Abstract
Multigene families encoding diverse secreted peptide hormones play important roles in plant development. A need exists to efficiently elucidate the structures and post-translational-modifications of these difficult-to-isolate peptide hormones in planta so that their biological functions can be determined. A mass spectrometry and bioinformatics approach was developed to comprehensively analyze the secreted peptidome of Medicago hairy root cultures and xylem sap. We identified 759 spectra corresponding to the secreted products of twelve peptide hormones including four CEP (C-TERMINALLY ENCODED PEPTIDE), two CLE (CLV3/ENDOSPERM SURROUNDING REGION RELATED) and six XAP (XYLEM SAP ASSOCIATED PEPTIDE) peptides. The MtCEP1, MtCEP2, MtCEP5 and MtCEP8 peptides identified differed in post-translational-modifications. Most were hydroxylated at conserved proline residues but some MtCEP1 derivatives were tri-arabinosylated. In addition, many CEP peptides possessed unexpected N- and C-terminal extensions. The pattern of these extensions suggested roles for endo- and exoproteases in CEP peptide maturation. Longer than expected, hydroxylated and homogeneously modified mono- and tri-arabinosylated CEP peptides corresponding to their in vivo structures were chemically synthesized to probe the effect of these post-translational-modifications on function. The ability of CEP peptides to elevate root nodule number was increased by hydroxylation at key positions. MtCEP1 peptides with N-terminal extensions or with tri-arabinosylation modification, however, were unable to impart increased nodulation. The MtCLE5 and MtCLE17 peptides identified were of precise size, and inhibited main root growth and increased lateral root number. Six XAP peptides, each beginning with a conserved DY sulfation motif, were identified including MtXAP1a, MtXAP1b, MtXAP1c, MtXAP3, MtXAP5 and MtXAP7. MtXAP1a and MtXAP5 inhibited lateral root emergence. Transcriptional analyses demonstrated peptide hormone gene expression in the root vasculature and tip. Since hairy roots can be induced on many plants, their corresponding root cultures may represent ideal source materials to efficiently identify diverse peptide hormones in vivo in a broad range of species.
A diversity of secreted plant peptides derived from over 15 multi-gene families play crucial roles as short and long range signaling molecules. These peptide hormones influence many important aspects of plant growth and development (1). Of these, CLE and CEP peptides affect the deployment pattern of the root system and root nodule formation through systemic and local interactions with their corresponding receptors (2–5), whereas the biological activity of XAP peptides (6) has not yet been elucidated.
To date, only a few laboratories have isolated and characterized a handful of secreted peptide hormones from plant materials and validated their in vivo forms and post-translational-modifications (PTMs)1 (5, 7–14). Notably, this represents only a small fraction of the ∼1000 predicted small peptide-encoding genes (15). Plant tissue cultures, xylem sap, and submerged seedlings, whole roots and hairy root cultures overexpressing peptide-hormone encoding genes have been used as source materials to isolate peptide hormones (5, 6, 9, 12, 14). Without knowledge of the in vivo forms of peptide hormones, the precise nature of the interaction of these putative ligands with their receptors cannot be determined. For example, IDA (INFLORESCENCE DEFICIENT IN ABSCISSION) peptides varying in length and hydroxyl modifications have substantial differences in binding affinity to their receptors and biological activity (16–18).
The very low native concentration of these peptides makes their isolation and proteomic identification difficult. The identification of peptide hormones in planta and the characterization of their structure and function requires (1) optimizing the natural production of peptides in the source tissue, (2) selecting a starting material that is most enriched in native peptide hormones, and (3) effective concentration and purification of the diversity of peptides from the sample. In addition, sensitive and accurate mass spectrometry (MS) detection and an effective bioinformatics strategy is required because peptide hormones are derived from undefined, native proteolytic processing activity. The ability to synthesize molecules identical to native peptide hormones is also necessary, particularly if some peptide PTMs are not amenable to synthesis through commercial facilities. Finally, the characterization of the phenotypic effect of the peptide on plants needs to be defined and, ideally, the strategy employed should be applicable to a wide range of plants. Therefore, there is a need to optimize methods and approaches to isolate peptide hormone so that they can be applied to a wide range of plants.
Thus far, the quantitative assessment of the natural abundance of peptide hormones, combined with the use of titration bioassays, has shown that the CEP and CLE peptides in planta are present in the mid pm to low nm range (9, 14). To identify in vivo peptides and/or boost low production, gene overexpression (12, 13), data-dependent MS (5) and the use of materials thought to be enriched in certain peptide hormones, such as xylem sap (6), have assisted the detection of several low abundance peptide hormones. An effective bioinformatics approach is also required to identify PTMs that are known to occur in plant peptide hormones in vivo (e.g. proline hydroxylation, tyrosine sulfation and glycosylation of hydroxylproline). In addition, bioinformatics settings need to be relaxed so that peptides of unexpected lengths can be detected and potential information on peptide maturation can be gathered. This approach enables a fuller appreciation of the diversity of peptide hormones to be determined.
Chemical purification combined with MS was used recently to identify several putative mobile peptide species in soybean xylem sap including CLE, CEP, and XAP peptides, but their biological activity was not assessed (6). Soybeans were also found to be a suitable host for overexpressing the Lotus RS2 gene in transgenic roots and this enabled identification of its tri-arabinosylated CLE product in the xylem sap collected from the shoot. The presence, however, of the corresponding RS2 CLE peptide in the xylem sap of Lotus, from which the gene was derived, was not demonstrated (13). Therefore, the broader utility of methods using xylem sap as the source material for isolating peptide hormones has, to date, not been established. Nevertheless, the fractionation of crude Arabidopsis xylem sap on a C18 trap column followed by nanoflow separation and targeted Orbitrap MS in multiple reaction monitoring mode was used recently to detect several CEP peptides (5). Recently, we used a similar targeted MS approach to identify and quantify nine different 15 amino acid variants of the MtCEP1 peptide in the culture medium of Agrobacterium rhizogenes transformed hairy roots overexpressing MtCEP1 (9). The MtCEP1 variants containing a 412 Da glycosylation modification of proline on position 11 (P11) were detected in very small amounts in MtCEP1 overexpressing Medicago hairy root cultures and this mass corresponds to that of tri-arabinose (9). Tri-arabinosylation is required for the maximal biological activity of CLE peptides (13, 19), but the significance of this modification is still unknown for CEP peptides, because glycosylated peptide standards are difficult to access synthetically.
Herein, we comprehensively characterized the secreted peptidome of M. truncatula to identify a plethora of native peptides that are secreted into hairy root cultures and the xylem sap. To maximize the ability to identify the full diversity of peptide hormones, we developed a tandem MS (MS/MS) and bioinformatics strategy that accommodated variations in the length of the secreted peptides as well as several PTMs. This generated 759 spectra that corresponded to peptides from the CEP, CLE and XAP gene families that varied in length and PTM, as well as the breakdown fragments of high molecular weight xylem sap proteins. The effect of these diverse CEP peptides on root architecture was tested to determine the biological relevance of the N- and C-terminal extensions as well as hydroxylation and tri-arabinosylation PTMs. The tri-arabinosylated derivative was prepared by solid phase peptide synthesis (SPPS) using a specially synthesized tri-arabinosylated hydroxyproline 'building block' (20) as this Fmoc-protected glycosylamino acid is not commercially available for peptide synthesis. The results indicated that MtCEP1 derivatives with N-terminal extensions or tri-arabinosylation had reduced or abolished biological activity especially in relation to promoting root nodule formation. Phylogenetics was used to classify XAP and XAP-like peptides into 5 distinct families, one of which was homologous to Arabidopsis (CASPARIAN STRIP INTEGRITY FACTOR) CIF peptides (21). Given the ease at which hairy roots can be induced on a multitude of plants (22), we propose that hairy root cultures combined with the methodologies used here will have wide utility in the simultaneous identification and characterization of multiple plant hormones in a broad range of species. In addition, the identification of typical xylem sap proteins in the secreted fraction of hairy root cultures suggest that this material partially reflects the composition of xylem sap, and the peptide hormones therein are, therefore, candidates for being long distance signals. This was supported by the localization of peptide hormone expression in root vascular cells where proximity to the xylem vessels would enable these secreted peptides to join the xylem stream. Finally, the diversity of peptides with different lengths in the exudate material provided information about the possible maturation process by which peptide hormones are derived and the results implicate roles for endo- as well as exoproteases.
