylation of this site (or these sites depending on the specific isoform and species) results in structural alterations within the sarcomeres that potentiate the magnitude and velocity of force production (47, 48). Consistently, we have previously shown that the age-related decrease in the phosphorylation of MLC-2F (at Ser14/15) correlates with a reduction in force production, shortening velocity, and power output in the fast-twitch skeletal muscle fibers of aging F344BN rats (17). Likewise, similar findings have been reported in muscles from aging humans (16).
Although phosphorylation of the cardiac isoform of TnI is well-established within the literature (49), phosphorylation of the fast- and slow-skeletal muscle-specific isoforms of TnI has not previously been reported. Thus, the functional significance of fs-/ss-TnI phosphorylation in skeletal muscle contractile function remains to be determined. Notably, despite the fact that the phosphorylation of TnI isoforms in fast- and slow-twitch skeletal muscles was unchanged with age (Figs. 4, 6A), the relative abundance of GSS-ssTnI, the glutathionylated proteoform of ssTnI, increased in the slow-skeletal muscles of aged F344BN rats (Fig. 5). This finding is particularly relevant given that oxidative protein damage is a well-established facet of the aging process (50). Moreover, evidence in the literature suggests that the oxidative modification of contractile proteins may negatively impact contractile function (14, 15, 51). Therefore, enhanced glutathionylation of amino acid residues vulnerable to oxidation in ssTnI may represent an adaptive modification that helps preserve contractile function in slow-twitch skeletal muscle.
Top-down proteomics analysis also uncovered alterations in the phosphorylation of Tpm isoforms in fast- but not slow-twitch skeletal muscles with age (Figs. 4–6). Specifically, the phosphorylation of αTpm in the fast-twitch skeletal muscles of aging rats was significantly increased at 36 months of age versus that in muscles from 6- and 24-month-old animals (Figs. 4, 6A). The phosphorylation of βTpm was not altered with age in the fast- or slow-twitch skeletal muscles of aging rats (Figs. 4, 6A). Although the site of phosphorylation in Tpm isoforms was not determined in this study, prior studies have shown that the sole site of phosphorylation in both α- and β-Tpm is the penultimate residue, Ser283 (52–54). Functionally, the phosphorylation of Tpm isoforms at Ser283 has been shown to increase the rigidity of the Tpm head-to-tail overlap domain (55). The increased stiffness of Tpm head-to-tail polymers enhances the responsiveness of the thin filament to intracellular Ca2+ concentrations (55). Therefore, the increase in Tpm isoform phosphorylation in fast-twitch skeletal muscle may be a compensatory change in response to age-related alterations in cellular Ca2+ handling (56).
Sarcopenia is Associated with Changes in the Phosphorylation of the Z-Disc Protein Cypher in Fast-Twitch, but Not Slow-Twitch, Skeletal Muscles
Recent studies have identified a variety of mutations in Cypher, a Z-disc protein that belongs to the Enigma subfamily of PDZ/LIM proteins, in patients with dilated cardiomyopathy, hypertrophic cardiomyopathy, and skeletal muscle myopathies, including myofibrillar myopathies (57). In this study, we were able to detect and identify two different splice isoforms of Cypher in the fast- and slow-twitch skeletal muscles of aging F344BN rats (supplemental Figs. S25–S27). One of these isoforms is analogous to the short isoform Cypher2s, which has previously been identified in the striated muscles of mice (UniProtKB/Swiss-Prot accession number A0A0G2JXR0; supplemental Tables S1, S3; supplemental Fig. S27) (40). Interestingly, the second short isoform of Cypher that we detected and characterized using top-down proteomics was determined to be an isoform that has, to the best of our knowledge, only been detected at the transcript level (UniProtKB/Swiss-Prot accession number Q5XIG1; supplemental Tables S1, S3, supplemental Fig. S26) (41). We have designated this isoform Cypher4s, following the naming convention established by Huang et al. wherein “s” denotes Cypher isoforms containing exons 5–7, which are only found in Cypher isoforms expressed in skeletal muscle (40). Of note, this isoform appears to be unique among skeletal muscle-specific isoforms of Cypher in that it does not contain exon 7, which is present in all other Cypher isoforms expressed in skeletal muscle (supplemental Table S4).
