Abstract
Glycosylation is perhaps the most common post-translational modification. Recently there has been growing interest in cataloging the glycan structures, glycoproteins, and specific sites modified and deciphering the biological functions of glycosylation. Although the results are piling up for N-glycosylation, O-glycosylation is seriously trailing behind. In our review we reiterate the difficulties researchers have to overcome in order to characterize O-glycosylation. We describe how an ingenious cell engineering method delivered exciting results, and what could we gain from “wild-type” samples. Although we refer to the biological role(s) of O-glycosylation, we do not provide a complete inventory on this topic.
To understand the biological role of glycosylation, in an ideal world one would study intact glycoproteins. Recent developments have made such analyses possible (1), but proteins modified at multiple sites, displaying significant macro- as well as microheterogeneity still represent a challenge. Mostly the chromatographic separation is lacking. One would have to separate the protein of interest from all other components, and somehow fractionate the isomeric structures. Even with relatively successful top-down experiments (1, 2), one also has to collect data using the “second best” solutions (1) studying the glycan pool that provides detailed information about the carbohydrate structures; (2) characterizing intact glycopeptides that provide information about the microheterogeneity; (3) gaining information about the unmodified sequences that may yield information about the macroheterogeneity.
The Different “Classes” of Extracellular O-Glycosylation
All O-glycopeptides feature a carbohydrate residue covalently linked to the hydroxyl group of an amino acid. Among the coded amino acids serine, threonine and tyrosine can be modified this way. The modifying sugar unit directly linked to the amino acid could be Fuc, Glc, GalNAc, GlcNAc, Man, and Xyl. The modifications are performed in the ER1 and the Golgi. Thus, they affect secreted proteins and the extracellular domain of membrane proteins. The lumenal side of ER, Golgi and certain vesicles are considered as such.
O-Fucosylation
α-linked O-fucose modification was originally considered EGF-domain specific (3). Its consensus sequence was determined as CXXGG(S/T)C and the observation of an elongated structure, NeuAcα2,6Galβ1,4GlcNAcβ1, 3Fuc has also been reported (Fig. 1A (4)). The presence of the 2 Gly residues N-terminal to the site of glycosylation is not a very strict requirement. For example, Thr-3103 of Versican core protein, preceded by an Ala instead of Gly, has been detected bearing a single Fuc, as well as di- and trisaccharides (5, 6). Thrombospondin type 1 repeats (TSRs) also may be O-fucosylated (7, 8). Presently, the CX2/3(S/T)CX2G sequence is considered as the consensus motif (9). Both consensus motifs are linked somewhat to the Cys-framework within the specified domains, and the enzymes performing the “deposition” of the core sugar unit are Protein O-fucosyl transferase-1 and −2 for EGF or TSR domains, respectively (8, 10). The glycan extension also follows two distinct pathways (11). Fucoses on the EGF domain can be elongated to the above mentioned tetrasaccharide, whereas in TSRs only a β1,3-linked Glc is added to the core unit. Our knowledge about the biological function(s) of O-fucosylation is quite limited, though it has been implicated in protein-protein interactions, intercellular signaling and protein folding (9, 12, 13). The O-fucosylation of the IgG1 light chain has also been reported. The modified sequence does not comply with any of the consensus requirements listed above, and it features only the α-linked Fuc (14). This observation suggests the existence of a different pathway.
Fig. 1.
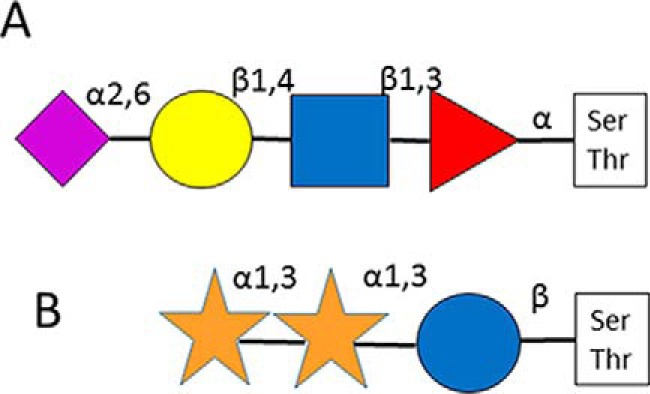
Detailed structures of O-fucosyl glycan (A) (4); and O-glucosyl glycan (B) (17).
O-Glucosylation
O-Glucosylation was first described on blood clotting Factors VII and IX (15, 16). The β-linked Glc residue was reported to be elongated with one or two α 1,3-linked Xyl moieties (Fig. 1B (17)). The modification seems to be EGF-domain-specific, Protein Z as well as thrombospondin have been reported as modified in their EGF domains (16, 18). Recently, both O-glucosylation and fucosylation has been reported on the EGF-like domain of AMACO, an extracellular matrix protein of unknown function (19). Already the first reports speculated that there is a consensus motif for O-glucosylation: CXSXPC (16). The enzyme responsible for the modification is O-glucosyltransferase Rumi (20). Interestingly, Rumi also may function as protein O-xylosyltransferase (21). The exact biological role of this modification has not been deciphered yet, but EGF-glucosylation seems to be essential for mouse embryonic development and Notch signaling (22).
O-GalNAcylation or Mucin-type Glycosylation
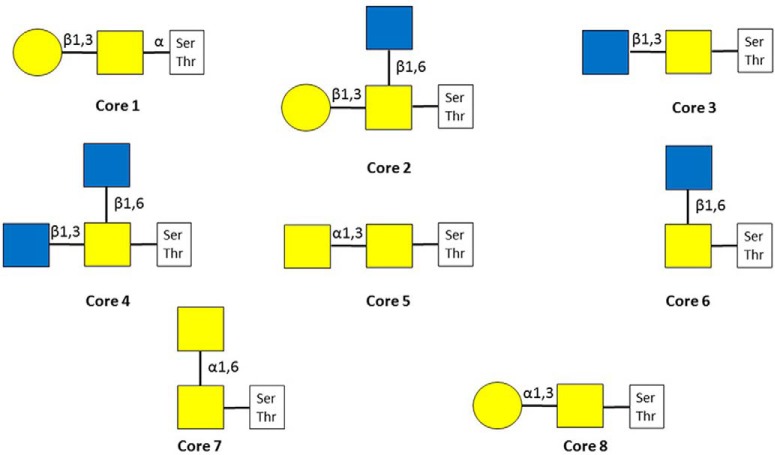
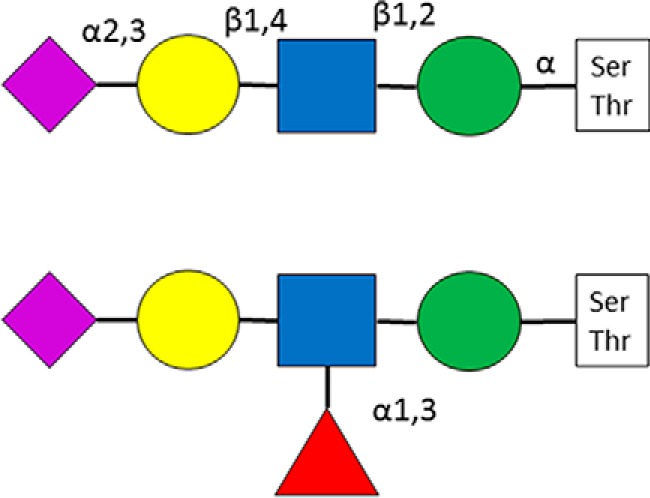
This is the most common mammalian O-glycosylation. It was named after a family of secreted and transmembrane proteins that feature heavily glycosylated repetitive peptide stretches, the so-called “variable number of tandem repeat” regions. More than 20 different GalNAc-transferases may perform the primary glycosylation step (23). As these glycosyltransferases display distinct but overlapping substrate specificities (24), there is no consensus motif for mucin-type O-glycosylation, although there are some tendencies. Thr residues are modified to much higher extent compared with Ser (25, 26). Pro, and to lesser extent, Ser, Thr, Ala, and Gly residues are overrepresented around modification sites (24–28). Eight different core structures exist (Fig. 2) (23). Core-1–4 structures may be considered common, core-1 and core-2 (that may be considered a branched core-1) glycans are the most frequently occurring ones in serum (29). The core-1 structure is the only type of glycan that has been identified on Tyr-residues unambiguously so far (30, 31). Cores 1 and 2 may be elongated into poly-N-acetyllactosamine chains and may feature antigens, such as blood-type determinants (23, 32, 33). Even some small O-linked structures are antigenic: the core GalNAc by itself or sialylated in the sixth position as well as the core-1 disaccharide, and these are termed as Tn, sialyl-Tn and T antigens, respectively (23). Cores 5–8 are rare, and have been detected only from specific sources (23). All cores may be modified with sialic acids - terminal Gal residues bear only α2,3-linked ones. Covalent modifications of these mucin-type glycans have been reported, such as the O-acetylation of neuraminic acid (1, 23, 34), and the sulfation of different Gal and GlcNAc units (23, 35, 36). Mucin-type O-glycosylation has been implicated in a wide variety of biological processes, such as interaction with pathogens (37, 38), cell adhesion (39–41), and proteolytic processing (40, 42–45). Altered glycosylation patterns have been linked to different diseases. For example, defective glycosylation of IgA1 has been reported in patients with IgA nephropathy and Henoch-Schoenlein purpura nephritis (46). It has also been documented that the O-glycosylation pattern is altered by cancer (24): higher Tn antigen levels increase its invasiveness (47, 48).
Fig. 2.
Mucin-type glycan core structures.
O-GlcNAcylation
It has been known for decades that Ser and Thr residues of nuclear and cytosolic proteins may be modified with a β-linked GlcNAc (49). O-GlcNAcylation fulfills signaling and regulatory functions within the cell (50). Both the enzyme performing the glycosylation as well as the glycosidase removing it have been identified (51–54). More recently, it has been reported that extracellular proteins also may feature this modification (55). The enzyme performing the modification within the ER lumen, EGF-domain-specific O-linked N-acetylglucosamine transferase has first been identified in Drosophila (56) and later in mammals (57). In Drosophila this modification has been implicated in cell-cell interaction (56). The biological role(s) of extracellular O-GlcNAcylation in mammals has not been determined yet, although multiple proteins have been found bearing O-GlcNAc in extracellular environment (5).
O-Mannosylation
O-mannosylation was originally categorized as a unique fungal type of O-glycosylation. Later it was discovered that mammalian dystroglycans also bear such modifications (Fig. 3 (58)), and their glycosylation deficiency leads to pathological changes (58). Cadherins were also identified as extensively modified with O-mannosyl glycans (59) and that O-mannosylation of E-cadherin is crucial for cell adhesion (60). The modification is initiated in the ER, and from various studies only a handful of additional glycoproteins were found O-mannosylated (6, 61–63). O-glycan studies revealed that mammalian brain is especially rich in such modifications (64). In this study, the existence of branched O-mannosyl glycan structures was also demonstrated (64), the core Man is modified with an additional, β1,6-linked GlcNAc that may be further elongated (65). It has been reported that branched O-mannosyl structures displaying HNK-1 epitopes (sulfoglucuronyl “capping”) influence cell-cell and cell-matrix interactions in the developing nervous system (66). Recently, protein modification with a single mannose residue has also been reported (67, 68).
Fig. 3.
The detailed structures of some O-mannosyl glycans (58).
O-Xylosylation
In a special class of glycoproteins, proteoglycans the core saccharide unit is xylose and it modifies Ser residues. Proteoglycans feature huge extended glycan chains: glycosaminoglycans (GAGs). This type of O-glycosylation will not be discussed in this paper, for an excellent summary on the structure and biosynthesis of proteoglycans see chapter 17 of (23).
Difficulties Encountered in O-Glycosylation Analysis
Despite the differences among the glycosylation processes, the resulting glycans and their biological roles, the different classes of O-glycosylation represent very similar analytical challenges.