EXPERIMENTAL PROCEDURES
Establishing Axenic Medicago Root Cultures
Axenic cultures of Medicago roots transformed with A. rhizogenes ARqua1 containing the empty vector (pK7WG2D.1) or pK7WG2D.1 with 35S:MtCEP1 were established (9, 23). Root cultures were grown in 150 ml liquid Fåhraeus medium containing 5 mm KNO3 and 1% sucrose in the dark for 14 days (9).
Xylem Sap Extraction
Medicago plants grown for 6 weeks on a potting mix were decapitated and xylem sap (∼600 μl/plant) was collected under normal root pressure for 8 h on ice (24). Sap was stored at −20 °C and the 20 ml cumulative volume was centrifuged to remove debris before processing. Seeds of Glycine max cultivar Williams 82 were germinated, and xylem sap was extracted from 8–10 week old plants (24).
Peptides Extraction and Isolation
In planta peptides in root cultures and xylem sap were concentrated for MS analysis using o-chlorophenol/acetone precipitation followed by size exclusion chromatography and concentration/desalting by reversed-phase chromatography using C18 micro-pipette tips (9). After lyophilization, samples were resuspended in 400 μl of 3% acetonitrile with 0.1% formic acid prior to nano-liquid chromatography-electrospray ionization (LC-ESI)-MS/MS analysis.
Nano-LC-ESI-MS/MS and Data Analysis
A comprehensive description of the strategy employed for MS is given in supplemental Methods. The raw files generated by nano-LC-ESI-MS/MS were analyzed with Proteome Discoverer 2.1 (Thermo Scientific) using SEQUEST against three different databases. The Medicago peptide databases contained about 225 sequences from known members of the peptide families for CLE, CEP, RGF, IDA, PSK, PSY, CEP-like, CIF, EPF, and XAP which were identified from a phylogenetic examination of the M. truncatula genome using the domain sequence and a BLAST approach (essentially as outlined in 26). The sequences for all the peptides were obtained from jcvi.org and NCBI. The raw files were also run against CEP database containing about 940 CEP peptide sequences from all the species (26) and Medicago truncatula whole protein database containing 57477 sequences (www.medicagogenome.org; Mt4.0v1_GenesProteinSeq_20130731_1800 released on 31–07-2013). The bioinformatics strategy screened for spectra corresponding to in planta peptides (selecting no enzyme cleavage) ranging in length from 6 to 150 amino acids to ensure that all possible larger than expected species were identified. The fragmentation type used was high energy collision dissociation (HCD), and the precursor and fragment mass tolerance were set at 10 ppm and 0.08 Da, respectively. The bioinformatics approach is detailed in Supplemental Methods and the stepwise flowchart of the strategy employed for in planta peptide identification is shown in supplemental Fig. S1. A maximum of 3 equal dynamic modifications were allowed per peptide for oxidation (i.e. hydroxylation) at M and P (mass shift of + 15.995 Da), sulfation of S, T and Y (mass shift of + 79.957 Da), pentosylation of P (mass shift of + 132.11 Da), tri-arabinosylation of P (mass shift of + 412.127 Da) and deamidation at N and Q (mass shift of + 0.984 Da).
Experimental Design and Statistical Rationale
The samples derived from root cultures containing vector control or 35S:MtCEP1 or xylem sap were subjected to nano-LC-MS/MS (Q Exactive™ Hybrid Quadrupole-Orbitrap™ Mass Spectrometer, Thermo Fisher Scientific, San Jose, CA) using 7 μl of sample per technical repeat (9). Two independent biological and two technical replicates were used for each sample and two blank runs were included among each sample to minimize carryover. Acceptable peptide spectrum matches had a delta Cn better than or equal to 0.05, and a target false discovery rate (FDR) equal to or less than 0.01. For the discovery of in planta peptides, unambiguous matches also satisfy the following criteria: m/z of the precursor and fragment ions within the specified precision range, a near continuous series of fragment ion annotations in the spectra and significant q-value as generated by the Sequest HT search algorithm. Representative unedited annotated spectra with a minimum of 30% MS/MS assignments are presented as a proof of identity. Further details on search settings are provided in supplemental Methods.
Synthetic peptides
All hydroxylated peptides used in the study were synthesized by GL BioChem Ltd., Shanghai, China at > 95% purity. Peptides were validated qualitatively and quantitatively by HPLC (CXTH LC3000, Beijing) and MS (SHIMADZU LCMS-2010EV and SHIMADZU LCMS-2020, Japan) prior to use. MtCEP1-D1 variants (HyP4,11, HyP4, MaP11 and TaP11) were synthesized through Fmoc-strategy SPPS as described in the supplemental Methods. Mono- and tri-arabinosylated MtCEP1-D1 peptides (MaP11 and TaP11) were produced using N-Fmoc-protected and per-O-acetylated β-l-arabinofuranosyl and β1,2-tri-l-arabinofuranosyl-hydroxyproline building blocks as described in the Supplemental Methods and outlined in supplemental Fig. S2. The glycosyl-hydroxyproline building blocks were prepared according to a method from Corcilius et al. (20). Peptides were purified by reversed-phase HPLC and validated by analytical HPLC, low-resolution ESI-MS and high-resolution ESI-MS (See supplemental Methods for detailed conditions and analytical data). Peptides were verified to be the bis(trifluoroacetate) salts through a combination of 1H and 19F quantitative NMR (QNMR) using an internal standard of α,α,α-trifluorotoluene (See supplemental Methods and supplemental Fig. S3–S5 for details).
Biological Activity of Peptides
Three-day-old M. truncatula A17 seedlings were transferred to N-free or 5 mm KNO3 Fåhraeus medium plates (25) with or without synthetic peptides (1 μm). Plants were scored for primary root length, lateral root growth and nodule formation (23) at day 7 and 14. Plants were grown at 25 °C with a 16 h photoperiod and a photon flux density of 100 μmol m−2 s−1 (26) in a Thermoline growth chamber (Climatron, Australia). All root measurements were repeated independently in triplicate.
Phylogenetic analysis of XAP genes
XAP genes in Medicago, Glycine max and Arabidopsis were identified by generating a representative domain motif and then scanning for that motif across proteome databases (27). Due to the variable lengths of XAP domains, initial XAP motifs were generated in the motif discovery tool GLAM2 (28) based on previously identified XAP sequences (6). GLAM2SCAN was used to search the generated XAP motif against protein databases for each species (28). Different XAP type motifs were refined once gene groups had been established using the genes identified in the initial database searches. The genetic distance among all identified XAP genes was estimated using the maximum-likelihood algorithm RAxML v8.0.0 (27, 29). A phylogenetic tree of identified XAP genes was generated using Dendroscope (30) to group XAP genes.
Transcript Expression Analysis
The expression profiles of MtCEP, MtCLE and MtXAP in different zones of the root tissue was done using qRT-PCR (25). The root tip (1 mm), the young root (next 1 mm) and the mature root (next 10 mm) were harvested from 8-day-old plants and RNA was Trizol-extracted and purified using the RNeasy Plant Miniprep kit (Qiagen, Melbourne, Australia). First strand cDNA was synthesized using oligo (dT) and Superscript III Reverse Transcriptase kit (Invitrogen, CA) (23). The gene-specific primers used are listed in supplemental Table S1. The reactions and data analysis were carried out as described previously (25) and the Ct values of the target genes were normalized to the Ct values of MtUBQ10 (MtGI accession no. TC161574). Three biological and three technical replicates were used for each root region.