Surprisingly, quantitative top-down proteomics identified age-related changes in the phosphorylation of Cypher isoforms in rat fast-twitch, but not slow-twitch, skeletal muscles (Figs. 4–6). Specifically, the phosphorylation of both Cypher isoforms (Cypher4s and Cypher2s) was significantly decreased in the fast-twitch GAS muscles of rats in the 24-month-old age group in comparison to that in muscles from 6-month-old animals (Figs. 4, 6A). Conversely, although the phosphorylation of Cypher2s did not differ in the GAS muscles of 6- and 36-month-old animals, Cypher4s phosphorylation was also significantly decreased at GAS muscles at 36 months of age relative to that in muscles from 6-month-old animals (Figs. 4, 6A).
Prior studies have shown that Cypher serves as a protein kinase A anchoring protein and can be phosphorylated by protein kinase A in vitro, as well as in vivo (58); however, the functional significance of Cypher phosphorylation remains enigmatic. Therefore, additional studies will be necessary to determine not only the functional significance of Cypher phosphorylation, but also the significance of age-related alterations in Cypher phosphorylation in fast-twitch skeletal muscles.
Identification of Novel Isoform of the PDZ/LIM Protein Enigma
Enigma is the prototypical member of the Enigma subfamily of PDZ/LIM proteins and, like all members of this protein subfamily, can produce a variety of isoforms via alternative splicing of the Enigma pre-mRNA. Although only a single sequence was present in the UniProtKB/Swiss-Prot database for Enigma in rat (accession number Q9Z1Z9; corresponds to Enigma isoform 1 in human), sequences for six different isoforms of Enigma were present in the database for human (supplemental Table S2). Surprisingly, top-down MS/MS analysis revealed that the Enigma isoform identified in this study did not correspond to any known Enigma isoforms in the UniProtKB/Swiss-Prot database. Therefore, the isoform identified herein represents a novel short isoform of the Enigma protein (comprised of exons 1–4 and 6–9) that contains the N-terminal PDZ domain but lacks the three C-terminal LIM domains (Fig. 3, supplemental Table S2, supplemental Fig. S17).
Originally identified in 1994 by Wu and Gill as an insulin receptor interacting protein (59), Enigma, like other members of its subfamily, is a scaffolding protein that plays a key role in the organization of macromolecular signaling complexes. In striated muscle, the PDZ domain of Enigma was shown to interact with the C terminus of βTpm, promoting localization of Enigma to the boundary of Z-discs and I bands (60). Although the function(s) of Enigma isoforms in skeletal muscle have yet to be determined, the localization of Enigma to the boundary of the Z-discs in striated muscles raises the possibility that Enigma may mediate important signals generated within the Z-discs or, potentially, may influence contractility via signaling to myofilaments. Also of note, recent evidence has shown that Enigma is a target of the ubiquitin ligase Nedd4–1, which plays an important role in skeletal muscle atrophy (61).
DATA AVAILABILITY
All MS and MS/MS data have been submitted to the MassIVE data repository and are available at ftp://massive.ucsd.edu/MSV000081157.
