Abstract
Despite recent efforts toward control and elimination, malaria remains a major public health problem worldwide. Plasmodium falciparum resistance against artemisinin, used in front line combination drugs, is on the rise, and the only approved vaccine shows limited efficacy. Combinations of novel and tailored drug and vaccine interventions are required to maintain the momentum of the current malaria elimination program. Current evidence suggests that strain-transcendent protection against malaria infection can be achieved using whole organism vaccination or with a polyvalent vaccine covering multiple antigens or epitopes. These approaches have been successfully applied to the human-infective sporozoite stage. Both systemic and tissue-specific pathology during infection with the human malaria parasite P. falciparum is caused by asexual blood stages. Tissue tropism and vascular sequestration are the result of specific binding interactions between antigens on the parasite-infected red blood cell (pRBC) surface and endothelial receptors. The major surface antigen and parasite ligand binding to endothelial receptors, PfEMP1 is encoded by about 60 variants per genome and shows high sequence diversity across strains. Apart from PfEMP1 and three additional variant surface antigen families RIFIN, STEVOR, and SURFIN, systematic analysis of the infected red blood cell surface is lacking. Here we present the most comprehensive proteomic investigation of the parasitized red blood cell surface so far. Apart from the known variant surface antigens, we identified a set of putative single copy surface antigens with low sequence diversity, several of which are validated in a series of complementary experiments. Further functional and immunological investigation is underway to test these novel P. falciparum blood stage proteins as possible vaccine candidates.
An estimated 3.2 billion people - nearly half the world's population - are at risk of contracting malaria (1). Although the mortality rate has decreased over the last decade, malaria remains an acute public health problem in many countries and regions. In 2015 alone, there was an estimated 214 million clinical cases of malaria leading to about 438,000 deaths, most of them children under the age of five (1).
Severe and fatal malaria cases can largely be attributed to one parasite species, Plasmodium falciparum. After an initial and asymptomatic liver cycle, parasite propagation takes place in host red blood cells (RBC)1. On RBC invasion, parasites start an active process of host cell remodeling. As mature RBCs lack organelles and intracellular membranes, parasite-derived trafficking machinery is introduced into the host cell cytosol to export parasite antigens to the RBC and its plasma membrane. Such antigens can establish new permeation pathways for nutrient uptake, induce adherence of parasitized RBCs (pRBC) to the vascular lining in the deep tissue (sequestration), bind unparasitized RBCs (rosetting), or evade immunity by antigenic variation (reviewed in (2)).
Most of the known pRBC surface antigens are encoded by large multi-gene families, collectively named variant surface antigens (VSA). VSAs include P. falciparum erythrocyte membrane protein (PfEMP1) (3–5), P. falciparum–encoded repetitive interspersed families of polypeptides (RIFIN)(6, 7), surface-associated interspersed gene family (SURFIN)(8), and possibly others such as sub-telomeric variable open reading frame (STEVOR)(9, 10). PfEMP1s are the best characterized VSA, and multiple paralogs have been demonstrated to mediate adhesion of pRBCs to various endothelial receptors present in the microvasculature, including CD36, ICAM-1, VCAM-1 and endothelial protein C receptor (EPCR) (reviewed in (11)). Sequestration of pRBCs in the microvasculature is a key pathophysiological feature of malaria infection, and several VSAs including PfEMP1 induce strong antibody-mediated immune responses (12). The RIFIN proteins belong to the largest VSA family, with roughly 150 paralogs. These are classified into the surface-exposed type A and type B that localize to Maurer's clefts (13, 14) - parasite-derived membranous structures underneath the RBC plasma membrane (15). Like PfEMP1, RIFINs are immunogenic and have recently been shown to be involved in sequestration and rosetting of blood group A RBCs (16). Recently the two paralogs of the P. falciparum Hyp8 family (17) have also been localized at the pRBC surface (18).
The importance of VSAs such as PfEMP1 and RIFINs as immune targets strongly supports their development as vaccine candidates (12). However, major challenges toward this goal are the significant sequence diversity and large genetic repertoires of VSAs, as well as variant expression patterns in the population. Hence, there is a strong rationale to identify conserved pRBC surface antigens (or at least epitopes) with minimal allelic variation that could induce strain-transcendent immunity. Two previous studies aiming at identifying parasite antigens at the pRBC surface have used surface biotinylation techniques followed by isolation of membranes and subsequent mass spectrometry (19, 20). In a third study, parasite antigens were metabolically labeled before isolation of membranes (21). These pioneering studies identified several antigens, and several of them have subsequently been validated as markers of the Maurer's clefts. However, none of the putative pRBC surface antigens have since been confirmed. Limited information on parasite antigen composition at the pRBC surface is likely because of low antigen solubility (because these are usually membrane-anchored proteins), as well as low antigen expression.
Here we present the most comprehensive proteomic investigation of the pRBC membrane so far. For this purpose, we used two complementary approaches: (1) surface shaving and proteomic analysis of shaved and unshaved pRBC membranes in combination with profiling of immunogenic surface antigens by protein array (here termed membrane proteomics), and (2) surface shaving and proteomic analysis of released protein ectodomains in the supernatant (here termed supernatant proteomics). Our data demonstrate the identification and validation of a series of nonvariant pRBC surface antigens, and provide a rational basis for novel vaccine approaches targeting blood stage malaria parasites.
MATERIALS AND METHODS
Whole Genome Sequencing of Pf2004 Strain
P. falciparum Pf2004 genomic DNA was sequenced with an Illumina HiSeq platform, using 101 base pair (bp) paired end reads from a small-insert (200 bp) library. The raw data have been deposited in the NCBI Sequence Read Archive under accession number SAMN02630800. Variants were called using GATK Unified Genotyper (22).
Parasite In Vitro Culture Conditions
Plasmodium falciparum strain Pf2004–164/TdTomato (23) was kept in continuous culture according to standard procedures (24) with O+ red blood cells (RBCs) at 3% hematocrit and 10% O+ type serum in buffered malaria culture medium (MCM). Parasites were kept in T-75 flasks, each containing 25 ml MCM. Parasites were synchronized with repeated treatments of 5% sorbitol for 10 min, and kept in a shaking incubator for all described experiments. Parasites were selected to express the PfEMP1 variant VAR2CSA, by repeated panning using chondroitin sulfate A, as previously described (25).
Sample Separation by SDS-PAGE for Western Blot and Coomassie Gels
Five μl of each sample (corresponding to 2.5 × 106 pRBC/lane) were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) using 4–12% Bis-Tris gradient gels (Invitrogen, Carlsbad, CA), and subjected to either Western blot or Coomassie staining. For Western blot analysis, after overnight wet transfer using PVDF membranes, the membranes were blocked with 3% Bovine Serum Albumin (BSA) and probed for one hour with anti-Glycophorin C antibodies (1:250, Sigma-Aldrich), anti-ATS 6H-1 antibodies (1:100), anti-Spectrin antibodies (1:500, Abcam, Cambridge, UK), or with sera from the immunized mice described below (1:500). Mouse sera were pre-absorbed before probing for one hour with unparasitized RBCs at a 20:1 dilution. Detection by near-infrared fluorescence was performed using the LI-COR system (Lincoln, NE) after a secondary probe of anti-mouse IRDye-800CW-coupled antibodies (1:3000, LI-COR) was added for one hour. Washes were conducted with Phosphate-buffered saline (PBS) supplemented with 0.05% Tween-20 (Sigma-Aldrich, St. Louis, MO).
Lysis Measurements
Five μl of packed unparasitized RBCs were incubated with PBS, PBS supplemented with sucrose (5–40%), or RPMI for 15, 30, or 60 min. As a positive lysis control, the same amount of RBCs was incubated with 0.1% Triton-X 100 in PBS for 15, 30, or 60 min. Samples were subsequently centrifuged, and the supernatants collected. The amount of released hemoglobin (Hb) was measured at 415 nm by a Nanodrop spectrophotometer (Nanodrop, Wilmington, DE). The amount of lysis in each sample was calculated as follows: Lysis (test sample) = Absorbance415 (test sample)/Absorbance415 (positive lysis control), and presented as percentage.
Sample Preparation and Mouse Immunization
Trophozoite stage Pf2004–164/TdT (28–36 h p.i.) was harvested by magnet-activated cell sorting (MACS) and 5–6 × 108 pRBCs were washed 3× in PBS and split into two fractions. The first fraction (+) was incubated with 450 μl of PBS supplemented with 10% sucrose, 1 mg/ml trypsin and 1 mg/ml chymotrypsin (PBS-S+T/C; Sigma-Aldrich), and the second fraction (−) was incubated with 10% sucrose in PBS only (PBS-S) for 1 h while rotating at 37 °C. Cells were then collected by centrifugation, washed twice in PBS supplemented with a protease inhibitor mixture (PBS-PI; Roche, Basel, Switzerland), and washed once in PBS alone. Each cell pellet was subsequently lysed in 2 ml ice-cold hypotonic lysis buffer (5 mm KH2PO4, pH 7.4) for 20 min with repeated vortexing. The cell lysates were then centrifuged for 15 min at 20,000 × g and 80% of the resulting supernatant removed. The remaining cell pellets were lysed two more times and 80% of the supernatant removed. The remaining membrane pellets were then transferred to a new tube and washed 3× in hypotonic lysis buffer and once in PBS by centrifugation for 15 min at 20,000 × g. The final membranes were mixed with TiterMax Gold Adjuvant (Sigma-Aldrich) at a 1:1 ratio and used for animal immunization. For each membrane preparation, a group of six female BALB/c mice from Harlan (Frederick, MD) were immunized three times with 100 μl of the membrane/adjuvant-mix over a course of 6 weeks at BioQual (Rockville, MD).
Experimental Design and Statistical Rationale for pRBC Membrane Proteomics
Three biological replicates (A, B, and C) of trophozoite stage Pf2004–164/TdT (28–32 h p.i.) pRBCs were harvested using magnetic cell sorting columns (MACS, Miltenyi Biotec, Auburn, CA) resulting in a yield of 4.8–5.3 × 108 pRBCs (88% parasitemia). The three samples were washed 3x in PBS and split into two fractions. The first fraction (+) was incubated with 450 μl PBS-S+T/C, and the second (−) with PBS-S for one hour while rotating at 37 °C. Cells were collected by centrifugation, washed twice in PBS-PI, and once in PBS alone. Membranes from each sample were subsequently prepared as above. For subsequent Western blots, all samples were normalized to contain the same number of cells (5 × 107 cells per 100 μl loading buffer).
Twenty-five microliters of membrane proteins isolated from pRBCs with or without trypsin shaving in SDS sample buffer were loaded to SDS-PAGE (4–12% bis-tris gel, 10 well, 1.5 mm) (Novex by Life Technologies, Carlsbad, CA) and run in MOPS buffer for 1.5 h. A total of 72 bands resulting from six samples were excised for subsequent LC/MS/MS analysis. The 72 samples were derived from three biological replicates, each with or without trypsin shaving, and 12 bands from each sample: four bands (ABCD) over 180 kDa region, four bands (EFGH) from 100–130 kDa region, one band (I) from 70 kDa region and three bands (JKL) around 25–35 kDa region. In-gel digestion of the 72 bands was performed in three batches using an Intavis DigestPro with 32 well set-up (Cologne, Germany). Briefly, the in-gel digestion method included gel band destaining (50:50 ammonium bicarbonate/acetonitrile), shrinking of gel pieces (acetonitrile), reduction and alkylation (dithiothreitol and iodoacetamide solutions in ammonium bicarbonate), washing, in-gel trypsin digestion (8 h, Porcine sequencing trypsin, Promega, Madison, WI) and finally extraction of digested peptides with 50 ml of 1% trifluoroacetic acid/60% acetonitrile solution three times. The resulting peptide mixtures were analyzed by nano LC/MS/MS. Separation was performed using an in-house packed nano column with Magic C18 beads (Bruker, Billerica, MA; 75 μm × 10 cm) on a nanoAcquity HPLC (Waters corporation, Milford MA) in conjunction with an Orbitrap Velos (Thermo, Waltham, MA) using a top 20 data dependent acquisition (DDA) MS/MS experiment in ion trap mode with survey MS at 60 K resolution.
