Abstract
Several studies have begun to elucidate the genetic and developmental processes underlying major vertebrate traits. Few of these traits have evolved repeatedly in vertebrates, preventing the analysis of molecular mechanisms underlying these traits comparatively. Electric organs have evolved multiple times among vertebrates, presenting a unique opportunity to understand the degree of constraint and repeatability of the evolutionary processes underlying novel vertebrate traits. As there is now a completed genome sequence representing South American electric eels, we were motivated to obtain genomic sequence from a linage that independently evolved electric organs to facilitate future comparative analyses of the evolution and development of electric organs. We report here the sequencing and de novo assembly of the genome of the mormyrid Paramormyrops kingsleyae using short-read sequencing. In addition, we have completed a somatic transcriptome from 11 tissues to construct a gene expression atlas of predicted genes from this assembly, enabling us to identify candidate housekeeping genes as well as genes differentially expressed in the major somatic tissues of the mormyrid electric fish. We anticipate that this resource will greatly facilitate comparative studies on the evolution and development of electric organs and electroreceptors.
Keywords: electric fish, mormyridae, electric organs, Paramormyrops
Introduction
Electric organs (EOs) have evolved to produce electric fields for the purposes of communication, navigation, and, in extreme cases, for predation and defense. In contrast with most other vertebrate traits, there have been six independent origins of electrogenesis represented among extant vertebrates. The taxonomic diversity of electrogenic fishes is so profound that Darwin considered the multiple origins of EOs as a “special difficulty” to reconcile with his newly minted theory of natural selection (Darwin 1859). Though it has been 150 years since the publication of The Origin of Species, we still know remarkably little about the “steps by which these wondrous organs have been produced,” despite their clear benefit as a model for understanding general principles of how complex vertebrate tissues may have repeatedly evolved (Gallant et al. 2014).
In every group that has evolved electrogenesis, EOs originate during development from skeletal muscle (SM) progenitor cells and undergo a series of developmental steps that disable the ability to contract in favor of extreme electrical excitability. Gallant et al. (2014) reported the recent successful sequencing and assembly of the first electric fish genome (from Electrophorus electricus) and multiple transcriptome assemblies representing several species that independently evolved EOs. Comparative analysis of expression data elucidated a relatively small set of genes with highly consistent patterns of expression across lineages that independently evolved EOs, motivating a “shared toolkit” hypothesis for the evolution of EOs. The relative paucity of genomic resources representing groups of electric fishes that have evolved EOs independently, as well as the lack of functional tools in these groups, have limited progress to test this hypothesis more stringently (Pitchers et al. 2016).
Beyond the needs for additional comparative resources, a mormyrid genome is of interest to biologists for several reasons. First, it should be useful to those interested broadly in vertebrate genomics, as osteoglossomorphs, the most ancestral group of teleost fishes, are poorly represented among available fish genomes, represented only by the Asian Arowana Scleropages formosus (Bian et al. 2016; Li et al. 2016). Second, mormyrids are a long-established model in systems neurobiology: they boast one of the most thorough descriptions of the neural basis of a vertebrate behavior (Heiligenberg 1991), and have contributed to understanding mechanisms for preservation and analysis of temporal information (Hopkins 1999; Kawasaki 1997), synaptic plasticity (Bell et al. 1997), and cerebellar function (Bell et al. 2008). The availability of genomic resources for mormyrid fishes should greatly facilitate molecular approaches to the continued dissection of neural circuits in this system (Carlson and Gallant 2013).
Finally, mormyrids are an emerging model in evolutionary biology for understanding the evolution of prezygotic isolation mechanisms (Arnegard et al. 2005, 2010b; Feulner et al. 2009; Gallant et al. 2011). African weakly electric fish (mormyriformes) are among one of the most rapidly speciating groups of ray-finned fishes (Rabosky et al. 2013) and much of this diversity comes from rapid radiations of mormyriformes in the genera Paramormyrops (Sullivan et al. 2002, 2004) and Campylomormyrus. Paramormyrops are morphologically cryptic, but distinct in their electrical signals. While the phylogenetic relationship among mormyrids is well understood, the origin of this species diversity is less well understood. Field and laboratory playback experiments reveal strong preferences for species-specific EO discharge (EOD) waveforms among weakly electric fishes (Hopkins and Bass 1981; Arnegard et al. 2006) and recent work has demonstrated that EOD diversification outpaces other forms of ecological and morphological diversification (Arnegard et al. 2010b). EODs are produced by well-characterized and discrete anatomical and physiological substrates related to the function of other electrically excitable tissues such as muscle and nerve (Bennett and Grundfest 1961; Arnegard et al. 2010a; Gallant et al. 2011). Comparative mormyrid genomics would greatly facilitate the ability to link molecular changes to the evolution of reproductive isolation, that is, speciation.