Supplementary Material
Footnotes
* Financial support was kindly provided by NIH F31 HL128086 (to Z.R.G.), NIH R01 HL109810 and R01 HL096971 (to Y.G.), and NIH R01 AG030423 (to J.M.A.). Y.G. would like to acknowledge NIH R01 GM117058 and high-end instrument grant S10OD018475.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- Tpm
- tropomyosin
- TnI
- troponin I
- TnT
- troponin T
- TnC
- troponin C
- MHC
- myosin heavy chain
- PTMs
- post-translational modifications
- MS
- mass spectrometry
- MS/MS
- tandem MS
- ECD
- electron capture dissociation
- F344BN
- Fisher 344 × Brown Norway F1 hybrid
- GAS
- gastrocnemius
- SOL
- soleus
- CSA
- cross-sectional area
- LC
- liquid chromatography
- q-TOF
- quadrupole-time-of-flight
- LTQ
- linear ion trap
- FT-ICR
- Fourier transform ion cyclotron resonance
- Ptotal
- total protein phosphorylation
- fsTnT
- fast skeletal troponin T
- fsTnI
- fast skeletal troponin I
- αTpm
- α-tropomyosin
- MLC-1F
- fast skeletal myosin essential light chain 1
- MLC-3F
- fast skeletal myosin essential light chain 3
- fsTnC
- fast skeletal troponin C
- MLC-2F
- fast skeletal isoform of the myosin regulatory light chain
- nEnigma
- novel isoform of the Enigma protein
- ssTnT1
- slow skeletal troponin T isoform 1
- ssTnI
- slow skeletal troponin I
- MLC-1S
- slow skeletal myosin essential light chain
- ssTnC
- slow skeletal troponin C
- MLC-1V
- slow skeletal/ventricular myosin essential light chain
- MLC-2S
- slow skeletal/ventricular myosin regulatory light chain
- βTpm
- β-tropomyosin
- pfsTnI
- phosphorylated fsTnI
- GSS-fsTnI
- glutathionylated fsTnI
- pαTpm
- phosphorylated αTpm
- pβTpm
- phosphorylated βTpm
- pMLC-2F
- mono-phosphorylated MLC-2F
- ppMLC-2F
- bis-phosphorylated MLC-2F
- pssTnT1
- mono-phosphorylated ssTnT1
- pssTnI
- mono-phosphorylated ssTnI
- GSS-ssTnI
- glutathionylated ssTnI
- pCypher2s
- mono-phosphorylated Cypher2s
- pCypher4s
- mono-phosphorylated Cypher4s.
REFERENCES
- 1. Cohen S., Nathan J. A., and Goldberg A. L. (2015) Muscle wasting in disease: molecular mechanisms and promising therapies. Nat. Rev. Drug Discov. 14, 58–74 [DOI] [PubMed] [Google Scholar]
- 2. Ryall J. G., Schertzer J. D., and Lynch G. S. (2008) Cellular and molecular mechanisms underlying age-related skeletal muscle wasting and weakness. Biogerontology 9, 213–228 [DOI] [PubMed] [Google Scholar]
- 3. Rolland Y., Czerwinski S., Abellan Van Kan G., Morley J. E., Cesari M., Onder G., Woo J., Baumgartner R., Pillard F., Boirie Y., Chumlea W. M., and Vellas B. (2008) Sarcopenia: its assessment, etiology, pathogenesis, consequences and future perspectives. J. Nutr. Health Aging 12, 433–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Janssen I., Heymsfield S. B., and Ross R. (2002) Low relative skeletal muscle mass (sarcopenia) in older persons is associated with functional impairment and physical disability. J. Am. Geriatr. Soc. 50, 889–896 [DOI] [PubMed] [Google Scholar]
- 5. Janssen I., Shepard D. S., Katzmarzyk P. T., and Roubenoff R. (2004) The healthcare costs of sarcopenia in the United States. J. Am. Geriatr. Soc. 52, 80–85 [DOI] [PubMed] [Google Scholar]
- 6. Wiener J. M., and Tilly J. (2002) Population ageing in the United States of America: implications for public programmes. Int. J. Epidemiol. 31, 776–781 [DOI] [PubMed] [Google Scholar]
- 7. Ciciliot S., Rossi A. C., Dyar K. A., Blaauw B., and Schiaffino S. (2013) Muscle type and fiber type specificity in muscle wasting. Int. J. Biochem. Cell Biol. 45, 2191–2199 [DOI] [PubMed] [Google Scholar]
- 8. Thompson L. V. (1994) Effects of age and training on skeletal muscle physiology and performance. Phys. Ther. 74, 71–81 [DOI] [PubMed] [Google Scholar]