Data base searching was performed against a combination of four databases containing 31,385 entries (5538 for P. falciparum 3D7, 5536 for P. falciparum Pf2004, 20,177 for Uniprot_human_rev and 128 for cRAP_proteomics) using Mascot search engine version 2.5.1 (Matrix Science Inc., Boston, MA). Database search parameters included fixed enzyme search with trypsin and maximum two missed cleavage events, fixed modification of cysteines with carbamidomethylation, variable modifications of methionine oxidation, asparagine and glutamine deamidation and N-terminal acetylation, mass tolerances of 10 ppm and 0.8 Da for MS and MS/MS respectively. Further filtering and summarization of the data was performed in Scaffold version 4.4.4 (Proteome Software, Portland, OR) with protein and peptide probability set at 95% with a minimum of 2 unique peptides. A false discovery rate (FDR) less than 0.5% for proteins was measured by the equation FDR = D/T where D was the number of counts of decoys and T was the target identification hits observed using the decoy database. The search results from six samples (3 biological replicates with/without trypsin shaving) of the same band were grouped as one experiment in Scaffold. Twelve sample protein reports with total unique peptide count information were exported from Scaffold and combined into one table. Single protein hits, which are only shown once in six samples or once in each arm, were removed from the table. Software R version 3.1.2 was used to calculate the average of total unique peptide counts for each protein of the treated and nontreated samples, fold changes, p values and FDR rates of an unpaired t test.
Experimental Design and Statistical Rationale for pRBC Supernatant Proteomics
Two biological replicates (A and B) of synchronized ring stage parasite cultures at 2–4 h post invasion (p.i.) were collected by centrifugation. Each replicate was resuspended in 50 ml PBS supplemented with 32.5 mg EZ-Link Sulfo-NHS-Biotin (Life Technologies) to label host (RBC) surface proteins. After room temperature incubation for 30 min on a rotating shaker, pRBCs were collected and washed twice in 100 mm glycine/PBS to neutralize the biotin, and twice in PBS alone. Each replicate was subsequently re-suspended in MCM and parasites allowed to continue their development as described above. The following day, trophozoite stage pRBCs (28–32 p.i.) were harvested by MACS, yielding 5.7 × 108 pRBCs (74% parasitemia, replicate A) and 7.1 × 108 pRBCs (91% parasitemia, replicate B).
Each replicate was subsequently split into two fractions. The first fraction (+) was incubated with 500 μl PBS+10% sucrose supplemented with 70 μg/ml mass-spectrometry grade Trypsin (Sigma-Aldrich), whereas the second fraction (−) was mock-treated (PBS and 10% sucrose alone). After a one-hour incubation at 37 °C while rotating, the four samples (A+ and A−, B+ and B-) were centrifuged for 8 min at 240 × g at room temperature to separate the supernatant containing surface shaved peptides and/or pRBC lysis products from intact pRBCs. For subsequent Western blots, 5 × 107 intact pRBCs from each sample were resuspended in 100 μl SDS loading buffer. The supernatants were first cleared of any remaining cell debris by centrifugation for 20 min at 14,000 × g at 4 °C. Next, the supernatants were cleared from “contaminating” host RBC surface proteins by incubating each supernatant (500 μl) with 70 μl of washed MagnaBind Strepatavidin Beads (Life Technologies) for 30 min while rotating. The beads were removed with the MACS magnet and the supernatants were centrifuged for 20 min as 14,000 × g to remove any remaining beads.
Next samples were prepared for quantitative mass spectrometry with iTRAQ. A diagram of the procedure is shown in supplemental Fig. S1. Protein content of each of the four samples (A+ and A−, B+ and B-) was measured using Pierce BCA protein assay. Twenty mm DTT was added to 100 μg of protein and samples were incubated for 30 min at 37 °C. Iodoacetamide was added at a final concentration of 50 mm and samples were incubated for 30 min in the dark at room temperature. Before trypsin digestion, urea concentration was diluted to less than 1 m by adding water and pH adjusted to 8 with 1 m Tris solution. 2 μg sequencing grade trypsin (Promega, Madison, WI) was added (1:50 enzyme to substrate ratio) and samples were incubated at 37 °C with shaking for 16 h. The reaction was stopped by addition of formic acid (FA) to a final concentration of 1% and the solution was desalted with a 1 cc (30 mg) Oasis HLB reverse phase cartridge (Waters, Milford, MA) conditioned with 3 × 500 μl acetonitrile (ACN), followed by 4 × 500 μl 0.1% FA. Samples were loaded onto the cartridges and washed with 3 × 500 μl 0.1% FA. Desalted peptides were eluted by 2 applications of 500 μl of 80% ACN/0.1% FA. Eluates were frozen, dried via vacuum centrifugation and stored at −80 °C before peptide fractionation.
For each sample the de-salted peptides were labeled with iTRAQ 4-plex reagents according to the manufacturer's instructions (AB Sciex, Foster City, CA). The differentially labeled peptides were mixed to form a single pooled sample and subsequently desalted on 500 mg SepPak columns. The pooled sample was fractionated by reverse phase chromatography into 30 fractions and analyzed by tandem LC/MS. Briefly, each of the 30 fractions was re-suspended in 8 μl of 3% ACN/0.5% FA before analysis using a Q Exactive mass spectrometer coupled to an EASY-nLC 1000 UHPLC (Thermo Scientific). A PicoFrit column (New Objective, Woburn, MA), with an inner diameter of 75 μm and packed with 20 cm of ReproSil-Pur C18 1.9 μm particles, was directly interfaced to the Q Exactive instrument equipped with a custom nano-electrospray ionization source. The peptide mixture (2 μg) from each of the 30 fractions were injected and separated by a 110-min gradient from 5–60% solvent B. MS/MS analysis settings for protein identification were as follows: one precursor MS scan at 70,000 resolution in profile mode was followed by data-dependent scans of the top 12 most abundant ions at low-resolution (17,500) in profile mode. Dynamic exclusion was enabled for duration of 20 s. MS/MS spectra were collected with normalized collision energy of 28 and an isolation width of 2.5 m/z. The extensive peptide fractionation coupled with in depth MS analysis allowed detection and identification of very low levels of peptides.
All mass spectra were processed using the Spectrum Mill software package Rev B.05.0 preRelease (Agilent Technologies, Santa Clara, CA). Precursor ion quantification was performed using extracted ion chromatograms for each precursor ion. The peak area for the extracted ion chromatograms of each precursor ion subjected to MS/MS was calculated automatically by the Spectrum Mill software in the intervening high-resolution MS1 scans of the LC-MS/MS runs using narrow windows around each individual member of the isotope cluster. Peak widths in both the time and m/z domains were dynamically determined based on MS scan resolution, precursor charge and m/z, subject to quality metrics on the relative distribution of the peaks in the isotope cluster versus theoretical. Similar MS/MS spectra acquired on the same precursor m/z in the same dissociation mode within ±60 s were merged. Precursor ion purity (PIP) was calculated for every precursor ion triggered for MS/MS from its corresponding MS1 spectrum as 100% × the intensity of the precursor ion isotopic envelope/total intensity in the precursor isolation window. MS/MS spectra with precursor charge >7 and poor quality MS/MS spectra, which failed the quality filter by not having a sequence tag length >1 (i.e. minimum of three masses separated by the in-chain mass of an amino acid) were excluded from searching.
For peptide identification MS/MS spectra were searched against two concatenated protein sequence databases, each containing UniProt Human (78,370 entries) and common laboratory contaminant proteins (73 entries), in addition to either P. falciparum 3D7 (5538 entries) or Pf2004 (5536 entries). Search parameters included: ESI QExactive scoring parameters, Trypsin_nonspecific_N_term enzyme specificity (allows semitryptic peptides ending in K or R), 40% minimum matched peak intensity, ±20 ppm precursor mass tolerance, ±20 ppm product mass tolerance, and carbamidomethylation of cysteines and iTRAQ labeling of lysines and peptide n-termini as fixed modifications. Allowed variable modifications were oxidationM, pyroGlu-Q, Deamidated-N, and pyroCarbamidomethyl-C with a precursor MH_ shift range of −18 to 64 Da. After the first round of autovalidation described below, a second round of searches was performed on the remaining MS/MS spectra with the variable modifications parameter revised to allow peptide N termini to be acetylated rather than iTRAQ labeled, and the sequence database search space limited to only proteins with a peptide matched in the first round of searches. The purpose of the two-step search was to identify peptides with a cleaved N-terminal PEXEL motif (RxLxD/E/Q), resulting in a mature N-terminal sequence xD/E/Q (26).
Peptide spectrum matches for individual spectra were automatically designated as confidently assigned using the Spectrum Mill autovalidation module to apply target-decoy based false-discovery rate (FDR) scoring threshold criteria via a three-step auto threshold strategy at the peptide and protein levels. First, peptide autovalidation was performed using an auto thresholds strategy with a minimum sequence length of 6, automatic variable range precursor mass filtering, and score and delta Rank1 - Rank2 score thresholds optimized to yield a spectral level FDR estimate for precursor charges 2 thru 6 of <1.2% for each precursor charge state in each LC-MS/MS run. Second, a round of manual validation was performed after the second round of searches to select the peptides with the cleaved PEXEL motif. This only increased the number of validated PSM's from 74,373 to 74,601. Third, protein polishing autovalidation was applied to further filter all the peptide-level validated spectra with the primary goal of eliminating peptides identified with low scoring peptide spectrum matches (PSM's) that represent proteins identified by a single peptide from a single patient, so-called one-hit wonders. The following parameters were used; minimum number of experiments protein group is observed in: 1, minimum protein score: 10. After assembling protein groups from the autovalidated peptides for an experiment, protein polishing removed PSM's were from the set obtained in the initial peptide-level autovalidation step if they contributed to protein groups that have protein scores at or below the minimum protein score. In the filtered results each identified protein was detected with multiple peptides unless a single excellent scoring peptide was the sole match. These autovalidation steps yielded FDR estimates of 0.78% at the spectrum level, 1.13% at the peptide level, and 0.08% at the protein level. Because the protein level FDR estimate neither explicitly requires a minimum number of distinct peptides per protein nor adjusts for the number of possible tryptic peptides per protein, it may underestimate false positive protein identifications for large proteins observed only on the basis of multiple low scoring PSM's. In calculating scores at the protein level and reporting the identified proteins, redundancy is addressed in the following manner: the protein score is the sum of the scores of distinct peptides. A distinct peptide is the single highest scoring instance of a peptide detected through an MS/MS spectrum. MS/MS spectra for a peptide may have been recorded multiple times, (i.e. as different precursor charge states, isolated from adjacent bRP fractions, modified by oxidation of Met) but are still counted as a single distinct peptide. When a peptide sequence >8 residues long is contained in multiple protein entries in the sequence database, the proteins are grouped together and the highest scoring one and its accession number are reported. In some cases when the protein sequences are grouped in this manner there are distinct peptides, which uniquely represent a lower scoring member of the group (isoforms or family members). Each of these instances spawns a subgroup and multiple subgroups are reported and counted toward the total number of proteins. Each protein level iTRAQ log2 ratio was calculated as the median of all PSM level log2 ratios contributing to the protein remaining after excluding those PSM's lacking an iTRAQ label, having a negative delta forward-reverse score (half of all false-positive identifications), or having a precursor ion purity < 50% (MS/MS has significant precursor isolation contamination from coeluting peptides). To identify significantly enriched surface proteins, we used the Limma package in the R environment to calculate moderated t test p values corrected by the Benjamini-Hochberg method.
In Silico Analysis of Candidate Antigens
The genetic diversity of candidate proteins was measured with the nonsynonymous pi statistic using custom scripts. Variation data for the nonsynonymous pi calculations derive from 45 culture adapted isolates from Senegal for which whole genome Illumina sequencing data were previously published (68). RNAseq data generation was performed using RNA extracted from patient samples from Senegal (Bei et al., unpublished data). Briefly, the samples were maintained in short-term ex vivo culture and then harvested at the late trophozoite to mid-schizont stage and lysed with TRI reagent BD (Sigma-Aldrich) before RNA extraction. RNA was sent to the Beijing Genomics Institute for library construction following poly(A) selection. Reads were trimmed with Trimmomatic, ends of reads were removed if phred quality was less than 15 (28), reads were aligned to the 3D7 reference genome with TopHat (29), transcript expression was measured with CuffLinks (30) and samples were compared with CuffDiff2 (31). Statistical analysis was performed using the fragments per kilobase per million (FPKM) metric values for each locus.
