Abstract
Among the chaperones and co-chaperones regulating the glucocorticoid receptor (GR), FK506 binding protein (FKBP) 51 is the most intensely investigated across different disciplines. This review provides an update on the role of the different co-chaperones of Hsp70 and Hsp90 in the regulation of GR function. The development leading to the focus on FKBP51 is outlined. Further, a survey of the vast literature on the mechanism and function of FKBP51 is provided. This includes its structure and biochemical function, its regulation on different levels—transcription, post-transcription, and post-translation—and its function in signaling pathways. The evidence portraying FKBP51 as a scaffolding protein organizing protein complexes rather than a chaperone contributing to the folding of individual proteins is collated. Finally, FKBP51’s involvement in physiology and disease is outlined, and the promising efforts in developing drugs targeting FKBP51 are discussed.
Keywords: chaperone, glucocorticoid receptor, FKBP51, FKBP5, signaling pathway, drug
1. Introduction
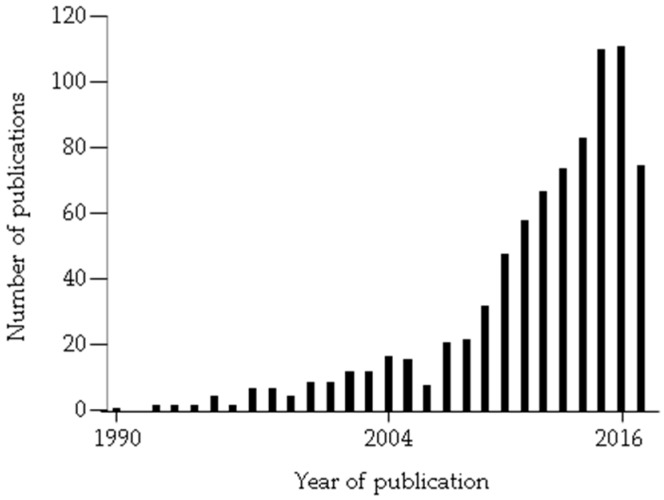
The original discovery of FK506 binding protein (FKBP) 51 was described more than 25 years ago alongside the analysis of steroid receptor complex components [1]. Accordingly, steroid receptors (SRs) and the glucocorticoid receptor (GR) in particular served as the main experimental systems for the initial investigations of FKBP51’s function and mechanism [2,3,4,5], while additional functions of FKBP51 emerged soon afterward [6]. Scientists in several laboratories characterized FKBP51 as a strong inhibitor of GR function [7,8,9,10]. Since GR is a crucial part of the stress hormone axis, also known as HPA (hypothalamus–pituitary–adrenals) axis, and since increasing evidence became available for dysregulated GR in stress-related diseases, such as depression [11], the finding of FKBP51 as an inhibitor of GR inspired a genotype association study on depression [12]. This study discovered that polymorphisms of the FKBP51-encoding gene FKBP5 were associated with stress hormone axis reactivity and response to antidepressants [12] and amplified interest in FKBP51 research worldwide (see Figure 1 for a summary of the number of FKBP51-publications per year since 1990), along with the discovery of several additional functions of FKBP51 [6].
Figure 1.
Number of publications on FKBP51 per year since 1990. The numbers are based on a Pubmed search that included FKBP5, FKBP51, FKBP54, FKBP 5, FKBP 51, and FKBP 54. The initial publications described FKBP51 as immunophilin p54. For 2017, citations were retrieved October 30 of this year.
Given the importance of GR in the initial characterization of FKBP51, we will first provide an update of the contribution of chaperones and co-chaperones to GR function (Section 2), with a focus on FKBP51 and a brief introduction into its structure and biochemical function (Section 3). This is followed by a survey of the literature on regulation and function of FKBP51 (Section 4 and Section 5) and on drug development efforts (Section 6) and by concluding remarks (Section 7).
2. Glucocorticoid Receptor Co-Chaperones, Focus on FKBP51
The glucocorticoid receptor (GR) belongs to the large superfamily of nuclear receptors (NRs), which typically act as transcriptional regulators in a ligand-dependent manner [13,14,15]. They share a common structure that became apparent already before any of their genomic sequence was known [16]: an N-terminal activator domain, a central DNA binding domain, and a C-terminal activator domain 2, which is also the domain of ligand binding [17]. NRs are grouped into seven subfamilies, NR0-6, based on their homology [18]. GR, designated as NR3C1, belongs to the NR3 subfamily, which encompasses all steroid hormone receptors.
GR is functionally involved in a broad spectrum of physiological processes, including the immune, cardiovascular, reproductive, nervous, and metabolic system [19]. This vast range of physiological functions requires intricate control of its activity [20]. In addition to molecular chaperones and co-chaperones, which predominantly are known as regulators of folding and conformation of GR in the cytosol, there is a vast array of cofactors that determine GR’s activity as a transcription factor on the chromatin. These are not the subject of this survey, excellent reviews have been published [20,21,22].
2.1. Complexity of GR Regulating Chaperones and Co-Chaperones
The first confirmed chaperone found associated with SRs was heat shock protein (Hsp) 90 [4,23,24]. Hsp90 is a highly abundant protein in eukaryotic cells making up 1–2% of the cytosolic protein content under non-stress conditions and up to 5% under stress conditions [25], now known as a key folding and assembly platform involved in numerous cellular processes [26,27,28]. The discovery of the core components of the GR-chaperone heterocomplex and their step-wise, ATP-dependent assembly has been described in several reviews [2,4,5]. Briefly, in vitro reconstitution experiments revealed that five chaperones and co-chaperones suffice to fold GR to a conformation with high affinity hormone binding competence: Hsp40, Hsp70, hop, Hsp90, and p23 [2,29]. The basic maturation step is thought to involve the initial recognition of GR by Hsp40, which leads to association with ATP-bound Hsp70 [30,31,32]. In the next step, Hsp70-Hsp90 organizing protein (hop) promotes the transfer of the partially folded GR to the Hsp90-based folding platform; hop leaves the folding complex at later stages. Hsp90 primarily acts on the hormone binding domain and keeps it in a hormone binding competent state. P23 is a cofactor of Hsp90 that keeps Hsp90 in the ATP-bound state, thus promoting receptor binding of Hsp90 [33]. More recently, the coordinated action of the Hsp70- and Hsp90-based cycles in the folding and maturation of GR has been analyzed in more detail [34].
While the studies in vitro as well as in yeast provided important mechanistic insight into the basic process of steroid receptor folding such as GR [2,4,35], the chaperone action on SRs in their homologous environment in mammalian cells is more complex. Both Hsp70 and Hsp90 are fine-tuned by a vast array of co-chaperones [26,36,37,38,39]. In addition, the functional outcome of chaperone and co-chaperone action also depends on the specific context. For example, p23 appears to have several functions as it is known to act as an Hsp90 co-chaperone, as an autonomous molecular chaperone, and as a prostaglandin E2 synthase [40,41,42,43,44]. With respect to its influence on GR activity, in vivo experiments in mammalian cells provided evidence for both stimulation and inhibition of GR-dependent transcription, possibly relating to experimental conditions including cellular context and expression levels of p23 [45,46,47].
2.2. Hsp70 and Hsp90 Cofactors
Of the cofactors of Hsp70 and Hsp90, only a small fraction has been analyzed for their potential effect on GR. To provide a glimpse of the complexity, there are 13 known Hsp70 genes in the human genome [48,49], and a genome-wide analysis has revealed the existence of 41 Hsp40 family members. Of these 41, 34 feature typical J domains that confer association with Hsp70, while 7 have partially conserved J-like domains [50,51]. Hsp70 cofactors with known effects on GR are listed in Table 1, along with a short description and references. More recently added was Hsp70 binding protein 1 (HSPBP1), which, like Bcl2-associated athanogene 1 (BAG1), is a nucleotide exchange factor of Hsp70 [52,53]. Similar to the longer BAG1 isoforms BAG1M and BAG1L [54,55,56,57], increased expression of HSPBP1 reduced GR-dependent transcriptional activity [58].
Table 1.
Cofactors of Hsp90 and Hsp70 with demonstrated influence on GR activity. For more details, the reader is referred to the original citations provided. (Cofactors of Hsp90 and Hsp70 relevant for GR activity).
| Name | Description | Citations |
|---|---|---|
| Hsp70 cofactors | ||
| BAG1 (Bcl2 Associated Athanogene 1) | Nucleotide exchange factor for Hsp70 proteins. L- and M-forms inhibit GR in mammalian cells. | [52,54,56,58] |
| Hip (Hsp70 interacting protein, ST13) | Stabilizes the ADP bound state of Hsp70, opposes the inhibitory effect of BAG1, enhances GR function in the yeast model, possibly also independently of Hsp70. | [59,60,61] |
| Hsp40 (Heat shock protein 40) | Family of J-domain proteins, enhances Hsp70’s ATP hydrolysis, one of the 5 components in vitro reconstitution system, forms the initial contact in SR folding. | [29,30,62] |
| Hsp110 (Heat shock protein 110) | Sse1 in yeast, related to and nucleotide exchange factor of Hsp70, enhances GR maturation in the yeast model. | [63,64] |
| HSPBP1 (Hsp70 binding protein 1) | Nucleotide exchange factor of Hsp70. Overexpression in mammalian cells decreases GR-dependent transcription. | [53,58] |
| Hsp90 cofactors | ||
| Aha1 (Activator of Hsp 90 ATPase activity 1) | Stimulates the ATPase activity of Hsp90. Enhances GR function in the yeast model and in mammalian cells. | [65,66,67] |
| Cns1 (cyclophilin seven suppressor) | TPR domain containing protein. Evidence from yeast suggests enhancement of GR function. | [68,69] |
| Cyp40 (Cyclophilin 40, Cpr6 and Cpr7 in yeast) | TPR domain containing protein and PPIase. Influences GR in some yeast models, no association and effect shown in mammalian cells. | [70,71,72,73] |
| FKBP51 (FK506 binding protein 51) | TPR domain containing protein and PPIase. Potent inhibitor of GR in mammalian cells. Decreases hormone affinity and delays nuclear translocation. | [7,8] |
| FKBP52 (FK506 binding protein 52) | TPR domain containing protein and PPIase. Potentiates GR activity in the yeast model. Context-dependency in mammalian cells. | [7,9,74,75] |
| FKBPL (FK506 binding protein like, WISp39) | TPR domain containing protein with PPIase-like domain. Affects levels, nuclear translocation, and transcriptional activity of GR in a cell line dependent manner. | [76] |
| P23 (also known as PTGES3, prostaglandin E synthase 3) | Part of the basal in vitro reconstitution system. Promotes Hsp90’s ATP-bound state and substrate-interaction. Context-dependent action on GR activity, chromatin effects. | [2,45,46,47,77,78] |
| PP5 (protein phosphatase 5, Ppt1 in yeast) | Contains TPR domain protein, dephosphorylates Hsp90. Enhances Hsp90 and GR activity in the yeast model. Overexpression reduces GR activity in mammalian cells. | [71,79,80,81,82] |
| Tah1 (TPR-containing protein associated with Hsp90) | TPR domain containing protein, in complex with Pih1. Evidence from yeast suggests stimulatory effect on GR. | [27] |
| SGT1 (Suppressor of G2 allele of Skp1) | Enhances Hsp90 chaperone cycle and GR activity in the yeast model. | [39] |
| XAP2 (HBV X- Associated Protein 2 = FKBP37) | TPR domain containing protein with PPIase-like domain. Interferes with GR activity upon overexpression in mammalian cells. | [83] |
| Cofactors for both Hsp70 and Hsp90 | ||
| CHIP (Carboxy- terminus of Hsp70- binding protein | TPR domain containing protein, has E3 ubiquitin ligase activity. Reduces GR protein levels and function in mammalian cells. | [71,84] |
| Hop (Hsp70–Hsp90 organizing protein) | Part of the basal in vitro foldosome reconstitution system. Mediates the transfer from the Hsp70- to the Hsp90-based folding platform. | [2,34,85] |
| TPR2 (TPR repeat protein 2 = DNAJC7) | Features TPR- and J-domain. Stimulates ATP hydrolysis and polypeptide binding by Hsp70. Excess inhibits GR folding in vitro and activity in vivo. | [71,86] |
| SGTA (Small Gluta- mine-rich TPR-con- taining Protein α) | TPR domain containing protein. Knock-down and ectopic expression in yeast and mammalian cells evidence an inhibitory role on GR function. | [87] |
Most of the known GR-regulating Hsp90 cofactors have been reviewed before [2,3,34,88]. Briefly, an important group of Hsp90 cofactors competes for access to Hsp90 and the associated hetero-complexes by binding to the N-terminal EEVD-motif of Hsp90 through their tetratricopeptide repeat (TPR) motif domain [5,71,89]. This combinatorial assembly creates a wide variety of compositions and regulatory possibilities. Some of the TPR-proteins bring additional biochemical functions to the Hsp90 complex, such as phosphatase activity in the case of PP5 or peptidylprolylisomerase (PPIase) activity in the case of most TPR-containing immunophilins. While clear evidence has been provided for the involvement of PP5’s phosphatase activity in GR function through changing both Hsp90 and GR phosphorylation [79,80,90], the role, if any, of the PPIase activity of GR-regulating immunophilins such as FKBP51 and FKBP52 remains unknown [6,74]. The PPIase domain, however, likely plays a crucial role in regulating GR, with effects on hormone binding affinity and nuclear translocation [7,9,74,75]. The competitive interaction of the TPR proteins with Hsp90 is an example to illustrate the dependency of the influence of a given TPR protein of the cellular context, as its effect is modulated by the abundance and nature of the other available TPR proteins. For example, while FKBP52 potentiates GR function in the yeast model [9,74], its overexpression in mammalian cells produces very little, if any, effect on GR, likely due to the presence of numerous other TPR cofactors that already promote GR function [7,71].
More recently described GR regulating TPR-containing Hsp90 cofactors are CNS1, FKBPL, SGTA, SGT1, Tah1, and XAP2 (Table 1). Both inhibitory and stimulatory effects on GR function have been described for these proteins, depending on their abundance as well as the cellular context [27,39,68,69,76,83,87]. Whether or not they are also involved in the regulation of intranuclear GR mobility and recycling, similarly to other chaperones and cochaperones, remains to be elucidated [45,91,92,93,94,95].
3. Structure and Biochemical Function of FKBP51
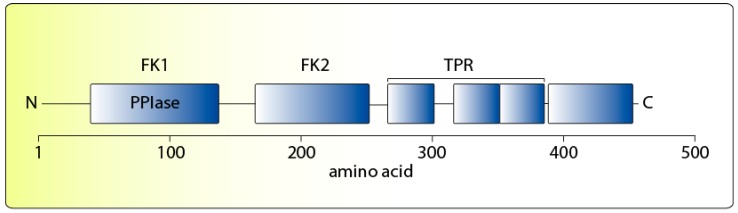
FKBP51 and FKBP52 share high homology (60%, similarity 70%) and the same domain structure [96] (see also Figure 2). They encompass two N-terminal domains with homology to FKBP12 (FK1 and FK2); only FK1 binds the immunosuppressive drug FK506 (also called tacrolimus) and is biochemically active in expediting the isomerization of peptidyl-prolyl bonds of model peptides with similar activity between FKBP51 and FKBP52 [97,98]. The C-terminal TPR domain endows the two FKBPs with the ability to interact with the EEVD motif at the C-terminal region of Hsp90 [7,89,99], in competition with all the other available TPR proteins in the cell [71,100]. Amino acids downstream of the TPR domain at the very C-terminal end of FKBP51 have also been reported to contribute to Hsp90 binding [101].
Figure 2.
Schematic representation of the FKBP51 protein domain structure. TPR = tetratricopeptide repeat domain conveying Hsp90 interaction.
The FK1 domain is the primary determinant for the divergent actions of FKBP51 and FKBP52 on GR [7,9]. However, while this PPIase domain is important, its enzymatic function is not. It has been proposed that the association of the FKBPs with Hsp90 in its “closed” conformation [26] enables the FK1 domain to contact the ligand binding domain of GR and other SRs, thereby influencing the conformation and hormone binding affinity [96]. The structures of the FK1s of the two immunophilins differ around the PPIase pocket by several residues of a proline-rich region [74,98,102], which likely causes differential protein interaction and the differential effects on GR activity [74,96]. With respect to GR function, it has not been resolved yet whether FKBP51 contributes a specific effect in inhibiting the receptor or merely competes with GR-stimulating TPR proteins such as FKBP52 out of the Hsp90-based heterocomplex.
FKBP51 has originally been characterized as chaperone and Hsp90 co-chaperone, i.e., involved in the process of folding individual proteins [97,103,104]. As outlined before, it is particularly challenging to prove the in vivo relevance of the PPIase activity and this has not yet been achieved, neither for FKBP51 nor for FKBP52 [6,74]. An increasing number of studies suggests that the most important molecular action of FKBP51 is its ability to serve as protein scaffolder that associates with several regulatory proteins, thus influencing a variety of signal transduction pathways (see Section 5 below for more details).