- 9. Thompson L. V. (2009) Age-related muscle dysfunction. Exp. Gerontol. 44, 106–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Johnson M. L., Robinson M. M., and Nair K. S. (2013) Skeletal muscle aging and the mitochondrion. Trends Endocrinol. Metab. 24, 247–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Miller M. S., Callahan D. M., and Toth M. J. (2014) Skeletal muscle myofilament adaptations to aging, disease, and disuse and their effects on whole muscle performance in older adult humans. Front. Physiol. 5, 369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Moss R. L., Diffee G. M., and Greaser M. L. (1995) Contractile properties of skeletal muscle fibers in relation to myofibrillar protein isoforms. Rev. Physiol. Biochem. Pharmacol. 126, 1–63 [DOI] [PubMed] [Google Scholar]
- 13. Schiaffino S., and Reggiani C. (1996) Molecular diversity of myofibrillar proteins: gene regulation and functional significance. Physiol. Rev. 76, 371–423 [DOI] [PubMed] [Google Scholar]
- 14. Callahan D. M., Miller M. S., Sweeny A. P., Tourville T. W., Slauterbeck J. R., Savage P. D., Maugan D. W., Ades P. A., Beynnon B. D., and Toth M. J. (2014) Muscle disuse alters skeletal muscle contractile function at the molecular and cellular levels in older adult humans in a sex-specific manner. J. Physiol. 592, 4555–4573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Prochniewicz E., Thomas D. D., and Thompson L. V. (2005) Age-related decline in actomyosin function. J. Gerontol. A Biol. Sci. Med. Sci. 60, 425–431 [DOI] [PubMed] [Google Scholar]
- 16. Miller M. S., Bedrin N. G., Callahan D. M., Previs M. J., Jennings M. E., Ades P. A., Maughan D. W., Palmer B. M., and Toth M. J. (2013) Age-related slowing of myosin actin cross-bridge kinetics is sex specific and predicts decrements in whole skeletal muscle performance in humans. J. Appl. Physiol. 115, 1004–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gregorich Z. R., Peng Y., Cai W., Jin Y., Wei L., Chen A. J., McKiernan S. H., Aiken J. M., Moss R. L., Diffee G. M., and Ge Y. (2016) Top-Down Targeted Proteomics Reveals Decrease in Myosin Regulatory Light-Chain Phosphorylation That Contributes to Sarcopenic Muscle Dysfunction. J. Proteome Res. 15, 2706–2716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Purves-Smith F. M., Sgarioto N., and Hepple R. T. (2014) Fiber typing in aging muscle. Exerc. Sport. Sci. Rev. 42, 45–52 [DOI] [PubMed] [Google Scholar]
- 19. Smith L. M., Kelleher N. L., and Proteomics C. (2013) f. T. D. Proteoform: a single term describing protein complexity. Nat. Methods 10, 186–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Siuti N., and Kelleher N. L. (2007) Decoding protein modifications using top-down mass spectrometry. Nat. Methods 4, 817–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gregorich Z. R., and Ge Y. (2014) Top-down proteomics in health and disease: challenges and opportunities. Proteomics 14, 1195–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang H., and Ge Y. (2011) Comprehensive analysis of protein modifications by top-down mass spectrometry. Circ. Cardiovasc. Genet. 4, 711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zubarev R. A., Horn D. M., Fridriksson E. K., Kelleher N. L., Kruger N. A., Lewis M. A., Carpenter B. K., and McLafferty F. W. (2000) Electron capture dissociation for structural characterization of multiply charged protein cations. Anal. Chem. 72, 563–573 [DOI] [PubMed] [Google Scholar]