We selected Paramormyrops kingsleyae for a first mormyrid genome sequence partly because it is among the best studied species of Paramormyrops (Hopkins and Bass 1981; Sullivan et al. 2002; 2004), and partly due to its geographically widespread distribution in West-Central Africa (Stiassny et al. 2007). In addition to this, recent work has identified P. kingsleyae as a “microcosm” of signal evolution in Paramormyrops (Gallant et al. 2011, 2017) due to polymorphic EOD signaling.
Materials and Methods
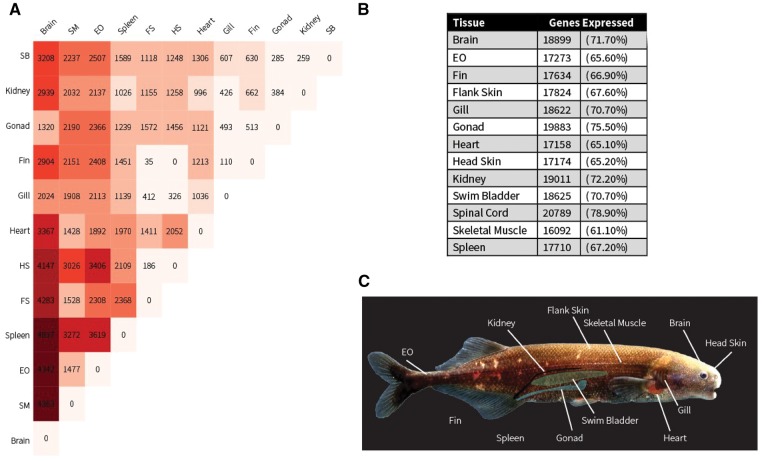
A complete description of methods utilized is detailed in the Supplementary Materials online, which we briefly summarize here. We sequenced a single individual juvenile P. kingsleyae (PKING1) to high coverage (90×) using short-insert and mate-pair libraries of 100 bp length on an Illumina HiSeq 2000. We assembled the draft genome of all sequences with ALLPATHS-LG (Gnerre et al. 2011) using default parameter settings, subjecting assembly input reads to quality control as detailed in ALLPATHS documentation. Finally, we obtained RNA-seq data from 11 tissues of one individual (PKING2), and using Trinity (Grabherr et al. 2011), assembled a PASA (Haas et al. 2003), merged, genome-guided and de novo transcriptome to aid in annotation of the genome. We used the MAKER pipeline (Cantarel et al. 2008) using the protocol suggested by Campbell et al. (2014) to perform gene annotation. We constructed a gene expression on this annotated gene using RNA-seq data obtained from 3 individuals (PKING1, PKING2, and PKING3) from 13 tissues (see fig 1C): brain, regressed gonads (two testis and one ovary, all sampled under non-breeding conditions), SM, spleen, EO, heart, kidney, swim bladder, gill, spinal cord, fin, electroreceptor enriched skin from the head (head skin), and electroreceptor impoverished skin from the flank (flank skin). We then examined between- and within-sample correlation of read counts as an ad hoc method of quality control for between sample tissue contamination, as some tissues were difficult to dissect “purely” from each organism. This approach excluded spinal cord and spleen from further analysis. We proceeded with examining expression profiles of each of 26,348 genes in each of the 11 tissues sampled. Based on expression profiles, we identified two sets of “housekeeping genes” with high stringency and low stringency criteria. Next, we computed highly significant (FDR-corrected P-value < 0.001) differentially expressed genes between all pairs of tissues. We compared gene expression and functional enrichment between EO and SM, motivated by our longstanding interest in differential expression of genes and gene ontology terms specifically between EO and SM (Gallant et al. 2012, 2014).