4. Regulation of FKBP51
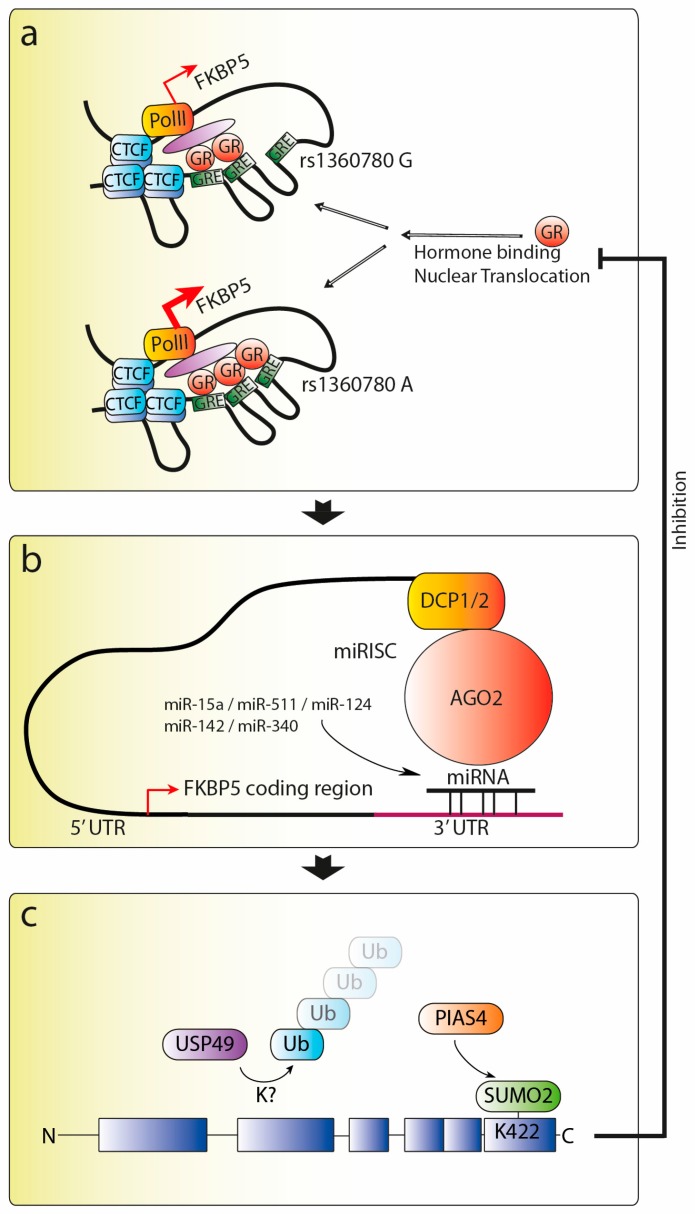
Not only is FKBP51 a multifunctional protein (see Section 5), but it is also regulated at multiple layers. Specifically, it can be transcriptionally regulated by several factors, genetic polymorphisms, and DNA methylation, ultimately determining the expression levels of the FKBP5 gene (see Section 4.1). A couple of microRNAs (miRNAs) have been recently proposed to target the FKBP5 mRNA, thereby regulating FKBP5’s mRNA stability and/or translation into a functional protein (see Section 4.2). A final layer of modulation takes place post-translationally, influencing the protein stability, and, as we will discuss below, the interaction between FKBP51 and the GR (see Section 4.3). A schematic representation of the different layers of FKBP51 regulation is provided in Figure 3. Altogether, this multitude of regulatory mechanisms allows for a complex modulation of the FKBP51-GR system in response to environmental stimuli and underlies its association with several pathological conditions, as discussed in the following sections.
Figure 3.
Model of FKBP51’s regulation at different levels. (a) Transcriptionally, FKBP5 is modulated by several single nucleotide polymorphisms, including the functional rs1360780 located within intron 2 and chromatin organizing proteins such as the CCCTC-binding factor. The scheme in Panel A is a hypothetical combination of two chromatin architecture studies [105,106]; (b) post-transcriptionally, microRNAs can interfere with the stability and translation of the FKBP5 mRNA molecules; (c) post-translationally, FKBP51 is regulated by post-translational modifications, such as ubiquitination (probably at lysines 352 und 376, www.nextprot.org/entry/NX_Q13451/proteomics) and SUMOylation. FKBP51 not only is engaged in various signaling pathways (see below), but also forms an ultrashort negative feedback loop by inhibiting its own production at the transcriptional level. Abbreviations: AGO2 = argonaute 2 (the catalytic subunit of the RISC complex); GR = glucocorticoid receptor; GRE = glucocorticoid response element; CTCF = CCCTC-binding factor; DCP = decapping protein; miRNA = microRNA; PIAS = protein inhibitor of activated STAT; PolII = RNA polymerase II; RISC = RNA-induced silencing complex; SUMO = small ubiquitin-like modifier USP = ubiquitin-specific peptidase; UTR = untranslated region.
4.1. Transcriptional Regulation
The gene encoding FKBP51 is FKBP5, which in humans is located at 6p21.31 and consists of 13 exons and 12 introns spanning more than 150 kb of genomic DNA [107]. The most important hallmark of the FKBP5 gene, at least with respect to GR regulation, is the presence of functional glucocorticoid responsive elements (GREs) constituting an intracellular short feedback loop [6,106,108]. It has been suggested that FKBP4 (the gene encoding FKBP52) and FKBP5 evolved from the same gene through duplication [109]. Nevertheless, glucocorticoid-dependent transcription has been described only for FKBP5. The structure of the FKBP5 gene is very similar to that of FKBP4, including their exon–intron boundaries, with the exception that FKBP5 presents three extra non-coding exons at its 3′ region [107]. Several polymorphisms have been identified in the FKBP5 gene, including functional ones with known implications in FKBP5 expression and function [110]. Around 613 single nucleotide polymorphisms (SNPs) and 57 indels (insertions/deletions), including a 3.3 kb deletion, have been discovered so far [107]. One SNP in particular, rs1360780, has been associated with significantly higher FKBP51 levels, as it is linked to differences in GR sensitivity [12]. This same SNP, which is in close proximity to a glucocorticoid-responsive element (GRE) in intron 2 of the FKBP5 gene, has also been shown to alter the extent of mRNA and protein induction following GR activation [105] (Figure 3a).
As previously mentioned, FKBP5 is a target for several transcription factors, among which the GR is the most commonly discussed in the context of stress. Chromatin-immunoprecipitations and reporter gene analyses identified GREs located several kilobases upstream and downstream of the transcriptional start site as determinants of the glucocorticoid response of the gene [106,108]. This entails long-range interactions and the involvement of chromatin organizing proteins such as CCCTC-binding factor (CTCF), a general determinant of chromatin domains [105,106,111] (Figure 3b). Interestingly, binding sites for several transcription factors are not primarily located in the promoter region; in fact, several relevant binding sites are located within intronic and distal regions of the FKBP5 gene [107]. These regions appear to have an even stronger glucocorticoid responsiveness compared to the promoter region itself, with some studies showing an absence or a very low response of the promoter sequence to glucocorticoids [108,112]. Importantly, some of these distal regions show promiscuous responsiveness to other hormone receptors in addition to GR, such as the androgen receptor (AR) and the progesterone receptor (PR) [113]. In particular, AR has been shown to selectively recruit cAMP response element-binding protein to a distal enhancer located 65 kb downstream of the transcription start site in the fifth intron of the FKBP5 gene, ultimately leading to chromatin remodeling and modulation of its expression [113].
As another means of regulation, binding of transcription and chromatin architecture factors is influenced by DNA methylation at the FKBP5 locus, which can directly mediate the effects of environmental stimuli on the gene. In mice, chronic (4 weeks) exposure with corticosterone decreases DNA methylation at distinct CpGs along with enhanced expression of Fkbp5 [114]. In this model, DNA methylation was reversible and reappeared after the cessation of corticosterone exposure. Several studies have provided evidence that DNA methylation across different regions of the FKBP5 gene can influence its expression and interact with specific polymorphisms. For instance, hypermethylation of the FKBP5 gene has been associated with a reduced expression of specific transcript variants in patients with MDD [115]. Indirect evidence for a role of DNA methylation in FKBP5 transcription is often provided by associations with physiological parameters. For example, a positive correlation between FKBP5 methylation with a thickness of the transverse frontopolar gyrus has been reported in MDD patients [116]. A landmark study has shown that childhood trauma can reduce DNA methylation specifically in the intron 7 of the FKBP5 gene in subjects presenting the risk allele of the rs1360780 SNP [105], suggesting a complex interplay between genotype, DNA methylation, and the effects of environment on the FKBP5 gene transcription. DNA methylation apparently also contributes to the upregulation of FKBP5 transcription with age [117].
Finally, the FKBP5 transcription has also been shown to be controlled in a tissue-specific manner, probably through a combination of the aforementioned main mechanisms. In particular, a study performed in mice showed that Fkbp5 is expressed throughout the entire brain, with some regions showing higher expression than others [109]. In particular, regions with a high baseline expression of Fkbp5, such as the hippocampus, the premammillary nucleus, and the motor nuclei of the nervus trigeminus and nervus facialis, tend to show a more modest induction of Fkbp5 expression in response to dexamethasone treatment (a synthetic glucocorticoid analog) [109]. In other words, dexamethasone treatment can induce a much more prominent induction of Fkbp5 expression in tissues where its baseline expression is typically low, such as the hypothalamic paraventricular nucleus or the central amygdala, in line with the reported role of FKBP51 as a negative regulator of GR sensitivity [109].
4.2. Post-Transcriptional Regulation
Different studies have suggested a post-transcriptional regulation of FKBP5 expression by means of modulating its mRNA stability. Independent studies indicate the role of specific miRNAs through interference with FKBP5 mRNA stability and translation by binding to the 3’UTR region. After the processing and biogenesis of mature miRNAs, these regulatory molecules are incorporated into a ribonuclear particle to form the RNA-induced gene silencing complex (RISC) as part of their translational repressor role [118]. An efficient search complex for specific target mRNAs is also facilitated by the association of miRNAs with argonaute (Ago) proteins [119]. Not only has FKBP5 mRNA been detected in the argonaute RISC catalytic component 2 (Ago2) complex [120], but it has also been shown to bind argonaute and the trinucleotide repeat-containing proteins, which are both part of miRNA-induced silencing complexes [107].
So far, five different miRNA molecules have been reported or predicted to bind to the FKBP5 mRNA: miR-124 [107], miR-15a [120], miR-142 [120], miR-340 [120], and miR-511 [121] (Figure 3b). Specifically, miR-124 was shown to bind to FKBP5’s exon 9 using data from photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation (PAR-CLIP) [107], although this finding has yet to be replicated. Additionally, by investigating the effects of chronic stress in mice, a recent study found that an increase in miR-15a levels in the amygdala-Ago2 complex was associated with a concomitant reduction in the levels of FKBP5 mRNA and FKBP51 protein [120]. The authors were able to confirm FKBP5 as a target of miR-15a by a luciferase assay and by showing that the seed sequence for this miRNA binding was highly conserved at the 3’UTR of FKBP5. Interestingly, this miR-15a-specific downregulation of FKBP5 expression was later suggested to underlie an important coping mechanism to chronic stress, which warrants further investigation [120].
Finally, by using multiple programs to predict potential miRNA candidates that could target FKBP5, three miRNA were identified with potential binding to this gene: miR-142, miR-340, and miR-511 [121]. Although all three miRNAs were shown to regulate FKBP5 mRNA expression in vitro, only miR-511 was found to robustly reduce FKBP51 protein levels as well [121]. miR-511 has been shown to suppress the glucocorticoid-induced upregulation of FKBP51 in cells and primary neurons, suggesting an important role of this miRNA in FKBP51’s modulatory function in GR activity [122]. In addition, this miR also contributes to age-dependent regulation of FKBP5 and neuronal development and differentiation [121], suggesting it as a potential target in the modulation of FKBP51.
4.3. Post-Translational Regulation
The final level of regulation comes with the modification of the FKBP51 protein itself by post-translational mechanisms. These include the chemical addition of functional groups or proteins to FKBP51, in addition to the cleavage or degradation of the protein or specific regulatory subunits. In general, the most common post-translational modifications (PTMs) include phosphorylation, glycosylation, ubiquitination, nitrosylation, methylation, acetylation, lipidation, and SUMOylation.
Overall, evidence for PTMs on FKBP51 is scarce. As opposed to FKBP52, FKBP51’s sequence lacks a phosphorylation site on its FK linker region (a seven- to nine-amino-acid loop that connects the FK1 and FK2 domains) [123]. Although several phosphorylation sites are known for FKBP51 (https://www.phosphosite.org), their functional relevance has not been revealed yet. However, the two peptide modifications ubiquitination and SUMOylation have been reported in the modulation of FKBP51 and its function [124]. SUMOylation of proteins involves the covalent addition of a member of the SUMO (small ubiquitin-like modifier) family of proteins to a protein target. Once bound, the SUMO can have one of three non-mutually exclusive effects: it can interfere with the binding of another protein, it can act as a recruiter of new interacting proteins, or it can lead to conformational changes in the SUMOylated substrate [125]. Ubiquitination also can have a number of different effects, depending on the type of chaining of several ubiquitin moieties, where marking proteins for proteasomal degradation is the prototypic case [126].
FKBP51 can be SUMOylated at Lys422 to allow for its association to the GR complex though interaction with Hsp90 [124]. Accordingly, FKBP51 mutants that could not be SUMOylated failed to interact with Hsp90 and GR, suggesting an important modification in FKBP51’s activity on GR signaling [124]. FKBP51 is further modified by the ubiquitin specific peptidase (UPS) 49. Association between USP49 and FKBP51 has been demonstrated, leading to deubiquitination and stabilization of FKBP51 and further consequences with respect to FKBP51-dependent Akt1 activity [127] (Figure 3c).
5. Functions of FKBP51
FKBP51 is a protein with many described functions in addition to its GR modulatory properties. Not only does it interact with several other co-chaperones to regulate SRs function, but it has also been suggested to modulate pathways related to the immune function, autophagy, epigenetic remodeling, apoptosis, cell growth, cytoskeleton dynamics, and metabolism, among others. In this section, we will briefly discuss some of its known functions in signaling pathways and their relevance for health and disease.
5.1. Regulation of Immune Pathways
The most common role of FKBP51 is linked to its regulatory effect on the action of GR, as previously discussed in this review. Given the known function of glucocorticoids and GR on inflammation [128,129], it is not surprising that FKBP51 has been shown to present direct and indirect effects on immune and inflammatory pathways as well. Specifically, FKBP51 is thought to influence the nuclear factor kappa-light-chain-enhancer of activated B cells (NFκB) system by several different mechanisms. This family of molecules acts as transcription factors that regulate immune and inflammatory responses, among other processes [128], and is considered one of the major regulators of the expression and production of inflammatory cytokines [130].
FKBP51 has been shown to physically associate with the inhibitor of κB kinase α (IKKα), IKKε, TGF-β activated kinase 1, and mitogen-activated protein kinase kinase kinase [131,132], all of which belong to the NFκB signaling pathway. The direct effect of FKBP51 on NFκB, however, is still under investigation. A preclinical study using an animal model of chronic constriction injury showed that FKBP51 knockdown reduced the production of proinflammatory cytokines in the dorsal root ganglion of rats [133], and the same study showed that the inhibition of FKBP51 significantly reduced the activation of NFκB in the same model [133]. One of the mechanisms by which FKBP51 may act here is by inhibiting calcineurin activity [134], which counteracts NFκB activation by dephosphorylating IκB and preventing its proteasomal degradation [131,135]. In addition, a role for FKBP51 in the innate immune response to viral infection has been proposed, which is mediated by the FKBP51 interaction with TNF receptor-associated factor proteins [136]. Finally, FKBP51 has also been shown to modulate the immunosuppressant effects of myeloid-derived suppressor cells by increasing inducible nitric oxide synthase, arginase-1, reactive oxygen species, and enhancing NFκB activity in vivo [137].
5.2. Regulation of the AKT Pathway
The AKT/protein kinase B signaling pathway is a pivotal player in the control of cellular function both in health and in pathological conditions [138]. Among several regulatory mechanisms, its activity can be modulated by the phosphorylation status of two sites, namely Ser473 and Thr308 [139]. FKBP51 has been shown to regulate AKT activity and to function by interfering with its phosphorylation through a scaffolding mechanism. Studies have shown that FKBP51 can interact with AKT at multiple domains independently of their phosphorylation status, either directly or indirectly via Hsp90 [140]. Briefly, FKBP51 can increase the interaction between AKT (which binds to the FK1 domain) and the pleckstrin homology domain leucine-rich repeat protein phosphatase (PHLPP), which binds to FKBP51’s C-terminus [139]. By doing so, FKBP51 facilitates the dephosphorylation of AKT at Ser473 by PHLPP. Finally, a recent study also showed that the FKBP51 expression status may determine whether and to which extent antidepressants alter the phosphorylation of AKT Set473, which is a key mechanism in the action of these drugs [141].
5.3. Regulation of Microtubule Dynamics
One of the roles proposed for FKBP51 is the regulation of the cytoskeleton, more specifically microtubule dynamics. Studies have shown that it can promote the stabilization of microtubules by interacting with tau in a complex with Hsp90 [142,143]. The mechanism proposed is that phosphorylation of tau leads to its association with FKBP51 and isomerization to a cis configuration; this eventually allows for its dephosphorylation by the phosphatase PP5 and its recycling and stabilization to microtubules [142,143]. Interestingly, as seen in the FKBP51 role on GR modulation, FKBP52 exerts the opposite effect. FKBP52 has been shown to directly interact with tubulin whether polymerized or not, ultimately preventing or inhibiting microtubule formation [144].
Of note, this microtubule-modulating property of FKBP51 and FKBP52 can have important effects on determining neurite length in neuronal cells, which might have further consequences in terms of cognitive and behavioral functions in animals. Accordingly, neurite outgrowth has been shown to reduce after the overexpression of FKBP52 in PC12 cells [144], and the balance between FKBP51 and FKBP52 has been suggested to play a key role during the early mechanisms of neuronal differentiation [145].
5.4. Modulation of Autophagy
Based on initial observations that FKBP51 could shift the cellular response to irradiation from apoptosis to autophagy [146], some recent studies have focused on the mechanisms by which FKBP51 could play a role in autophagy and related mechanisms [141,147]. Autophagy is an important and highly regulated cellular process responsible for targeting damaged proteins, lipids, glycogens, and organelles from the cytosol to lysosomes for destruction [148]. The hypothesis that FKBP51 could control autophagy was also supported by evidence of glucocorticoid-induced autophagy [149] and that the autophagy regulators AKT and Beclin1 were among the proteins controlled by Hsp90 and/or its co-chaperone FKBP51 [103].
Accordingly, FKBP51 has been shown to bind to Beclin1 and alter its phosphorylation status, ultimately promoting the induction of autophagic pathways [141]. It does so by interacting with PHLPP and AKT and thereby favoring AKT’s dephosphorylation at Ser473, which leads to Beclin1 recruitment and dephosphorylation [141]. Interestingly, antidepressants have been shown to act in a synergistic way with FKBP51 in the regulation of autophagy, and the baseline FKBP51 levels actually correlated with the potential of antidepressants to induce autophagy [141]. Based on recent evidence of an association between autophagy and neurotransmission [150], these FKBP51-mediated processes might eventually underlie pathological mechanisms in depression and other neuropsychiatric conditions, as discussed in Section 5.7.