Recombinant Protein Expression, Purification, and Antibody Generation
Recombinant ectodomain constructs for PfJ23, PIESP2, PF3D7_1310500, and PF3D7_1352500 were designed by removing predicted native signal peptide and transmembrane domain portions of the protein. Constructs with a predicted PEXEL motif were truncated to remove sequence N-terminal to the PEXEL cleavage site (26). This resulted in the following constructs: PfJ23(90–221), PIESP2(46–361), PF3D7_1310500(24–154), and PF3D7_1352500(23–166). Each construct was appended with an N-terminal mammalian signal sequence (METDTLLLWVLLLWVPGSTG) for secretory expression and a C-terminal 10xHis tag for immobilized metal affinity column (IMAC) purification. Constructs underwent codon harmonization for synthesis (GeneArt, Regensburg, Germany) and were cloned into the pcDNA3.1 plasmid (Invitrogen) for heterologous expression in mammalian cells. The fully automated mammalian cell secretory overexpression system PEPP (Protein Expression and Purification Platform; GNF, San Diego, CA) was used to express and purify the recombinant proteins. In brief, 35 ml of cultures seeded with 1.4 × 106 HEK293 F cells (Invitrogen, Cat. R79007) were transfected with 35 μg of plasmid DNA using the Polyethylenimine HCI MAX transfection reagent per the manufacturers protocol (Polysciences Inc., Warminster, PA). Cells were incubated at 37 °C in 8% CO2 gas environment for 5 days. Culture supernatants were harvested and applied to Ni-NTA agarose (Qiagen) for gravity flow purification. Proteins were desalted using Nap-10 sephadex (GE Healthcare, Little Chalfont, UK) and eluted with 1X DPBS (Invitrogen) resulting in a final storage solution of phosphate buffered saline, pH 7.4 for each protein sample. For each construct, protein from 10 culture wells was pooled and concentrated to ∼1.0 mg/ml for animal immunization and antibody generation.
Antisera were generated by immunization of three rats per construct using standard protocols (GenScript, Piscataway NJ). Preimmune serum samples were harvested for each study animal on the day prior (d −1) to immunization. On day zero, each animal was immunized by intraperitoneal injection (i.p.) with a primary dose of 50 μg of recombinant protein in Freund's Complete Adjuvant. This was followed by three boosts of 25 μg protein in Freund's Incomplete Adjuvant administered i.p. on days 14, 25, and 56. Antisera were collected on day 63 and tested by ELISA for reactivity against the recombinant protein relative to preimmune control sera.
Validation of Candidates by Western Blot and Flow Cytometry
Western blots for the purpose of candidate validation were performed as described above. For flow cytometry experiments, trophozoite stage Pf2004–164/TdT were harvested by MACS, washed 3x in PBS, subjected to one hour incubation with either PBS-S+T/C or PBS-S, and washed twice in PBS-PI, and once in PBS alone. Candidate polyclonal antibodies (all produced in rat) were diluted to test concentrations (1:25, 1:50, 1:100) in PBS supplemented with 1% fetal bovine serum (FBS) and pre-absorbed for one hour with unparasitized RBCs at a 1:1 dilution to remove antibodies nonspecifically binding RBCs. Pre-absorbed antibodies were then incubated with trypsin- or mock-treated pRBCs at 0.4% hematocrit. An SBP-1 N-terminal rabbit antibody, kindly provided by Tobias Spielmann, was used as a control. Surface binding was detected using Alexa Fluor® 488 anti-rat IgG dye (Thermo Fisher Scientific; 1:500) or anti-rabbit in the case of SBP-1. Washes were conducted with PBS supplemented with 1% FBS. To distinguish unparasitized and parasitized RBCs, samples were incubated with Vybrant® DyeCycle™ Violet Stain (Thermo Fisher Scientific; 1:500) diluted in Hank's Buffered Salt Solution (Thermo Fisher Scientific). Cells were ultimately resuspended in 200 μl Hank's buffer and analyzed on a MACSQuant® VYB (Miltenyi Biotec, Bergisch Gladbach, Germany) or LSRFORTESSA X-20 (BD Biosciences, San Jose, CA).
Validation of Candidates by Fluorescence Microscopy
For immunofluorescence microscopy, trophozoite stage Pf2004–164/TdT at 5–8% parasitemia were washed 3x in PBS, subjected to one-hour incubation with either PBS-S+T/C or PBS-S, and washed twice in PBS-PI, and once in PBS alone. Cells were fixed for 40 min in 3% paraformaldehyde/0.01% glutaraldehyde, washed 3× in PBS, permeabilized for 10 min using 0.1% Triton-X and washed 3×. As described above, candidate polyclonal antibodies were pre-absorbed for one hour with unparasitized RBCs. Antibodies (diluted in PBS supplemented with 3% BSA) or SBP-1 antibody were then incubated with trypsin- or mock-treated pRBCs at 0.4% hematocrit, and surface binding was detected using Alexa Fluor® 488 anti-IgG dye (Thermo Fisher Scientific; 1:500). Washes were conducted with PBS supplemented with 3% BSA. Cells were mounted with Vectashield with DAPI (Vector Labs, Burlingame, CA).
High resolution multicolor microscopy was conducted on two microscopes. We used a Zeiss Elyra PS.1 microscope equipped with a high resolution AXIO Observer Z1 inverted microscope stand with transmitted (HAL), UV (HBO) and laser illumination sources. The objective used for imaging was an α-Plan-Apochromat 100 × 1.46 NA DIC VIS immersion oil lens, and four laser lines (405, 488, 561 and 642 nm) as well as BF and DIC. The microscope was equipped with an XL TIRF Dark S1 incubator. We also used a Zeiss observer Z1 spinning disc confocal microscope, equipped with a Yokogawa CSU-X1 filter wheel and spinning disc unit, a Photometrics Evolve 512 delta EM-CCD camera, four laser lines (405, 488, 561, and 642 nm) and an α-Plan-Apochromat 100 × 1.46 NA DIC VIS immersion oil lens.
Images from both microscopes were acquired using the Zen 2012 software. To avoid any cross talk among the channels, images were collected in line sequential mode with a dwell time of 1.1 μs and z-increments of 0.19 μm. Images were further processed using Huygens deconvolution software and analyzed for pixel intensity spatial correlation analysis, using the imaging software Fiji. Results were expressed as Pearson's correlation coefficient, and Maders split coefficients were also obtained.
RESULTS
Membrane Proteomics: An Immune Profiling Approach to Identify P. falciparum Surface Antigens
In a first series of experiments, we performed a combination of immune profiling using sera from mice immunized with shaved or unshaved pRBC membranes and proteomics of pRBC membranes, with the aim to identify pRBC surface antigens (Fig. 1A). To evaluate the surface proteome of sequestering P. falciparum parasites, we used the Pf2004 parasite strain, as it has previously been used for cytoadherence studies (32, 33). To ensure that our Pf2004 strain expressed surface antigens capable of cytoadhering, we selected the parasites to express the chondroitin sulfate A adhering PfEMP1 variant VAR2CSA (34), resulting in Pf2004_CSA.
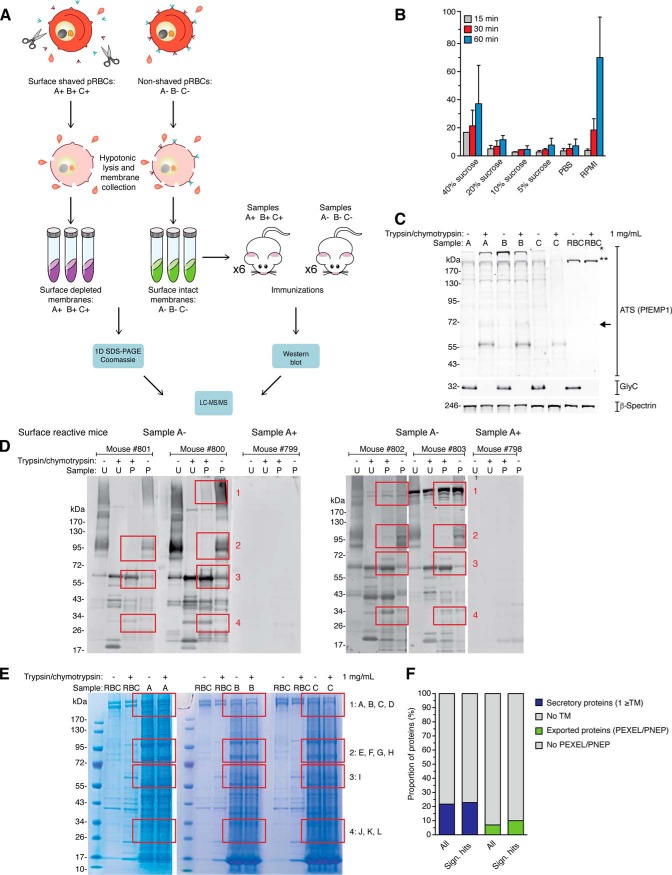
Fig. 1.
Membrane proteomics. A, Overview of the membrane proteomics process: biological replicates (A, B, and C) of purified and intact pRBCs (28–34 h post infection) were either surface-shaved with trypsin and chymotrypsin (+), or mock-treated with sucrose-supplemented PBS (−). B, Lysis optimization. Five μl of RBCs were lysed with either PBS, PBS supplemented with 5–20% sucrose, or RPMI, for 30–60 min. Lysis is presented as percentage of hemoglobin in each sample and normalized to a positive lysis control with 0.1% Triton X-100 in PBS, which was set to 100%. C, Validation of proteomics samples. Membranes were collected for Western blot analysis for each of the six samples (A±, B±, and C±). Membranes were probed with anti-Glycophorin C or anti-ATS antibodies. The anti-ATS antibody detects both full-length PfEMP1 (>300 kDa, marked with an asterisk), as well as the surface truncated PfEMP1 (70–95 kDa, box), which mainly consists of the cytoplasmic C-terminal ATS region of PfEMP1. Full-length PfEMP1 migrates very closely to cross-reactive RBC spectrins (marked with a double asterisk). Anti-Spectrin antibodies were used as a loading control. Each lane represents protein extract from 2.5 × 106 pRBC. D, Membranes from the six samples (A±, B±, and C±) were analyzed by Western blot using surface reactive sera from the six mice immunized with mock-treated pRBCs. Four areas of interest (1–4) where bands are present only in untreated cells or bands are truncated on treatment are marked with red boxes. U = unparasitized RBCs, p = pRBCs. E, Membranes from the six samples (A±, B±, and C±) were separated by SDS-PAGE and stained with Coomassie blue. Areas of interest were divided into multiple bands (A-L) for in-gel digestion. U = unparasitized RBCs, p = pRBCs). F, Bar charts describing distribution of transmembrane (TM, blue) and exported (green) proteins, comparing proteins significantly enriched in the untreated sample (Sign. hits) with all proteins identified (All).
First, to optimize the preparation of surface-shaved pRBCs, we assessed whether addition of sucrose (as used in surface shaving of Gram-positive bacteria (35, 36)) could reduce RBC lysis previously seen with PBS alone. Specifically, we monitored the degree of lysis over time in unparasitized RBC samples by measuring hemoglobin absorbance at 415 nm (37). Six digestion conditions were tested: PBS supplemented with 0, 5, 10, 20, and 40% sucrose, and RPMI. High levels of sucrose (40%), as used for surface shaving of bacteria, resulted in slightly more lysis, hence we decided to use PBS supplemented with 10% sucrose for the surface-shaving step in subsequent experiments (Fig. 1B). Next, three biological replicates (A, B, and C) of purified and intact pRBCs (28–34 h post infection) were surface-shaved with a standard mixture of 1 mg/ml each of trypsin and chymotrypsin (+)(e.g. (38)), or mock-treated with sucrose-supplemented PBS (−). Cells from these 6 samples were collected by centrifugation, and membranes purified by multiple rounds of hypotonic pRBC lysis. To determine the surface shaving efficiency, we monitored the truncation of PfEMP1 and the RBC surface marker Glycophorin C in the trypsin- and mock-treated cell pellets by Western blot (Fig. 1C). The anti-ATS antibody, which detects both full-length (>300 kDa) and truncated membrane-anchored PfEMP1 (70–95 kDa), was used to monitor the shaving efficiency of PfEMP1 (39). Glycophorin C and PfEMP1 were intact in the mock-treated samples (A-, B-, and C-), whereas the molecules were absent (Glycophorin C) or truncated (C-terminal tail of PfEMP1 after proteolytic cleavage) in the surface-shaved samples (A+, B+ and C+), demonstrating efficient surface shaving (Fig. 1C).