Reporter Gene Assays
Promoter sequences (2–2.5 kb) derived from MtCEP5, MtCEP8, MtCLE5, or MtCLE17 were cloned, fused upstream to the GUS reporter gene, transformed into the pKGWFS7 destination vector and introduced into A. rhizogenes K599 (23). The gene-specific primers used for making the GUS-fusions are listed in supplemental Table S1. Medicago roots were transformed with K599 harboring the vectors. GUS staining was performed and vibratome root sections were prepared (1000 Plus; Vibratome Company, St Louis, MO) (31). Tissues and sections were imaged using a stereomicroscope (Leica Microsystems Inc., Deerfield, IL). Staining and sectioning were performed three times, each with five independently transformed plants.
RESULTS
The Medicago Peptidome Contains Several Peptide Hormones and Xylem Sap Protein Breakdown Products
MS/MS analysis of the culture exudates of Medicago hairy roots demonstrated unequivocal matches to peptide sequences that in all cases included the predicted regulatory peptide domains of specific members of the CEP, CLE, or XAP gene families (Table I). Analysis of the 104 peptide groups showed unambiguous matches to the peptide domains encoded by MtCEP1, MtCEP2, MtCEP5, MtCEP8, MtCLE5, MtCLE17, MtXAP1a, MtXAP1b, and MtXAP5 (Fig. 1–3; summarized in Table I). All the confident matches were identified with mass precision of 2 ppm and highly significant q-values (supplemental File S1). By contrast, MS/MS analysis of Medicago xylem sap identified several XAP1b, XAP1c, XAP3 and XAP7 peptides (Table I, supplemental File S1). Members of other peptide families were not detected. In addition, hairy root cultures and xylem sap both contained peptides that corresponded to typical high molecular weight xylem sap proteins (e.g. peroxidases, endoglycanases/chitinases and β-glucosidases; supplemental File S2), that are known to be secreted into xylem sap of a diversity of plants (24). As expected, SDS-PAGE confirmed the presence of a suite of typical high molecular weight proteins in Medicago xylem sap (supplemental Fig. S6) (24). To benchmark the sensitivity of our procedures and techniques, soybean xylem sap was isolated and analyzed. Twelve CEP, CLE, and XAP peptide species were identified in as little as 5 ml of xylem sap which included domain 1 of GmCEP3 and GmCEP21, domain 2 of GmCEP13, GmXAP1a, and GmXAP1b, which have not been described previously (supplemental Table S2).
Table I. Secreted peptides found in M. truncatula root cultures and/or xylem sap. MtCEP1, MtCEP2, MtCEP5, MtCEP8, MtCLE5, MtCLE17, MtXAP1a, and MtXAP5 were present in root cultures only. MtXAP1c, MtXAP 3, and MtXAP 7 were present in xylem sap only, whereas MtXAP1b was present in both root cultures and xylem sap. D1 = domain 1; D2 = domain 2; Nm = no modification. 1The verified or 2predicted domain sequences of the peptide species. *Peptides present only in Medicago root cultures expressing 35S:MtCEP1. #Peptide present only in Medicago root cultures expressing empty vector. HyP: hydroxylated proline (numbering indicates the position of PTM in the proposed final domain); TaP: tri-arabinosylation at proline; Y2S: sulfation at tyrosine at position 2.
| Accession number (% coverage) | Peptide and proposed final domain | Type and position of PTMs | Size of peptide variants found | Unique peptides (Number of spectra) |
|---|---|---|---|---|
| Medtr5g030490.1 | MtCEP1-D1 | Nm | 10–41 | 43 (446) |
| XP_003612914.2 (64) | AFQPTTPGNSPGVGH1 | HyP11 | ||
| HyP4,11 | ||||
| HyP7,11 | ||||
| Hy4,7,11 | ||||
| TaP11 | ||||
| HyP4,TaP11 | ||||
| HyP7,TaP11 | ||||
| HyP4,7,TaP11 | ||||
| MtCEP1-D2 | Nm | 9–27 | ||
| EFQKTNPGHSPGVGH1 | HyP11 | |||
| HyP7,11 | ||||
| AFK45165.1 (24) | MtCEP2 | HyP7,11 | 15–20 | 2 (13) |
| AFRPTTPGHSPGIGH1 | HyP4,7,11 | |||
| Medtr5g017710.1 | MtCEP5 | HyP11 | 14–21 | 4 (84) |
| XP_003611775.1 (25) | AFRPTTPGHSPGVGH1 | HyP7,11 | ||
| HyP4,7,11 | ||||
| AC233112_1014.1 (15) | MtCEP8* | HyP11 | 15 | 1 (4) |
| AFRPTTPGNSPGVGH1 | HyP4,11 | |||
| HyP7,11 | ||||
| HyP4,7,11 | ||||
| Medtr1g100733.1 | MtCLE5 | HyP4,7 | 12 | 1 (10) |
| XP_013469691.1 (14) | HVVPSGPNPLHN1 | |||
| Medtr5g085990.1 | MtCLE17* | Nm | 12 | 1 (3) |
| XP_003616944.1 (17) | RKVPTGSNPLHN1 | |||
| Medtr7g086770.1 | MtXAP1a# | HyP16 | 16–17 | 2 (25) |
| XP_003624724.1 (18) | DYGGTGANTDHEPKPPR2 | Y2S,HyP16 | ||
| Medtr6g027480.1 | MtXAP1b | HyP16 | 17–19 | 3 (119) |
| XP_013451515.1 (22) | DYEGPGANKEHNPKSPGNG2 | Y2S | ||
| Y2S,HyP16 | ||||
| Y2S,HyP5,13 | ||||
| Medtr6g027470.1 | MtXAP1c | Y2S,HyP16 | 17–21 | 2 (44) |
| XP_013451514.1 (24) | DYPGTGPNHHHDPKSPG2 | |||
| Medtr8g037800.1 | MtXAP3 | HyP9 | 15 | 3 (6) |
| XP_003627749.1 (29) | DYGRYDPSPTFSKPP2 | |||
| Medtr3g491850.1 | MtXAP5* | Y2S | 18 | 1 (2) |
| XP_013461431.1 (23) | DYDEAGPNPKHSKKPGKG2 | |||
| Medtr4g108700.1 | MtXAP7 | Y2S,HyP4,13 | 13 | 1 (3) |
| XP_013457905.1 (16) | DYAPSGANGRHTP2 |
Fig. 1.
MS/MS spectra of MtCEP peptides identified in M. truncatula hairy root cultures. A, MtCEP2 peptide with a 5-amino acid N-terminal extension and hydroxylation at P12, 16 was identified at 19.93 min (m/z 698.32532). B, MtCEP5 peptide with a 5-amino acid N-terminal, 1-amino acid C-terminal extension and hydroxylation at P9, 12, 16, was identified at 15.27 min (m/z 718.66510). C, A MtCEP8 peptide domain hydroxylated at P4, 7, 11 was identified at 10.69 min (m/z 772.36017).
Fig. 2.
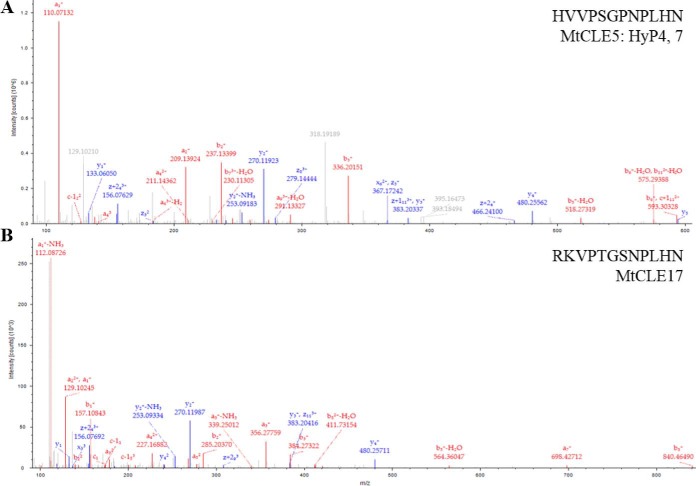
MS/MS spectra of MtCLE peptides identified in M. truncatula hairy root cultures. A, MtCLE5 peptide domain identified at 17.03 min (m/z 433.88617) was hydroxylated at P4, 7. B, MtCLE17 peptide domain was identified at 12.86 min (m/z 440.57791).
