Abstract
Amoebae have been considered as a genetic “melting pot” for its symbionts, facilitating genetic exchanges of the bacteria that co-inhabit the same host. To test the “melting pot” hypothesis, we analyzed six genomes of amoeba endosymbionts within Rickettsiales, four of which belong to Holosporaceae family and two to Candidatus Midichloriaceae. For the first time, we identified plasmids in obligate amoeba endosymbionts, which suggests conjugation as a potential mechanism for lateral gene transfers (LGTs) that underpin the “melting pot” hypothesis. We found strong evidence of recent LGTs between the Rickettsiales amoeba endosymbionts, suggesting that the LGTs are continuous and ongoing. In addition, comparative genomic and phylogenomic analyses revealed pervasive and recurrent LGTs between Rickettsiales and distantly related amoeba-associated bacteria throughout the Rickettsiales evolution. Many of these exchanged genes are important for amoeba–symbiont interactions, including genes in transport system, antibiotic resistance, stress response, and bacterial virulence, suggesting that LGTs have played important roles in the adaptation of endosymbionts to their intracellular habitats. Surprisingly, we found little evidence of LGTs between amoebae and their bacterial endosymbionts. Our study strongly supports the “melting pot” hypothesis and highlights the role of amoebae in shaping the Rickettsiales evolution.
Keywords: lateral gene transfer, Rickettsiales, amoeba endosymbionts
Introduction
Amoebae are one of the most ubiquitous protists and are widespread in diverse habitats such as soil, water, air, and engineered environments (Schmitz-Esser, et al. 2008). As predators, they feed on various bacteria and exhibit a great impact in controlling the microbial communities (Clarholm 1981). Amoebae are known to harbor a wide range of intracellular bacteria belonging to dispersed phylogenetic lineages (Moliner, et al. 2010; Schmitz-Esser, et al. 2008). Some of these bacteria, including members from Rickettsiales (Birtles, et al. 2000; Fritsche, et al. 1999; Horn, et al. 1999), Bacteroidetes (Horn, et al. 2001), and Chlamydiae (Amann, et al. 1997; Birtles, et al. 1997; Fritsche, et al. 2000; Horn, et al. 2000), are obligate amoeba endosymbionts, while others can cause disease in human and animals (Albert-Weissenberger, et al. 2007; Thomas and McDonnell 2007). Because macrophages are amoeboid cells, it has been suggested that amoebae serve as an evolutionary “training ground” for the emergence of these specialized bacterial pathogens (Molmeret, et al. 2005).
The “melting pot” hypothesis proposes that amoebae serve as a fertile ground allowing genetic exchanges among intra-amoebal bacteria (Bertelli and Greub 2012; Moliner, et al. 2010). It can be considered as a special case of the “intracellular arena” hypothesis that genetic material can be exchanged between bacteria that co-localize the same intracellular environment (Bordenstein and Wernegreen 2004). Multiple lines of evidence support such lateral gene transfers (LGTs). For example, phylogenetic analyses by Ogata et al. suggested that the whole tra cluster of Rickettsia bellii encoding the type IV secretion system (T4SS) was closely related to that of Candidatus Protochlamydiae amoebophila, both of which are capable of infecting amoebae (Ogata, et al. 2006). The analysis of the genome of Candidatus Amoebophilus asiaticus (Caa), an amoeba symbiont, revealed 37 genes of likely foreign origins (Schmitz-Esser, et al. 2010). Not only can amoebae serve as a place of gene exchanges for the microorganisms living within them, they can also participate in such exchanges. For example, phylogenetic analyses suggested one Sec7 domain-containing protein homologous to RalF was likely first transferred from eukaryotes to bacteria and then between Legionella and Rickettsia (Cox, et al. 2004; Ogata, et al. 2006). Genome sequencing of Legionella drancourtii revealed that it contains both a keto acid dehydrogenase and a sterol reductase gene most closely related to the amoebal homologs (Gimenez, et al. 2011). And it has been suggested that mimivirus, a virus that grows in amoebae and possesses one of the largest viral genomes (Raoult, et al. 2004), acquired 10% of its genes from amoebae (Filee, et al. 2008; Moreira and Brochier-Armanet 2008).
The “melting pot” hypothesis predicts that partners engaged in LGTs with amoeba endosymbionts should be mostly amoeba-associated bacteria. Rickettsiales represents an excellent model for testing the “melting pot” hypothesis. Rickettsiales is a deep-branched order of α-proteobacteria consisting of obligate intracellular bacteria in four distinct families: Rickettsiaceae, Anaplasmataceae, Holosporaceae, and Candidatus Midichloriaceae (although one recent study proposed Holosporaceae to be an independent order (Szokoli, et al. 2016). Members of Holosporaceae and Candidatus Midichloriaceae are mostly endosymbionts of unicellular protists such as Paramecium and Acanthamoeba, while their relatives in Rickettsiaceae and Anaplasmataceae (e.g., Rickettsia, Ehrlichia, and Anaplasma) are mostly notable specialized pathogens in a broad range of multicellular eukaryotic hosts, such as arthropods, nematodes, and mammals. The “melting pot” hypothesis predicts that members of Holosporaceae and Candidatus Midichloriaceae should participate in extensive LGTs with other amoeba-associated bacteria.
Although many members of Rickettsiaceae and Anaplasmataceae have been sequenced, few genomes in Holosporaceae and Candidatus Midichloriaceae are available. Recently, six Rickettsiales amoeba endosymbionts were sequenced, four of which belong to the Holosporaceae family (Candidatus Odyssella thessalonicensis [Cot] [Georgiades, et al. 2011], Candidatus Caedibacter acanthamobae [Cca], Candidatus Paracaedibacter acanthamoebae [Cpa], Candidatus Paracaedibacter symbiosus [Cps] [Wang and Wu 2014, 2015]) and two belong to the Candidatus Midichloriaceae family (Candidatus Jidaibacter acanthamoeba [Cja] [Schulz, et al. 2016] and Endosymbiont of Acanthamoeba UWC8 [Eau] [Wang and Wu 2014, 2015]).
With the much improved taxonomic representation, we tested the “melting pot” hypothesis using Rickettsiales endosymbionts of amoeba as our model system. First, we identified possible mechanisms of LGT from genomes of the amoeba endosymbionts. We then performed comparative genomic and phylogenomic analyses and tested the “melting pot” hypothesis by identifying partners involved in the LGTs with these endosymbionts. We extended our analyses to other lineages of Rickettsiales to assess the impact of intra-amoebal LGTs on the evolution of Rickettsiales.