Fig. 1—
Atlas of gene expression based on 11 tissues from 3 P. kingsleyae specimens. (A) Heatmap matrix of differentially expressed genes between pairs of tissues (numbers indicated in each box). Warmer colors indicate larger numbers of differentially expressed genes between tissue pairs. (B) List of number of genes considered “expressed” (TPM reads > 1) in each tissue, and expressed as a percent of total genes expressed in at least one tissue. (C) Photograph of Paramormyrops kingsleyae overlayed with a colorized diagram indicating sources of tissues for transcriptome sequencing. HS, head skin; FS, flank skin.
Results and Discussion
Paramormyrops kingsleyae Genome Assembly and De Novo Transcriptome Assembly
The combined gDNA sequence reads were assembled using ALLPATHS-LG to a total base size of 735 Mb. The assembly was made up of a total of 4,668 scaffolds with an N50 scaffold length of over 1.7 Mb (N50 contig length was 37.6 kb). We refer to this assembly as the PKINGS_0.1 assembly. In the PKINGS_0.1 assembly, the largest scaffold was over 7 Mb in length. We estimated total repetitive content to be 26% (kmer = 25 scale) and GC composition to be 43%. The assembly statistics are compared to those obtained from a previous assembly of the E. electricus genome in Table 1. Using CEGMA (Parra et al. 2007), we were able to locate 435 of the 458 genes included in the CEGMA core set (94.9%). Of the 248 most conserved genes defined by Parra et al. (2007), 91.94% were identified as full length and 94.34% were identified as either full or partial, indicating a high degree of completeness of the protein coding content of the genome.
Table 1.
Comparison of E. electricus and P. kingsleyae (v. 01) Assemblies
| Confirmed | E. electricus | P. kingsleyae |
|---|---|---|
| Genome size | 720 Mb | 880 Mb |
| Coverage | 55x | 83x |
| CEGMA core set representation | 97% | 95% |
| Full length | 88% | 91% |
| Partial | 99% | 94% |
| n contigs | 340,589 | 4,496 |
| Contig N50 | 104 kb | 37.6 kb |
| Scaffold N50 | 632 kb | 1.7 Mb |
| GC content | 42.50% | 43% |
| Genes Predicted | 22,000 | 27,677 |
We obtained two transcriptome assemblies of all reads from the 13 somatic tissues of PKING2: a genome-guided assembly consisting of 430,693 contigs and a de novo assembly consisting of 432,197 contigs. The two assemblies were merged using PASA for a final transcriptome assembly consisting of 418,012 non-redundant contigs used for annotation purposes.
Genome Annotation
The MAKER-standard gene set consisted of 27,677 predicted genes. We note that this is an unusually high number of genes for a teleost fish: the only other osteoglossiform genome available, S. formosus, predicted a maximum of 22,016 genes (Bian et al. 2016; Li et al. 2016). While it may be possible that Mormyrid fishes have a larger number of genes than other osteoglossiforms, our qualitative inspection of gene annotation has identified instances where gene annotations for a single gene may be “split.” We are currently working to systematically identify potentially split gene annotations and improve the annotation over time. Despite this, annotation edit distance (AED) score distributions (figure S1, Supplementary Material online) indicate that the majority of annotations are supported by either protein or RNA-seq assembly evidence: 90% of genes have an AED of 0.5 or lower (Campbell et al. 2014), indicating a relatively high quality of most gene annotations. As part of the annotation process, all predicted genes were functionally annotated based on homology to the UniProtKB/Swissprot database (November 2015) and for recognizable PFAM-A (release 29.0) domains using InterPro Scan.
Analysis of Gene Expression and Function
RNA-seq data collected in this study is summarized in supplementary file S1, Supplementary Material online. Of the 27,677 predicted genes, RNA-seq analysis supports expression of 26,438 (95.1%) of the genes identified in this set in at least one tissue. We counted the number of genes expressed in each tissue (fig. 1B) which ranged from 61.1% to 78.9% of the total transcriptome.
We performed analysis of differential expression using the edgeR (Robinson et al. 2010) package in “classic mode,” computing pairwise differential expression between all sets of tissues for each of the 26,438 expressed genes. We detected that 13,054 genes were differentially expressed genes in at least one tissue. The number of pairwise differences between tissues is shown as a heatmap in figure 1A. A list of differentially expressed genes, together with average and standard deviation of expression values across replicates, is provided as supplemental file S2, Supplementary Material online.