Fig. 3.
MS/MS spectra of MtXAP peptides identified in M. truncatula hairy root cultures. A, MtXAP1a peptide identified at 15.50 min (m/z 636.59821) was sulfated and hydroxylated at Y2 and P16, respectively. B, MtXAP1b peptide identified at 12.78 min (m/z 1032.41846) was sulfated and hydroxylated at Y2 and P16, respectively. C, MtXAP5 peptide identified at 9.17 min (m/z 501.98227) was sulfated at Y2.
Peptides Identified in Root Cultures Possess Highly Variable PTM Patterns and Length
Hydroxylation of proline residues was the most common PTM found in CEP, CLE and XAP peptides (Table I). In addition, some MtCEP1 spectra showed evidence of the presence of a glycosyl PTM consistent with the mass of tri-arabinose (412 Da). XAP peptides were also deduced to be modified at tyrosine by sulfation (Table I) as the N-terminal DY motif is a common sulfation signal in plants (32, 33). Thirteen spectra of the 759 examined (i.e. two corresponding to D1 and eight corresponding to D2 of MtCEP1, and three corresponding to MtCLE17) contained no PTMs (Table I). GmCEP/CLE/XAP peptides were also identified with hydroxylprolination, tri-arabinosylation and sulfation PTMs (supplemental Table S2). The length of MtCEP1, MtCEP2 and MtCEP5 peptides was highly variable, whereas MtCEP8 and the two MtCLE peptides corresponded precisely to the expected 15 and 12 amino acid domains, respectively (Table I; Fig. 1, 2). Similarly, the length of MtXAP1 and MtXAP1b ranged from 16 to 19 amino acids, whereas MtXAP5 consisted of 18 amino acids only (Table I; Fig. 3). Variability in the termini of most GmCEP/CLE/XAP peptides was also found (supplemental Table S2).
CEP Peptide Maturation Involves N- and C-terminal Trimming
The majority of the MtCEP1 peptides identified (79 of D1; Fig. 4A, 4B and 46 of D2; Table I; Fig. 4A, 4C) corresponded to the conserved 15 amino acid CEP domain size (27). The hairy root cultures, however, contained peptide variants of different lengths that were larger than the expected 15 amino acid CEP domain size that extended to the N- and C-terminal flanking sequences of the predicted domains (Fig. 4A; Fig. 5). These longer peptides collectively spanned 60% of the MtCEP1 propeptide (Fig. 4A). The largest MtCEP1-D1 fragment possessed 41 amino acids that extended from a dibasic (KK) site in the MtCEP1 propeptide to an R residue flanking the D1 right border (Fig. 4A; Fig. 5A). A 35 amino acid variant appeared to be a predominant intermediate in MtCEP1 maturation as it triggered 63 individual MS/MS spectra (Fig. 4A, 4B). This pattern of N-terminal extensions (Fig. 4A) is consistent with exoprotease-like trimming activities. Interestingly, no fragment was detected that encompassed both domains of MtCEP1 (Fig. 4A). No peptide that matched solely to the variable region of any peptide encoding gene was found (Fig. 4A). Larger than expected variants of MtCEP2 and MtCEP5 were also found (Fig. 1).
Fig. 4.
Diversity of MtCEP1-D1 and D2 peptides isolated from Medicago hairy root cultures. A, Twenty-eight MtCEP1-D1 and 19 MtCEP1-D2 peptide variants ranging in length from 10–41 and 9–27 amino acids were identified, respectively. Black bar represents signal peptide and white bars represent variable region (VR). The dashed line depicts the overall coverage of the propeptide by the longest peptide variants of D1 and D2. B and C, The number of spectra relative to peptide length is shown for D1 (B) and D2 (C), respectively. The gold, magenta and green sequences in (A), and bars in (B) and (C) represent some of the longer peptide variants with low abundance; representative spectra shown in Fig. 5. Red and blue bars and amino acid sequences represent the predominant 15 amino acid D1 and D2 peptide domains, respectively.
Fig. 5.
MS/MS spectra of the extended MtCEP1 peptides. A, The D1 extended peptide consisting of 41 amino acids (D1–41). Nano-LC-MS/MS spectrum at 26.96 min (m/z 867.61340) showed a signature ion fragment at y6 indicating hydroxylation at P36. B, Identification of the D1 extended peptide consisting of 23 amino acids (D1–23). Nano-MS/MS spectrum at 13.55 min (m/z 799.69806) showed signature ion fragments peaks at y6, y10, and y13 indicating hydroxylation at P11, 14, 18. C, Identification of the D2 extended peptide consisting of 20 amino acids (D2–20). MS/MS spectrum at 15.73 min (m/z 1063.48047) showed signature ion peak y6 indicating hydroxylation at P15.
Biological Activity of MtCEP1-D1 is Influenced by N- or C-terminal Extensions, PTM, and Amino Acid Substitution
It is known from studies of INFLORESCENCE DEFICIENT IN ABSCISSION (IDA) peptides that N-terminal extensions greatly affect biological activity and receptor binding efficiency (16–18). Therefore, the biological effects of longer MtCEP1 variants on the formation of lateral roots and nodules were assessed by synthesizing four MtCEP1-D1 peptides with identical hydroxyproline residues at P4 and 11, but with different N- or C-terminal amino acid extensions (Fig. 6A). The structure of these synthesized peptides mirrored those found in vivo. The peptides with N-terminal extensions were unable to increase root nodule number but retained the ability to decrease the number of emerged lateral roots (Fig. 6B). The variant with one amino acid C-terminal extension did not affect biological activity (Fig. 6B).
Fig. 6.

Biological activity of the extended and modified MtCEP1-D1 peptides. A and C, Sequence of extended and modified MtCEP1-D1 variants, respectively (red = hydroxylated P, green = mono-arabinosylated P; Blue = tri-arabinosylated P). B and D, The number of lateral roots (LR) and nodules were scored on plants at 14 days (n ≥ 21) for extended (B) and modified (D) MtCEP1-D1 variants. To assess root nodule number, plants were grown on N-free medium containing 1 μm peptide, and inoculated with S. meliloti WSM 1022. Lateral roots were scored on plants grown on 1 μm peptide supplemented N-rich medium. Statistically significant differences indicated by letters were determined with ANOVA, followed by Tukey's test, p < 0.05. Error bars = standard error.
The spectra of four MtCEP1-D1 variants in the 35S:MtCEP1 root cultures indicated that these 15 amino acid peptides were modified by tri-arabinose at P11 (Table I). Therefore, to assess the significance of this modification, derivatives of MtCEP1 containing either hydroxylation at P4 (HyP4) or hydroxylation at P4 and 11 (HyP4,11) or mono-arabinose at P11 (MaP11) or tri-arabinose at P11 (TaP11) were synthesized using SPPS (Fig. 6C) and tested for biological activity. These derivatives were identical in structure except for the modifications at P11. Surprisingly, tri-arabinosylation abolished the biological activity of the peptide, whereas peptides lacking hydroxylation at P11 or possessing mono-arabinose at P11 retained biological activity (Fig. 6D).