The Lack of Consensus Motif
Although N-glycosylation features a consensus motif, and the potential sites can usually be isolated in individual peptides upon proteolytic treatment, the same is not true for O-glycopeptides. Because so many different glycosyltransferases are involved in the primary protein modification, not surprisingly, there is no universal consensus motif. Multiple potential modification sites are present in most of the peptides irrespective of the proteolytic enzyme used, and not infrequently these sites are clustered. Only O-glucosylation and O-fucosylation are different, as these are linked to specific domains, and the modified amino acids are in a specific position in reference to the disulfide bridges (see above).
However, as described earlier the existence of a fucosyl-transferase of different specificity cannot be excluded (14).
Significant Macroheterogeneity
N-glycosites are believed mostly highly, if not fully occupied—although because the glycopeptides are usually selectively enriched the available information may not be entirely reliable (69, 70). O-linked glycosylation sites may feature a lot more significant macroheterogeneity - most available data seem to suggest this (25, 71). However, there are reports that indicate the full occupancy of certain modification sites (72, 73). At the same time, assessing the occupancy rates for O-glycopeptides even in a single protein is extremely challenging because the ionization efficiency of the unmodified and differently glycosylated sequences is not the same and cannot be calculated or predicted. Identification of lower abundance glycoforms is also hampered by the chromatographic behavior of differently modified glycopeptides (see later).
Microheterogeneity
Although N-linked glycosylation sites may feature 50+ different glycan structures (6), the glycan repertoire of individual O-glycosylation sites seems to be more limited (6, 74). At the same time, the O-glycan-pool analyses of gastric mucins revealed an impressive array of oligosaccharide structures (32, 33). However, this diversity has not yet been “translated” to site-specific modifications. Nevertheless, it has been reported that the same sites can be modified by glycans of different mucin-type core structures (75, 76), or mannosyl as well as GalNAc-core glycans (6, 77). It has also been reported that the mucin-type core structure may be altered during recycling (78). Assessing the macro- and microheterogeneity is complicated by the presence of multiple and (frequently) clustered modification sites whose differential occupancy may yield numerous isomeric structures. Complexity is also increased by rampant proteolytic digestion in body fluids (25, 79, 80).
Chromatographic Behavior
The vast majority of proteomics data is acquired using reversed-phase separation of the sample components before MS/MS analysis. Increasing acidity leads to longer reversed phase retention times for N-glycopeptides when weak acids, such as formic or acetic acid are used as ion pairing agents, i.e. in most LC-MS/MS experiments (34). The addition of a few “neutral” sugar units, for example, an extra antenna, does not alter the retention times significantly, and different N-glycoforms of the same acidity usually coelute (6, 34, 81). Thus, from the accurately measured masses of the coeluting components additional glycoforms can be identified even if no MS/MS data were acquired or relatively poor MS/MS spectra resulted from the analyses (6). O-glycopeptides are somewhat different in this aspect. Acidic O-glycopeptides also elute later than their neutral counterparts (34). At the same time, the addition of just a disaccharide at a different position, i.e. multiple modification has an even more profound effect on their chromatographic behavior. It has been documented that because of such alteration the retention time might be shortened by as much as 5 min when a relatively shallow LC gradient was applied (80). Thus, finding all O-glycoforms represents a bigger challenge even in single protein analysis than the characterization of N-glycosylation.
MS/MS Behavior
Mass spectrometry has become the method of choice for all kind of PTM analyses. It has been essential for the characterization of glycosylation as well. Here we present what information can be gleaned from spectra acquired using different MS/MS techniques. All our examples represent mucin-type glycosylation. However, the few existing/available data on the other type of O-glycopeptides indicate that the rules of fragmentation are very similar for the other cores as well (see references for Fuc (5, 6, 14); Glc (82); Man (6, 83); and GlcNAc (84)).
Collision-induced dissociation is the most widely used MS/MS activation method. It was reported more than two decades ago that the favored process in the CID analysis of O-glycosylation is the gas-phase loss of the glycan via a rearrangement reaction without leaving a telltale sign on the previously modified residue (85). Since then, numerous papers and reviews discussed the CID fragmentation pattern of O-glycopeptides and also the possibility and limitations of peptide sequence and modification site assignments (86, 87).
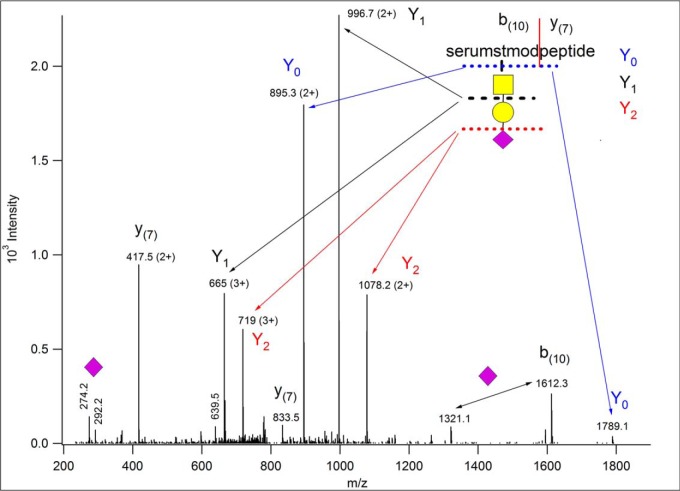
Fig. 4 represents the “typical” ion trap CID fragmentation of an O-glycopeptide decorated with a simple oligosaccharide. These data were acquired from a tryptic digest, in LC/MS/MS mode on an LTQ-Orbitrap XL mass spectrometer (Thermo Fisher Scientific, Waltham, MA). Although the precursor ions were measured with high resolution, the CID acquisition was performed in the linear ion trap. Ion trap CID is a resonance-activation process leading mostly to single bond cleavages. Glycosidic bond fragmentation clearly dominates the spectrum. Not surprisingly very abundant fragments were detected with charge retention at the reducing end (more precisely on the peptide connected to it). These are the so-called Y ions, numbered from the reducing end of the glycan (for nomenclature see (88)). Such sugar-unit-loss ions may be formed with the same charge as the precursor ion, but also at lower charge states. The only clearly visible nonreducing end fragments in this spectrum represent the terminal neuraminic acid at m/z 292 and 274, i.e. the B1 fragment and an ion formed via water loss from it, respectively. The B3 fragment (NeuAcHexHexNAc, at m/z 657) was also detected, however, practically at the noise level. Among the fragments unaccounted for there is one that seems to be present both as singly and doubly charged (remember, the spectrum was recorded with low resolution and low mass accuracy). One could guess that it is most likely a peptide y fragment formed in front of a Pro residue. O-glycopeptides frequently contain prolines (27, 28), and this imino acid usually yields an abundant y ion. In addition, the other half of the peptide, i.e. the corresponding b fragment may also be unusually abundant. This is the case for this peptide, the corresponding b ion was indeed detected at m/z 1612 (yi+bn-i = MH++1), still bearing the trisaccharide. Interestingly, this fragment was also detected without the terminal sialic acid at m/z 1321 indicating secondary fragmentation, unexpected in ion trap CID. To sum it up, from these data one can deduce the composition of the glycan: NeuAcHexHexNAc, the linearity of its structure, and the mass of the peptide. One can speculate that it may be about 17 residues long, and a Pro residue may be present within the sequence, and the glycosylation occurs N-terminally from this Pro. Measuring the fragments in the Orbitrap would confirm the charge states guessed, and deliver masses with high accuracy, but the amino acid sequence and the modification site still would not be identified whereas some of the weaker ions might be lost because of the lower sensitivity of the analyzer.
Fig. 4.
Ion trap CID data of m/z 815.731(3+). From these data one can deduce the composition of the glycan: NeuAcHexHexNAc, the linearity of its structure, and the mass of the peptide. One can speculate that it may be about 17 residues long, and a Pro residue may be present within the sequence, and the glycosylation occurs N-terminally from this Pro. The peptide fragment numbering in parantheses indicates that the lengths of the corresponding sequence stretches were guessed from the m/z values.
Beam-type CID performed in collision cells (Q-TOF instruments or HCD (higher-energy C-trap dissociation) in Thermo mass spectrometers) may provide much needed information on the identity of the peptide at the expense of the glycan characterization. The beam-type CID spectrum of the same glycopeptide (Fig. 5) is full of informative peptide fragments, the modified sequence can readily be identified as AVGAQVLESTPPPHVMR [682–698] of Uniprot F1MNW4, bovine ITIH2 protein. However, one can only guess/calculate the glycan mass and structure because of the (usually) complete loss of the glycan structure upon this type of activation. The oxonium ions clearly indicate the presence of neuraminic acid (m/z 274 and 292) and GalNAc (m/z 204). This latter fragment ion is diagnostic for both GlcNAc and GalNAc, but the distribution of further fragments clearly establishes GalNAc on this peptide (89). One can then consider the simplest mucin-type sialylated structure, NeuAcGalGalNAc and the presence of the doubly charged Y0 at m/z 895.49(2+) confirms our hunch. (Because the ion trap CID data revealed the mass and linearity of the glycan as well as the mass of the peptide, using ion trap and beam-type CID data combined may deliver more reliable and faster results. Except we do not know of any software that enables one to perform O-glycopeptide analysis doing so.) A database search will deliver the peptide identification once the proper glycan structure is permitted as variable modification. However, site assignments usually cannot be accomplished from CID/HCD data. In our example either Ser-10 or Thr-11 bears the trisaccharide. Thus, the C-terminal fragments starting from y8 or y9 should feature the appropriate mass shift, except each was detected without the modification, and that is rather typical for beam-type CID spectra. Sometimes abundant peptide fragments (typically representing Xxx-Pro bond cleavages) may be observed both unmodified and partially or fully retaining the glycan. One certainly could try to lower the collision energy to prevent sugar losses. Acquiring data at different collision energies may provide sufficient information for both sequence identification and site assignment. However, the extent of fragmentation depends on a lot of factors, and finding the optimal conditions for most components in automated LC/MS/MS settings may be a daunting task.
Fig. 5.
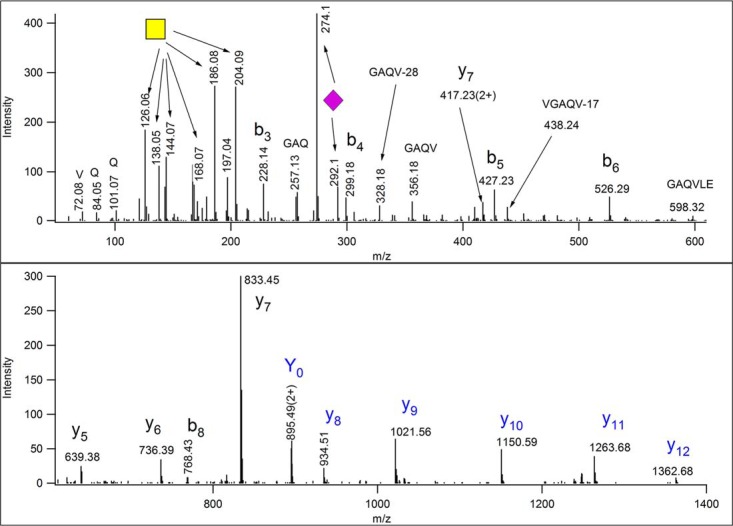
Beam-type (Q-TOF) CID data of m/z 815.73(3+). From these data one could identify the modified peptide as AVGAQVLESTPPPHVMR ([682–698] of bovine ITIH2 protein (Uniprot ID: F1MNW4)). The glycan composition can be “guessed” from the oxonium ions of HexNAc and Neu5Ac, and from the presence of m/z 895.49(2+) as Neu5AcHexHexNAc. Intensity pattern of the fragments of the HexNAc oxonium ion (m/z 204) ascertain its identity as GalNAc (89). Thus, the sugar structure is most likely Neu5AcGalGalNAc. Its linearity or branching cannot be determined, because larger Y fragments were not detected, nor were m/z 454.16 or 495.18 observed, indicating sialylation of Gal or GalNAc, respectively. The site of modification cannot be assigned. All ions that were gas-phase deglycosylated, and thus, were detected unmodified are printed in blue. Fragment y8 belongs to this series only if Thr-10 is the modification site.
