Abstract
Normal pancreatic islet β-cells (PBCs) abundantly secrete insulin in response to elevated blood glucose levels, in order to maintain an adequate control of energy balance and glucose homeostasis. However, the molecular mechanisms underlying the insulin secretion are unclear. Improving our understanding of glucose-stimulated insulin secretion (GSIS) mechanisms under normal conditions is a prerequisite for developing better interventions against diabetes. Here, we aimed at identifying novel signaling pathways involved in the initial release of insulin from PBCs after glucose stimulation using quantitative strategies for the assessment of phosphorylated proteins and sialylated N-linked (SA) glycoproteins.
Islets of Langerhans derived from newborn rats with a subsequent 9–10 days of maturation in vitro were stimulated with 20 mm glucose for 0 min (control), 5 min, 10 min, and 15 min. The isolated islets were subjected to time-resolved quantitative phosphoproteomics and sialiomics using iTRAQ-labeling combined with enrichment of phosphorylated peptides and formerly SA glycopeptides and high-accuracy LC-MS/MS. Using bioinformatics we analyzed the functional signaling pathways during GSIS, including well-known insulin secretion pathways. Furthermore, we identified six novel activated signaling pathways (e.g. agrin interactions and prolactin signaling) at 15 min GSIS, which may increase our understanding of the molecular mechanism underlying GSIS. Moreover, we validated some of the regulated phosphosites by parallel reaction monitoring, which resulted in the validation of eleven new phosphosites significantly regulated on GSIS. Besides protein phosphorylation, alteration in SA glycosylation was observed on several surface proteins on brief GSIS. Interestingly, proteins important for cell-cell interaction, cell movement, cell-ECM interaction and Focal Adhesion (e.g. integrins, semaphorins, and plexins) were found regulated at the level of sialylation, but not in protein expression. Collectively, we believe that this comprehensive Proteomics and PTMomics survey of signaling pathways taking place during brief GSIS of primary PBCs is contributing to understanding the complex signaling underlying GSIS.
The prevalence of hyperglycemia is steadily increased over the last decades and globally developed along with rising availability of high-energy foods (1, 2). The progression of glucose toxicity leads to all symptoms of cardiovascular disease, coronary heart disease, stroke, obesity, and diabetes mellitus, however the underlying pathophysiological- and molecular mechanisms in the pancreatic endocrine cells are poorly understood (3, 4). Pancreatic islet β-cells (PBCs)1 are among the most important endocrine cells, constituting around ∼80% of the cell type percentage within the rodent islets of Langerhans. The physiological function of PBCs is to sense the blood glucose concentration and subsequently regulate the insulin secretory response to maintain an adequate control of energy balance and glucose homeostasis in the body (5). The presence of high blood glucose markedly increases insulin release from PBCs through biphasic insulin granule biogenesis, trafficking, and exocytosis in normal PBCs through a variety of cellular signaling mechanisms (5, 6). However, chronic hyperglycemia causes deterioration of PBCs functions resulting in profound dysregulation of the insulin secretion processes and impairment in PBCs sensitivity to glucose (7, 8). Although substantial studies have showed epidemic and physiological evidence for the glucose-stimulated insulin secretion (GSIS) process in PBCs, the molecular mechanism underlying GSIS and hyperglycemia regulated insulin action is still unclear. The common GSIS model of action (9), involving glucose metabolism, closure of ATP sensitive potassium channels leading to depolarization of the plasma membrane and subsequent opening of Calcium (Ca2+) channels, Ca2+ influx and activation of exocytotic pathways leading to emptying of insulin granules, have been challenged as incomplete (10). Contributions from other ion channels to the depolarization processes identified through PBCs lacking ATP sensitive potassium channels open for novel molecular mechanisms underlying GSIS.
Conversely, improving our understanding of high glucose-regulated signaling pathways underlying insulin sensitivity/secretion can contribute to the restoration of PBCs function by functional activation or inhibition of potential therapeutic targets. For example, GLP-1-based therapies are recommended to control the level of blood glucose through enhancing insulin secretory response signaling in patients with type 2 diabetes (T2D) (11–13).
Despite the enormous effort put into β-cell biology to understanding the mechanisms of the dynamic time-dependent GSIS and subsequent insulin release from PBCs, only the overall signaling pathways have been characterized. One of the major problems in characterizing GSIS is the multitude of complex signaling pathways, which participate in such a highly controlled process as release of insulin, as most of the body is relying on this molecule for proper function. Many of the signaling pathways are highly controlled and regulated by dynamic, reversible post-translational modifications (PTMs), such as protein phosphorylation, which are notoriously difficult to molecularly characterize. Nonetheless, recent advances in large-scale proteomics technologies have unraveled novel signaling pathways coupled with quantities of PTMs of the islets cell systems. Although previous large-scale studies showed the quantitative analysis of the proteome or phosphoproteome in the rat insulinoma β-cell line (INS1-E) or islets responding to glucose (14–16), there is still a lack of experimental evidence on changes in phosphosites related to transient exposure of primary PBCs to glucose. In addition, little is presented to elucidate the dynamic relationships of cellular signaling mechanisms between the temporal regulation of protein expression and PTMs such as phosphorylation and especially glycosylation.
In the present study, we investigated signaling pathways associated with GSIS in PBCs derived from isolated rat islets of Langerhans. PTMomics can be defined as the global study of PTMs and is a relatively new research area aiming at understanding the function and consequences of PTMs of proteins in a cell. Specifically, our aim was to reveal which phosphosites and N-linked sialylated glycosylation sites that were reversible regulated on brief glucose stimulation and link these to cellular signaling pathways using bioinformatics tools. We have recently developed a novel quantitative proteomics/PTMomics strategy targeting both the proteome and selected PTMs such as phosphorylation, and N-linked sialylation (17–20). We applied a modified TiSH protocol (19, 21), to enrich and fractionate N-linked sialylated glycopeptides, phosphopeptides, and unmodified peptides from PBCs derived from isolated islets of newborn rats matured over 9 days (∼1500 islets per condition), with low and high glucose stimulation over an initial temporal range from 0 to 15 min. We analyzed the functional significance of signaling pathways during the insulin release β-cells and found that the molecular events occurring are closely associated with known insulin signaling and calcium signaling pathway. We also identified new interactions network in various signaling pathways and biological context among regulated proteins and PTMs that link extracellular integrin signaling with the nuclear lumen including nuclear pore complex proteins. Especially the regulation of protein sialylation is completely novel in relation to brief GSIS and could relate to nonconventional GSIS signaling mechanisms. We highlight activated or inhibited cellular signaling pathways, based on regulated proteins/phosphopeptides/sialylated glycopeptides, which were of high relevance to increase our understanding of GSIS functions. In addition, we performed Parallel Reaction Monitoring (PRM) using heavy labeled synthetic phosphopeptides to validate the regulation of a subset of phosphopeptides and found that the validation correlated very well with the initial iTRAQ quantitative analysis, verifying the quality of the initial analysis. Moreover, we confirmed new regulators on compensatory response to high glucose stimulation compared with control using PRM and found that eleven previous unknown phosphosites (Psen1;S368, Fam135a;S40, Arfgef3;S348 and S1829, Atp2b1;S1216, Arhgef11;S51, Sphkap;S1167, R3hdm2;S872, RGD1562037;S930, C17orf59;S43, and Dennd4c;S935) were significantly phosphorylated/dephosphorylated at 10 min GSIS. Taken together, we believe this comprehensive Proteomics and PTMomics analysis will be useful to help further decipher the molecular networks underlying the temporal regulation of GSIS.
EXPERIMENTAL PROCEDURES
Isolation and Culture of Rat Islets of Langerhans
Neonatal rat pancreatic islets were isolated from 4 to 5-days-old Wistar rat pups (Taconic, NY) essentially as previously described (22). Pups were killed by decapitation and pancreases were dissected and placed in ice-cold serum-free Hanks balanced salt solution (HBSS; Lonza, Switzerland). The isolated Pancreas were distributed into tubes containing collagenase (Roche; final concentration 0.44 mg/ml), and partially dissociated by manually shaking the tubes for 8 min. Subsequently, islets were washed 4 times with ice-cold HBSS containing 10% newborn calf serum (NCS; Biological industries) and isolated using Histopaque (Sigma; 1.077 g/ml density) gradients for 20 min at 300 g without brake. Islets were collected from the interphase, washed 3 times with ice-cold HBSS/10% NCS and handpicked under a stereo microscope (Leica). The isolated islets were cultured for 9–10 days in vitro (DIV) in RPMI 1640 with glutamax and supplemented with 10% NCS, 100 U/ml penicillin and 100 μg/ml streptomycin (Gibco Thermo Fisher Scientific, MA) in 100 mm Petri dishes (Nunc; approx. 500 islets/dish) at 5% CO2 and 37 °C. The medium was changed the day after culture set-up and after 7 DIV.
Glucose-stimulated Insulin Secretion, Islet Collection, and Insulin ELISA
After 9–10 DIV islets were stimulated with glucose for 0 min (control), 5 min, 10 min, or 15 min in Krebs-Ringer HEPES buffer (KRHB; 115 mm NaCl, 4.7 mm KCl, 2.6 mm CaCl2, 1.2 mm KH2PO4, 1.2 mm MgSO4, 20 mm HEPES, 2 mm glutamine, 5 mm NaHCO3, 1% (v/v) PenStrep and 0.2% (w/v) BSA; pH 7.4). Glucose stimulation was performed in 100 mm Petri dishes for islets for the proteomics experiment whereas islets used to measure insulin secretion were stimulated in 24-well plates (Nunc, Thermo Fisher Scientific, MA) containing 20 islets/well. Before the glucose stimulation, islets were incubated in KRHB containing 0.5 mm glucose (low glucose) for 90 min at 5% CO2 and 37 °C. Subsequently, the islets were stimulated with fresh KRHB (low glucose) or 20 mm glucose for 5, 10, or 15 min and islets incubated for 5 min in KRHB containing 0.5 mm glucose served as controls. In the 24-well plates KRHB was collected before and after glucose stimulation for insulin measurement. For the proteomics part KRHB was removed and islets collected in ice-cold PBS, centrifuged 1 min at 300 × g and washed once in PBS followed by snap-freezing in liquid nitrogen. Approximately 1500 islets were pooled per condition in low binding safeseal microcentrifuge tubes (Sorenson Bioscience Inc., UT) to minimize loss of modified peptides binding to generic plastic tubes in the following digestion. Islets were stored at −80 °C until further processing. Insulin levels in KRHB before and after glucose stimulation were determined by ELISA (Mercodia, Uppsala, Sweden) using rat insulin as standard. The ELISA was performed according to the manufacturer's instructions.
Cell lysis, Protein Purification, and Digestion
Islets for the proteomics workflow were lysed, reduced, and predigested in 6 m Urea, 2 m Thiourea, containing 10 mm DTT (all Sigma, Germany) and 2 μl Lys-C (Wako, VA) supplemented with PhosSTOP phosphatase inhibitor (Roche) for 2 h at RT. Thereafter, the lysates were diluted 10 times using 20 mm Triethylammonium bicarbonate buffer (TEAB; pH adjusted to 7.5) and tip-sonicated for 2 × 20 s on ice. Then, samples were alkylated by 20 mm iodacetamide (IAA) for 20 min in the dark before digestion with 2% (w/w) trypsin (Promega, WI) overnight (ON) at 37 °C.
