Abstract
Neanderthals contributed genetic material to modern humans via multiple admixture events. Initial admixture events presumably occurred in Western Asia shortly after humans migrated out of Africa. Despite being a focal point of admixture, earlier studies indicate lower Neanderthal introgression rates in some Western Asian populations as compared with other Eurasian populations. To better understand the genome-wide and phenotypic impact of Neanderthal introgression in the region, we sequenced whole genomes of nine present-day Europeans, Africans, and the Western Asian Druze at high depth, and analyzed available whole genome data from various other populations, including 16 genomes from present-day Turkey. Our results confirmed previous observations that contemporary Western Asian populations, on an average, have lower levels of Neanderthal-introgressed DNA relative to other Eurasian populations. Modern Western Asians also show comparatively high variability in Neanderthal ancestry, which may be attributed to the complex demographic history of the region. We further replicated the previously described depletion of putatively functional sequences among Neanderthal-introgressed haplotypes. Still, we find dozens of common Neanderthal-introgressed haplotypes in the Turkish sample associated with human phenotypes, including anthropometric and metabolic traits, as well as the immune response. One of these haplotypes is unusually long and harbors variants that affect the expression of members of the CCR gene family and are associated with celiac disease. Overall, our results paint a complex first picture of the genomic impact of Neanderthal introgression in the Western Asian populations.
Keywords: Anatolia, celiac disease, malaria, metabolism, immunity, genetic anthropology
Introduction
Recent studies have shown that archaic hominins contributed genetic material to modern humans (Green et al. 2010; Hammer et al. 2011; Reich et al. 2010; Xu et al. 2017). The origin and impact of these introgressions vary geographically and happened through multiple instances. For example, Neanderthals contributed genetic material to all Eurasian peoples, possibly through a single introgression event in the Middle East during the out-of-Africa migrations ∼50,000–60,000 years ago (Green et al. 2010). Follow-up analyses have shown evidence for a second, smaller introgression event affecting only Asian groups (Wall et al. 2013). More recent studies have also identified additional geography-specific introgression events from other archaic hominins to ancestors of contemporary human populations (Meyer et al. 2012; Hsieh et al. 2016; Vernot et al. 2016). As such, all modern humans from outside-of-Africa are estimated to carry 1–3% Neanderthal DNA in their genomes (Green et al. 2010; Meyer et al. 2012; Prüfer et al. 2014). Moreover, haplotype level scrutinization of these admixture events revealed a significant depletion of coding sequences among haplotypes admixed from Neanderthals, suggesting strong negative selection acting on these sequences (Sankararaman et al. 2014; Vernot and Akey 2014; Harris and Nielsen 2016; Juric et al. 2016). In contrast, a small but measurable number of introgressed haplotypes have been adaptively maintained in the human population (Huerta-Sánchez et al. 2014; Dannemann et al. 2016; Gittelman et al. 2016; Quach et al. 2016; Racimo et al. 2017). In addition, recent studies have now shown that Neanderthal-introgressed haplotypes might affect the expression levels of multiple genes (Dannemann et al. 2017; McCoy et al. 2017).
Based on the current data, it has been suggested that the first and most significant introgression from Neanderthals occurred in Western Asia (Green et al. 2010; Prüfer et al. 2014). Specifically, contemporary East and West Eurasian genomes share more Neanderthal alleles than sub-Saharan African genomes do (Vernot and Akey 2014). This observation is consistent with an introgression event in Western Asia, after the population ancestral to contemporary West and East Eurasians migrated out of Africa, but had not yet split into two isolated populations. However, little is known about the distribution and functional significance of Neanderthal introgression in contemporary Western Asian populations, except for a handful of studies (reviewed in Taskent and Gokcumen 2017). Notably, a recent study found lower proportions of Neanderthal ancestry in ancient genomes from the Middle East (Lazaridis et al. 2016). This study further identified high levels of basal Eurasian ancestry (Lazaridis et al. 2014) in these ancient West Asian genomes, which was negatively correlated with Neanderthal ancestry, suggesting that the hypothetical basal Eurasian lineage carried lower levels of Neanderthal ancestry than other ancestral Eurasian lineages (Lazaridis et al. 2016). The degree of basal Eurasian ancestry could also explain variation in Neanderthal ancestry among present-day West Eurasian genomes. For instance, a high level of basal Eurasian or sub-Saharan African ancestry could underlie the observation that there is a relatively low proportion of Neanderthal ancestry in a present-day Qatari Bedouin population as compared with European and some other Middle Eastern populations (Rodriguez-Flores et al. 2016).
In this study, we investigate whether the unique population history of the region affected the distribution of Neanderthal introgression among Western Asian populations, concentrating on the Druze, a small, and ethnically homogenous closed population of the Levant, and the more diverse population of modern-day Turkey. Western Asia is especially interesting as the region has been central to major demographic events since the early Neolithic (Yunusbayev et al. 2015; Kılınç et al. 2016). Moreover, the distribution of genetic variation in Western Asia may be different than what has repeatedly been observed for European populations, that is, a strong correlation with geography consistent with an isolation-by-distance model (Novembre et al. 2008). Specifically, recent inbreeding, migration, and local isolation may create deviations in the clinal distribution of genetic variation in this region (Gokcumen et al. 2011; Scott et al. 2016), which may give rise to considerable heterogeneity in the levels of Neanderthal introgression among populations.