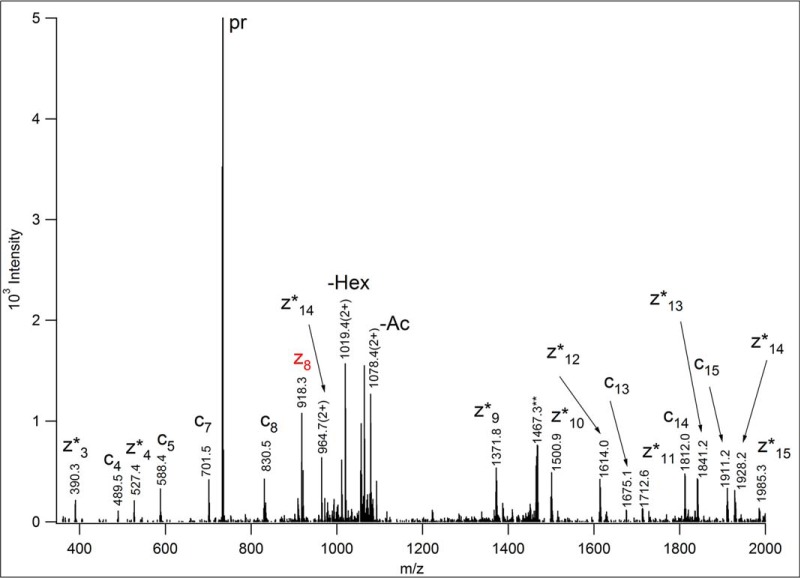
Electron capture and electron transfer dissociation (ECD and ETD, respectively), two recent MS/MS activation methods, lead to the formation of radical precursor ions that undergo fragmentation mostly yielding peptide backbone fragments via cleavages between the amino group and the alpha carbon, while retaining the amino acid side chains and their modifications intact (90, 91). Thus, efficient ECD/ETD fragmentation enables both the identification of the modified peptide and the assignment of the site of modification. Indeed, practically all recent successful O-glycosylation studies used radical based fragmentation to decipher modification sites (6, 25, 26, 74, 79, 80, 92–95). ECD is performed in FT-ICR mass spectrometers, whereas ETD works in ion traps and more efficiently. Thus, we included an ETD spectrum to illustrate the advantages and limitations of the method. Fig. 6 shows the ETD spectrum of the human homologue of the earlier described ITIH2 peptide. This spectrum was acquired from m/z 733.692(3+) precursor ion, in the linear ion trap of an LTQ-Orbitrap Elite mass spectrometer (Thermo Fisher Scientific). Because in ETD the amino acid side chains mostly remain intact, one has to make assumptions about the glycans present, and has to provide a list of variable modifications during the database search. It is practical to start with the most common structures or with the targeted glycans if a selective enrichment was performed. In our case the human serum sample was treated with neuraminidase, thus, only HexNAc (representing GalNAc) and HexHexNAc (representing GalGalNAc) modifications were considered. From the spectrum presented AQGSQVLES(HexNAcHex)TPPPHVMR [682–698] of Uniprot P19823, human ITIH2 protein was identified with high confidence. However, the modification site assignment had to be confirmed manually. Ser-4 could be excluded as modification site by the presence of unmodified N-terminal fragment ion series c4-c8. However, the other potential sites are in adjacent positions, and the observation of c9 and/or z8 was necessary to make an unambiguous site assignment. As indicated with its red color, only z8 was detected, unmodified, and thus, Ser-9 was assigned as the glycosylation site. Interestingly, most of the C-terminal fragments were observed as z+1 ions, the products of hydrogen migration (96), and some carbohydrate fragmentation, reported earlier for O-glycopeptides (92, 97) was also detected. The ETD fragments also could have been measured in the Orbitrap, with high resolution and mass accuracy making the assignments more reliable, but unfortunately at the expense of sensitivity.
Fig. 6.
ETD spectrum of m/z 733.692(3+), acquired in the linear ion trap of an LTQ-Orbitrap Elite mass spectrometer (Thermo Fisher Scientific). AQGSQVLES(HexNAcHex)TPPPHVMR [682–698] of human ITIH2 protein (Uniprot ID: P19823) was identified from these data. In order to identify the peptide modified, common O-glycan structures were listed as variable modifications in the database search. The identity or the linkage of the glycans cannot be established from these data. The fragments were measured with low resolution, thus, the charge state of the ions was “determined” from the potential matches. Asterisks indicate the products of hydrogen migration, i.e. z+1 fragments. Although Ser-4 easily could be excluded as the glycosylation site, the C-terminal fragment printed in red provided the decisive information for the assignment of Ser-9 for that role, because it indicates that Thr-10 is unmodified. Some carbohydrate fragmentation was also detected: the loss of 42 Da from the N-acetylhexosamine, and some terminal hexose loss, both from the charge-reduced precursor ion. The precursor ion is labeled as “pr.” The **-labeled ion is the charge-reduced ion of a coeluting (2+) component.
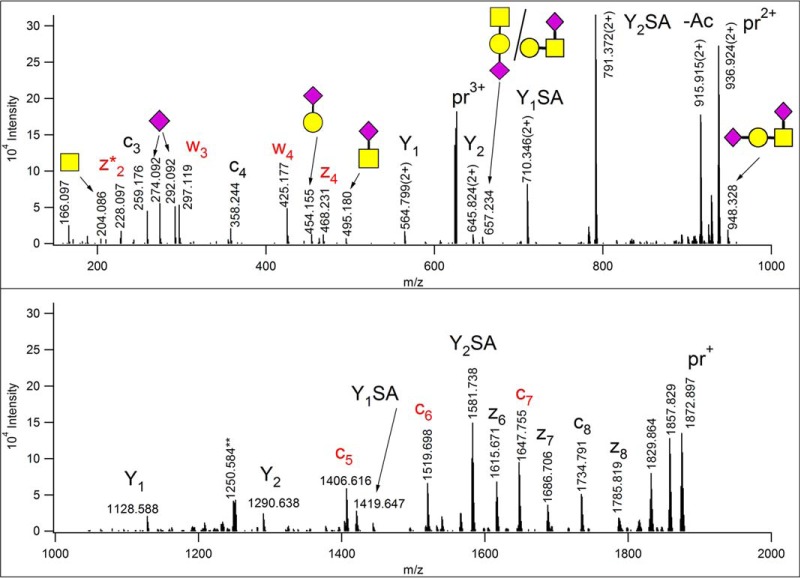
The Thermo Tribrid mass spectrometers (Thermo Fisher Scientific), with new design features dramatically improved the sensitivity of ETD analysis. Thus, measuring the fragments in the Orbitrap analyzer does not translate into significant information loss. In addition, a new MS/MS activation option, EThcD has been made available. In this hybrid technique ETD activation is performed first, then the entire “ion package” is subjected to mild HCD activation to achieve higher sequence coverage (98). Recently, EThcD has been promoted for the more efficient analysis of post-translationally modified sequences, including glycopeptides (2). An EThcD spectrum is presented in Fig. 7. These data were acquired on an Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo Fisher Scientific) from a precursor ion at m/z 624.950(3+) (with the default NCE value of 15%), the fragments were measured at high resolution and with high mass accuracy in the Orbitrap.
Fig. 7.
EThcD spectrum of m/z 624.950(3+), acquired on a Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo Fisher Scientific) (at NCE = 15%). The modified sequence was identified from these data as AVAVTLQSH [342–350] of human YIPF3 protein (Uniprot ID: Q9GZM5). The glycan is most likely the disialo mucin-type core-1 structure, one of the glycans listed as potential variable modifications of Ser and Thr residues. Extensive HexNAc fragmentation was not observed, thus, the GalNAc's identity cannot be established unambiguously from these data. In the reducing end fragment ion Y2SA, the sialic acid could be linked either to the GalNAc or the Gal. The modification site was identified as Thr-5, fragments printed in red are unique to this positional variant. No fragment ions indicated modification on the Ser residue. The precursor ion and its charge-reduced versions are labeled as “pr.”
This activation combines the advantages of collision-induced and radical-driven dissociation. First, we have plenty of information on the modified amino acid sequence in form of ETD-derived backbone cleavages. Interestingly, the collisional activation of some radical C-terminal ETD fragments led to C-C bond cleavages in some amino acid side chains yielding two w ions (for nomenclature see (99)), a phenomenon already reported for the differentiation of the isomeric Ile/Leu pair (100). Otherwise, no additional peptide fragmentation was detected, rather the glycosidic bonds were cleaved. An almost complete oxonium ion series was detected for this glycopeptide. In contrast to “normal” HCD, the single unit oxonium ions are weaker, the fragments distinguishing between GlcNAc and GalNAc were not observed, m/z 366 representing HexHexNAc is missing, but all the larger sialylated fragments are present, even the intact tetrasaccharide. Because the low mass range is also covered, and abundant reducing end fragments are also present and the ions are measured within a few ppm, this technique seems to offer more information about the glycans than ion trap CID.
In summary, we have to point out that usually none of the above mentioned techniques provide “full glycan, amino acid sequence and modification site assignment” coverage. All our examples were singly modified glycopeptides and EThcD delivered the best results. However, peptides bearing multiple, different O-glycan structures depending on their amino acid and glycan sequences, carbohydrate/peptide size ratio, and charge density still may yield very limited information from all aspects.
Data Interpretation
Search engines were developed for the identification of unmodified, mostly tryptic peptides. Covalent modifications that are stable on MS/MS activation and are specific for any given amino acid make the assignment of such modifications, including site determination, relatively straightforward.
However, in O-glycosylation analysis we must deal with a wide variety of glycans, and also with their potential coexistence within the same peptide. Thus, even single protein characterization can turn into a very complicated process, and high throughput glycopeptide analysis remains extraordinarily challenging. Unless a specific enrichment strategy was followed (and even such protocols may produce surprising results (6)), multiple glycan structures must be listed as potential variable modifications. Thus, in a way, a protein and a glycan database search must be performed simultaneously. In addition, the peptides may be decorated with multiple, sometimes different glycans. Furthermore, both peptide and glycan fragmentation must be considered. Activation-dependent peptide fragmentation scoring and the probability-based evaluation of the reliability thereof has been established for unmodified sequences. Glycopeptide glycan fragmentation assignment is like an “add-on” feature. Glycopeptide fragmentation has not been studied and evaluated yet extensively, especially for O-glycosylation. We simply did not have sufficiently large data sets available. What also complicates the matter, that search strategies have been developed to handle data from a single activation method. However, as discussed earlier, for glycopeptide characterization it would be better to use the information delivered by the different MS/MS strategies combined, sometimes adding retention time information and mass measurement data to the “mixture.” Thus, we would need other “mining tools,” not only humans, combining all these data. For the lack of better solutions, there are studies, even of high throughput nature, where data interpretation was/is performed from combined MS/MS data, mostly manually (93–95). At the same time, there are two search engines that are regularly used for O-glycopeptide identification from ETD or HCD data, and these are Byonic (101) and Protein Prospector (97, 102). Byonic is a commercial product, whereas Protein Prospector is available freely on the web and can also be downloaded and installed in-house. These search engines display a comparable performance and very similar shortcomings. Because Ser and Thr residues are frequently located in clusters, even ETD spectra may not reveal which amino acid bears the glycan. Protein Prospector features a built-in site assignment evaluation score, SLIP (site localization in peptide) score (103), and it either will identify the glycosite confidently, or list the potential modification sites whenever there is not sufficient information for site determination. As for HCD data the glycan modification should be specified as “neutral loss” for Protein Prospector (because of the preferential elimination of the glycan on HCD activation, see MS/MS behavior section). Byonic does not require such “special treatment,” and it labels not only peptide fragments but also glycan oxonium ions as well as some of the sugar losses. Unfortunately, it also will “identify” a modification site even when there is no supportive evidence in the spectrum that is usually the case. In addition, characterizing peptides modified by multiple different O-glycans has not been reliably solved yet, although both search engines permit specifying such search parameters. For example, a mass addition of 1312 Da may indicate the presence of two GalGalNAc structures and two sialic acids. However, this composition may correspond to a single hexasaccharide, or two monosialo-structures, where the sialic acids might be bond to either the Gal or the core GalNAc, or we may have a combination of a disialo tetrasaccharide and an “uncapped” disaccharide structure. EThcD may address some of these issues, because it may contain sufficient information for glycan size determination, peptide identification as well as site assignment. However, to use reliably all the information included in the spectra requires significant software development.