5.5. Regulation of DNA Methylation
Many stress-related conditions and neuropsychiatric disorders show alterations in DNA methylation markers and related mechanisms. Early-life and lifetime stresses have also been consistently shown to induce epigenetic alterations and long-lasting DNA methylation changes, which suggests the GR-FKBP51 system as a player in the establishment and modulation of these processes. In fact, FKBP51 has been recently shown to control the phosphorylation and activity of DNA methyltransferase 1 (DNMT1), a key epigenetic enzyme responsible for the maintenance of DNA methylation [151].
DNMT1 can be regulated by the action of several different molecular interactions and posttranslational modifications [152]. Among its regulatory mechanisms, the phosphorylation of Ser154 by cyclin-dependent kinases (CDKs) is of particular importance [153]. An interesting model has been proposed in which FKBP51 and FKBP52 compete for the binding to CDK5 based on their structural similarity [151]. When overexpressed, FKBP51 was shown to displace FKBP52 from CDK5, which decreases the interaction between CDK5 with DNMT1 [151]. This was shown to reduce DNMT1 phosphorylation at Ser154 and ultimately its enzymatic activity, leading to a global DNA demethylation in vitro [151]. Altogether, these results suggest a key role for FKBP51 in modulating DNA methylation. As discussed in the previous sections, the FKBP5 gene itself can also be regulated by methylation, suggesting another modulatory feedback loop with potential impact in health and disease [6]. For example, epigenetic upregulation of FKBP5 transcription with age has been reported to impair stress-resiliency [117]. This mechanism, together with age-dependent miR-511 activity [121], may also contribute to the genetic association of FKBP5 with cognitive function in aged individuals [154]. In line with a role in aging, elevated levels of FKBP5 transcripts have been correlated with shorter telomere length in depression [155]. However, it should be noted that FKBP51 has been reported to enhance human telomerase reverse transcriptase activity [156], so that the mechanistic basis requires further investigation.
5.6. Regulation of Metabolism
Dysregulation of the HPA axis in obesity has been reported in several studies. Not coincidently, there is emerging literature on the role of FKBP51 in metabolism regulation and obesity-related conditions [157]. In fact, glucocorticoids can induce FKBP5 expression in the adipose tissue, where higher FKBP5 expression has been associated with markers of insulin resistance, such as insulin, homeostatic model assessment of insulin resistance, subcutaneous adipocyte diameter [158], and lower levels of plasma HDL (high-density lipoprotein)-cholesterol [158]. Accordingly, FKBP5 SNPs have been associated with type 2 diabetes [158], insulin resistance, and elevated serum triglycerides [159]. It is likely that the dysregulated HPA axis activity seen in carriers of the FKBP5 risk variants may mediate lipolysis and insulin resistance, hindering vulnerable subjects that require a rigorous glycemic and lipid control as part of their routine management [159]. In addition, FKBP5 variants have also been associated with significantly lower weight loss after bariatric surgery [160].
In mice, Fkbp5 expression was found to be responsive to a high-fat diet and chronic social defeat stress in the hypothalamus and hippocampus, respectively, and hypothalamic Fkbp5 expression was found to be related to an increased body weight [161]. Work on mice and rats demonstrated that fasting induces FKBP51 in ventromedial, paraventricular, and arcuate hypothalamic nuclei as well [162]. In addition, virus-mediated overexpression of FKBP51 in the hypothalamus led to an elevated body weight in mice on a high-fat diet [162]. Conversely, mice lacking FKBP51 are resistant to diet-induced obesity [163], consistent with the finding that FKBP51 controls adipogenesis [164,165]. Altogether, these findings suggest a novel role of FKBP51 as an important target in metabolic regulation.
5.7. Implications in Diseases: Focus on Depression
Because of the plethora of functions and means of regulation of FKBP51, it is not surprising that this system has been associated with several medical conditions. The most common of those are related to acute or chronic exposure to stress, such as most neuropsychiatric disorders, mostly due to FKBP51’s stress inducibility and possibly to its key role in controlling GR function. With different degrees of involvement, FKBP51 has been previously associated with substance abuse [166,167], autism [168], bipolar disorder [169,170], depression [12,116], schizophrenia [171], post-traumatic stress disorder (PTSD) [172,173], suicide [174], attention-deficit/hyperactivity disorder [175], anxiety [176], and the effects of early trauma and stress in general [177,178,179].
Among all of these conditions, depression is the most studied and has been consistently associated with FKBP51 in multiple independent populations. Accordingly, major depressive disorder (MDD) has been linked to a dysfunction in the HPA axis by several studies [180,181,182], and a landmark study from 2004 showed the role of FKBP5 in antidepressant response causing the initial boom in the study of FKBP51 in clinical populations [12].
A summary of clinical studies investigating the association between depression and FKBP51 is provided in Table 2. As can be seen, the vast majority of studies have focused on associations between depression and the FKBP5 genotype by investigating multiple single nucleotide polymorphisms (SNPs) throughout the gene (with a particular focus on the initially reported rs1360780). In general, it is currently hypothesized that depressed patients have increased basal FKBP51 levels [155,183,184] (although not observed in all populations [115,185]) that may be leading to a GR resistance [186,187]; alternatively, the basal promoter activity may be less relevant than the stress reactivity of the promoter that is shaped by genotype and epigenotype [6]. Moreover, the FKBP5 genotype has been reported to interact with the MDD diagnosis to predict structural neuroanatomical changes [116,188,189,190], as well as with prior lifetime trauma and/or stress to increase the risk for depression [191,192,193,194]. A recent meta-analysis performed with 26,582 subjects has also confirmed a significant association between both rs1360780 and rs3800373 with MDD [195].
Table 2.
Studies investigating the association between FKBP51 and depression.
| Ref. | FKBP51 Measure | Sample | Finding |
|---|---|---|---|
| [183] | Protein levels in blood | Patients with MDD (n = 30) and healthy controls (n = 35) | Higher FKBP51 levels in the cytoplasm of blood cells from depressed patients. Significant correlation between cytoplasmic GR and FKBP5 levels. |
| [186] | mRNA levels in blood after dexamethasone administration | Two cohorts: (1) n = 18 cases/18 controls; (2) n = 11 cases/13 controls | Lower FKBP5 induction after dexamethasone in patients compared to controls. |
| [185] | mRNA levels in the hippocampus (cornu ammonis and dentate gyrus) | MDD patients (n = 43) and healthy controls (n = 43) | No difference between groups. No association between FKBP5 expression and hippocampal volumes. |
| [206] | mRNA levels in blood | Pregnant women with a lifetime history of mood or anxiety disorders (n = 106) | Effect of gestation trimester in upregulating FKBP5 expression. Reduced magnitude of upregulation of FKBP5 across pregnancy in depressed women. |
| [155] | mRNA levels in the hippocampus | MDD patients (n = 10) and controls (n = 10) | Increased FKBP5 mRNA levels in patients compared to controls. |
| [184] | Protein and mRNA levels in the frontal cortex | Controls (n = 12), MDD patients (n = 12), MDD with psychosis (n = 12), HIV+ (n = 11), and HIV+ with MDD (n = 11) | Elevated FKBP51 at both the transcript and protein levels correlated with MDD. Higher frequency of the rs3800373 CC genotype in the MDD and MDD/psychosis groups. |
| [207] | mRNA levels in blood and genotype (rs3800373) | MDD patients at first visit and euthymic at second visit (n = 56); euthymic patients at first visit and MDD at second visit (n = 30) | Correlation of change in the severity of depressive mood with FKBP5 gene expression in individuals homozygous for GG of the SNP rs3800373. |
| [115] | DNA methylation and mRNA levels in PBMCs | Controls (n = 20), MDD patients with (n = 14) or without serious suicidal ideation (n = 10) | Hypermethylation of FKBP5 gene in MDD patients with concomitant reduced expression of FKBP5 transcript variants 1, 2, and 3. Further analyses showed that differences were primarily seen in the MDD-suicide group compared to controls. |
| [116] | Genotype (rs1360780) and DNA methylation | MDD patients (n = 114) and healthy controls (n = 88) | Interaction between the FKBP5 genotype and MDD diagnosis on gray matter volumes of several brain regions. Allele-specific positive correlation of the FKBP5 gene methylation with thickness of the transverse frontopolar gyrus. |
| [191] | Genotype (rs1360780, rs9470080, rs9394309) | N = 1431 participants with available data on early life stress and depressive symptoms at midlife | FKBP5 SNPs interacted with early life stress exposure, but not with recent stressful life events, in predicting self-reported depressive symptoms in midlife. |
| [208] | Genotype (rs1360780) | N = 131 outpatients with moderate to severe depression | The FKBP5 rs1360780 T allele was significantly associated with antidepressant treatment increasing suicidal ideation, with a partly drug-specific pattern of association. |
| [209] | Genotype (rs1360780) | N = 922 hospital staff members (depressive state, n = 309; non-depressive state, n = 613) | Significant association of the FKBP5 SNP as main effect on depressive state. No significant association between the depressive state and the SNP x stressful life events interaction. |
| [190] | Genotype (several SNPs) | Inpatients with unipolar depression (n = 268) and controls (n = 284) | Association between SNPs (especially rs3800373 and rs4713916) in the FKBP5 gene with vulnerability to unipolar depression. Association of the FKBP5 genotype with HPA axis activity after citalopram treatment, as well as with right hippocampal volume. |
| [210] | Genotype, mRNA expression and DNA methylation in blood | Patients with remitted major depression (n = 61) and healthy controls (n = 55) | Genotype-dependent plasma cortisol response to psychosocial stress exposure in controls (not in patients). rs1360780 T-carrier controls responded with a blunted FKBP5 mRNA expression after psychosocial stress. Depression- and genotype-specific differences in FKBP5 methylation in intron 7. |
| [211] | Genotype (rs1360780) | N = 489 children | Significant three-way interaction between FKBP5 genotype, victimization, and child sex predicting depressive symptoms. |
| [212] | Genotype (rs1360780) | Children of mothers with (n = 81) and without (n = 81) a history of depression | Children of depressed mothers who carried the reactive genotype of FKBP5 rs1360780 exhibited less sustained attention to sad faces and more sustained attention to happy faces (information-processing bias). |
| [188] | Genotype (rs1360780) | N = 115 monozygotic twin pairs discordant or concordant for depression | Additive effect of right hippocampal connectivity alterations and FKBP5 genotype on depression risk (CC genotype carriers who have low nodal strength in the right hippocampus show higher depression risk). |
| [213] | Genotype (several SNPs) | Patients with MDD (n = 218) and controls (n = 742) | Association between five FKBP5 SNPs (rs1360780, rs9470080, rs4713916, rs9296158, and rs9394309) and MDD. Two haplotype combinations were significantly more frequent in MDD than in controls. |
| [214] | Genotype (rs1360780) | Patients with a first primary cancer diagnosis (n = 7,320) | No association between FKBP5 genetic variant and use of antidepressants or hospital contact for depression after diagnosis of cancer. |
| [215] | Genotype (rs1360780, rs9296158, rs3800373, rs9470080) | N = 361 pregnant women | No association between FKBP5 genotypes or haplotypes with depressive symptoms during and after pregnancy. |
| [187] | Genotype (rs1360780) | Inpatients in a depressive episode (n = 68) and healthy controls (n = 87) | Significant interaction between MDD and T allele on GR-stimulated FKBP5 mRNA expression (reduced induction of FKBP5 mRNA in T carriers). GR resistance in T carrier patients. |
| [216] | Genotype (several SNPs) | N = 2928 participants with genetic data and information about depressive symptoms | Significant association between minor alleles of 4 FKBP5 SNPs (rs9470080, rs9394309, rs7748266, and rs1360780) with decreased cortisol area under the curve and increased risk of depressive symptoms. |
| [217] | Genotype (rs1360780) | N = 344 outpatients with chronic hepatitis C initiating IFN-alpha and ribavirin therapy | No association between FKBP5 genetic variants and IFN-induced depression. |
| [192] | Genotype (rs3800373, rs1360780, rs4713916, rs9296158, rs9470080) | N = 884 participants (community sample) | No genetic main effect on major depressive episode. Interactions between the five SNPs and traumatic (but not separation) events (especially severe trauma). Homozygosity for the minor allele of selected FKBP5 SNPs suggested as a risk genotype for the development of a major depressive episode in subjects with prior trauma exposure. |
| [218] | Genotype (rs1360780, rs4713916, rs3800373) | N = 271 children/adolescents whose mothers had experiences at least two major depressive episodes and N = 165 controls | No differences in the frequency of the genotypes between the two groups. No association between FKBP5 genotypes and child and adolescent depression scores. |
| [12] | Genotype (several SNPs) and protein levels in blood | N = 294 depressed individuals and N = 339 controls | Individuals with the TT genotype (rs1360780) experienced more than twice as many depressive episodes before the index episode than the other genotypes. |
| [219] | Genotype (rs1360780, rs3800373, rs9296158, rs9470080) | N = 131 adult patients who have received a kidney transplant | FKBP5 rare alleles at three out of four SNPs in FKBP5 (rs1360780, rs9296158, and rs9470080) were associated with increased depressive symptoms. |
| [220] | Genotype (rs3800373) | N = 106 school-aged children | Attachment security was negatively associated with depressive symptoms among children with two minor alleles of the FKBP5 SNP. Maternal attachment anxiety was positively associated with depressive symptoms in these children. |
| [221] | Genotype (rs1360780) | N = 311 physically healthy subjects | Dysfunctional attitudes predisposing to depression were significantly higher in the group with the T-allele than in that without this allele (particularly in achievement and self-control). |
| [189] | Genotype (rs1360780) | Adult patients with MDD (n = 40) and healthy controls (n = 20) | T-allele carriers had a later onset of disease compared with CC homozygous patients. Patients expressing the T-allele exhibited functional and structural differences in areas involved in emotional perception and inhibition. Interaction between the risk allele and higher CTQ scores mediates structural alterations. |
| [222] | Genotype (rs1360780, rs4713916) | Depressed unrelated inpatients with a major depressive episode (n = 412) and controls (n = 634) | No difference in FKBP5 genotypes between groups. |
| [223] | Genotype (rs1360780) | Patients with depression (n = 457) and healthy controls (n = 2286) | T-allele and TT genotype were overrepresented in depression for men. |
| [176] | Genotype (rs7757037, rs1360780, rs4713902) | Patients with MDD (n = 657) and healthy controls (n = 462) | T-allele of rs1360780 was more frequent among patients with MDD with a comorbidity of anxiety disorders, compared to those without. |
| [193] | Genotype (rs1360780) | N = 2157 Caucasian subjects | Significant interaction of physical abuse with the TT genotype of rs1360780 was found to increase depressive symptoms. |
| [194] | Genotype (rs3800373, rs9296158, rs1360870, rs9470080) | N = 236 high-risk, low-income women | FKBP5 moderation of the indirect effects of maltreatment on depression and dissociation via limbic irritability. |
| [224] | Genotype (rs1360780) | N = 246 Alzheimer’s disease patients with or without major depressive disorder | Significant association of FKBP5 with MDD in Alzheimer’s disease. C-allele was associated with a higher risk of depression. |
| [195] | Genotype (rs1360780, rs4713916, rs3800373, rs755658) | Meta-analysis with a total sample size of 26,582, including 14,491 MDD patients and 14,091 controls | No significant association between any of the FKBP5 SNPs and MDD susceptibility when all samples were pooled. After removing one heterogeneous study, both the rs1360780 T-allele and the rs3800373 C-allele were significantly associated with MDD. |
| [225] | Genotype (rs1360780, rs3800373, rs9470080) | Meta-analysis with a total sample of 15,109 participants. | Individuals who carry T-allele of rs1360780, C-allele of rs3800373 or T-allele of rs9470080 exposed to early-life trauma had higher risks for depression or PTSD. |
GR = glucocorticoid receptor; IFN = interferon; MDD = major depressive disorder; PBMC = peripheral blood mononuclear cells; PTSD = post-traumatic stress disorder; SNP = single nucleotide polymorphism; STAR*D = Sequenced Treatment Alternatives to Relieve Depression.
Finally, 10 studies have investigated the role of FKBP5 in the response to treatment among depressed patients. As seen in Table 3, eight studies have confirmed this association [12,196,197,198,199,200,201], while two failed to show a role for the FKBP5 genotype on treatment response [202,203]. A meta-analysis from 2013 was also able to confirm this association and its SNP and ethnicity specificity [204]. The proposed role for FKBP51 in MDD treatment response is also supported by several studies showing the importance and dependence of FKBP51 function in the mechanism of actions of antidepressants, as discussed in Section 5.2, Section 5.4 and Section 6.1 [141,151,205].
Table 3.
Studies investigating the role of FKBP51 in antidepressant response.
| Ref. | FKBP5 Measure | Sample | Finding |
|---|---|---|---|
| [196] | mRNA levels in blood | Patients with MDD (n = 74) and healthy controls (n = 34) | Association between successful antidepressant response and reduction of FKBP5 mRNA levels after 8 weeks of treatment with escitalopram or nortriptyline. |
| [202] | Genotype (rs1360780, rs3800373) | N = 246 geriatric patients with depression | No association between FKBP5 and clinical outcomes after 8 weeks of treatment with paroxetine and mirtazapine. |
| [226] | Genotype (rs1360780) | Depressive outpatients (n = 159) and controls (n = 96) | T allele carriers showed a 2.10 increased risk for non-responding at 4th week to 12-week citalopram treatment (almost significant tendency). |
| [197] | Genoytpe (rs3800373, rs1360780) | N = 304 depressive inpatients with unipolar or bipolar depression | Carriers of the FKBP5 variants had a trend toward a higher chance to response (mainly in those treated with antidepressant drug combinations or with venlafaxine) |
| [203] | Genotype (rs1360780) | N = 125 outpatients with major depression (n = 119) or dysthymic disorders (n = 6) | No association between rs1360780 and short-term antidepressant treatment response (fluoxetine; 20 mg/day) or lifetime depressive episodes. |
| [12] | Genotype (several SNPs) | N = 233 depressed inpatients | Association between three SNPs in FKBP5 (rs1360780, rs1334894 and rs755658) with antidepressant response. In a replication sample, rs1360780 showed a significant association and rs3800373 showed a trend for an association with response to antidepressants. |
| [198] | Genotype (rs1360780) | N = 1953 STAR*D outpatients and N = 275 MARS in patients | Interaction between the TT genotype of FKBP5 rs1360780 and the GG genotype of GRIK4 (rs12800734) in the prediction of antidepressant response. |
| [199] | Genotype (rs1360780) | N = 298 inpatients with MDD | Interaction between FKBP5 genotype at rs1360780 and treatment mode. C-allele carriers had a significantly worse outcome when treated naturalistically. TT-genotype subjects showed a superior treatment response across both SSTR and TAU treatment conditions. |
| [200] | Genotype (several SNPs) | N = 529 MDD patients from the Mayo clinic and n = 96 from the STAR*D study | Association between the SNP rs352428 with 8-week SSRI treatment response in the Mayo study and 6-week treatment response in the STAR*D replication study. |
| [201] | Genotype (rs1360780, rs4713916) | Outpatients with non-psychotic MDD (n = 1809) and controls (n = 729) | rs1360780 was significantly associated with MDD in White non-Hispanics. Significant association between rs4713916 and remission. |
| [204] | Genotype (rs1360780, rs3800373) | Meta-analysis with a total sample of 2,194 subjects for rs1360780 and 2,049 for rs3800373 | rs1360780: In Caucasians T-allele carriers showed a marginal evidence of better response, while in the other/mixed ethnic subgroup a better response was seen in C/C homozygous subjects. rs3800373: In Caucasians C-allele carriers showed a trend of better response. |
MARS = Munich Antidepressant Response Signature; MDD = major depressive disorder; SSTR = structured, stepwise treatment algorithm; STAR*D = Sequenced Treatment Alternatives to Relieve Depression; TAU = treatment as usual.