The 15 amino acid MtCEP8, MtCEP5 and MtCEP2 domain sequences differed from MtCEP1-D1 by one (Q to R), two (Q to R, N to H) and three amino acids (Q to R, N to H and V to I), respectively (Fig. 7A), and they either contained identical or differing hydroxylation patterns to MtCEP1-D1 (Table I). These amino acid substitutions represent natural variation at these three positions and the biological significance of these modifications is not known. To test the influence of these amino acid residues and differing PTM patterns on biological activity, we synthesized the peptide domains corresponding to the MtCEP2, MtCEP5 and MtCEP8 species found in the root cultures. In addition, the biological activity of synthesized MtCEP2, MtCEP5, and MtCEP8 derivatives with fixed hydroxylations at P4 and P11 were compared with MtCEP1-D1 with hydroxylation at P4 and P11, which is the most biologically active variant of MtCEP1 (9). For root nodule formation, the results showed that MtCEP1-D1, MtCEP5, and MtCEP8 hydroxylations at P4 and 11, and MtCEP8 hydroxylation at P4, 7, and 11 increased nodule number (Fig. 7B). No other derivatives positively affect nodule number. For lateral roots, all derivatives considerably decreased lateral root number except MtCEP2 hydroxylated at P4, 7 and 11, and MtCEP8 hydroxylated at P11 (Fig. 7C).
Fig. 7.

Biological activity of synthetic MtCEP peptides. A, Amino acid sequences of MtCEP1, 2, 5, and 8. The red residues inside the box represent different amino acids to MtCEP1-D1. B and C, The effect of peptide structure and proline hydroxylation were assessed on nodule number (B) and lateral root number (C). Plants were grown identically to those in Fig. 6 (n ≥ 21). Statistically significant differences indicated by letters were determined with ANOVA, followed by Tukey's test, p < 0.05. Error bars = standard error.
MtCLE5 is Homologous to Tracheary Element Differentiation Inhibitory Factor (TDIF) but Inhibits Main Root Growth
The biological activity of MtCLE5 and MtCLE17 peptides was assessed on Medicago roots. MtCLE5 resembles AtCLE41 and AtCLE42 peptides in amino acid sequence at key residues (34) (Fig. 8A). AtCLE41 and AtCLE42 are TDIF CLE peptides and have not been shown to affect main or lateral root growth in any plant tested so far (14, 35). MtCLE5, however, conferred classic non-TDIF CLE peptide biological activities (36) by inhibiting primary root length and promoting lateral root number. Given the sequence similarity of MtCLE17 to AtCLE20 (37) and OsCLE302 (38), this peptide inhibited main root growth and promoted lateral root number, as expected (Fig. 8A, 8B).
Fig. 8.

Biological activity of synthetic MtCLE and MtXAP peptides. A, Amino acid sequence homology of MtCLE5 with AtCLE41 and AtCLE42 (TDIF peptides in Arabidopsis), and MtCLE17 with AtCLE20 and OsCLE302. AtCLE41 sequence is identical to MtCLE65 (33) which is nomenclatured as MtCLE06 at NCBI. The red residues inside the box represent the dissimilar amino acids. B and C, Differential effects on lateral root and primary root (PR) length of MtCLE peptides (B) and MtXAP peptides (C). Plants were grown on Fåhraeus medium containing 5 mm KNO3 and scored at day 14 (n ≥ 21). *p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001 (Two tail Student's t test). Error bars = standard error.
MtXAP1a and MtXAP5 Decrease Lateral Root Number
MtXAP1a and MtXAP5 affected the lateral roots negatively but induced no other root phenotype, whereas MtXAP1b did not influence root architecture (Fig. 8C). Since many small peptides are 12–15 amino acids in length and terminate at H or N and these terminal amino acids have been shown to enhance interactions with conserved RxR motifs in the corresponding receptor (32), we synthesized shorter MtXAP peptide derivatives terminating in either N or H. All these XAP derivatives were biologically inactive (supplemental Fig. S7).
XAP and XAP-like Peptides Form Five Groups
The phylogenetic relationship between XAP and XAP-like peptides that begin with the DY sulfation motif are unclear as is the length of the final biologically active peptide. Therefore, a consensus XAP motif was generated and used to identify other XAP peptides in Medicago, soybean and Arabidopsis (supplemental Fig. S8A). A total of 46 XAP peptide hormones starting with a DY motif were identified across the three species and their phylogenetic relatedness was determined (supplemental Fig. S9). The N-termini of XAP peptides were highly consistent as there was no evidence for any N-terminal extension, whereas their C-termini were variable. The predicted RGF, XAP, CIF, PSY and PSK gene products share homology both within the regulatory motif (supplemental Fig. S8B–S8F) as well as to the N-terminal side of the conserved DY motif, suggesting a common origin. There was evidence for sequence conservation to the N-terminal side of the common DY motif (underlined sequences in supplemental Fig. S8). The phylogenetic analysis also shows that MtXAP3 is a CIF peptide as these clearly form one family.
Transcript Analysis and Reporter Gene Assays Show Preference for Root Vascular Expression
To determine the in situ expression pattern of the peptide hormone genes, qRT-PCR of MtCEP, MtCLE and MtXAP was examined in root tissues and in the root tip region. The expression in young root and root tip was normalized against the mature root. MtCEP1 and MtCEP8 show maximum expression in young roots whereas MtCEP2 and MtCEP5 express more prominently in root tips (Fig. 9A). GUS fusions to the promoter regions of MtCEP5 and MtCEP8 showed that they express in root vasculature and in the tip (supplemental Fig. S10). MtCLE5 mainly expressed in mature root whereas MtCLE17 expressed predominantly in the root tip (Fig. 9B). Reporter gene assays showed that MtCLE5 and MtCEP17 are expressed in the vasculature, root epidermal cells and the root tip (supplemental Fig. S10). Transcript expression of MtXAP1a, MtXAP1b, and MtXAP5 are higher in mature root and young root, and lowest in the root tip (Fig. 9B).
Fig. 9.

Transcript analysis of peptide encoding genes in different root zones. A and B, Expression of MtCEP (A), MtCLE and MtXAP (B) genes were studied in 8-day-old Medicago plants grown on N-free media. (n = 9). Statistically significant differences indicated by letters were determined with ANOVA, followed by Tukey's test, p < 0.05. Error bars = standard error.
DISCUSSION
Here, we provide the precise methodology and bioinformatics approach necessary to enrich, purify and characterize the peptidome of secreted plant fluids and identify a plethora of peptide hormones. From an analysis of 759 matching spectra, we identified the mature secreted products of twelve M. truncatula peptide hormone genes (MtCEP1, MtCEP2, MtCEP5, MtCEP8, MtCLE5, MtCLE17, MtXAP1a, MtXAP1b, MtXAP1c, MtXAP3, MtXAP5, and MtXAP7), and defined their PTMs. A minimum of 30% assignment of the predicted MS/MS cleavages along with identification of the parental ion within 2 ppm of the expected mass, combined with iterative spectra and significant q-values found in multiple samples enabled confident identifications to be made. The comprehensive bioinformatic approach identified in vivo peptide hormones with a wide variation in length. In addition, from an analysis of as little as 5 ml of soybean xylem sap from cv. Williams 82, we identified twelve G. max peptide hormones. These G. max peptides overlapped with the seven species found in cv. Enrei (6).
We also found that hairy root culture medium and the xylem fluids of M. truncatula and G. max contained peptide breakdown products corresponding to well-characterized high molecular weight xylem sap proteins (24). This suggests that hairy root culture fluid is enriched in xylem sap proteins and peptide hormones. This is consistent with the expression of several genes in the root vascular tissue where peptide hormone secretion in proximity to the xylem vessels would enable them to join the xylem stream and be potential long distance signals. There is evidence for the long-distance movement of CLE, CEP, and XAP peptides (7, 21). The initial circa 10 mm root seeded into the hairy root culture medium is severed from existing transgenic roots and therefore, as the root system branches from this piece to form an extensive root mass, any xylem fluid in the proliferating roots would be free to leak from the severed end into the medium on a continuous basis. Given the relative ease at which axenic hairy roots can be established and continuously cultured using A. rhizogenes mediated transformation of diverse plants, the strategy for peptide hormone extraction, identification and characterization outlined here should find wide utility. For example, hairy roots can form on multiple members of the Solanaceae, Rosaceae, Fabaceae, Crassulaceae, Caesalpiniaceae, Brassicaceae, Polygonaceae, and Asteraceae families (22).