- 24. Zhang J., Zhang H., Ayaz-Guner S., Chen Y. C., Dong X., Xu Q., and Ge Y. (2011) Phosphorylation, but not alternative splicing or proteolytic degradation, is conserved in human and mouse cardiac troponin T. Biochemistry 50, 6081–6092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dong X., Sumandea C. A., Chen Y. C., Garcia-Cazarin M. L., Zhang J., Balke C. W., Sumandea M. P., and Ge Y. (2012) Augmented phosphorylation of cardiac troponin I in hypertensive heart failure. J. Biol. Chem. 287, 848–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Peng Y., Gregorich Z. R., Valeja S. G., Zhang H., Cai W., Chen Y. C., Guner H., Chen A. J., Schwahn D. J., Hacker T. A., Liu X., and Ge Y. (2014) Top-down proteomics reveals concerted reductions in myofilament and Z-disc protein phosphorylation after acute myocardial infarction. Mol. Cell. Proteomics 13, 2752–2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang J., Guy M. J., Norman H. S., Chen Y. C., Xu Q., Dong X., Guner H., Wang S., Kohmoto T., Young K. H., Moss R. L., and Ge Y. (2011) Top-down quantitative proteomics identified phosphorylation of cardiac troponin I as a candidate biomarker for chronic heart failure. J. Proteome Res. 10, 4054–4065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Medzihradszky K. F., Zhang X., Chalkley R. J., Guan S., McFarland M. A., Chalmers M. J., Marshall A. G., Diaz R. L., Allis C. D., and Burlingame A. L. (2004) Characterization of Tetrahymena histone H2B variants and posttranslational populations by electron capture dissociation (ECD) Fourier transform ion cyclotron mass spectrometry (FT-ICR MS). Mol. Cell. Proteomics 3, 872–886 [DOI] [PubMed] [Google Scholar]
- 29. Ge Y., Rybakova I. N., Xu Q., and Moss R. L. (2009) Top-down high-resolution mass spectrometry of cardiac myosin binding protein C revealed that truncation alters protein phosphorylation state. Proc. Natl. Acad. Sci. U.S.A. 106, 12658–12663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ansong C., Wu S., Meng D., Liu X., Brewer H. M., Deatherage Kaiser B. L., Nakayasu E. S., Cort J. R., Pevzner P., Smith R. D., Heffron F., Adkins J. N., and Pasa-Tolic L. (2013) Top-down proteomics reveals a unique protein S-thiolation switch in Salmonella Typhimurium in response to infection-like conditions. Proc. Natl. Acad. Sci. U.S.A. 110, 10153–10158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pesavento J. J., Mizzen C. A., and Kelleher N. L. (2006) Quantitative analysis of modified proteins and their positional isomers by tandem mass spectrometry: human histone H4. Anal. Chem. 78, 4271–4280 [DOI] [PubMed] [Google Scholar]
- 32. Steen H., Jebanathirajah J. A., Rush J., Morrice N., and Kirschner M. W. (2006) Phosphorylation analysis by mass spectrometry: myths, facts, and the consequences for qualitative and quantitative measurements. Mol. Cell. Proteomics 5, 172–181 [DOI] [PubMed] [Google Scholar]
- 33. Chamot-Rooke J., Mikaty G., Malosse C., Soyer M., Dumont A., Gault J., Imhaus A. F., Martin P., Trellet M., Clary G., Chafey P., Camoin L., Nilges M., Nassif X., and Duménil G. (2011) Posttranslational modification of pili upon cell contact triggers N. meningitidis dissemination. Science 331, 778–782 [DOI] [PubMed] [Google Scholar]
- 34. Hintz C. S., Coyle E. F., Kaiser K. K., Chi M. M., and Lowry O. H. (1984) Comparison of muscle fiber typing by quantitative enzyme assays and by myosin ATPase staining. J. Histochem. Cytochem. 32, 655–660 [DOI] [PubMed] [Google Scholar]