Materials and Methods
For this study, we used data from 1000 Genomes Project Phase 1 (1000 Genomes Project Consortium 2012), Turkish Genome Research Project (Alkan et al. 2014) and Human Origins data sets (Lazaridis et al. 2014). In addition, we compiled a dataset of ten samples. Specifically, we sequenced 9 genomes: two individuals from Finnish population (with European ancestry), one individual from CEPH population (Utah residents with Northern and Western European ancestry), three individuals from Druze population (from Lebanon and Syria, with Western Asian ancestry), and three individuals from Mbuti Pygmy population (with Central African ancestry), all of which were purchased from Coriell Institute for Medical Research (https://www.coriell.org/). In addition, we also included previously sequenced CEPH sample NA12878 in our variant calling pipeline and compiled a vcf file from all 10 genomes. (https://www.coriell.org/; last accessed November 13, 2017). The samples and the populations used in this study are summarized in supplementary table S1, Supplementary Material online.
The library preparation was conducted by New York Genome Center core facility using standard procedures (TruSeqDNA Nano). Each sample was sequenced to ∼30× with 150-bp paired-end sequences using an Illumina HiSeq X platform. Duplicate reads were removed by Picard and BWA was used to map the sequences to Hg19 (Ghr37). The variation calls were done by GATK (McKenna et al. 2010). All mapping and variant calling was done by standard procedures in New York Genome Center. BAM files are available at Short Read Archive and the joint genotyping vcf file can be found at https://gokcumenlab.org/.
The expression quantitative loci information for rs10540 was downloaded from GTEX (The GTEx Consortium 2015). All the bioinformatics analyses were done using publically available software and custom scripts, which can be found here (https://github.com/taskent/W.Asia-Neanderthal-Introgression/blob/master/Sstar.py; last accessed November 13, 2017). The details of our methodology is described in supplementary methods, Supplementary Material online.
Results
Variation in Neanderthal Ancestry within and among Western Asian Populations
To determine the Neanderthal introgression patterns in Western Asian populations, we first set out to determine relative Neanderthal ancestry levels in two distinct populations: the Druze and the population of present-day Turkey, as compared with other Eurasian populations. To do this, we first analyzed the Human Origins (HO) data set (Lazaridis et al. 2014) and calculated D-statistics of the form D(HO Druze or Turkish, HO Eurasian or African; Neanderthal, Chimpanzee) for various population combinations.
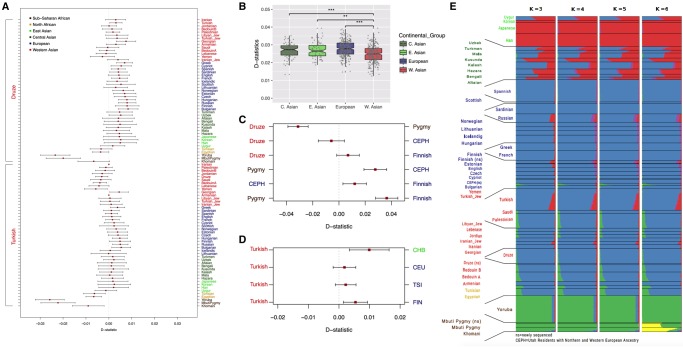
Our results showed that these two present-day Western Asian populations consistently carry lower proportions of Neanderthal introgression relative to Europeans, Central Asians, and East Asians (fig. 1A). This observation is concordant with the trends documented globally in present-day populations (Mallick et al. 2016) as well as using ancient genomes (Lazaridis et al. 2016). We found the same trend comparing individual Neanderthal ancestry estimates of the form D(Test, Yoruba; Neanderthal, Chimp) among West Asian and other Eurasian groups. The difference in estimated admixture proportions between Western Asians and other Eurasians is systematic and significantly lower for Western Asians (P < 0.01, Wilcoxon Rank Sum Test, fig. 1B).
Fig. 1.
—Neanderthal ancestry proportions and population structure in Eurasian human populations. The results were color coded according to the geographic regions of origin for all the panels: Africa: Dark Brown; North Africa: Light Brown; East Asia: Light Green; Central Asia: Dark Green; Europe: Dark Blue; Western Asia: Red. (A) Distribution of D-statistics for the Human Origins (HO) data set calculated in the form D(Test, Druze; Neanderthal, Chimpanzee), and D(Test, Turkish; Neanderthal, Chimpanzee). Results show the Neanderthal ancestry proportions in the Druze and Turkish populations relative to various Test populations included in the Human Origins (HO) data set (Lazaridis et al. 2014). Approximately 30,000 single nucleotide polymorphisms that are derived in the Neanderthal genome were used in each comparison (supplementary table S1, Supplementary Material online). Results for other comparisons can be found in supplementary table S1, Supplementary Material online. (B) Comparison of the differences in D-statistics between continental groups. The data in each boxplot are D-statistics calculated for each sample in that continental group available from the HO data set. The D-statistics were calculated in the form D(Test, Yoruba; Neanderthal, Chimpanzee). (C) D-statistics calculated for the Turkish samples sequenced in Turkish Genome Project as compared with Eurasian populations sequenced in 1,000 Genomes Project. Distribution of D-statistics calculated in the form D(Test, TGP; Neanderthal, Chimpanzee). Approximately 100,000 polymorphic transversions that are derived in the Neanderthal genome were used in each of these comparisons (supplementary table S1, Supplementary Material online). Error bars show two standard deviations around the mean (Z = 2). (D) The D-statistic comparisons between the samples sequenced in this study. The sequenced samples are from individuals of Druze (n = 3), Pigmy (n = 3), Fin (n = 2), and Central European (n = 2) ancestry (n = 10). D-statistics were calculated in the form D(Test1, Test2; Neanderthal, Chimpanzee). Approximately 140,000 polymorphic transversions that are derived in Neanderthal genome were used in each of these comparisons (supplementary table S1, Supplementary Material online). Error bars show two standard deviations around the mean (Z = 2). (E) ADMIXTURE analysis results calculated using the Human Origins data set (Lazaridis et al. 2014). Each row shows ancestry components estimated using different (k = 3, 4, 5, 6) number of clusters. Ancestry proportions were calculated for 746 individuals included in the 43 populations in the Human Origins data set.