6. Drugs and FKBP51
FKBP51 is being explored for possibilities for drug development, like other steroid receptor-associated immunophilins [88,227]. In general, two roles for FKBP51 in drug treatment emerge from the literature: (1) FKBP51 organizes signaling pathways that are relevant for the effects of other drugs. Thus, its status needs to be considered when targeting other proteins that link to FKBP51-influenced pathways; (2) FKBP51 may serve as direct drug target. Obviously, targeting FKBP51 directly also has the potential to impact drug effects that use FKBP51-dependent pathways.
6.1. FKBP51 Influences the Effects of Other Drugs
The multiple roles of FKBP51 in signaling transduction include pathways that are targeted by drugs. Most notable are anti-cancer drugs and antidepressants. Owing to the observation that the role of FKBP51 may depend on the type of cancer, its contribution to cancer drug treatment cannot be generalized. As reviewed elsewhere, while FKBP51 is upregulated in several human cancers, it is downregulated in others [228]. This may relate to differential importance of growth-regulatory pathways in different cancer types. For example, enhanced expression of FKBP51 can stimulate AR in prostate cancer [229,230] and thus drive malignancy, while as a negative regulator of Akt1 FKBP51 is important for the action of chemotherapeutic agents in pancreatic cancer cells [231].
The dependency of antidepressant drug treatment on the FKBP51 levels has been shown in several studies employing human cell lines, mouse models, and peripheral blood cells from depressed patients and healthy controls [12,141,147,151,205]. More specifically, the effect of antidepressants on mouse behavior and on the GSK3β pathway, global and local (BDNF) DNA methylation, and autophagy depend on FKBP51. Furthermore, the extent of the antidepressant effect on these FKBP51-directed pathways in patient-derived blood cells was predictive of clinical treatment success [141,151,205]. Even though the molecular mechanisms discovered are very plausible, causality has not yet been established.
6.2. FKBP51 as A Drug Target
As a member of the immunophilin protein family, FKBP51 binds to the immune suppressive drug FK506 (also called tacrolimus) [232]. FK506 binds to the catalytic pocket thereby blocking the PPIase activity of the FKBPs. Its physiological effects, however, are explained by the fact that it is a large molecule protruding out of the PPIase pocket [233], leading to changes in the interaction profile of the FKBPs. The best investigated example is the FK506-dependent interaction of FKBP12 and other FKBPs with calcineurin, which leads to the inhibition of this phosphatase and thus affects the activity of the transcription factor NFAT (nuclear factor of activated T-cells) as the basis of the immune suppressive effect of FK506 [234]. Since FK506 binds to all members of the FKBP family, other drugs need to be developed to achieve selectivity for FKBP51. It should be mentioned, though, that some of the effects of FK506 have been ascribed specifically to the inhibition of FKBP51, in particular by blocking the AR and thereby inhibiting prostate cancer cell growth [229,230].
Other efforts have been aimed at disrupting the interaction between Hsp90 and TPR proteins, including FKBP51. For example, the coumermycin-A1-related compound novobiocin has been reported as a C-terminal inhibitor of Hsp90 that prevents binding to FKBP51 and other TPR proteins at millimolar concentrations [235]. Furthermore, the small molecule SM145 (which binds between the N and middle domain of Hsp90) reduces interaction between Hsp90 and FKBP51, FKBP52 and other TPR proteins, which also leads to lower levels of FKBP51, FKBP52, and GR [236]. In contrast to N-terminal inhibitors of Hsp90, such as geldanamycin, this C-terminal inhibitor does not mount a heat shock response. Nevertheless, these compounds cannot be considered as specific to FKBP51.
When the structural details of the TPR–Hsp90 interaction were revealed, it was suggested that this interaction might serve as drug target to disrupt the interaction of Hsp90 with TPR proteins, including immunophilins [89,99]. In fact, a hop-derived TPR peptide has been designed, fused to the cell-penetrating antennapedia homeodomain sequence (Antp) [237]. This peptide has been shown to disrupt the interaction of Hsp90 with Hop, but not with PP5 or with FKBP51, and to induce cell death in several cancer cell lines [238,239]. Whether a similar strategy will be successful for targeting the Hsp90-FKBP51 interaction remains an open question. Specificity would be a particular challenge, also when devising EEVD-peptides, and this strategy would be designed to target Hsp90-dependent functions of FKBP51 while leaving others intact. For many of FKBP51’s cellular actions the Hsp90-dependency is not known. In addition, to use GR regulation as an example, it is not known whether FKBP51 exerts a specific action or merely acts by displacing other TPR proteins from Hsp90. In fact, the overexpression of PP5’s TPR domain produced a strong GR-inhibitory effect [81]. Thus, even if inhibitors of FKBP51’s association with Hsp90 could be designed, they probably would act as promoters of some actions of FKBP51, by inhibiting GR on the one hand, and by freeing up FKBP51 from Hsp90 complexes possibly enhancing its Hsp90-independent effects. Nevertheless, very recently, X-ray crystallography and computational modeling have been used for a detailed analysis of the binding of the Hsp90 C-terminal peptide binding to FKBP51 as the basis for a structure-based drug design targeting this interaction [240].
In recent years, ligands for FKBP51 have been synthesized that are selective in the sense that they do not bind to FKBP52 [241,242]. These ligands are often referred to as “inhibitors,” because they block FKBP51’s PPIase activity [241]. However, they may not inhibit all functions of FKBP51 (see above). Nevertheless, these compounds are very promising as they show antidepressant-like, anxiolytic and pain-relieving effects in mouse models [241,243,244]. While these compounds have been carefully designed to bind FKBP51, but not FKBP52, currently it cannot be excluded that binding to other FKBPs contributes to some of their physiological effects. The development of these FKBP51 ligands has greatly benefitted from high resolution crystal structure analyses [245,246]. Crystal structures reveal a particular conformational state, which represents one of many other possible conformations a molecule can typically adopt in solution. Therefore, insights from molecular dynamics simulation, binding free energy calculation, conformational dynamics, and unbinding pathway analysis may contribute to further improvement in the development of these and other FKBP51 ligands [247,248]. In addition, in light of the manifold pathway involvements of FKBP51, it appears useful to determine which of them are actually affected by these drugs, positively or negatively. Whenever possible, rationally designing drugs that affect a subset of FKBP51’s pathway actions is particularly desirable.
7. Conclusions
The important role of the FKBP51 system in the regulation of SR activity has led to a series of discoveries in the field of stress-related conditions. Important findings have contributed to the understanding of not only pathophysiological mechanisms of several illnesses (in particular depression and other affective disorders), but also treatment response toward pharmacogenomics approaches and personalized medicine. In parallel, as discussed in Section 6, advancements in chemical and drug design are finally able to pharmacologically target FKBP51 with relatively good precision, with promising opportunities for the treatment of stress-related mental disorders and possibly other diseases.
While this particular focus on FKBP51 research is undeniably important, the preclinical investigation of FKBP51 regulation has led to the discovery of several novel GR-independent functions and signaling pathways involving this protein. Even if they are also somewhat connected to neuronal mechanisms and the response to antidepressants, these novel mechanisms provide the background for innovative investigations of the FKBP51 system in conditions other than the commonly studied neuropsychiatric disorders.
In addition to the growing body of evidence of associations between FKBP51 and medical conditions, it is also noteworthy that this system also seems to influence specific parameters in healthy controls that may or may not have an effect on disease risk. In particular, several studies have reported associations between the FKBP5 genotype and brain alterations in non-clinical populations. Variants of the rs1360780 have been associated with altered volumes of the dorsal anterior and posterior cingulate cortexes [249], right amygdala [250], left amygdala [251], and the right middle and inferior orbitofrontal regions [250]. Moreover, FKBP5 polymorphisms have been associated with an altered resting-state activity in the frontotemporal–parietal network in controls [252]. The significance of these findings in increasing the risk for diseases still needs to be investigated, primarily by longitudinal studies.
In summary, accumulating evidence suggests a key role for FKBP51 in multiple cellular functions with potential impact in brain anatomy, metabolism, cognition, risk for diseases, and response to treatments. The multilayered regulation of FKBP51 from its transcription to protein stability provides the basis for the development of innovative molecules exploiting different mechanisms to target FKBP51 and its pathways, with the ultimate goal of improving the lives and prognoses of patients and subjects at risk. To achieve this goal, a more in-depth mechanistic understanding of the multifaceted actions of FKBP51 is required, including the different factors acting synergistically or in an opposing manner.
Acknowledgments
Nils C. Gassenwas supported in part by a Young Investigator NARSAD (National Alliance for Research on Schizophrenia and Depression) grant.
Abbreviations
| BAG1 | Bcl2-associated athanogene 1 |
| CDK | cyclin-dependent kinase |
| DNMT1 | DNA methyltransferase 1 |
| FKBP | FK506 binding protein |
| GR | glucocorticoid receptor |
| GRE | glucocorticoid response element |
| HPA | hypothalamus–pituitary–adrenals |
| HSP | heat shock protein |
| HSPBP1 | Hsp70 binding protein 1 |
| MDD | major depressive disorder |
| NFκB | nuclear factor kappa-light-chain-enhancer of activated B cells |
| NR | nuclear receptor |
| PHLPP | pleckstrin homology domain leucine-rich repeat protein phosphatase |
| PPIase | peptidylprolylisomerase |
| PTM | post translational modification |
| RISC | RNA-induced gene silencing complex |
| SNP | single nucleotide polymorphisms |
| SR | steroid receptor |
| SUMO | small ubiquitin-like modifier |
| TPR | tetratricopeptide repeat |
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Smith D.F., Faber L.E., Toft D.O. Purification of unactivated progesterone receptor and identification of novel receptor-associated proteins. J. Biol. Chem. 1990;265:3996–4003. [PubMed] [Google Scholar]
- 2.Grad I., Picard D. The glucocorticoid responses are shaped by molecular chaperones. Mol. Cell. Endocrinol. 2007;275:2–12. doi: 10.1016/j.mce.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 3.Echeverria P.C., Picard D. Molecular chaperones, essential partners of steroid hormone receptors for activity and mobility. Biochim. Biophys. Acta. 2010;1803:641–649. doi: 10.1016/j.bbamcr.2009.11.012. [DOI] [PubMed] [Google Scholar]
- 4.Pratt W.B., Toft D.O. Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocr. Rev. 1997;18:306–360. doi: 10.1210/er.18.3.306. [DOI] [PubMed] [Google Scholar]
- 5.Pratt W.B., Galigniana M.D., Harrell J.M., DeFranco D.B. Role of hsp90 and the hsp90-binding immunophilins in signalling protein movement. Cell. Signal. 2004;16:857–872. doi: 10.1016/j.cellsig.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 6.Rein T. FK506 binding protein 51 integrates pathways of adaptation: FKBP51 shapes the reactivity to environmental change. Bioessays. 2016;38:894–902. doi: 10.1002/bies.201600050. [DOI] [PubMed] [Google Scholar]
- 7.Wochnik G.M., Rüegg J., Abel G.A., Schmidt U., Holsboer F., Rein T. FK506-binding proteins 51 and 52 differentially regulate dynein interaction and nuclear translocation of the glucocorticoid receptor in mammalian cells. J. Biol. Chem. 2005;280:4609–4616. doi: 10.1074/jbc.M407498200. [DOI] [PubMed] [Google Scholar]
- 8.Denny W.B., Valentine D.L., Reynolds P.D., Smith D.F., Scammell J.G. Squirrel monkey immunophilin FKBP51 is a potent inhibitor of glucocorticoid receptor binding. Endocrinology. 2000;141:4107–4113. doi: 10.1210/endo.141.11.7785. [DOI] [PubMed] [Google Scholar]
- 9.Riggs D.L., Roberts P.J., Chirillo S.C., Cheung-Flynn J., Prapapanich V., Ratajczak T., Gaber R., Picard D., Smith D.F. The Hsp90-binding peptidylprolyl isomerase FKBP52 potentiates glucocorticoid signaling in vivo. EMBO J. 2003;22:1158–1167. doi: 10.1093/emboj/cdg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scammell J.G., Denny W.B., Valentine D.L., Smith D.F. Overexpression of the FK506-binding immunophilin FKBP51 is the common cause of glucocorticoid resistance in three New World primates. Gen. Comp. Endocrinol. 2001;124:152–165. doi: 10.1006/gcen.2001.7696. [DOI] [PubMed] [Google Scholar]
- 11.Holsboer F. The corticosteroid receptor hypothesis of depression. Neuropsychopharmacology. 2000;23:477–501. doi: 10.1016/S0893-133X(00)00159-7. [DOI] [PubMed] [Google Scholar]
- 12.Binder E.B., Salyakina D., Lichtner P., Wochnik G.M., Ising M., Pütz B., Papiol S., Seaman S., Lucae S., Kohli M.A., et al. Polymorphisms in FKBP5 are associated with increased recurrence of depressive episodes and rapid response to antidepressant treatment. Nat. Genet. 2004;36:1319–1325. doi: 10.1038/ng1479. [DOI] [PubMed] [Google Scholar]
- 13.Gustafsson J.A. Historical overview of nuclear receptors. J. Steroid Biochem. Mol. Biol. 2016;157:3–6. doi: 10.1016/j.jsbmb.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 14.Gronemeyer H., Gustafsson J.A., Laudet V. Principles for modulation of the nuclear receptor superfamily. Nat. Rev. Drug Discov. 2004;3:950–964. doi: 10.1038/nrd1551. [DOI] [PubMed] [Google Scholar]
- 15.Sladek F.M. What are nuclear receptor ligands? Mol. Cell. Endocrinol. 2011;334:3–13. doi: 10.1016/j.mce.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wrange O., Gustafsson J.A. Separation of the hormone- and DNA-binding sites of the hepatic glucocorticoid receptor by means of proteolysis. J. Biol. Chem. 1978;253:856–865. [PubMed] [Google Scholar]
- 17.Bain D.L., Heneghan A.F., Connaghan-Jones K.D., Miura M.T. Nuclear receptor structure: Implications for function. Annu. Rev. Physiol. 2007;69:201–220. doi: 10.1146/annurev.physiol.69.031905.160308. [DOI] [PubMed] [Google Scholar]
- 18.Germain P., Staels B., Dacquet C., Spedding M., Laudet V. Overview of nomenclature of nuclear receptors. Pharmacol. Rev. 2006;58:685–704. doi: 10.1124/pr.58.4.2. [DOI] [PubMed] [Google Scholar]
- 19.Garabedian M.J., Harris C.A., Jeanneteau F. Glucocorticoid receptor action in metabolic and neuronal function. F1000 Res. 2017;6:1208. doi: 10.12688/f1000research.11375.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weikum E.R., Knuesel M.T., Ortlund E.A., Yamamoto K.R. Glucocorticoid receptor control of transcription: Precision and plasticity via allostery. Nat. Rev. Mol. Cell Biol. 2017;18:159–174. doi: 10.1038/nrm.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caratti G., Matthews L., Poolman T., Kershaw S., Baxter M., Ray D. Glucocorticoid receptor function in health and disease. Clin. Endocrinol. 2015;83:441–448. doi: 10.1111/cen.12728. [DOI] [PubMed] [Google Scholar]
- 22.King H.A., Trotter K.W., Archer T.K. Chromatin remodeling during glucocorticoid receptor regulated transactivation. Biochim. Biophys. Acta. 2012;1819:716–726. doi: 10.1016/j.bbagrm.2012.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Picard D., Khursheed B., Garabedian M.J., Fortin M.G., Lindquist S., Yamamoto K.R. Reduced levels of hsp90 compromise steroid receptor action in vivo. Nature. 1990;348:166–168. doi: 10.1038/348166a0. [DOI] [PubMed] [Google Scholar]
- 24.Sanchez E.R., Toft D.O., Schlesinger M.J., Pratt W.B. Evidence that the 90-kDa phosphoprotein associated with the untransformed L-cell glucocorticoid receptor is a murine heat shock protein. J. Biol. Chem. 1985;260:12398–12401. [PubMed] [Google Scholar]