Materials and Methods
LGTs between Rickettsiales and Other Amoeba-Associated Bacteria
For each gene of the six genomes of Rickettsiales endosymbionts of amoeba (Cca, Cpa, Cps, Eau, Cot, and Cja), a BLASTP search was performed against 2,461 bacterial, 144 archaeal, and 109 viral genomes. To include LGTs that happened prior to and during the divergence of the Rickettsiales lineages, we excluded the BLASTP hits in Rickettsiales and identified the query sequences that have non α-proteobacterial best hits as candidate genes for LGTs (evalue cutoff 1e-7). We performed the same analysis for 13 other representative Rickettsiales species that are not amoeba endosymbionts (Rickettsia prowazekii str. Madrid E, Rickettsia conorii str. Malish 7, Rickettsia massiliae MTU5, Rickettsia bellii RML369-C, Rickettsia felis URRWXCal2, Rickettsia felis LSU-Lb, Rickettsia buchneri ISO7, Wolbachia wMel, Orientia tsutsugamushi str. Boryong, Anaplasma phagocytophilum str. HZ, Ehrlichia canis str. Jake, Neorickettsia risticii str. Illinois, and Candidatus Midichloria mitochondrii). For functional annotation, the candidate genes were classified into COGs by hidden Markov model search using HMMer3 (Eddy 1998).
To focus on bacterial species that are more likely engaged in LGTs with Rickettsiales, we ranked the non α-proteobacterial species by the number of times that they showed up as the best hit in the BLASTP search and then performed phylogenetic analysis for candidate genes with a best hit in the top 10 ranked species. Genes were clustered into families using the Markov Cluster Algorithm with all-against-all BLASTP e-value cutoff of 1e-15 (Enright, et al. 2002). For each gene family, its homologs from all complete bacterial and archaeal genomes were retrieved by BLASTP search (e-value cutoff 1e-15). Protein sequences were aligned using MAFFT (Katoh, et al. 2002) and ambiguously aligned columns were trimmed using ZORRO with a probability cutoff of 0.4 as suggested (Wu, et al. 2012). Phylogenetic trees were constructed using RAxML with the best model selected by the program (Stamatakis 2014). Gene families with spurious alignment (e.g. families with Ankyrin or Sel1 repeats) were excluded from the phylogenetic analysis. When possible, each individual tree was rooted using three different rooting methods, rooting with Archaea or Deinococcus as the outgroup or midpoint rooting. Each of the rooted trees was scanned for a bipartition in which the Rickettsiales genes clustered with homologs in the 10 amoeba-associated bacteria using a customized perl script. The species tree of Rickettsiales and the non-α-proteobacterial LGT partners was reconstructed using the 31 bacterial universal marker genes in AMPHORA2 (supplementary fig. S1, Supplementary Material online) (Wu and Scott 2012). LGT events were inferred by reconciling the gene tree with the species tree to minimize the number of the LGT events (maximum parsimony). The same phylogenetic analysis pipeline was applied to identify LGTs between the 19 Rickettsiales genomes and 109 viral genomes.
LGTs between Rickettsiales Endosymbionts and the Amoeba Hosts
We employed a similar phylogenetic analysis to identify LGTs between the Rickettsiales amoeba endosymbionts and the hosts. For each gene of the six endosymbiont genomes, a BLASTP search was performed against all complete bacterial and archaeal genomes and genomes of 12 amoeba species in the supergroups Amoebozoa and Excavata (Dictyostelium discoideum, Dictyostelium fasciculatum, Dictyostelium purpureum, Naegleria fowleri, Naegleria gruberi, Entamoeba dispar, Entamoeba histolytica, Entamoeba invadens, Entamoeba moshkovskii, Entamoeba nuttalli, Polysphondylium pallidum, and Acanthamoeba castellanii) and 27 other eukaryotic representatives selected to represent a broad range of phylogenetic diversity (Wang and Wu 2014, 2015) (supplementary table S1, Supplementary Material online), to infer the timing of LGT events and distinguish LGTs that are specific to amoebae from other bacteria–eukaryote LGTs (e.g. ancient bacteria–eukaryote LGTs via mitochondria). A bacterial gene was retained for further phylogenetic analysis if its top five hits contained an amoebal sequence (e-value cutoff 1e-7). Bacterial genes passing the initial BLASTP search were then clustered into families using the Markov Cluster Algorithm (Enright, et al. 2002), and homologs from all complete bacterial, archaeal, viral, and eukaryotic genomes were retrieved by BLASTP (e-value cutoff 1e-15). Protein sequences were aligned using MAFFT (Katoh, et al. 2002) and trimmed using ZORRO with a probability cutoff of 0.4 as suggested (Wu, et al. 2012). Phylogenetic trees were constructed using RAxML with the best model selected by the program (Stamatakis 2014) and were scanned for a bipartition where the endosymbiont sequences clustered with the amoeba homologs.
Results and Discussion
Amoeba Endosymbiont Genomes Exhibit High Level of Plasticity
Major features of the six genomes are summarized in table 1. In addition to the main chromosomes, the Cca genome has four plasmids, and both Cpa and Cps have one plasmid. The size and coding capacity of the draft Cps genome are comparable with Cca, Cpa, and Cot in the same family with all the 31 bacterial universal marker genes identified (table 1) (Wu and Eisen 2008), indicating that it is unlikely to miss many genes. Remarkably, genomes of Cca, Cpa, and Cps exhibit a high level of plasticity indicated by the presence of numerous mobile genetic elements, transposases, and repetitive regions (table 1). Such mobile genetic elements and repeats likely facilitate the recombination between plasmids and the chromosome, and the integration of laterally acquired plasmid genes into the chromosome.
Table 1.