Even though lower Neanderthal ancestry is a consistent trend for Western Asian populations, it also shows noticeable variation. To determine whether this variation originates from within- or between-population differences, or both, we calculated D-statistics for each individual within a given population D(Individual [Test], Yoruba; Neanderthal, Chimp) (supplementary fig. S1, Supplementary Material online). Indeed, we observed that both within- and between population variation of D-statistics were higher among Western Asian populations as compared with other continental groups. For example, the Druze has lower levels of Neanderthal ancestry as compared with European, Central Asian, and East Asian populations. However, this population also shows a higher level of within-population variation in Neanderthal ancestry. We further noticed two distinct groups of Western Asian populations with respect to Neanderthal ancestry that they carry, populations associated with the Arabian peninsula showed conspicuously low Neanderthal ancestry as compared with other Eurasian populations, whereas those from northern and eastern regions of Western Asia showed only slightly lower levels of Neanderthal ancestry as compared with other Eurasians (supplementary fig. S1, Supplementary Material online). This is concordant with the recent observation that the Qatari populations of Bedouin ancestry, but not all Middle Easterners, showed lower levels of Neanderthal ancestry as compared with European, Central Asian, and East Asian populations (Rodriguez-Flores et al. 2016). Overall, our results showed that D-statistics between pairs of Western Asian populations are more variable than D-statistics between pairs of European or Asian populations (supplementary table S1, Supplementary Material online). We further quantified these observations and calculated an ∼42% increase in the standard deviation of D-statistic values among Western Asian populations, compared with the same values calculated among European populations, a significant difference as assessed by random permutations (P < 0.001, supplementary fig. S2, Supplementary Material online).
To validate our findings with higher power, we used two whole genome sequencing data sets (1000 Genomes Project Consortium 2012; Alkan et al. 2014). First, we calculated similar D-statistics with published Turkish genomes of high coverage, which we merged with the 1000 Genomes Phase I data set. This indicated lower Neanderthal ancestry in the Turkish population relative to other Eurasian populations (fig. 1C), as we had observed earlier in Turkish genomes in Human Origins data set. Second, we sequenced three Druze, three European, and three Pygmy genomes all with high coverage (>30×) using (∼150 bp) paired-end reads. The library preparation, sequencing platform, mapping, and calling algorithms for these genomes are identical to avoid any technical biases (see supplementary methods, Supplementary Material online). Unexpectedly, the sequenced Druze individuals were found to have similar levels of Neanderthal ancestry as European individuals (fig. 1D), in contrast to lower levels found using the 39 independent Druze individuals of the Human Origins data set.
To rule out sample mix up and further investigate the source of this variation in Druze, we merged the data sets using common variants genotyped in our data set and the Human Origins data set. ADMIXTURE analysis on this merged data set confirmed that the three Druze individuals sequenced had similar ancestry components as the Druze in the Human Origins data set (fig. 1E). Using the same merged data set, we also conducted individual Neanderthal ancestry estimate comparisons (supplementary fig. S3A, Supplementary Material online). This analysis showed that the D-statistics from three Druze individuals that we sequenced in this study actually falls within the distribution of the variation of D-statistics observed in Druze from the Human Origins data set (supplementary fig. S3B, Supplementary Material online). Based on these, we conclude that sampling bias is the main source of the observed disparity between data sets, which marks the higher level of variation in Neanderthal ancestry within and among Western Asian populations.
It is plausible that immigration, isolation, and inbreeding (Scott et al. 2016) have created genetic structure and heterogeneity within and across Western Asian populations. Heterogeneity especially with regards to varying levels of sub-Saharan or basal Eurasian ancestry may explain the elevated variation in allele sharing with Neanderthals among Western Asian genomes. This scenario would be consistent with the results of a previous study that found different levels of Neanderthal ancestry among three Qatari populations (Rodriguez-Flores et al. 2016), possibly due to differences in their levels of sub-Saharan and basal Eurasian ancestries (Scott et al. 2016).