Quantitative Proteomics Using Peptide Labeling Approach
Peptide concentration was measured by Qubit® Fluorometric quantification (Thermo Scientific) according to the manufacturer's instructions. The total amount of protein for 1500 islets was ∼135 μg in each group. A total of 120 μg was aliquoted from all samples and lyophilized before labeling with iTRAQ 4plex (AB Sciex). Two biological replicates were made and labeling was performed as follows: control 114, 5 min glucose 115, 10 min glucose 116, and 15 min glucose 117. The labeling was performed according to the manufacturer's protocol and complete labeling was validated by running combined aliquots on MALDI MS (Bruker Daltonics, Germany). The samples were mixed in a 1:1:1:1 ratio and stored at −20 °C until PTM enrichment.
Enrichment of phosphorylated peptides and sialylated (SA) N-linked glycopeptides
The purification of phosphopeptides and SA N-linked glycopeptides was performed according to a slightly modified TiSH protocol (21, 23), in which nonmodified peptides are first separated from these two modified peptide species using TiO2. Hereafter the multi- and monophosphorylated peptides are separated from the formerly SA N-linked glycopeptides after a deglycosylation step, using the Sequential elution from IMAC (SIMAC) procedure. Briefly, the lyophilized iTRAQ labeled sample was made up to 1 ml loading buffer (1 m glycolic acid, 80% ACN, 5% TFA) and added with TiO2 beads (GL Sciences, Japan) at 0.6 mg/100 μg (bead/peptide), and incubated at RT for 10 min. The suspension was centrifuged for 15 s in a table centrifuge and the supernatant loaded onto a second batch of TiO2 (containing half the amount of TiO2 as initially used) and incubated at RT for 15 mins. The two batches of TiO2 were washed with 100 μl of washing buffer 1 (80% ACN, 1% TFA) and centrifuged for 15 s in a table centrifuge. The supernatant was removed, and the beads were washed with 100 μl washing buffer 2 (10% ACN, 0.1% TFA) and centrifuged for 15 s in a table centrifuge. The supernatant was removed, and the beads were dried in a vacuum centrifuge for 5 min. The bound peptides were eluted with 100 μl of 1% ammonium hydroxide for 15 min and then centrifuged at 1000 g for 1 min. The eluted peptides were passed over a C8 stage tip (3 mTM EmporeTM Bioanalytical Technologies, SigmaAldrich, MO) (24) to retain the TiO2 beads and dried by vacuum centrifugation to produce the enriched phosphopeptide/sialylated glycopeptide fraction. The flow through from the initial loading buffer (containing nonmodified peptides) and washes were combined and dried by vacuum centrifugation to produce the nonmodified peptide fraction. The nonmodified peptide fraction was acidified with TFA and desalted on a R3 stage tip column before HILIC fractionation.
Deglycosylation
To remove the glycan structures from the enriched SA N-linked glycopeptides the sample was resuspended in 20 mm TEAB, pH 8.0 and treated with 2 μl N-glycosidase F (New England Biolabs) and 0.5 μl Sialidase A (ProZyme, CA) ON at 37 °C (25).
SIMAC
The deglycosylated samples were resuspended in SIMAC loading buffer (50% ACN and 0.1% TFA) and pH was adjusted with 10% TFA within an interval of 1.6–1.8. PhosSelect IMAC beads (Sigma Aldrich) were equilibrated with loading buffer and combined with the samples and incubated for 1 h under gentle rotation. After incubation, the samples were centrifuged and the supernatant recovered. The beads were transferred to a constricted GELoader tip (Eppendorf® AG, Germany) and the column was washed with loading buffer, collecting the FT in the same tube as the previous supernatant. Mono-phosphorylated and formerly SA N-linked glycopeptides were eluted from the column with an acidic solution (20% ACN, 1% TFA). The multi-phosphorylated peptides were subsequently eluted from the column with a basic solution (1% ammonium hydroxide solution, pH 11.3). The basic elution containing multiphosphopeptides was acidified before desalting on a R3 column and the eluted peptides were lyophilized before LC-MS/MS. The supernatants and acidic eluates collected after SIMAC were subjected to another round of TiO2 purification to separate mono-phosphopeptides from formerly SA N-linked glycopeptides. The samples were adjusted to 70% ACN and 2% TFA and the TiO2 beads were added to the solution and incubated on a shaker as described above. The incubation round was performed twice with half amount of beads the second time. The beads were pelleted and the supernatant was recovered (containing formerly N-linked glycopeptides). The two pellets of TiO2 were pooled with 50% ACN, 0.1% TFA, the beads were pelleted and the supernatant was recovered. The beads were dried and bound phosphopeptides were eluted from the beads with 1% ammonium hydroxide solution, pH 11.3. The phosphopeptide elution was acidified with 100% formic acid and 10% TFA and desalted using a R3 column before HILIC fractionation. The formerly SA N-linked glycopeptides were lyophilized before desalting.
Sample Desalting
Samples were desalted before HILIC fractionation or direct LC-MS/MS analysis (multi-phosphopeptides). The desalting columns were self-made by inserting a small plug of C18 (3 mTM EmporeTM Bioanalytical Technologies) material into the constricted end of a 200 μl tip and packed with Poros R3 or a mixture of R2 and R3 reversed-phase resin (Applied Biosystems) applying manual air pressure with a syringe, followed by an optimized desalting procedure (26). Briefly, the samples were acidified before loading onto the columns (equilibrated with 0.1% TFA), followed by washing with 0.1% TFA, and peptides were eluted using 60% ACN, 0.1% TFA and were lyophilized before further processing.
Hydrophilic Interaction Liquid Chromatography (HILIC)
The mono-phosphorylated, deglycosylated and the nonmodified peptide samples were subjected to fractionation using HILIC. Briefly, these samples were resuspended in 90% ACN, 0.1% TFA (Solvent B) and loaded onto a 450 μm OD x 320 μm ID x 17 cm micro-capillary column packed with TSK Amide-80 resin material (Tosoh Bioscience, PA) using an Agilent 1200 Series HPLC (Agilent, CA). Peptides were separated using a gradient from 100–60% Solvent B (Solvent A: 0.1% TFA) running for 30 min at a flow-rate of 6 μl/min. Fractions were collected every 1 min and combined into 12–15 final fractions based on the UV chromatogram and subsequently dried by vacuum centrifugation.
Reversed-phase nanoLC-ESI-MS/MS
The samples were resuspended in 0.1% formic acid (FA) and loaded onto an EASY-nLC system (Thermo Scientific). The samples were loaded onto a two-column system containing a 3 cm pre-column and a 17 cm analytic column both consisting of fused silica capillary (75 μm inner diameter) packed with ReproSil - Pur C18 AQ 3 μm reversed-phase material (Dr. Maisch). The peptides were eluted with an organic solvent gradient from 100% phase A (0.1% FA) to 34% phase B (95% ACN, 0.1% FA) at a constant flowrate of 250 nL/min. Depending on the samples, based on the HILIC, the gradient was from 1 to 34% solvent B in 60 min or 90 min, 34% to 50% solvent B in 10min, 50–100% solvent B in 5 min and 8 min at 100% solvent B. The nLC was online connected to a Q-Exactive HF Mass Spectrometer (Thermo Fisher Scientific) operated at positive ion mode with data-dependent acquisition. The Orbitrap acquired the full MS scan with an automatic gain control (AGC) target value of 3 × 106 ions and a maximum fill time of 100ms. Each MS scan was acquired at high-resolution (60,000 full width half maximum (FWHM)) at m/z 200 in the Orbitrap with a mass range of 400–1400 Da. The 12 most abundant peptide ions were selected from the MS for higher energy collision-induced dissociation (HCD) fragmentation (collision energy: 32V). Fragmentation was performed at high resolution (30,000 FWHM) for a target of 1 × 105 and a maximum injection time of 60 ms using an isolation window of 1.2 m/z and a dynamic exclusion. All raw data were viewed in Xcalibur v3.0 (Thermo Fisher Scientific).
MS Data Processing and Statistical Analysis
The raw MS data sets were processed for protein/peptide identification using the MSGF+ (v9979, 07/16/2014) (27) combined MASIC (28) pipeline with a peptide mass tolerance of ±10 ppm, reporter ion m/z tolerance half width of 2 mDa, and a false discovery rate (FDR) of 1% for proteins and peptides. All peak lists were searched against both the UniProtKB/Swiss-Prot database (2015_08, 7928 entries) of rat sequences with decoy and mouse sequences (2013_06, 16,613 entries) with decoy using the parameters as follows: enzyme, trypsin; maximum missed cleavages, 2; fixed modification, carbamidomethylation (C), iTRAQ tags (K, peptide N termini); variable modifications, oxidation (M) and phosphorylation (S,T,Y). Data sets with raw MS values were filtered to remove potential errors using several criteria. For relative protein quantification, the output tsv file from MSGF+ combine MASIC pipeline was imported into Microsoft Excel and filtered as follows: elimination of contaminants and reversed sequences for each accession number. For sialylated peptides, the N-X-S/T consensus sequence is required to be considered a deglycopeptide. Protein/Phosphopeptide/Sialylated-glycopeptides relative expression values from the respective accession number/unique peptide were calculated by summing all reporter ion intensity of each channel and normalized to the number of total reporter ion intensity of each channel estimating the relative amounts of the different accession number within relative sample. The resulting ratios were logarithmized (base = 2) to achieve a normal distribution. Ratios were averaged and an accession number/unique peptide with modification was accepted as differentially expressed proteins/peptides with ratio values (5 min/0 min, 10 min/0 min, and 15 min/0 min) beyond p < 0.1 in normal distribution. To further assess the individual statistical significance of the expression level differentially expressed protein, phosphorylation, and sialylation that had a same direction of alteration change (positive or negative) in all experiments were selected. Fragment ion masses (b- and y-type ions) with the modification were checked at the peptide backbone in the MS/MS data sets using ScanRanker and PhosphoRS (29, 30). Clustering analysis was performed using GProX (31), using expression changes for regulated proteins/phosphopeptides or z-scores (Inhibition < −2 or 2 < Activation) for canonical pathways. Gene Ontology annotation enrichment analysis was performed using the DAVID Bioinformatics Resource (v6.8) (32). Ingenuity Pathway analysis (IPA, Qiagen, CA) was used to functionally annotate genes implicated in canonical pathways, using a Fisher's exact test with a p value threshold of 0.05.
Network Analysis for the Regulated Proteome/PTMome
The regulated proteins/phosphorylation/sialylation were searched against the STRING database version 10.0 (33) for protein-protein interactions. STRING defines a metric called “confidence score” to define interaction confidence; we selected all interactions for our regulated data set that had a confidence score ≥0.9 (the highest confidence).