Main Features of the Six Amoeba Endosymbiont Genomes in This Study
| Cca | Cpa | Cps | Eau | Cot | Cja | |
|---|---|---|---|---|---|---|
| Genome size (bp) | 2,175,773 | 2,485,110 | 2,723,225 | 1,615,277 | 2,848,788 | 2,370,652 |
| Number of contigs | 5 | 2 | 148 | 1 | 20 | 249 |
| GC % | 37.9 | 41.0 | 41.0 | 34.7 | 41.9 | 33.7 |
| Plasmids | 4 | 1 | ≥1 | 0 | NAa | NAa |
| Predicted ORFs | 2,332 | 2,439 | 2,383 | 1,608 | 2,673 | 2,070 |
| ORFs with assigned functions (%) | 56.1 | 56.5 | 53.3 | 65.2 | 50.0 | 59.8 |
| Average ORF length (bp) | 808 | 864 | 793 | 898 | 893 | 934 |
| Percent of genome that is coding | 86.4 | 85.2 | 71.1 | 89.4 | 84.0 | 82.1 |
| Ribosomal RNA operon | 1 | 2 | 2 | 1 | 1 | 1 |
| Transfer RNA | 42 | 42 | 41 | 36 | 40 | 36 |
| Transposase | 17 | 12 | 15 | 9 | 20 | 21 |
| Mobile genetic element | 98 | 140 | 63 | 1 | 33 | 46 |
| Phage genes | 19 | 11 | 9 | 19 | 8 | 5 |
| Percent of genome that is repetitiveb | 8.9 | 4.6 | 4.3 | 1.1 | 0.04 | 6.5 |
| Proteins with eukaryotic domain (Ankyrin, LRR, SEL1, F- or U-Box) | 57 | 68 | 93 | 60 | 118 | 187 |
| Flagellar genes | 5 | 27 | 28 | 29 | 28 | 35 |
a NA due to the incomplete nature of genome assembly.
b Repetitive regions were defined as fragments of at least 50 bp with at least 97% sequence identity.
Conjugation Is a Potential Mechanism for LGTs between the Amoeba Endosymbionts
One prominent feature of the three amoeba endosymbionts (Cca, Cpa, and Cps) is the presence of plasmids in their genomes. Plasmids have been identified in many Rickettsia species such as R. felis (Ogata, et al. 2005), R. buchneri (Gillespie, et al. 2012), R. massiliae (Blanc, et al. 2007), and Rickettsia peacockii (Felsheim, et al. 2009). However, to our knowledge, no plasmid has been reported for obligate amoeba endosymbionts sequenced to date. Finding of the plasmids in these endosymbiont genomes is significant because it suggests that conjugation can potentially mediate LGTs between bacterial species living inside amoebae, as it does in those Rickettsia species.
The chromosome of Cca encodes two complete sets of genes for the F-like tra T4SS, the molecular machinery required for conjugation (supplementary fig. S2A, Supplementary Material online). These genes are located in two syntenic and highly similar gene clusters (82.2% DNA sequence identity), indicative of a recent duplication. Furthermore, the four plasmids of Cca encode several additional proteins involved in conjugation. For example, one gene with a traD domain was found in all four plasmids and was duplicated in pCca2. Also, the four plasmids encode a total of five mobA genes, the relaxase essential for the initiation of plasmid conjugation (Bhattacharjee and Meyer 1993). Accordingly, the origin of transfer (oriT) region was found adjacent to the mobA gene in all four plasmids. We also identified in pCca4 one ORF homologous to traG that is involved in F-pilus assembly (Firth and Skurray 1992). Together with the two complete F-like T4SSs encoded on the main chromosome, we think the four Cca plasmids are most likely conjugative. This provides a mechanistic basis for LGTs between Cca and other amoeba symbionts.
In comparison, the chromosomes of both Cpa and Cps possess a partial tra system with several components missing (supplementary fig. S2A, Supplementary Material online). Based on the gene content, it is not immediately clear whether plasmids in Cpa and Cps are conjugative or not. Phylogenetic analysis of tra genes placed the amoeba endosymbionts with other Rickettsiales as a sister clade to Candidatus Protochlamydia amoebophila and Simkania negevensis (both from phylum Chlamydiae), suggesting an ancient intra-amoebal LGT (supplementary fig. S3, Supplementary Material online).
The plasmids in Cca most likely contribute to the endosymbiont–host interaction, as three of the four plasmids each encodes an operon of the P-like vir T4SS. Unlike F-like T4SS that is known to transfer DNAs only (Lawley, et al. 2003), P-like T4SS is also capable of translocating protein effectors into the host cell (Cascales and Christie 2003; Siamer and Dehio 2015). The three Cca vir T4SS operons exhibit high sequence similarity to each other (≥81.3% DNA sequence identity), indicating that they are products of recent duplications. The operon structures are highly similar to that of the archetypal vir T4SS in Agrobacterium tumefaciens Ti plasmid but different from those of vir T4SSs found previously in other Rickettsiales species [the Rickettsiales vir homolog (rvh) T4SS] (Gillespie, et al. 2016) (supplementary fig. S2B, Supplementary Material online). Consistently, phylogenetic analysis of concatenated vir genes placed Cca as a sister clade to Magnetospirillum and away from other Rickettsiales (supplementary fig. S4, Supplementary Material online). It is possible that Cca retained the ancestral α-proteobacterial P-T4SS, which was subsequently lost in other Rickettsiales and replaced by the rvh operon. Alternatively, Cca could have acquired the vir T4SS via conjugative transfer from non-Rickettsiales species instead of from the common ancestor of Rickettsiales.
In addition to conjugation, we also found signature of possible LGTs by transduction. This is supported by numerous phage genes identified in the amoeba endosymbionts (table 1).
Evidence of Recent Lateral Gene Transfers between Rickettsiales Endosymbionts of Amoeba
There is strong evidence supporting recent LGTs between amoeba endosymbionts. We identified four DNA fragments greater than 2 kb with at least 90% DNA sequence identity among three amoeba endosymbionts in Rickettsiales (Cpa, Cps, and Cot), suggesting that genetic exchange likely occurred very recently among them (table 2). Of them, two large fragments of 8,686 bp and 7,619 bp were highly similar between the chromosomes of Cpa and Cps, with 97.5% and 95.0% DNA sequence identity, respectively, which are significantly higher than the average DNA sequence identity of 87.6% between the homologous regions of the two genomes (one-sample T-test P < 1e-4). At synonymous sites, these two fragments are 95.4% and 87.9% identical, respectively. Using 0.9% DNA sequence divergence at synonymous sites per million years (Ochman, et al. 1999), it is estimated that they diverged ∼5.2 and ∼14.2 Ma. Notably, both fragments are part of integrative and conjugative elements flanked by tRNA genes and mobile genetic elements. Cpa and Cps are currently living in different species of Acanthamoeba (UWC9 and UWET39, respectively). The simplest explanation is that Cpa and Cps exchanged genes when they once coexisted within the same host cell, although it is also possible that they acquired these genes separately through third parties.