To investigate this notion, we studied ancestral components in the Human Origins data set genomes using ADMIXTURE (Alexander et al. 2009). This revealed conspicuous heterogeneity among Western Asian populations (fig. 1E), with varying levels of African and Asian contributions within and among Western Asian populations (e.g., higher East Asian ancestry in Turkish genomes, and higher African ancestry in Palestinian genomes, compared with the Druze). We then tested the hypothesis that Neanderthal ancestry estimated for Western Asian individuals would be negatively correlated with their estimated sub-Saharan ancestral components. Testing this within each population, we found no significant negative correlation except for Yemenis (P < 0.05, Spearman nonparametric correlation test, supplementary table S2, Supplementary Material online). However, when we pooled data for each continental group and compared the sub-Saharan component across all genomes within that group with the estimated Neanderthal ancestry in the same genomes, we found a highly significant negative correlation for Western Asia (rho=−0.234, P < 0.0002; supplementary table S2, Supplementary Material online). We were able to see the same significant trend when we compared the average sub-Saharan component in each Western Asian population with the average Neanderthal ancestry in the same population across populations (rho=−0.739, P < 0.003, supplementary fig. S4 and table S2, Supplementary Material online). As such, our results support the notion that varying levels of sub-Saharan ancestry among Western Asian genomes contributed to the observed differences in Neanderthal ancestry among these populations.
Neanderthal-Introgressed Haplotypes in the Turkish Population
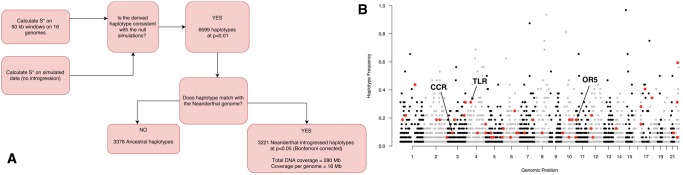
Next, we aimed to delineate the genomic impact of Neanderthal-introgressed haplotypes in Western Asian populations. As a case example, we focused on the 16 present-day Turkish individuals for whom we have access to high-quality genomes (due to sample size restrictions we could not include the Druze individuals in this analysis). We followed the S*-based pipeline recently fine-tuned for identifying introgressed haplotypes (Vernot and Akey 2014). Briefly, we determined putatively introgressed haplotypes in 16 Turkish genomes (i.e., 32 sets of phased chromosomes), as well as in 16 Western European and in 16 East Asian genomes for comparative purposes (fig. 2A and Supplementary Material online). Briefly, using a combination of empirical analyses and simulation-based modeling, this pipeline identifies stretches of DNA carrying unusually high numbers of proximate, derived single nucleotide variants present in Eurasian genomes, but not found in sub-Saharan African genomes (see supplementary methods, Supplementary Material online, for details).
Fig. 2.
—S* statistics workflow and haplotypes. (A) S* analysis workflow. The boxes show each major computational step to determine Neanderthal-introgressed haplotypes. The number of haplotypes that we found in our pipeline at each step was indicated within the respective boxes. (B) Distribution of Neanderthal-introgressed haplotypes detected by S* over the Turkish genomes (n = 16) sequenced for the Turkish Genome Project (TGP) (Alkan et al. 2014). The x-axis shows the chromosomal locations of the haplotypes. Y-axis indicates the allele frequency observed in the Turkish sample (n = 16 individuals). Haplotypes labeled by red show haplotypes carrying GWAS variants where the Neanderthal carries the derived allele. Arrows show the haplotypes carrying C-C motif chemokine receptor (CCR) family, toll-like receptor (TLR-1, TLR-6, TLR-10), and olfactory receptor-5 (OR5) family genes.
Using our pipeline, we identified 6,599 derived haplotypes in 16 Turkish genomes with an average size of ∼72 kb (supplementary table S3, Supplementary Material online). Because the S* statistic does not consider information from Neanderthal genomes, we further refined our data set by probabilistically categorizing the derived haplotypes into those that harbor higher than expected numbers of derived Neanderthal alleles (introgressed), and those do not (ancestral) (see Supplementary Material online for details). Not surprisingly, we found that 3,378 (∼51%) of these divergent haplotypes are not enriched in Neanderthal derived alleles and are thus likely remnants of the ancestral genetic structure in Africa, rather than an introgression from Neanderthals (Yang et al. 2012).
The remaining 3,221 (∼49%) derived haplotypes were likely introgressed from Neanderthals (FDR < 0.05), of which 1,790 (∼56%) are found in more than one copy (supplementary table S3, Supplementary Material online). The 3221 putatively Neanderthal-introgressed haplotypes observed in 16 Turkish genomes cover close to 280 Mb (∼9%) of sequence, corresponding on an average to 18 Mb of Neanderthal-introgressed DNA in each genome. Among these, 1,431 (∼42%) are singletons, that is, observed in 1 out of 32 copies, whereas the rest of the haplotypes are observed at least twice. As expected, Neanderthal-introgressed haplotypes are larger but less frequent than ancestral haplotypes (fig. 2B and supplementary fig. S5, Supplementary Material online).
Of the common Neanderthal-introgressed haplotypes, 584 (33%) were not found in previous studies (Vernot and Akey 2014) and could be region- or population-specific. This observation is consistent with the observation that the frequencies of Neanderthal-introgressed haplotypes can be highly variable both within and among Eurasian populations (Sankararaman et al. 2014; Vernot et al. 2016). In our further analyses, we focused on understanding the genomic and evolutionary impact of the Neanderthal introgression in the Turkish population using the ancestral derived haplotypes as an internal control.