- 25.Welch W.J., Feramisco J.R. Purification of the major mammalian heat shock proteins. J. Biol. Chem. 1982;257:14949–14959. [PubMed] [Google Scholar]
- 26.Schopf F.H., Biebl M.M., Buchner J. The HSP90 chaperone machinery. Nat. Rev. Mol. Cell Biol. 2017;18:345–360. doi: 10.1038/nrm.2017.20. [DOI] [PubMed] [Google Scholar]
- 27.Zhao R., Davey M., Hsu Y.C., Kaplanek P., Tong A., Parsons A.B., Krogan N., Cagney G., Mai D., Greenblatt J., et al. Navigating the chaperone network: An integrative map of physical and genetic interactions mediated by the hsp90 chaperone. Cell. 2005;120:715–727. doi: 10.1016/j.cell.2004.12.024. [DOI] [PubMed] [Google Scholar]
- 28.Taipale M., Krykbaeva I., Koeva M., Kayatekin C., Westover K.D., Karras G.I., Lindquist S. Quantitative analysis of HSP90-client interactions reveals principles of substrate recognition. Cell. 2012;150:987–1001. doi: 10.1016/j.cell.2012.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pratt W.B., Dittmar K.D. Studies with Purified Chaperones Advance the Understanding of the Mechanism of Glucocorticoid Receptor-hsp90 Heterocomplex Assembly. Trends Endocrinol. Metab. 1998;9:244–252. doi: 10.1016/S1043-2760(98)00059-9. [DOI] [PubMed] [Google Scholar]
- 30.Hernandez M.P., Chadli A., Toft D.O. HSP40 binding is the first step in the HSP90 chaperoning pathway for the progesterone receptor. J. Biol. Chem. 2002;277:11873–11881. doi: 10.1074/jbc.M111445200. [DOI] [PubMed] [Google Scholar]
- 31.Kimura Y., Yahara I., Lindquist S. Role of the protein chaperone YDJ1 in establishing Hsp90-mediated signal transduction pathways. Science. 1995;268:1362–1365. doi: 10.1126/science.7761857. [DOI] [PubMed] [Google Scholar]
- 32.Laufen T., Mayer M.P., Beisel C., Klostermeier D., Mogk A., Reinstein J., Bukau B. Mechanism of regulation of hsp70 chaperones by DnaJ cochaperones. Proc. Natl. Acad. Sci. USA. 1999;96:5452–5457. doi: 10.1073/pnas.96.10.5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheung J., Smith D.F. Molecular chaperone interactions with steroid receptors: An update. Mol. Endocrinol. 2000;14:939–946. doi: 10.1210/mend.14.7.0489. [DOI] [PubMed] [Google Scholar]
- 34.Kirschke E., Goswami D., Southworth D., Griffin P.R., Agard D.A. Glucocorticoid receptor function regulated by coordinated action of the Hsp90 and Hsp70 chaperone cycles. Cell. 2014;157:1685–1697. doi: 10.1016/j.cell.2014.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caplan A.J. Yeast molecular chaperones and the mechanism of steroid hormone action. Trends Endocrinol. Metab. 1997;8:271–275. doi: 10.1016/S1043-2760(97)00079-9. [DOI] [PubMed] [Google Scholar]
- 36.Bracher A., Verghese J. The nucleotide exchange factors of Hsp70 molecular chaperones. Front Mol. Biosci. 2015;2:10. doi: 10.3389/fmolb.2015.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Craig E.A., Marszalek J. How Do J-Proteins Get Hsp70 to Do So Many Different Things? Trends Biochem. Sci. 2017;42:355–368. doi: 10.1016/j.tibs.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duncan E.J., Cheetham M.E., Chapple J.P., van der S.J. The role of HSP70 and its co-chaperones in protein misfolding, aggregation and disease. Subcell. Biochem. 2015;78:243–273. doi: 10.1007/978-3-319-11731-7_12. [DOI] [PubMed] [Google Scholar]
- 39.Sahasrabudhe P., Rohrberg J., Biebl M.M., Rutz D.A., Buchner J. The Plasticity of the Hsp90 Co-chaperone System. Mol. Cell. 2017;67:947–961. doi: 10.1016/j.molcel.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 40.Felts S.J., Toft D.O. p23, a simple protein with complex activities. Cell Stress Chaperones. 2003;8:108–113. doi: 10.1379/1466-1268(2003)008<0108:PASPWC>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Young J.C., Hartl F.U. Polypeptide release by Hsp90 involves ATP hydrolysis and is enhanced by the co-chaperone p23. EMBO J. 2000;19:5930–5940. doi: 10.1093/emboj/19.21.5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bose S., Weikl T., Bugl H., Buchner J. Chaperone function of Hsp90-associated proteins. Science. 1996;274:1715–1717. doi: 10.1126/science.274.5293.1715. [DOI] [PubMed] [Google Scholar]
- 43.Freeman B.C., Toft D.O., Morimoto R.I. Molecular chaperone machines: Chaperone activities of the cyclophilin Cyp-40 and the steroid aporeceptor-associated protein p23. Science. 1996;274:1718–1720. doi: 10.1126/science.274.5293.1718. [DOI] [PubMed] [Google Scholar]
- 44.Tanioka T., Nakatani Y., Semmyo N., Murakami M., Kudo I. Molecular identification of cytosolic prostaglandin E2 synthase that is functionally coupled with cyclooxygenase-1 in immediate prostaglandin E2 biosynthesis. J. Biol. Chem. 2000;275:32775–32782. doi: 10.1074/jbc.M003504200. [DOI] [PubMed] [Google Scholar]
- 45.Freeman B.C., Yamamoto K.R. Disassembly of transcriptional regulatory complexes by molecular chaperones. Science. 2002;296:2232–2235. doi: 10.1126/science.1073051. [DOI] [PubMed] [Google Scholar]
- 46.Freeman B.C., Felts S.J., Toft D.O., Yamamoto K.R. The p23 molecular chaperones act at a late step in intracellular receptor action to differentially affect ligand efficacies. Genes Dev. 2000;14:422–434. [PMC free article] [PubMed] [Google Scholar]
- 47.Wochnik G.M., Young J.C., Schmidt U., Holsboer F., Hartl F.U., Rein T. Inhibition of GR-mediated transcription by p23 requires interaction with Hsp90. FEBS Lett. 2004;560:35–38. doi: 10.1016/S0014-5793(04)00066-3. [DOI] [PubMed] [Google Scholar]
- 48.Daugaard M., Rohde M., Jaattela M. The heat shock protein 70 family: Highly homologous proteins with overlapping and distinct functions. FEBS Lett. 2007;581:3702–3710. doi: 10.1016/j.febslet.2007.05.039. [DOI] [PubMed] [Google Scholar]
- 49.Kominek J., Marszalek J., Neuveglise C., Craig E.A., Williams B.L. The complex evolutionary dynamics of Hsp70s: A genomic and functional perspective. Genome Biol. Evol. 2013;5:2460–2477. doi: 10.1093/gbe/evt192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qiu X.B., Shao Y.M., Miao S., Wang L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell Mol. Life Sci. 2006;63:2560–2570. doi: 10.1007/s00018-006-6192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Powers E.T., Balch W.E. Diversity in the origins of proteostasis networks—A driver for protein function in evolution. Nat. Rev. Mol. Cell. Biol. 2013;14:237–248. doi: 10.1038/nrm3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Höhfeld J., Jentsch S. GrpE-like regulation of the hsc70 chaperone by the anti-apoptotic protein BAG-1. EMBO J. 1997;16:6209–6216. doi: 10.1093/emboj/16.20.6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shomura Y., Dragovic Z., Chang H.C., Tzvetkov N., Young J.C., Brodsky J.L., Guerriero V., Hartl F.U., Bracher A. Regulation of Hsp70 function by HspBP1: Structural analysis reveals an alternate mechanism for Hsp70 nucleotide exchange. Mol. Cell. 2005;17:367–379. doi: 10.1016/j.molcel.2004.12.023. [DOI] [PubMed] [Google Scholar]
- 54.Schmidt U., Wochnik G.M., Rosenhagen M.C., Young J.C., Hartl F.U., Holsboer F., Rein T. Essential role of the unusual DNA-binding motif of BAG-1 for inhibition of the glucocorticoid receptor. J. Biol. Chem. 2003;278:4926–4931. doi: 10.1074/jbc.M212000200. [DOI] [PubMed] [Google Scholar]
- 55.Schmidt U., Holsboer F., Rein T. Role of the hsp70 cochaperone BAG1 in glucocorticoid receptor function and stress-related diseases. Proc. Natl. Acad. Sci. USA. 2008;105:E101. doi: 10.1073/pnas.0809354105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schneikert J., Hubner S., Langer G., Petri T., Jaattela M., Reed J., Cato A.C. Hsp70-RAP46 interaction in downregulation of DNA binding by glucocorticoid receptor. EMBO J. 2000;19:6508–6516. doi: 10.1093/emboj/19.23.6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schneikert J., Hubner S., Martin E., Cato A.C. A nuclear action of the eukaryotic cochaperone RAP46 in downregulation of glucocorticoid receptor activity. J. Cell Biol. 1999;146:929–940. doi: 10.1083/jcb.146.5.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Knapp R.T., Wong M.J., Kollmannsberger L.K., Gassen N.C., Kretzschmar A., Zschocke J., Hafner K., Young J.C., Rein T. Hsp70 cochaperones HspBP1 and BAG-1M differentially regulate steroid hormone receptor function. PLoS ONE. 2014;9:e85415. doi: 10.1371/journal.pone.0085415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Höhfeld J., Minami Y., Hartl F.U. Hip, a novel cochaperone involved in the eukaryotic Hsc70/Hsp40 reaction cycle. Cell. 1995;83:589–598. doi: 10.1016/0092-8674(95)90099-3. [DOI] [PubMed] [Google Scholar]
- 60.Kanelakis K.C., Murphy P.J., Galigniana M.D., Morishima Y., Takayama S., Reed J.C., Toft D.O., Pratt W.B. Hsp70 interacting protein Hip does not affect glucocorticoid receptor folding by the hsp90-based chaperone machinery except to oppose the effect of BAG-1. Biochemistry. 2000;39:14314–14321. doi: 10.1021/bi001671c. [DOI] [PubMed] [Google Scholar]
- 61.Nelson G.M., Prapapanich V., Carrigan P.E., Roberts P.J., Riggs D.L., Smith D.F. The heat shock protein 70 cochaperone hip enhances functional maturation of glucocorticoid receptor. Mol. Endocrinol. 2004;18:1620–1630. doi: 10.1210/me.2004-0054. [DOI] [PubMed] [Google Scholar]
- 62.Dittmar K.D., Banach M., Galigniana M.D., Pratt W.B. The role of DnaJ-like proteins in glucocorticoid receptor.hsp90 heterocomplex assembly by the reconstituted hsp90.p60.hsp70 foldosome complex. J. Biol. Chem. 1998;273:7358–7366. doi: 10.1074/jbc.273.13.7358. [DOI] [PubMed] [Google Scholar]
- 63.Mandal A.K., Gibney P.A., Nillegoda N.B., Theodoraki M.A., Caplan A.J., Morano K.A. Hsp110 chaperones control client fate determination in the hsp70-Hsp90 chaperone system. Mol. Biol. Cell. 2010;21:1439–1448. doi: 10.1091/mbc.E09-09-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu X.D., Morano K.A., Thiele D.J. The yeast Hsp110 family member, Sse1, is an Hsp90 cochaperone. J. Biol. Chem. 1999;274:26654–26660. doi: 10.1074/jbc.274.38.26654. [DOI] [PubMed] [Google Scholar]
- 65.Harst A., Lin H., Obermann W.M. Aha1 competes with Hop, p50 and p23 for binding to the molecular chaperone Hsp90 and contributes to kinase and hormone receptor activation. Biochem. J. 2005;387:789–796. doi: 10.1042/BJ20041283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lotz G.P., Lin H., Harst A., Obermann W.M. Aha1 binds to the middle domain of Hsp90, contributes to client protein activation, and stimulates the ATPase activity of the molecular chaperone. J. Biol. Chem. 2003;278:17228–17235. doi: 10.1074/jbc.M212761200. [DOI] [PubMed] [Google Scholar]
- 67.Panaretou B., Siligardi G., Meyer P., Maloney A., Sullivan J.K., Singh S., Millson S.H., Clarke P.A., Naaby-Hansen S., Stein R., et al. Activation of the ATPase activity of hsp90 by the stress-regulated cochaperone aha1. Mol. Cell. 2002;10:1307–1318. doi: 10.1016/S1097-2765(02)00785-2. [DOI] [PubMed] [Google Scholar]
- 68.Marsh J.A., Kalton H.M., Gaber R.F. Cns1 is an essential protein associated with the hsp90 chaperone complex in Saccharomyces cerevisiae that can restore cyclophilin 40-dependent functions in cpr7Delta cells. Mol. Cell. Biol. 1998;18:7353–7359. doi: 10.1128/MCB.18.12.7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tesic M., Marsh J.A., Cullinan S.B., Gaber R.F. Functional interactions between Hsp90 and the co-chaperones Cns1 and Cpr7 in Saccharomyces cerevisiae. J. Biol. Chem. 2003;278:32692–32701. doi: 10.1074/jbc.M304315200. [DOI] [PubMed] [Google Scholar]
- 70.Banerjee A., Periyasamy S., Wolf I.M., Hinds T.D., Jr., Yong W., Shou W., Sanchez E.R. Control of glucocorticoid and progesterone receptor subcellular localization by the ligand-binding domain is mediated by distinct interactions with tetratricopeptide repeat proteins. Biochemistry. 2008;47:10471–10480. doi: 10.1021/bi8011862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schülke J.P., Wochnik G.M., Lang-Rollin I., Gassen N.C., Knapp R.T., Berning B., Yassouridis A., Rein T. Differential impact of tetratricopeptide repeat proteins on the steroid hormone receptors. PLoS ONE. 2010;5:e11717. doi: 10.1371/journal.pone.0011717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ratajczak T., Ward B.K., Minchin R.F. Immunophilin chaperones in steroid receptor signalling. Curr. Top. Med. Chem. 2003;3:1348–1357. doi: 10.2174/1568026033451934. [DOI] [PubMed] [Google Scholar]
- 73.Duina A.A., Marsh J.A., Kurtz R.B., Chang H.C., Lindquist S., Gaber R.F. The peptidyl-prolyl isomerase domain of the CyP-40 cyclophilin homolog Cpr7 is not required to support growth or glucocorticoid receptor activity in Saccharomyces cerevisiae. J. Biol. Chem. 1998;273:10819–10822. doi: 10.1074/jbc.273.18.10819. [DOI] [PubMed] [Google Scholar]
- 74.Riggs D.L., Cox M.B., Tardif H.L., Hessling M., Buchner J., Smith D.F. Noncatalytic role of the FKBP52 peptidyl-prolyl isomerase domain in the regulation of steroid hormone signaling. Mol. Cell. Biol. 2007;27:8658–8669. doi: 10.1128/MCB.00985-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Galigniana M.D., Radanyi C., Renoir J.M., Housley P.R., Pratt W.B. Evidence that the peptidylprolyl isomerase domain of the hsp90-binding immunophilin FKBP52 is involved in both dynein interaction and glucocorticoid receptor movement to the nucleus. J. Biol. Chem. 2001;276:14884–14889. doi: 10.1074/jbc.M010809200. [DOI] [PubMed] [Google Scholar]
- 76.McKeen H.D., McAlpine K., Valentine A., Quinn D.J., McClelland K., Byrne C., O’Rourke M., Young S., Scott C.J., McCarthy H.O., et al. A novel FK506-like binding protein interacts with the glucocorticoid receptor and regulates steroid receptor signaling. Endocrinology. 2008;149:5724–5734. doi: 10.1210/en.2008-0168. [DOI] [PubMed] [Google Scholar]
- 77.Hutchison K.A., Stancato L.F., Owens-Grillo J.K., Johnson J.L., Krishna P., Toft D.O., Pratt W.B. The 23-kDa acidic protein in reticulocyte lysate is the weakly bound component of the hsp foldosome that is required for assembly of the glucocorticoid receptor into a functional heterocomplex with hsp90. J. Biol. Chem. 1995;270:18841–18847. doi: 10.1074/jbc.270.32.18841. [DOI] [PubMed] [Google Scholar]
- 78.Grad I., McKee T.A., Ludwig S.M., Hoyle G.W., Ruiz P., Wurst W., Floss T., Miller C.A., III, Picard D. The Hsp90 cochaperone p23 is essential for perinatal survival. Mol. Cell. Biol. 2006;26:8976–8983. doi: 10.1128/MCB.00734-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Soroka J., Wandinger S.K., Mausbacher N., Schreiber T., Richter K., Daub H., Buchner J. Conformational switching of the molecular chaperone Hsp90 via regulated phosphorylation. Mol. Cell. 2012;45:517–528. doi: 10.1016/j.molcel.2011.12.031. [DOI] [PubMed] [Google Scholar]
- 80.Wandinger S.K., Suhre M.H., Wegele H., Buchner J. The phosphatase Ppt1 is a dedicated regulator of the molecular chaperone Hsp90. EMBO J. 2006;25:367–376. doi: 10.1038/sj.emboj.7600930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen M.S., Silverstein A.M., Pratt W.B., Chinkers M. The tetratricopeptide repeat domain of protein phosphatase 5 mediates binding to glucocorticoid receptor heterocomplexes and acts as a dominant negative mutant. J. Biol. Chem. 1996;271:32315–32320. doi: 10.1074/jbc.271.50.32315. [DOI] [PubMed] [Google Scholar]
- 82.Pazdrak K., Straub C., Maroto R., Stafford S., White W.I., Calhoun W.J., Kurosky A. Cytokine-Induced Glucocorticoid Resistance from Eosinophil Activation: Protein Phosphatase 5 Modulation of Glucocorticoid Receptor Phosphorylation and Signaling. J. Immunol. 2016;197:3782–3791. doi: 10.4049/jimmunol.1601029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Laenger A., Lang-Rollin I., Kozany C., Zschocke J., Zimmermann N., Rüegg J., Holsboer F., Hausch F., Rein T. XAP2 inhibits glucocorticoid receptor activity in mammalian cells. FEBS Lett. 2009;583:1493–1498. doi: 10.1016/j.febslet.2009.03.072. [DOI] [PubMed] [Google Scholar]
- 84.Connell P., Ballinger C.A., Jiang J., Wu Y., Thompson L.J., Höhfeld J., Patterson C. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat. Cell. Biol. 2001;3:93–96. doi: 10.1038/35050618. [DOI] [PubMed] [Google Scholar]
- 85.Alvira S., Cuellar J., Rohl A., Yamamoto S., Itoh H., Alfonso C., Rivas G., Buchner J., Valpuesta J.M. Structural characterization of the substrate transfer mechanism in Hsp70/Hsp90 folding machinery mediated by Hop. Nat. Commun. 2014;5:5484. doi: 10.1038/ncomms6484. [DOI] [PubMed] [Google Scholar]