There was considerable variation in the PTMs of the peptide hormones identified. The proline moieties of CEP, CLE and XAP peptides identified in M. truncatula were variously hydroxylated. GmCLE32 and some MtCEP1 derivatives were glycosylated in a manner consistent with tri-arabinosylation. Since the conserved DY amino acids at the N-terminus is a well-recognized sulfation signal (32, 33), we concluded that the tyrosine moieties of MtXAPs were sulfated, and the sulfated versions of XAP1a and MtXAP5 that we synthesized had biological activity.
We also found natural variation in the length of N- and C-terminal sequences flanking the CEP domains in both M. truncatula and G. max. By contrast, the four related groups of MtXAP peptides identified in M. truncatula and G. max (XAP1/2, XAP3, XAP5 and XAP7) had defined N-termini but mostly variable C-termini, whereas CLE peptides appeared to be more accurately excised from the propeptide in M. truncatula.
The pattern of secreted CEP products indicates that endoproteolytic activity occurs at some distance from the N-terminal borders of the conserved domains. Since the MtCEP1 peptide has two domains and no peptide was found that spanned MtCEP1-D1 and MtCEP1-D2, this implies that an efficient endoproteolytic cleavage occurs between D1 and D2. In case of XAP peptides, it is possible that the motifs underlined in supplemental Fig. S5 could form part of an endoproteolytic cleavage site that potentially would enable the precise processing required to yield a product starting with the DY motif.
A model for plant peptide hormone maturation is presented in Fig. 10. The signal peptide cleavage triggers the propeptide to enter the secretion pathway followed by endoproteolytic and PTM activity. There is evidence for secretion pathway localization of several PTM enzymes (39–41). In addition, aminopeptidase-like and carboxypeptidase-like activity was evident in CEP maturation, and carboxypeptidase-like activity was implicated in XAP maturation. Carboxypeptidases appear to have a more definitive role in peptide hormone maturation (42), but it is not clear if this exoprotease activity is extracellular or this occurs before CEP and XAP secretion to the apoplast (Fig 10). There is some evidence for plant plasma membrane located amino peptidase activity (43), but little evidence for aminopeptidase secretion via the ER/Golgi pathway (44). Some evidence for extracellular trimming of peptide hormones has been reported (16, 24, 45, 46). Given that the xylem protein breakdown peptides also appeared to be subjected to aminopeptidase-like and carboxypeptidase-like activity, this supports their locality in the extracellular fluid. Similarly, these types of N- and C-terminal trimming activities are also found in the maturation of animal peptide hormones such as angiotensin and neuropeptides, which greatly affects their activity (47–49). Recently, the length of N-terminal extensions of IDA peptides has been shown to greatly affect the affinity for its receptor and its biological activity (17, 18, 50), and crystallography studies show specific interactions of peptide ligands with precise sites in their corresponding receptor (32) which would be affected by N- or C-terminal extensions. It is possible that N- and C-terminal trimming may play an important role in influencing peptide hormone maturation and/or receptor affinity.
Fig. 10.

Model for peptide hormone maturation. Signal peptide removal from the CEP, CLE and XAP prepropeptides enables their entry into the ER/Golgi-dependent secretion pathway. Further endoprotolytic processing and PTMs of proline and tyrosine would most likely occur during the secretion process, with more than one endoprotease being required for multidomain cleavages. Peptide hormone gene expression in the vascular tissue in proximity to the xylem vessels would enable the secreted product to join the xylem stream. The N- and C-terminal trimming activity may occur in the apoplast or on the cell surface or during secretion, and either trim larger less-biologically-active secreted products to a more biologically active size or be responsible for the turnover of secreted proteins. Evidence for the N- and C-terminal trimming of the peptides resulting from the breakdown of the high molecular weight xylem sap proteins (the xylem sap protein “degradome”) supports the latter hypothesis.
The biological activity of CEPs was distinct from CLE and some XAP peptides. We synthesized CEP peptides identical to those found in vivo and determined the effects of variations in PTMs, length and amino acid composition on their ability to influence M. truncatula root nodule and lateral root deployment and number. Surprisingly, only few CEP peptides were able to promote nodule number, but most CEP peptides negatively affected lateral root deployment. The implications of this variable activity on lateral organ formation on the interaction of CEPs with its putative receptor, CRA2 (51) is unknown.
We had previously shown that MtCEP1-D1 is further glycosylated with a three pentose oligosaccharide at P11 (9). By analogy with known glycopeptide hormones (AtCLV3, SlCLV3, LjCLERS1/2 and PSY) and cell wall extensin glycoproteins, this glycan most likely corresponds to the β-O-linked β1,2-tri-l-arabinofuranose oligomer (13, 19, 32, 52, 53). To probe the effect of glycosylation, we generated suitably protected derivatives of hydroxyproline containing β-O-linked monoarabinofuranose and β1,2-triarabinofuranose modifications and used these to synthesize glycosylated MtCEP1-D1 variants (mono- and tri-arabinose at P11), which were subsequently bioassayed along with the nonglycosylated form. Unexpectedly, tri-arabinosylation at P11 diminished activity, whereas mono-arabinosylation did not inhibit biological activity. This contrasts with most structurally characterized CLE peptide hormones where tri-arabinosylation appears essential for maximal biological activity (13, 53, 54). The strong biological activity of the dominant 15 amino acid MtCEP1-D1 peptide hydroxylated at P4 and 11 strongly suggests that it corresponds to the most biologically active mature functional peptide hormone.
The MtCLE17 peptide identified has the typical biological activity of CLE peptides in Arabidopsis as it reduces apical dominance and promotes lateral root formation in Medicago. MtCLE5 appears orthologous to the TDIF-like peptides (34), however, unlike TDIF-like peptides it also suppressed main root growth and promoted lateral root growth. In this regard, MtCLE5 is like a recently published synthetic hybrid CLE peptide (36). Therefore, MtCLE5 could represent a naturally occurring hybrid CLE peptide. Its expression was localized primarily to the root vasculature. XAP peptides are of uncertain mature size because of variability in their C-termini. We tested the biological activity of short and longer versions of the XAP peptides and showed that only the longer derivatives inhibit root growth, thus concluding that these peptides are indeed 17 or 18 amino acids long.
Transcript analysis suggested that several peptide hormone genes express predominantly in the vasculature including MtCEP5, MtCEP8, MtCLE5, and MtCLE17. Therefore, they could be directed to the xylem stream and be potential long-distance peptide hormones. The results of Tabata et al. and Okamoto et al. (5, 6) are consistent with those presented here. In addition, MtXAP1b/c, MtXAP3 (a member of the CIF family) and MtXAP7 were identified in Medicago xylem sap, and therefore are also candidates to be long distance peptides (6, 21).
The role of plant peptides in controlling important aspects of development is an exciting area of plant biology. Plants encode circa 1000 genes predicted to generate small secreted signaling peptides, however, the lack of in vivo validation of small secreted signaling expression and knowledge of the amino acid sequences of the mature peptide together with their PTMs has hampered their analysis. These peptides will most likely interact with a variety of uncharacterized surface receptors to trigger a wide array of developmental and defense responses through local and systemic circuits. The efficient isolation and identification of these secreted peptides from secreted fluids such as hairy root culture medium or xylem sap is a necessary step to determine their structure and biological activity, identify the receptor partner(s) and decipher these signaling circuits. The use of hairy root cultures combined with the detailed methods and approaches outlined here offers a way to identify multiple root-derived peptides in a single experiment in an axenic environment free of plant hormones and free of any possible feedback inhibition on peptide production that is controlled by the shoot. The lack of feedback inhibition may enable some peptides to accumulate to higher levels than in xylem fluid. In addition, hairy roots are readily formed on a variety of plants and can be easily exposed, for example, to different growth regimes or plant phytohormones to identify condition-specific production of root peptide hormones. In addition, the vast diversity of different sized peptide hormones found and the variations in their PTMs provides an insight into their processing and maturation. In total, we identified twelve M. truncatula peptide hormones from M. truncatula secreted fluids. The results suggest that peptide maturation may be more complicated than previously thought and involve endo- and exoproteolytic activities as well as other PTMs. Given that the hairy root culture fluids are xylem-sap-like, new long distance signaling peptides may be identified. These methods should enable multiple endogenous peptide hormones at mid pm to low nm levels to be detected routinely in hairy root cultures and xylem fluids in a wide variety of plants.