Neanderthal Introgression Shapes Variation Related to Innate Immunity and Immune-Mediated Disorders in Turkish Populations
To determine the functional relevance of Neanderthal-introgressed haplotypes in the Turkish samples, we first analyzed the annotated functional sequences that overlap with Neanderthal-introgressed haplotypes. Neanderthal haplotypes found in the Turkish population are depleted for exonic sequences, where we observe that only 1.1% of the Neanderthal-introgressed haplotypes are covered by exons, as compared with the expected 1.5% for the similar-sized regions (P < 10−15, chi-squared test). This result is consistent with the previously reported notion that there is widespread negative selection against functional Neanderthal alleles (Harris and Nielsen 2016; Juric et al. 2016).
Earlier work has also shown that a small number of introgressed Neanderthal haplotypes with phenotypic and biomedical effects have reached high frequencies in non-African populations. For example, a previously reported (Dannemann et al. 2016) Neanderthal-introgressed haplotype that is common in Eurasia and overlaps multiple Toll-like receptor genes (supplementary fig. S6, Supplementary Material online) is found in 31% of the sampled Turkish chromosomes. Looking at the tag variants for this haplotype we found that it has 15% frequency in the Western European populations, but reaches 50% in the East Asian populations. It is possible that this Neanderthal-introgressed haplotype increased in frequency in the Turkish population due to recent Asian migrations into Anatolia (Di Benedetto et al. 2001; Berkman et al. 2008).
To further investigate putatively functional introgressed Neanderthal haplotypes in the Turkish population, we searched for introgressed haplotypes that harbor single nucleotide variants associated with human traits in genome-wide association studies (GWAS) (MacArthur et al. 2017). Specifically, we searched in the GWAS database for derived single nucleotide variants within the introgressed haplotype regions that are also derived in the Altai Neanderthal genome. We found 55 such GWAS variants (supplementary table S4, Supplementary Material online), 42 of which were observed among at least 2 Turkish chromosomes (≥6.25% allele frequency).
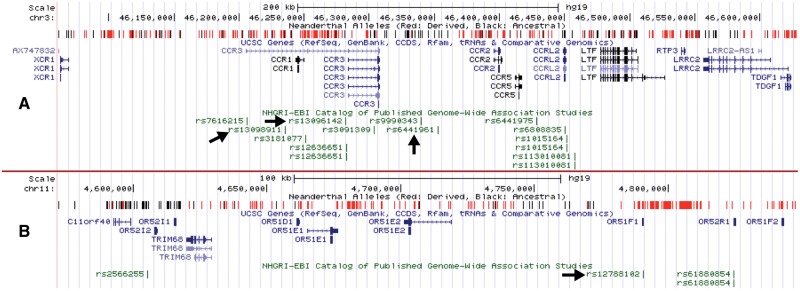
Concordant with previous observations (Khrameeva et al. 2014; Deschamps et al. 2016), 18 of the 41 common GWAS variants on Neanderthal-derived haplotypes in the Turkish population were immune- and metabolism-related. One of these haplotypes is also the largest (>500 kb) among all Neanderthal haplotypes we found in our analysis (fig. 3A). It is found in n = 3 (9%) of the Turkish chromosomes and has remained fully intact in one individual, which appears unlikely assuming at least 55,000 years, or 2,200 generations since admixture and given the recombination rate of this region (P < 10−15; see Supplementary Material online). The haplotype harbors rs13098911, a variant associated with celiac disease (Dubois et al. 2010). This haplotype overlaps with the C-C motif chemokine receptor (CCR) gene family with a known role in HIV infection (Choe et al. 1996). Intriguingly, other variants within the same region, which are not linked to the Neanderthal haplotype, were associated with Behcet’s disease (Kirino et al. 2013), an autoimmune disorder highly common in Western Asia. Collectively, its functional relevance and unusual size raise the possibility that this haplotype may have been maintained in Western Asian populations through adaptive forces, making it an ideal candidate for future studies.
Fig. 3.
—Neanderthal-introgressed haplotypes with putative functional effects. The graphs show genome-browser snapshots of genomic regions where we detected Neanderthal-introgressed haplotypes harboring GWAS variants. The upper ruler indicates a scale for the region. The “Neanderthal Alleles” track denotes the variants used in our S* pipeline and the colors indicate whether the variants are derived (red) in the Neanderthal genome or not (black). The “UCSC Genes” track shows the location of exons (thick blue sticks) and introns (thin blue sticks). (A) Neanderthal-introgressed haplotype carrying GWAS variants associated with celiac disease. The haplotype carries multiple C-C motif chemokine receptor (CCR) genes. The track on the bottom shows the GWAS variants in the region. The arrows show the GWAS variants, which are linked with celiac disease and linked with the Neanderthal haplotype. (B) Neanderthal-introgressed haplotype carrying multiple olfactory receptor 5 (OR5) genes. The haplotype also carries a GWAS variant (rs12788102) associated with the severity of malaria infections (Band et al. 2013). The arrow shows the GWAS variant associated with the severity of malaria infections.
We found another similarly large (>250 kb) Neanderthal-derived haplotype, which harbors rs12788102, a variant strongly associated with severity of malaria (Band et al. 2013) (fig. 3B). This haplotype overlaps with multiple olfactory receptor 5 (OR5) subfamily genes and is found at 20% frequency among the 32 sets of Turkish chromosomes investigated. To our knowledge, this is the first time this haplotype has been highlighted to be introgressed from Neanderthals into humans. Overall, Neanderthal-introgressed haplotypes may have observable effects on immunity-related variation within the Turkish population.