Table 2.
DNA Fragments with Evidence of Recent LGTs between the Rickettsiales Endosymbionts of Amoebae
| Sequence Length (bp) | LGT Species | Start Position in Cpa Genome (bp) | DNA Sequence Identity (%) | Genes Encoded (Copy Number) |
|---|---|---|---|---|
| 8,686 | Cpa-Cps | 995, 818 | 97.49 | Phosphopantetheinyl transferase (1), FabD (2), hypothetical protein (1) |
| 7,619 | Cpa-Cps | 389, 575 | 94.99 | LysR-type transcriptional regulator (1), Glu/Leu/Phe/Val dehydrogenase (1), O-methyltransferase (1), low-specificity L-threonine aldolase (1), hypothetical protein (2) |
| 2,850 | Cpa-Cot | 818, 798, 1, 701, 879 | 98.46 | Putative cell wall-associated hydrolase (1) |
| 2,505 | Cpa-Cps | 1, 759, 225 | 98.48 | Patatin-like phospholipase protein (2) |
Lateral Gene Transfers Are Predominantly between Amoeba-Associated Bacteria
The strong evidence of LGT between the Rickettsiales endosymbionts of amoeba prompted us to search LGTs between them and other amoeba-associated bacteria and test the “melting pot” hypothesis. To this end, we performed a BLASTP search of the genomes of six Rickettsiales endosymbionts of acanthamoeba against all complete bacterial and archaeal genomes and identified LGT genes as those that had non α-proteobacterial homologs as their best hits. To include LGTs that happened prior to and during the divergence of the Rickettsiales lineages, we excluded hits belonging to Rickettsiales. As a comparison, we also performed the same analysis for genomes of 13 other Rickettsiales representatives that infect human and animals. We excluded genes with α-proteobacterial best hits because the majority of them will likely be vertically inherited. We note that our approach will miss potential LGTs between Rickettsiales endosymbionts of amoeba and α-proteobacteria.
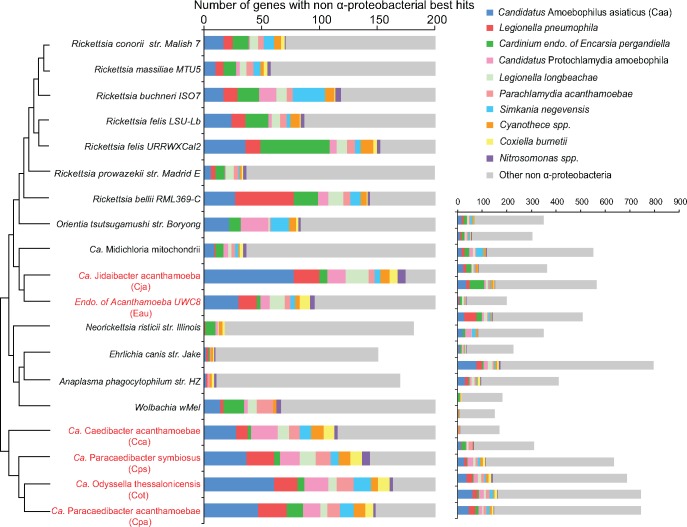
Among the Rickettsiales genomes surveyed, Cja had the largest number of genes that had non α-proteobacterial best hits (796 genes), followed by Cpa (744 genes), Cot (744 genes), Cps (688 genes), and Cca (635 genes) (fig. 1). All of them are endosymbionts of amoebae. A large number of such genes were also identified in R. felis URRWXCal2 (564 genes), R. buchneri (551 genes), and R. bellii (508 genes). Although R. bellii is known to infect amoebae (Ogata, et al. 2006), R. felis and R. buchneri are not. In comparison, a much smaller set of LGT genes were found in genomes of Neorickettsia (182 genes), Ehrlichia (151 genes), Anaplasma (170 genes), and the tick endosymbiont Candidatus Midichloria mitochondrii (226 genes). We ranked the non-α-proteobacterial species by the number of times that they showed up as the best hit in the BLASTP search. Remarkably, we found that the top 10 ranked bacteria are either amoeba-associated symbionts or free-living bacteria that share the same habitats with amoebae, whether we used the six Rickettsiales endosymbionts of amoeba genomes or all 19 Rickettsiales genomes as the query in the BLASTP search (fig. 1, supplementary tables S2 and S3, Supplementary Material online). Five of the top six non-α-proteobacterial species are all amoeba symbionts (in descending order): Caa (Bacteroidetes), Legionella pneumophila (γ-proteobacteria), Candidatus Protochlamydia amoebophila (Chlamydiae), Legionella longbeachae (γ-proteobacteria), and Parachlamydia acanthamoebae (Chlamydiae). Many of them have been suggested to engage in LGTs with members of Rickettsia (e.g. Merhej, et al. 2011; Ogata, et al. 2006; Schmitz-Esser, et al. 2010). Cardinium endosymbiont of Encarsia pergandiella (Bacteroidetes) is closely related to Caa. Interestingly, comparative genomic analysis suggested that their last common ancestor (LCA) was likely an amoeba symbiont (Penz, et al. 2012). Among the four remaining species, S. negevensis (Chlamydiae) and Coxiella burnetii (γ-proteobacteria) are obligate intracellular pathogens capable of infecting amoebae (Kahane, et al. 2001; La Scola and Raoult 2001). Cyanotheces spp. (Cyanobacteria) and Nitrosomonas spp. (β-proteobacteria) are free-living bacteria widely distributed in aquatic habitats such as wastewater in which amoebae are abundant. The predominance of amoeba-associated symbionts or free-living bacteria that cohabit with amoebae as the best hits of the six Rickettsiales endosymbionts of amoeba genomes (fig. 1, supplementary table S2, Supplementary Material online) supports the prediction of the “melting pot” hypothesis that LGTs are mostly between amoeba-associated bacteria.
Fig. 1.
—Predominance of amoeba-associated bacteria in the best hits of Rickettsiales genes. The histogram on the bottom right represents the taxonomic distribution of the non α-proteobacterial best hits (x axis) for genes in the six amoeba endosymbionts (highlighted in red) and the 13 Rickettsiales representatives (y axis). The top 10 ranked non-α-proteobacterial species with the largest number of best hits are shown. The histogram is enlarged and shown at the center.