- 35. Cai W., Guner H., Gregorich Z. R., Chen A. J., Ayaz-Guner S., Peng Y., Valeja S. G., Liu X., and Ge Y. (2015) MASH Suite Pro: a comprehensive software tool for top-down proteomics. Mol. Cell. Proteomics [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu X., Sirotkin Y., Shen Y., Anderson G., Tsai Y. S., Ting Y. S., Goodlett D. R., Smith R. D., Bafna V., and Pevzner P. A. (2012) Protein identification using top-down. Mol. Cell. Proteomics 11, M111.008524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lushaj E. B., Johnson J. K., McKenzie D., and Aiken J. M. (2008) Sarcopenia accelerates at advanced ages in Fisher 344xBrown Norway rats. J. Gerontol. A Biol. Sci. Med. Sci. 63, 921–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lipman R. D., Chrisp C. E., Hazzard D. G., and Bronson R. T. (1996) Pathologic characterization of brown Norway, brown Norway x Fischer 344, and Fischer 344 x brown Norway rats with relation to age. J. Gerontol. A Biol. Sci. Med. Sci. 51, B54–B59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Staron R. S., Kraemer W. J., Hikida R. S., Fry A. C., Murray J. D., and Campos G. E. (1999) Fiber type composition of four hindlimb muscles of adult Fisher 344 rats. Histochem. Cell Biol. 111, 117–123 [DOI] [PubMed] [Google Scholar]
- 40. Huang C., Zhou Q., Liang P., Hollander M. S., Sheikh F., Li X., Greaser M., Shelton G. D., Evans S., and Chen J. (2003) Characterization and in vivo functional analysis of splice variants of cypher. J. Biol. Chem. 278, 7360–7365 [DOI] [PubMed] [Google Scholar]
- 41. Strausberg R. L., Feingold E. A., Grouse L. H., Derge J. G., Klausner R. D., Collins F. S., Wagner L., Shenmen C. M., Schuler G. D., Altschul S. F., Zeeberg B., Buetow K. H., Schaefer C. F., Bhat N. K., Hopkins R. F., Jordan H., Moore T., Max S. I., Wang J., Hsieh F., Diatchenko L., Marusina K., Farmer A. A., Rubin G. M., Hong L., Stapleton M., Soares M. B., Bonaldo M. F., Casavant T. L., Scheetz T. E., Brownstein M. J., Usdin T. B., Toshiyuki S., Carninci P., Prange C., Raha S. S., Loquellano N. A., Peters G. J., Abramson R. D., Mullahy S. J., Bosak S. A., McEwan P. J., McKernan K. J., Malek J. A., Gunaratne P. H., Richards S., Worley K. C., Hale S., Garcia A. M., Gay L. J., Hulyk S. W., Villalon D. K., Muzny D. M., Sodergren E. J., Lu X., Gibbs R. A., Fahey J., Helton E., Ketteman M., Madan A., Rodrigues S., Sanchez A., Whiting M., Young A. C., Shevchenko Y., Bouffard G. G., Blakesley R. W., Touchman J. W., Green E. D., Dickson M. C., Rodriguez A. C., Grimwood J., Schmutz J., Myers R. M., Butterfield Y. S., Krzywinski M. I., Skalska U., Smailus D. E., Schnerch A., Schein J. E., Jones S. J., Marra M. A., Team M. G. C. P. (2002) Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc. Natl. Acad. Sci. U.S.A. 99, 16899–16903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lexell J., Taylor C. C., and Sjöström M. (1988) What is the cause of the ageing atrophy? Total number, size and proportion of different fiber types studied in whole vastus lateralis muscle from 15- to 83-year-old men. J. Neurol. Sci. 84, 275–294 [DOI] [PubMed] [Google Scholar]
- 43. Lexell J. (1995) Human aging, muscle mass, and fiber type composition. J. Gerontol. A Biol. Sci. Med. Sci. 50 Spec No, 11–16 [DOI] [PubMed] [Google Scholar]
- 44. Alway S. E., and Siu P. M. (2008) Nuclear apoptosis contributes to sarcopenia. Exerc. Sport Sci. Rev. 36, 51–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gannon J., Doran P., Kirwan A., and Ohlendieck K. (2009) Drastic increase of myosin light chain MLC-2 in senescent skeletal muscle indicates fast-to-slow fibre transition in sarcopenia of old age. Eur. J. Cell Biol. 88, 685–700 [DOI] [PubMed] [Google Scholar]
- 46. Szczesna D. (2003) Regulatory light chains of striated muscle myosin. Structure, function and malfunction. Curr. Drug Targets Cardiovasc. Haematol. Disord. 3, 187–197 [DOI] [PubMed] [Google Scholar]