To specifically identify immunogenic pRBC surface antigens, we immunized two sets of BALB/c mice with either surface-shaved or intact pRBCs. The rationale was that antibodies against immunogenic pRBC surface antigens are only present in mice immunized with intact pRBCs, and therefore these antigens are only detected in antibody-based detection methods when intact pRBCs are probed. Probing preparations of surface-shaved or intact pRBCs by Western blot with mouse sera demonstrated that only mice immunized with intact pRBC membranes (A−, B−, C−) elicited a detectable immune response against pRBC membranes (Fig. 1D). Therefore, we focused our further analysis on sera from mice immunized with intact pRBC membranes (i.e. surface reactive sera). Using these sera in Western blot analyses, we identified a series of bands in four areas (1–4) that were only present when either intact or surface-shaved pRBCs were probed with the surface reactive sera (e.g. proteins disappearing or emerging as truncated versions on pRBC surface shaving) (Fig. 1D). To isolate areas of interest, we separated membrane proteins by SDS-PAGE, stained the gels with Coomassie Blue and excised the four regions of interest for subsequent in-gel digestion and mass spectrometry analysis (Fig. 1E).
Analysis of Immunogenic Surface Candidates
For proteomic analysis of membranes, all six samples (A±, B±, C±) were separated by SDS-PAGE, and a total of 12 bands per sample covering the four areas of interest were excised. After in-gel digestion and processing, samples were analyzed by nanoflow liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS). To potentially generate a more extensive template for analysis of peptide sequences, we also performed whole genome sequencing of Pf2004_CSA by Illumina sequencing. Reads were mapped onto the fully sequenced and assembled P. falciparum 3D7 reference strain. Peptides and proteins were identified by searching the MS/MS spectra against the translated genomes of Pf2004_CSA as well as the reference strain 3D7.
The peptide and protein identification reports of 12 bands, exported from Scaffold software, are shown in supplemental Tables S1 and S2, respectively. Analysis of the samples by LC-MS/MS resulted in the identification of a total of 917 P. falciparum proteins. Thirty-six of these were detected uniquely identified using the 3D7 database, 95 detected using the Pf2004_CSA database and the remaining proteins detected by both databases (supplemental Tables S1 and S2). Importantly, 251 proteins were identified with significant enrichment in the untreated sample (supplemental Table S3). Forty-five of these were also significantly enriched in a lower molecular weight gel band in the treated sample, further suggesting possible surface exposure. Out of the 251 hits, 54 (22%) were predicted to be secretory proteins, i.e. containing a signal sequence and/or transmembrane domain (TM) according to Plasmodium genomics resource (www.PlasmoDB.org) (Fig. 1F, supplemental Table S3). Finally, a total of 26 out of the 54 secretory proteins had an export motif, a characteristic N-terminal amino acid motif required for proteolytic cleavage of parasite proteins exported into the host cell (17, 26, 40–42). We detected the PfEMP1 variant expressed by the parasite strain analyzed, VAR2CSA, as well as a type A-RIFIN variant among the exported antigen hits (supplemental Table S3). In addition, several known Maurer's clefts proteins (e.g. PfSBP1 (43), MAHRP1 (44)) and soluble exported proteins were identified. PIESP2 was also found with significant enrichment in the untreated sample and at lower molecular weight in the treated sample. PIESP2 was previously identified at the pRBC surface by surface biotinylation (20). In addition, we identified an exported protein previously termed PfJ23 (21) or Hyp16 (17) with a signal sequence and C-terminal transmembrane domain. This protein was identified in P. falciparum ghost membranes and localized to Maurer's clefts; however, surface localization was not investigated (21). Finally, PHISTc protein PFI1780w, a highly polymorphic parasite antigen (45) that was recently localized to the pRBC membrane (46), was also observed in our data. Altogether, following the traditional approach of proteomic analysis of ghost membranes combined with surface shaving, we detected the two major known variant surface antigen families, PfEMP1 and RIFINs, in addition to PIESP2, PfJ23, PFI1780w, several additional known exported proteins and eight membrane proteins with unknown localization (supplemental Table S3).
Supernatant Proteomics: An Orthogonal Surface Shaving Approach to Identify P. falciparum Antigens
Membrane surface proteins are notoriously difficult to detect because of their low yield at the surface. We therefore developed an alternative surface shaving method that was initially described for profiling of surface-exposed peptide epitopes in the Gram-positive bacterium Staphylococcus aureus (36). Here, surface shaving is based on proteolytic treatment of whole cells to release surface exposed peptides for their subsequent identification by LC-MS/MS (Fig. 2A). The pRBC surface was biotinylated shortly after host cell invasion to specifically label host proteins. The goal was to allow the intraerythrocytic parasite to mature into a trophozoite and express its own repertoire of antigens on the surface of the intact pRBC, before shaving and analysis by LC-MS/MS.
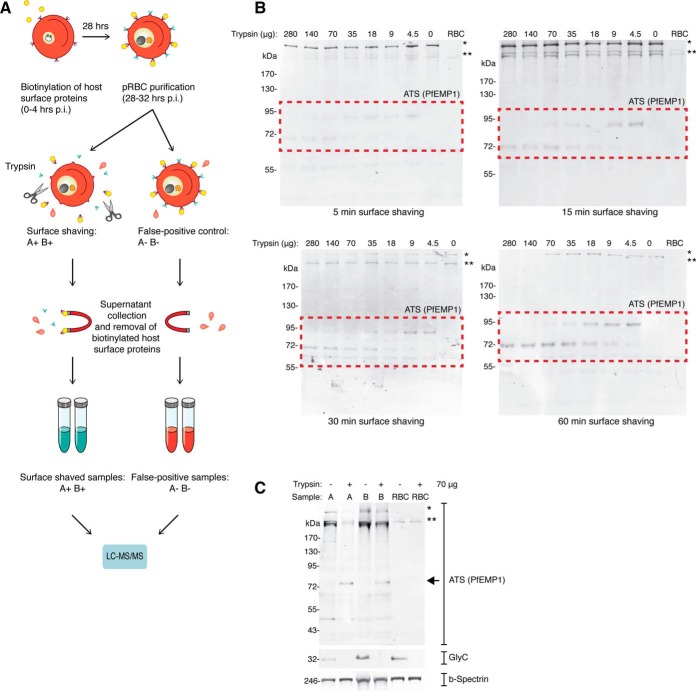
Fig. 2.
Supernatant proteomics. A, Overview of supernatant proteomics process. B, Surface shaving optimization. Intact and live trophozoite stage pRBC were surface shaved with a titration of trypsin, and analyzed by Western blot. Each lane represents protein extract from 2.5 × 106 pRBC. RBC refers to unparasitized RBCs. C, Validation of proteomics samples. After supernatants were collected for surface proteomics, the remaining cell pellet for each of the four samples were collected for Western blot analysis (A−, A+, B−, and B+). To validate the surface shaving method, membranes were probed with anti-Glycophorin C and anti-ATS antibodies. Anti-Spectrin antibodies were used as a loading control. Each lane represents protein extract from 2.5 × 106 pRBC.
Proteolysis conditions were initially investigated and optimized to minimize contamination of lysis products in the released material. The degree of surface shaving was monitored by incubating purified intact pRBCs with different concentrations of trypsin for 5, 15, 30, and 60 min, and analyzing the resulting cell pellets by Western blot (Fig. 2B). Again, the anti-ATS antibody was used to monitor the shaving efficiency. Truncated PfEMP1 molecules representing the C-terminal tail after proteolytic cleavage could be observed even after a short 5 min shaving procedure and using a low concentration of trypsin. However, more efficient surface shaving could be observed after 60 min of digestion at higher concentrations of trypsin. Importantly, the two highest trypsin concentrations used (140 and 280 μg/ml) resulted in a reduction of the full-length PfEMP1 signal, suggesting cell lysis and exposure of internal PfEMP1 under these conditions. We therefore decided to use 70 μg/ml of trypsin for further experiments, which appeared to be as efficient but less harmful to the intact pRBCs.
Two biological replicates (A and B) of purified and intact pRBCs (28–34 h post infection) were incubated with 70 μg/ml trypsin (+), or mock-treated with sucrose-supplemented PBS (−) for 60 min. Cells were removed by centrifugation and the remaining supernatants containing surface released peptides were collected and subjected to trypsin digestion for further analysis. Surface shaving efficiency of the pRBC samples was monitored by Western blot of truncated PfEMP1 and the RBC surface marker Glycophorin C in the trypsin- and mock-treated cell pellets, as described above. Glycophorin C and PfEMP1 were intact in the mock-treated samples (A− and B−), but absent or truncated in the trypsin-treated samples (A+ and B+), demonstrating efficient surface shaving (Fig. 2C).
Revisiting the P. falciparum Surface Proteome Using Quantitative Supernatant Proteomics
To obtain more precise quantification of peptides derived from shaved and control samples we used the iTRAQ (Isobaric Tags for Relative and Absolute Quantification) chemical labeling strategy (reviewed in (47)), in combination with LC-MS/MS (supplemental Fig. S1). Surface proteins may present only a small ectodomain with few trypsin cleavage sites at the surface and may consequently be underrepresented in complex surface supernatants. We therefore allowed for proteins to be identified by a single peptide and searched for peptides with N-terminal PEXEL cleavage sites. The Plasmodium EXport ELement is an N-terminal sequence motif with the consensus sequence RxLxD/E/Q required for export of parasite proteins into the host cell (42). The motif is cleaved by the aspartic protease Plasmepsin V, resulting in a mature acetylated sequence with the consensus xD/E/Q (26, 40, 48).
LC-MS/MS analysis of the four samples (A−, A+, B−, B+) resulted in the identification of 2737 P. falciparum proteins (supplemental Tables S4–S7). Most of the proteins were identified based on observation of multiple tryptic peptides, but we also allowed for identification of proteins based on single tryptic peptides (120 with 3D7 database and 118 with Pf2004 database). In addition, a small proportion of proteins (28 with 3D7 database and 35 with Pf2004 database) were identified (exclusively or in addition) based on the observation of single peptides formed by the characteristic cleavage within the PEXEL motif of exported parasite proteins and N-acetylation of the newly formed N terminus (supplemental Table S8). Most importantly, 381 proteins showed significantly different abundance profiles between shaved and control samples (Fig. 3A and supplemental Tables S9 and S10). Of those 381 proteins, 96 were at least 1.5-fold more abundant in the shaved sample compared with control, and we termed these surface-enriched proteins (Fig. 3A and supplemental Table S11).
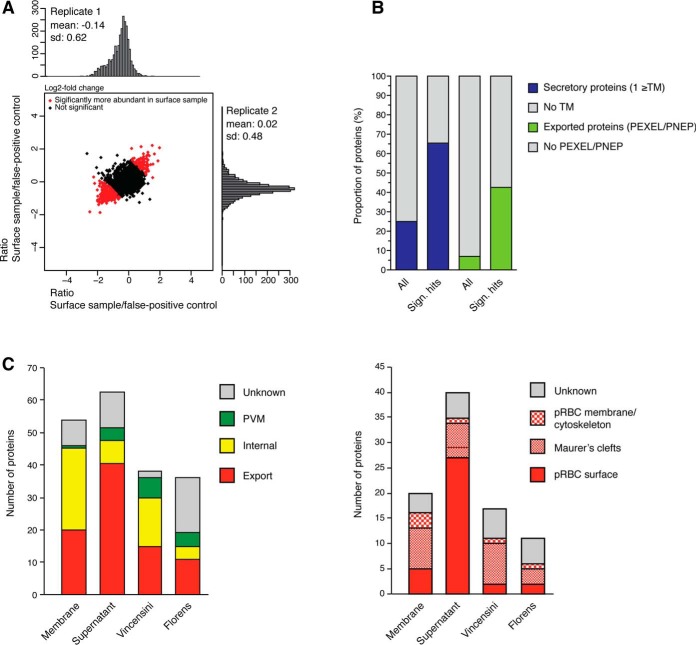
Fig. 3.