DATA AVAILABILITY
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD007614.
Supplementary Material
Acknowledgments
We thank Nijat Imin and Huw Ogilvie for advice in assembling the XAP phylogenetic tree.
Footnotes
* This work was supported by ARC grants to MAD: DP150104050 and LP150100826. NP was partly supported by an Endeavor Fellowship. NAMR was supported by an ANU Ph.D. scholarship supported by DP120101893. AI was supported by an Australian Post-graduate Award and an AW Howard Memorial Award. LC was supported by the Bruce-Veness Chandler and the John A. Lamberton research scholarships.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- PTMs
- post-translational-modifications
- Mt
- Medicago truncatula
- D1
- domain 1
- D2
- domain 2
- Gm
- Glycine max
- MS
- mass spectrometry.
REFERENCES
- 1. Czyzewicz N., Yue K., Beeckman T., and De Smet I. (2013) Message in a bottle: small signalling peptide outputs during growth and development. J. Exp. Bot. 64, 5281–5296 [DOI] [PubMed] [Google Scholar]
- 2. Araya T., Miyamoto M., Wibowo J., Suzuki A., Kojima S., Tsuchiya Y. N., Sawa S., Fukuda H., von Wiren N., and Takahashi H. (2014) CLE-CLAVATA1 peptide-receptor signaling module regulates the expansion of plant root systems in a nitrogen-dependent manner. Proc. Natl. Acad. Sci. U.S.A. 111, 2029–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Delay C., Imin N., and Djordjevic M. A. (2013) Regulation of Arabidopsis root development by small signaling peptides. Front. Plant Sci. 4, 352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Djordjevic M. A., Mohd-Radzman N. A., and Imin N. (2015) Small-peptide signals that control root nodule number, development, and symbiosis. J. Exp. Bot. 66, 5171–5181 [DOI] [PubMed] [Google Scholar]
- 5. Tabata R., Sumida K., Yoshii T., Ohyama K., Shinohara H., and Matsubayashi Y. (2014) Perception of root-derived peptides by shoot LRR-RKs mediates systemic N-demand signaling. Science 346, 343–346 [DOI] [PubMed] [Google Scholar]
- 6. Okamoto S., Suzuki T., Kawaguchi M., Higashiyama T., and Matsubayashi Y. (2015) A comprehensive strategy for identifying long-distance mobile peptides in xylem sap. Plant J. 84, 611–620 [DOI] [PubMed] [Google Scholar]
- 7. Fukuda H., and Higashiyama T. (2011) Diverse functions of plant peptides: entering a new phase. Plant Cell Physiol. 52, 1–4 [DOI] [PubMed] [Google Scholar]
- 8. Matsubayashi Y., Hanai H., Hara O., and Sakagami Y. (1996) Active fragments and analogs of the plant growth factor, phytosulfokine: structure-activity relationships. Biochem. Biophys. Res. Commun. 225, 209–214 [DOI] [PubMed] [Google Scholar]
- 9. Mohd-Radzman N. A., Binos S., Truong T. T., Imin N., Mariani M., and Djordjevic M. A. (2015) Novel MtCEP1 peptides produced in vivo differentially regulate root development in Medicago truncatula. J. Exp. Bot. 66, 5289–5300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Roberts I., Smith S., Stes E., De Rybel B., Staes A., van de Cotte B., Njo M. F., Dedeyne L., Demol H., Lavenus J., Audenaert D., Gevaert K., Beeckman T., and De Smet I. (2016) CEP5 and XIP1/CEPR1 regulate lateral root initiation in Arabidopsis. J. Exp. Bot. 67, 4889–4899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Whitford R., Fernandez A., Tejos R., Perez A. C., Kleine-Vehn J., Vanneste S., Drozdzecki A., Leitner J., Abas L., Aerts M., Hoogewijs K., Baster P., De Groodt R., Lin Y. C., Storme V., Van de Peer Y., Beeckman T., Madder A., Devreese B., Luschnig C., Friml J., and Hilson P. (2012) GOLVEN secretory peptides regulate auxin carrier turnover during plant gravitropic responses. Dev. Cell 22, 678–685 [DOI] [PubMed] [Google Scholar]
- 12. Ohyama K., Ogawa M., and Matsubayashi Y. (2008) Identification of a biologically active, small, secreted peptide in Arabidopsis by in silico gene screening, followed by LC-MS-based structure analysis. Plant J. 55, 152–160 [DOI] [PubMed] [Google Scholar]
- 13. Okamoto S., Shinohara H., Mori T., Matsubayashi Y., and Kawaguchi M. (2013) Root-derived CLE glycopeptides control nodulation by direct binding to HAR1 receptor kinase. Nat. Commun. 4, 2191. [DOI] [PubMed] [Google Scholar]
- 14. Ito Y., Nakanomyo I., Motose H., Iwamoto K., Sawa S., Dohmae N., and Fukuda H. (2006) Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science 313, 842–845 [DOI] [PubMed] [Google Scholar]
- 15. Lease K. A., and Walker J. C. (2006) The Arabidopsis unannotated secreted peptide database, a resource for plant peptidomics. Plant Physiol. 142, 831–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schardon K., Hohl M., Graff L., Pfannstiel J., Schulze W., Stintzi A., and Schaller A. (2016) Precursor processing for plant peptide hormone maturation by subtilisin-like serine proteinases. Science 354, 1594–1597 [DOI] [PubMed] [Google Scholar]
- 17. Santiago J., Brandt B., Wildhagen M., Hohmann U., Hothorn L. A., Butenko M. A., and Hothorn M. (2016) Mechanistic insight into a peptide hormone signaling complex mediating floral organ abscission. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Butenko M. A., Wildhagen M., Albert M., Jehle A., Kalbacher H., Aalen R. B., and Felix G. (2014) Tools and Strategies to Match Peptide-Ligand Receptor Pairs. Plant Cell 26, 1838–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ohyama K., Shinohara H., Ogawa-Ohnishi M., and Matsubayashi Y. (2009) A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nat. Chem. Biol. 5, 578–580 [DOI] [PubMed] [Google Scholar]
- 20. Corcilius L., Hastwell A. H., Zhang M., Williams J., Mackay J. P., Gresshoff P. M., Ferguson B. J., and Payne R. J. (2017) Arabinosylation Modulates the Growth-Regulating Activity of the Peptide Hormone CLE40a from Soybean. Cell Chem. Biol. 24, 1347–1355 [DOI] [PubMed] [Google Scholar]
- 21. Doblas V. G., Smakowska-Luzan E., Fujita S., Alassimone J., Barberon M., Madalinski M., Belkhadir Y., and Geldner N. (2017) Root diffusion barrier control by a vasculature-derived peptide binding to the SGN3 receptor. Science 355, 280–284 [DOI] [PubMed] [Google Scholar]
- 22. Porter J. R. (1991) Host Range and Implications of Plant Infection by Agrobacterium-Rhizogenes. Crit. Rev. Plant Sci. 10, 387–421 [Google Scholar]
- 23. Imin N., Mohd-Radzman N. A., Ogilvie H. A., and Djordjevic M. A. (2013) The peptide-encoding CEP1 gene modulates lateral root and nodule numbers in Medicago truncatula. J. Exp. Bot. 64, 5395–5409 [DOI] [PubMed] [Google Scholar]
- 24. Djordjevic M. A., Oakes M., Li D. X., Hwang C. H., Hocart C. H., and Gresshoff P. M. (2007) The glycine max xylem sap and apoplast proteome. J. Proteome Res. 6, 3771–3779 [DOI] [PubMed] [Google Scholar]
- 25. Kusumawati L., Imin N., and Djordjevic M. A. (2008) Characterization of the secretome of suspension cultures of Medicago species reveals proteins important for defense and development. J. Proteome Res. 7, 4508–4520 [DOI] [PubMed] [Google Scholar]