Targeted Quantification Using Parallel-Reaction Monitoring (PRM) and Data Analysis
Our LC/PRM-MS assay was developed for research use, so the experimental design aligns with the goals of a Tier-2 assay (34). PRM assay was carried out using heavy isotope-labeled synthetic phosphopeptides (JPT Peptide Technologies, Germany) and a Q-Exactive™ HF mass spectrometer (Thermo Fisher Scientific). Rat pancreatic islets samples from control (no glucose stimulation) and 10 min glucose stimulation were used for validation. Heavy-labeled (K or R) phosphopeptides corresponding to endogenous phosphopeptides were spiked with peptide extracts as internal standards before performing enrichment of phosphopeptides using TiO2 as described above. The PRM method consisted of a full MS scan configured as above followed by targeted MS/MS scans as defined by a time-scheduled inclusion list. The parameters were set as follows; MS: 120K resolution, MS/MS: 15K resolution, 3e6 AGC target, 15 ms maximum injection, 1.1 m/z isolation window, and 29% normalized collision energy for fragmentation. We used Skyline (35) (version 3.5.0.9319) for quantitative data analysis. Individual endogenous peptides peak areas were extracted from the precursor ions (M and M+1), normalized using standard deviation (0 min: 0.9972 and 10 min: 1.0028) measured of summing all precursor ion areas of heavy-labeled peptides from 0 min to 10 min, and log2 transformed for comparing 0 min (control) and 10 min for relative quantification of 13 target phosphopeptides. We assessed phosphorylation localization with PhosphoRS probability (>0.99) of iTRAQ data set and required at least one diagnostic fragment ion of PRM results. We further confirmed high correlation of retention time (Pearson correlation of 0.9997) between 0 and 10 min from endogenous and heavy peptides (supplemental Table S1). The Skyline files were uploaded into Panorama Public (https://panoramaweb.org/labkey/UPTjTd.url).
Experimental Design and Statistical Rationale
In this study, we used freshly isolated islets of Langerhans from newborn rats. Islets were serum starved then stimulated with 20 mm glucose for 0 min (control), 5 min, 10 min, and 15 min. Two biological replicates were performed. All the regulated proteins/phosphopeptides/sialylated peptides (z-score for p value <0.1) correlated (Pearson correlation of 0.77 to 0.79) with time scales between the two biological replicates. The PRM assay was performed using one biological replicate consisting of pool of islets derived from 20 pubs for each condition. The PRM assay was performed using several technical analyses. We chose to perform PRM assay for novel phosphosites by analyzing the sub-data set relative from the canonical insulin secretory-associated pathways to uncover new biological insights.
RESULTS AND DISCUSSION
Temporal Quantification of the PBC Proteome, Phosphoproteome, and Sialiome
To characterize the initial mechanisms underlying glucose stimulated insulin secretion (GSIS) in PBCs, we applied a modification of our TiSH quantitative proteomics and PTMomics (nonmodification, phosphorylation, and N-linked sialylation) approach (17, 19, 36) with iTRAQ labeling using newborn rat derived islets (∼1,500 islets per condition), which had been matured for 9–10 days in vitro (DIV). We used 20 mm glucose short-term stimulation over a period from 0 to 15 min (Fig. 1A). By ELISA we confirmed significantly increased insulin release in response to high glucose stimulation for 10 and 15 min compared with controls (low glucose) (Fig. 2). We conducted a MS-GF+ database search of the biological duplicate experiment and identified a total of 8491, 4027, and 1011 proteins, and 61284, 11534, and 1942 peptides (FDR < 1%) in the nonmodified, phosphorylated, and formerly N-linked sialylated fractions, respectively. For the further analysis we considered only the peptides and proteins that were found in both biological replicates (Fig. 1B). PTM analyses require markedly more material than “normal” quantitative proteomics therefore only two biological replicates were conducted here, because of the low amount of islets obtained from each rat.
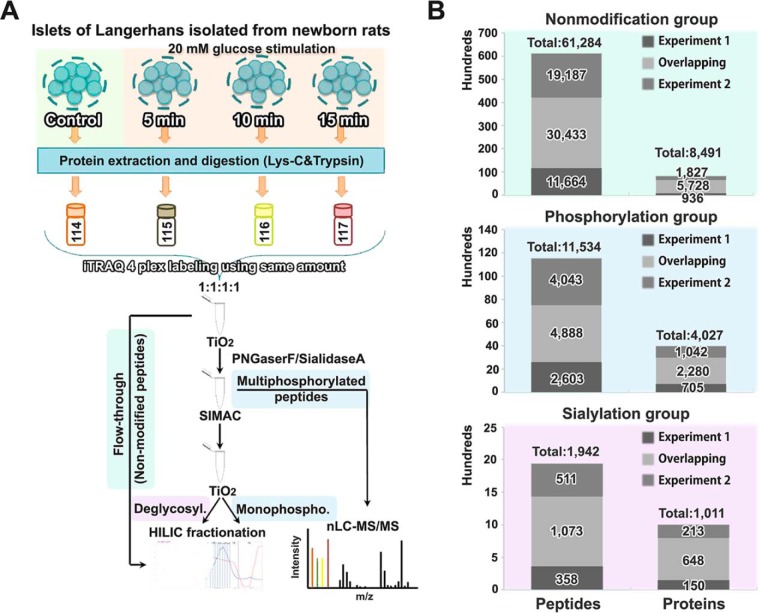
Fig. 1.
Experimental set-up and identification overview. A, Rat pancreatic islets of Langerhans were stimulated with high glucose (20 mm) for 5 min, 10 min or 15 min and nontreated islets served as control. Islets were digested using Lys-C and trypsin, and the resulting peptides were labeled with iTRAQ 4-plex and combined in a 1:1:1:1 ratio. Phosphorylated peptides and sialylated N-linked glycosylated peptides were enriched by a modified TiSH protocol, combining TiO2 with enzymatic deglycosylation, SIMAC and HILIC. Finally, bioinformatic analysis was performed. B, Summary of the numbers of identified proteins and peptides (FDR < 1%) in the nonmodified, phosphorylated, and N-linked sialylated protein/peptide group, respectively. The overlapping proteins and peptides between the two replicates were used for further analysis.
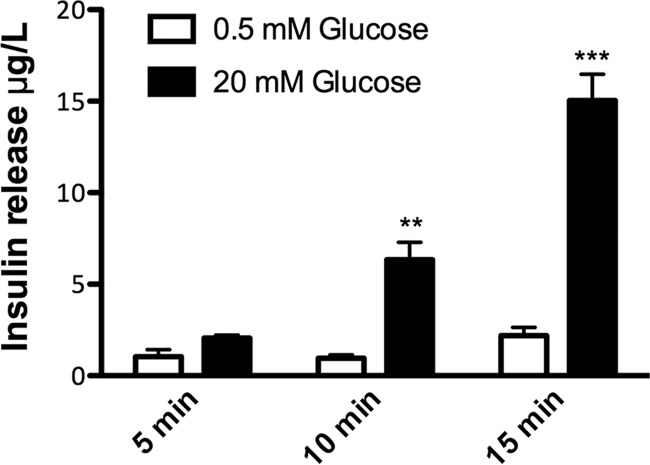
Fig. 2.
Glucose-stimulated insulin release from islets. Rat pancreatic islets grown for 9–10 days in vitro were pre-incubated with low glucose (0.5 mm) before stimulation with high glucose (20 mm) for 5 min, 10 min, or 15 min in Krebs-Ringer HEPES buffer (KRHB). Insulin levels in KRHB before and after high glucose stimulation were determined by ELISA using rat insulin as standard. Data are expressed as mean ± the standard error of the mean (S.E., n = 3–5, two independent experiments). Statistical analysis was performed with one-way ANOVA followed by Bonferroni's multiple comparison test; **p < 0.01; ***p < 0.001.
To identify the high glucose-regulated proteins, phosphorylated peptides, and formerly N-linked sialylated glycopeptides, we adopted statistical significance (p < 0.1) from z-scores with the same direction of alteration (positive or negative) in both experiments. Correlation data showed high reproducibility of the large-scale quantification between the two biological replicates with the expression changes of regulated proteins/phosphopeptides/sialylated glycopeptides (Pearson correlation of 0.77 to 0.79) (Fig. 3A). In the data from the nonmodified group, 97 proteins were differentially expressed in the time course data (Fig. 3A). In the phosphorylation group, the phosphorylation levels of 264 phosphosites were significantly altered in the temporal data responding to high glucose stimulus (Fig. 3A). In the formerly N-linked sialylation group, the sialylation level of 88 N-linked glycosites was substantially modulated in the temporal data responding to high glucose stimulation (Fig. 3A). The high glucose-regulated proteins and PTM peptides with phosphorylation or formerly N-linked sialylation are listed in supplemental Table S2. In the data set most regulation is observed at the phosphorylation level as we expected, because of the phosphorylation-dependent signaling pathways involved in glucose metabolism, calcium signaling, exocytosis, and endocytosis mechanisms observed in PBCs.
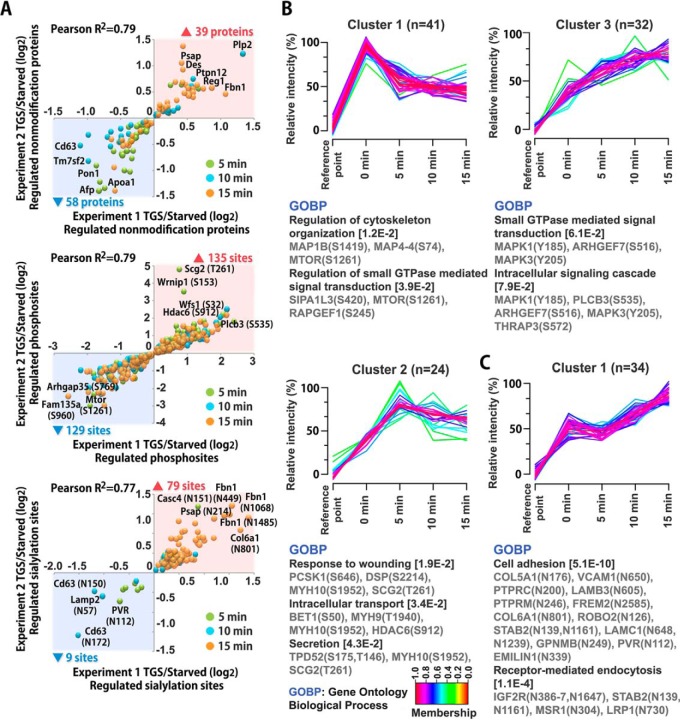
Fig. 3.
Correlation of regulated proteins/peptides and cluster analysis. A, Rat pancreatic islets were stimulated with high glucose for 5 min, 10 min or 15 min and compared with untreated controls. Scatter plots are showing correlation of all significantly regulated proteins (nonmodified), phosphopeptides and sialylated peptides from two independent experiments (Pearson correlation of 0.77 to 0.79). The numbers of up- and downregulated proteins/peptides are indicated by ▴ and ▾ respectively, and the color labels show the time point with the most pronounced regulation of the individual protein/peptide. TGS, temporal glucose stimulation; starved, no glucose stimulation. B, C, Fuzzy c-means clustering analysis showing temporal profile of phosphopeptides (B) and sialylated peptides (C) and their associated biological processes from DAVID gene ontology analysis.