Supernatant proteomics analysis and overall comparison. A, Log2fold change scatter plot of the 2737 identified proteins from the two biological replicates. The 381 proteins with significantly different peptide abundance between surface shaved and lysis control samples are marked in red (corrected p values ≤ 0.05), B, Bar charts show distribution of transmembrane (TM) and PEXEL proteins, comparing proteins significantly enriched in surface shaving sample (Significant hits; blue/green, 100 proteins) and all proteins identified (gray, 2730). C, A comparison of total number and localization of secretory proteins (left graph) and exported proteins (right graph) across our two proteomics approaches and two previously published methods (20, 21).
We observed enrichment of secretory proteins in the surface-enriched fraction. Specifically, 62 of the 96 (65%) surface-enriched proteins had a signal sequence or at least one TM (compared with 25% of all 2737 proteins identified), i.e. were predicted to be secretory proteins (Fig. 3B). Out of these 62 proteins, 40 (65%) had a signal sequence and/or PEXEL motif (compared with 7% of all proteins identified) or other evidence of export, demonstrating enrichment of exported proteins. Finally, 27 of the 40 exported proteins were members of the RIFIN, PfEMP1, SURFIN, and MC-2TM families. Apart from seven known Maurer's cleft markers (e.g. SBP1, MAHRP1, we also identified several putative surface antigens. These included GEXP07 (18, 49), PIESP2 (20, 21), and PfJ23 (21). GEXP07 was recently identified as a parasite-derived pRBC surface receptor for the adhesive chemokine CX3CL1 (18). In addition, we identified 11 membrane proteins with no known localization. Interestingly, seven out of the 54 secreted proteins identified with the membrane proteomic approach were also identified in the analysis of supernatants. These shared proteins included a type A RIFIN, PfEMP1, PfJ23, PIESP2, PFI1780w and the two Maurer's clefts markers SBP1 and MAHRP1 (supplemental Tables S3 and S11, and supplemental Fig. S2).
We also compared the subset of secretory and exported antigens identified by supernatant proteomics or membrane proteomics approaches with two previous studies using ghost membranes with (20) or without a surface biotinylation enrichment step (21) (Fig. 3C). This comparison demonstrated that our supernatant proteomics approach yielded by far the greatest number of known surface antigens, followed by our membrane proteomics approach. In contrast, neither of the two previous studies identified any variant surface antigens. These data demonstrate that our refined methodology has improved enrichment for surface antigens compared with the two previous studies. Finally, SBP1, PIESP2, and PfJ23 were the only hits that were identified in at least three out of the four studies, with PIESP2 being detected in all four studies.
In Vivo Expression and Sequence Diversity of Significant Hits
The rationale for this study was to identify invariant antigens exposed at the pRBC surface of a cytoadherent parasite strain, as a basis for vaccine candidate prioritization. We therefore analyzed the expression profile and sequence polymorphism of the potential surface antigen hits identified by our two complementary approaches; namely the 54 secretory antigens identified from membrane proteomics and the 62 secretory surface-enriched proteins identified from supernatant proteomics. The two major pRBC surface antigen families, PfEMP1 and RIFIN, undergo antigenic variation and therefore only a subset of variants is expressed within the parasite population during infection. Such a mechanism of immune evasion makes these antigens poor candidates for vaccine development. To determine expression levels of our candidate surface antigens during infection, we performed expression analysis on three parasite samples obtained directly from malaria-infected patients from an endemic area in Senegal. Because many secreted parasite antigens show high levels of allelic sequence variation, we used an RNAseq approach to avoid known mapping and hybridization issues experienced with traditional microarray methods. Not surprisingly, var gene variants (encoding PfEMP1), as well as rifin and MC-2TM variants showed low expression levels in each individual patient compared with single copy genes such as PIESP2 and PfJ23 (Fig. 4).
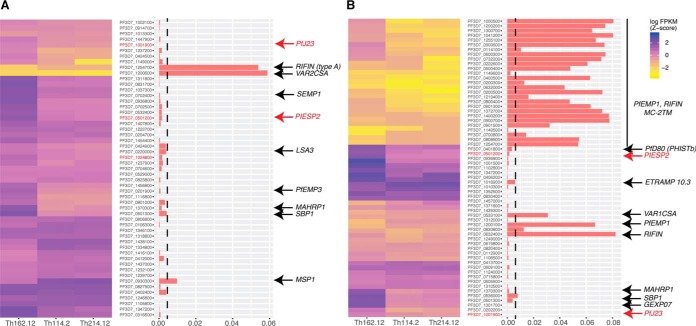
Fig. 4.
In vivo expression and allelic diversity for secretory proteins from membrane proteomics (A) and supernatant proteomics (B). Both graphs show transcriptional expression of all hits (A: n = 54, B: n = 65) in three patient samples from Senegal (left) and the ratio of nonsynonymous to synonymous SNPs across all hits based on a set of 64 parasite genomes from Senegal (right). Hits proposed for further validation (Table I) are shown in bold. The subset of hits validated by specific antibodies is marked in red. Vertical dashed line indicates our diversity threshold used for selection of candidates (πΝ < 0.004).
A second mechanism of immune evasion is allelic variation, and many parasite proteins, including early vaccine candidates, are highly polymorphic across strains (50). To quantify sequence diversity of the hits we measured their mean pairwise nonsynonymous diversity (πΝ) across 64 parasite isolates from Senegal. Collectively var genes, rifin and MC-2TMs showed the highest levels of sequence polymorphisms whereas many single genes showed very low levels, including PIESP2 and PfJ23. Altogether these data support the hypothesis that single copy genes encoding putative surface antigens, such as PIESP2 and PfJ23, may be potential vaccine candidates that could induce strain-transcending immunity.
Based on the combined data from the two complementary surface proteomics experiments and the in silico analysis, we have assembled a shortlist of candidate surface antigens for prioritization as vaccine candidates (Table I). For this purpose, we have further filtered the combined lists of transmembrane proteins identified by our two complementary approaches (supplemental Tables S3 and S11). After removal of variant antigens (PfEMP1, RIFIN, SURFIN), soluble proteins (signal sequence but no predicted TM), proteins with validated localization inside the pRBC (mostly Maurer's clefts) and corresponding genes with high levels of amino acid polymorphism (πΝ > 0.004), we propose to prioritize a shortlist of 11 candidate surface antigens for further investigation (Table I).
Table I. Candidate surface antigens prioritized for further validation. Shown are the eleven candidate proteins with key features derived from PlasmoDB, including number of transmembrane domains (TM), presence of a signal peptide (SP) and an export motif. We also included previously reported localization, isolation method by which the candidate was identified, and genetic diversity. Proteins validated for surface expression are marked in bold.
| Accession ID | TM | SP | Export motif | Protein description | Previously described localizations | Detection method | Polymorphism level (πN) |
|---|---|---|---|---|---|---|---|
| PF3D7_0112900 | 2 | YES | YES | Plasmodium exported protein | Supernatant | 0.00049 | |
| PF3D7_0220000 | 2 | YES | YES | LSA3 | Membranes | 0.0039 | |
| PF3D7_0501200 | 3 | YES | YES | PIESP2 | MC (21), Surface (20) | Membranes, Supernatant | 0.0016 |
| PF3D7_0619800 | 6 | NO | NO | Conserved Plasmodium membrane protein | Supernatant | 0.0014 | |
| PF3D7_0820400 | 2 | NO | NO | Conserved Plasmodium protein | Supernatant | 0.00040 | |
| PF3D7_1001900 | 3 | YES | YES | PfJ23 | MC (21) | Membranes/Supernatant | 0.000096 |
| PF3D7_1024800 | 2 | YES | NO | Conserved Plasmodium protein | Membranes | 0.0017 | |
| PF3D7_1249000 | 4 | NO | NO | Conserved Plasmodium membrane protein | Supernatant | 0.00053 | |
| PF3D7_1301700 | 3 | YES | YES | GEXP07 | Supernatant | 0.000061 | |
| PF3D7_1310500 | 2 | YES | NO | Conserved Plasmodium protein | Supernatant | 0.00040 | |
| PF3D7_1439300 | 3 | YES | NO | Conserved Plasmodium protein | Supernatant | 0.00055 |
Validation of Surface Exposure for a Subset of Candidates
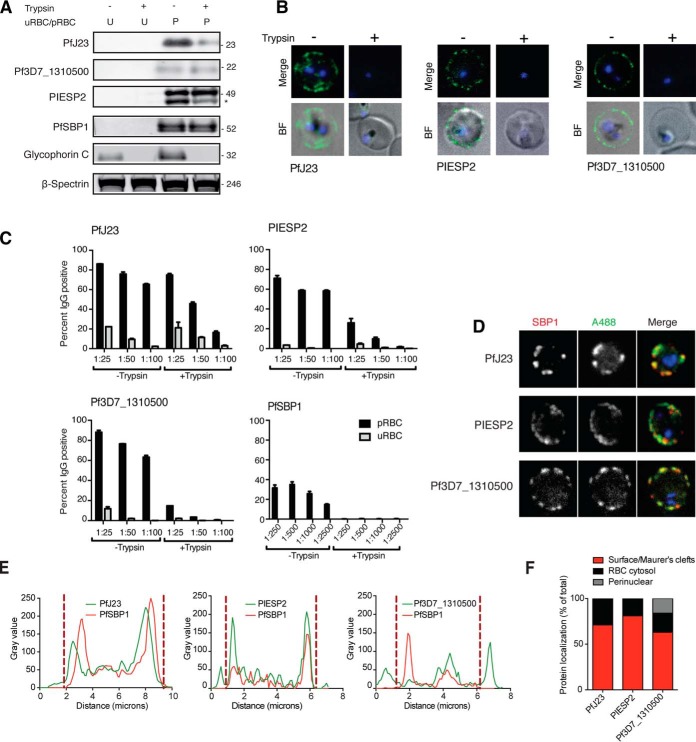
In order to validate a subset of our 11 top candidates as pRBC surface antigens, we generated polyclonal antibodies against three of them to be able to confirm their subsequent subcellular localization. We selected PfJ23 and PIESP2, as these are the only proteins that have been detected in both of our studies. Notably, PIESP2 has also been detected by both previous proteomics studies (20, 21). In addition, we selected a so far uncharacterized secretory antigen (Pf3D7_1310500).
Antibodies were first tested for specificity by Western blot analysis, and they all showed a specific band at the expected size of the mature protein, whereas no signal was detected when unparasitized RBCs were probed (Fig. 5A and supplemental Fig. S3). For PfJ23 and PIESP2, a band at the expected size (23 kDa for PfJ23 and 46 kDa for PIESP2) was trypsin-sensitive, indicating that at least a portion of the protein is surface exposed. Next, purified live pRBCs ± pre-treatment with trypsin/chymotrypsin were incubated with antigen-specific antibodies and pRBC surface recognition was detected by live microscopy (Fig. 5B). All three antibodies showed complete loss of signal on the pRBC surface on trypsin treatment. Parallel experiments using flow cytometry readout demonstrated minimal recognition of unparasitized RBCs and reduced recognition under + trypsin/chymotrypsin conditions compared with - trypsin/chymotrypsin conditions (Fig. 5C). Anti-Pf3D7_1310500 antibodies labeled more than 60% of surface-intact pRBCs at all concentrations tested, with this reactivity almost completely abolished after the addition of trypsin. In contrast, the Maurer's cleft marker PfSBP1 showed no evidence for surface localization either by Western blot or flow cytometry, as expected (Fig. 5A, 5B). Finally, we tested each antibody by immunofluorescence microscopy to confirm subcellular localizations of the proteins. As expected, all three proteins showed partial colocalization with SBP-1, and additional more peripheral labeling near the plasma membrane (Fig. 5D, 5E). Across all pRBCs examined, >60% protein localization was at the surface/Maurer's clefts for PfJ23 and PIESP2 (Fig. 5F). In contrast, Pf3D7_1310500 showed less colocalization with PfSBP1, as well as higher background signal and a higher percentage of cells showing perinuclear localization within the parasite (Fig. 5F). Taken together, these results provide strong evidence for pRBC surface expression of PfJ23 and PIESP2 and they are also suggestive of a smaller pool of Pf3D7_1310500 at the pRBC surface.