In order to facilitate qPCR analysis in mormyrid electric fish, particularly for systems neuroscience applications, we wished to identify putative “housekeeping genes” that would provide reliable references for relative quantification approaches. These genes ought to be (1) ubiquitously expressed in all tissues examined, (2) not differentially expressed between any tissues greater than 2-fold, and (3) had relatively little variance in expression across tissue replicates. We determined two stringency criteria (high and low) for these determinations. Using our high-stringency criteria, we identified 21 housekeeping genes, and using our low stringency criteria, we detected 1134 genes. The identities of these genes are listed in supplemental file S3, Supplementary Material online. Heatmaps of log-transformed expression values for both the high and low stringency sets of “housekeeping genes” are shown in figures S2 and S3, Supplementary Material online, respectively, and indicate the ubiquity of expression and the low variance between tissues of these genes, particularly among the high stringency set.
Examination of the SM and EO Transcriptomes
Our analysis identified 1,478 differentially expressed genes (FDR < 0.001; CPM > 2) between EO and SM. Of these, 702 were upregulated in EO and 775 were upregulated in SM. The identities and expression values in all tissues are reported in supplemental file S4, Supplementary Material online. These differentially expressed genes were examined for significantly (FDR < 0.05) over- and underrepresented GO terms in each tissue type. In SM, we found 57 overrepresented GO terms, and no underrepresented GO terms. In EO, we found three overrepresented GO terms and five underrepresented GO terms. The under- and overrepresented GO terms for each tissue, along with associated gene accession numbers, are reported in supplemental file S5, Supplementary Material online.
The results of these analyses represent the first comprehensive portrait of gene expression in the EO of mormyrids. Enriched GO terms in the SM set were highly redundant, but associated with the function of sarcomeres, which have been shown to be downregulated in EOs across lineages, including the mormyrid species Brienomyrus brachyistius and among the genus Campylomormyrus (Gallant et al. 2012, 2014; Lamanna et al. 2015).
Perhaps unsurprisingly, among the most abundant genes in the EO are those involved in ion transport. One particularly highly expressed protein is the calcium binding protein s100 (PKINGS_0.1_G033944), which has been noted to be highly expressed in the EOs of B. brachyistius (Gallant et al. 2012). In addition, the sodium channel gene scn4aa (PKINGS_0.1_G046371), potassium channel gene kcna7a (PKINGS_0.1_G046371), inward rectifying potassium channel Kcnj2 (PKINGS_0.1_G051417), and sodium-potassium transporting ATPse beta 1 subunit (PKINGS_0.1_G020465) atp1b1 are among the 50 most abundant genes expressed in the EO. The importance of scn4aa in mormyridae is well documented elsewhere (Zakon et al. 2006; Arnegard et al. 2010c); however, potassium channel genes have more recently gained attention in mormyrids (Swapna et al. 2017). Nagel et al. (2017) noted the strong expression of the inward rectifying potassium channel kcnj2 in Campylomormyrus previously.
In addition to the abundant expression of these specific ion channels and key transcription factors, gene–ontology analysis indicates enriched representation of plasma membrane–associated proteins in the EO, which have hitherto received very little attention. Given the rather prolific diversity of membrane morphology in mormyrid electrocytes (Bennett and Grundfest 1961; Hopkins 1999; Gallant et al. 2011; Sullivan et al. 2002, 2004) and its role in EOD signal diversity (Bennett and Grundfest 1961; Gallant et al. 2011), these genes are likely an important area of focus in future studies.
Also among the top 50 most highly abundant, differentially expressed transcripts in the EO is the transcription factor mef2a (PKINGS_0.1_G007371), which was also detected to be highly upregulated in the EOs of B. brachyistius (Gallant et al. 2012). mef2a is not consistently upregulated across all independent origins of electrogenesis (Gallant et al. 2014), but seems to be characteristic of mormyrid EOs. The retention of relatively high expression of an “early” muscle regulatory factors in an adult, fully differentiated tissue is striking, and may yield clues into understanding the steps by which the developmental program that normally yields muscle might be modified to produce EOs.