Gene Ontology Analysis of Post-translationally Modified Proteins Differentially Phosphorylated or Sialylated After High Glucose Stimulation
To further characterize the temporal associated biological processes of differentially regulated phosphorylated peptides and formerly sialylated N-linked glycopeptides in an unbiased manner, we employed integration of unsupervised fuzzy c-means clustering analysis of relative intensity (%) and DAVID gene ontology analysis. In Fig. 3B, we showed three clusters for genes with regulated phosphorylation, involved in the regulation of cytoskeleton organization, regulation of small GTPase mediated signal transduction, intracellular signaling cascade/transport, and secretion. The results for modulated sialylation revealed significant differences between proteins associated with cell adhesion and receptor-mediated endocytosis (Fig. 3C).
High Glucose Stimulation Regulates the Canonical Pathways for GSIS
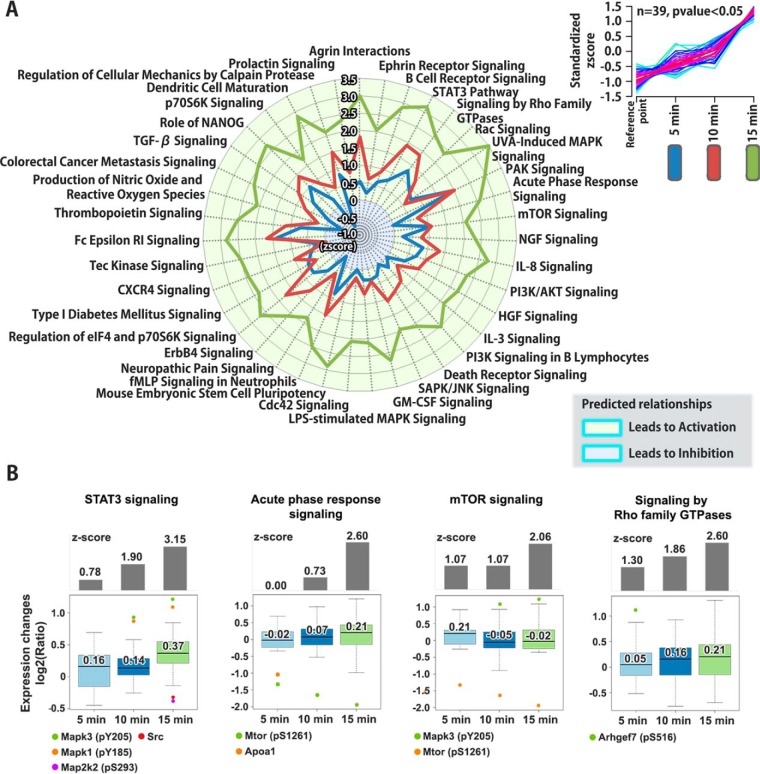
We evaluated whether known signaling pathways responsible for insulin release were associated with the molecular events occurring in our temporal large-scale data set. We analyzed the pathways most related to canonical signaling pathways responding to high glucose stimulation using statistical significance (z-score and p value < 0.05) through IPA analysis. We also predicted the dynamics of activated or inhibited canonical pathways by using significant z-scores (Inhibition < −2 or > 2 Activation) and clustering analysis. Three different clustering patterns (illustrated in the upper right corner of Fig. 4A and supplemental Fig. S1A and S2A) for the signaling pathways with activation or inhibition z-scores, were found at the three time points, 5min GSIS, 10 min GSIS and 15 min GSIS, for the nonmodification group and both PTM groups (phosphorylation and sialylation), respectively (Fig. 4A and supplemental Fig. S1A and S2A). We further examined the regulated molecules in four representative signaling pathways with outlier ratio values, based on a statistical significance of interquartile range (IQR) test (less than quartile 1 − 1.5 × IQR, or greater than quartile 3 + 1.5 × IQR) (Fig. 4B and supplemental Fig. S1B and S2B). In several outliers of the IQR test, phosphorylation of a protein was known or predicted, but no data on the precise site of modification have previously been published. In Fig. 4, four signaling pathways (STAT3 signaling, acute phase response signaling, mTOR signaling, and signaling by Rho family GTPases) showed a significant increased level (activation) at 15 min of GSIS. In STAT3 signaling, phosphosites in the mitogen-activated protein kinase 3 (Mapk3) and mitogen-activated protein kinase 1 (Mapk1) at Y205 and Y185 were upregulated, whereas dual specificity mitogen-activated protein kinase 2 (Map2k2) at S293 and proto-oncogene tyrosine-protein kinase (Src) were detected with significant downregulated phosphosites on proteins at 15 min of GSIS. Activated STAT3 phosphorylation and signaling enhanced GSIS in PBCs (37). The Rho family of small GTPase-mediated mTOR signaling is well known to be activated by glucose (38), and in the acute phase response signaling implicated in type 2 diabetes and the dysfunction of insulin secretion (39), but it is not known that acute phase response signaling in PBCs itself is critical to the GSIS process. Further, our data suggest that the activation of acute phase response signaling after 15 min of high glucose stimulation is considered to play an important role for the acute (first-phase) insulin secretory response to high glucose. In supplemental Fig. S1, four signaling pathways (Integrin signaling, phospholipase C signaling, calcium signaling, and IL-6 signaling) displayed an elevated level (activation) at 10 and 15 min of GSIS. In Integrin signaling, the phosphosites in Rho guanine nucleotide exchange factor 7 (Arhgef7), microtubule-associated serine/threonine-protein kinase 4 (Mast4), Rap guanine nucleotide exchange factor 1 (Rapgef1), Mapk3, and Mapk1 at S516, S1203, S245, Y205, and Y185, respectively, were observed. Interestingly, these signaling pathways are interrelated and closely linked to the activation of insulin secretion (40). Furthermore, cytosolic Ca2+-dependent signaling is well known as a major signal transduction pathway mediator in GSIS (41).
Fig. 4.
Mapping of affected canonical pathways by glucose stimulated insulin release from β-cells. A, Affected canonical signaling pathways in rat pancreatic islets after 5 min, 10 min or 15 min high glucose stimulation on the proteome, phosphoproteome and sialylome using Ingenuity Pathway Analysis (IPA) software are shown as a radar chart. The pathways shown in the radar chart are based on one of three dynamic pathway patterns found from the activation or inhibition z-scores. The pattern is illustrated as a cluster in the right corner (the two other patterns are found in supplemental Fig. S1 and S2). B, The regulated molecules of four representative signaling pathways were evaluated using interquartile range (IQR) test with significant z-scores (Inhibition < −2 or 2 < Activation).
Interestingly, in this study the glucose-regulated proteins were mostly downregulated after 5 min of stimulation, whereas most of the upregulated proteins were found after 10 and 15 min of glucose stimulation. Putative regulators after 5 min of high glucose stimulation might be linked with intracellular Ca2+ homeostasis, because dynamic changes in Ca2+ concentration was observed in the nucleus and the mitochondrion of rat β-cells (INS-1E) during the first 2–5 min of high glucose stimulation (42, 43). However, the roles of nuclear proteins have not yet been examined in a large-scale study. Thus, subcellular localization-dependent studies could investigate new mechanisms that participate in the function of GSIS. An alternate splicing of the protein Atp2b1 has been found, and its function is importantly implicated in the Ca2+ homeostasis as it is required for optimal cellular activity and insulin secretion (44). In our data, we found that the phosphorylation of Atp2b1 at S1216 was validated to be significantly upregulated under GSIS conditions; however, we could not distinguish the different isoforms by the proteomics data. This new data provides new clues to elucidate the Ca2+ homeostasis-mediated dynamic phosphorylation changes during GSIS processes.
In supplemental Fig. S2, four signaling pathways (Protein kinase A signaling, ERK/MAPK signaling, cAMP-mediated signaling, and CREB signaling) showed a suppressed level (inhibition) at 10 min and activation or reverting to baseline function after 15 min GSIS. In protein kinase A signaling, three phosphosites of Plcb3, Mapk3, and Mapk1 at S535, Y205, and Y185, respectively, were significantly upregulated. Notably, PKA/cAMP/ERK/MAPK/CREB signaling is the key component of GSIS and GLP-1 action (45).
Newly Identified and Highly Activated Signaling Pathways at 15 min GSIS
We also provide new findings with six proposed signaling pathways; agrin interactions, prolactin signaling, tec kinase signaling, actin nucreation by the ARP-WASP complex, MIF-mediated glucocorticoid, and ciliary neurotrophic factor (CNTF) signaling. These are mostly activated at 15 min GSIS, associated with a high degree of insulin secretion. A functional role for the six signaling pathways has not yet been elucidated in GSIS, even though they may play a role in PBCs biology and neuronal functions. For example, Bezakova et al. descripted that agrin interactions have critical roles in the organization of the post-synaptic density, the formation of the synapse, the activation of the Rho-family of small GTPases, the aggregation of lipid rafts, and the organization of the cytoskeleton (46). Furthermore, Auffret et al. demonstrated that prolactin signaling is essential for PBCs development and pancreas ontogenesis (47). Here we showed that these pathways are highly dynamic in the initial 15 min of GSIS, even with significant statistical variations at the three time-points covering the initial biphasic insulin-release. These data suggest that activation of the new potential signaling pathways may increase the insulin secretion and thus may contributes to the mechanism of GSIS. The proteins involved in significant activation/inhibition pathways are listed in supplemental Table S3. Future studies on the six new signaling pathways will more precisely uncover their roles in GSIS.