Fig. 5.
Validation of candidates. A, Unparasitized RBC and intact and live trophozoite stage pRBC (± pre-treatment with trypsin/chymotrypsin) analyzed by Western blot, and membranes were probed with polyclonal antibodies targeting candidate. A trypsin sensitive band at the expected size was detected for pRBCs probed with PfJ23 and PIESP2, whereas detection of Pf3D7_1310500 did not appear to be trypsin dependent. PfSBP1, Glycophorin C, and β-spectrin were included as controls. Each lane represents protein extract from 2.5 × 106 pRBC. Full Western blots are provided in supplemental Fig. S3. B, Surface reactivity of candidate antibodies to purified pRBCs (± pre-treatment with trypsin/chymotrypsin) was detected by live microscopy. Cells were stained with the nuclear dye Vybrant violet and the surface labeled using AlexaFluor488 goat anti-rat secondary antibody. Antibody signal is only detectable on the pRBC surface without prior trypsin treatment of the cells. C, Surface reactivity of candidate antibodies to purified pRBCs (± pre-treatment with trypsin/chymotrypsin) was detected by flow cytometry. Cells were gated for live cells and single cells, and uRBCs and pRBCs were subsequently separately gated based on Vybrant Violet fluorescence and surface reactivity measured using AlexaFluor488 goat anti-rat secondary antibody. Surface reactivity was measured as percent of cells positive for AlexaFluor488. Surface reactivity of all antibodies tested decreased with trypsin treatment, but this change was most significant for PIESP2, Pf3D7_1310500 and the lowest dilution (1:100) of PfJ23. D, Immunofluorescence analysis of the localization of PfJ23, PIESP2 and Pf3D7_1310500 (detected with specific sera) in fixed and permeabilized trophozoite pRBCs shows partial colocalization in the cytosol of pRBC with the Maurer's cleft resident protein PfSBP1, as well as pRBC surface localization. E, Fluorescence plot profile of PfJ23, PIESP2 and Pf3D7_1310500 localization confirms colocalization with PfSBP1 in addition to surface localization. Dashed lines mark the boundary between Maurer's cleft and peripheral/surface labeling based on SBP1 distribution. F, Immunofluorescence-based analysis of protein localization across multiple cells classified by fluorescence intensity percentage, shows that up to 70% of PfJ23, 90% of PIESP2, and 69% of Pf3D7_1310500, respectively, localizes to the pRBC surface or Maurer's clefts, whereas the remaining localization is cytosolic (and perinuclear in the case of Pf3D7_1310500).
DISCUSSION
Malaria parasites remodel the host RBC including its surface, to interact with the environment and adhere to the vascular endothelium. Several variants of the immunodominant antigen at the pRBC surface, PfEMP1, have been characterized in detail. For example, VAR2CSA—the most conserved PfEMP1 variant, an antigen responsible for placental sequestration and pregnancy-associated complications during malaria infection—is currently being evaluated as a blood stage vaccine candidate (51). Apart from PfEMP1, several other variant antigens have been identified at the pRBC surface, including RIFIN, SURFIN, and potentially STEVOR. However, a comprehensive characterization of the P. falciparum pRBC membrane has so far been limited to two proteomic studies published more than ten years ago (20, 21). Importantly, only one of these two studies (20) focused specifically on the pRBC surface. With a better understanding of the full surface antigen repertoire of the pRBC, we can identify conserved and constitutively expressed antigens, which we hypothesize represent better vaccine candidates than the variant antigens that have been explored to date. Here we applied two complementary surface shaving approaches to systematically map the pRBC surface. Key to both approaches was the shaving of the pRBC for subsequent differential analysis of hits between shaved and unshaved samples.
Surface shaving is a well-established method to investigate mechanisms of merozoite invasion into RBCs (52) and it has previously been suggested for evaluation of the pRBC surface (19, 53). In the case of supernatant proteomics, contamination with cytoplasmic proteins is a likely issue caused by cell lysis during the surface shaving procedure (53, 54). Prepared samples for supernatant proteomics are also expected to contain a fraction of mature schizonts that rupture during the surface shaving procedure, resulting in the release of free merozoites. As a consequence, the supernatants collected after surface shaving will not only contain proteins that are exposed on the pRBC surface (true positive surface proteins), but also soluble cytoplasmic proteins and proteins exposed on the merozoite surface (false positive surface proteins)(53). We attempted to control for these issues by introducing a false positive control in both approaches, where intact pRBCs were incubated without the presence of trypsin, as has previously been described in surface proteome studies of bacteria (55). Notably, surface shaving with the subsequent collection of cleaved ectodomains in the supernanant without prior depletion of host proteins has successfully been applied to the rodent malaria parasite, Plasmodium berghei (53). Our approach, however, has some critical improvements, including (1) selective host biotinylation before surface shaving for subsequent host protein depletion, and (2) quantitative mass spectrometry using iTRAQ to address the issue of internal protein leakage.
Overall, supernatant proteomics yielded 62 secretory proteins whereas membrane proteomics yielded 54 secretory proteins. In comparison, Vincensini et al. and Florens et al. detected 38 and 36 secretory proteins, respectively (20, 21). Supernatant proteomics also yielded by far the largest number of known surface antigens among the secretory proteins, suggesting that this approach is currently the most efficient method for pRBC surface antigen detection. With both supernatant proteomics and membrane proteomics, we were able to identify multiple variants of RIFIN as well as the PfEMP1 variant VAR2CSA, demonstrating that both methods indeed detect true positive surface antigens. This is important, as neither of the two previous studies detected a single paralog of these two variant surface antigen families (20, 21). Apart from solubility issues, low expression levels combined with limited sensitivity are likely reasons for the lack of detection of these two variant surface antigen families in previous work. Interestingly, the previously published studies by Florens et al. and Vincensini et al. and our work presented here have yielded a small set of overlapping Maurer's clefts proteins, including MAHRP1 (44), PfSBP1 (43), Rex1/2 (56), PfJ23, PIESP2 and PfD80 (21). Although PfSBP1 and MAHRP1 are well-characterized structural components of Maurer's clefts (43, 44, 57, 58), some of the other proteins may be en route to the pRBC surface. Indeed, Maurer's clefts are trafficking platforms for deposition of parasite antigens such as PfEMP1, RIFIN and STEVOR at the pRBC surface (15, 59). We hypothesized that a subset of our hits that have previously been localized to Maurer's clefts, including PfJ23 and PIESP2, may represent a pool of surface localized protein.
We were interested in identifying and validating invariant surface antigens with low allelic diversity, as a starting point for their evaluation for vaccine development. Therefore, we evaluated ex vivo transcriptional profiles and genetic diversity of all secretory hits from both the membrane-and the supernatant proteomics approaches. As expected, paralogs from variant antigen families were both the most polymorphic as well as the least expressed among all hits measured. However, we also identified a subset of hits with high expression levels and low sequence polymorphism, which are key features for the development of a vaccine with strain-transcending efficacy. Based on the combined proteomics data and the ex vivo expression and diversity information, we have compiled a shortlist of candidates that we propose for further evaluation. This list includes previously described antigens PIESP2, PfJ23, LSA3, and GEXP07, as well as one additional protein predicted to be exported (PF3D7_0112900) and six membrane proteins with no known localization. Notably four of these antigens (PIESP2, LSA3, GEXP07 and a membrane protein with unknown localization, PF3D7_01024800) are included in a protein array of immunogenic P. falciparum proteins that is widely used to measure human antibody responses to malaria infection. Using this platform, a recent study has demonstrated that responses to LSA3 correlate with protective immunity in a cohort in Kenya (60). We have validated three of the candidate antigens experimentally and demonstrated partial pRBC surface expression for all three using three complementary methods. In contrast, we did not detect any PfSBP1 signal on the pRBC surface, which is expected because it is a resident protein of Maurer's clefts. It has previously been shown that only a fraction of PfEMP1 is surface-exposed whereas the majority is present internally (59). Indeed, our data presented in Fig. 5 support a model where most of the surface antigen pool is localized within the cell, rather than surface-exposed. The functional significance of such a model remains to be determined.
There is a renewed interest in the global health community for the eventual elimination and eradication of malaria, but also recognition that new tools will be needed to achieve this goal. The development of an effective malaria vaccine has long been a goal and there are numerous vaccine development programs underway. These programs fall into three general classes - pre-erythrocytic vaccines, vaccines targeting the symptomatic blood stages, and vaccines specifically designed to block development and transmission in the mosquito. Until recently, most of these efforts were based on proteins identified in an initial series of discovery studies performed more than three decades ago (e.g. (61–63)). For example, many blood stage vaccine targets currently in trials and development were selected in the early 1980s based on protein abundance and measured human immune responses from patients to the parasite antigens (64–66). Vaccines developed from such targets have met with limited success or outright failure (67–70), in part because of the extreme sequence diversity of the chosen proteins (71, 72). Here we present the most comprehensive analysis of the pRBC surface since these early discovery studies. We have identified and partially validated a short list of putative pRBC surface antigens with high expression in patients and low sequence diversity. Based on this work, we propose further functional and immunological investigation of these antigens for prioritization as novel P. falciparum blood stage vaccine candidates.
DATA AVAILABILITY
The original mass spectra have been deposited in the public proteomics repository MassIVE and are accessible at ftp://massive.ucsd.edu/MSV000081541. Annotated MS/MS spectra for all peptides with N-terminal PEXEL cleavage sites and spectra corresponding to proteins to be identified by a single peptide are available at http://msviewer.ucsf.edu/prospector/cgi-bin/msform.cgi?form=msviewer. Use MS-Viewer search key: jed26hegyp (3D7 search results) and d9d1qtpg0f (Pf2004 search results).
Supplementary Material
Acknowledgments
We thank Dr. Seth Redmond and Allison Griggs for data analysis and generation of graphs shown in Fig. 5. Thanks also to Dr. Michael Duffy at Royal Melbourne Hospital, Melbourne, Australia for providing the 6H-1 anti-ATS antibody and Dr. Tobias Spielmann at Bernhard-Nocht-Institut für Tropenmedizin for providing the SBP-1 antibody.
Footnotes
* This work was supported by a Sanofi Pasteur Discovery award. SNB was supported by postdoctoral fellowships from the American Heart Association and the Swedish Society for Medical Research (SSMF), and KD was supported by a Herchel Smith graduate fellowship. MDN was supported by an SNF Early Postdoc Mobility Fellowship. MM was supported by a career development award from the Burroughs Wellcome Fund.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- RBC
- red blood cell
- pRBC
- parasite-infected red blood cell
- PfEMP1
- plasmodium falciparum erythrocyte membrane protein 1
- Stevor
- sub-telomeric variable open reading frame
- Rifin
- repetitive interspersed families of polypeptides
- Surfin
- surface-associated interspersed gene family
- VSA
- variant surface antigen
- EPCR
- endothelial protein C receptor
- CD36
- cluster of differentiation 36
- ICAM-1
- intercellular adhesion molecule 1
- VCAM-1
- vascular cell adhesion molecule 1
- MCM
- malaria Culture Medium
- SDS-PAGE
- sodium dodecyl sulfate polyacrylamide gel electrophoresis
- PVDF
- polyvinylidene fluoride
- BSA
- bovine serum albumin
- PBS
- phosphate buffered saline
- RPMI
- Roswell Park Memorial Institute
- Hb
- hemoglobin
- MACS
- magnet-activated cell sorting
- LS-MS/MS
- liquid chromatography tandem mass-spectrometry
- DDA
- data dependent acquisition
- FDR
- false discovery rate
- iTRAQ
- isobaric tag for relative and absolute quantification
- DTT
- dithiothreitol
- ACN
- acetonitrile
- PIP
- precursor ion purity
- PEXEL
- plasmodium export element
- PSM
- peptide spectrum match
- RNA
- ribonucleid acid
- FKPM
- fragments per kilobase per million
- IMAC
- immobilized metal affinity column
- PEPP
- protein expression and purification platform
- FBS
- fetal bovine serum
- BF
- bright field
- DIC
- differential interference contrast
- CSA
- chondroitin sulfate A
- MAHRP1
- membrane-associated histidine-rich protein 1
- SBP1
- skeleton binding protein 1
- PHIST
- plasmodium helical interspersed sub-telomeric family
- PIESP2
- plasmodium falciparum infected erythrocyte surface protein 1
- PfMC-2TM
- plasmodium falciparum Maurer's clefts 2 transmembrane domaine protein.