Data Availability
The MAKER standard annotation file for PKINGS_0.1 and the PASA-merged transcriptome assembly together with the expression data described in this manuscript are available through the EFISHGENOMICS web portal (http://efishgenomics.integrativebiology.msu.edu) for comparative data analysis with other electric fish.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by the National Science Foundation (1455405: J.R.G., PI). The authors declare no competing financial interests. The authors thank Monica Lucas and Katherine Shaw with technical assistance in isolating RNA from Paramormyrops and for fish care. They also thank Carl Hopkins, Bruce Carlson, and Matthew Arnegard, Roger Afene, Jean Danielle Mbega, and Marie-Francois Eva for assistance with fish collection in Gabon. They also thank the MSU RTSF Genomics core facility with assistance in RNA-seq analysis and troubleshooting library preparation.
Literature Cited
- Arnegard ME, Zwickl DJ, Lu Y, Zakon HH.. 2010a. Old gene duplication facilitates origin and diversification of an innovative communication system—twice. PNAS 10751:22172–22177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnegard ME, Bogdanowicz SM, Hopkins CD.. 2005. Multiple cases of striking genetic similarity between alternate electric fish signal morphs in sympatry. Evolution. 592:324–343.http://dx.doi.org/10.1111/j.0014-3820.2005.tb00993.x [PubMed] [Google Scholar]
- Arnegard ME, Jackson BS, Hopkins CD.. 2006. Time-domain signal divergence and discrimination without receptor modification in sympatric morphs of electric fishes. J Exp Biol. 209(Pt 11):2182–2198. [DOI] [PubMed] [Google Scholar]
- Arnegard ME, et al. 2010b. Sexual signal evolution outpaces ecological divergence during electric fish species radiation. Evol Int J Org Evol. 1763:335–356. [DOI] [PubMed] [Google Scholar]
- Arnegard ME, Zwickl DJ, Lu Y, Zakon HH.. 2010c. Old gene duplication facilitates origin and diversification of an innovative communication system–twice. Proc Natl Acad Sci U S A. 10751:22172–22177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell CC, Han V, Sawtell NB.. 2008. Cerebellum-like structures and their implications for cerebellar function. Annu Rev Neurosci. 31:1–24.http://dx.doi.org/10.1146/annurev.neuro.30.051606.094225 [DOI] [PubMed] [Google Scholar]
- Bell CC, Han VZ, Sugawara Y, Grant K.. 1997. Synaptic plasticity in a cerebellum-like structure depends on temporal order. Nature. 3876630:278–281. [DOI] [PubMed] [Google Scholar]
- Bennett MVL, Grundfest H.. 1961. Studies on morphology and electrophysiology of electric organs III. Electrophysiology of electric organs in mormyrids. In: Chagas C, Paes de Carvalho A, editors. Bioelectrogenesis. New York: Elsevier. p. 113-135.
- Bian C, et al. 2016. The Asian arowana (Scleropages formosus) genome provides new insights into the evolution of an early lineage of teleosts. Sci Rep. 6:24501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MS, Holt C, Moore B, Yandell M.. 2014. Genome annotation and curation using MAKER and MAKER-P. Curr Protoc Bioinformatics. 48:4.11.11–39. [DOI] [PMC free article] [PubMed]
- Cantarel BL, et al. 2008. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 181:188–196.http://dx.doi.org/10.1101/gr.6743907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson BA, Gallant JR.. 2013. From sequence to spike to spark: evo-devo-neuroethology of electric communication in mormyrid fishes. J Neurogenet. 273:106–129.http://dx.doi.org/10.3109/01677063.2013.799670 [DOI] [PubMed] [Google Scholar]
- Darwin C. 1859. On the origin of species by means of natural selection. London: J. Murray. [Google Scholar]
- Feulner PG, Plath M, Engelmann J, Kirschbaum F, Tiedemann R.. 2009. Magic trait electric organ discharge (EOD): dual function of electric signals promotes speciation in African weakly electric fish. Commun Integr Biol. 24:329–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallant JR, Arnegard ME, Sullivan JP, Carlson BA, Hopkins CD.. 2011. Signal variation and its morphological correlates in Paramormyrops kingsleyae provide insight into the evolution of electrogenic signal diversity in mormyrid electric fish. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. doi: 10.1007/s00359-011-0643-8. [DOI] [PubMed]