The data interpretation is not finished when the search engine delivers a list of the identified glycopeptides, however high the confidence level may be. As discussed earlier, certain rules apply, such as where O-glycosylation is performed, what kind of glycans can be deposited on the amino acid side chains etc. Thus, we must consider the cellular localization of the proteins on our list as well as their topology. Finding extracellular glycosylation on—predicted to be—cytosolic domains may overwrite topology predictions (6). Similarly, allegedly cytosolic and nuclear proteins detected with extracellular glycan structures might have been missassigned or may have a secreted isoform. In addition, with the ever-improving analytical tools one may discover novel, unexpected glycan structures or modification sites. For example, mucin-type glycosylation has been reported for Tyr (30, 31) and GlcNAcylation of Cys has been observed (104). HLA peptides have been discovered featuring elongated GlcNAc residues (105, 106). These neoantigens may serve as targets for immunotherapy.
O-Glycosylation Studies
Single Protein Analysis
The analysis of isolated proteins led to the discovery and characterization of the different type of O-glycans. In the not so distant past proteases of different specificity, glycosidases, and even Edman degradation had to be used for such jobs. For example, the glycosylation of human casein was studied using such methods in 1980 (107). A decade later mass spectrometry was introduced into the protein analytical repertoire and it holds a unique position there ever since. All classes of O-glycosylation have been described in single protein analysis using mass spectrometry. (For references see: O-fucosylation (3, 4, 14, 19, 109); O-glucosylation (15, 18, 19); mucin-type glycosylation (30, 71, 109–114); O-mannosylation (60, 62, 66)). Some proteins feature multiple types of O-glycosylation, such as Coagulation Factor VII (115), Notch 1 (116), or dystroglycan (83). Single protein analysis is still the best approach for the comprehensive characterization of PTMs.
Recombinant Proteins
Presently, the majority of approved biopharmaceuticals are glycoproteins. These represent a specific case of single protein characterization. As the physicochemical, pharmacokinetic, and immunologic properties of these proteins may be altered by glycosylation changes, in-depth glycoprofiling of these proteins is essential. To display glycan pattern similar to that of human, these proteins are usually expressed in eukaryotic cell lines such as Chinese hamster ovary (CHO), myeloma (NS0), or hybridoma (SP2/0). However, depending on the culture medium and the physiological status of the host cells, the glycosylation pattern of these proteins may vary batch-to-batch. Although therapeutic efficacy, immunogenicity, and circulation half-life have clearly been linked to N-glycosylation (117–120), the role of O-glycosylation is less explored. Possible function of O-glycans may be masking antigenic sites of the proteins as evidenced by elevated antigenecity of O-glycosylation deficient human recombinant granulocyte stimulating factor (121). Among several glycan-related issues linked to N-glycosylation, sialic acid O-acetylation, and the presence of the nonhuman NeuGc may have implications toward O-linked structures as well. The presence of these sialic acid variants have been demonstrated on O-linked structures in wild-type samples (1, 34).
The largest and best-selling therapeutic proteins are recombinant immunoglobulins (mAbs) applied widely for cancer and autoimmune diseases. Proper N-glycosylation of IgGs is indispensable for proper Fc-effector functions and serum half-life. On the other hand, O-glycosylation has not been observed on the most abundant IgG subclass, IgG1, although O-glycosylation of the less abundant IgG2 and IgG3 subclasses has been reported (122, 123). Therefore, presently there are no O-glycosylation issues related to this class of biologics.
However, there are important biologics O-glycosylated. Erythropoietin (EPO) is used for prevention of anemia in cancer patients and restoration of hemoglobin level in patients with renal failure. Site-specific N- and O-glycosylation of this 30 kDa protein have been extensively studied (1, 124–128). The human chorionic gonadotrophin, used for over 30 years for treating female infertility, is a 26 kDa glycoprotein with 4 N- and 4 O-glycosylation sites. In a recent study, ten different samples from urinary- and recombinant hCGs were analyzed and compared in a site-specific and quantitative manner (129).
Therapeutic fusion proteins which are created from different proteins using recombinant DNA technology may also belong here. An example is Etanercept, a TNFα inhibitor used for treatment of rheumatoid arthritis. The linker region of this protein is heavily O-glycosylated, 12 Ser/Thr residues have been shown to be modified with neutral, mono- and disialylated core-1 type structures (73).
High Throughput Data
Extracellular O-glycosylation data acquired from complex samples are rather limited. The dynamic range of protein concentration is overwhelming in body fluids or tissue samples—for example, concentration of individual proteins spans 11 orders of magnitude in human plasma (130), and the vast majority of these proteins are not- or just N-glycosylated. To characterize O-glycosylation in complex samples, some enrichment is essential. Basically, three enrichment strategies have been applied for this purpose: (i) hydrophilic interaction liquid chromatography (HILIC), (ii) periodate oxidation-hydrazide capture, and (iii) lectin affinity chromatography.
HILIC is suitable for the enrichment of glycopeptides with relatively high glycan/peptide ratio including N-glycopeptides and multiply modified O-glycopeptides. Until now few O-glycopeptides were characterized form such mixtures (131). However, some O-glycosylated short peptide sequences, such as those generated by aspecific/broad specificity proteases may be isolated separately from N-glycopeptides as demonstrated by a human plasma study incubating the samples with Proteinase K before the chromatography (95). Though we would like to point out that automated interpretation of short peptide sequences generated by aspecific/broad specificity proteases is usually quite unreliable.
The hydrazide capture and release approach exploits that vicinal cis diols can be easily and selectively oxidized using periodate and the resulting aldehydes can be isolated on immobilized hydrazide functionalities. This method has been developed for N-glycoprotein isolation (132) and further optimized for enrichment of sialic acid containing glycoproteins (133). The unmodified part of the protein is removed by tryptic digestion followed by mild acidic release of the glycopeptides. The glycosidic bond linking the terminal sialic acid and the penultimate saccharide unit is hydrolyzed during this process, information on the original intact glycan structure is therefore lost. Similarly, to HILIC, both N- and O-glycosylated molecules are isolated by this method. Peptides and proteins with N-terminal Ser and Thr are also readily oxidized by periodate (134), therefore this method is more suitable for protein-level enrichment. The hydrazide capture-and release method has been used for studying O-glycosylation in human urine (93) and cerebrospinal fluid (94).
Finally, there are several lectins affording O-glycopeptide enrichment. Jacalin, a plant lectin from Artocarpus integrifolia shows preference toward Galβ1,3GalNAc with unsubstituted GalNAc C6-OH (135), therefore it is an attractive option for samples such as human plasma/serum where mucin core-1 type O-glycosylation is dominant (136) keeping in mind that the disialylated mucin core-1 structure that comprises some 20% of human plasma O-glycans (29) cannot be captured by this lectin. Glycopeptides displaying core-2 O-glycans will not be isolated either as the GlcNAc moiety is in 1,6 linkage with the core GalNAc. Peanut agglutinin also binds the Galβ1,3GalNAc structure, but sialylation inhibits the binding (137). Vicia villosa and soybean agglutinin show preference toward GalNAc enabling enrichment of O-glycosylated proteins or peptides from samples where elongation of mucin-type O-glycosylation is inhibited (see Simple Cell technology below). Finally, broad-specificity lectins such as wheat germ agglutinin (138) can also be exploited for O-glycopeptide enrichment. Although lectins show varying affinity toward their preferred substrates, high nonspecific background is very typical. This feature may partly be attributed to agarose frequently used as solid support for lectin affinity chromatography. Immobilization of lectins on other supports withstanding more stringent washing conditions such as POROS (139) may improve this situation.
Numerous O-glycosylation studies rely on lectin affinity isolation. Jacalin has been used for O-glycosylation characterization of fetal calf serum (25, 92), human plasma (140) and serum (79). Peanut agglutinin has been used for characterization of protein O-glycosylation of human plasma, platelets and endothelial cells (26) yielding the largest data set for protein O-glycosylation to date. Wheat germ agglutinin has been used in studies reporting O-glycosylation of the mouse synaptosome (6) and liver (74), and this lectin also proved to be useful for characterization of human serum O-glycosylation (79, 80).
Interestingly, although HILIC, sialic-acid-based isolation as well as affinity chromatography with wheat germ agglutinin are suitable for the enrichment of some core mannosyl and fucosyl structures, only a few such glycopeptides have been identified in high-throughput studies (6). Thus, we must assume that the occurrence of these modifications is much lower than that of the mucin-type structures, and more targeted enrichment strategies have to be developed in order to find the differently O-glycosylated proteins.
SimpleCell Technology
Although the above examples represented the analysis of wild-type samples, the most remarkable breakthrough in O-glycosylation analysis has been delivered by the Clausen-group introducing an innovative new approach, SimpleCell technology. First, mucin-type O-glycosylation has been extensively studied on genetically engineered cell lines, where O-glycan extension is blocked (31, 141–143). This approach has been extended to probe the specificity of different GalNAc-transferases (144, 145). The latter studies revealed that there is significant redundancy in the O-GalNAc transferase system, and most of the sites are “covered” by multiple enzymes. However, the different GalNAc transferases have specific substrate subsets that are linked to distinct cellular processes. This suggests that some GalNAc transferases may regulate these processes through their specific substrate proteins. The technology was also applied successfully to an entirely different glycosylation pathway, O-mannosylation (59). Although very impressive results were gathered, the method suffers from certain shortcomings. The first results were gained from cancer cell lines. Because it is believed that in cancer the equilibrium between core glycosylation and elongation is broken (24), one would expect a higher degree of glycosylation in such samples than in healthy tissues, although we do not have any proof of that (31). However, the authors themselves pointed out that higher density glycosylation may result from the prevention of elongation (141, 146). They also suggested that the increased glycan density may adversely affect the proteolytic digestion as well as mass spectrometry detection and MS/MS-based identification. On the other hand, in a recent study, King et al. reported that there were no substantial differences between site distribution and localization in glycoengineered immortalized cell lines and in native samples (26). Distinguishing the two N-acetylhexosamines used by human cells also represents a challenge for glycopeptides with truncated glycan structures. Luckily, it has been reported that GalNAc and GlcNAc produce a characteristic fragmentation pattern in beam-type collisional activation (89). However, the reliability of such assignments has not been tested on a large enough data set yet. Obviously, the biggest issue with SimpleCell technology is the loss of information on structural diversity. Still, this is a very powerful tool, and there are numerous promising projects where this technique will deliver significant new knowledge, and will point us to the right direction.
Biological Significance
Cataloguing all proteins expressed within a given cell type is not a straightforward task even in a qualitative manner, despite the numerous claims of “comprehensive” characterization of different proteomes. Compiling the information about O-glycosylation is on its way. Characterizing protein populations, i.e. deciphering which modifications coexist has just started for glycoproteins (71). Intact protein fractionation and mass spectrometry seem to work better for N-glycoproteins (147), but the newest results are very promising for more complex species as well (2). Elucidating the biological role of these modifications, and how the macro- and microheterogeneity modulate these functions is another important question. We listed some observations about the potential or proven function of these PTMs above while describing the different classes of O-glycosylation.
In addition, we want to point out that there are some exciting possibilities for an “interplay” among different extracellular post-translational modifications of Ser/Thr/Tyr residues. It is widely accepted that intracellular GlcNAcylation of Ser/Thr residues and the phosphorylation of the same sites or positions nearby may represent a regulatory switch (148). We speculated that such interactions may not be restricted to the intracellular space. Thus, we performed a preliminary investigation how frequently these modifications occur on the same residue or in close proximity (within 5 amino acids). We investigated glycoproteins identified in the first SimpleCell study (31), and considered glycosylation sites reported then or later by the same group. Most of these data are listed on PhosphoSitePlus, where we also found information about the phosphorylation sites. In the Steentoft paper (31) ∼350 O-glycosylation sites in 122 proteins were reported. We found that 43 proteins featured 142 “overlapping” phosphorylation and glycosylation sites. In most cases (113 instances) the very same residues were assigned as modification sites in both processes (supplemental Table S1). Similarly to the intercellular GlcNAc/phosphorylation interplay (139), the degree of overlapping may not be statistically significant in general, a conclusion also reported along with the largest mucin-type glycosylation data set (26). However, we believe for certain proteins a strong correlation among these modifications may exist, similarly to intracellular regulative processes (148). For example, it has been reported that the secretion level of FGF23 is controlled by a delicate balance between phosphorylation of Ser-180 and mucin-type glycosylation of Thr-178 (149); and glycosylation defect in osteopontin led to increased phosphorylation (41).