- 86.Brychzy A., Rein T., Winklhofer K.F., Hartl F.U., Young J.C., Obermann W.M. Cofactor Tpr2 combines two TPR domains and a J domain to regulate the Hsp70/Hsp90 chaperone system. EMBO J. 2003;22:3613–3623. doi: 10.1093/emboj/cdg362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Paul A., Garcia Y.A., Zierer B., Patwardhan C., Gutierrez O., Hildenbrand Z., Harris D.C., Balsiger H.A., Sivils J.C., Johnson J.L., et al. The cochaperone SGTA (small glutamine-rich tetratricopeptide repeat-containing protein alpha) demonstrates regulatory specificity for the androgen, glucocorticoid, and progesterone receptors. J. Biol. Chem. 2014;289:15297–15308. doi: 10.1074/jbc.M113.535229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ratajczak T., Cluning C., Ward B.K. Steroid Receptor-Associated Immunophilins: A Gateway to Steroid Signalling. Clin. Biochem. Rev. 2015;36:31–52. [PMC free article] [PubMed] [Google Scholar]
- 89.Scheufler C., Brinker A., Bourenkov G., Pegoraro S., Moroder L., Bartunik H., Hartl F.U., Moarefi I. Structure of TPR domain-peptide complexes: Critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell. 2000;101:199–210. doi: 10.1016/S0092-8674(00)80830-2. [DOI] [PubMed] [Google Scholar]
- 90.Hinds T.D., Jr., Stechschulte L.A., Cash H.A., Whisler D., Banerjee A., Yong W., Khuder S.S., Kaw M.K., Shou W., Najjar S.M., et al. Protein phosphatase 5 mediates lipid metabolism through reciprocal control of glucocorticoid receptor and peroxisome proliferator-activated receptor-gamma (PPARgamma)1. J. Biol. Chem. 2011;286:42911–42922. doi: 10.1074/jbc.M111.311662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liu J., DeFranco D.B. Chromatin recycling of glucocorticoid receptors: Implications for multiple roles of heat shock protein 90. Mol. Endocrinol. 1999;13:355–365. doi: 10.1210/mend.13.3.0258. [DOI] [PubMed] [Google Scholar]
- 92.Haché R.J., Tse R., Reich T., Savory J.G., Lefebvre Y.A. Nucleocytoplasmic trafficking of steroid-free glucocorticoid receptor. J. Biol. Chem. 1999;274:1432–1439. doi: 10.1074/jbc.274.3.1432. [DOI] [PubMed] [Google Scholar]
- 93.Tago K., Tsukahara F., Naruse M., Yoshioka T., Takano K. Regulation of nuclear retention of glucocorticoid receptor by nuclear Hsp90. Mol. Cell. Endocrinol. 2004;213:131–138. doi: 10.1016/j.mce.2003.10.057. [DOI] [PubMed] [Google Scholar]
- 94.Elbi C., Walker D.A., Romero G., Sullivan W.P., Toft D.O., Hager G.L., DeFranco D.B. Molecular chaperones function as steroid receptor nuclear mobility factors. Proc. Natl. Acad. Sci. USA. 2004;101:2876–2881. doi: 10.1073/pnas.0400116101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Stavreva D.A., Müller W.G., Hager G.L., Smith C.L., McNally J.G. Rapid glucocorticoid receptor exchange at a promoter is coupled to transcription and regulated by chaperones and proteasomes. Mol. Cell. Biol. 2004;24:2682–2697. doi: 10.1128/MCB.24.7.2682-2697.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Storer C.L., Dickey C.A., Galigniana M.D., Rein T., Cox M.B. FKBP51 and FKBP52 in signaling and disease. Trends Endocrinol. Metab. 2011;22:481–490. doi: 10.1016/j.tem.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pirkl F., Buchner J. Functional analysis of the Hsp90-associated human peptidyl prolyl cis/trans isomerases FKBP51, FKBP52 and Cyp40. J. Mol. Biol. 2001;308:795–806. doi: 10.1006/jmbi.2001.4595. [DOI] [PubMed] [Google Scholar]
- 98.Sinars C.R., Cheung-Flynn J., Rimerman R.A., Scammell J.G., Smith D.F., Clardy J. Structure of the large FK506-binding protein FKBP51, an Hsp90-binding protein and a component of steroid receptor complexes. Proc. Natl. Acad. Sci. USA. 2003;100:868–873. doi: 10.1073/pnas.0231020100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Brinker A., Scheufler C., Von Der M.F., Fleckenstein B., Herrmann C., Jung G., Moarefi I., Hartl F.U. Ligand discrimination by TPR domains. Relevance and selectivity of EEVD-recognition in Hsp70 x Hop x Hsp90 complexes. J. Biol. Chem. 2002;277:19265–19275. doi: 10.1074/jbc.M109002200. [DOI] [PubMed] [Google Scholar]
- 100.Ebong I.O., Beilsten-Edmands V., Patel N.A., Morgner N., Robinson C.V. The interchange of immunophilins leads to parallel pathways and different intermediates in the assembly of Hsp90 glucocorticoid receptor complexes. Cell Discov. 2016;2:16002. doi: 10.1038/celldisc.2016.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Barent R.L., Nair S.C., Carr D.C., Ruan Y., Rimerman R.A., Fulton J., Zhang Y., Smith D.F. Analysis of FKBP51/FKBP52 chimeras and mutants for Hsp90 binding and association with progesterone receptor complexes. Mol. Endocrinol. 1998;12:342–354. doi: 10.1210/mend.12.3.0075. [DOI] [PubMed] [Google Scholar]
- 102.Wu B., Li P., Liu Y., Lou Z., Ding Y., Shu C., Ye S., Bartlam M., Shen B., Rao Z. 3D structure of human FK506-binding protein 52: Implications for the assembly of the glucocorticoid receptor/Hsp90/immunophilin heterocomplex. Proc. Natl. Acad. Sci. USA. 2004;101:8348–8353. doi: 10.1073/pnas.0305969101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Taipale M., Jarosz D.F., Lindquist S. HSP90 at the hub of protein homeostasis: Emerging mechanistic insights. Nat. Rev. Mol. Cell. Biol. 2010;11:515–528. doi: 10.1038/nrm2918. [DOI] [PubMed] [Google Scholar]
- 104.Taipale M., Tucker G., Peng J., Krykbaeva I., Lin Z.Y., Larsen B., Choi H., Berger B., Gingras A.C., Lindquist S. A quantitative chaperone interaction network reveals the architecture of cellular protein homeostasis pathways. Cell. 2014;158:434–448. doi: 10.1016/j.cell.2014.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Klengel T., Mehta D., Anacker C., Rex-Haffner M., Pruessner J.C., Pariante C.M., Pace T.W., Mercer K.B., Mayberg H.S., Bradley B., et al. Allele-specific FKBP5 DNA demethylation mediates gene-childhood trauma interactions. Nat. Neurosci. 2013;16:33–41. doi: 10.1038/nn.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Paakinaho V., Makkonen H., Jaaskelainen T., Palvimo J.J. Glucocorticoid receptor activates poised FKBP51 locus through long-distance interactions. Mol. Endocrinol. 2010;24:511–525. doi: 10.1210/me.2009-0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pelleymounter L.L., Moon I., Johnson J.A., Laederach A., Halvorsen M., Eckloff B., Abo R., Rossetti S. A novel application of pattern recognition for accurate SNP and indel discovery from high-throughput data: Targeted resequencing of the glucocorticoid receptor co-chaperone FKBP5 in a Caucasian population. Mol. Genet. Metab. 2011;104:457–469. doi: 10.1016/j.ymgme.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hubler T.R., Scammell J.G. Intronic hormone response elements mediate regulation of FKBP5 by progestins and glucocorticoids. Cell. Stress Chaperones. 2004;9:243–252. doi: 10.1379/CSC-32R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Galat A. A note on clustering the functionally-related paralogues and orthologues of proteins: A case of the FK506-binding proteins (FKBPs) Comput. Biol. Chem. 2004;28:129–140. doi: 10.1016/j.compbiolchem.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 110.Scharf S.H., Liebl C., Binder E.B., Schmidt M.V., Müller M.B. Expression and regulation of the Fkbp5 gene in the adult mouse brain. PLoS ONE. 2011;6:e16883. doi: 10.1371/journal.pone.0016883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ong C.T., Corces V.G. CTCF: An architectural protein bridging genome topology and function. Nat. Rev. Genet. 2014;15:234–246. doi: 10.1038/nrg3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shen L., Oshida T., Miyauchi J., Yamada M., Miyashita T. Identification of novel direct transcriptional targets of glucocorticoid receptor. Leukemia. 2004;18:1850–1856. doi: 10.1038/sj.leu.2403516. [DOI] [PubMed] [Google Scholar]
- 113.Magee J.A., Chang L.W., Stormo G.D., Milbrandt J. Direct, androgen receptor-mediated regulation of the FKBP5 gene via a distal enhancer element. Endocrinology. 2006;147:590–598. doi: 10.1210/en.2005-1001. [DOI] [PubMed] [Google Scholar]
- 114.Lee R.S., Tamashiro K.L., Yang X., Purcell R.H., Harvey A., Willour V.L., Huo Y., Rongione M., Wand G.S., Potash J.B. Chronic corticosterone exposure increases expression and decreases deoxyribonucleic acid methylation of Fkbp5 in mice. Endocrinology. 2010;151:4332–4343. doi: 10.1210/en.2010-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Roy B., Shelton R.C., Dwivedi Y. DNA methylation and expression of stress related genes in PBMC of MDD patients with and without serious suicidal ideation. J. Psychiatr. Res. 2017;89:115–124. doi: 10.1016/j.jpsychires.2017.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Han K.M., Won E., Sim Y., Kang J., Han C., Kim Y.K., Kim S.H., Joe S.H., Lee M.S., Tae W.S., et al. Influence of FKBP5 polymorphism and DNA methylation on structural changes of the brain in major depressive disorder. Sci. Rep. 2017;7:42621. doi: 10.1038/srep42621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sabbagh J.J., O’Leary J.C., III, Blair L.J., Klengel T., Nordhues B.A., Fontaine S.N., Binder E.B., Dickey C.A. Age-associated epigenetic upregulation of the FKBP5 gene selectively impairs stress resiliency. PLoS ONE. 2014;9:e107241. doi: 10.1371/journal.pone.0107241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Fries G.R., Carvalho A.F., Quevedo J. The miRNome of bipolar disorder. J. Affect. Disord. 2017 doi: 10.1016/j.jad.2017.09.025. in press. [DOI] [PubMed] [Google Scholar]
- 119.Klein M., Chandradoss S.D., Depken M., Joo C. Why Argonaute is needed to make microRNA target search fast and reliable. Semin. Cell Dev. Biol. 2017;65:20–28. doi: 10.1016/j.semcdb.2016.05.017. [DOI] [PubMed] [Google Scholar]
- 120.Volk N., Pape J.C., Engel M., Zannas A.S., Cattane N., Cattaneo A., Binder E.B., Chen A. Amygdalar MicroRNA-15a Is Essential for Coping with Chronic Stress. Cell Rep. 2016;17:1882–1891. doi: 10.1016/j.celrep.2016.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zheng D., Sabbagh J.J., Blair L.J., Darling A.L., Wen X., Dickey C.A. MicroRNA-511 Binds to FKBP5 mRNA, Which Encodes a Chaperone Protein, and Regulates Neuronal Differentiation. J. Biol. Chem. 2016;291:17897–17906. doi: 10.1074/jbc.M116.727941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Aghajan M., Jonai N., Flick K., Fu F., Luo M., Cai X., Ouni I., Pierce N., Tang X., Lomenick B., et al. Chemical genetics screen for enhancers of rapamycin identifies a specific inhibitor of an SCF family E3 ubiquitin ligase. Nat. Biotechnol. 2010;28:738–742. doi: 10.1038/nbt.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Cox M.B., Riggs D.L., Hessling M., Schumacher F., Buchner J., Smith D.F. FK506-binding protein 52 phosphorylation: A potential mechanism for regulating steroid hormone receptor activity. Mol. Endocrinol. 2007;21:2956–2967. doi: 10.1210/me.2006-0547. [DOI] [PubMed] [Google Scholar]
- 124.Antunica-Noguerol M., Budziñski M.L., Druker J., Gassen N.C., Proto-Cassina L., Senin S., Aprile-Garcia F., Holsboer F., Rein T., Liberman A.C., et al. The activity of the glucocorticoid receptor is regulated by SUMO conjugation to FKBP51. Cell Death Differ. 2016;23:1579–1591. doi: 10.1038/cdd.2016.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wilkinson K.A., Henley J.M. Mechanisms, regulation and consequences of protein SUMOylation. Biochem. J. 2010;428:133–145. doi: 10.1042/BJ20100158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ikeda F., Dikic I. Atypical ubiquitin chains: New molecular signals. ‘Protein Modifications: Beyond the Usual Suspects’ review series. EMBO Rep. 2008;9:536–542. doi: 10.1038/embor.2008.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Luo K., Li Y., Yin Y., Li L., Wu C., Chen Y., Nowsheen S., Hu Q., Zhang L., Lou Z., et al. USP49 negatively regulates tumorigenesis and chemoresistance through FKBP51-AKT signaling. EMBO J. 2017;36:1434–1446. doi: 10.15252/embj.201695669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bekhbat M., Rowson S.A., Neigh G.N. Checks and balances: The glucocorticoid receptor and NFkB in good times and bad. Front. Neuroendocrinol. 2017;46:15–31. doi: 10.1016/j.yfrne.2017.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Newton R., Shah S., Altonsy M.O., Gerber A.N. Glucocorticoid and cytokine crosstalk: Feedback, feedforward, and co-regulatory interactions determine repression or resistance. J. Biol. Chem. 2017;292:7163–7172. doi: 10.1074/jbc.R117.777318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lawrence T. The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harb. Perspect. Biol. 2009;1:a001651. doi: 10.1101/cshperspect.a001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Romano S., Mallardo M., Romano M.F. FKBP51 and the NF-kappaB regulatory pathway in cancer. Curr. Opin. Pharmacol. 2011;11:288–293. doi: 10.1016/j.coph.2011.04.011. [DOI] [PubMed] [Google Scholar]
- 132.Bouwmeester T., Bauch A., Ruffner H., Angrand P.O., Bergamini G., Croughton K., Cruciat C., Eberhard D., Gagneur J., Ghidelli S., et al. A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway. Nat. Cell Biol. 2004;6:97–105. doi: 10.1038/ncb1086. [DOI] [PubMed] [Google Scholar]
- 133.Yu H.M., Wang Q., Sun W.B. Silencing of FKBP51 alleviates the mechanical pain threshold, inhibits DRG inflammatory factors and pain mediators through the NF-kappaB signaling pathway. Gene. 2017;627:169–175. doi: 10.1016/j.gene.2017.06.029. [DOI] [PubMed] [Google Scholar]
- 134.Li T.K., Baksh S., Cristillo A.D., Bierer B.E. Calcium- and FK506-independent interaction between the immunophilin FKBP51 and calcineurin. J. Cell Biochem. 2002;84:460–471. doi: 10.1002/jcb.10026. [DOI] [PubMed] [Google Scholar]
- 135.Giraudier S., Chagraoui H., Komura E., Barnache S., Blanchet B., LeCouedic J.P., Smith D.F., Larbret F., Taksin A.L., Moreau-Gachelin F., et al. Overexpression of FKBP51 in idiopathic myelofibrosis regulates the growth factor independence of megakaryocyte progenitors. Blood. 2002;100:2932–2940. doi: 10.1182/blood-2002-02-0485. [DOI] [PubMed] [Google Scholar]
- 136.Akiyama T., Shiraishi T., Qin J., Konno H., Akiyama N., Shinzawa M., Miyauchi M., Takizawa N., Yanai H., Ohashi H., et al. Mitochondria-nucleus shuttling FK506-binding protein 51 interacts with TRAF proteins and facilitates the RIG-I-like receptor-mediated expression of type I IFN. PLoS ONE. 2014;9:e95992. doi: 10.1371/journal.pone.0095992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kim Y.S., Kim Y.J., Lee J.M., Kim E.K., Park Y.J., Choe S.K., Ko H.J., Kang C.Y. Functional changes in myeloid-derived suppressor cells (MDSCs) during tumor growth: FKBP51 contributes to the regulation of the immunosuppressive function of MDSCs. J. Immunol. 2012;188:4226–4234. doi: 10.4049/jimmunol.1103040. [DOI] [PubMed] [Google Scholar]
- 138.Manning B.D., Toker A. AKT/PKB Signaling: Navigating the Network. Cell. 2017;169:381–405. doi: 10.1016/j.cell.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wang L. FKBP51 regulation of AKT/protein kinase B phosphorylation. Curr. Opin. Pharmacol. 2011;11:360–364. doi: 10.1016/j.coph.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Fabian A.K., Marz A., Neimanis S., Biondi R.M., Kozany C., Hausch F. InterAKTions with FKBPs--mutational and pharmacological exploration. PLoS ONE. 2013;8:e57508. doi: 10.1371/journal.pone.0057508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Gassen N.C., Hartmann J., Zschocke J., Stepan J., Hafner K., Zellner A., Kirmeier T., Kollmannsberger L., Wagner K.V., Dedic N., et al. Association of FKBP51 with Priming of Autophagy Pathways and Mediation of Antidepressant Treatment Response: Evidence in Cells, Mice, and Humans. PLoS Med. 2014;11:e1001755. doi: 10.1371/journal.pmed.1001755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Jinwal U.K., Koren J., III, Borysov S.I., Schmid A.B., Abisambra J.F., Blair L.J., Johnson A.G., Jones J.R., Shults C.L., O’Leary J.C., III, et al. The Hsp90 cochaperone, FKBP51, increases Tau stability and polymerizes microtubules. J. Neurosci. 2010;30:591–599. doi: 10.1523/JNEUROSCI.4815-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Cioffi D.L., Hubler T.R., Scammell J.G. Organization and function of the FKBP52 and FKBP51 genes. Curr. Opin. Pharmacol. 2011;11:308–313. doi: 10.1016/j.coph.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Chambraud B., Belabes H., Fontaine-Lenoir V., Fellous A., Baulieu E.E. The immunophilin FKBP52 specifically binds to tubulin and prevents microtubule formation. FASEB J. 2007;21:2787–2797. doi: 10.1096/fj.06-7667com. [DOI] [PubMed] [Google Scholar]