- Gallant JR, Hopkins CD, Deitcher DL.. 2012. Differential expression of genes and proteins between electric organ and skeletal muscle in the mormyrid electric fish Brienomyrus brachyistius. J Exp Biol. 215(Pt 14):2479–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallant JR, et al. 2017. Variation in electric signals among Paramormyorps kingsleyae is due to substantial population structure imposed by geographic isolation. bioRxiv 154047. doi: https://doi.org/10.1101/154047
- Gallant JR, et al. 2014. Genomic basis for the convergent evolution of electric organs. Science 3446191:1522–1525.http://dx.doi.org/10.1126/science.1254432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnerre S, et al. 2011. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc Natl Acad Sci USA. 1084:1513–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 297:644–652.http://dx.doi.org/10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, et al. 2003. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 3119:5654–5666.http://dx.doi.org/10.1093/nar/gkg770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiligenberg W. 1991. Neural nets in electric fish. Cambridge, MA: MIT Press. [Google Scholar]
- Hopkins CD. 1999. Design features for electric communication. J Exp Biol. 202(Pt 10):1217–1228. [DOI] [PubMed] [Google Scholar]
- Hopkins CD, Bass A.. 1981. Temporal coding of species recognition signals in an electric fish. Science. 2124490:85–87. [DOI] [PubMed] [Google Scholar]
- Kawasaki M. 1997. Sensory hyperacuity in the jamming avoidance response of weakly electric fish. Curr Opin Neurobiol. 74:473–479.http://dx.doi.org/10.1016/S0959-4388(97)80025-6 [DOI] [PubMed] [Google Scholar]
- Lamanna F, Kirschbaum F, Waurick I, Dieterich C, Tiedemann R.. 2015. Cross-tissue and cross-species analysis of gene expression in skeletal muscle and electric organ of African weakly-electric fish (Teleostei; Mormyridae). BMC Genomics. 16:668.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, et al. 2016. A chromosome-level genome assembly of the Asian arowana, Scleropages formosus. Sci Data. 3:160105.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel R, Kirschbaum F, Tiedemann R.. 2017. Electric organ discharge diversification in mormyrid weakly electric fish is associated with differential expression of voltage-gated ion channel genes. J Comp Physiol A. 2033:183–195. [DOI] [PubMed] [Google Scholar]
- Parra G, Bradnam K, Korf I.. 2007. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 239:1061–1067.http://dx.doi.org/10.1093/bioinformatics/btm071 [DOI] [PubMed] [Google Scholar]
- Pitchers WR, Constantinou SJ, Losilla M, Gallant JR.. 2016. Electric fish genomics: progress, prospects, and new tools for neuroethology. J Physiol Paris. 1103: 259–272. [DOI] [PubMed] [Google Scholar]
- Rabosky DL, et al. 2013. Rates of speciation and morphological evolution are correlated across the largest vertebrate radiation. Nat. Commun. 4:1958. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK.. 2010. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 261:139–140.http://dx.doi.org/10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiassny MLJ, Teugels GG, Hopkins CD.. 2007. Poissons d'eaux douces et saumâtres de basse Guinée, ouest de l'Afrique centrale = The fresh and brackish water fishes of Lower Guinea, West-Central Africa. Paris, France, Tervuren, Belgique: Institut de recherche pour le développement: Muséum national d'histoire naturelle; Musée royal de l'Afrique centrale.
- Sullivan JP, Lavoue S, Arnegard ME, Hopkins CD.. 2004a. AFLPs resolve phylogeny and reveal mitochondrial introgression within a species flock of African electric fish (Mormyroidea: Teleostei). Evolution. 58:825–841. [DOI] [PubMed] [Google Scholar]
- Sullivan JP, Lavoue S, Hopkins CD.. 2002. Discovery and phylogenetic analysis of a riverine species flock of African electric fishes (Mormyridae: Teleostei). Evolution. 563:597–616.http://dx.doi.org/10.1111/j.0014-3820.2002.tb01370.x [DOI] [PubMed] [Google Scholar]
- Swapna I, et al. 2017. Electrostatic Tuning of a Potassium Channel in Electric Fish. bioRxiv 206243 [DOI] [PMC free article] [PubMed]
- Zakon HH, Lu Y, Zwickl D, Hillis D.. 2006. Sodium channel genes and the evolution of diversity in communication signals of electric fishes: convergent molecular evolution. Proc Natl Acad Sci USA. 10310:3675–3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MAKER standard annotation file for PKINGS_0.1 and the PASA-merged transcriptome assembly together with the expression data described in this manuscript are available through the EFISHGENOMICS web portal (http://efishgenomics.integrativebiology.msu.edu) for comparative data analysis with other electric fish.