- 26. Holmes P., Goffard N., Weiller G. F., Rolfe B. G., and Imin N. (2008) Transcriptional profiling of Medicago truncatula meristematic root cells. BMC Plant Biol. 8, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ogilvie H. A., Imin N., and Djordjevic M. A. (2014) Diversification of the C-TERMINALLY ENCODED PEPTIDE (CEP) gene family in angiosperms, and evolution of plant-family specific CEP genes. BMC Genomics 15, 870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Frith M. C., Saunders N. F., Kobe B., and Bailey T. L. (2008) Discovering sequence motifs with arbitrary insertions and deletions. PLoS Comput. Biol. 4, e1000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stamatakis A. (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 [DOI] [PubMed] [Google Scholar]
- 30. Huson D. H., and Scornavacca C. (2012) Dendroscope 3: an interactive tool for rooted phylogenetic trees and networks. Syst. Biol. 61, 1061–1067 [DOI] [PubMed] [Google Scholar]
- 31. Saur I. M., Oakes M., Djordjevic M. A., and Imin N. (2011) Crosstalk between the nodulation signaling pathway and the autoregulation of nodulation in Medicago truncatula. New Phytol. 190, 865–874 [DOI] [PubMed] [Google Scholar]
- 32. Amano Y., Tsubouchi H., Shinohara H., Ogawa M., and Matsubayashi Y. (2007) Tyrosine-sulfated glycopeptide involved in cellular proliferation and expansion in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 104, 18333–18338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Song W., Liu L., Wang J., Wu Z., Zhang H., Tang J., Lin G., Wang Y., Wen X., Li W., Han Z., Guo H., and Chai J. (2016) Signature motif-guided identification of receptors for peptide hormones essential for root meristem growth. Cell Res. 26, 674–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hastwell A. H., de Bang T. C., Gresshoff P. M., and Ferguson B. J. (2017) CLE peptide-encoding gene families in Medicago truncatula and Lotus japonicus, compared with those of soybean, common bean and Arabidopsis. Sci. Rep. 7, 9384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Djordjevic M. A., Oakes M., Wong C. E., Singh M., Bhalla P., Kusumawati L., and Imin N. (2011) Border sequences of Medicago truncatula CLE36 are specifically cleaved by endoproteases common to the extracellular fluids of Medicago and soybean. J. Exp Bot. 62, 4649–4659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hirakawa Y., Shinohara H., Welke K., Irle S., Matsubayashi Y., Torii K. U., and Uchida N. (2017) Cryptic bioactivity capacitated by synthetic hybrid plant peptides. Nat. Commun. 8, 14318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Meng L., and Feldman L. J. (2010) CLE14/CLE20 peptides may interact with CLAVATA2/CORYNE receptor-like kinases to irreversibly inhibit cell division in the root meristem of Arabidopsis. Planta 232, 1061–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kinoshita A., Nakamura Y., Sasaki E., Kyozuka J., Fukuda H., and Sawa S. (2007) Gain-of-function phenotypes of chemically synthetic CLAVATA3/ESR-related (CLE) peptides in Arabidopsis thaliana and Oryza sativa. Plant Cell Physiol. 48, 1821–1825 [DOI] [PubMed] [Google Scholar]
- 39. Ogawa-Ohnishi M., Matsushita W., and Matsubayashi Y. (2013) Identification of three hydroxyproline O-arabinosyltransferases in Arabidopsis thaliana. Nat. Chem. Biol. 9, 726–730 [DOI] [PubMed] [Google Scholar]
- 40. Hanai H., Nakayama D., Yang H., Matsubayashi Y., Hirota Y., and Sakagami Y. (2000) Existence of a plant tyrosylprotein sulfotransferase: novel plant enzyme catalyzing tyrosine O-sulfation of preprophytosulfokine variants in vitro. FEBS Lett. 470, 97–101 [DOI] [PubMed] [Google Scholar]
- 41. Yuasa K., Toyooka K., Fukuda H., and Matsuoka K. (2005) Membrane-anchored prolyl hydroxylase with an export signal from the endoplasmic reticulum. Plant J. 41, 81–94 [DOI] [PubMed] [Google Scholar]
- 42. Tamaki T., Betsuyaku S., Fujiwara M., Fukao Y., Fukuda H., and Sawa S. (2013) SUPPRESSOR OF LLP1 1-mediated C-terminal processing is critical for CLE19 peptide activity. Plant J. 76, 970–981 [DOI] [PubMed] [Google Scholar]
- 43. Murphy A., and Taiz L. (1999) Localization and characterization of soluble and plasma membrane aminopeptidase activities in Arabidopsis seedlings. Plant Physiol. Bioch. 37, 431–443 [Google Scholar]
- 44. Matsui M., Fowler J. H., and Walling L. L. (2006) Leucine aminopeptidases: diversity in structure and function. Biol. Chem. 387, 1535–1544 [DOI] [PubMed] [Google Scholar]
- 45. Ni J., and Clark S. E. (2006) Evidence for functional conservation, sufficiency, and proteolytic processing of the CLAVATA3 CLE domain. Plant Physiol. 140, 726–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ni J., Guo Y., Jin H., Hartsell J., and Clark S. E. (2011) Characterization of a CLE processing activity. Plant Mol. Biol. 75, 67–75 [DOI] [PubMed] [Google Scholar]
- 47. Hildebrand D., Merkel P., Eggers L. F., and Schluter H. (2013) Proteolytic processing of angiotensin-I in human blood plasma. PLoS ONE 8, e64027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hook V., Funkelstein L., Lu D., Bark S., Wegrzyn J., and Hwang S. R. (2008) Proteases for processing proneuropeptides into peptide neurotransmitters and hormones. Annu. Rev. Pharmacol. Toxicol. 48, 393–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Velez J. C., Ierardi J. L., Bland A. M., Morinelli T. A., Arthur J. M., Raymond J. R., and Janech M. G. (2012) Enzymatic processing of angiotensin peptides by human glomerular endothelial cells. Am. J. Physiol. Renal Physiol 302, F1583–F1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schaller A., Stintzi A., and Graff L. (2012) Subtilases - versatile tools for protein turnover, plant development, and interactions with the environment. Physiol. Plant 145, 52–66 [DOI] [PubMed] [Google Scholar]
- 51. Mohd-Radzman N. A., Laffont C., Ivanovici A., Patel N., Reid D., Stougaard J., Frugier F., Imin N., and Djordjevic M. A. (2016) Different Pathways Act Downstream of the CEP Peptide Receptor CRA2 to Regulate Lateral Root and Nodule Development. Plant Physiol. 171, 2536–2548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Akiyama Y., and Kato K. (1977) Structure of Hydroxyproline-Arabinoside from Tobacco Cells. Agr Biol. Chem. Tokyo 41, 79–81 [Google Scholar]
- 53. Xu C., Liberatore K. L., MacAlister C. A., Huang Z., Chu Y. H., Jiang K., Brooks C., Ogawa-Ohnishi M., Xiong G., Pauly M., Van Eck J., Matsubayashi Y., van der Knaap E., and Lippman Z. B. (2015) A cascade of arabinosyltransferases controls shoot meristem size in tomato. Nat. Genet. 47, 784–792 [DOI] [PubMed] [Google Scholar]
- 54. Kaeothip S., Ishiwata A., and Ito Y. (2013) Stereoselective synthesis of Arabidopsis CLAVATA3 (CLV3) glycopeptide, unique protein post-translational modifications of secreted peptide hormone in plant. Org. Biomol. Chem. 11, 5892–5907 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD007614.