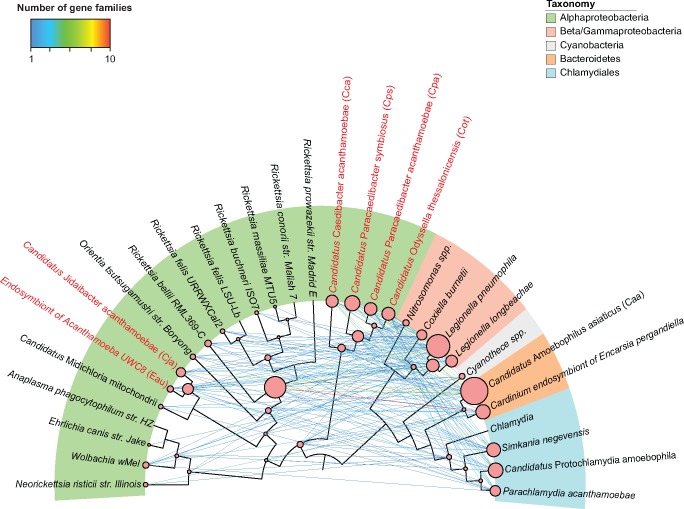
To further investigate the timing of the intra-amoebal LGTs, we reconstructed phylogenetic trees of genes whose best hits were in the top 10 amoeba-associated bacteria. We inferred LGT events by reconciling the gene trees with the species tree of Rickettsiales using maximum parsimony to minimize the number of LGT events. From 274 gene families, we identified a total of 320 LGT events between Rickettsiales and the 10 amoeba-associated bacteria ( supplementary table S4, Supplementary Material online), suggesting intra-amoebal LGTs were pervasive and likely occurred throughout the evolutionary history of Rickettsiales (fig. 2). For example, at least nine gene families, including the ATP/ADP translocase that has been well characterized as a LGT candidate (Schmitz-Esser, et al. 2004), were found to be laterally transferred between the LCA of all Rickettsiales and members of Bacteroidetes, Legionellales, and Chlamydiales. This is consistent with the notion that LCA of Rickettsiales was likely an amoeba endosymbiont and LGTs occurred at a time when it shared a common host with ancestors of Legionella or Chlamydiales (Greub and Raoult 2003). Forty-five LGT events were inferred between the LCA of Rickettsia and other amoeba-associated bacteria, suggesting the LCA of Rickettsia was likely also an amoeba endosymbiont.
Fig. 2.
—Gene transfer network illustrating LGTs between Rickettsiales and distantly related amoeba-associated bacteria. Each edge represents an LGT route between the two nodes. Edge color corresponds to the number of gene families transferred between the two nodes. The size of the node is proportional to the number of laterally transferred gene families involving the nodes. The six amoeba endosymbionts are highlighted in red.
At the species level, four amoeba endosymbionts in the Holosporaceae family, Cps, Cpa, Cot, and Cca, had the largest number of LGT events, with each having at least 23 LGTs with the 10 amoeba-associated bacteria. Sixteen and nine LGTs were found in Cja and Eau in the Candidatus Midichloriaceae family, respectively. In Rickettsiaceae and Anaplasmataceae, R. bellii had the largest number of LGTs (10). In comparison, much fewer LGTs were found involving other Rickettsia species (fig. 2), including R. buchneri and R. felis that have been reported to undergo frequent LGTs (Gillespie, et al. 2012, 2014). Our results and those of Gillespie et al. (2012, 2014) are consistent as our results indicate that most of the LGTs detected in Rickettsia species occurred in their LCA, likely an amoeba endosymbiont, although it is also possible that individual Rickettsia species have exchanged genes with other partners in non-amoeba hosts.
Among the distantly related LGT partners, Caa was the most frequently involved species with 62 LGT events, followed by L. pneumophila and Candidatus Protochlamydia amoebophila. Our results suggest that recurrent LGTs with distantly related amoeba-associated bacteria have significantly shaped the Rickettsiales genomes throughout their evolutionary history.
Laterally Transferred Genes Are Enriched in Amoeba-Symbiont Interactions
We next examined the functions of the genes exchanged with the 10 amoeba-associated bacteria to further understand how LGTs have shaped the biology of Rickettsiales. Functional breakdown of the LGT genes into Cluster of Orthologous Groups (COGs) revealed a significant enrichment of genes important for amoeba–symbiont interactions. Of the 210 gene families with COG assignments, at least 71 families (33.8%) are related to the transport system, antibiotic resistance, stress response, cell signaling (i.e. kinases), or bacterial virulence factors (i.e. hydrolases, lipoproteins) (supplementary table S4, Supplementary Material online). In comparison, very few LGT genes are involved in bacterial central metabolisms such as energy biogenesis. Bacterial transporters are important for the symbionts to exploit host cells and uptake nutrients. We identified a sodium/proline transporter (COG0591), a branched-chain amino acid permease (COG1114), a sodium/alanine transporter (COG1115), a sodium/glutamate symporter (COG1301), and a thiamine transporter (COG3840) that are likely essential for the survival of the endosymbionts because they all depend on the host for amino acids due to the absence of complete de novo amino acid biosynthesis pathways in their genomes. In addition, a large proportion of LGT genes are involved in antibiotic resistance, including a fosA fosfomycin resistance protein (COG0346), a MATE family efflux transporter (COG0534), a rarD drug/metabolite transporter (COG0697), an acrF RND efflux transporter (COG0841), a mexH multi-drug efflux transporter (COG0845), a mccF microcin C7 resistance protein (COG1619), a blaD class D beta-lactamase (COG2602), a bcr2 bicyclomycin resistance protein (COG2814), and an ampD enzyme for beta-lactamase regulation (COG3023). Genetic exchange of these antibiotic resistant genes could be important for the survival of the symbionts within amoebae who are likely exposed to antibiotics in soil and water. We also found evidence of intra-amoebal LGTs for numerous genes related to bacterial virulence and stress responses, such as a patatin-like phospholipase (COG3621), a capM glycosyltransferase (COG0438), and several heat shock proteins (e.g., COG0071 and COG0265). LGT is often thought to confer selective advantages to the recipient organism. The enrichment of genes related to amoeba–symbiont interactions suggests that there is selective pressure favoring a functional gene transfer within amoebae, in which the symbionts frequently exchange strategies of host cell interactions with each other to quickly adapt to their common intracellular habitats.