- 145.Quinta H.R., Maschi D., Gomez-Sanchez C., Piwien-Pilipuk G., Galigniana M.D. Subcellular rearrangement of hsp90-binding immunophilins accompanies neuronal differentiation and neurite outgrowth. J. Neurochem. 2010;115:716–734. doi: 10.1111/j.1471-4159.2010.06970.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Romano S., D’Angelillo A., Pacelli R., Staibano S., De L.E., Bisogni R., Eskelinen E.L., Mascolo M., Cali G., Arra C., et al. Role of FK506-binding protein 51 in the control of apoptosis of irradiated melanoma cells. Cell Death. Differ. 2010;17:145–157. doi: 10.1038/cdd.2009.115. [DOI] [PubMed] [Google Scholar]
- 147.Gassen N.C., Hartmann J., Schmidt M.V., Rein T. FKBP5/FKBP51 enhances autophagy to synergize with antidepressant action. Autophagy. 2015;11:578–580. doi: 10.1080/15548627.2015.1017224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Laane E., Tamm K.P., Buentke E., Ito K., Kharaziha P., Oscarsson J., Corcoran M., Bjorklund A.C., Hultenby K., Lundin J., et al. Cell death induced by dexamethasone in lymphoid leukemia is mediated through initiation of autophagy. Cell Death. Differ. 2009;16:1018–1029. doi: 10.1038/cdd.2009.46. [DOI] [PubMed] [Google Scholar]
- 149.Zhi X., Feng W., Rong Y., Liu R. Anatomy of autophagy: From the beginning to the end. Cell. Mol. Life Sci. 2017:1–17. doi: 10.1007/s00018-017-2657-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Vijayan V., Verstreken P. Autophagy in the presynaptic compartment in health and disease. J. Cell Biol. 2017;216:1895–1906. doi: 10.1083/jcb.201611113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Gassen N.C., Fries G.R., Zannas A.S., Hartmann J., Zschocke J., Hafner K., Carrillo-Roa T., Steinbacher J., Preissinger S.N., Hoeijmakers L., et al. Chaperoning epigenetics: FKBP51 decreases the activity of DNMT1 and mediates epigenetic effects of the antidepressant paroxetine. Sci. Signal. 2015;8:ra119. doi: 10.1126/scisignal.aac7695. [DOI] [PubMed] [Google Scholar]
- 152.Lyko F. The DNA methyltransferase family: A versatile toolkit for epigenetic regulation. Nat. Rev. Genet. 2017 doi: 10.1038/nrg.2017.80. [DOI] [PubMed] [Google Scholar]
- 153.Lavoie G., St-Pierre Y. Phosphorylation of human DNMT1: Implication of cyclin-dependent kinases. Biochem. Biophys. Res. Commun. 2011;409:187–192. doi: 10.1016/j.bbrc.2011.04.115. [DOI] [PubMed] [Google Scholar]
- 154.Fujii T., Ota M., Hori H., Hattori K., Teraishi T., Matsuo J., Kinoshita Y., Ishida I., Nagashima A., Kunugi H. The common functional FKBP5 variant rs1360780 is associated with altered cognitive function in aged individuals. Sci. Rep. 2014;4:6696. doi: 10.1038/srep06696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Mamdani F., Rollins B., Morgan L., Myers R.M., Barchas J.D., Schatzberg A.F., Watson S.J., Akil H., Potkin S.G., Bunney W.E., et al. Variable telomere length across post-mortem human brain regions and specific reduction in the hippocampus of major depressive disorder. Transl. Psychiatr. 2015;5:e636. doi: 10.1038/tp.2015.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lagadari M., Zgajnar N.R., Gallo L.I., Galigniana M.D. Hsp90-binding immunophilin FKBP51 forms complexes with hTERT enhancing telomerase activity. Mol. Oncol. 2016;10:1086–1098. doi: 10.1016/j.molonc.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 157.Zannas A.S., Balsevich G., Gassen N.C. The emerging role of FKBP5 in the regulation of metabolism and body weight. Surg. Obes. Relat. Dis. 2016;12:1560–1561. doi: 10.1016/j.soard.2016.05.016. [DOI] [PubMed] [Google Scholar]
- 158.Pereira M.J., Palming J., Svensson M.K., Rizell M., Dalenback J., Hammar M., Fall T., Sidibeh C.O., Svensson P.A., Eriksson J.W. FKBP5 expression in human adipose tissue increases following dexamethasone exposure and is associated with insulin resistance. Metabolism. 2014;63:1198–1208. doi: 10.1016/j.metabol.2014.05.015. [DOI] [PubMed] [Google Scholar]
- 159.Fichna M., Krzysko-Pieczka I., Zurawek M., Skowronska B., Januszkiewicz-Lewandowska D., Fichna P. FKBP5 polymorphism is associated with insulin resistance in children and adolescents with obesity. Obes. Res. Clin. Pract. 2016 doi: 10.1016/j.orcp.2016.11.007. in press. [DOI] [PubMed] [Google Scholar]
- 160.Hartmann I.B., Fries G.R., Bucker J., Scotton E., von D.L., Kauer-Sant’Anna M. The FKBP5 polymorphism rs1360780 is associated with lower weight loss after bariatric surgery: 26 months of follow-up. Surg. Obes. Relat. Dis. 2016;12:1554–1560. doi: 10.1016/j.soard.2016.04.016. [DOI] [PubMed] [Google Scholar]
- 161.Balsevich G., Uribe A., Wagner K.V., Hartmann J., Santarelli S., Labermaier C., Schmidt M.V. Interplay between diet-induced obesity and chronic stress in mice: Potential role of FKBP51. J. Endocrinol. 2014;222:15–26. doi: 10.1530/JOE-14-0129. [DOI] [PubMed] [Google Scholar]
- 162.Yang L., Isoda F., Yen K., Kleopoulos S.P., Janssen W., Fan X., Mastaitis J., Dunn-Meynell A., Levin B., McCrimmon R., et al. Hypothalamic Fkbp51 is induced by fasting, and elevated hypothalamic expression promotes obese phenotypes. Am. J. Physiol. Endocrinol. Metab. 2012;302:E987–E991. doi: 10.1152/ajpendo.00474.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Stechschulte L.A., Qiu B., Warrier M., Hinds T.D., Jr., Zhang M., Gu H., Xu Y., Khuder S.S., Russo L., Najjar S.M., et al. FKBP51 Null Mice Are Resistant to Diet-Induced Obesity and the PPARgamma Agonist Rosiglitazone. Endocrinology. 2016;157:3888–3900. doi: 10.1210/en.2015-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Stechschulte L.A., Hinds T.D., Jr., Khuder S.S., Shou W., Najjar S.M., Sanchez E.R. FKBP51 controls cellular adipogenesis through p38 kinase-mediated phosphorylation of GRalpha and PPARgamma. Mol. Endocrinol. 2014;28:1265–1275. doi: 10.1210/me.2014-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zhang L., Qiu B., Wang T., Wang J., Liu M., Xu Y., Wang C., Deng R., Williams K., Yang Z., et al. Loss of FKBP5 impedes adipocyte differentiation under both normoxia and hypoxic stress. Biochem. Biophys. Res. Commun. 2017;485:761–767. doi: 10.1016/j.bbrc.2017.02.126. [DOI] [PubMed] [Google Scholar]
- 166.Dogan M.V., Lei M.K., Beach S.R., Brody G.H., Philibert R.A. Alcohol and tobacco consumption alter hypothalamic pituitary adrenal axis DNA methylation. Psychoneuroendocrinology. 2016;66:176–184. doi: 10.1016/j.psyneuen.2016.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Levran O., Randesi M., Li Y., Rotrosen J., Ott J., Adelson M., Kreek M.J. Drug addiction and stress-response genetic variability: Association study in African Americans. Ann. Hum. Genet. 2014;78:290–298. doi: 10.1111/ahg.12064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Patel N., Crider A., Pandya C.D., Ahmed A.O., Pillai A. Altered mRNA Levels of Glucocorticoid Receptor, Mineralocorticoid Receptor, and Co-Chaperones (FKBP5 and PTGES3) in the Middle Frontal Gyrus of Autism Spectrum Disorder Subjects. Mol. Neurobiol. 2016;53:2090–2099. doi: 10.1007/s12035-015-9178-2. [DOI] [PubMed] [Google Scholar]
- 169.Seifuddin F., Pirooznia M., Judy J.T., Goes F.S., Potash J.B., Zandi P.P. Systematic review of genome-wide gene expression studies of bipolar disorder. BMC Psychiatry. 2013;13:213. doi: 10.1186/1471-244X-13-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Fries G.R., Moreno M.P.V., Gubert C., Santos B.T.M.Q., Sartori J., Eisele B., Ferrari P., Fijtman A., Rüegg J., Gassen N.C., et al. Hypothalamic-pituitary-adrenal axis dysfunction and illness progression in bipolar disorder. Int. J. Neuropsychopharmacol. 2014 doi: 10.1093/ijnp/pyu043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Misiak B., Stramecki F., Gaweda L., Prochwicz K., Sasiadek M.M., Moustafa A.A., Frydecka D. Interactions Between Variation in Candidate Genes and Environmental Factors in the Etiology of Schizophrenia and Bipolar Disorder: A Systematic Review. Mol. Neurobiol. 2017:1–26. doi: 10.1007/s12035-017-0708-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Fani N., King T.Z., Shin J., Srivastava A., Brewster R.C., Jovanovic T., Bradley B., Ressler K.J. Structural and functional connectivity in posttraumatic stress disorder: Associations with fkbp5. Depress. Anxiety. 2016;33:300–307. doi: 10.1002/da.22483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Watkins L.E., Han S., Harpaz-Rotem I., Mota N.P., Southwick S.M., Krystal J.H., Gelernter J., Pietrzak R.H. FKBP5 polymorphisms, childhood abuse, and PTSD symptoms: Results from the National Health and Resilience in Veterans Study. Psychoneuroendocrinology. 2016;69:98–105. doi: 10.1016/j.psyneuen.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 174.De la Cruz-Cano E. Association between FKBP5 and CRHR1 genes with suicidal behavior: A systematic review. Behav. Brain Res. 2017;317:46–61. doi: 10.1016/j.bbr.2016.09.032. [DOI] [PubMed] [Google Scholar]
- 175.Isaksson J., Allen M., Nilsson K.W., Lindblad F. Polymorphisms in the FK506 binding protein 5 gene are associated with attention deficit hyperactivity disorder and diurnal cortisol levels. Acta Paediatr. 2015;104:910–915. doi: 10.1111/apa.13056. [DOI] [PubMed] [Google Scholar]
- 176.Minelli A., Maffioletti E., Cloninger C.R., Magri C., Sartori R., Bortolomasi M., Congiu C., Bignotti S., Segala M., Giacopuzzi M., et al. Role of allelic variants of FK506-binding protein 51 (FKBP5) gene in the development of anxiety disorders. Depress. Anxiety. 2013;30:1170–1176. doi: 10.1002/da.22158. [DOI] [PubMed] [Google Scholar]
- 177.Ising M., Depping A.M., Siebertz A., Lucae S., Unschuld P.G., Kloiber S., Horstmann S., Uhr M., Müller-Myhsok B., Holsboer F. Polymorphisms in the FKBP5 gene region modulate recovery from psychosocial stress in healthy controls. Eur. J. Neurosci. 2008;28:389–398. doi: 10.1111/j.1460-9568.2008.06332.x. [DOI] [PubMed] [Google Scholar]
- 178.Yeo S., Enoch M.A., Gorodetsky E., Akhtar L., Schuebel K., Roy A., Goldman D. The influence of FKBP5 genotype on expression of FKBP5 and other glucocorticoid-regulated genes, dependent on trauma exposure. Genes Brain Behav. 2017;16:223–232. doi: 10.1111/gbb.12342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Lessard J., Holman E.A. FKBP5 and CRHR1 polymorphisms moderate the stress-physical health association in a national sample. Health Psychol. 2014;33:1046–1056. doi: 10.1037/a0033968. [DOI] [PubMed] [Google Scholar]
- 180.Maric N.P., Adzic M. Pharmacological modulation of HPA axis in depression—New avenues for potential therapeutic benefits. Psychiatr. Danub. 2013;25:299–305. [PubMed] [Google Scholar]
- 181.Dean J., Keshavan M. The neurobiology of depression: An integrated view. Asian J. Psychiatry. 2017;27:101–111. doi: 10.1016/j.ajp.2017.01.025. [DOI] [PubMed] [Google Scholar]
- 182.Pariante C.M., Lightman S.L. The HPA axis in major depression: Classical theories and new developments. Trends Neurosci. 2008;31:464–468. doi: 10.1016/j.tins.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 183.Lukic I., Mitic M., Soldatovic I., Jovicic M., Maric N., Radulovic J., Adzic M. Accumulation of Cytoplasmic Glucocorticoid Receptor Is Related to Elevation of FKBP5 in Lymphocytes of Depressed Patients 3573. J. Mol. Neurosci. 2014;55:951–958. doi: 10.1007/s12031-014-0451-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Tatro E.T., Everall I.P., Masliah E., Hult B.J., Lucero G., Chana G., Soontornniyomkij V., Achim C.L. Differential expression of immunophilins FKBP51 and FKBP52 in the frontal cortex of HIV-infected patients with major depressive disorder. J. Neuroimmune Pharmacol. 2009;4:218–226. doi: 10.1007/s11481-009-9146-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Frodl T., Carballedo A., Frey E.M., O’Keane V., Skokauskas N., Morris D., Gill M., Hughes M.M., Harkin A., Connor T. Expression of glucocorticoid inducible genes is associated with reductions in cornu ammonis and dentate gyrus volumes in patients with major depressive disorder. Dev. Psychopathol. 2014;26:1209–1217. doi: 10.1017/S0954579414000972. [DOI] [PubMed] [Google Scholar]
- 186.Menke A., Arloth J., Putz B., Weber P., Klengel T., Mehta D., Gonik M., Rex-Haffner M., Rubel J., Uhr M., et al. Dexamethasone stimulated gene expression in peripheral blood is a sensitive marker for glucocorticoid receptor resistance in depressed patients. Neuropsychopharmacology. 2012;37:1455–1464. doi: 10.1038/npp.2011.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Menke A., Klengel T., Rubel J., Bruckl T., Pfister H., Lucae S., Uhr M., Holsboer F., Binder E.B. Genetic variation in FKBP5 associated with the extent of stress hormone dysregulation in major depression. Genes Brain Behav. 2013;12:289–296. doi: 10.1111/gbb.12026. [DOI] [PubMed] [Google Scholar]
- 188.Cordova-Palomera A., de Reus M.A., Fatjo-Vilas M., Falcon C., Bargallo N., van den Heuvel M.P., Fananas L. FKBP5 modulates the hippocampal connectivity deficits in depression: A study in twins. Brain Imaging Behav. 2017;11:62–75. doi: 10.1007/s11682-015-9503-4. [DOI] [PubMed] [Google Scholar]
- 189.Tozzi L., Carballedo A., Wetterling F., McCarthy H., O’Keane V., Gill M., Morris D., Fahey C., Meaney J., Frodl T. Single-Nucleotide Polymorphism of the FKBP5 Gene and Childhood Maltreatment as Predictors of Structural Changes in Brain Areas Involved in Emotional Processing in Depression. Neuropsychopharmacology. 2016;41:487–497. doi: 10.1038/npp.2015.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Zobel A., Schuhmacher A., Jessen F., Hofels S., Von W.O., Metten M., Pfeiffer U., Hanses C., Becker T., Rietschel M., et al. DNA sequence variants of the FKBP5 gene are associated with unipolar depression. Int. J. Neuropsychopharmacol. 2010;13:649–660. doi: 10.1017/S1461145709991155. [DOI] [PubMed] [Google Scholar]
- 191.Lahti J., la-Mikkula H., Kajantie E., Haljas K., Eriksson J.G., Raikkonen K. Associations Between Self-Reported and Objectively Recorded Early Life Stress, FKBP5 Polymorphisms, and Depressive Symptoms in Midlife. Biol. Psychiatry. 2016;80:869–877. doi: 10.1016/j.biopsych.2015.10.022. [DOI] [PubMed] [Google Scholar]
- 192.Zimmermann P., Brückl T., Nocon A., Pfister H., Binder E.B., Uhr M., Lieb R., Moffitt T.E., Caspi A., Holsboer F., et al. Interaction of variants in the FKBP5 gene and adverse life events in predicting the first depression onset: Results from a ten-year prospective community study. Am. J. Psychiatry. 2011;168:1107–1116. doi: 10.1176/appi.ajp.2011.10111577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Appel K., Schwahn C., Mahler J., Schulz A., Spitzer C., Fenske K., Stender J., Barnow S., John U., Teumer A., et al. Moderation of adult depression by a polymorphism in the FKBP5 gene and childhood physical abuse in the general population. Neuropsychopharmacology. 2011;36:1982–1991. doi: 10.1038/npp.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Dackis M.N., Rogosch F.A., Oshri A., Cicchetti D. The role of limbic system irritability in linking history of childhood maltreatment and psychiatric outcomes in low-income, high-risk women: Moderation by FK506 binding protein 5 haplotype 3714. Dev. Psychopathol. 2012;24:1237–1252. doi: 10.1017/S0954579412000673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Rao S., Yao Y., Ryan J., Li T., Wang D., Zheng C., Xu Y., Xu Q. Common variants in FKBP5 gene and major depressive disorder (MDD) susceptibility: A comprehensive meta-analysis. Sci. Rep. 2016;6:32687. doi: 10.1038/srep32687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Cattaneo A., Gennarelli M., Uher R., Breen G., Farmer A., Aitchison K.J., Craig I.W., Anacker C., Zunsztain P.A., McGuffin P., et al. Candidate genes expression profile associated with antidepressants response in the GENDEP study: Differentiating between baseline ‘predictors’ and longitudinal ‘targets’. Neuropsychopharmacology. 2013;38:377–385. doi: 10.1038/npp.2012.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Kirchheiner J., Lorch R., Lebedeva E., Seeringer A., Roots I., Sasse J., Brockmoller J. Genetic variants in FKBP5 affecting response to antidepressant drug treatment. Pharmacogenomics. 2008;9:841–846. doi: 10.2217/14622416.9.7.841. [DOI] [PubMed] [Google Scholar]
- 198.Horstmann S., Lucae S., Menke A., Hennings J.M., Ising M., Roeske D., Müller-Myhsok B., Holsboer F., Binder E.B. Polymorphisms in GRIK4, HTR2A, and FKBP5 show interactive effects in predicting remission to antidepressant treatment. Neuropsychopharmacology. 2010;35:727–740. doi: 10.1038/npp.2009.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Stamm T.J., Rampp C., Wiethoff K., Stingl J., Mossner R., Malley O., Ricken R., Seemüller F., Keck M., Fisher R., et al. The FKBP5 polymorphism rs1360780 influences the effect of an algorithm-based antidepressant treatment and is associated with remission in patients with major depression. J. Psychopharmacol. 2016;30:40–47. doi: 10.1177/0269881115620459. [DOI] [PubMed] [Google Scholar]