Site-specific glycan heterogeneity may represent a unique kind of PTM-interplay. We need more information even to guess the biological significance for displaying mucin-type glycosylation versus mannosylation on the same site (6), or glucosylation and fucosylation in close proximity (19); or for altering the mucin-type core during membrane protein recycling (78).
CONCLUSIONS
The glycosylation of isolated proteins, especially recombinant biologicals have been studied for decades using a wide variety of analytical tools, including mass spectrometry. The analysis of intact proteins, the novel activation methods, ETD and EThcD have been quickly introduced into the analytical “tool set,” and made single protein characterization more comprehensive, more successful. At the same time, high throughput site-specific characterization of O-glycosylation is still the exception not the norm, although there has been significant progress in the large-scale analysis of protein O-glycosylation in the last decade. Several enrichment methods facilitating effective glycopeptide isolation have been developed. High sensitivity and high mass accuracy mass spectrometers equipped with ETD enable site-specific characterization of O-glycosylation. Bioinformatic tools for reliable automated data interpretation are under progressive development. Presently, integrative interpretation of MS/MS data acquired with multiple fragmentation techniques is lacking, although this approach would certainly lead to faster and more reliable glycopeptide identifications. Thus, careful, critical inspection/evaluation of the automated assignments is still highly recommended.
The vast majority of O-glycosylation data, both for single protein- and large-scale analysis, was acquired on mucin type O-glycosylation. The only other type of O-glycosylation targeted by large-scale studies is O-mannosylation. Although the Simple Cell technology yielded many mannosylation sites, the only other large-scale study revealed a single O-mannosylated protein (the same study also reported an O-fucosylated protein) (6). Single-protein studies have provided additional information on protein O-mannosylation, fucosylation and glucosylation.
The largest mucin-type O-glycopeptide data set published to date (26) identified over 1000 O-glycosylation sites in ∼650 glycoproteins. Based on this amount of data we may start to speculate about potential novel functions of protein O-glycosylation, as the authors indeed tried to identify shared features and made certain predictions. As other studies will deliver large scale O-glycosylation data from different sources, new hypotheses will emerge and this process eventually will lead to better understanding of the biological role(s) of O-glycosylation—but there is a long way to go.
Supplementary Material
Footnotes
* This work was supported by the following grants: Hungarian Scientific Research Fund 105611 (to Z. Darula), and the Economic Development and Innovation Operative Programmes GINOP-2.3.2-15-2016-00001 and GINOP-2.3.2-15-2016-00020 from the Ministry for National Economy.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- ER
- endoplasmic reticulum
- ECD
- electron capture dissociation
- EGF
- epidermal growth factor
- ETD
- electron transfer dissociation
- Fuc
- fucose
- Gal
- galactose
- GalNAc
- N-acetylgalactosamine
- Glc
- glucose
- GlcNAc
- N-acetylglucosamine
- Hex
- hexose
- HexNAc
- N-acetylhexosamine
- HILIC
- hydrophilic interaction liquid chromatography
- Man
- mannose
- NeuAc
- N-acetylneuraminic acid (sialic acid)
- NeuGc
- N-glycolylneuraminic acid
- PTM
- post-translational modification
- TSR
- thrombospondin type 1 repeat
- Xyl
- xylose.
REFERENCES
- 1. Yang Y., Liu F., Franc V., Halim L. A., Schellekens H., and Heck A. J. (2016) Hybrid mass spectrometry approaches in glycoprotein analysis and their usage in scoring biosimilarity. Nat. Commun. 7, 13397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Franc V., Yang Y., and Heck A. J. (2017) Proteoform Profile Mapping of the Human Serum Complement Component C9 Revealing Unexpected New Features of N-, O-, and C-Glycosylation. Anal. Chem. 89, 3483–3491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Harris R. J., Leonard C. K., Guzzetta A. W., and Spellman M. W. (1991) Tissue plasminogen activator has an O-linked fucose attached to threonine-61 in the epidermal growth factor domain. Biochemistry 30, 2311–2314 [DOI] [PubMed] [Google Scholar]
- 4. Harris R. J., and Spellman M. W. (1993) O-linked fucose and other post-translational modifications unique to EGF modules. Glycobiology 3, 219–224 [DOI] [PubMed] [Google Scholar]
- 5. Alfaro J. F., Gong C. X., Monroe M. E., Aldrich J. T., Clauss T. R., Purvine S. O., Wang Z., Camp D. G. 2nd, Shabanowitz J., Stanley P., Hart G. W., Hunt D. F., Yang F., and Smith R. D. (2012) Tandem mass spectrometry identifies many mouse brain O-GlcNAcylated proteins including EGF domain-specific O-GlcNAc transferase targets. Proc. Natl. Acad. Sci. U.S.A. 109, 7280–7285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Trinidad J. C., Schoepfer R., Burlingame A. L., and Medzihradszky K. F. (2013) N- and O-glycosylation in the murine synaptosome. Mol. Cell. Proteomics 12, 3474–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hofsteenge J., Huwiler K. G., Macek B., Hess D., Lawler J., Mosher D. F., and Peter-Katalinic J. (2001) C-mannosylation and O-fucosylation of the thrombospondin type 1 module. J. Biol. Chem. 276, 6485–6498 [DOI] [PubMed] [Google Scholar]
- 8. Luo Y., Koles K., Vorndam W., Haltiwanger R. S., and Panin V. M. (2006) Protein O-fucosyltransferase 2 adds O-fucose to thrombospondin type 1 repeats. J. Biol. Chem. 281, 9393–9399 [DOI] [PubMed] [Google Scholar]
- 9. Leonhard-Melief C., and Haltiwanger R. S. (2010) O-fucosylation of thrombospondin type 1 repeats. Methods Enzymol. 480, 401–416 [DOI] [PubMed] [Google Scholar]
- 10. Wang Y., Lee G. F., Kelley R. F., and Spellman M. W. (1996) Identification of a GDP-L-fucose:polypeptide fucosyltransferase and enzymatic addition of O-linked fucose to EGF domains. Glycobiology 6, 837–842 [DOI] [PubMed] [Google Scholar]
- 11. Luo Y., Nita-Lazar A., and Haltiwanger R. S. (2006) Two distinct pathways for O-fucosylation of epidermal growth factor-like or thrombospondin type 1 repeats. J. Biol. Chem. 281, 9385–9392 [DOI] [PubMed] [Google Scholar]
- 12. Luther K. B., and Haltiwanger R. S. (2009) Role of unusual O-glycans in intercellular signaling. Int. J. Biochem. Cell Biol. 41, 1011–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Takeuchi H., and Haltiwanger R. S. (2014) Significance of glycosylation in Notch signaling. Biochem. Biophys. Res. Commun. 453, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Valliere-Douglass J. F., Brady L. J., Farnsworth C., Pace D., Balland A., Wallace A., Wang W., Treuheit M. J., and Yan B. (2009) O-fucosylation of an antibody light chain: characterization of a modification occurring on an IgG1 molecule. Glycobiology 19, 144–152 [DOI] [PubMed] [Google Scholar]
- 15. Hase S., Kawabata S., Nishimura H., Takeya H., Sueyoshi T., Miyata T., Iwanaga S., Takao T., Shimonishi Y., and Ikenaka T. (1988) A new trisaccharide sugar chain linked to a serine residue in bovine blood coagulation factors VII and IX. J. Biochem. 104, 867–868 [DOI] [PubMed] [Google Scholar]
- 16. Nishimura H., Kawabata S. I., Kisiel W., Hase S., Ikenaka T., Takao T., Shimonishi Y., and Iwanaga S. (1989) Identification of a disaccharide (Xyl-Glc) and a trisaccharide (Xyl2-Glc) O-glycosidically linked to a serine residue in the first epidermal growth factor-like domain of human factors VII and IX and protein Z and bovine protein Z. J. Biol. Chem. 264, 20320–20325 [PubMed] [Google Scholar]
- 17. Hase S., Nishimura H., Kawabata S., Iwanaga S., and Ikenaka T. (1990) The structure of (xylose)2glucose-O-serine 53 found in the first epidermal growth factor-like domain of bovine blood clotting factor IX. J. Biol. Chem. 265, 1858–1861 [PubMed] [Google Scholar]
- 18. Nishimura H., Yamashita S., Zeng Z., Walz D. A., and Iwanaga S. (1992) Evidence for the existence of O-linked sugar chains consisting of glucose and xylose in bovine thrombodpondin. J. Biochem. 111, 460–464 [DOI] [PubMed] [Google Scholar]
- 19. Gebauer J. M., Müller S., Hanisch F. G., Paulsson M., and Wagener R. (2008) O-glucosylation and O-fucosylation occur together in close proximity on the first epidermal growth factor repeat of AMACO (VWA2 protein). J. Biol. Chem. 283, 17846–17854 [DOI] [PubMed] [Google Scholar]
- 20. Acar M., Jafar-Nejad H., Takeuchi H., Rajan A., Ibrani D., Rana N. A., Pan H., Haltiwanger R. S., and Bellen H. J. (2008) Rumi is a CAP10 domain glycosyltransferase that modifies Notch and is required for Notch signaling. Cell. 132, 247–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Takeuchi H., Fernández-Valdivia R. C., Caswell D. S., Nita-Lazar A., Rana N. A., Garner T. P., Weldeghiorghis T. K., Macnaughtan M. A., Jafar-Nejad H., and Haltiwanger R. S. (2011) Rumi functions as both a protein O-glucosyltransferase and a protein O-xylosyltransferase. Proc. Natl. Acad. Sci. U.S.A. 108, 16600–16605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fernandez-Valdivia R., Takeuchi H., Samarghandi A., Lopez M., Leonardi J., Haltiwanger R. S., and Jafar-Nejad H. (2011) Regulation of mammalian Notch signaling and embryonic development by the protein O-glucosyltransferase Rumi. Development 138, 1925–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Varki A., Cummings R. D., Esko J. D., Freeze H. H., Stanley P., Bertozzi C. R., Hart G. W., and Etzler M. E. (2009) Essentials of Glycobiology, 2nd Ed., Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press: [PubMed] [Google Scholar]
- 24. Gill D. J., Clausen H., and Bard F. (2011) Location, location, location: new insights into O-GalNAc protein glycosylation. Trends Cell Biol. 21, 149–158 [DOI] [PubMed] [Google Scholar]
- 25. Darula Z., Sherman J., and Medzihradszky K. F. (2012) How to dig deeper? Improved enrichment methods for mucin core-1 type glycopeptides. Mol. Cell. Proteomics 11, O111.016774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. King S. L., Joshi H. J., Schjoldager K. T., Halim A., Madsen T. D., Dziegiel M. H., Woetmann Vakhrushev S. Y., and Wandall H. H. (2017) Characterizing the O-glycosylation landscape of human plasma, platelets, and endothelial cells. Blood Advances 1, 429–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wilson I. B., Gavel Y., and von Heijne G. (1991) Amino acid distributions around O-linked glycosylation sites. Biochem. J. 275, 529–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Christlet T. H., and Veluraja K. (2001) Database analysis of O-glycosylation sites in proteins. Biophys. J. 80, 952–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yabu M., Korekane H., and Miyamoto Y. (2014) Precise structural analysis of O-linked oligosaccharides in human serum. Glycobiology 24, 542–553 [DOI] [PubMed] [Google Scholar]