Analysis of the GSIS Interaction Network
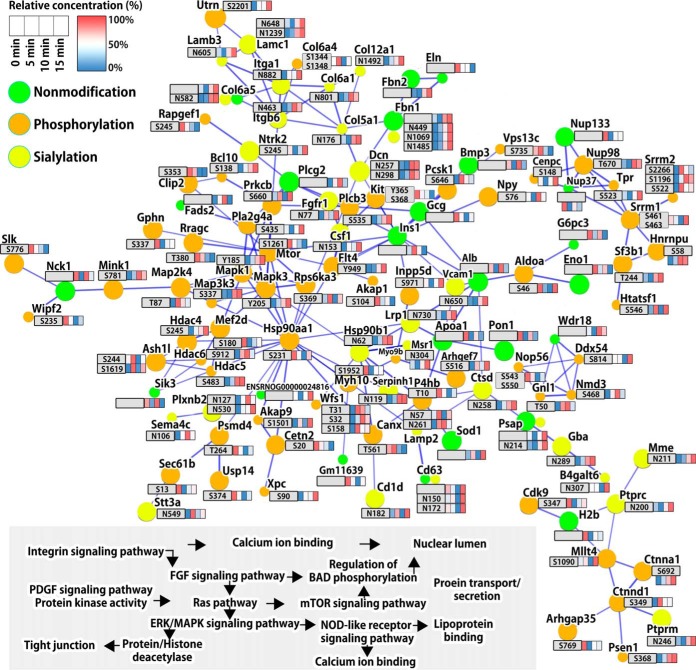
To systematically reveal protein-protein interaction networks among significantly regulated proteins, phosphoproteins, and formerly N-linked sialylated proteins, we used a combination of STRING (Search Tool for the Retrieval of Interacting Genes/Proteins), DAVID (Gene Ontology annotation enrichment analysis), and KEGG pathway analyses on the temporal large-scale data set. In the global protein-protein signaling network, we classified several complexes and biological context forming prominent, tightly connected and/or overlapping nodes (Fig. 5). We show that causal relationships and individual molecular interaction with regulated PTMs drives fifteen both known and poorly defined functional annotation and signaling pathways such as integrin signaling, FGF signaling, Ras pathway, mTOR signaling, ERK/MAPK signaling, regulation of BAD phosphorylation, regulation of nuclear lumen including the nuclear pore complex, regulation of acetylation of histone and other proteins (Fig. 5). After high glucose stimulation, glucose-mediated signals facilitated the alteration of members of cell adhesion molecule signaling (integrin signaling and FGF signaling) with sialylation across the cell surface. The phosphorylation of members of kinase activation signaling (Ras signaling, mTOR signaling, and ERK/MAPK signaling) was regulated in response to cell surface signaling pathways with altered sialylation. As downstream molecular events, the regulations of nuclear lumen, nuclear pore complex, and acetylation of histone proteins may induce the nuclear proteins translocation and transcription in nuclear environment for insulin biosynthesis and secretion. These data provide potential mechanisms that extracellular high glucose levels affected the systemic signal transduction pathways from the cell surface to the nucleus in PBCs, both involving protein phosphorylation and sialylation.
Fig. 5.
Time-resolved map of protein-protein interaction network of regulated proteins. STRING subnetworks among significantly regulated proteins, phosphorylation, and N-linked sialylation after high glucose stimulation in PBCs. The quantitative information for each node is shown as relative concentration in a box format described in the upper left corner. In the global protein-protein signaling network several complexes and biological context formed prominent, tightly connected and/or overlapped nodes. Fifteen functional annotations and signaling pathways were found interconnected and interdependent, which are outlined at the lower left part of the figure and organized in accordance with the interaction network. STRING confidence score ≥ 0.9 (the highest confidence).
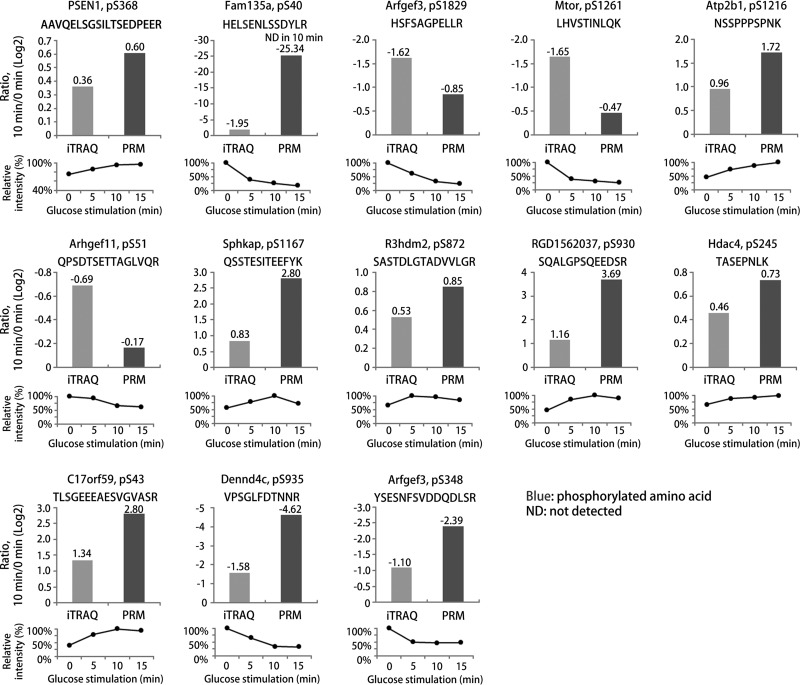
Validation of Highly Regulated Phosphopeptides with Parallel Reaction Monitoring (PRM)
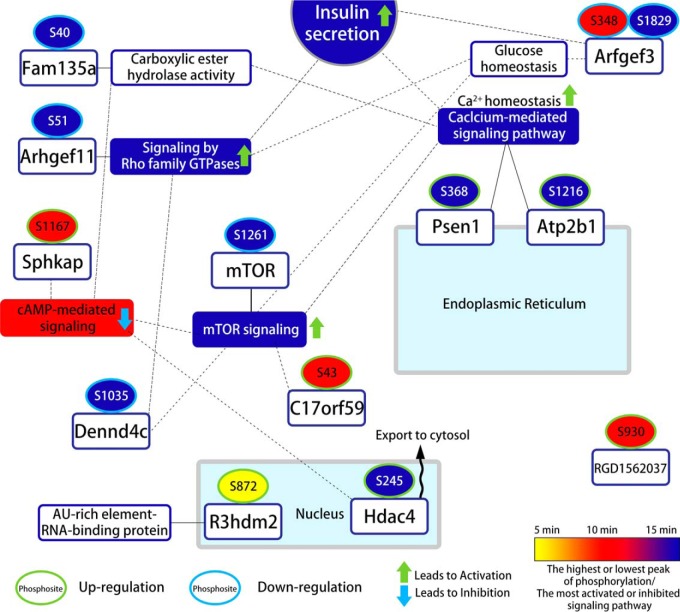
Among regulated phosphopeptides, we validated thirteen differentially regulated phosphosites identified in our iTRAQ-based quantitative analysis using PRM assays with synthetic phosphopeptides (Fig. 6 and 7). As shown in Fig. 6, we verified thirteen phosphopeptides in twelve proteins in the response to GSIS, which occurred in PBCs between the control (0 min) and 10 min GSIS. The PRM results showed a good correlation with the iTRAQ data (supplemental Table S1). The phosphorylation levels of presenilin 1 (Psen1;S368), plasma membrane Ca2+-transporting ATPase (Atp2b1;S1216), SPHK1-interactor and AKAP domain-containing protein (Sphkap;S1167), R3H domain-containing protein 2 (R3hdm2;S872), protein RGD1562037 (S930), histone deacetylase 4 (Hdac4;S245), and BLOC-1-related complex subunit 6 (C17orf59;S43) increased significantly after 10 min GSIS compared with 0 min GSIS. However, protein FAM135A (S40), brefeldin A-inhibited guanine nucleotide-exchange protein 3 (Arfgef3;S348 and S1829), serine/threonine-protein kinase mTOR (Mtor;S1261), rho guanine nucleotide exchange factor 11 (Arhgef11;S51) and DENN domain-containing protein 4C (Dennd4c;S935) decreased significantly in the same samples. We found in the iTRAQ data that the phosphorylation level of mTOR at S1261 decreased rapidly after 15 min GSIS when compared with nonstimulated PBCs. Activated mTORC1 signaling causes ER stress and an increase of the unfolded protein response, thereby inhibiting the mTORC1 positive effector S6K1 (48–50). TSC/Rheb signaling promotes mTOR S1261 phosphorylation with mTORC1 activation, as well as cell growth and cell cycle progression during glucose stimulated conditions, but it has not been demonstrated that insulin or glucose regulates the destabilization of the mTOR-raptor interaction (51–54). Thus, these results suggest that mTOR dephosphorylation at S1261 may modulate insulin production/secretion or glucose homeostasis in PBCs. Interestingly, we also observed an increased level of phosphorylation of C17orf59 at S43 in GSIS condition of PBCs. Consistently, the association of C17orf59 and mTOR phosphorylation is an essential factor in inhibiting mTORC1 (55), and this is dependent on high glucose stimulation. These data suggest that dephosphorylation of mTOR at S1261 and phosphorylation of C17orf59 at S43 may act to inhibit mTORC1 glucose metabolism, resulting in insulin secretion at high glucose conditions. In addition, histone deacetylase proteins (HDACs) are well known regulators of chromatin structure and gene transcription in the nucleus, and they are the key modulators of fatty acid and glucose metabolism in obesity and T2D (56, 57). Our data show increased phosphorylation levels of HDACs under high glucose stimulated conditions, such as Hdac4;S245, Hdac5;S483, and Hdac6;S912. Interestingly, Ozcan et al. reported that Camk2 modulates the phosphorylation (S465 and S629) of Hdac4 and blocks nuclear translocation of Hdac4 to the cytoplasm of hepatocytes in livers from obese mice, which promotes the PERK-TRB3 pathway and defective insulin signaling (58). In addition, Luan et al. demonstrated that cAMP-mediated Hdac4 activation is associated with insulin sensitivity and energy balance in obesity (59). Our data suggests a link between the phosphorylation of HDACs and control of glucose uptake and insulin processing in the GSIS cellular events in PBCs. Arfgef3 is an important protein associated with reduced insulin granule biogenesis and insulin secretion in PBCs (60). Recent studies have shown that in Arfgef3-knockout mice, secretory insulin granule biogenesis and glucagon secretion were highly increased in PBCs and pancreatic islet alpha cells, respectively (61, 62). Here, we provide the first evidence that Arfgef3 was dephosphorylated at S348 and S1829 under GSIS conditions, phosphosites that have not previously been identified. It suggests that dephosphorylation of Arfgef3 contributes to improve the islet functions by altering the cellular signaling mechanisms involved in GSIS, possibly acting to enhance biphasic insulin granule biogenesis and glucagon secretion in the two cell types, respectively. We also found a decrease in the phosphorylation of Dennd4c at S935, which is known to be a potential key regulator of glucose transporter (Glut4) translocation (63, 64). Knockdown of Dennd4c significantly suppressed Glut4 translocation, thereby triggering the activation of a GTP-bound form of Rabs (63, 64). Our results suggest that the novel phosphosite of Dennd4c may modulate the accumulation of Glut4 transport vesicles. These results provide the first molecular characterization of GSIS mediated alterations in phosphorylation with high accuracy, using our quantitative strategy on PBCs from isolated pancreatic islets. A summary of the validated phosphosites and their potential involvement in cellular signaling and PBCs functions is illustrated in Fig. 7. These data provide substantial evidence that the regulations of these phosphosites could be accounted for fundamental GSIS mechanisms from the cell membrane to the nucleus. It remains to be examined whether this signal transduction may be directly or indirectly to increase glucose-mediated (prepro)insulin biosynthesis, insulin secretory granules, or insulin secretion in the intra-islet environment.
Fig. 6.
Parallel reaction monitoring (PRM) validation of selected phosphopeptides. Thirteen phosphopeptides of 12 proteins showing regulation in the iTRAQ-based phosphoproteome data in response to high glucose stimulation for 10 min as compared with controls were validation by PRM. The PRM phosphorylation levels of all phosphopeptides correlated well with the iTRAQ analysis. All histograms show the Log2-ratios of 10 min high glucose compared with controls in the iTRAQ and PRM analysis, whereas the lower graphs show the time-resolved regulation found in the iTRAQ data at 0, 5, 10, and 15 min high glucose stimulation shown as average percent expression from two independent experiments (the highest expression set at 100%).