Neanderthal-Introgressed Haplotypes Affect Multiple Metabolism Genes in the Turkish Population
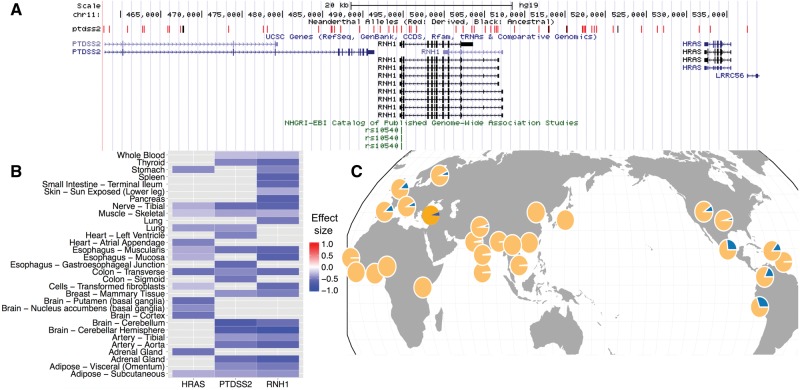
Previous studies have shown that the genes related to metabolism may have been particularly affected by Neanderthal-introgressed haplotypes (Khrameeva et al. 2014). Here, we identified two independent Neanderthal-variants (rs13201877, rs10540) common in this Turkish sample (at ∼9% and ∼6% frequency, respectively) and associated with Body Mass Index (BMI). Among these, the haplotype block that carries rs10540 is >80 kb and harbors three genes: PTDSS2, RNH1, and HRAS (fig. 4). The haplotype is defined by hundreds of alleles and, consequently, the causal variant(s) that lead to the change in BMI remain unknown. The haplotype is intact (i.e., not recombined) in >5,000 human haplotypes phased in the 1000 Genomes data set (1000 Genomes Project Consortium 2012). In this data set, the haplotype’s allele frequency is between 7% and 11% in European populations. It is absent in sub-Saharan African populations and found only rarely in some East Asian populations.
Fig. 4.
—A Neanderthal-introgressed haplotype with putative metabolism-related effects. (A) A Neanderthal-introgressed haplotype carrying an obesity-associated GWAS variant. The graphs show genome-browser snapshots of genomic regions where we detected Neanderthal-introgressed haplotypes harboring a GWAS variant. The upper ruler indicates a scale for the region. The “Neanderthal Alleles” track denotes the variants used in our S* pipeline and the colors indicate whether the variants are derived (red) in Neanderthal genome or not (black). The “UCSC Genes” track shows the location of exons (thick blue sticks) and introns (thin blue sticks). The GWAS variant is shown in green. (B) Genotype tissue expression profile of the GWAS variant rs10540. The range for this heatmap is from red (positive impact on expression) to blue (negative effect on expression). Note that the impact of this haplotype is negative on all three genes. (C) The frequency distribution of the Neanderthal-introgressed haplotype among world populations. The blue section of the pie shows the allele frequency of the introgressed haplotype and the yellow shows all other haplotypes.
We then investigated the cis-regulatory influence of this haplotype using the GTEx data set (The GTEx Consortium 2015). This investigation revealed a general inhibitory effect (supplementary table S4, Supplementary Material online): PTDSS2, RNH1, and HRAS, three highly expressed genes in multiple adult tissues that overlap with this haplotype, are all significantly downregulated in the tissues we could assess (fig. 4). RNH1 is an RNAse inhibitor and has been shown to regulate angiogenin (Lee et al. 1988). HRAS is a well-studied proto-oncogene (Krontiris et al. 1985; Bos 1989). It is developmentally important with variants associated with the Costello syndrome (Aoki et al. 2005), a drastic developmental disorder and myopathy with excess of muscle spindles (van der Burgt et al. 2007). Finally, PTDSS2 is the main enzyme in the biosynthesis of phosphatidylserine, which accounts for up to 10% of cell membrane phospholipids (Tomohiro et al. 2009). In summary, this Neanderthal-introgressed haplotype has broad effects on the expression of multiple genes and a well-established association to BMI. The adaptive significance of these biological effects (or lack thereof) remains to be investigated.
Conclusion
The nature and biological impact of gene flow from Neanderthals to modern human ancestors have lately attracted major interest in anthropological genomics. In this study, we measured the levels of Neanderthal introgression in Western Asian populations, and mapped, for the first time, the haplotype-level impact of Neanderthal introgression in Turkish genomes. Our study provides a first look at the functional impact of Neanderthal introgression in a Western Asian population, paving the way for future population comparisons.
We replicated previous studies (Lazaridis et al. 2016; Rodriguez-Flores et al. 2016) showing that contemporary Western Asian populations have similar or lower levels of Neanderthal introgression than other Eurasian populations. The presence of sub-Saharan African ancestry and possible ancestry from a basal Eurasian lineage with lower (or no) signatures of Neanderthal introgression are the most parsimonious explanations for this observation. We also find considerable variation in the levels of Neanderthal introgression among Western Asian populations (even between one sample-set to another from the Druze population), which we also attribute to variable sub-Saharan African ancestry. As such, it is important here to note that Western Asia has a complex population history and the currently available genome data may not be fully representative of the region’s population diversity.