- 30. Halim A., Brinkmalm G., Rüetschi U., Westman-Brinkmalm A., Portelius E., Zetterberg H., Blennow K., Larson G., and Nilsson J. (2011) Site-specific characterization of threonine, serine, and tyrosine glycosylations of amyloid precursor protein/amyloid beta-peptides in human cerebrospinal fluid. Proc. Natl. Acad. Sci. U.S.A. 108, 11848–11853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Steentoft C., Vakhrushev S. Y., Vester-Christensen M. B., Schjoldager K. T., Kong Y., Bennett E. P., Mandel U., Wandall H., Levery S. B., and Clausen H. (2011) Mining the O-glycoproteome using zinc-finger nuclease-glycoengineered SimpleCell lines. Nat. Methods 8, 977–982 [DOI] [PubMed] [Google Scholar]
- 32. Jin C., Kenny D. T., Skoog E. C., Padra M., Adamczyk B., Vitizeva V., Thorell A., Venkatakrishnan V., Lindén S. K., and Karlsson N. G. (2017) Structural diversity of human gastric mucin glycans. Mol. Cell. Proteomics pii: mcp.M117.067983 [DOI] [PubMed] [Google Scholar]
- 33. Rossez Y., Maes E., Lefebvre Darroman T., Gosset P., Ecobichon C., Joncquel Chevalier Curt M., Boneca I. G., Michalski J. C., and Robbe-Masselot C. (2012) Almost all human gastric mucin O-glycans harbor blood group A, B or H antigens and are potential binding sites for Helicobacter pylori. Glycobiology 22, 1193–1206 [DOI] [PubMed] [Google Scholar]
- 34. Medzihradszky K. F., Kaasik K., and Chalkley R. J. (2015) Characterizing sialic acid variants at the glycopeptide level. Anal. Chem. 87, 3064–3071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Solecka B. A., Weise C., Laffan M. A., and Kannicht C. (2016) Site-specific analysis of von Willebrand factor O-glycosylation. J. Thromb. Haemost. 14, 733–746 [DOI] [PubMed] [Google Scholar]
- 36. Xia B., Royall J. A., Damera G., Sachdev G. P., and Cummings R. D. (2005) Altered O-glycosylation and sulfation of airway mucins associated with cystic fibrosis. Glycobiology 15, 747–775 [DOI] [PubMed] [Google Scholar]
- 37. Ricciuto J., Heimer S. R., Gilmore M. S., and Argüeso P. (2008) Cell surface O-glycans limit Staphylococcus aureus adherence to corneal epithelial cells. Infect. Immun. 76, 5215–5220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Guzman-Aranguez A., and Argüeso P. (2010) Structure and biological roles of mucin-type O-glycans at the ocular surface. Ocul. Surf. 8, 8–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lasky L. A., Singer M. S., Dowbenko D., Imai Y., Henzel W. J., Grimley C., Fennie C., Gillett N., Watson S. R., and Rosen S. D. (1992) An endothelial ligand for L-selectin is a novel mucin-like molecule. Cell 69, 927–938 [DOI] [PubMed] [Google Scholar]
- 40. Pan Y., Yago T., Fu J., Herzog B., McDaniel J. M., Mehta-D”Souza P., Cai X., Ruan C., McEver R. P., West C., Dai K., Chen H., and Xia L. (2014) Podoplanin requires sialylated O-glycans for stable expression on lymphatic endothelial cells and for interaction with platelets. Blood. 124, 3656–3665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kariya Y., Kanno M., Matsumoto-Morita K., Konno M., Yamaguchi Y., and Hashimoto Y. (2014) Osteopontin O-glycosylation contributes to its phosphorylation and cell-adhesion properties. Biochem. J. 463, 93–102 [DOI] [PubMed] [Google Scholar]
- 42. Peng J., Jiang J., Wang W., Qi X., Sun X. L., and Wu Q. (2011) Glycosylation and processing of pro-B-type natriuretic peptide in cardiomyocytes. Biochem. Biophys. Res. Commun. 411, 593–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Leuenberger B., Hahn D., Pischitzis A., Hansen M. K., and Sterchi E. E. (2003) Human meprin beta: O-linked glycans in the intervening region of the type I membrane protein protect the C-terminal region from proteolytic cleavage and diminish its secretion. Biochem. J. 369, 659–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Goth C. K., Halim A., Khetarpal S. A., Rader D. J., Clausen H., and Schjoldager K. T. (2015) A systematic study of modulation of ADAM-mediated ectodomain shedding by site-specific O-glycosylation. Proc. Natl. Acad. Sci. U.S.A. 112, 14623–14628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Goth C. K., Tuhkanen H. E., Khan H., Lackman J. J., Wang S., Narimatsu Y., Hansen L. H., Overall C. M., Clausen H., Schjoldager K. T., and Petäjä-Repo U. E. (2017) Site-specific O-glycosylation by polypeptide N-Acetylgalactosaminyltransferase 2 (GalNAc-transferase T2) co-regulates β1-adrenergic receptor N-terminal cleavage. J. Biol. Chem. 292, 4714–4726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Allen A. C., Willis F. R., Beattie T. J., and Feehally J. (1998) Abnormal IgA glycosylation in Henoch-Schönlein purpura restricted to patients with clinical nephritis. Nephrol. Dial. Transplant. 13, 930–934 [DOI] [PubMed] [Google Scholar]
- 47. Chou C. H., Huang M. J., Chen C. H., Shyu M. K., Huang J., Hung J. S., Huang C. S., and Huang M. C. (2015) Up-regulation of C1GALT1 promotes breast cancer cell growth through MUC1-C signaling pathway. Oncotarget. 6, 6123–6135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hofmann B. T., Schlüter L., Lange P., Mercanoglu B., Ewald F., Fölster A., Picksak A. S., Harder S., El Gammal A. T., Grupp K., Güngör C., Drenckhan A., Schlüter H., Wagener C., Izbicki J. R., Jücker M., Bockhorn M., and Wolters-Eisfeld G. (2015) COSMC knockdown mediated aberrant O-glycosylation promotes oncogenic properties in pancreatic cancer. Mol. Cancer 14, 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Torres C. R., and Hart G. W. (1984) Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes. Evidence for O-linked GlcNAc. J. Biol. Chem. 259, 3308–3317 [PubMed] [Google Scholar]
- 50. Hardivillé S., and Hart G. W. (2014) Nutrient regulation of signaling, transcription, and cell physiology by O-GlcNAcylation. Cell. Metab. 20, 208–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Haltiwanger R. S., Blomberg M. A., and Hart G. W. (1992) Glycosylation of nuclear and cytoplasmic proteins. Purification and characterization of a uridine diphospho-N-acetylglucosamine:polypeptide beta-N-acetylglucosaminyltransferase. J. Biol. Chem. 267, 9005–9013 [PubMed] [Google Scholar]
- 52. Kreppel L. K., Blomberg M. A., and Hart G. W. (1997) Dynamic glycosylation of nuclear and cytosolic proteins. Cloning and characterization of a unique O-GlcNAc transferase with multiple tetratricopeptide repeats. J. Biol. Chem. 272, 9308–9315 [DOI] [PubMed] [Google Scholar]
- 53. Dong D. L., and Hart G. W. (1994) Purification and characterization of an O-GlcNAc selective N-acetyl-beta-D-glucosaminidase from rat spleen cytosol. J. Biol. Chem. 269, 19321–19330 [PubMed] [Google Scholar]
- 54. Gao Y., Wells L., Comer F. I., Parker G. J., and Hart G. W. (2001) Dynamic O-glycosylation of nuclear and cytosolic proteins: cloning and characterization of a neutral, cytosolic beta-N-acetylglucosaminidase from human brain. J. Biol. Chem. 276, 9838–9845 [DOI] [PubMed] [Google Scholar]
- 55. Matsuura A., Ito M., Sakaidani Y., Kondo T., Murakami K., Furukawa K., Nadano D., Matsuda T., and Okajima T. (2008) O-linked N-acetylglucosamine is present on the extracellular domain of notch receptors. J. Biol. Chem. 283, 35486–35495 [DOI] [PubMed] [Google Scholar]
- 56. Sakaidani Y., Nomura T., Matsuura A., Ito M., Suzuki E., Murakami K., Nadano D., Matsuda T., Furukawa K., and Okajima T. (2011) O-linked-N-acetylglucosamine on extracellular protein domains mediates epithelial cell-matrix interactions. Nat. Commun. 2, 583. [DOI] [PubMed] [Google Scholar]
- 57. Sakaidani Y., Ichiyanagi N., Saito C., Nomura T., Ito M., Nishio Y., Nadano D., Matsuda T., Furukawa K., and Okajima T. (2012) O-linked-N-acetylglucosamine modification of mammalian Notch receptors by an atypical O-GlcNAc transferase Eogt1. Biochem. Biophys. Res. Commun. 419, 14–19 [DOI] [PubMed] [Google Scholar]
- 58. Chiba A., Matsumura K., Yamada H., Inazu T., Shimizu T., Kusunoki S., Kanazawa I., and Kobata A. (1997) Structures of sialylated O-linked oligosaccharides of bovine peripheral nerve alpha-dystroglycan. The role of a novel O-mannosyl-type oligosaccharide in the binding of alpha-dystroglycan with laminin. J. Biol. Chem. 272, 2156–2162 [DOI] [PubMed] [Google Scholar]
- 59. Vester-Christensen M. B., Halim A., Joshi H. J., Steentoft C., Bennett E. P., Levery S. B., Vakhrushev S. Y., and Clausen H. (2013) Mining the O-mannose glycoproteome reveals cadherins as major O-mannosylated glycoproteins. Proc. Natl. Acad. Sci. U.S.A. 110, 21018–21023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lommel M., Winterhalter P. R., Willer T., Dahlhoff M., Schneider M. R., Bartels M. F., Renner-Müller I., Ruppert T., Wolf E., and Strahl S. (2013) Protein O-mannosylation is crucial for E-cadherin-mediated cell adhesion. Proc. Natl. Acad. Sci. U.S.A. 110, 21024–21029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Martinez T., Pace D., Brady L., Gerhart M., and Balland A. (2007) Characterization of a novel modification on IgG2 light chain. Evidence for the presence of O-linked mannosylation. J. Chromatogr. A 1156, 183–187 [DOI] [PubMed] [Google Scholar]
- 62. Bleckmann C., Geyer H., Lieberoth A., Splittstoesser F., Liu Y., Feizi T., Schachner M., Kleene R., Reinhold V., and Geyer R. (2009) O-glycosylation pattern of CD24 from mouse brain. Biol. Chem. 390, 627–645 [DOI] [PubMed] [Google Scholar]
- 63. Pacharra S., Hanisch F. G., and Breloy I. (2012) Neurofascin 186 is O-mannosylated within and outside of the mucin domain. J. Proteome Res. 11, 3955–3964 [DOI] [PubMed] [Google Scholar]
- 64. Chai W., Yuen C. T., Kogelberg H., Carruthers R. A., Margolis R. U., Feizi T., and Lawson A. M. (1999) High prevalence of 2-mono- and 2,6-di-substituted manol-terminating sequences among O-glycans released from brain glycopeptides by reductive alkaline hydrolysis. Eur. J. Biochem. 263, 879–888 [DOI] [PubMed] [Google Scholar]
- 65. Inamori K., Endo T., Gu J., Matsuo I., Ito Y., Fujii S., Iwasaki H., Narimatsu H., Miyoshi E., Honke K., and Taniguchi N. (2004) N-acetylglucosaminyltransferase IX acts on the GlcNAcβ1,2-Manα1-Ser/Thr moiety, forming a 2,6-branched structure in brain O-mannosyl glycan. J. Biol. Chem. 279, 2337–2340 [DOI] [PubMed] [Google Scholar]
- 66. Abbott K. L., Matthews R. T., and Pierce M. (2008) Receptor tyrosine phosphatase beta (RPTPbeta) activity and signaling are attenuated by glycosylation and subsequent cell surface galectin-1 binding. J. Biol. Chem. 283, 33026–33035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Winterhalter P. R., Lommel M., Ruppert T., and Strahl S. (2013) O-glycosylation of the non-canonical T-cadherin from rabbit skeletal muscle by single mannose residues. FEBS Lett. 587, 3715–3721 [DOI] [PubMed] [Google Scholar]