Fig. 7.
Time-resolved overview of regulated and validated phosphosites related to insulin secretion pathways in pancreatic islet β-cells using PRM with synthetic phosphopeptides.
One additional interesting finding in our data set is the strong regulation of the S646 in Neuroendocrine convertase 1. This enzyme, also referred to as prohormone convertase 1 (PC1) is, together with prohormone convertase 2 (PC2), responsible for the cleavage of the proinsulin into the mature form for insulin in the endoplasmic reticulum (ER). The regulation of the activity of this enzyme is poorly understood, but our data suggest that it is regulated by phosphorylation caused by activation of an ER specific kinase after short time glucose stimulation of PBCs. The action of PC1/2 is essential for the generation of functional mature insulin from PBCs and therefore the identification of this phosphosite as a potential activator of this protein is an important finding.
Alteration in Sialylation of Membrane Proteins Associated with GSIS
Alteration of sialic acids on membrane proteins was unexpected because the time of stimulation was very short and therefore membrane protein degradation or new synthesis and processing of membrane proteins could be neglected. However, several membrane-associated proteins were found to change specifically in the sialylation level, judged by the selective binding of sialylated glycopeptides to TiO2, as previously described (17) (supplemental Table S2). In order to obtain an overview of the protein networks that was associated with the changes, we imported the proteins showing altered sialylation in all three time points into the STRING network interaction program. The result is illustrated in supplemental Fig. S3. Here several distinct networks of proteins are observed, including proteins involved in cell-cell interaction, Focal adhesion, Semaphorin signaling, Integrin signaling and lysosomal degradation. Many of the protein networks are interconnected, illustrating a substantial modulation of sialylation of the cell surface important for the biological outcome of glucose stimulation of PBCs. One very important group of proteins is the Semaphorins, which are membrane bound or secreted proteins that binds to the group of receptor proteins called plexins. Each group of semaphorins has their own plexin receptor. All plexin proteins have a cytoplasmic domain that encode a GTPase-activating protein for Ras-related protein (R-Ras) and a GTPase binding domain (65), that are capable of associate with other signaling molecules and thereby initiate signal transduction pathways in the cell. For example, the Semaphorin 4C and 3E, which changed in sialylation after short GSIS, bind to the plexin B2 and A1, respectively. Both plexins also show change in sialylation after brief GSIS. The activation of these two plexins results in downstream activation of RHOA and subsequent changes of the dynamic of the actin cytoskeleton. In addition, activation of R-Ras is known to promote cell adhesion, by regulation of integrin activation, cell mobility, migration and polarity (66, 67) and it is involved in the activation of the extracellular signal-regulated kinase (ERK)/mitogen-activated protein kinase (MAPK) pathway, involved in protein synthesis and cell differentiation (68). Several of the signaling molecules downstream from the plexins identified in this study showed changes in phosphorylation, including phosphosites known to control the enzyme activity, e.g. ERK/MAPK, mTOR, PI3K (phosphatidylinositol 3-kinase), AKT (RAC-alpha kinase), and S6K (p70 ribosomal S6 Kinase)(68).
An interesting observation, linking protein sialylation with phosphorylation directly in a PTM cross-talk, taking place during GSIS, is the regulation of the interaction between integrin's and ECM proteins in relation to various processes in the cell that determine the highly dynamic link between the cytoskeleton and its connection with the ECM, defined as focal adhesion. Several integrin's and their ligands were found to change specifically in sialylation on distinct N-linked glycosites after brief GSIS (supplemental Fig. S3). Activated integrin's promote focal adhesion kinase autophosphorylation on a tyrosine residue, which lead to its activation and phosphorylation of downstream proteins, i.e. Paxillin, which are involved in modulation of the cytoskeleton in the cell. In the present study we found from IPA analysis of the phosphorylation data, that actin cytoskeleton, Paxillin, RAS, RAC, and PAK signaling pathways were systematically activated after high glucose stimulation. In addition, we identified a 1.8-fold upregulation of the Y118 in Paxillin, but only in one of the replicates. The autophosphorylation site in FAK at Y397 is located on a large very acidic tryptic peptide which makes it difficult to detect by LC-MS/MS. Besides we identify alteration in phosphorylation of several guanine nucleotide exchange factors, PLCβ, SH2 domain proteins, Synaptopodin-2, Actin filament-associated protein 1 and MAPK, all known to be involved in focal adhesion. Interestingly, previous reports have shown that focal adhesion is crucial for GSIS and involves the activation of FAK and Paxillin (69), which is involved in heavily modulation of the dynamic of the F-actin in the cell, to recruit insulin granules to the plasma membrane shortly after glucose stimulation of PBCs (70). Blocking the interaction between the ECM and β1 integrin, with a specific antibody for β1 integrin was shown to inhibit the phosphorylation of FAK (Y397), Paxillin (Y118), and ERK1/2 (T202/Y204) (70), strongly indicating that integrin modulation on the surface of PBCs are highly important for the subsequent release of insulin in GSIS. The interaction between ECM proteins and Integrin's is believed to be depending on the glycosylation, especially sialylation, status of integrin's (e.g. (71)), which is known to modulate its conformation (72). For example, addition of α2–6-linked sialic acid to β1 integrin alters its binding activity to several of the ligands including fibronectin (72), Laminin (73), and collagens (74). Our observation that many integrin's and their ligands are changing in sialylation after short GSIS, is linking the Ca2+ mediated release of insulin from PBCs with a substantial modulation of the surface protein interactions probably catalyzed by alteration in sialylation. This fast alteration in sialylation of surface proteins is peculiar, as most of the N-linked glycosites are increasing in sialylation, placing sialyltransferases central in this process. It is well known that Neuraminidases are active on the cell surface but very little has been reported on sialyltransferases outside of the Golgi apparatus. Exactly which sialyltransferases that mediates the glucose stimulation signals to the surface proteins in PBCs and how these sialyltransferases are activated in response to increased glucose levels is currently unknown and will require a substantial focus in the future to fully understand the GSIS process. However, it is evident from our study, that surface protein sialylation is an important, very much overlooked PTM involved in GSIS.
Interestingly, our data showed the increased level of sialylation on four receptor proteins (IGF2R;insulin-like growth factor 2 receptor, STAB2;stabilin-2, MSR1;scavenger receptor type A, and LRP1;prolow-density lipoprotein receptor-related protein 1) associated with receptor-mediated endocytosis at 10 and 15 min after high glucose stimulation, in concert with the regulation of small GTPase mediated signal transduction and intracellular signaling cascade/transport in the formed cluster 3 of Fig. 3B. G-protein-coupled-receptors (GPCR)-mediated signal transduction can promote a reversible redistribution of receptors from the plasma membrane to intracellular membrane through endocytosis within seconds to minutes after agonist-induced activation (75). Moreover, these receptors are involved in regulation of cholesterol and/or glucose efflux and transport in the cell. Our data suggest that the increased sialylation of receptors (IGF2R (N386–7 and N1647), STAB2 (N139 and N1161), MSR1 (N304), and LRP1 (N730)) may play a role of regulation of receptor trafficking, thereby inhibit the import of glucose or cholesterol into the intracellular space in PBCs.
Concluding remarks
We here demonstrate the first comprehensive temporal identification and quantification of the proteome and selected PTMs (phosphoproteins and N-linked sialylated glycoproteins) of PBCs in response to brief glucose stimulation. Our optimized workflow for the low amount of material allowed us to effectively investigate alterations in protein expression, phosphorylation and N-linked sialylation implicated in insulin secretion-related signal transduction pathways. The data presented here confirmed many signaling pathways already known to be involved in GSIS, such as pathways related to Ca2+ mediated signaling, exocytosis, endocytosis and insulin granule docking. However, our study identified many novel regulatory phosphosites important for these pathways and also novel phosphosites on proteins not previously thought to be involved in the GSIS process. Among the novel finding, beside selected phosphosites and novel signaling pathways, we for the first time showed alteration in sialylation of surface proteins, such as integrins, integrin ligands, semaphorins and plexins in response to brief glucose stimulation of PBCs. The alteration in sialylation is highly connected to focal adhesion, which is known to be critical in GSIS, however, the exact biological role for the sialylation is presently not known. The identification of intracellular signaling pathways that are involved in insulin secretion on GSIS of PBCs has lead us to a better understanding of the molecular mechanisms underlying the GSIS event. This is highly important to understand the pathology underlying metabolic diseases such as type 1 and 2 diabetes and to identify novel targets for the treatment of these diseases. We believe that the data presented here will be useful as a blueprint for future studies of the mechanisms of high glucose-regulated hyper insulin production/secretion in primary PBCs.
DATA AVAILABILITY
Data are available via ProteomeXchange (http://proteomecentral.proteomexchange.org/cgi/GetDataset) with identifier PXD007689, allow viewing of results using PRIDE Inspector (76, 77). Skyline files from the PRM analysis were uploaded into Panorama Public (https://panoramaweb.org/labkey/UPTjTd.url).
Supplementary Material
Acknowledgments
This study was supported by the Danish Medical Research Council (Grant number: 1331-00338B), The Novo Nordisk Foundation (Grant number: NNF16OC0023448) and the Villum Center for Bioanalytical Sciences at SDU.
Footnotes
* This study was supported by the Danish Medical Research Council (grant number: 1331-00338B), The Novo Nordisk Foundation (Grant number: NNF16OC0023448) and the Villum Center for Bioanalytical Sciences at SDU.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- PBCs
- pancreatic islet β-cells
- GSIS
- glucose stimulated insulin secretion
- HILIC
- hydrophilic liquid chromatography
- SA
- sialylated
- PRM
- parallel reaction monitoring
- PTMs
- posttranslational modifications
- SIMAC
- sequential elution from IMAC
- IQR
- inter-quartile range
- T2D
- Type 2 Diabetes
- HDACs
- histone deacetylase proteins
- PSEN1
- presenilin 1
- ATP2B1
- plasma membrane Ca2+-transporting ATPase
- SPHKAP
- SPHK1-interactor and AKAP domain-containing protein
- R3HDM2
- R3H domain-containing protein 2
- RGD1562037
- protein RGD1562037
- HDAC4
- histone deacetylase 4
- C17orf59
- BLOC-1-related complex subunit 6
- FAM135A
- protein FAM135A
- ARFGEF3
- brefeldin A-inhibited guanine nucleotide-exchange protein 3
- MTOR
- serine/threonine-protein kinase mTOR
- ARHGEF11
- rho guanine nucleotide exchange factor 11
- DENND4C
- DENN domain-containing protein 4C
- R-RAS
- Ras-related protein
- ERK
- extracellular signal-regulated kinase
- MAPK
- mitogen-activated protein kinase
- IGF2R
- insulin-like growth factor 2 receptor
- STAB2
- stabilin-2
- MSR1
- scavenger receptor type A
- LRP1
- prolow-density lipoprotein receptor-related protein 1.