- 47. Patel J. R., Diffee G. M., Huang X. P., and Moss R. L. (1998) Phosphorylation of myosin regulatory light chain eliminates force-dependent changes in relaxation rates in skeletal muscle. Biophys. J. 74, 360–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhi G., Ryder J. W., Huang J., Ding P., Chen Y., Zhao Y., Kamm K. E., and Stull J. T. (2005) Myosin light chain kinase and myosin phosphorylation effect frequency-dependent potentiation of skeletal muscle contraction. Proc. Natl. Acad. Sci. U.S.A. 102, 17519–17524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Layland J., Solaro R. J., and Shah A. M. (2005) Regulation of cardiac contractile function by troponin I phosphorylation. Cardiovasc. Res. 66, 12–21 [DOI] [PubMed] [Google Scholar]
- 50. Finkel T., and Holbrook N. J. (2000) Oxidants, oxidative stress and the biology of ageing. Nature 408, 239–247 [DOI] [PubMed] [Google Scholar]
- 51. Moen R. J., Klein J. C., and Thomas D. D. (2014) Electron paramagnetic resonance resolves effects of oxidative stress on muscle proteins. Exerc. Sport Sci. Rev. 42, 30–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Heeley D. H. (2013) Phosphorylation of tropomyosin in striated muscle. J. Muscle Res. Cell Motil. 34, 233–237 [DOI] [PubMed] [Google Scholar]
- 53. Peng Y., Yu D., Gregorich Z., Chen X., Beyer A. M., Gutterman D. D., and Ge Y. (2013) In-depth proteomic analysis of human tropomyosin by top-down mass spectrometry. J Muscle Res. Cell Motil. 34, 199–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Peng Y., Chen X., Zhang H., Xu Q., Hacker T. A., and Ge Y. (2013) Top-down targeted proteomics for deep sequencing of tropomyosin isoforms. J. Proteome Res. 12, 187–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lehman W., Medlock G., Li X. E., Suphamungmee W., Tu A. Y., Schmidtmann A., Ujfalusi Z., Fischer S., Moore J. R., Geeves M. A., and Regnier M. (2015) Phosphorylation of Ser283 enhances the stiffness of the tropomyosin head-to-tail overlap domain. Arch. Biochem. Biophys. 571, 10–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Delbono O., O'Rourke K. S., and Ettinger W. H. (1995) Excitation-calcium release uncoupling in aged single human skeletal muscle fibers. J. Membr. Biol. 148, 211–222 [DOI] [PubMed] [Google Scholar]
- 57. Sheikh F., Bang M. L., Lange S., and Chen J. (2007) “Z”eroing in on the role of Cypher in striated muscle function, signaling, and human disease. Trends Cardiovasc. Med. 17, 258–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lin C., Guo X., Lange S., Liu J., Ouyang K., Yin X., Jiang L., Cai Y., Mu Y., Sheikh F., Ye S., Chen J., Ke Y., and Cheng H. (2013) Cypher/ZASP is a novel A-kinase anchoring protein. J. Biol. Chem. 288, 29403–29413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wu R. Y., and Gill G. N. (1994) LIM domain recognition of a tyrosine-containing tight turn. J. Biol. Chem. 269, 25085–25090 [PubMed] [Google Scholar]
- 60. Guy P. M., Kenny D. A., and Gill G. N. (1999) The PDZ domain of the LIM protein enigma binds to beta-tropomyosin. Mol. Biol. Cell 10, 1973–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. D'Cruz R., Plant P. J., Pablo L. A., Lin S., Chackowicz J., Correa J., Bain J., and Batt J. (2016) PDLIM7 is a novel target of the ubiquitin ligase Nedd4–1 in skeletal muscle. Biochem. J. 473, 267–276 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All MS and MS/MS data have been submitted to the MassIVE data repository and are available at ftp://massive.ucsd.edu/MSV000081157.