- 68. Bartels M. F., Winterhalter P. R., Yu J., Liu Y., Lommel M., Möhrlen F., Hu H., Feizi T., Westerlind U., Ruppert T., and Strahl S. (2016) Protein O-mannosylation in the murine brain: occurrence of mono-O-mannosyl glycans and identification of new substrates. PLoS ONE 11:e0166119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhou H., Froehlich J. W., Briscoe A. C., and Lee R. S. (2013) The GlycoFilter: A simple and comprehensive sample preparation platform for proteomics, N-glycomics and glycosylation site assignment. Mol. Cell. Proteomics 12, 2981–2991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Clerc F., Reiding K. R., Jansen B. C., Kammeijer G. S., Bondt A., and Wuhrer M. (2016) Human plasma protein N-glycosylation. Glycoconj. J. 33, 309–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Windwarder M., and Altmann F. (2014) Site-specific analysis of the O-glycosylation of bovine fetuin by electron-transfer dissociation mass spectrometry. J. Proteomics 108, 258–268 [DOI] [PubMed] [Google Scholar]
- 72. Kato K., Jeanneau C., Tarp M. A., Benet-Pagès A., Lorenz-Depiereux B., Bennett E. P., Mandel U., Strom T. M., and Clausen H. (2006) Polypeptide GalNAc-transferase T3 and familial tumoral calcinosis. Secretion of fibroblast growth factor 23 requires O-glycosylation. J. Biol. Chem. 281, 18370–18377 [DOI] [PubMed] [Google Scholar]
- 73. Houel S., Hilliard M., Yu Y. Q., McLoughlin N., Martin S. M., Rudd P. M., Williams J. P., and Chen W. (2014) N- and O-glycosylation analysis of etanercept using liquid chromatography and quadrupole time-of-flight mass spectrometry equipped with electron-transfer dissociation functionality. Anal. Chem. 86, 576–584 [DOI] [PubMed] [Google Scholar]
- 74. Medzihradszky K. F., Kaasik K., and Chalkley R. J. (2015) Tissue-specific glycosylation at the glycopeptide level. Mol. Cell. Proteomics 14, 2103–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Engelmann K., Kinlough C. L., Müller S., Razawi H., Baldus S. E., Hughey R. P., and Hanisch F. G. (2015) Transmembrane and secreted MUC1 probes show trafficking-dependent changes in O-glycan core profiles. Glycobiology 15, 1111–1124 [DOI] [PubMed] [Google Scholar]
- 76. Prakobphol A., Thomsson K. A., Hansson G. C., Rosen S. D., Singer M. S., Phillips N. J., Medzihradszky K. F., Burlingame A. L., Leffler H., and Fisher S. J. (1998) Human low-molecular-weight salivary mucin expresses the sialyl lewisx determinant and has L-selectin ligand activity. Biochemistry 37, 4916–4927 [DOI] [PubMed] [Google Scholar]
- 77. Gomez Toledo A., Raducu M., Cruces J., Nilsson J., Halim A., Larson G., Rüetschi U., and Grahn A. (2012) O-Mannose and O-N-acetyl galactosamine glycosylation of mammalian α-dystroglycan is conserved in a region-specific manner. Glycobiology 22, 1413–1423 [DOI] [PubMed] [Google Scholar]
- 78. Razawi H., Kinlough C. L., Staubach S., Poland P. A., Rbaibi Y., Weisz O. A., Hughey R. P., and Hanisch F. G. (2013) Evidence for core 2 to core 1 O-glycan remodeling during the recycling of MUC1. Glycobiology 23, 935–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Darula Z., Sarnyai F., and Medzihradszky K. F. (2016) O-glycosylation sites identified from mucin core-1 type glycopeptides from human serum. Glycoconj. J. 33, 435–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Pap A., Medzihradszky K. F., and Darula Z. (2017) Using “spectral families” to assess the reproducibility of glycopeptide enrichment: human serum O-glycosylation revisited. Anal. Bioanal. Chem. 409, 539–550 [DOI] [PubMed] [Google Scholar]
- 81. Medzihradszky K. F. (2005) Characterization of protein N-glycosylation. Methods Enzymol. 405, 116–138 [DOI] [PubMed] [Google Scholar]
- 82. Rana N. A., Nita-Lazar A., Takeuchi H., Kakuda S., Luther K. B., and Haltiwanger R. S. (2011) O-glucose trisaccharide is present at high but variable stoichiometry at multiple sites on mouse Notch1. J. Biol. Chem. 286, 31623–31637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Nilsson J., Nilsson J., Larson G., and Grahn A. (2010) Characterization of site-specific O-glycan structures within the mucin-like domain of alpha-dystroglycan from human skeletal muscle. Glycobiology 20, 1160–1169 [DOI] [PubMed] [Google Scholar]
- 84. Vosseller K., Trinidad J. C., Chalkley R. J., Specht C. G., Thalhammer A., Lynn A. J., Snedecor J. O., Guan S., Medzihradszky K. F., Maltby D. A., Schoepfer R., and Burlingame A. L. (2006) O-linked N-acetylglucosamine proteomics of postsynaptic density preparations using lectin weak affinity chromatography and mass spectrometry. Mol. Cell. Proteomics 5, 923–934 [DOI] [PubMed] [Google Scholar]
- 85. Medzihradszky K. F., Gillece-Castro B. L., Settineri C. A., Townsend R. R., Masiarz F. R., and Burlingame A. L. (1990) Structure determination of O-linked glycopeptides by tandem mass spectrometry. Biomed. Environ. Mass Spectrom. 19, 777–781 [DOI] [PubMed] [Google Scholar]
- 86. Peter-Katalinić J. (2005) Methods in enzymology: O-glycosylation of proteins. Methods Enzymol. 405, 139–171 [DOI] [PubMed] [Google Scholar]
- 87. Zauner G., Kozak R. P., Gardner R. A., Fernandes D. L., Deelder A. M., and Wuhrer M. (2012) Protein O-glycosylation analysis. Biol. Chem. 393, 687–708 [DOI] [PubMed] [Google Scholar]
- 88. Domon B., and Costello C. E. (1988) A systematic nomenclature for carbohydrate fragmentations in FAB-MS/MS spectra of glycoconjugates. Glycoconjugate J. 5, 397–409 [Google Scholar]
- 89. Halim A., Westerlind U., Pett C., Schorlemer M., Rüetschi U., Brinkmalm G., Sihlbom C., Lengqvist J., Larson G., and Nilsson J. (2014) Assignment of saccharide identities through analysis of oxonium ion fragmentation profiles in LC-MS/MS of glycopeptides. J. Proteome Res. 13, 6024–6032 [DOI] [PubMed] [Google Scholar]
- 90. Mirgorodskaya E., Roepstorff P., and Zubarev R. A. (1999) Localization of O-glycosylation sites in peptides by electron capture dissociation in a Fourier transform mass spectrometer. Anal. Chem. 71, 4431–4436 [DOI] [PubMed] [Google Scholar]
- 91. Mikesh L. M., Ueberheide B., Chi A., Coon J. J., Syka J. E., Shabanowitz J., and Hunt D. F. (2006) The utility of ETD mass spectrometry in proteomic analysis. Biochim. Biophys. Acta 1764, 1811–1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Darula Z., and Medzihradszky K. F. (2009) Affinity enrichment and characterization of mucin core-1 type glycopeptides from bovine serum. Mol. Cell. Proteomics 8, 2515–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Halim A., Nilsson J., Rüetschi U., Hesse C., and Larson G. (2012) Human urinary glycoproteomics; attachment site-specific analysis of N- and O-linked glycosylations by CID and ECD. Mol. Cell. Proteomics doi: 10.1074/mcp.M111.013649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Halim A., Rüetschi U., Larson G., and Nilsson J. (2013) LC-MS/MS characterization of O-glycosylation sites and glycan structures of human cerebrospinal fluid glycoproteins. J. Proteome Res. 12, 573–584 [DOI] [PubMed] [Google Scholar]
- 95. Hoffmann M., Marx K., Reichl U., Wuhrer M., and Rapp E. (2016) Site-specific O-glycosylation analysis of human blood plasma proteins. Mol. Cell. Proteomics 15, 624–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Bakken V., Helgaker T., and Uggerud E. (2004) Models of fragmentations induced by electron attachment to protonated peptides. Eur. J. Mass Spectrom. 10, 625–638 [DOI] [PubMed] [Google Scholar]
- 97. Darula Z., Chalkley R. J., Baker P., Burlingame A. L., and Medzihradszky K. F. (2010) Mass spectrometric analysis, automated identification and complete annotation of O-linked glycopeptides. Eur. J. Mass Spectrom. (Chichester) 16, 421–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Frese C. K., Altelaar A. F., van den Toorn H., Nolting D., Griep-Raming J., Heck A. J., and Mohammed S. (2012) Toward full peptide sequence coverage by dual fragmentation combining electron-transfer and higher-energy collision dissociation tandem mass spectrometry. Anal. Chem. 84, 9668–9673 [DOI] [PubMed] [Google Scholar]
- 99. Biemann K. (1990) Appendix 5. Nomenclature for peptide fragment ions (positive ions). Methods Enzymol. 193, 886–887 [DOI] [PubMed] [Google Scholar]
- 100. Zhokhov S. S., Kovalyov S. V., Samgina T. Y., and Lebedev A. T. (2017) An EThcD-Based Method for Discrimination of Leucine and Isoleucine Residues in Tryptic Peptides. J. Am. Soc. Mass Spectrom. 28, 1600–1611 [DOI] [PubMed] [Google Scholar]
- 101. Bern M., Kil Y. J., and Becker C. (2012) Byonic: advanced peptide and protein identification software. Curr. Protoc. Bioinformatics Chapter 13:Unit13.20. doi: 10.1002/0471250953.bi1320s40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Clauser K. R., Baker P., and Burlingame A. L. (1999) Role of accurate mass measurement (+/− 10 ppm) in protein identification strategies employing MS or MS/MS and database searching. Anal. Chem. 71, 2871–2882 [DOI] [PubMed] [Google Scholar]
- 103. Baker P. R., Trinidad J. C., and Chalkley R. J. (2011) Modification site localization scoring integrated into a search engine. Mol. Cell. Proteomics doi: 10.1074/mcp.M111.008078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Maynard J. C., Burlingame A. L., and Medzihradszky K. F. (2016) Cysteine S-linked N-acetylglucosamine (S-GlcNAcylation), A new post-translational modification in mammals. Mol. Cell. Proteomics 15, 3405–3411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Marino F., Bern M., Mommen G. P. M., Leney A. C., van Gaans-van den Brink J. A. M., Bonvin A. M. J. J., Becker C., van Els C. A. C. M., and Heck A. J. R. (2015) Extended O-GlcNAc on HLA class-I-bound peptides. J. Am. Chem. Soc. 137, 10922–10925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Malaker S. A., Penny S. A., Steadman L. G., Myers P. T., Loke J. C., Raghavan M., Bai D. L., Shabanowitz J., Hunt D. F., and Cobbold M. (2017) Identification of glycopeptides as posttranslationally modified neoantigens in leukemia. Cancer Immunol. Res. 5, 376–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Fiat A. M., Jollès J., Aubert J. P., Loucheux-Lefebvre M. H., and Jollès P. (1980) Localisation and importance of the sugar part of human casein. Eur. J. Biochem. 111, 333–339 [DOI] [PubMed] [Google Scholar]
- 108. Verbij F. C., Stokhuijzen E., Kaijen P. H., van Alphen F., Meijer A. B., and Voorberg J. (2016) Identification of glycans on plasma-derived ADAMTS13. Blood 128, e51–e58 [DOI] [PubMed] [Google Scholar]
- 109. Agarwala K. L., Kawabata S., Takao T., Murata H., Shimonishi Y., Nishimura H., and Iwanaga S. (1994) Activation peptide of human factor IX has oligosaccharides O-glycosidically linked to threonine residues at 159 and 169. Biochemistry 33, 5167–5171 [DOI] [PubMed] [Google Scholar]
- 110. Medzihradszky K. F., Gillece-Castro B. L., Hardy M. R., Townsend R. R., and Burlingame A. L. (1996) Structural elucidation of O-linked glycopeptides by high energy collision-induced dissociation. J. Am. Soc. Mass Spectrom. 7, 319–328 [DOI] [PubMed] [Google Scholar]