- 200.Ellsworth K.A., Moon I., Eckloff B.W., Fridley B.L., Jenkins G.D., Batzler A., Biernacka J.M., Abo R., Brisbin A., Ji Y., et al. FKBP5 genetic variation: Association with selective serotonin reuptake inhibitor treatment outcomes in major depressive disorder. Pharmacogenet. Genom. 2013;23:156–166. doi: 10.1097/FPC.0b013e32835dc133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Lekman M., Laje G., Charney D., Rush A.J., Wilson A.F., Sorant A.J., Lipsky R., Wisniewski S.R., Manji H., McMahon F.J., et al. The FKBP5-gene in depression and treatment response—An association study in the Sequenced Treatment Alternatives to Relieve Depression (STAR*D) Cohort. Biol. Psychiatry. 2008;63:1103–1110. doi: 10.1016/j.biopsych.2007.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Sarginson J.E., Lazzeroni L.C., Ryan H.S., Schatzberg A.F., Murphy G.M., Jr. FKBP5 polymorphisms and antidepressant response in geriatric depression. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2010;153B:554–560. doi: 10.1002/ajmg.b.31019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Tsai S.J., Hong C.J., Chen T.J., Yu Y.W. Lack of supporting evidence for a genetic association of the FKBP5 polymorphism and response to antidepressant treatment 26. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2007;144B:1097–1098. doi: 10.1002/ajmg.b.30246. [DOI] [PubMed] [Google Scholar]
- 204.Niitsu T., Fabbri C., Bentini F., Serretti A. Pharmacogenetics in major depression: A comprehensive meta-analysis. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2013;45:183–194. doi: 10.1016/j.pnpbp.2013.05.011. [DOI] [PubMed] [Google Scholar]
- 205.Gassen N.C., Hartmann J., Zannas A.S., Kretzschmar A., Zschocke J., Maccarrone G., Hafner K., Zellner A., Kollmannsberger L.K., Wagner K.V., et al. FKBP51 inhibits GSK3beta and augments the effects of distinct psychotropic medications. Mol. Psychiatry. 2016;21:277–289. doi: 10.1038/mp.2015.38. [DOI] [PubMed] [Google Scholar]
- 206.Katz E.R., Stowe Z.N., Newport D.J., Kelley M.E., Pace T.W., Cubells J.F., Binder E.B. Regulation of mRNA expression encoding chaperone and co-chaperone proteins of the glucocorticoid receptor in peripheral blood: Association with depressive symptoms during pregnancy. Psychol. Med. 2012;42:943–956. doi: 10.1017/S0033291711002121. [DOI] [PubMed] [Google Scholar]
- 207.Tatro E.T., Nguyen T.B., Bousman C.A., Masliah E., Grant I., Atkinson J.H., Everall I.P. Correlation of major depressive disorder symptoms with FKBP5 but not FKBP4 expression in human immunodeficiency virus-infected individuals. J. Neurovirol. 2010;16:399–404. doi: 10.3109/13550284.2010.504248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Perroud N., Bondolfi G., Uher R., Gex-Fabry M., Aubry J.M., Bertschy G., Malafosse A., Kosel M. Clinical and genetic correlates of suicidal ideation during antidepressant treatment in a depressed outpatient sample. Pharmacogenomics. 2011;12:365–377. doi: 10.2217/pgs.10.189. [DOI] [PubMed] [Google Scholar]
- 209.Shimasaki A., Kondo K., Saito T., Esaki K., Otsuka Y., Mano K., Ikeda M., Iwata N. A genetic variant in 12q13, a possible risk factor for bipolar disorder, is associated with depressive state, accounting for stressful life events. PLoS ONE. 2014;9:e115135. doi: 10.1371/journal.pone.0115135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Höhne N., Poidinger M., Merz F., Pfister H., Bruckl T., Zimmermann P., Uhr M., Holsboer F., Ising M. FKBP5 Genotype-Dependent DNA Methylation and mRNA Regulation after Psychosocial Stress in Remitted Depression and Healthy Controls. Int. J. Neuropsychopharmacol. 2014;18 doi: 10.1093/ijnp/pyu087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.VanZomeren-Dohm A.A., Pitula C.E., Koss K.J., Thomas K., Gunnar M.R. FKBP5 moderation of depressive symptoms in peer victimized, post-institutionalized children. Psychoneuroendocrinology. 2015;51:426–430. doi: 10.1016/j.psyneuen.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Owens M., Harrison A.J., Burkhouse K.L., McGeary J.E., Knopik V.S., Palmer R.H., Gibb B.E. Eye tracking indices of attentional bias in children of depressed mothers: Polygenic influences help to clarify previous mixed findings. Dev. Psychopathol. 2016;28:385–397. doi: 10.1017/S0954579415000462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Szczepankiewicz A., Leszczynska-Rodziewicz A., Pawlak J., Narozna B., Rajewska-Rager A., Wilkosc M., Zaremba D., Maciukiewicz M., Twarowska-Hauser J. FKBP5 polymorphism is associated with major depression but not with bipolar disorder. J. Affect. Disord. 2014;164:33–37. doi: 10.1016/j.jad.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 214.Suppli N.P., Bukh J.D., Moffitt T.E., Caspi A., Johansen C., Tjonneland A., Kessing L.V., Dalton S.O. Genetic variants in 5-HTTLPR, BDNF, HTR1A, COMT, and FKBP5 and risk for treated depression after cancer diagnosis. Depress. Anxiety. 2017;34:845–855. doi: 10.1002/da.22660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Schneider M., Engel A., Fasching P.A., Haberle L., Binder E.B., Voigt F., Grimm J., Faschingbauer F., Eichler A., Dammer U., et al. Genetic variants in the genes of the stress hormone signalling pathway and depressive symptoms during and after pregnancy. Biomed. Res. Int. 2014;2014:469278. doi: 10.1155/2014/469278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Velders F.P., Kuningas M., Kumari M., Dekker M.J., Uitterlinden A.G., Kirschbaum C., Hek K., Hofman A., Verhulst F.C., Kivimaki M., et al. Genetics of cortisol secretion and depressive symptoms: A candidate gene and genome wide association approach. Psychoneuroendocrinology. 2011;36:1053–1061. doi: 10.1016/j.psyneuen.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Udina M., Navines R., Egmond E., Oriolo G., Langohr K., Gimenez D., Valdes M., Gomez-Gil E., Grande I., Gratacos M., et al. Glucocorticoid Receptors, Brain-Derived Neurotrophic Factor, Serotonin and Dopamine Neurotransmission are Associated with Interferon-Induced Depression. Int. J. Neuropsychopharmacol. 2016;19 doi: 10.1093/ijnp/pyv135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Lewis G., Collishaw S., Harold G., Rice F., Thapar A. Maternal depression and child and adolescent depression symptoms: An exploratory test for moderation by CRHR1, FKBP5 and NR3C1 gene variants. Behav. Genet. 2012;42:121–132. doi: 10.1007/s10519-011-9482-1. [DOI] [PubMed] [Google Scholar]
- 219.Shinozaki G., Jowsey S., Amer H., Biernacka J., Colby C., Walker D., Black J., Rundell J., Stegall M., Mrazek D.A. Relationship between FKBP5 polymorphisms and depression symptoms among kidney transplant recipients. Depress. Anxiety. 2011;28:1111–1118. doi: 10.1002/da.20879. [DOI] [PubMed] [Google Scholar]
- 220.Borelli J.L., Smiley P.A., Rasmussen H.F., Gomez A., Seaman L.C., Nurmi E.L. Interactive effects of attachment and FKBP5 genotype on school-aged children’s emotion regulation and depressive symptoms. Behav. Brain Res. 2017;325:278–289. doi: 10.1016/j.bbr.2016.07.035. [DOI] [PubMed] [Google Scholar]
- 221.Suzuki A., Matsumoto Y., Sadahiro R., Enokido M., Goto K., Otani K. Relationship of the FKBP5 C/T polymorphism with dysfunctional attitudes predisposing to depression. Compr. Psychiatry. 2014;55:1422–1425. doi: 10.1016/j.comppsych.2014.04.019. [DOI] [PubMed] [Google Scholar]
- 222.Sarubin N., Hilbert S., Naumann F., Zill P., Wimmer A.M., Nothdurfter C., Rupprecht R., Baghai T.C., Buhner M., Schule C. The sex-dependent role of the glucocorticoid receptor in depression: Variations in the NR3C1 gene are associated with major depressive disorder in women but not in men. Eur. Arch. Psychiatry Clin. Neurosci. 2017;267:123–133. doi: 10.1007/s00406-016-0722-5. [DOI] [PubMed] [Google Scholar]
- 223.Lavebratt C., Aberg E., Sjoholm L.K., Forsell Y. Variations in FKBP5 and BDNF genes are suggestively associated with depression in a Swedish population-based cohort. J. Affect. Disord. 2010;125:249–255. doi: 10.1016/j.jad.2010.02.113. [DOI] [PubMed] [Google Scholar]
- 224.Arlt S., Demiralay C., Tharun B., Geisel O., Storm N., Eichenlaub M., Lehmbeck J.T., Wiedemann K., Leuenberger B., Jahn H. Genetic risk factors for depression in Alzheimer’s disease patients. Curr. Alzheimer Res. 2013;10:72–81. doi: 10.2174/156720513804871435. [DOI] [PubMed] [Google Scholar]
- 225.Wang Q., Shelton R.C., Dwivedi Y. Interaction between early-life stress and FKBP5 gene variants in major depressive disorder and post-traumatic stress disorder: A systematic review and meta-analysis. J. Affect. Disord. 2018;225:422–428. doi: 10.1016/j.jad.2017.08.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Papiol S., Arias B., Gastó C., Gutiérrez B., Catalán R., Fañanás L. Genetic variability at HPA axis in major depression and clinical response to antidepressant treatment. J. Affect. Disord. 2007;104:83–90. doi: 10.1016/j.jad.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 227.Ratajczak T. Steroid Receptor-Associated Immunophilins: Candidates for Diverse Drug-Targeting Approaches in Disease. Curr. Mol. Pharmacol. 2015;9:66–95. doi: 10.2174/1874467208666150519113639. [DOI] [PubMed] [Google Scholar]
- 228.Mazaira G.I., Camisay M.F., De L.S., Erlejman A.G., Galigniana M.D. Biological relevance of Hsp90-binding immunophilins in cancer development and treatment. Int. J. Cancer. 2016;138:797–808. doi: 10.1002/ijc.29509. [DOI] [PubMed] [Google Scholar]
- 229.Periyasamy S., Hinds T., Jr., Shemshedini L., Shou W., Sanchez E.R. FKBP51 and Cyp40 are positive regulators of androgen-dependent prostate cancer cell growth and the targets of FK506 and cyclosporin A. Oncogene. 2010;29:1691–1701. doi: 10.1038/onc.2009.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Ni L., Yang C.S., Gioeli D., Frierson H., Toft D.O., Paschal B.M. FKBP51 promotes assembly of the Hsp90 chaperone complex and regulates androgen receptor signaling in prostate cancer cells. Mol. Cell. Biol. 2010;30:1243–1253. doi: 10.1128/MCB.01891-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Pei H., Li L., Fridley B.L., Jenkins G.D., Kalari K.R., Lingle W., Petersen G., Lou Z., Wang L. FKBP51 affects cancer cell response to chemotherapy by negatively regulating Akt. Cancer Cell. 2009;16:259–266. doi: 10.1016/j.ccr.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Kang C.B., Hong Y., Dhe-Paganon S., Yoon H.S. FKBP family proteins: Immunophilins with versatile biological functions. Neurosignals. 2008;16:318–325. doi: 10.1159/000123041. [DOI] [PubMed] [Google Scholar]
- 233.Van Duyne G.D., Standaert R.F., Karplus P.A., Schreiber S.L., Clardy J. Atomic structure of FKBP-FK506, an immunophilin-immunosuppressant complex. Science. 1991;252:839–842. doi: 10.1126/science.1709302. [DOI] [PubMed] [Google Scholar]
- 234.Liu J., Farmer J.D., Jr., Lane W.S., Friedman J., Weissman I., Schreiber S.L. Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP-FK506 complexes. Cell. 1991;66:807–815. doi: 10.1016/0092-8674(91)90124-H. [DOI] [PubMed] [Google Scholar]
- 235.Allan R.K., Mok D., Ward B.K., Ratajczak T. Modulation of chaperone function and cochaperone interaction by novobiocin in the C-terminal domain of Hsp90: Evidence that coumarin antibiotics disrupt Hsp90 dimerization. J. Biol. Chem. 2006;281:7161–7171. doi: 10.1074/jbc.M512406200. [DOI] [PubMed] [Google Scholar]
- 236.McConnell J.R., Alexander L.A., McAlpine S.R. A heat shock protein 90 inhibitor that modulates the immunophilins and regulates hormone receptors without inducing the heat shock response. Bioorg. Med. Chem. Lett. 2014;24:661–666. doi: 10.1016/j.bmcl.2013.11.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Kabouridis P.S. Biological applications of protein transduction technology. Trends Biotechnol. 2003;21:498–503. doi: 10.1016/j.tibtech.2003.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Horibe T., Torisawa A., Kohno M., Kawakami K. Molecular mechanism of cytotoxicity induced by Hsp90-targeted Antp-TPR hybrid peptide in glioblastoma cells. Mol. Cancer. 2012;11:59. doi: 10.1186/1476-4598-11-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Horibe T., Kohno M., Haramoto M., Ohara K., Kawakami K. Designed hybrid TPR peptide targeting Hsp90 as a novel anticancer agent. J. Transl. Med. 2011;9:8. doi: 10.1186/1479-5876-9-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Kumar R., Moche M., Winblad B., Pavlov P.F. Combined X-ray crystallography and computational modeling approach to investigate the Hsp90 C-terminal peptide binding to FKBP51. Sci. Rep. 2017;7:14288. doi: 10.1038/s41598-017-14731-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Gaali S., Kirschner A., Cuboni S., Hartmann J., Kozany C., Balsevich G., Namendorf C., Fernandez-Vizarra P., Sippel C., Zannas A.S., et al. Selective inhibitors of the FK506-binding protein 51 by induced fit. Nat. Chem. Biol. 2015;11:33–37. doi: 10.1038/nchembio.1699. [DOI] [PubMed] [Google Scholar]
- 242.Feng X., Pomplun S., Hausch F. Recent Progress in FKBP Ligand Development. Curr. Mol. Pharmacol. 2015;9:27–36. doi: 10.2174/1874467208666150519113313. [DOI] [PubMed] [Google Scholar]
- 243.Hartmann J., Wagner K.V., Gaali S., Kirschner A., Kozany C., Ruhter G., Dedic N., Hausl A.S., Hoeijmakers L., Westerholz S., et al. Pharmacological Inhibition of the Psychiatric Risk Factor FKBP51 Has Anxiolytic Properties. J. Neurosci. 2015;35:9007–9016. doi: 10.1523/JNEUROSCI.4024-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Maiaru M., Tochiki K.K., Cox M.B., Annan L.V., Bell C.G., Feng X., Hausch F., Geranton S.M. The stress regulator FKBP51 drives chronic pain by modulating spinal glucocorticoid signaling. Sci. Transl. Med. 2016;8:325ra19. doi: 10.1126/scitranslmed.aab3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Pomplun S., Wang Y., Kirschner A., Kozany C., Bracher A., Hausch F. Rational design and asymmetric synthesis of potent and neurotrophic ligands for FK506-binding proteins (FKBPs) Angew. Chem. Int. Ed. Engl. 2015;54:345–348. doi: 10.1002/anie.201408776. [DOI] [PubMed] [Google Scholar]
- 246.Bracher A., Kozany C., Thost A.K., Hausch F. Structural characterization of the PPIase domain of FKBP51, a cochaperone of human Hsp90. Acta Crystallogr. D Biol. Crystallogr. 2011;67:549–559. doi: 10.1107/S0907444911013862. [DOI] [PubMed] [Google Scholar]
- 247.Shi D., Bai Q., Zhou S., Liu X., Liu H., Yao X. Molecular dynamics simulation, binding free energy calculation and unbinding pathway analysis on selectivity difference between FKBP51 and FKBP52: Insight into the molecular mechanism of isoform selectivity. Proteins. 2017 doi: 10.1002/prot.25401. [DOI] [PubMed] [Google Scholar]
- 248.LeMaster D.M., Hernandez G. Conformational Dynamics in FKBP Domains: Relevance to Molecular Signaling and Drug Design. Curr. Mol. Pharmacol. 2015;9:5–26. doi: 10.2174/1874467208666150519113146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Fujii T., Ota M., Hori H., Hattori K., Teraishi T., Sasayama D., Higuchi T., Kunugi H. Association between the common functional FKBP5 variant (rs1360780) and brain structure in a non-clinical population. J. Psychiatr. Res. 2014;58:96–101. doi: 10.1016/j.jpsychires.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 250.Hirakawa H., Akiyoshi J., Muronaga M., Tanaka Y., Ishitobi Y., Inoue A., Oshita H., Aizawa S., Masuda K., Higuma H., et al. FKBP5 is associated with amygdala volume in the human brain and mood state: A voxel-based morphometry (VBM) study. Int. J. Psychiatry Clin. Pract. 2016;20:106–115. doi: 10.3109/13651501.2016.1144772. [DOI] [PubMed] [Google Scholar]
- 251.Holz N.E., Buchmann A.F., Boecker R., Blomeyer D., Baumeister S., Wolf I., Rietschel M., Witt S.H., Plichta M.M., Meyer-Lindenberg A., et al. Role of FKBP5 in emotion processing: Results on amygdala activity, connectivity and volume. Brain Struct. Funct. 2015;220:1355–1368. doi: 10.1007/s00429-014-0729-5. [DOI] [PubMed] [Google Scholar]
- 252.Bryant R.A., Felmingham K.L., Liddell B., Das P., Malhi G.S. Association of FKBP5 polymorphisms and resting-state activity in a frontotemporal-parietal network. Transl. Psychiatry. 2016;6:e925. doi: 10.1038/tp.2016.149. [DOI] [PMC free article] [PubMed] [Google Scholar]