REFERENCES
- 1. WHO. (2016) WHO Malaria Report 2016
- 2. Maier A. G., Cooke B. M., Cowman A. F., and Tilley L. (2009) Malaria parasite proteins that remodel the host erythrocyte. Nature Rev. Microbiology 7, 341–354 [DOI] [PubMed] [Google Scholar]
- 3. Su X. Z., Heatwole V. M., Wertheimer S. P., Guinet F., Herrfeldt J. A., Peterson D. S., Ravetch J. A., and Wellems T. E. (1995) The large diverse gene family var encodes proteins involved in cytoadherence and antigenic variation of Plasmodium falciparum-infected erythrocytes. Cell 82, 89–100 [DOI] [PubMed] [Google Scholar]
- 4. Smith J. D., Chitnis C. E., Craig A. G., Roberts D. J., Hudson-Taylor D. E., Peterson D. S., Pinches R., Newbold C. I., and Miller L. H. (1995) Switches in expression of Plasmodium falciparum var genes correlate with changes in antigenic and cytoadherent phenotypes of infected erythrocytes. Cell 82, 101–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baruch D. I., Pasloske B. L., Singh H. B., Bi X., Ma X. C., Feldman M., Taraschi T. F., and Howard R. J. (1995) Cloning the P. falciparum gene encoding PfEMP1, a malarial variant antigen and adherence receptor on the surface of parasitized human erythrocytes. Cell 82, 77–87 [DOI] [PubMed] [Google Scholar]
- 6. Fernandez V., Hommel M., Chen Q., Hagblom P., and Wahlgren M. (1999) Small, clonally variant antigens expressed on the surface of the Plasmodium falciparum-infected erythrocyte are encoded by the rif gene family and are the target of human immune responses. J. Exp. Med. 190, 1393–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kyes S. A., Rowe J. A., Kriek N., and Newbold C. I. (1999) Rifins: a second family of clonally variant proteins expressed on the surface of red cells infected with Plasmodium falciparum. Proc. Natl. Acad. Sci. U.S.A. 96, 9333–9338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Winter G., Kawai S., Haeggstrom M., Kaneko O., von Euler A., Kawazu S., Palm D., Fernandez V., and Wahlgren M. (2005) SURFIN is a polymorphic antigen expressed on Plasmodium falciparum merozoites and infected erythrocytes. J. Exp. Med. 201, 1853–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Niang M., Yan Yam X., and Preiser P. R. (2009) The Plasmodium falciparum STEVOR multigene family mediates antigenic variation of the infected erythrocyte. PLoS Pathogens 5, e1000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Niang M., Bei A. K., Madnani K. G., Pelly S., Dankwa S., Kanjee U., Gunalan K., Amaladoss A., Yeo K. P., Bob N. S., Malleret B., Duraisingh M. T., and Preiser P. R. (2014) STEVOR is a Plasmodium falciparum erythrocyte binding protein that mediates merozoite invasion and rosetting. Cell Host Microbe 16, 81–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rowe J. A., Claessens A., Corrigan R. A., and Arman M. (2009) Adhesion of Plasmodium falciparum-infected erythrocytes to human cells: molecular mechanisms and therapeutic implications. Expert Rev. Mol. Med. 11, e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chan J. A., Fowkes F. J., and Beeson J. G. (2014) Surface antigens of Plasmodium falciparum-infected erythrocytes as immune targets and malaria vaccine candidates. Cell Mol. Life Sci. 71, 3633–3657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Petter M., Haeggstrom M., Khattab A., Fernandez V., Klinkert M. Q., and Wahlgren M. (2007) Variant proteins of the Plasmodium falciparum RIFIN family show distinct subcellular localization and developmental expression patterns. Mol. Biochem. Parasitol. 156, 51–61 [DOI] [PubMed] [Google Scholar]
- 14. Joannin N., Abhiman S., Sonnhammer E. L., and Wahlgren M. (2008) Sub-grouping and sub-functionalization of the RIFIN multi-copy protein family. BMC Genomics 9, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mundwiler-Pachlatko E., and Beck H. P. (2013) Maurer's clefts, the enigma of Plasmodium falciparum. Proc. Natl. Acad. Sci. U.S.A. 110, 19987–19994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Goel S., Palmkvist M., Moll K., Joannin N., Lara P., Akhouri R. R., Moradi N., Ojemalm K., Westman M., Angeletti D., Kjellin H., Lehtio J., Blixt O., Idestrom L., Gahmberg C. G., Storry J. R., Hult A. K., Olsson M. L., von Heijne G., Nilsson I., and Wahlgren M. (2015) RIFINs are adhesins implicated in severe Plasmodium falciparum malaria. Nat. Med. 21, 314–317 [DOI] [PubMed] [Google Scholar]
- 17. Sargeant T. J., Marti M., Caler E., Carlton J. M., Simpson K., Speed T. P., and Cowman A. F. (2006) Lineage-specific expansion of proteins exported to erythrocytes in malaria parasites. Genome Biol. 7, R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hermand P., Ciceron L., Pionneau C., Vaquero C., Combadiere C., and Deterre P. (2016) Plasmodium falciparum proteins involved in cytoadherence of infected erythrocytes to chemokine CX3CL1. Sci. Reports 6, 33786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sharling L., Sowa K. M., Thompson J., Kyriacou H. M., and Arnot D. E. (2007) Rapid and specific biotin labelling of the erythrocyte surface antigens of both cultured and ex-vivo Plasmodium parasites. Malar J. 6, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Florens L., Liu X., Wang Y., Yang S., Schwartz O., Peglar M., Carucci D. J., Yates JR. 3rd, Wub Y. (2004) Proteomics approach reveals novel proteins on the surface of malaria-infected erythrocytes. Mol. Biochem. Parasitol. 135, 1–11 [DOI] [PubMed] [Google Scholar]
- 21. Vincensini L., Richert S., Blisnick T., Van Dorsselaer A., Leize-Wagner E., Rabilloud T., and Braun Breton C. (2005) Proteomic analysis identifies novel proteins of the Maurer's clefts, a secretory compartment delivering Plasmodium falciparum proteins to the surface of its host cell. Mol. Cell. Proteomics 4, 582–593 [DOI] [PubMed] [Google Scholar]
- 22. DePristo M. A., Banks E., Poplin R., Garimella K. V., Maguire J. R., Hartl C., Philippakis A. A., del Angel G., Rivas M. A., Hanna M., McKenna A., Fennell T. J., Kernytsky A. M., Sivachenko A. Y., Cibulskis K., Gabriel S. B., Altshuler D., and Daly M. J. (2011) A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Brancucci N. M., Goldowitz I., Buchholz K., Werling K., and Marti M. (2015) An assay to probe Plasmodium falciparum growth, transmission stage formation and early gametocyte development. Nat. Protocols 10, 1131–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Trager W., and Jensen J. B. (1976) Human malaria parasites in continuous culture. Science 193, 673–675 [DOI] [PubMed] [Google Scholar]
- 25. Buffet P. A., Gamain B., Scheidig C., Baruch D., Smith J. D., Hernandez-Rivas R., Pouvelle B., Oishi S., Fujii N., Fusai T., Parzy D., Miller L. H., Gysin J., and Scherf A. (1999) Plasmodium falciparum domain mediating adhesion to chondroitin sulfate A: a receptor for human placental infection. Proc. Natl. Acad. Sci. U.S.A. 96, 12743–12748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Boddey J. A., Hodder A. N., Gunther S., Gilson P. R., Patsiouras H., Kapp E. A., Pearce J. A., de Koning-Ward T. F., Simpson R. J., Crabb B. S., and Cowman A. F. (2010) An aspartyl protease directs malaria effector proteins to the host cell. Nature 463, 627–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Park D. J., Lukens A. K., Neafsey D. E., Schaffner S. F., Chang H. H., Valim C., Ribacke U., Van Tyne D., Galinsky K., Galligan M., Becker J. S., Ndiaye D., Mboup S., Wiegand R. C., Hartl D. L., Sabeti P. C., Wirth D. F., and Volkman S. K. (2012) Sequence-based association and selection scans identify drug resistance loci in the Plasmodium falciparum malaria parasite. Proc. Natl. Acad. Sci. U.S.A. 109, 13052–13057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lohse M., Bolger A. M., Nagel A., Fernie A. R., Lunn J. E., Stitt M., and Usadel B. (2012) RobiNA: a user-friendly, integrated software solution for RNA-Seq-based transcriptomics. Nucleic Acids Res. 40, W622–W627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., and Salzberg S. L. (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Trapnell C., Williams B. A., Pertea G., Mortazavi A., Kwan G., van Baren M. J., Salzberg S. L., Wold B. J., and Pachter L. (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Trapnell C., Hendrickson D. G., Sauvageau M., Goff L., Rinn J. L., and Pachter L. (2013) Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31, 46–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Avril M., Cartwright M. M., Hathaway M. J., and Smith J. D. (2011) Induction of strain-transcendent antibodies to placental-type isolates with VAR2CSA DBL3 or DBL5 recombinant proteins. Malar J. 10, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hommel M., Elliott S. R., Soma V., Kelly G., Fowkes F. J., Chesson J. M., Duffy M. F., Bockhorst J., Avril M., Mueller I., Raiko A., Stanisic D. I., Rogerson S. J., Smith J. D., and Beeson J. G. (2010) Evaluation of the antigenic diversity of placenta-binding Plasmodium falciparum variants and the antibody repertoire among pregnant women. Infection Immunity 78, 1963–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Salanti A., Dahlback M., Turner L., Nielsen M. A., Barfod L., Magistrado P., Jensen A. T., Lavstsen T., Ofori M. F., Marsh K., Hviid L., and Theander T. G. (2004) Evidence for the involvement of VAR2CSA in pregnancy-associated malaria. J. Exp. Med. 200, 1197–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dreisbach A., van der Kooi-Pol M. M., Otto A., Gronau K., Bonarius H. P., Westra H., Groen H., Becher D., Hecker M., and van Dijl J. M. (2011) Surface shaving as a versatile tool to profile global interactions between human serum proteins and the Staphylococcus aureus cell surface. Proteomics 11, 2921–2930 [DOI] [PubMed] [Google Scholar]
- 36. Rodriguez-Ortega M. J., Norais N., Bensi G., Liberatori S., Capo S., Mora M., Scarselli M., Doro F., Ferrari G., Garaguso I., Maggi T., Neumann A., Covre A., Telford J. L., and Grandi G. (2006) Characterization and identification of vaccine candidate proteins through analysis of the group A Streptococcus surface proteome. Nat. Biotechnol. 24, 191–197 [DOI] [PubMed] [Google Scholar]
- 37. Zijlstra W. G., Buursma A., and Meeuwsen-van der Roest W. P. (1991) Absorption spectra of human fetal and adult oxyhemoglobin, de-oxyhemoglobin, carboxyhemoglobin, and methemoglobin. Clin. Chem. 37, 1633–1638 [PubMed] [Google Scholar]