In this study, we also describe the first haplotype-level Neanderthal introgression map for a Western Asian population. Despite a general depletion of functional sequences among Neanderthal-introgressed haplotypes, we still identified dozens of haplotypes common within our Turkish sample and previously associated with phenotypic variations, including multiple immunity- and metabolism-related variants. These included a haplotype >500 kb that harbors multiple CCR genes, which appears to have remained intact solely through neutral processes given the recombination rate in this region. This haplotype has been associated with celiac disease (Dubois et al. 2010). Moreover, variants within this haplotype have been discussed within the context of HIV and plague resistance (Galvani and Novembre 2005), as well as Behcet’s disease, an immune-mediated disorder especially common in the Turkish population (Kirino et al. 2013). Some of these variants could have been maintained adaptively, although this notion needs to be further scrutinized using larger data sets than those used here. Irrespective of their adaptive nature, our results indicate that Neanderthal-derived alleles have contributed to functional variation in Western Asia.
In addition to functionally annotated and common variants, we found hundreds of putatively functional (e.g., overlapping coding sequence) Neanderthal-introgressed haplotypes in the Turkish population that could be region- or population-specific. Since few genome-wide association studies have been conducted in Turkey and the Middle East, we are yet to discern the exact phenotypic impact of these population-specific haplotypes. This premise is exciting for future studies, as population-specific genomewide association studies in various populations have been identifying Neanderthal variants of functional significance, including variants linked to metabolic disorders (SIGMA Type 2 Diabetes Consortium et al. 2014). We, therefore, argue that the next step toward understanding the phenotypic impact of Neanderthal introgression is to focus on the impact of population-specific variants inherited from Neanderthals.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
The authors thank Marie Saito and Skyler Resendez for critical readings of previous versions of this manuscript. We thank Gulsah Kilinc for data curation. This study is primarily funded by O.G.’s start-up funds. This project is also supported partially by National Science Foundation under Grant No. 1714867 to O.G. M.S. was supported by a TÜBA GEBİP award. The authors declare that there is no conflict of interest with organization regarding to the materials discussed in this article.
Literature Cited
- 1000 Genomes Project Consortium. 2012. An integrated map of genetic variation from 1,092 human genomes. Nature 491:56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander DH, Novembre J, Lange K.. 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 199:1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkan C, et al. 2014. Whole genome sequencing of Turkish genomes reveals functional private alleles and impact of genetic interactions with Europe, Asia and Africa. BMC Genomics 15:963.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki Y, et al. 2005. Germline mutations in HRAS proto-oncogene cause Costello syndrome. Nat Genet. 3710:1038–1040. [DOI] [PubMed] [Google Scholar]
- Band G, et al. 2013. Imputation-based meta-analysis of severe malaria in three African populations. PLoS Genet. 95:e1003509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkman CC, Dinc H, Sekeryapan C, Togan I.. 2008. Alu insertion polymorphisms and an assessment of the genetic contribution of Central Asia to Anatolia with respect to the Balkans. Am J Phys Anthropol. 1361:11–18. [DOI] [PubMed] [Google Scholar]
- Bos JL. 1989. ras oncogenes in human cancer: a review. Cancer Res. 4917:4682–4689. [PubMed] [Google Scholar]
- Choe H, et al. 1996. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell 857:1135–1148. [DOI] [PubMed] [Google Scholar]
- Dannemann M, Andrés AM, Kelso J.. 2016. Introgression of Neandertal- and Denisovan-like haplotypes contributes to adaptive variation in human toll-like receptors. Am J Hum Genet. 981:22–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannemann M, Prüfer K, Kelso J.. 2017. Functional implications of Neandertal introgression in modern humans. Genome Biol. 181:61.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschamps M, et al. 2016. Genomic signatures of selective pressures and introgression from archaic hominins at human innate immunity genes. Am J Hum Genet. 981:5–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Benedetto G, et al. 2001. DNA diversity and population admixture in Anatolia. Am J Phys Anthropol. 1152:144–156. [DOI] [PubMed] [Google Scholar]
- Dubois PCA, et al. 2010. Multiple common variants for celiac disease influencing immune gene expression. Nat Genet. 424:295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand EY, Patterson N, Reich D, Slatkin M.. 2011. Testing for ancient admixture between closely related populations. Mol Biol Evol. 288:2239–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvani AP, Novembre J.. 2005. The evolutionary history of the CCR5-Delta32 HIV-resistance mutation. Microbes Infect. 72:302–309. [DOI] [PubMed] [Google Scholar]
- Gittelman RM, et al. 2016. Archaic hominin admixture facilitated adaptation to out-of-Africa environments. Curr Biol. 2624:3375–3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokcumen Ö, et al. 2011. Biological ancestries, kinship connections, and projected identities in four central Anatolian settlements: insights from culturally contextualized genetic anthropology. Am Anthropol. 1131:116–131. [DOI] [PubMed] [Google Scholar]