REFERENCES
- 1. Danaei G., Finucane M. M., Lu Y., Singh G. M., Cowan M. J., Paciorek C. J., Lin J. K., Farzadfar F., Khang Y. H., Stevens G. A., Rao M., Ali M. K., Riley L. M., Robinson C. A., Ezzati M., and Global Burden of Metabolic Risk Factors of Chronic Diseases Collaborating, G. (2011) National, regional, and global trends in fasting plasma glucose and diabetes prevalence since 1980: systematic analysis of health examination surveys and epidemiological studies with 370 country-years and 2.7 million participants. Lancet 378, 31–40 [DOI] [PubMed] [Google Scholar]
- 2. Collaboration N. C. D. R. F. (2016) Worldwide trends in diabetes since 1980: a pooled analysis of 751 population-based studies with 4.4 million participants. Lancet 387, 1513–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Laakso M. (1999) Hyperglycemia and cardiovascular disease in type 2 diabetes. Diabetes 48, 937–942 [DOI] [PubMed] [Google Scholar]
- 4. Grundy S. M., Benjamin I. J., Burke G. L., Chait A., Eckel R. H., Howard B. V., Mitch W., Smith S. C. Jr, and Sowers J. R. (1999) Diabetes and cardiovascular disease: a statement for healthcare professionals from the American Heart Association. Circulation 100, 1134–1146 [DOI] [PubMed] [Google Scholar]
- 5. MacDonald P. E., Joseph J. W., and Rorsman P. (2005) Glucose-sensing mechanisms in pancreatic beta-cells. Philos. Trans. R. Soc. Lond. B Biol. Sci. 360, 2211–2225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rorsman P., and Renstrom E. (2003) Insulin granule dynamics in pancreatic beta cells. Diabetologia 46, 1029–1045 [DOI] [PubMed] [Google Scholar]
- 7. Marshak S., Leibowitz G., Bertuzzi F., Socci C., Kaiser N., Gross D. J., Cerasi E., and Melloul D. (1999) Impaired beta-cell functions induced by chronic exposure of cultured human pancreatic islets to high glucose. Diabetes 48, 1230–1236 [DOI] [PubMed] [Google Scholar]
- 8. Chiu K. C., Cohan P., Lee N. P., and Chuang L. M. (2000) Insulin sensitivity differs among ethnic groups with a compensatory response in beta-cell function. Diabetes Care 23, 1353–1358 [DOI] [PubMed] [Google Scholar]
- 9. Henquin J. C. (2000) Triggering and amplifying pathways of regulation of insulin secretion by glucose. Diabetes 49, 1751–1760 [DOI] [PubMed] [Google Scholar]
- 10. Henquin J. C., Nenquin M., Ravier M. A., and Szollosi A. (2009) Shortcomings of current models of glucose-induced insulin secretion. Diabetes Obes. Metab. 11, 168–179 [DOI] [PubMed] [Google Scholar]
- 11. Marre M., Shaw J., Brandle M., Bebakar W. M., Kamaruddin N. A., Strand J., Zdravkovic M., Le Thi T. D., Colagiuri S., and group, L-Ss (2009) Liraglutide, a once-daily human GLP-1 analogue, added to a sulphonylurea over 26 weeks produces greater improvements in glycaemic and weight control compared with adding rosiglitazone or placebo in subjects with Type 2 diabetes (LEAD-1 SU). Diabet. Med. 26, 268–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Natalicchio A., Biondi G., Marrano N., Labarbuta R., Tortosa F., Spagnuolo R., D'Oria R., Carchia E., Leonardini A., Cignarelli A., Perrini S., Laviola L., and Giorgino F. (2016) Long-term exposure of pancreatic beta-cells to palmitate results in SREBP-1C-dependent decreases in GLP-1 receptor signaling via CREB and AKT and insulin secretory response. Endocrinology 157, 2243–2258 [DOI] [PubMed] [Google Scholar]
- 13. Cohen O., Filetti S., Castaneda J., Maranghi M., and Glandt M. (2016) When intensive insulin therapy (MDI) fails in patients with Type 2 diabetes: switching to GLP-1 receptor agonist versus insulin pump. Diabetes Care 39, S180–S186 [DOI] [PubMed] [Google Scholar]
- 14. Sacco F., Humphrey S. J., Cox J., Mischnik M., Schulte A., Klabunde T., Schafer M., and Mann M. (2016) Glucose-regulated and drug-perturbed phosphoproteome reveals molecular mechanisms controlling insulin secretion. Nat. Commun. 7, 13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Han D., Moon S., Kim Y., Ho W. K., Kim K., Kang Y., Jun H., and Kim Y. (2012) Comprehensive phosphoproteome analysis of INS-1 pancreatic beta-cells using various digestion strategies coupled with liquid chromatography-tandem mass spectrometry. J. Proteome Res. 11, 2206–2223 [DOI] [PubMed] [Google Scholar]
- 16. Li J., Li Q., Tang J., Xia F., Wu J., and Zeng R. (2015) Quantitative phosphoproteomics revealed glucose-stimulated responses of islet associated with insulin secretion. J. Proteome Res. 14, 4635–4646 [DOI] [PubMed] [Google Scholar]
- 17. Palmisano G., Lendal S. E., Engholm-Keller K., Leth-Larsen R., Parker B. L., and Larsen M. R. (2010) Selective enrichment of sialic acid-containing glycopeptides using titanium dioxide chromatography with analysis by HILIC and mass spectrometry. Nat. Protoc. 5, 1974–1982 [DOI] [PubMed] [Google Scholar]
- 18. Thingholm T. E., Jensen O. N., Robinson P. J., and Larsen M. R. (2008) SIMAC (sequential elution from IMAC), a phosphoproteomics strategy for the rapid separation of monophosphorylated from multiply phosphorylated peptides. Mol. Cell. Proteomics 7, 661–671 [DOI] [PubMed] [Google Scholar]
- 19. Engholm-Keller K., Birck P., Storling J., Pociot F., Mandrup-Poulsen T., and Larsen M. R. (2012) TiSH–a robust and sensitive global phosphoproteomics strategy employing a combination of TiO2, SIMAC, and HILIC. J. Proteomics 75, 5749–5761 [DOI] [PubMed] [Google Scholar]
- 20. Larsen M. R., Thingholm T. E., Jensen O. N., Roepstorff P., and Jorgensen T. J. (2005) Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol. Cell. Proteomics 4, 873–886 [DOI] [PubMed] [Google Scholar]
- 21. Engholm-Keller K., and Larsen M. R. (2016) Improving the Phosphoproteome Coverage for Limited Sample Amounts Using TiO2-SIMAC-HILIC (TiSH) Phosphopeptide Enrichment and Fractionation. Methods Mol. Biol. 1355, 161–177 [DOI] [PubMed] [Google Scholar]
- 22. Brunstedt J. (1980) Rapid isolation of functionally intact pancreatic islets from mice and rats by percollTM gradient centrifucation. Diabete Metabolisme 6, 87–89 [PubMed] [Google Scholar]
- 23. Engholm-Keller K., Birck P., Storling J., Pociot F., Mandrup-Poulsen T., and Larsen M. R. (2012) TiSH - a robust and sensitive global phosphoproteomics strategy employing a combination of TiO(2), SIMAC, and HILIC. J. Proteomics 75, 5749–5761 [DOI] [PubMed] [Google Scholar]
- 24. Thingholm T. E., and Larsen M. R. (2009) The use of titanium dioxide micro-columns to selectively isolate phosphopeptides from proteolytic digests. Methods Mol. Biol. 527, 57–66, xi [DOI] [PubMed] [Google Scholar]
- 25. Larsen M. R., Jensen S. S., Jakobsen L. A., Heegaard N. H. (2007) Exploring the sialiome using titanium dioxide chromatography and mass spectrometry. Mol. Cell. Proteomics 6, 1778–1787 [DOI] [PubMed] [Google Scholar]
- 26. Thingholm T. E., Jorgensen T. J., Jensen O. N., and Larsen M. R. (2006) Highly selective enrichment of phosphorylated peptides using titanium dioxide. Nat. Protoc. 1, 1929–1935 [DOI] [PubMed] [Google Scholar]
- 27. Kim S., and Pevzner P. A. (2014) MS-GF+ makes progress towards a universal database search tool for proteomics. Nat. Commun. 5, 5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Monroe M. E., Shaw J. L., Daly D. S., Adkins J. N., and Smith R. D. (2008) MASIC: a software program for fast quantitation and flexible visualization of chromatographic profiles from detected LC-MS(/MS) features. Computational Biol. Chem. 32, 215–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ma Z. Q., Chambers M. C., Ham A. J., Cheek K. L., Whitwell C. W., Aerni H. R., Schilling B., Miller A. W., Caprioli R. M., and Tabb D. L. (2011) ScanRanker: Quality assessment of tandem mass spectra via sequence tagging. J. Proteome Res. 10, 2896–2904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Taus T., Kocher T., Pichler P., Paschke C., Schmidt A., Henrich C., and Mechtler K. (2011) Universal and confident phosphorylation site localization using phosphoRS. J. Proteome Res. 10, 5354–5362 [DOI] [PubMed] [Google Scholar]
- 31. Rigbolt K. T., Vanselow J. T., and Blagoev B. (2011) GProX, a user-friendly platform for bioinformatics analysis and visualization of quantitative proteomics data. Mol. Cell. Proteomics 10, O110 007450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Huang da W, Sherman B. T., and Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 33. Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K. P., Kuhn M., Bork P., Jensen L. J., and von Mering C. (2015) STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447–D452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Carr S. A., Abbatiello S. E., Ackermann B. L., Borchers C., Domon B., Deutsch E. W., Grant R. P., Hoofnagle A. N., Huttenhain R., Koomen J. M., Liebler D. C., Liu T., MacLean B., Mani D. R., Mansfield E., Neubert H., Paulovich A. G., Reiter L., Vitek O., Aebersold R., Anderson L., Bethem R., Blonder J., Boja E., Botelho J., Boyne M., Bradshaw R. A., Burlingame A. L., Chan D., Keshishian H., Kuhn E., Kinsinger C., Lee J. S., Lee S. W., Moritz R., Oses-Prieto J., Rifai N., Ritchie J., Rodriguez H., Srinivas P. R., Townsend R. R., Van Eyk J., Whiteley G., Wiita A., and Weintraub S. (2014) Targeted peptide measurements in biology and medicine: best practices for mass spectrometry-based assay development using a fit-for-purpose approach. Mol. Cell. Proteomics 13, 907–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. MacLean B., Tomazela D. M., Shulman N., Chambers M., Finney G. L., Frewen B., Kern R., Tabb D. L., Liebler D. C., and MacCoss M. J. (2010) Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Palmisano G., Parker B. L., Engholm-Keller K., Lendal S. E., Kulej K., Schulz M., Schwammle V., Graham M. E., Saxtorph H., Cordwell S. J., and Larsen M. R. (2012) A novel method for the simultaneous enrichment, identification, and quantification of phosphopeptides and sialylated glycopeptides applied to a temporal profile of mouse brain development. Mol. Cell. Proteomics 11, 1191–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xi Y., Liu S., Bettaieb A., Matsuo K., Matsuo I., Hosein E., Chahed S., Wiede F., Zhang S., Zhang Z. Y., Kulkarni R. N., Tiganis T., and Haj F. G. (2015) Pancreatic T cell protein-tyrosine phosphatase deficiency affects beta cell function in mice. Diabetologia 58, 122–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Inoki K., Li Y., Xu T., and Guan K. L. (2003) Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 17, 1829–1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Festa A., D'Agostino R. Jr, Tracy R. P., Haffner S. M., and Insulin Resistance Atherosclerosis Study (2002) Elevated levels of acute-phase proteins and plasminogen activator inhibitor-1 predict the development of type 2 diabetes: the insulin resistance atherosclerosis study. Diabetes 51, 1131–1137 [DOI] [PubMed] [Google Scholar]