Lateral Gene Transfers between Amoeba Endosymbionts and Viruses
Giant viruses such as Mimivirus, Megavirus, and Marseillevirus reside within amoebae and have been shown to be involved in the LGTs with intra-amoebal bacteria and the hosts (Boyer, et al. 2009; Filee, et al. 2008; Koonin and Yutin 2010). We performed a similar phylogenomic analysis to identify potential LGTs between the Rickettsiales amoeba endosymbionts and 109 viral genomes. BLASTP search identified a total of 34 genes from 11 Rickettsiales genomes, including 5 amoeba endosymbionts, with best hits in 11 viral genomes (supplementary fig. S5A and table S5, Supplementary Material online). Of them, Cja and Orientia tsutsunamushi had the largest number of best hits to viruses (12 and 10, respectively). From the viral side, Acanthamoeba polyphaga mimivirus has the most number of best hits to Rickettsiales (nine genes). Phylogenetic analysis identified an endonuclease and three hypothetical proteins that were likely laterally transferred between Cca, Cpa, and Cot and two viral lineages (supplementary fig. S5B and table S5, Supplementary Material online).
LGTs between Amoeba Host and Endosymbionts
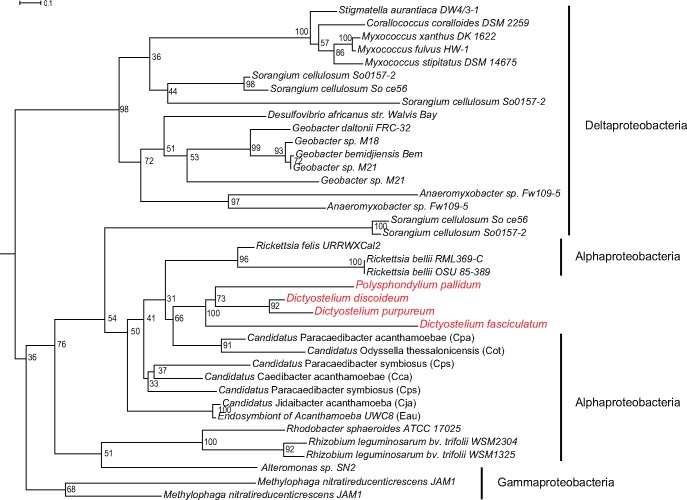
Not only can amoebae serve as a fertile ground where endosymbionts exchange genetic materials with each other, they could also participate in such LGTs (Moliner, et al. 2009). This is because foreign bacterial DNAs are also readily accessible to genomes of amoeba, unlike in animals where sequestered germline cells present an obstacle to LGT, as proposed in the weak-link model of LGT (Huang 2013). To test this hypothesis, we searched for potential inter-kingdom LGT events between the Rickettsiales endosymbionts and their amoeba hosts using a phylogenomic approach. We found three gene families with evidence of LGTs between the Rickettsiales symbionts and their host: a ferritin-like protein and two hypothetical proteins. Ferritins are a family of iron storage proteins that organisms use to maintain iron homeostasis. They remove excess ferrous ions from the cytoplasm in prevention of cellular damage caused by oxidative stress (Harrison and Arosio 1996). Ferritins are found in almost all living organisms, and bacterial ferritins (ferritin-like proteins) are homologous but distantly related to ferritins found in eukaryotes. Phylogenetic analysis of ferritin gene placed four amoebae of the Dictyosteliidae family (D. discoideum, D. purpureum, D. fasciculatum, and P. pallidum) within Rickettsiales as a sister clade with Cpa and Cps (fig. 3), indicating a LGT from the endosymbionts to the host, although the exact donor of the gene could not be unambiguously determined. Interestingly, none of the four amoebae possesses the eukaryotic version of ferritin gene, although the gene is found in several other amoeba genomes such as N. gruberi and A. castellanii. This suggests an orthologous gene replacement event where the host ferritin gene in the LCA of Dictyosteliidae has been replaced by the endosymbiont bacterial ferritin gene. No intron sequence was found for the ferritin gene in the four amoebae. For the two hypothetical genes showing LGT evidence, one contains a small MutS-related (smr) domain and is exclusively shared by Cca, Caa, and N. gruberi (BLASTP e-value ≤ 3e-48). It is not immediately clear whether the protein is functional in DNA mismatch repair. The other gene contains a NUC1 endonuclease domain and is exclusively shared by Cps and D. purpureum (BLASTP e-value = 7e-53).
Fig. 3.
—Maximum likelihood phylogeny for the ferritin-like protein showing LGT between Rickettsiales endosymbionts and four amoeba species. The tree was rooted using midpoint rooting. Bootstrap values for internal nodes are shown beside them. The four amoeba species are highlighted in red.
Given that amoeba’s genomes are much more accessible to bacterial DNAs than those of multicellular eukaryotes, it is remarkable that very few bacteria-to-amoebae LGTs were identified. Several factors could contribute to the deficiency of amoeba–symbiont LGTs. First, the transcription and translation machinery differs considerably between bacteria and eukaryotes, making it difficult for the transferred genes to be functionally integrated. In principle, a successfully transferred bacterial gene needs to acquire a nuclear promoter before it can be transcribed and retained in the host genome. Acquisition of eukaryotic features such as promoter and intron has been observed for genes transferred from Buchnera to the pea aphid host (Nikoh, et al. 2010). Second, eukaryotic genes are usually part of a larger and more complex network than their bacterial counterparts. Known as the complexity hypothesis, it has been shown that such genes with higher network connectivity are more recalcitrant to LGTs (Cohen, et al. 2011). Also, the scarce sampling of eukaryotic genomes and removal of sequences highly similar to bacteria prior to eukaryotic genome assembly could contribute to the paucity of amoeba–symbiont LGTs detected (Danchin 2016). Lastly, because we only focused on LGTs between Rickettsiales endosymbionts and eukaryotes in this study, we excluded LGTs from other bacterial lineages to eukaryotes, such as those reported in (Alsmark, et al. 2013).
Conclusion
In this study, we tested the “melting pot” hypothesis using the Rickettsiales as our model system. As predicted by the “melting pot” hypothesis, we showed that Rickettsiales symbionts living inside amoebae participated in extensive LGTs with other distantly related amoeba-associated bacteria. We also showed that intra-amoebal LGT genes were enriched in amoeba–symbiont interactions and likely conferred fitness benefits to the endosymbionts. Our results provide strong support to the “melting pot” hypothesis and show that LGTs have and continue to play an important role in shaping the Rickettsiales evolution.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This research was funded by an award from Thomas F. & Kate Miller Jeffress Memorial Trust to MW.