- 111. Müller S., Alving K., Peter-Katalinic J., Zachara N., Gooley A. A., and Hanisch F. G. (1999) High density O-glycosylation on tandem repeat peptide from secretory MUC1 of T47D breast cancer cells. J. Biol. Chem. 274, 18165–18172 [DOI] [PubMed] [Google Scholar]
- 112. Perdivara I., Petrovich R., Allinquant B., Deterding L. J., Tomer K. B., and Przybylski M. (2009) Elucidation of O-glycosylation structures of the beta-amyloid precursor protein by liquid chromatography-mass spectrometry using electron transfer dissociation and collision induced dissociation. J. Proteome Res. 8, 631–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Takahashi K., Wall S. B., Suzuki H., Smith AD 4th, Hall S., Poulsen K., Kilian M., Mobley J. A., Julian B. A., Mestecky J., Novak J., and Renfrow M. B. (2010) Clustered O-glycans of IgA1: defining macro- and microheterogeneity by use of electron capture/transfer dissociation Mol. Cell. Proteomics 9, 2545–2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Takahashi K., Smith A. D., Poulsen K., Kilian M., Julian B. A., Mestecky J., Novak J., and Renfrow M. B. (2012) Naturally occurring structural isomers in serum IgA1 O-glycosylation. J. Proteome Res. 11, 692–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Fenaille F., Groseil C., Ramon C., Riandé S., Siret L., Chtourou S., and Bihoreau N. (2008) Mass spectrometric characterization of N- and O-glycans of plasma-derived coagulation factor VII. Glycoconj. J. 25, 827–842 [DOI] [PubMed] [Google Scholar]
- 116. Moloney D. J., Shair L. H., Lu F. M., Xia J., Locke R., Matta K. L., and Haltiwanger R. S. (2000) Mammalian Notch1 is modified with two unusual forms of O-linked glycosylation found on epidermal growth factor-like modules. J. Biol. Chem. 275, 9604–9611 [DOI] [PubMed] [Google Scholar]
- 117. Walsh G., and Jefferis R. (2006) Post-translational modifications in the context of therapeutic proteins. Nat. Biotechnol. 24, 1241–1252 [DOI] [PubMed] [Google Scholar]
- 118. Hossler P., Khattak S. F., and Li Z. J. (2009) Optimal and consistent protein glycosylation in mammalian cell culture. Glycobiology 19, 936–949 [DOI] [PubMed] [Google Scholar]
- 119. Albrecht S., Hilliard M., and Rudd P. (2014) Therapeutic proteins: facing the challenges of glycobiology. JHPOR 1, 12–17 [Google Scholar]
- 120. Costa A. R., Rodrigues M. E., Henriques M., Oliveira R., and Azeredo J. (2014) Glycosylation: impact, control and improvement during therapeutic protein production. Crit. Rev. Biotechnol. 34, 281–299 [DOI] [PubMed] [Google Scholar]
- 121. Gribben J. G., Devereux S., Thomas N. S., Keim M., Jones H. M., Goldstone A. H., and Linch D. C. (1990) Development of antibodies to unprotected glycosylation sites on recombinant human GM-CSF. Lancet 335, 434–437 [DOI] [PubMed] [Google Scholar]
- 122. Kim H., Yamaguchi Y., Masuda K., Matsunaga C., Yamamoto K., Irimura T., Takahashi N., Kato K., and Arata Y. (1994) O-glycosylation in hinge region of mouse immunoglobulin G2b. J. Biol. Chem. 269, 12345–12350 [PubMed] [Google Scholar]
- 123. Plomp R., Dekkers G., Rombouts Y., Visser R., Koeleman C. A., Kammeijer G. S., Jansen B. C., Rispens T., Hensbergen P. J., Vidarsson G., and Wuhrer M. (2015) Hinge-region O-glycosylation of human immunoglobulin G3 (IgG3). Mol. Cell. Proteomics 14, 1373–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Gong B., Burnina I., Stadheim T. A., and Li H. (2013) Glycosylation characterization of recombinant human erythropoietin produced in glycoengineered Pichia pastoris by mass spectrometry. J. Mass Spectrom. 48, 1308–1317 [DOI] [PubMed] [Google Scholar]
- 125. Giménez E., Ramos-Hernan R., Benavente F., Barbosa J., and Sanz-Nebot V. (2012) Analysis of recombinant human erythropoietin glycopeptides by capillary electrophoresis electrospray-time of flight-mass spectrometry. Anal. Chim. Acta 709, 81–90 [DOI] [PubMed] [Google Scholar]
- 126. Jiang J., Tian F., Cai Y., Qian X., Costello C. E., and Ying W. (2014) Site-specific qualitative and quantitative analysis of the N- and O-glycoforms in recombinant human erythropoietin. Anal. Bioanal. Chem. 406, 6265–6274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Byeon J., Lim Y. R., Kim H. H., and Suh J. K. (2015) Structural identification of a non-glycosylated variant at Ser126 for O-glycosylation site from EPO BRP, human recombinant erythropoietin by LC/MS analysis. Mol. Cells 38, 496–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Stübiger G., Marchetti M., Nagano M., Grimm R., Gmeiner G., Reichel C., and Allmaier G. (2005) Characterization of N- and O-glycopeptides of recombinant human erythropoietins as potential biomarkers for doping analysis by means of microscale sample purification combined with MALDI-TOF and quadrupole IT/RTOF mass spectrometry. J. Sep. Sci. 28, 1764–1778 [DOI] [PubMed] [Google Scholar]
- 129. Zhu H., Qiu C., Ruth A. C., Keire D. A., and Ye H. (2017) A LC-MS all-in-one workflow for site-specific location, identification and quantification of N-/O- glycosylation in human chorionic gonadotropin drug products. AAPS J. doi: 10.1208/s12248-017-0062-z [DOI] [PubMed] [Google Scholar]
- 130. Anderson N. L., and Anderson N. G. (2002) The human plasma proteome: history, character, and diagnostic prospects. Mol. Cell. Proteomics 1, 845–867 [DOI] [PubMed] [Google Scholar]
- 131. Hägglund P., Matthiesen R., Elortza F., Højrup P., Roepstorff P., Jensen O. N., and Bunkenborg J. (2007) An enzymatic deglycosylation scheme enabling identification of core fucosylated N-glycans and O-glycosylation site mapping of human plasma proteins. J. Proteome Res. 6, 3021–3031 [DOI] [PubMed] [Google Scholar]
- 132. Zhang H., Li X. J., Martin D. B., and Aebersold R. (2003) Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat. Biotechnol. 21, 660–666 [DOI] [PubMed] [Google Scholar]
- 133. Nilsson J., Rüetschi, Halim U. A., Hesse C., Carlsohn E., Brinkmalm G., and Larson G. (2009) Enrichment of glycopeptides for glycan structure and attachment site identification. Nat. Methods 6, 809–811 [DOI] [PubMed] [Google Scholar]
- 134. Geoghegan K. F., and Stroh J. G. (1992) Site-directed conjugation of nonpeptide groups to peptides and proteins via periodate oxidation of a 2-amino alcohol. Application to modification at N-terminal serine. Bioconjug. Chem. 3, 138–146 [DOI] [PubMed] [Google Scholar]
- 135. Tachibana K., Nakamura S., Wang H., Iwasaki H., Tachibana K., Maebara K., Cheng L., Hirabayashi J., and Narimatsu H. (2006) Elucidation of binding specificity of Jacalin toward O-glycosylated peptides: quantitative analysis by frontal affinity chromatography. Glycobiology 16, 46–53 [DOI] [PubMed] [Google Scholar]
- 136. Yamada K., Hyodo S., Kinoshita M., Hayakawa T., and Kakehi K. (2010) Hyphenated technique for releasing and MALDI MS analysis of O-glycans in mucin-type glycoprotein samples. Anal. Chem. 82, 7436–7443 [DOI] [PubMed] [Google Scholar]
- 137. Reisner Y., Linker-Israeli M., and Sharon N. (1976) Separation of mouse thymocytes into two subpopulations by the use of peanut agglutinin. Cell. Immunol. 25, 129–134 [DOI] [PubMed] [Google Scholar]
- 138. Chang C. F., Pan J. F., Lin C. N., Wu I. L., Wong C. H., and Lin C. H. (2011) Rapid characterization of sugar-binding specificity by in-solution proximity binding with photosensitizers. Glycobiology 21, 895–902 [DOI] [PubMed] [Google Scholar]
- 139. Trinidad J. C., Barkan D. T., Gulledge B. F., Thalhammer A., Sali A., Schoepfer R., and Burlingame A. L. (2012) Global identification and characterization of both O-GlcNAcylation and phosphorylation at the murine synapse. Mol. Cell. Proteomics 11, 215–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Bai X., Li D., Zhu J., Guan Y., Zhang Q., and Chi L. (2015) From individual proteins to proteomic samples: characterization of O-glycosylation sites in human chorionic gonadotropin and human-plasma proteins. Anal. Bioanal. Chem. 407, 1857–1869 [DOI] [PubMed] [Google Scholar]
- 141. Steentoft C., Vakhrushev S. Y., Joshi H. J., Kong Y., Vester-Christensen M. B., Schjoldager K. T., Lavrsen K., Dabelsteen S., Pedersen N. B., Marcos-Silva L., Gupta R., Bennett E. P., Mandel U., Brunak S., Wandall H. H., Levery S. B., and Clausen H. (2013) Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 32, 1478–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Vakhrushev S. Y., Steentoft C., Vester-Christensen M. B., Bennett E. P., Clausen H., and Levery S. B. (2013) Enhanced mass spectrometric mapping of the human GalNAc-type O-glycoproteome with SimpleCells. Mol. Cell. Proteomics 12, 932–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Yang Z., Halim A., Narimatsu Y., Jitendra Joshi H., Steentoft C., Schjoldager K. T., Alder Schulz M., Sealover N. R., Kayser K. J., Paul Bennett E., Levery S. B., Vakhrushev S. Y., and Clausen H. (2014) The GalNAc-type O-Glycoproteome of CHO cells characterized by the SimpleCell strategy. Mol. Cell. Proteomics 13, 3224–3235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Schjoldager K. T., Vakhrushev S. Y., Kong Y., Steentoft C., Nudelman A. S., Pedersen N. B., Wandall H. H., Mandel U., Bennett E. P., Levery S. B., and Clausen H. (2012) Probing isoform-specific functions of polypeptide GalNAc-transferases using zinc finger nuclease glycoengineered SimpleCells. Proc. Natl. Acad. Sci. U.S.A. 109, 9893–9898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Schjoldager K. T., Joshi H. J., Kong Y., Goth C. K., King S. L., Wandall H. H., Bennett E. P., Vakhrushev S. Y., and Clausen H. (2015) Deconstruction of O-glycosylation–GalNAc-T isoforms direct distinct subsets of the O-glycoproteome. EMBO Rep. 16, 1713–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Bennett E. P., Mandel U., Clausen H., Gerken T. A., Fritz T. A., and Tabak L. A. (2012) Control of mucin-type O-glycosylation: a classification of the polypeptide GalNAc-transferase gene family. Glycobiology 22, 736–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Bush D. R., Zang L., Belov A. M., Ivanov A. R., and Karger B. L. (2016) High resolution CZE-MS quantitative characterization of intact biopharmaceutical proteins: proteoforms of interferon-β1. Anal. Chem. 88, 1138–1146 [DOI] [PubMed] [Google Scholar]
- 148. Hart G. W., Slawson C., Ramirez-Correa G., and Lagerlof O. (2011) Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu. Rev. Biochem. 80, 825–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Tagliabracci V. S., Engel J. L., Wiley S. E., Xiao J., Gonzalez D. J., Nidumanda Appaiah H., Koller A., Nizet V., White K. E., and Dixon J. E. (2014) Dynamic regulation of FGF23 by Fam20C phosphorylation, GalNAc-T3 glycosylation, and furin proteolysis. Proc. Natl. Acad. Sci. U.S.A. 111, 5520–5525 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.