- Green RE, et al. 2010. A draft sequence of the Neandertal genome. Science 3285979:710–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer MF, et al. 2011. Genetic evidence for archaic admixture in Africa. PNAS 10837:15123–15128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris K, Nielsen R.. 2016. The genetic cost of Neanderthal introgression. Genetics 2032:881–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh P, et al. 2016. Model-based analyses of whole-genome data reveal a complex evolutionary history involving archaic introgression in Central African Pygmies. Genome Res. 265:291–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Sánchez E, et al. 2014. Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA. Nature 512:194–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juric I, Aeschbacher S, Coop G.. 2016. The strength of selection against Neanderthal introgression. PLoS Genet. 1211:e1006340.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khrameeva EE, et al. 2014. Neanderthal ancestry drives evolution of lipid catabolism in contemporary Europeans. Nat Commun. 5:3584.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kılınç GM, et al. 2016. The demographic development of the first farmers in Anatolia. Curr Biol. 26:2659–2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirino Y, et al. 2013. Genome-wide association analysis identifies new susceptibility loci for Behçet’s disease and epistasis between HLA-B*51 and ERAP1. Nat Genet. 452:202–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krontiris TG, DiMartino NA, Colb M, Parkinson DR.. 1985. Unique allelic restriction fragments of the human Ha-ras locus in leukocyte and tumour DNAs of cancer patients. Nature 3136001:369–374. [DOI] [PubMed] [Google Scholar]
- Lazaridis I, et al. 2014. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature 5137518:409–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaridis I, et al. 2016. Genomic insights into the origin of farming in the ancient Near East. Nature 5367617:419–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee FS, Fox EA, Zhou HM, Strydom DJ, Vallee BL.. 1988. Primary structure of human placental ribonuclease inhibitor. Biochemistry 2723:8545–8553. [DOI] [PubMed] [Google Scholar]
- MacArthur J, et al. 2017. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 45(D1):D896–D901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallick S, et al. 2016. The Simons Genome Diversity Project: 300 genomes from 142 diverse populations. Nature 5387624:201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy RC, Wakefield J, Akey JM.. 2017. Impacts of Neanderthal-introgressed sequences on the landscape of human gene expression. Cell 1685:916–927.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, et al. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 209:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M, et al. 2012. A high-coverage genome sequence from an archaic Denisovan individual. Science 3386104:222–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novembre J, et al. 2008. Genes mirror geography within Europe. Nature 4567218:98–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N, et al. 2012. Ancient admixture in human history. Genetics 1923:1065–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prüfer K, et al. 2014. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 5057481:43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 813:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quach H, et al. 2016. Genetic adaptation and Neandertal admixture shaped the immune system of human populations. Cell 1673:643–656.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racimo F, et al. 2017. Archaic adaptive introgression in TBX15/WARS2. Mol Biol Evol. 343:509–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D, et al. 2010. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature 4687327:1053–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Flores JL, et al. 2016. Indigenous Arabs are descendants of the earliest split from ancient Eurasian populations. Genome Res. 262:151–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankararaman S, et al. 2014. The genomic landscape of Neanderthal ancestry in present-day humans. Nature 5077492:354–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott EM, et al. 2016. Characterization of Greater Middle Eastern genetic variation for enhanced disease gene discovery. Nat Genet. 489:1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SIGMA Type 2 Diabetes Consortium, Williams AL, et al. 2014. Sequence 60 variants in SLC16A11 are a common risk factor for type 2 diabetes in Mexico. Nature 506:97–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taskent RO, Gokcumen O.. 2017. The multiple histories of Western Asia: Perspectives from ancient, and modern genomes. Human Biology 892:doi: 10.13110/humanbiology.89.2.01. [DOI] [PubMed] [Google Scholar]
- The GTEx Consortium. 2015. The Genotype-Tissue Expression (GTEx) pilot analysis: multi tissue gene regulation in humans. Science 348:648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomohiro S, Kawaguti A, Kawabe Y, Kitada S, Kuge O.. 2009. Purification and characterization of human phosphatidylserine synthases 1 and 2. Biochem J. 418:421–429. [DOI] [PubMed] [Google Scholar]
- van der Burgt I, et al. 2007. Myopathy caused by HRAS germline mutations: implications for disturbed myogenic differentiation in the presence of constitutive HRas activation. J Med Genet. 447:459–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernot B, Akey JM.. 2014. Resurrecting surviving Neandertal lineages from modern human genomes. Science 3436174:1017–1021. [DOI] [PubMed] [Google Scholar]
- Vernot B, et al. 2016. Excavating Neandertal and Denisovan DNA from the genomes of Melanesian individuals. Science 3526282:235–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall JD, et al. 2013. Higher levels of neanderthal ancestry in East Asians than in Europeans. Genetics 194:199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Lachance J, Tishkoff SA, Hey J, Xing J.. 2013. Apparent variation in Neanderthal admixture among African populations is consistent with gene flow from Non-African populations. Genome Biol Evol. 511:2075–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, et al. 2017. Archaic hominin introgression in Africa contributes to functional salivary MUC7 genetic variation. Mol Biol Evol. 3410:2704–2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang MA, Malaspinas A-S, Durand EY, Slatkin M.. 2012. Ancient structure in Africa unlikely to explain Neanderthal and non-African genetic similarity. Mol Biol Evol. 2910:2987–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yunusbayev B, et al. 2015. The genetic legacy of the expansion of Turkic-speaking nomads across Eurasia. PLoS Genet. 114:e1005068. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.