Literature Cited
- Albert-Weissenberger C, Cazalet C, Buchrieser C.. 2007. Legionella pneumophila—a human pathogen that co-evolved with fresh water protozoa. Cell Mol Life Sci. 644:432–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsmark C, et al. 2013. Patterns of prokaryotic lateral gene transfers affecting parasitic microbial eukaryotes. Genome Biol. 142:R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann R, et al. 1997. Obligate intracellular bacterial parasites of acanthamoebae related to Chlamydia spp. Appl Environ Microbiol. 631:115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertelli C, Greub G.. 2012. Lateral gene exchanges shape the genomes of amoeba-resisting microorganisms. Front Cell Infect Microbiol. 2:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee MK, Meyer RJ.. 1993. Specific binding of MobA, a plasmid-encoded protein involved in the initiation and termination of conjugal DNA transfer, to single-stranded oriT DNA. Nucleic Acids Res. 2119:4563–4568.http://dx.doi.org/10.1093/nar/21.19.4563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birtles RJ, et al. 2000. ′Candidatus Odyssella thessalonicensis′ gen. nov., sp. nov., an obligate intracellular parasite of Acanthamoeba species. Int J Syst Evol Microbiol. 50 Pt 1:63–72. [DOI] [PubMed] [Google Scholar]
- Birtles RJ, Rowbotham TJ, Storey C, Marrie TJ, Raoult D.. 1997. Chlamydia-like obligate parasite of free-living amoebae. Lancet 3499056:925–926. [DOI] [PubMed] [Google Scholar]
- Blanc G, et al. 2007. Lateral gene transfer between obligate intracellular bacteria: evidence from the Rickettsia massiliae genome. Genome Res. 1711:1657–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordenstein SR, Wernegreen JJ.. 2004. Bacteriophage flux in endosymbionts (Wolbachia): infection frequency, lateral transfer, and recombination rates. Mol Biol Evol. 2110:1981–1991. [DOI] [PubMed] [Google Scholar]
- Boyer M, et al. 2009. Giant Marseillevirus highlights the role of amoebae as a melting pot in emergence of chimeric microorganisms. Proc Natl Acad Sci U S A. 10651:21848–21853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascales E, Christie PJ.. 2003. The versatile bacterial type IV secretion systems. Nat Rev Microbiol. 12:137–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarholm M. 1981. Protozoan grazing of bacteria in soil-impact and importance. Microb Ecol. 74:343–350. [DOI] [PubMed] [Google Scholar]
- Cohen O, Gophna U, Pupko T.. 2011. The complexity hypothesis revisited: connectivity rather than function constitutes a barrier to horizontal gene transfer. Mol Biol Evol. 284:1481–1489. [DOI] [PubMed] [Google Scholar]
- Cox R, Mason-Gamer RJ, Jackson CL, Segev N.. 2004. Phylogenetic analysis of Sec7-domain-containing Arf nucleotide exchangers. Mol Biol Cell. 154:1487–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danchin EG. 2016. Lateral gene transfer in eukaryotes: tip of the iceberg or of the ice cube? BMC Biol. 141:101.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. 1998. Profile hidden Markov models. Bioinformatics 149:755–763.http://dx.doi.org/10.1093/bioinformatics/14.9.755 [DOI] [PubMed] [Google Scholar]
- Enright AJ, Van Dongen S, Ouzounis CA.. 2002. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 307:1575–1584.http://dx.doi.org/10.1093/nar/30.7.1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsheim RF, Kurtti TJ, Munderloh UG.. 2009. Genome sequence of the endosymbiont Rickettsia peacockii and comparison with virulent Rickettsia rickettsii: identification of virulence factors. PLoS One 412:e8361.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filee J, Pouget N, Chandler M.. 2008. Phylogenetic evidence for extensive lateral acquisition of cellular genes by Nucleocytoplasmic large DNA viruses. BMC Evol Biol. 8:320.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth N, Skurray R.. 1992. Characterization of the F plasmid bifunctional conjugation gene, traG. Mol Genet. 2321:145–153. [DOI] [PubMed] [Google Scholar]
- Fritsche TR, et al. 1999. In situ detection of novel bacterial endosymbionts of Acanthamoeba spp. phylogenetically related to members of the order Rickettsiales. Appl Environ Microbiol. 651:206–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsche TR, et al. 2000. Phylogenetic diversity among geographically dispersed Chlamydiales endosymbionts recovered from clinical and environmental isolates of Acanthamoeba spp. Appl Environ Microbiol. 666:2613–2619.http://dx.doi.org/10.1128/AEM.66.6.2613-2619.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiades K, Madoui MA, Le P, Robert C, Raoult D.. 2011. Phylogenomic analysis of Odyssella thessalonicensis fortifies the common origin of Rickettsiales, Pelagibacter ubique and Reclimonas americana mitochondrion. PLoS One 69:e24857.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie JJ, et al. 2014. Genomic diversification in strains of Rickettsia felis Isolated from different arthropods. Genome Biol Evol. 71:35–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie JJ, et al. 2012. A Rickettsia genome overrun by mobile genetic elements provides insight into the acquisition of genes characteristic of an obligate intracellular lifestyle. J Bacteriol. 1942:376–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie JJ, et al. 2016. The Rickettsia type IV secretion system: unrealized complexity mired by gene family expansion. Pathog Dis. 74:doi: 10.1093/femspd/ftw058http://dx.doi.org/10.1093/femspd/ftw058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez G, et al. 2011. Insight into cross-talk between intra-amoebal pathogens. BMC Genomics 12:542.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greub G, Raoult D.. 2003. History of the ADP/ATP-translocase-encoding gene, a parasitism gene transferred from a Chlamydiales ancestor to plants 1 billion years ago. Appl Environ Microbiol. 699:5530–5535.http://dx.doi.org/10.1128/AEM.69.9.5530-5535.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison PM, Arosio P.. 1996. The ferritins: molecular properties, iron storage function and cellular regulation. Biochim Biophys Acta. 12753:161–203. [DOI] [PubMed] [Google Scholar]
- Horn M, Fritsche TR, Gautom RK, Schleifer KH, Wagner M.. 1999. Novel bacterial endosymbionts of Acanthamoeba spp. related to the Paramecium caudatum symbiont Caedibacter caryophilus. Environ Microbiol. 14:357–367. [DOI] [PubMed] [Google Scholar]