- 40. Clark E. A., and Brugge J. S. (1995) Integrins and signal transduction pathways: the road taken. Science 268, 233–239 [DOI] [PubMed] [Google Scholar]
- 41. Ahren B. (2009) Islet G protein-coupled receptors as potential targets for treatment of type 2 diabetes. Nat. Rev. Drug Discov. 8, 369–385 [DOI] [PubMed] [Google Scholar]
- 42. Merglen A., Theander S., Rubi B., Chaffard G., Wollheim C. B., and Maechler P. (2004) Glucose sensitivity and metabolism-secretion coupling studied during two-year continuous culture in INS-1E insulinoma cells. Endocrinology 145, 667–678 [DOI] [PubMed] [Google Scholar]
- 43. Sileno S., D'Oria V., Stucchi R., Alessio M., Petrini S., Bonetto V., Maechler P., Bertuzzi F., Grasso V., Paolella K., Barbetti F., and Massa O. (2014) A possible role of transglutaminase 2 in the nucleus of INS-1E and of cells of human pancreatic islets. J. Proteomics 96, 314–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kamagate A., Herchuelz A., Bollen A., and Van Eylen F. (2000) Expression of multiple plasma membrane Ca(2+)-ATPases in rat pancreatic islet cells. Cell Calcium 27, 231–246 [DOI] [PubMed] [Google Scholar]
- 45. MacDonald P. E., El-Kholy W., Riedel M. J., Salapatek A. M., Light P. E., and Wheeler M. B. (2002) The multiple actions of GLP-1 on the process of glucose-stimulated insulin secretion. Diabetes 51, S434–S442 [DOI] [PubMed] [Google Scholar]
- 46. Bezakova G., and Ruegg M. A. (2003) New insights into the roles of agrin. Nat. Rev. Mol. Cell Biol. 4, 295–308 [DOI] [PubMed] [Google Scholar]
- 47. Auffret J., Freemark M., Carre N., Mathieu Y., Tourrel-Cuzin C., Lombes M., Movassat J., and Binart N. (2013) Defective prolactin signaling impairs pancreatic beta-cell development during the perinatal period. Am. J. Physiol. Endocrinol. Metab. 305, E1309–E1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ozcan U., Ozcan L., Yilmaz E., Duvel K., Sahin M., Manning B. D., and Hotamisligil G. S. (2008) Loss of the tuberous sclerosis complex tumor suppressors triggers the unfolded protein response to regulate insulin signaling and apoptosis. Mol. Cell 29, 541–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Polak P., and Hall M. N. (2009) mTOR and the control of whole body metabolism. Curr. Opin. Cell Biol. 21, 209–218 [DOI] [PubMed] [Google Scholar]
- 50. Laplante M., and Sabatini D. M. (2012) mTOR signaling in growth control and disease. Cell 149, 274–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Acosta-Jaquez H. A., Keller J. A., Foster K. G., Ekim B., Soliman G. A., Feener E. P., Ballif B. A., and Fingar D. C. (2009) Site-specific mTOR phosphorylation promotes mTORC1-mediated signaling and cell growth. Mol. Cell. Biol. 29, 4308–4324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ekim B., Magnuson B., Acosta-Jaquez H. A., Keller J. A., Feener E. P., and Fingar D. C. (2011) mTOR kinase domain phosphorylation promotes mTORC1 signaling, cell growth, and cell cycle progression. Mol. Cell. Biol. 31, 2787–2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Swenne I. (1982) The role of glucose in the in vitro regulation of cell cycle kinetics and proliferation of fetal pancreatic B-cells. Diabetes 31, 754–760 [DOI] [PubMed] [Google Scholar]
- 54. Kim D. H., Sarbassov D. D., Ali S. M., King J. E., Latek R. R., Erdjument-Bromage H., Tempst P., and Sabatini D. M. (2002) mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110, 163–175 [DOI] [PubMed] [Google Scholar]
- 55. Schweitzer L. D., Comb W. C., Bar-Peled L., and Sabatini D. M. (2015) Disruption of the Rag-Ragulator Complex by c17orf59 Inhibits mTORC1. Cell Rep. 12, 1445–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ye J. (2013) Improving insulin sensitivity with HDAC inhibitor. Diabetes 62, 685–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mathias R. A., Guise A. J., and Cristea I. M. (2015) Post-translational modifications regulate class IIa histone deacetylase (HDAC) function in health and disease. Mol. Cell. Proteomics 14, 456–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ozcan L., Ghorpade D. S., Zheng Z., de Souza J. C., Chen K., Bessler M., Bagloo M., Schrope B., Pestell R., and Tabas I. (2016) Hepatocyte DACH1 Is Increased in Obesity via Nuclear Exclusion of HDAC4 and Promotes Hepatic Insulin Resistance. Cell Rep. 15, 2214–2225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Luan B., Goodarzi M. O., Phillips N. G., Guo X., Chen Y. D., Yao J., Allison M., Rotter J. I., Shaw R., and Montminy M. (2014) Leptin-mediated increases in catecholamine signaling reduce adipose tissue inflammation via activation of macrophage HDAC4. Cell Metab. 19, 1058–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Li H., Wei S., Cheng K., Gounko N. V., Ericksen R. E., Xu A., Hong W., and Han W. (2014) BIG3 inhibits insulin granule biogenesis and insulin secretion. EMBO Rep. 15, 714–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Liu T., Li H., Gounko N. V., Zhou Z., Xu A., Hong W., and Han W. (2014) Detection of insulin granule exocytosis by an electrophysiology method with high temporal resolution reveals enlarged insulin granule pool in BIG3-knockout mice. Am. J. Physiol. Endocrinol. Metab. 307, E611–E618 [DOI] [PubMed] [Google Scholar]
- 62. Li H., Liu T., Lim J., Gounko N. V., Hong W., and Han W. (2015) Increased biogenesis of glucagon-containing secretory granules and glucagon secretion in BIG3-knockout mice. Mol. Metab. 4, 246–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sadacca L. A., Bruno J., Wen J., Xiong W., and McGraw T. E. (2013) Specialized sorting of GLUT4 and its recruitment to the cell surface are independently regulated by distinct Rabs. Mol. Biol. Cell 24, 2544–2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sano H., Peck G. R., Kettenbach A. N., Gerber S. A., and Lienhard G. E. (2011) Insulin-stimulated GLUT4 protein translocation in adipocytes requires the Rab10 guanine nucleotide exchange factor Dennd4C. J. Biol. Chem. 286, 16541–16545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wang Y., Pascoe H. G., Brautigam C. A., He H., and Zhang X. (2013) Structural basis for activation and non-canonical catalysis of the Rap GTPase activating protein domain of plexin. Elife 2, e01279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wozniak M. A., Kwong L., Chodniewicz D., Klemke R. L., and Keely P. J. (2005) R-Ras controls membrane protrusion and cell migration through the spatial regulation of Rac and Rho. Mol. Biol. Cell 16, 84–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ada-Nguema A. S., Xenias H., Hofman J. M., Wiggins C. H., Sheetz M. P., and Keely P. J. (2006) The small GTPase R-Ras regulates organization of actin and drives membrane protrusions through the activity of PLCepsilon. J. Cell Sci. 119, 1307–1319 [DOI] [PubMed] [Google Scholar]
- 68. Mendoza M. C., Er E. E., and Blenis J. (2011) The Ras-ERK and PI3K-mTOR pathways: cross-talk and compensation. Trends Biochem. Sci. 36, 320–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Rondas D., Tomas A., and Halban P. A. (2011) Focal adhesion remodeling is crucial for glucose-stimulated insulin secretion and involves activation of focal adhesion kinase and paxillin. Diabetes 60, 1146–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rondas D., Tomas A., Soto-Ribeiro M., Wehrle-Haller B., and Halban P. A. (2012) Novel mechanistic link between focal adhesion remodeling and glucose-stimulated insulin secretion. J. Biol. Chem. 287, 2423–2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Suzuki O., Abe M., and Hashimoto Y. (2015) Sialylation and glycosylation modulate cell adhesion and invasion to extracellular matrix in human malignant lymphoma: Dependency on integrin and the Rho GTPase family. Int. J. Oncol. 47, 2091–2099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Pretzlaff R. K., Xue V. W., and Rowin M. E. (2000) Sialidase treatment exposes the beta1-integrin active ligand binding site on HL60 cells and increases binding to fibronectin. Cell Adhes. Commun. 7, 491–500 [DOI] [PubMed] [Google Scholar]
- 73. Pochec E., Litynska A., Amoresano A., and Casbarra A. (2003) Glycosylation profile of integrin alpha 3 beta 1 changes with melanoma progression. Biochim. Biophys. Acta 1643, 113–123 [DOI] [PubMed] [Google Scholar]
- 74. Shaikh F. M., Seales E. C., Clem W. C., Hennessy K. M., Zhuo Y., and Bellis S. L. (2008) Tumor cell migration and invasion are regulated by expression of variant integrin glycoforms. Exp. Cell Res. 314, 2941–2950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Tsao P., Cao T., and von Zastrow M. (2001) Role of endocytosis in mediating downregulation of G-protein-coupled receptors. Trends Pharmacol. Sci. 22, 91–96 [DOI] [PubMed] [Google Scholar]
- 76. Perez-Riverol Y., Xu Q. W., Wang R., Uszkoreit J., Griss J., Sanchez A., Reisinger F., Csordas A., Ternent T., Del-Toro N., Dianes J. A., Eisenacher M., Hermjakob H., and Vizcaino J. A. (2016) PRIDE inspector toolsuite: moving toward a universal visualization tool for proteomics data standard formats and quality assessment of ProteomeXchange data sets. Mol. Cell. Proteomics 15, 305–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Deutsch E. W., Csordas A., Sun Z., Jarnuczak A., Perez-Riverol Y., Ternent T., Campbell D. S., Bernal-Llinares M., Okuda S., Kawano S., Moritz R. L., Carver J. J., Wang M., Ishihama Y., Bandeira N., Hermjakob H., and Vizcaino J. A. (2017) The ProteomeXchange consortium in 2017: supporting the cultural change in proteomics public data deposition. Nucleic Acids Res. 45, D1100–D1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available via ProteomeXchange (http://proteomecentral.proteomexchange.org/cgi/GetDataset) with identifier PXD007689, allow viewing of results using PRIDE Inspector (76, 77). Skyline files from the PRM analysis were uploaded into Panorama Public (https://panoramaweb.org/labkey/UPTjTd.url).