- 38. Melcher M., Muhle R. A., Henrich P. P., Kraemer S. M., Avril M., Vigan-Womas I., Mercereau-Puijalon O., Smith J. D., and Fidock D. A. (2010) Identification of a role for the PfEMP1 semi-conserved head structure in protein trafficking to the surface of Plasmodium falciparum infected red blood cells. Cell. Microbiol. 12, 1446–1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rug M., Prescott S. W., Fernandez K. M., Cooke B. M., and Cowman A. F. (2006) The role of KAHRP domains in knob formation and cytoadherence of P falciparum-infected human erythrocytes. Blood 108, 370–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Russo I., Babbitt S., Muralidharan V., Butler T., Oksman A., and Goldberg D. E. (2010) Plasmepsin V licenses Plasmodium proteins for export into the host erythrocyte. Nature 463, 632–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hiller N. L., Bhattacharjee S., van Ooij C., Liolios K., Harrison T., Lopez-Estrano C., and Haldar K. (2004) A host-targeting signal in virulence proteins reveals a secretome in malarial infection. Science 306, 1934–1937 [DOI] [PubMed] [Google Scholar]
- 42. Marti M., Good R. T., Rug M., Knuepfer E., and Cowman A. F. (2004) Targeting malaria virulence and remodeling proteins to the host erythrocyte. Science 306, 1930–1933 [DOI] [PubMed] [Google Scholar]
- 43. Blisnick T., Morales Betoulle M. E., Barale J. C., Uzureau P., Berry L., Desroses S., Fujioka H., Mattei D., and Braun Breton C. (2000) Pfsbp1, a Maurer's cleft Plasmodium falciparum protein, is associated with the erythrocyte skeleton. Mol. Biochem. Parasitol. 111, 107–121 [DOI] [PubMed] [Google Scholar]
- 44. Spycher C., Rug M., Klonis N., Ferguson D. J., Cowman A. F., Beck H. P., and Tilley L. (2006) Genesis of and trafficking to the Maurer's clefts of Plasmodium falciparum-infected erythrocytes. Mol. Cell. Biol. 26, 4074–4085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chookajorn T., and Hartl D. L. (2006) Position-specific polymorphism of Plasmodium falciparum Stuttering motif in a PHISTc PFI1780w. Exp. Parasitol. 114, 126–128 [DOI] [PubMed] [Google Scholar]
- 46. Oberli A., Slater L. M., Cutts E., Brand F., Mundwiler-Pachlatko E., Rusch S., Masik M. F., Erat M. C., Beck H. P., and Vakonakis I. (2014) A Plasmodium falciparum PHIST protein binds the virulence factor PfEMP1 and comigrates to knobs on the host cell surface. FASEB J. 28, 4420–4433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rauniyar N., and Yates J. R. 3rd. (2014) Isobaric labeling-based relative quantification in shotgun proteomics. J. Proteome Res. 13, 5293–5309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chang H. H., Falick A. M., Carlton P. M., Sedat J. W., DeRisi J. L., and Marletta M. A. (2008) N-terminal processing of proteins exported by malaria parasites. Mol. Biochem. Parasitol. 160, 107–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Silvestrini F., Lasonder E., Olivieri A., Camarda G., van Schaijk B., Sanchez M., Younis Younis S., Sauerwein R., and Alano P. (2010) Protein export marks the early phase of gametocytogenesis of the human malaria parasite Plasmodium falciparum. Mol. Cell. Proteomics 9, 1437–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Volkman S. K., Hartl D. L., Wirth D. F., Nielsen K. M., Choi M., Batalov S., Zhou Y., Plouffe D., Le Roch K. G., Abagyan R., and Winzeler E. A. (2002) Excess polymorphisms in genes for membrane proteins in Plasmodium falciparum. Science 298, 216–218 [DOI] [PubMed] [Google Scholar]
- 51. Fried M., and Duffy P. E. (2015) Designing a VAR2CSA-based vaccine to prevent placental malaria. Vaccine 33, 7483–7488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wright G. J., and Rayner J. C. (2014) Plasmodium falciparum erythrocyte invasion: combining function with immune evasion. PLoS Pathogens 10, e1003943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Fonager J., Pasini E. M., Braks J. A., Klop O., Ramesar J., Remarque E. J., Vroegrijk I. O., van Duinen S. G., Thomas A. W., Khan S. M., Mann M., Kocken C. H., Janse C. J., and Franke-Fayard B. M. (2012) Reduced CD36-dependent tissue sequestration of Plasmodium-infected erythrocytes is detrimental to malaria parasite growth in vivo. J. Exp. Med. 209, 93–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Olaya-Abril A., Jimenez-Munguia I., Gomez-Gascon L., and Rodriguez-Ortega M. J. (2014) Surfomics: shaving live organisms for a fast proteomic identification of surface proteins. J. Proteomics 97, 164–176 [DOI] [PubMed] [Google Scholar]
- 55. Solis N., Larsen M. R., and Cordwell S. J. (2010) Improved accuracy of cell surface shaving proteomics in Staphylococcus aureus using a false-positive control. Proteomics 10, 2037–2049 [DOI] [PubMed] [Google Scholar]
- 56. Spielmann T., Hawthorne P. L., Dixon M. W., Hannemann M., Klotz K., Kemp D. J., Klonis N., Tilley L., Trenholme K. R., and Gardiner D. L. (2006) A cluster of ring stage-specific genes linked to a locus implicated in cytoadherence in Plasmodium falciparum codes for PEXEL-negative and PEXEL-positive proteins exported into the host cell. Mol. Biol. Cell 17, 3613–3624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Spycher C., Rug M., Pachlatko E., Hanssen E., Ferguson D., Cowman A. F., Tilley L., and Beck H. P. (2008) The Maurer's cleft protein MAHRP1 is essential for trafficking of PfEMP1 to the surface of Plasmodium falciparum-infected erythrocytes. Mol. Microbiol. 68, 1300–1314 [DOI] [PubMed] [Google Scholar]
- 58. Cooke B. M., Buckingham D. W., Glenister F. K., Fernandez K. M., Bannister L. H., Marti M., Mohandas N., and Coppel R. L. (2006) A Maurer's cleft-associated protein is essential for expression of the major malaria virulence antigen on the surface of infected red blood cells. J. Cell Biol. 172, 899–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kriek N., Tilley L., Horrocks P., Pinches R., Elford B. C., Ferguson D. J., Lingelbach K., and Newbold C. I. (2003) Characterization of the pathway for transport of the cytoadherence-mediating protein, PfEMP1, to the host cell surface in malaria parasite-infected erythrocytes. Mol. Microbiol. 50, 1215–1227 [DOI] [PubMed] [Google Scholar]
- 60. Dent A. E., Nakajima R., Liang L., Baum E., Moormann A. M., Sumba P. O., Vulule J., Babineau D., Randall A., Davies D. H., Felgner P. L., and Kazura J. W. (2015) Plasmodium falciparum protein microarray antibody profiles correlate with protection from symptomatic malaria in Kenya. J. Infect. Dis. 212, 1429–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gwadz R. W., Cochrane A. H., Nussenzweig V., and Nussenzweig R. S. (1979) Preliminary studies on vaccination of rhesus monkeys with irradiated sporozoites of Plasmodium knowlesi and characterization of surface antigens of these parasites. Bull. World Health Organ. 57, 165–173 [PMC free article] [PubMed] [Google Scholar]
- 62. Saul A., Maloy W. L., Rock E. P., and Howard R. J. (1988) A portion of the Pf155/RESA antigen of Plasmodium falciparum is accessible on the surface of infected erythrocytes. Immunol. Cell Biol. 66 (Pt 4), 269–276 [DOI] [PubMed] [Google Scholar]
- 63. Cochrane A. H., Santoro F., Nussenzweig V., Gwadz R. W., and Nussenzweig R. S. (1982) Monoclonal antibodies identify the protective antigens of sporozoites of Plasmodium knowlesi. Proc. Natl. Acad. Sci. U.S.A. 79, 5651–5655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Richards J. S., and Beeson J. G. (2009) The future for blood-stage vaccines against malaria. Immunol. Cell Biol. 87, 377–390 [DOI] [PubMed] [Google Scholar]
- 65. Kemp D. J., Coppel R. L., Cowman A. F., Saint R. B., Brown G. V., and Anders R. F. (1983) Expression of Plasmodium falciparum blood-stage antigens in Escherichia coli: detection with antibodies from immune humans. Proc. Natl. Acad. Sci. U.S.A. 80, 3787–3791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Coppel R. L., Cowman A. F., Anders R. F., Bianco A. E., Saint R. B., Lingelbach K. R., Kemp D. J., and Brown G. V. (1984) Immune sera recognize on erythrocytes Plasmodium falciparum antigen composed of repeated amino acid sequences. Nature 310, 789–792 [DOI] [PubMed] [Google Scholar]
- 67. Aide P., Aponte J. J., Renom M., Nhampossa T., Sacarlal J., Mandomando I., Bassat Q., Manaca M. N., Leach A., Lievens M., Vekemans J., Dubois M. C., Loucq C., Ballou W. R., Cohen J., and Alonso P. L.. Safety, immunogenicity and duration of protection of the RTS, S/AS02(D) malaria vaccine: one year follow-up of a randomized controlled phase I/IIb trial. PloS One 5, e13838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Thera M. A., Doumbo O. K., Coulibaly D., Laurens M. B., Ouattara A., Kone A. K., Guindo A. B., Traore K., Traore I., Kouriba B., Diallo D. A., Diarra I., Daou M., Dolo A., Tolo Y., Sissoko M. S., Niangaly A., Sissoko M., Takala-Harrison S., Lyke K. E., Wu Y., Blackwelder W. C., Godeaux O., Vekemans J., Dubois M. C., Ballou W. R., Cohen J., Thompson D., Dube T., Soisson L., Diggs C. L., House B., Lanar D. E., Dutta S., Heppner D. G. Jr., and Plowe C. V.. A field trial to assess a blood-stage malaria vaccine. N Engl. J. Med. 365, 1004–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Genton B., Al-Yaman F., Betuela I., Anders R. F., Saul A., Baea K., Mellombo M., Taraika J., Brown G. V., Pye D., Irving D. O., Felger I., Beck H. P., Smith T. A., and Alpers M. P. (2003) Safety and immunogenicity of a three-component blood-stage malaria vaccine (MSP1, MSP2, RESA) against Plasmodium falciparum in Papua New Guinean children. Vaccine 22, 30–41 [DOI] [PubMed] [Google Scholar]
- 70. Roussilhon C., Oeuvray C., Muller-Graf C., Tall A., Rogier C., Trape J. F., Theisen M., Balde A., Perignon J. L., and Druilhe P. (2007) Long-term clinical protection from falciparum malaria is strongly associated with IgG3 antibodies to merozoite surface protein 3. PLoS Med. 4, e320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Volkman S. K., H. D., Wirth D. F., Nielsen K. M., Choi M., Batalov S., Zhou Y., Plouffe D., Le Roch K. G., Abagyan R., and Winzeler E. A. (2002) Excess polymorphisms in genes for membrane proteins in Plasmodium falciparum. Science 298, 216–218 [DOI] [PubMed] [Google Scholar]
- 72. Volkman S. K., Sabeti P. C., DeCaprio D., Neafsey D. E., Schaffner S. F., Milner D. A., Daily J. P., Sarr O., Ndiaye D., Ndir O., Mboup S., Duraisingh M. T., Lukens A., Derr A., Stange-Thomann N., Waggoner S., Onofrio R., Ziaugra L., Mauceli E., Gnerre S., Jaffe D. B., Zainoun J., Wiegand R. C., Birren B. W., Hartl D. L., Galagan J. E., Lander E. S., and Wirth D. F. (2007) A genome-wide map of diversity in Plasmodium falciparum. Nat. Genet. 39, 113–119 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original mass spectra have been deposited in the public proteomics repository MassIVE and are accessible at ftp://massive.ucsd.edu/MSV000081541. Annotated MS/MS spectra for all peptides with N-terminal PEXEL cleavage sites and spectra corresponding to proteins to be identified by a single peptide are available at http://msviewer.ucsf.edu/prospector/cgi-bin/msform.cgi?form=msviewer. Use MS-Viewer search key: jed26hegyp (3D7 search results) and d9d1qtpg0f (Pf2004 search results).