- Horn M, et al. 2001. Members of the cytophaga-flavobacterium-bacteroides phylum as intracellular bacteria of acanthamoebae: proposal of ′Candidatus Amoebophilus asiaticus′. Environ Microbiol. 37:440–449. [DOI] [PubMed] [Google Scholar]
- Horn M, et al. 2000. Neochlamydia hartmannellae gen. nov., sp. nov. (Parachlamydiaceae), an endoparasite of the amoeba Hartmannella vermiformis. Microbiology 1465:1231–1239. [DOI] [PubMed] [Google Scholar]
- Huang J. 2013. Horizontal gene transfer in eukaryotes: the weak-link model. Bioessays 3510:868–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahane S, Dvoskin B, Mathias M, Friedman MG.. 2001. Infection of Acanthamoeba polyphaga with Simkania negevensis and S. negevensis survival within amoebal cysts. Appl Environ Microbiol. 6710:4789–4795.http://dx.doi.org/10.1128/AEM.67.10.4789-4795.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T.. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 3014:3059–3066.http://dx.doi.org/10.1093/nar/gkf436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Yutin N.. 2010. Origin and evolution of eukaryotic large nucleo-cytoplasmic DNA viruses. Intervirology 535:284–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Scola B, Raoult D.. 2001. Survival of Coxiella burnetii within free-living amoeba Acanthamoeba castellanii. Clin Microbiol Infect. 72:75–79.http://dx.doi.org/10.1046/j.1469-0691.2001.00193.x [DOI] [PubMed] [Google Scholar]
- Lawley TD, Klimke WA, Gubbins MJ, Frost LS.. 2003. F factor conjugation is a true type IV secretion system. FEMS Microbiol Lett. 2241:1–15. [DOI] [PubMed] [Google Scholar]
- Merhej V, Notredame C, Royer-Carenzi M, Pontarotti P, Raoult D.. 2011. The rhizome of life: the sympatric Rickettsia felis paradigm demonstrates the random transfer of DNA sequences. Mol Biol Evol. 2811:3213–3223. [DOI] [PubMed] [Google Scholar]
- Moliner C, Fournier PE, Raoult D.. 2010. Genome analysis of microorganisms living in amoebae reveals a melting pot of evolution. FEMS Microbiol Rev. 343:281–294. [DOI] [PubMed] [Google Scholar]
- Moliner C, Raoult D, Fournier PE.. 2009. Evidence of horizontal gene transfer between amoeba and bacteria. Clin Microbiol Infect. 15:178–180. [DOI] [PubMed] [Google Scholar]
- Molmeret M, Horn M, Wagner M, Santic M, Abu Kwaik Y.. 2005. Amoebae as training grounds for intracellular bacterial pathogens. Appl Environ Microbiol. 711:20–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira D, Brochier-Armanet C.. 2008. Giant viruses, giant chimeras: the multiple evolutionary histories of Mimivirus genes. BMC Evol Biol. 8:12.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikoh N, et al. 2010. Bacterial genes in the aphid genome: absence of functional gene transfer from Buchnera to its host. PLoS Genet. 62:e1000827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H, Elwyn S, Moran NA.. 1999. Calibrating bacterial evolution. Proc Natl Acad Sci U S A. 9622:12638–12643.http://dx.doi.org/10.1073/pnas.96.22.12638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata H, et al. 2006. Genome sequence of Rickettsia bellii illuminates the role of amoebae in gene exchanges between intracellular pathogens. PLoS Genet. 25:e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata H, et al. 2005. The genome sequence of Rickettsia felis identifies the first putative conjugative plasmid in an obligate intracellular parasite. PLoS Biol. 38:e248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penz T, et al. 2012. Comparative genomics suggests an independent origin of cytoplasmic incompatibility in Cardinium hertigii. PLoS Genet. 810:e1003012.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raoult D, et al. 2004. The 1.2-megabase genome sequence of Mimivirus. Science 3065700:1344–1350. [DOI] [PubMed] [Google Scholar]
- Schmitz-Esser S, et al. 2004. ATP/ADP translocases: a common feature of obligate intracellular amoebal symbionts related to Chlamydiae and Rickettsiae. J Bacteriol. 1863:683–691.http://dx.doi.org/10.1128/JB.186.3.683-691.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Esser S, et al. 2010. The genome of the amoeba symbiont “Candidatus Amoebophilus asiaticus” reveals common mechanisms for host cell interaction among amoeba-associated bacteria. J Bacteriol. 1924:1045–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Esser S, et al. 2008. Diversity of bacterial endosymbionts of environmental acanthamoeba isolates. Appl Environ Microbiol. 7418:5822–5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz F, et al. 2016. A Rickettsiales symbiont of amoebae with ancient features. Environ Microbiol. 188:2326–2342. [DOI] [PubMed] [Google Scholar]
- Siamer S, Dehio C.. 2015. New insights into the role of Bartonella effector proteins in pathogenesis. Curr Opin Microbiol. 23:80–85. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 309:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szokoli F, et al. 2016. Disentangling the taxonomy of rickettsiales and description of two novel symbionts (“Candidatus Bealeia paramacronuclearis” and “Candidatus Fokinia cryptica”) Sharing the Cytoplasm of the Ciliate Protist Paramecium biaurelia. Appl Environ Microbiol. 8224:7236–7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas V, McDonnell G.. 2007. Relationship between mycobacteria and amoebae: ecological and epidemiological concerns. Lett Appl Microbiol. 454:349–357. [DOI] [PubMed] [Google Scholar]
- Wang Z, Wu M.. 2015. An integrated phylogenomic approach toward pinpointing the origin of mitochondria. Sci Rep. 5:7949.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Wu M.. 2014. Phylogenomic reconstruction indicates mitochondrial ancestor was an energy parasite. PLoS One 910:e110685.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Chatterji S, Eisen JA.. 2012. Accounting for alignment uncertainty in phylogenomics. PLoS One 71:e30288.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Eisen JA.. 2008. A simple, fast, and accurate method of phylogenomic inference. Genome Biol. 910:R151.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Scott AJ.. 2012. Phylogenomic analysis of bacterial and archaeal sequences with AMPHORA2. Bioinformatics 287:1033–1034. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.